Abstract
lynch syndrome (LS), an autosomal dominantly inherited disease previously known as hereditary non-polyposis colorectal cancer (HNPCC), leads to a high risk of colorectal cancer (CRC) as well as malignancy at certain sites including endometrium, ovary, stomach, and small bowel (Hampel et al., 2008; Lynch et al., 2009). Clinically, LS is considered the most common hereditary CRC-predisposing syndrome, accounting for about 3% of all CRC cases (Popat et al., 2005). LS is associated with mutations of DNA mismatch repair (MMR) genes such as MLH1, MSH2, MSH6, PMS2, and EPCAM (Ligtenberg et al., 2009; Lynch et al., 2009), which can trigger a high frequency of replication errors in both microsatellite regions and repetitive sequences in the coding regions of various cancer-related genes. Immunohistochemistry (IHC) tests followed by genetic analysis of these mutations play a significant role in diagnosis, treatment determination, and therapeutic response prediction of LS (Lynch et al., 2009; Alex et al., 2017; Ryan et al., 2017). Here, we report substitution of one base-pair in exon 1 of MLH3 (c.1397C>A) and a frameshift mutation in exon 19 of MLH1 (c.2250_2251ins AA) in a 43-year-old Chinese male with an LS pedigree.
Keywords: Lynch syndrome, DNA mismatch repair, Frameshift mutation
Lynch syndrome (LS), an autosomal dominantly inherited disease previously known as hereditary non-polyposis colorectal cancer (HNPCC), leads to a high risk of colorectal cancer (CRC) as well as malignancy at certain sites including endometrium, ovary, stomach, and small bowel (Hampel et al., 2008; Lynch et al., 2009). Clinically, LS is considered the most common hereditary CRC-predisposing syndrome, accounting for about 3% of all CRC cases (Popat et al., 2005). LS is associated with mutations of DNA mismatch repair (MMR) genes such as MLH1, MSH2, MSH6, PMS2, and EPCAM (Ligtenberg et al., 2009; Lynch et al., 2009), which can trigger a high frequency of replication errors in both microsatellite regions and repetitive sequences in the coding regions of various cancer-related genes. Immunohistochemistry (IHC) tests followed by genetic analysis of these mutations play a significant role in diagnosis, treatment determination, and therapeutic response prediction of LS (Lynch et al., 2009; Alex et al., 2017; Ryan et al., 2017). Here, we report substitution of one base-pair in exon 1 of MLH3 (c.1397C>A) and a frameshift mutation in exon 19 of MLH1 (c.2250_2251ins AA) in a 43-year-old Chinese male with an LS pedigree.
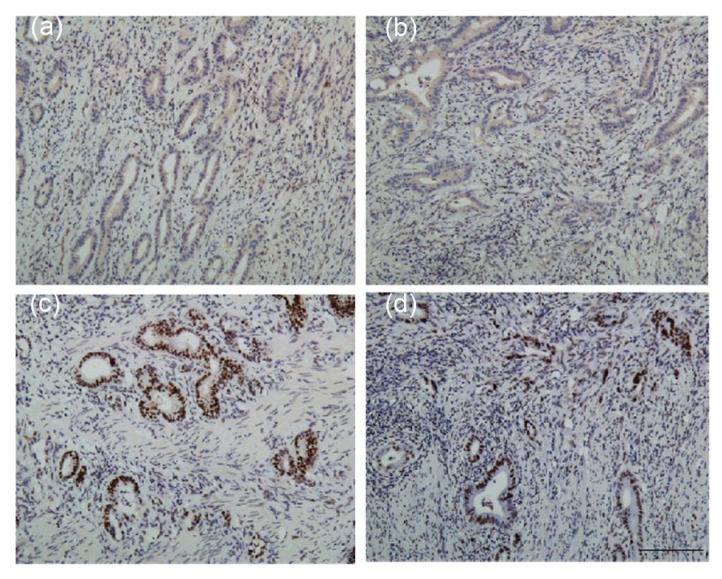
The patient was diagnosed with ascending colon cancer, receiving a laparoscopic right hemicolectomy at the age of 43 years in our hospital. His mother was diagnosed with endometrial cancer at the age of 58 years and his maternal grandmother suffered from colorectal cancer and died in her 60 years. His maternal uncle was diagnosed with colon cancer at the age of 50 years. As his family history fulfilled the Amsterdam II criteria for LS, we took a sample of his tumor for IHC analysis, which revealed defects in MLH1 and PMS2 proteins (Figs. 1a and 1b).
