Abstract
Purpose
Magnetic resonance imaging (MRI) provides non-invasive evaluation of patient’s anatomy without using ionizing radiation. Due to this and the high soft-tissue contrast, MRI use has increased and has potential for use in longitudinal studies where changes in patients’ anatomy or tumors at different time points are compared. Deformable image registration can be useful for these studies. Here we describe two data sets that can be used to evaluate the registration accuracy of systems for MR images, as it cannot be assumed to be the same as that measured on CT images.
Acquisition and Validation Methods
Two sets of images were created to test registration accuracy. (1) A porcine phantom was created by placing ten 0.35 mm gold markers into porcine meat. The porcine phantom was immobilized in a plastic container with movable dividers. T1-weighted, T2-weighted, and CT images were acquired with the porcine phantom compressed in 4 different ways. The markers were not visible on the MR images, due to the selected voxel size, so they did not interfere with the measured registration accuracy, while the markers were visible on the CT images and were used to identify the known deformation between positions. (2) Synthetic images were created using 28 head and neck squamous cell carcinoma patients who had MR scans pre-, mid-, and post-radiotherapy treatment. An inter- and intra-patient variation model was created using these patient scans. Four synthetic pre-treatment images were created using the inter-patient model, and four synthetic post-treatment images were created for each of the pre-treatment images using the intra-patient model.
Data Format and Usage Notes
The T1-weighted, T2-weighted, and CT scans of the porcine phantom in the four positions are provided. Four T1-weighted synthetic pre-treatment images each with four synthetic post-treatment images, and four T2-weighted synthetic pre-treatment images each with four synthetic post-treatment images are provided. Additionally, the applied deformation vector fields to generate the synthetic post-treatment images are provided. The data is available on TCIA under the collection MRI-DIR.
Potential Applications
The proposed database provides two sets of images (one acquired and one computer generated) for use in evaluating deformable image registration accuracy. T1- and T2-weighted images are available for each technique as the different image contrast in the two types of images may impact the registration accuracy.
II. Introduction
Magnetic resonance imaging (MRI) allows for non-invasive assessment of patients without ionizing radiation, which conventional imaging techniques, i.e. digital radiography and computed tomography (CT), do not allow. Additionally, MRI provides high soft-tissue contrast. Due to these advantages, MRI use has more than tripled in the United States over the past 20 years with similar trends in other developed countries 1. The advantages of MRI and its increased use put it in a position to be used for longitudinal studies, a study of the same patients over a period of time 2. If deformable registration is used to compare these images, the deformable image registration accuracy should be high as large errors could produce misinterpretation of the data, or misdiagnosis or mistreatment of the patient.
Many image registration software systems have been evaluated using CT images 3–8. The image contrast and intensity histograms are typically very different between CT and MRI. Additionally, MRI has geometric distortion and often artifacts are present at interfaces of air and bone. Due to these differences the registration accuracy of a system must be measured separately for each imaging modality. There have been several in-house systems that have been evaluated using MR images for different anatomic sites, including liver 9,10, prostate 11–13, and breast 14,15. However, there has been limited reporting on commercial systems’ accuracy with MR images. There are published databases with MRI longitudinal patient data, but they often do not contain anatomical T1- and T2-weighted images and do not have markers within the patients to measure registration accuracy with 16–23. These databases are mostly for predicting response which often assumes an accurate MR image registration or separate contours on each image.
This dataset contains two different sets of images to test MR image registration accuracy. The lack of MR image registration studies compared to CT may be due to the lack of available datasets. This dataset can help with this need and facilitate testing commercial and in-house systems’ registration accuracy. Additionally, this data set can aid in verifying registration systems which can then be used for the longitudinal databases that already exist for prediction. The dataset fills a need in a baseline step that should not be overlooked before proceeding with higher level studies that utilize deformable image registration with MR images.
