Summary
The plasticity of developing tissues relies on the adjustment of cell survival and growth rate to environmental cues. This includes the effect of mechanical cues on cell survival. Accordingly, compaction of an epithelium can lead to cell extrusion and cell death. This process was proposed to contribute to tissue homeostasis but also to facilitate the expansion of pretumoral cells through the compaction and elimination of the neighboring healthy cells. However, we know very little about the pathways that can trigger apoptosis upon tissue deformation, and the contribution of compaction-driven death to clone expansion has never been assessed in vivo. Using the Drosophila pupal notum and a new live sensor of ERK, we show first that tissue compaction induces cell elimination through the downregulation of epidermal growth factor receptor/extracellular signal regulated kinase (EGFR/ERK) pathway and the upregulation of the pro-apoptotic protein Hid. Those results suggest that the sensitivity of EGFR/ERK pathway to mechanics could play a more general role in the fine tuning of cell elimination during morphogenesis and tissue homeostasis. Second, we assessed in vivo the contribution of compaction-driven death to pretumoral cell expansion. We found that the activation of the oncogene Ras in clones can downregulate ERK and activate apoptosis in the neighboring cells through their compaction, which eventually contributes to Ras clone expansion. The mechanical modulation of EGFR/ERK during growth-mediated competition for space may contribute to tumor progression.
Keywords: mechanotransduction, apoptosis, cell competition, EGFR, ERK, Hid, epithelium, Drosophila
Graphical Abstract
Highlights
-
•
Caspase activity in Drosophila pupal notum is regulated by EGFR/ERK and hid
-
•
EGFR/ERK can be activated or downregulated by tissue stretching or compaction
-
•
Cell compaction near fast-growing clones downregulates ERK and triggers cell death
-
•
Compaction-driven ERK downregulation promotes fast-growing clone expansion
Moreno et al. show that cell elimination in the Drosophila pupal notum can be locally adjusted by tissue deformation through the modulation of EGFR/ERK pathway and the pro-apoptotic gene hid. Compaction-driven ERK downregulation also occurs near fast-growing clones and promotes clone expansion through neighboring cell elimination.
Introduction
Developing tissues can cope with perturbations, including stress from the environment or abnormal behaviors of a subset of cells. This robustness relies on the plasticity of cell behavior and/or fate, which can adjust to changes in the tissue environment. Modulation of cell proliferation and cell death can be driven by contact-dependent communication, extracellular diffusive factors, and/or mechanical inputs [1]. Accordingly, mechanical cues have been proposed to adjust the local rate of cell death and cell division to regulate tissue final size or to maintain it during homeostasis [2, 3, 4].
The regulation of cell death by mechanical inputs is well documented in epithelia. Epithelial compaction can induce cell extrusion and cell death in MDCK cells, zebrafish epidermis [5, 6, 7], and in the midline region of the Drosophila pupal notum (a single layer epithelium; Figure 1A) [8]. Recently, we showed that compaction-driven cell elimination in the pupal notum relies on caspase activation, which is required for and precedes every extrusion event [9]. Thus, some pathways must be sensitive to tissue deformations and trigger and/or modulate caspase activation. However, we could not find a clear contribution of known mechanosensitive pathways to midline cell elimination, including p53 [7], the JNK pathway [10], or the Hippo Yap/Taz pathway [9, 11]. Moreover, it also suggested that cells could have differential sensitivity to compaction depending on their sensitivity to apoptosis. Accordingly, activation of Ras in clones led to the preferential compaction and elimination of the neighboring wild-type (WT) cells [9]. Similarly, the high levels of p53 in mutant MDCK cells for the polarity gene scribble increase their sensitivity to compaction and trigger their elimination when surrounded by WT MDCK cells [7, 12]. Those eliminations have been proposed to promote the expansion of pretumoral cells through a so-called mechanical cell competition [7, 9, 13, 14]. However, the molecular pathway triggering cell death during mechanical cell competition in vivo was not yet identified, and it was not yet clear whether such elimination could significantly promote pretumoral clone expansion.