Fig. 1.
IHC examination of MMR protein expression in tumor sections
IHC examination revealed loss of MLH1 (a) and PMS2 (b) expression in tumor cells, while no loss of MSH2 (c) or MSH6 (d) expression was observed. Scale bar=200 μm. IHC: immunohistochemistry; MMR: mismatch repair
We then undertook genetic analysis of genes related to hereditary CRC to further characterize the mutations. Next generation sequencing was performed by polymerase chain reaction (PCR)-direct sequencing analysis, targeting genes including APC, AXIN2, EPCAM, MLH1, MLH3, MSH2, MSH6, MUTYH, PMS1, PMS2, STK11, PTEN, SMAD4, and BMPR1. Results were compared with the “1000 Genomes” browser (https://www.ncbi.nlm.nih.gov/variation/tools/1000genomes) and the pathogenicity of mutations was classified according to the recommendations of the American College of Medical Genetics and Genomics (ACMG; http://www.acmg.net). After excluding promoter methylation, the analysis showed a heterozygous substitution of one base-pair (c.1397C>A) in exon 1 of MLH3, whose transcript variant (p.Ser466Ter) leads to a protein truncated at codon 465 instead of codon 1429 in the wild type. Also, a heterozygous frameshift mutation in exon 19 of MLH1 (c.2250_2251insAA) was detected, which created a new reading frame at Val 752, encountering a premature stop codon at the following 32nd position (p.Val752L-ysfs*32) and eventually a protein of 782 codons instead of 757 in the wild type (Fig. S1). Both mutations were found to be dominant. Comparisons with previous findings from the “1000 Genomes” browser and ACMG recommendations showed that the mutation in MLH3 had not been previously reported. Therefore, we considered it to be a suspected pathogenic mutation. Although not previously reported as pathogenic, similar MLH1 nucleotides, detected in a South American LS pedigree (c.2252_2253dup-AA) (Dominguez-Valentin et al., 2013) and a Chinese pedigree (Sheng et al., 2008), were recognized as variants of unclassified significance (VUS).
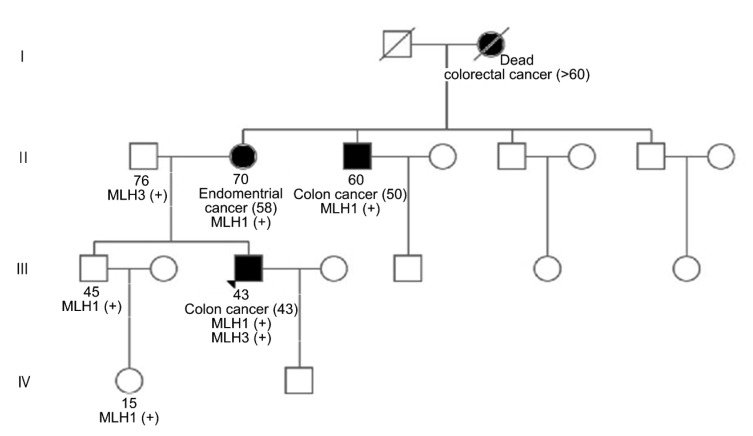
To confirm the germline mutation in those genes as well as the pathogenic effect of the MLH3 variants, we provided genetic counseling for the patient’s family members via Sanger sequencing, and recommended genetic analysis. The analysis showed that the germline mutation in MLH1 was carried by the patient’s brother, niece, and mother (Fig. S2a), while the same MLH3 variant was detected only in the patient’s father who was never diagnosed with a tumor (Fig. S2b). These results suggested that the mutation in MLH1 weighed more in the LS etiology of the proband. Genetic analysis of the maternal uncle with colon cancer was not carried out due to personal refusal. The pedigree chart is shown in Fig. 2.