III. Acquisition and Validation Methods
A. Porcine Phantom
A porcine shoulder cut was selected as it contained muscle, fat, and bone. Ten 0.35 mm gold markers were implanted in the porcine meat. The porcine meat with markers was then placed in a plastic container and secured with movable dividers (United States Plastic Company, Lima, OH). The container is 27.6 cm long with 7 notches, 21.0 cm wide with 5 notches, and 12.7 cm tall. The notches are 3.25 cm apart and 1.6 cm from the corner in the width, and 1.8cm from the corner in the length. There were two sets of dividers: long dividers that went into the notches on the width of the container and short dividers that went into the notches on the length of the container. The porcine meat was placed in four different positions within the container and imaged on CT and MRI in each position. Figure 1 shows the porcine phantom and dividers. The four positions consisted of: (1) one long divider in the first notch, (2) one long divider in the first notch and one short divider in the first notch, (3) one long divider in the first notch, one long divider in the last notch, and one short divider in the first notch, and (4) one long divider in the first notch, one long divider in the last notch, one short divider in the first notch, and one short divider in the last notch. Table 1 has the locations of the gold markers within the image and Table 2 has the applied deformations.
Figure 1. Porcine Phantom Representative Deformation.
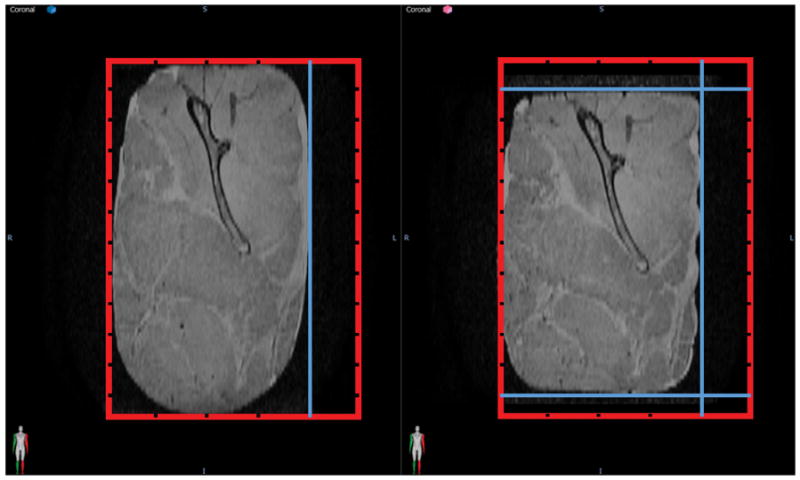
A deformation was applied to the porcine phantom by moving the dividers. The red box represents the container with the grooves for the movable dividers and the position of the dividers is shown in blue. The original position is shown on the left and a deformed position using more movable dividers to secure the phantom in place is shown on the right. (Reproduced with permission from Wiley 24.)
Table 1.
Locations of Gold Markers for Each Position Imaged
| Position 1 | Position 2 | Position 3 | Position 4 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Axial | Coronal | Sagittal | Axial | Coronal | Sagittal | Axial | Coronal | Sagittal | Axial | Coronal | Sagittal | |
| Marker 1 | −57.1 | 6.7 | −5 | −46.3 | 7.1 | −26.9 | −46.9 | 10.6 | −29.3 | −45.5 | 3.2 | −17.2 |
| Marker 2 | −52.1 | −2.6 | 19.1 | −44.6 | −3.6 | −1.1 | −41.3 | 6 | −2.4 | −42.9 | −7.7 | 8.6 |
| Marker 3 | −32.9 | −59.8 | −0.9 | −25.1 | −59.7 | −19.5 | −21.9 | −57.6 | −10.6 | −29.1 | −60.6 | −12.7 |
| Marker 4 | 7.9 | −10.9 | 21.6 | 13.9 | −11.5 | 2.8 | 12.9 | −2.2 | 1.2 | 15.4 | −14.9 | 11.4 |
| Marker 5 | 8.6 | 45.2 | 43.3 | 18.1 | 43.8 | 20.3 | 14.6 | 56 | 11 | 23 | 37.1 | 29.7 |
| Marker 6 | 28.4 | −43.9 | 2.2 | 32.6 | −43.6 | −17.3 | 31 | −43 | −15.8 | 31.5 | −45.3 | −11.2 |
| Marker 7 | 42.4 | 2.8 | −55.2 | 49.6 | 1.2 | −77.9 | 44.4 | −7.5 | −78.4 | 48.4 | −3.1 | −71 |
| Marker 8 | 45.5 | 44.2 | 44.4 | 52.9 | 43.1 | 23.2 | 47.1 | 53.6 | 12.2 | 56.6 | 35.5 | 28.8 |
| Marker 9 | 58.5 | −21.3 | 14.7 | 62.2 | −20.6 | −5.3 | 54.4 | −15.4 | −7.6 | 62 | −23 | 1.7 |
| Marker 10 | 74.1 | 28.5 | −14.6 | 79 | 28.2 | −36.9 | 68.2 | 28.9 | −47 | 79.1 | 24.2 | −31.3 |
Table 2.