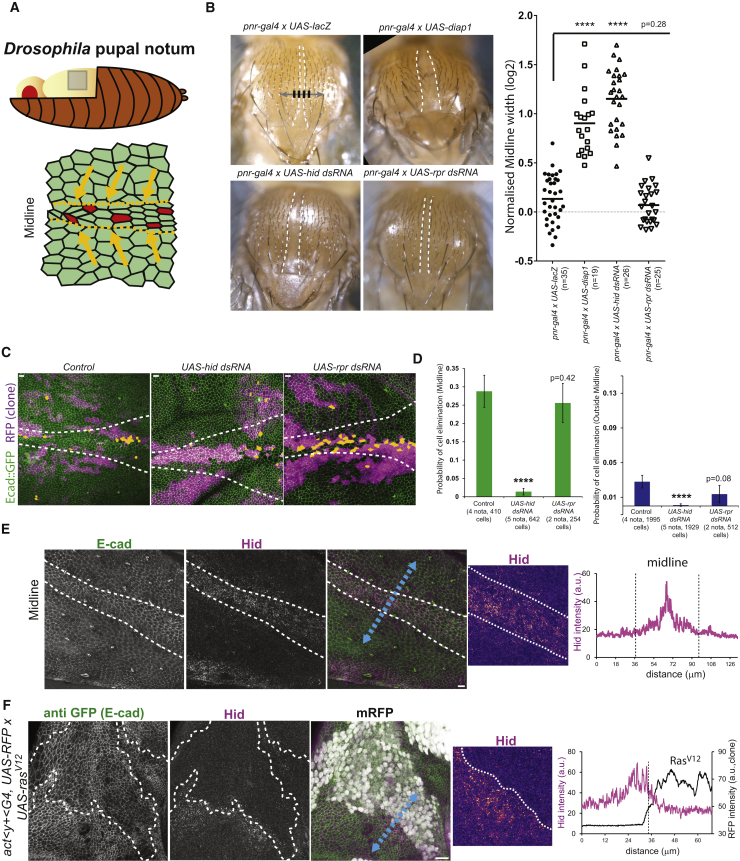
Figure 1.
Hid Is Required for Cell Elimination
(A) Schematic of the pupal notum and the midline (bottom). Orange arrows, compaction; red cells, caspase-activated cells.
(B) Adult thorax upon perturbation of cell death in the pnr domain. White dashed lines, midline. Black lines are used to measure the relative midline width (see STAR Methods). Right graph: normalized midline width is shown (log2 scale; one point = 1 thorax); t test with control; ∗∗∗∗p < 10−4.
(C) Live pupal nota expressing ubi-Ecad::GFP (green) with Gal4-expressing clones (RFP, magenta) in controls (ayG4 alone) or expressing UAS-hid dsRNA or UAS-rpr dsRNA (white dashed lines: midline). Orange cells: clonal cells that will die. Scale bars represent 10 μm.
(D) Probability of cell elimination in clones in the midline (left) and outside the midline (right). Fisher exact test with the control; ∗∗∗∗p < 10−4. Error bars indicate 95% confidence interval.
(E) Immunostaining of a pupal notum, z-projection of anti-E-cad (green), and anti-Hid (magenta) in the midline (white dashed line; 7/7 nota). Close-up view of Hid intensity in the midline in pseudocolor is shown in the right panel. Right graph: intensity profile of Hid along the blue dashed line (magenta) is shown. Scale bar represents 10 μm.
(F) Immunostaining of a pupal notum showing z-projection of anti-GFP (E-cad::GFP, green), anti-Hid (magenta), and upstream activating sequence (UAS)-nlsRFP signal (white) in vicinity of a clone where Ras was conditionally activated (UAS-rasV12; 24 hr activation at 29°C; white dashed lines: clone boundaries); 4/4 nota. Right: close up view of Hid intensity near the clone in pseudocolor is shown. Right graph: intensity profile of Hid along the blue dashed line (magenta) and RFP (clone, black line) is shown. Scale bar indicates 10 μm.