Fig. 2.
Pedigree chart of the family with Lynch syndrome
Black symbols indicate individuals diagnosed with cancers, with slashes indicating death. Ages upward mean the current age (years), while ages in the brackets indicate diagnosed age (years). A plus symbol indicates the mutation in the mismatch repair (MMR) genes. The triangle indicates the proband. Square: male; Circle: female
Genetic linkage analysis showed that 80% of LS cases are related to germline mutations in MSH2 and MLH1 (Alex et al., 2017), whereas the mutation we found in MLH1, previously reported in only one pedigree of a South American, was predicted to be a VUS by multiple in silico tools (Dominguez-Valentin et al., 2013). Germline mutation was revealed in the maternal pedigree, especially in patients with LS involving malignancy, which supports its etiological role according to the law of segregation. Also, deficiencies in both MLH1 and PMS2 were proved by IHC tests in the proband. We believe that an abnormal C-terminus in the MLH1 protein caused by such a mutation could provide an explanation. It was reported that the heterodimer of MLH1 combined with PMS2 serves as a major effector of MMR function in vivo, and the participation of the carboxy terminal homology (CTH) domain (residues 500–756 amino acids in MLH1) in the structure is crucial for its stabilization (Mohd et al., 2006). Moreover, mutations truncating or expanding MLH1 peptide are related to the loss of such domains (Kim et al., 2009), theoretically triggering the absence of MLH1 and PMS2. For these reasons, the MLH1 variant carried by the proband, extending the peptide and affecting the structure of the CTH domain, was supposed to destroy the stability of the heterodimer, causing MMR deficiency and eventually LS. Nevertheless, previous studies reported that patients with a defective MMR had a mean age of 45 years (Lynch et al., 2009), while patients in our study carrying only the MLH1 mutation were diagnosed after their 50 years. Further studies concerning the significance of similar variants are required.
We had suspected that the mutation in MLH3 plays an important role in tumorigenicity, but the results from genetic analysis suggested the contrary. The father lived tumor-free as a carrier of MLH3 mutation alone, which did not support individual pathogenicity. Clinically, there have been few reports concerning tumor predisposition with only MLH3 mutations (Hienonen et al., 2003; Mohd et al., 2006), providing little evidence. Also, Korhonen et al. (2008) functionally characterized seven missense mutations of MLH3, and disproved their individual function of interfering with MMR, describing them as low-risk mutations for LS.
Although the mutation in MLH1 was considered pathogenic in our case, we still believed that MLH3 variant made a difference. The proband was diagnosed with colon cancer at the age of 43 years, much younger than the other patients in his family. In addition, malignancy has not yet been diagnosed in his 45-year-old brother carrying MLH1 variant. Therefore, it is possible that MLH3 deficiency enhanced the tumorigenicity of MLH1 mutation and accelerated the occurrence of the tumor. According to the study of Chen et al. (2005), MLH3, following PMS2, takes part in tumor suppression in mice with MLH1. Also, Chen et al. (2008) showed that a loss of MLH3 accelerates the progression of gastrointestinal tumors on the basis of PMS2 deficiency. We believe that the compound heterozygote of these germline mutations is more likely to trigger LS. Further studies are needed to provide more conclusive evidence, as this is the first time that this phenomenon has been detected.
In conclusion, we identified a mutation in exon 19 of MLH1 and a novel germline mutation in exon 1 of MLH3. The mutation in MLH1, although described previously as a VUS, was detected in the maternal pedigree and possibly resulted in LS. MLH3 variant was not detected among other patients of the maternal pedigree, but we still believe it may have enhanced tumorigenicity in this case.
List of electronic supplementary materials
Sequencing of the proband
Sequencing of the patients of the Lynch syndrome pedigree
Footnotes
Contributors: Qiao-qi SUI and Wu JIANG wrote this report and collected references. Xiao-dan WU drew the figure of genetic reports and pedigree chart. Yi-hong LING diagnosed defective MMR and drew the figure of IHC examination. Pei-rong DING wrote an outline. Zhi-zhong PAN checked and approved the final version.