Deformations of Gold Marker Locations between Positions
| Position 1 to 2 | Position 1 to 3 | Position 1 to 4 | Position 2 to 3 | Position 2 to 4 | Position 3 to 4 | ||
|---|---|---|---|---|---|---|---|
| Marker 1 | Axial | 10.8 | 10.2 | 11.6 | −0.6 | 0.8 | 1.4 |
| Coronal | 0.4 | 3.9 | −3.5 | 3.5 | −3.9 | −7.4 | |
| Sagittal | −21.9 | −24.3 | −12.2 | −2.4 | 9.7 | 12.1 | |
|
|
|||||||
| Marker 2 | Axial | 7.5 | 10.8 | 9.2 | 3.3 | 1.7 | −1.6 |
| Coronal | −1 | 8.6 | −5.1 | 9.6 | −4.1 | −13.7 | |
| Sagittal | −20.2 | −21.5 | −10.5 | −1.3 | 9.7 | 11 | |
|
|
|||||||
| Marker 3 | Axial | 7.8 | 11 | 3.8 | 3.2 | −4 | −7.2 |
| Coronal | 0.1 | 2.2 | −0.8 | 2.1 | −0.9 | −3 | |
| Sagittal | −18.6 | −9.7 | −11.8 | 8.9 | 6.8 | −2.1 | |
|
|
|||||||
| Marker 4 | Axial | 6 | 5 | 7.5 | −1 | 1.5 | 2.5 |
| Coronal | −0.6 | 8.7 | −4 | 9.3 | −3.4 | −12.7 | |
| Sagittal | −18.8 | −20.4 | −10.2 | −1.6 | 8.6 | 10.2 | |
|
|
|||||||
| Marker 5 | Axial | 9.5 | 6 | 14.4 | −3.5 | 4.9 | 8.4 |
| Coronal | −1.4 | 10.8 | −8.1 | 12.2 | −6.7 | −18.9 | |
| Sagittal | −23 | −32.3 | −13.6 | −9.3 | 9.4 | 18.7 | |
|
|
|||||||
| Marker 6 | Axial | 4.2 | 2.6 | 3.1 | −1.6 | −1.1 | 0.5 |
| Coronal | 0.3 | 0.9 | −1.4 | 0.6 | −1.7 | −2.3 | |
| Sagittal | −19.5 | −18 | −13.4 | 1.5 | 6.1 | 4.6 | |
|
|
|||||||
| Marker 7 | Axial | 7.2 | 2 | 6 | −5.2 | −1.2 | 4 |
| Coronal | −1.6 | −10.3 | −5.9 | −8.7 | −4.3 | 4.4 | |
| Sagittal | −22.7 | −23.2 | −15.8 | −0.5 | 6.9 | 7.4 | |
|
|
|||||||
| Marker 8 | Axial | 7.4 | 1.6 | 11.1 | −5.8 | 3.7 | 9.5 |
| Coronal | −1.1 | 9.4 | −8.7 | 10.5 | −7.6 | −18.1 | |
| Sagittal | −21.2 | −32.2 | −15.6 | −11 | 5.6 | 16.6 | |
|
|
|||||||
| Marker 9 | Axial | 3.7 | −4.1 | 3.5 | −7.8 | −0.2 | 7.6 |
| Coronal | 0.7 | 5.9 | −1.7 | 5.2 | −2.4 | −7.6 | |
| Sagittal | −20 | −22.3 | −13 | −2.3 | 7 | 9.3 | |
|
|
|||||||
| Marker 10 | Axial | 4.9 | −5.9 | 5 | −10.8 | 0.1 | 10.9 |
| Coronal | −0.3 | 0.4 | −4.3 | 0.7 | −4 | −4.7 | |
| Sagittal | −22.3 | −32.4 | −16.7 | −10.1 | 5.6 | 15.7 | |
The CT images were captured on a GE Discovery CT 750HD (GE Healthcare, Waukesha, WI) in helical mode. The protocol for all four images was 120 kVp, 150 mA, 0.516 pitch, 0.8-s rotation time, 0.625-mm slice thickness, 0.625-mm spacing between slices, pixel size 0.39 mm × 0.39 mm, and CTDIvol 41.07 mGy.