See also Video S1 and STAR Methods.
Here, we show that tissue compaction induces cell elimination in the pupal notum through the downregulation of epidermal growth factor receptor/extracellular signal regulated kinase (EGFR/ERK) pathway and the upregulation of the pro-apoptotic protein Hid (head involution defective). Using a new Drosophila live sensor of ERK activity, we demonstrate that local tissue stretching or compaction transiently upregulate or downregulate ERK activity, hence increasing or decreasing cell survival. Moreover, we show that compaction-driven ERK downregulation near Ras-activated clones controls cell elimination and promotes clone expansion. The sensitivity of EGFR/ERK pathway to mechanics and its role in the fine tuning of cell elimination could play a more general role during tissue homeostasis and tumor progression.
Results
Cell Elimination in the Pupal Notum Is Regulated by Hid
We previously showed that a deletion covering the three pro-apoptotic genes hid, grim, and reaper (H99 deletion) strongly downregulated cell extrusion in the pupal notum [9]. Downregulation of hid by RNAi in the pupal notum (using pnr-gal4 driver) led to a significant widening of the midline in the adult fly thorax (a zone with a high rate of cell elimination) [8, 9, 15], similar to apoptosis downregulation through diap1 overexpression (Figure 1B). Moreover, downregulation of hid in clones strongly reduced their rate of cell extrusion inside or outside the midline (Figures 1C and 1D; Video S1), phenocopying the defects observed with the H99 deletion [9]. Accordingly, rpr downregulation had no significant effect on thorax morphology and clone survival (Figures 1B–1D; Video S1). Thus, hid must be responsible for the defects observed in H99 deletion. Finally, Hid protein also accumulated in regions showing a high rate of cell elimination, including the midline (Figure 1E) and in crowded regions near RasV12 clones (Figure 1F). Altogether, this suggests that Hid is a central regulator of cell death in the pupal notum and that it can be upregulated in crowded regions.
z projections of 16–18 hr APF pupal nota expressing ubi-Ecad::GFP (green) and Gal4 UAS-RFP clones (magenta) in control (UAS-lacZ, left), upon Hid downregulation in clones (UAS-hid dsRNA, middle), Rpr downregulation in clones (UAS-rpr dsRNA, right), overexpressing Bantam sponge (UAS-bantam sponge), an active from of PI3K (UAS-pi3kCA, middle), an active form of Raf (UAS-rafCA, right), EGFR downregulation in clones (UAS-egfr dsRNA) and upon overexpression of hEGFR::GFP (UAS-EGFR::GFP, clones are marked by the strong membrane GFP signal, note that the movie was filtered to obtain similar intensity ranges for E-cad and EGFR::GFP in the clones). The midline is encompassed by the white lines and the extruding cells in the clone are marked in yellow at the beginning of the movie. Anterior on the left, posterior on the right. Scale bars, 10 μm.