Electronic supplementary materials: The online version of this article (https://doi.org/10.1631/jzus.B1800105) contains supplementary materials, which are available to authorized users
Compliance with ethics guidelines: Qiao-qi SUI, Wu JIANG, Xiao-dan WU, Yi-hong LING, Zhi-zhong PAN, and Pei-rong DING declare that they have no conflict of interest.
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2008 (5). Informed consent was obtained from all patients for being included in the study. Additional informed consent was obtained from all patients for whom identifying information is included in this article.
References
- 1.Alex AK, Siqueira S, Coudry R, et al. Response to chemotherapy and prognosis in metastatic colorectal cancer with DNA deficient mismatch repair. Clin Colorectal Cancer. 2017;16(3):228–239. doi: 10.1016/j.clcc.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Chen PC, Dudley S, Hagen W, et al. Contributions by mutl homologues Mlh3 and Pms2 to DNA mismatch repair and tumor suppression in the mouse. Cancer Res. 2005;65(19):8662–8670. doi: 10.1158/0008-5472.CAN-05-0742. [DOI] [PubMed] [Google Scholar]
- 3.Chen PC, Kuraguchi M, Velasquez J, et al. Novel roles for MLH3 deficiency and TLE6-like amplification in DNA mismatch repair-deficient gastrointestinal tumorigenesis and progression. PLoS Genet. 2008;4(6):e1000092. doi: 10.1371/journal.pgen.1000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dominguez-Valentin M, Nilbert M, Wernhoff P, et al. Mutation spectrum in south american lynch syndrome families. Hered Cancer Clin Pract. 2013;11(1):18. doi: 10.1186/1897-4287-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hampel H, Frankel WL, Martin E, et al. Feasibility of screening for lynch syndrome among patients with colorectal cancer. J Clin Oncol. 2008;26(35):5783–5788. doi: 10.1200/JCO.2008.17.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hienonen T, Laiho P, Salovaara R, et al. Little evidence for involvement of MLH3 in colorectal cancer predisposition. Int J Cancer. 2003;106(2):292–296. doi: 10.1002/ijc.11218. [DOI] [PubMed] [Google Scholar]
- 7.Kim KH, Kim JY, Oh SI, et al. A novel germline mutation of hMLH1 in a korean hereditary non-polyposis colorectal cancer family. Int J Oncol. 2009;34(5):1313–1318. doi: 10.3892/ijo_00000258. [DOI] [PubMed] [Google Scholar]
- 8.Korhonen MK, Vuorenmaa E, Nyström M. The first functional study of MLH3 mutations found in cancer patients. Genes Chromosomes Cancer. 2008;47(9):803–809. doi: 10.1002/gcc.20581. [DOI] [PubMed] [Google Scholar]
- 9.Ligtenberg MJL, Kuiper RP, Chan TL, et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3' exons of TACSTD1 . Nat Genet. 2009;41(1):112–117. doi: 10.1038/ng.283. [DOI] [PubMed] [Google Scholar]
- 10.Lynch HT, Lynch PM, Lanspa SJ, et al. Review of the lynch syndrome: history, molecular genetics, screening, differential diagnosis, and medicolegal ramifications. Clin Genet. 2009;76(1):1–18. doi: 10.1111/j.1399-0004.2009.01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohd AB, Palama B, Nelson SE, et al. Truncation of the C-terminus of human MLH1 blocks intracellular stabilization of PMS2 and disrupts DNA mismatch repair. DNA Repair. 2006;5(3):347–361. doi: 10.1016/j.dnarep.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23(3):609–618. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 13.Ryan E, Sheahan K, Creavin B, et al. The current value of determining the mismatch repair status of colorectal cancer: a rationale for routine testing. Crit Rev Oncol Hematol. 2017;116:38–57. doi: 10.1016/j.critrevonc.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 14.Sheng JQ, Fu L, Sun ZQ, et al. Mismatch repair gene mutations in Chinese HNPCC patients. Cytogenet Genome Res. 2008;122(1):22–27. doi: 10.1159/000151312. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequencing of the proband
Sequencing of the patients of the Lynch syndrome pedigree