T1-weighted and T2-weighted MR images were acquired on a 3.0T Discovery 750 MRI scanner with an 8US TORSOPA coil (GE Healthcare). The T1-weighted scans used a three-dimensional spoiled gradient recalled echo sequence with the following acquisition parameters: flip angle 12°, repetition time 4.96 ms, echo time 2.1 ms, effective number of excitations 2, pixel bandwidth 325 Hz, field of view 25.6 cm, slice thickness 1 mm, and pixel size 1 mm × 1 mm. The T2-weighted scans used a two-dimensional fast spin echo sequence with the following acquisition parameters: flip angle 90°, repetition time 5884 ms, echo time 98 ms, effective number of excitations 2, pixel bandwidth 325 Hz, field of view 25.6 cm, slice thickness 2.5 mm, gap 1.5 mm, acquisition pixel size 1 mm × 1 mm, and zero filling interpolation × 2 (producing an image pixel size of 0.5 mm × 0.5 mm). The voxel size selected for both T1 and T2 weighted sequences were large enough for the gold markers to not be visible, therefore not interfering/affecting/biasing the results of the registration algorithms.
B. Synthetic Images
Twenty-eight patients with human papillomavirus-positive oropharyngeal squamous cell carcinoma who were treated with definitive chemoradiotherapy were selected from a prospective trial under a protocol approved by the institutional review board at MD Anderson Cancer Center with study-specific informed consent. Patients were imaged with T1- and T2-weighted MRI sequences at pre-treatment (within 1 week before treatment), mid-treatment (3–4 weeks after initiation of treatment), and post-treatment (6–8 weeks after conclusion of treatment). The first 28 patients to complete all three time point scans were selected. Patients selected for the generation of the synthetic images were scanned between December 2013 and October 2015. The patient median age was 57 (range 46–70), with 26 men and 2 women. Organ segmentation was performed on each patient’s T1-weighted images by the treating physician. The median left parotid volume was 30.9 cm3 (range 22.4–47.1 cm3), median right parotid volume was 33.5 cm3 (range 18.0–46.7 cm3), median left submandibular volume was 9.6 cm3 (range 5.7–19.7 cm3), median right submandibular volume was 9.4 cm3 (range 4.3–17.5 cm3), and median sublingual volume was 4.8 cm3 (range 1.6–9.4 cm3).
The T1- and T2-weighted scans were acquired on a 3.0T Discovery 750 MRI with a 6-element flex coil and flat insert table (GE Healthcare). The scans each had thirty slices with a 25.6cm axial field of view and covered the region encompassing the palatine process region cranially to the cricoid cartilage caudally in the superior-inferior direction with 4 mm slice thickness. The T1-weighted scans used a three-dimensional spoiled gradient recalled echo sequence with the following acquisition parameters: flip angle 15°, repetition time 3.6 ms, echo time 1 ms, effective number of excitations 0.7 (GE terminology, number of averages = percent sampling * number of signal averages/number of excitations), pixel bandwidth 325 Hz, acquisition pixel size 2 mm × 2mm, and zero filling interpolation × 2. This resulted in reconstructed voxel sizes of 1 mm × 1 mm × 4 mm. The T2-weighted scans used a two-dimensional fast spin echo multi-slice sequence with the following acquisition parameters: flip angle 90°, repetition time ~3600 ms, echo time ~100 ms, effective number of excitations 1, pixel bandwidth 195 Hz, slice thickness 2.5 mm, gap 1.5 mm, acquisition pixel size 1 mm × 1 mm, and zero filling interpolation × 2. This resulted in reconstructed voxel sizes of 0.5 mm × 0.5 mm × 4 mm.
T1- and T2-weighted synthetic image models were created separately. Each model was built using the following steps. Each patient’s mid- and post-treatment images were registered to his/her pre-treatment image. These deformations were used to create an intra-patient variation model. A template patient was selected from the 28 patients based on median age and median salivary gland volume. Each patient’s pre-treatment image was deformed to the template patient’s pre-treatment image. An inter-patient variation model was created using these deformations. All deformations were conducted using an in-house demons-based deformable image registration tool 5,6. Ninety-five percent of the variation was included in both the intra- and inter-patient variation models.