Cell Survival in the Midline Is Regulated by EGFR/Ras/Raf/ERK Pathway
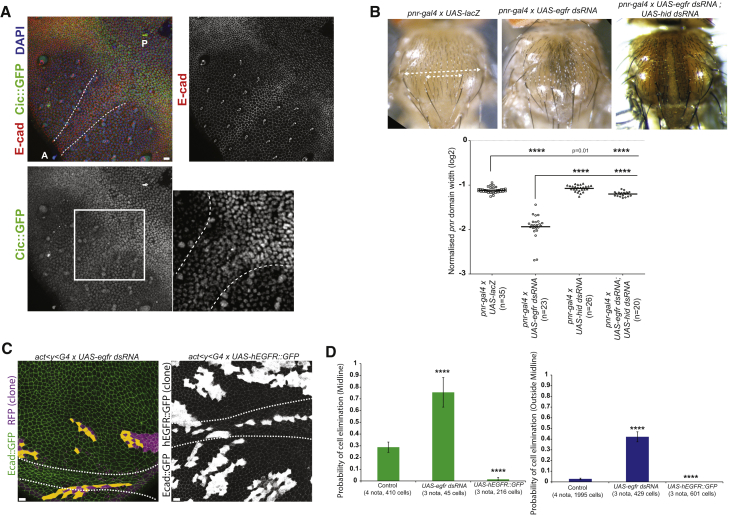
We next tried to identify the regulators of Hid in the midline of the notum. Hid can be upregulated by the steroid hormone ecdysone [16], downregulated by the pro-survival microRNA bantam [17], and downregulated by the EGFR/Ras/Raf/ERK pathway [18, 19]. There was no accumulation of ecdysone receptor (Figure S1A) or a downregulation of bantam in the midline (Figure S1B). Moreover, overexpression of a Bantam sponge in clones (which sequesters bantam microRNA [17] and reduces bantam activity; Figure S1C) did not affect cell elimination (Figures S1D and S1E; Video S1). Thus, ecdysone and bantam are not responsible for the upregulation of Hid in the midline. Interestingly, we observed a relative downregulation of ERK activity in the midline (Figures S1F and 2A). As such, we tried to identify the upstream signal regulating ERK activity in the midline. EGFR downregulation led to a narrowing of the pnr domain in the adult thorax (Figure 2B), which could be rescued by downregulation of hid (Figure 2B). Therefore, EGFR is required for cell survival in the pupal notum through hid downregulation. This effect was specific of EGFR, as we did not observe such narrowing upon Pvr/PDGF, FAK, Src42A, and Src64B downregulation (Figure S2A), other putative regulators of ERK [21, 22, 23]. We then tested the impact of EGFR on cell survival in the notum. Downregulation of EGFR in clones led to a strong increase of cell elimination everywhere in the notum, and overexpression of a chimeric EGFR (hEGFR::GFP, which can activate ERK independently of extracellular ligands) [24] nearly abolished cell elimination (Figures 2C and 2D; Video S1; ERK activity shown in Figure 3C). EGFR/Ras could promote cell survival through the phosphatidylinositol 3-kinase (PI3K) pathway or the Raf/mitogen-activated protein kinase (MAPK) pathway [25]. However, activation of PI3K in clones had little impact on the rate of cell elimination (Figure S2B; Video S1) and overexpression of an active form of Raf strongly abolished cell elimination (Figure S2C; Video S1). Altogether, we conclude that EGFR/Ras/Raf/ERK pathway is a central regulator of cell survival in the pupal notum.
Figure 2.
EGFR/ERK Downregulation Is Necessary and Sufficient for Cell Elimination
(A) Anti-E-cad (red), DAPI (blue), and anti-GFP (cic-Cic::GFP, BAC clone, green, negatively regulated by ERK) [20] in the midline (white dashed line; 7/7 nota); higher magnification shown on the right (white square region). 5/5 nota are shown. Scale bar represents 10 μm. A, anterior; P, posterior.
(B) Adult thorax in control (pnr-gal4 × UAS-lacZ) upon EGFR downregulation (pnr-gal4 × UAS-EGFR dsRNA) and downregulation of EGFR and Hid (pnr-gal4 × UAS-EGFR dsRNA; UAS-hid dsRNA). White dashed lines: pnr domain width and total thorax width (see STAR Methods). (Bottom graph) Normalized pnr domain width is shown (log2 scale; one point = 1 thorax); t tests; ∗∗∗∗p < 10−4.
(C) Live pupal nota expressing ubi-Ecad::GFP (green) with Gal4-expressing clones where EGFR is downregulated (UAS-egfr dsRNA, left; clones in magenta) or upregulated (UAS-hEGFR::GFP; H. sapiens extracellular domain and Drosophila intracellular domain; clones marked in white). White dashed lines: midline. Orange cells: clonal cells that will die. See Figure 1C for control. Scale bars represent 10 μm.
(D) Probability of cell elimination in the clones in the midline (top) and outside the midline (bottom). Fisher exact test with the control condition (same as Figures 1C and 1D); ∗∗∗∗p < 10−4. Error bars indicate 95% confidence interval.
See also Figures S1 and S2, Video S1, and STAR Methods.
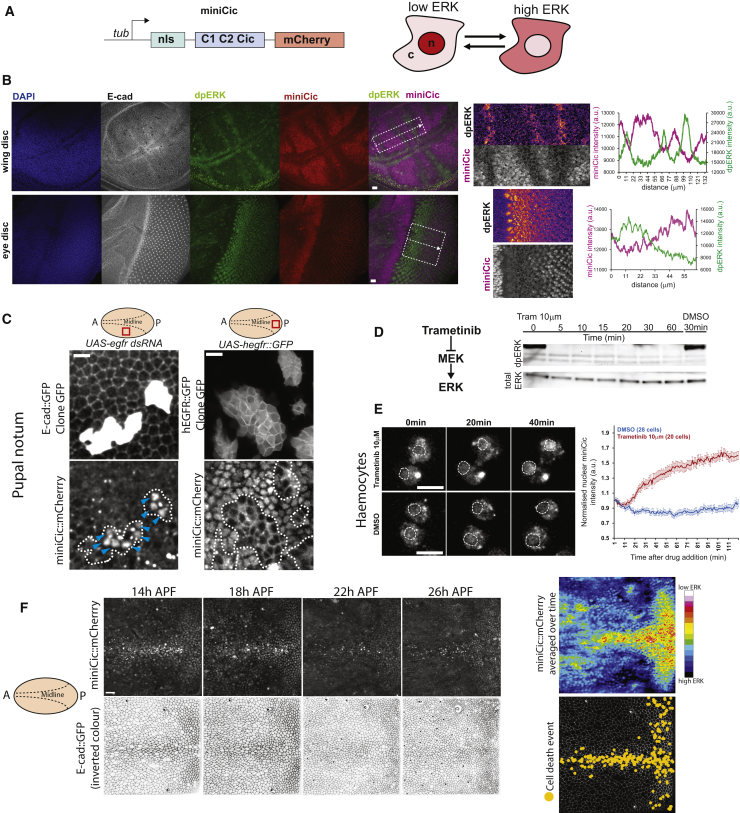
Figure 3.
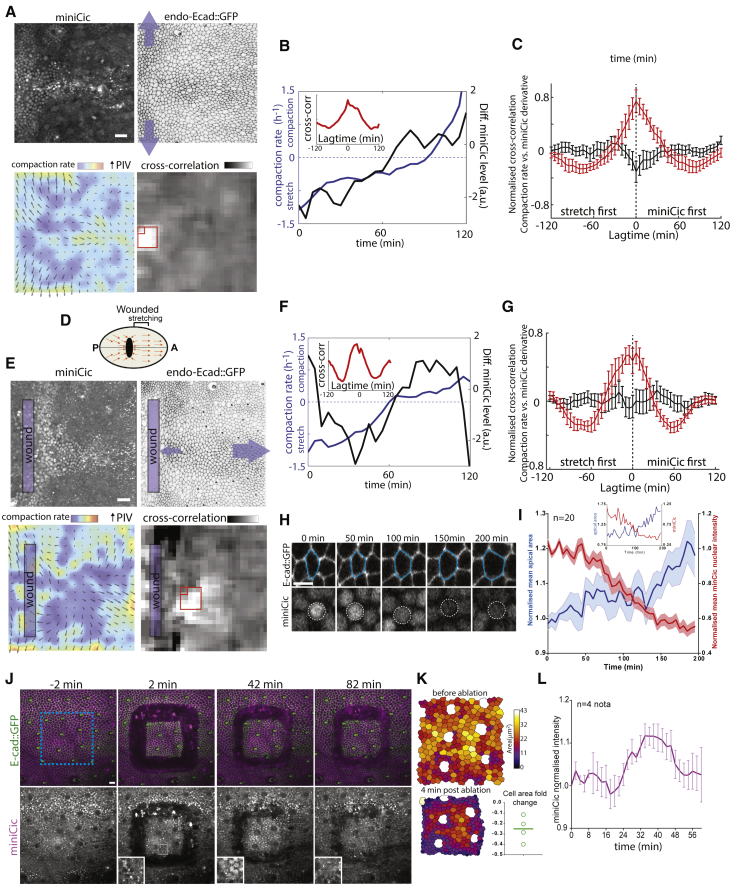
A New Live Sensor of ERK
(A) Schematic of the miniCic construct. Right cartoon: expected localization of miniCic at low ERK activity (accumulation in nucleus, n) and high ERK activity (exclusion in the cytoplasm, c) is shown.