The intra- and inter-patient variation model consistent of a range of values and use a random number generator to pull from the distribution, therefore each time the model is run to create a synthetic image, a different image is created. The template patient’s pre-treatment image was deformed using the inter-patient variation model to create synthetic pre-treatment images. The synthetic pre-treatment image was deformed using the intra-patient variation model to create synthetic post-treatment images. The inter-patient variation model was run 4 times to create 4 distinct synthetic pre-treatment images. For each synthetic pre-treatment image, the intra-patient variation model was run 4 times to create 4 distinct synthetic post-treatment images. This process is demonstrated in Figure 2. Due to differences in scan extent between patients, the number of usable slices for image registration accuracy assessment was reduced to 14 slices for T1-weighted images and 18 slices for T2-weighted images. More information on the intra- and inter-patient variation models is described by Yu et al. 25.
Figure 2. Workflow of Synthetic Image Generation.
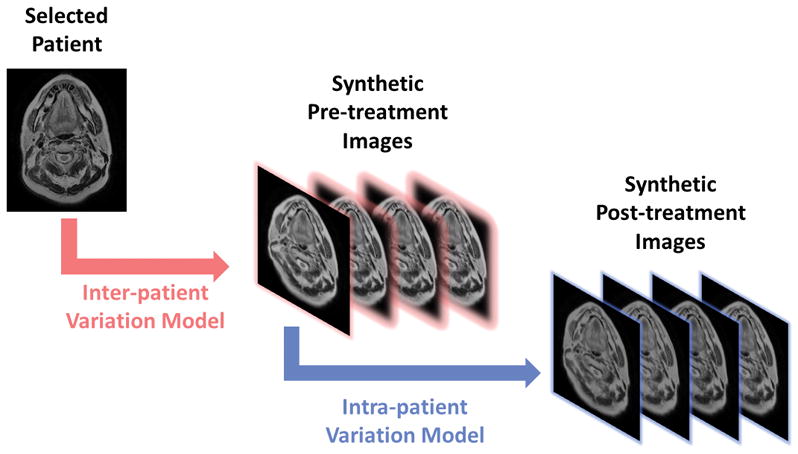
The generation of synthetic images is represented visually. The selected patient’s pre-treatment image is deformed by the inter-patient variation model to generate the synthetic pre-treatment images (highlighted in pink). For each of the synthetic pre-treatment images, four synthetic post-treatment images (highlighted in blue) are generated by deforming the synthetic pre-treatment image using the intra-patient variation model. (Reproduced with permission from Wiley 24.)
C. Computational Tools
The synthetic images are generated using computational tools that are in-house. The methodology of the in-house tool is described in-depth by Yu et al. 25. The deformation vector field used to generate the synthetic post-treatment images is in binary format with an accompanied text file describing the basic information of the vector field. The deformation field data has been decomposed in MATLAB (MathWorks, Natick, MA) for previous studies with synthetic images 24,25 and the MATLAB code for this decomposition has been included for ease of use. There are separate MATLAB scripts for the T1- and T2-weighted images included in the data set.
D. Data Validation
The T1- and T2-weighted MR sequences used for the porcine phantom were derived from patient T1- and T2-weighted sequences used in dynamic contrast enhanced MRI studies conducted at MD Anderson 26,27. These were chosen as they produce typical signal to noise ratios as seen in patient images, thus providing a more accurate estimate of deformable image registration software accuracy in practice.
The synthetic images were created using an established and verified methodology 25. The demons-based registration algorithm, which is a central component to the method, was shown to be accurate for MR images 24.
IV. Data Format and Usage Notes
The data is available on The Cancer Imaging Archive (TCIA) under the collection MRI-DIR (http://dx.doi.org/10.7937/K9/TCIA.2018.3f08iejt) (Data Citation 1). The data upload is separated by porcine phantom and synthetic images. The porcine phantom data is organized by the position of the dividers as described in Section IIIa. The series/study/acquisition date is set to January 1, 1990 for the first position, February 2, 1990 for the second position, March 3, 1990 for the third position, and April 4, 1990 for the fourth position. The series description is set to CT, T1, or T2 to identify the image. TCIA images can have two different structures: (1) ..\DOI\PatientID\StudyInstanceUID\SeriesInstanceUID\xxxxxx.dcm where xxxxxx starts at 000000.dcm and increments by 1 for each image within a series if downloaded from the TCIA wiki page or (2) ..\CollectionName\PatientID\StudyDate-StudyDescription\SeriesDescription\xxxxxx.dcm if downloaded from the Search function within TCIA.
The synthetic image data is similarly organized. The pre-treatment images have a series/study/acquisition date of January 1, 1990 and the post-treatment images have a series/study/acquisition date of February 2, 1990. The series description of the pre-treatment scans is set to T1Pre or T2Pre corresponding to the MR sequence. The post-treatment scans series description is set to T1Post1, T1Post2, T1Post3, and T1Post4 or T2Post1, T2Post2, T2Post3, T2Post4 corresponding to the MR sequence and the synthetic post-treatment image. The patient name and ID are set to T1_1, T1_2, T1_3, T1_4, T2_1, T2_2, T2_3, or T2_4, corresponding to the MR sequence and which synthetic image series is created. There is a DICOM RT structure file for each of the synthetic pre-treatment images located with pre-treatment images. This file contains 5 regions of interest (ROIs) that are identified: left/right parotid gland, left/right submandibular gland, and sublingual gland. The images and DICOM RT structure files when downloaded from TCIA have the same structure as described for the porcine phantom.
There are two deformation vector field data folders labeled T1DVFData and T2DVFData that correspond to the deformation vector field data for the synthetic T1- and T2-weighted images, respectively. Inside each of the deformation vector field folders are folders labeled Post1, Post2, Post3, and Post4. These folders contain a field and header file. There are two sets, designated as D2T or T2D. The D2T files are mapping the pre-treatment image to the post-treatment image (pre-treatment image is the secondary image) and the T2D files are mapping the post-treatment image to the pre-treatment image (pre-treatment image is the primary image). The header file contains the x, y, z dimension of the field file, the x, y, and z voxel dimension (unit: centimeters), the x, y, and z starting point of the reference and secondary location used for the rigid central alignment before deformation is applied (unit: centimeters). The field file stores all deformations along x direction (dx) followed by all deformations along y direction (dy) and all deformation along z direction (dz). In each deformation field block (dx, dy, or dz), the field data is arranged by scanning x direction first, then y direction, and finally z direction. The unit of the field data is the number of voxels. There is a MATLAB file supplied to decompose the T1 and T2 DVF data in their respective folders. The field and header file store data in centimeters while typically deformation vector fields are exported in millimeters from DICOM images. This has been corrected for in the MATLAB file.
V. Discussion
This data set provides two image sets to verify deformable image registration accuracy when using MR images. All images are in DICOM format, therefore, they can be imported to any image registration software that allows this format, e.g. all medical image registration software. These images can be used as a reference check for accuracy of image registration software using MR images. This was done for Velocity (Varian Medical Systems) and an in-house demons-based algorithm 24. Many commercial and in-house systems have been verified using CT images, but public repositories of MR images for testing are sparse. These two different image sets can act as an easily accessible option for researchers when deformable image registration software (whether in-house or commercial) accuracy needs to be checked with MR images before proceeding with other experiments.
A limitation of this data set is that all images were acquired under a specific protocol. The protocol for the patients and porcine phantom was very similar. Changes in acquisition parameters, such as number of averages, and changes in the image noise and artifacts could impact registration accuracy. There are several limitations specific to the patient data set. Firstly, it contains only head and neck patients and had displacements that were usually less than the slice thickness. Therefore, extending the results from the synthetic data alone to another site must be done with caution. Also, only the salivary glands were contoured for this data set. Additionally, the inter- and intra-patient variation models were created using an in-house demons-based algorithm. Other algorithms similar to the one used to create the inter- and intra-patient variation models may be biased in the results of the registration accuracy when using the synthetic images.
There are several limitations pertaining to the porcine phantom: the phantom did not shrink and the deformations are larger than those seen in patients. Not all sites of interest shrink or grow with time, so the lack of volume change in the meat does not impact all evaluations. The deformations that could be applied were limited due to the setup and thus can provide an extreme registration scenario to test registration accuracy with.
VI. Conclusion
The data set contains two separate techniques, synthetic and acquired images, to evaluate deformable image registration accuracy. Each technique contains T1- and T2-weighted images as registration accuracy may differ between the MR image types.
Acknowledgments
Rachel Ger is supported by the Rosalie B. Hite Graduate Fellowship in Cancer Research and the American Legion Auxiliary Fellowship awarded by The University of Texas MD Anderson Cancer Center UTHealth Graduate School of Biomedical Sciences. Carlos Cardenas has been funded by the George M. Stancel PhD Fellowship in the Biomedical Sciences. Dr. Clifton Fuller is a Sabin Family Foundation Fellow. Dr. Fuller receives direct funding and salary support from the National Institutes of Health (NIH), including: the National Institute for Dental and Craniofacial Research Award (1R01DE025248-01/R56DE025248-01); a National Science Foundation (NSF), Division of Mathematical Sciences, Joint NIH/NSF Initiative on Quantitative Approaches to Biomedical Big Data (QuBBD) Grant (NSF 1557679); the NIH Big Data to Knowledge (BD2K) Program of the National Cancer Institute (NCI) Early Stage Development of Technologies in Biomedical Computing, Informatics, and Big Data Science Award (1R01CA214825-01); NCI Early Phase Clinical Trials in Imaging and Image-Guided Interventions Program (1R01CA218148-01); an NIH/NCI Cancer Center Support Grant (CCSG) Pilot Research Program Award from the UT MD Anderson CCSG Radiation Oncology and Cancer Imaging Program (P30CA016672) and an NIH/NCI Head and Neck Specialized Programs of Research Excellence (SPORE) Developmental Research Program Award (P50 CA097007-10). Dr. Fuller receives direct industry grant support and has received travel funding from Elekta AB.
Footnotes
Data Citation
1. Rachel B. Ger, Jinzhong Yang, Yao Ding, Megan C. Jacobsen, Carlos E. Cardenas, Clifton D. Fuller, Rebecca M. Howell, Heng Li, R. Jason Stafford, Shouhao Zhou, Laurence E. Court (2018). Data from Synthetic and Phantom MR Images for Determining Deformable Image Registration Accuracy (MRI-DIR). The Cancer Imaging Archive. http://dx.doi.org/10.7937/K9/TCIA.2018.3f08iejt
References
- 1.Organization for Economic Co-operation and Development. Magnetic resonance imaging (MRI) exams. https://data.oecd.org/healthcare/magnetic-resonance-imaging-mri-exams.htm.
- 2.Caruana EJ, Roman M, Hernandez-Sanchez J, Solli P. Longitudinal studies. Journal of thoracic disease. 2015 Nov;7(11):E537–540. doi: 10.3978/j.issn.2072-1439.2015.10.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohamed AS, Ruangskul MN, Awan MJ, et al. Quality assurance assessment of diagnostic and radiation therapy-simulation CT image registration for head and neck radiation therapy: anatomic region of interest-based comparison of rigid and deformable algorithms. Radiology. 2015 Mar;274(3):752–763. doi: 10.1148/radiol.14132871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirby N, Chuang C, Ueda U, Pouliot J. The need for application-based adaptation of deformable image registration. Medical physics. 2013 Jan;40(1):011702. doi: 10.1118/1.4769114. [DOI] [PubMed] [Google Scholar]
- 5.Wang H, Dong L, Lii MF, et al. Implementation and validation of a three-dimensional deformable registration algorithm for targeted prostate cancer radiotherapy. International journal of radiation oncology, biology, physics. 2005 Mar 01;61(3):725–735. doi: 10.1016/j.ijrobp.2004.07.677. [DOI] [PubMed] [Google Scholar]
- 6.Wang H, Dong L, O’Daniel J, et al. Validation of an accelerated ‘demons’ algorithm for deformable image registration in radiation therapy. Physics in medicine and biology. 2005 Jun 21;50(12):2887–2905. doi: 10.1088/0031-9155/50/12/011. [DOI] [PubMed] [Google Scholar]
- 7.Rigaud B, Simon A, Castelli J, et al. Evaluation of deformable image registration methods for dose monitoring in head and neck radiotherapy. BioMed research international. 2015;2015:726268. doi: 10.1155/2015/726268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castillo R, Castillo E, Guerra R, et al. A framework for evaluation of deformable image registration spatial accuracy using large landmark point sets. Physics in medicine and biology. 2009 Apr 07;54(7):1849–1870. doi: 10.1088/0031-9155/54/7/001. [DOI] [PubMed] [Google Scholar]
- 9.Brock KK, Sharpe MB, Dawson LA, Kim SM, Jaffray DA. Accuracy of finite element model-based multi-organ deformable image registration. Medical physics. 2005 Jun;32(6):1647–1659. doi: 10.1118/1.1915012. [DOI] [PubMed] [Google Scholar]
- 10.Kaus MR, Brock KK, Pekar V, Dawson LA, Nichol AM, Jaffray DA. Assessment of a model-based deformable image registration approach for radiation therapy planning. International journal of radiation oncology, biology, physics. 2007 Jun 01;68(2):572–580. doi: 10.1016/j.ijrobp.2007.01.056. [DOI] [PubMed] [Google Scholar]
- 11.Brock KK, Nichol AM, Menard C, et al. Accuracy and sensitivity of finite element model-based deformable registration of the prostate. Medical physics. 2008 Sep;35(9):4019–4025. doi: 10.1118/1.2965263. [DOI] [PubMed] [Google Scholar]
- 12.Nichol AM, Brock KK, Lockwood GA, et al. A magnetic resonance imaging study of prostate deformation relative to implanted gold fiducial markers. International journal of radiation oncology, biology, physics. 2007 Jan 01;67(1):48–56. doi: 10.1016/j.ijrobp.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 13.Lian J, Xing L, Hunjan S, et al. Mapping of the prostate in endorectal coil-based MRI/MRSI and CT: a deformable registration and validation study. Medical physics. 2004 Nov;31(11):3087–3094. doi: 10.1118/1.1806292. [DOI] [PubMed] [Google Scholar]
- 14.Bruckner T, Lucht R, Brix G. Comparison of rigid and elastic matching of dynamic magnetic resonance mammographic images by mutual information. Medical physics. 2000 Oct;27(10):2456–2461. doi: 10.1118/1.1288241. [DOI] [PubMed] [Google Scholar]
- 15.Schnabel JA, Tanner C, Castellano-Smith AD, et al. Validation of nonrigid image registration using finite-element methods: application to breast MR images. IEEE transactions on medical imaging. 2003 Feb;22(2):238–247. doi: 10.1109/TMI.2002.808367. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Abramson RG, Arlinghaus LR, et al. Data From QIN-Breast. The Cancer Imaging Archive. 2016 [Google Scholar]
- 17.Shah N, Feng X, Lankerovich M, Puchalski RB, Keogh B. The Cancer Imaging Archive. 2016. Data from Ivy GAP. [Google Scholar]
- 18.Newitt D, Hylton N Team obotI-SNaAT. The Cancer Imaging Archive. 2016. Multi-center breast DCE-MRI data and segmentations from patients in the I-SPY 1/ACRIN 6657 trials. [Google Scholar]
- 19.Huang W, Tudorica A, Chui S, et al. Variations of dynamic contrast-enhanced magnetic resonance imaging in evaluation of breast cancer therapy response: a multicenter data analysis challenge. The Cancer Imaging Archive. 2014 doi: 10.1593/tlo.13838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang W, Ryan C, Beckett B, et al. The Cancer Imaging Archive. 2016. Data From QIN-SARCOMA. [Google Scholar]
- 21.Barboriak D. The Cancer Imaging Archive. 2015. Data From RIDER_NEURO_MRI. [Google Scholar]
- 22.Marcus DS, Fotenos AF, Csernansky JG, Morris JC, Buckner RL. Open access series of imaging studies: longitudinal MRI data in nondemented and demented older adults. Journal of cognitive neuroscience. 2010 Dec;22(12):2677–2684. doi: 10.1162/jocn.2009.21407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malone IB, Cash D, Ridgway GR, et al. MIRIAD--Public release of a multiple time point Alzheimer’s MR imaging dataset. NeuroImage. 2013 Apr 15;70:33–36. doi: 10.1016/j.neuroimage.2012.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ger RB, Yang J, Ding Y, et al. Accuracy of deformable image registration on magnetic resonance images in digital and physical phantoms. Medical physics. 2017 Jun 16; doi: 10.1002/mp.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu ZH, Kudchadker R, Dong L, et al. Learning anatomy changes from patient populations to create artificial CT images for voxel-level validation of deformable image registration. Journal of applied clinical medical physics. 2016;17(1):5888. doi: 10.1120/jacmp.v17i1.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooperative JHaNR-MD. Dynamic contrast-enhanced MRI detects acute radiotherapy-induced alterations in mandibular microvasculature: prospective assessment of imaging biomarkers of normal tissue injury. Scientific reports. 2016;6:29864. doi: 10.1038/srep29864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ger RB, Mohamed ASR, Awan MJ, et al. A Multi-Institutional Comparison of Dynamic Contrast-Enhanced Magnetic Resonance Imaging Parameter Calculations. Scientific reports. 2017 Sep 11;7(1):11185. doi: 10.1038/s41598-017-11554-w. [DOI] [PMC free article] [PubMed] [Google Scholar]