(B) z-projection of immunostainings of a wing imaginal disc (top) and an eye imaginal disc (bottom) at the L3 wandering stage (10/10 tissues for each) stained for DAPI (blue), E-cad (white), dpERK (green), and miniCic (anti-mCherry, red; purple on the overlay). Scale bars represent 10 μm. Close-up views (dashed white rectangles) are shown on the right (single plane; dpERK pseudocolor; miniCic greyscale); graphs: intensity line profiles of dpERK (green) and miniCic (purple) along white dashed rectangles.
(C) Live pupal nota expressing miniCic and endo-Ecad::GFP with EGFR-depleted clones (left, green GFP; UAS-egfr dsRNA) or clones overexpressing hEGFR (right, UAS-hegfr::GFP). Top schemes: localization of the clones in the notum is shown (red rectangles; A, anterior; P, posterior; dashed lines, midline; zone of high ERK activity on the left and low ERK activity for the right). Blue arrows: ectopic nuclear accumulation. White dashed lines: clone boundaries. Scale bars represent 10 μm.
(D) Western blot of dpERK (top) and total ERK (bottom) in S2 cells upon inhibition of ERK phosphorylation by trametinib (10 μM; a potent inhibitor of MEK, left scheme) at different time after drug treatment (in minutes). Control band: DMSO treatment (30 min after adding DMSO). Similar results were obtained at 1 μM.
(E) Live imaging of larval haemocytes primary culture expressing tub-miniCic upon treatment with 10 μM trametinib (top) or DMSO (bottom). Time (minutes) after drug deposition is shown. Dotted circles: nuclei (detected by transmitted light). Scale bars represent 10 μm. (Right graph) Mean normalized miniCic nuclear intensity after drug treatment (two independent experiments) is shown. Error bars are SEM.
(F) Snapshots of a live pupal notum expressing endo-Ecad::GFP and miniCic (local z-projection) at different time after pupal formation (APF). Anterior: left; posterior: right; midline in the center (see left scheme). Scale bar represents 20 μm. Right heatmap: averaged miniCic signal over the full movie is shown; cell death events over the full movie are shown (bottom right picture; one dot = one elimination).
Development of a New Live Sensor of ERK
We then asked whether ERK downregulation was playing an instructive role for caspase activation rather than a purely permissive function. To do so, we developed a new live sensor of ERK activity. Phosphorylation of the transcriptional repressor Capicua (Cic) by ERK leads to its removal from the nucleus [20]. We used a minimal domain of Cic lacking its DNA binding domain and containing the phosphorylation and docking site of ERK [20] fused to a nuclear localization sequence (NLS) and mCherry under the control of the tubulin promoter (Figure 3A; hereafter named miniCic). We expected to observe a nuclear accumulation of miniCic upon ERK downregulation and an exclusion from the nucleus upon ERK activation. miniCic nuclear intensity was indeed anticorrelated with the endogenous levels of active ERK in the wing and eye imaginal discs (Figure 3B; note the gradient in the eye disc) [26] and in the embryos (Figure S3A) with a clear exclusion from the nucleus in active regions (Figure 3B, right). Thus, miniCic is suitable for quantitative measurements of endogenous levels of ERK activity. MiniCic also accurately reported modulations of EGFR/ERK activity in the pupal notum by expressing EGFR double-stranded RNA (dsRNA) or hEGFR::GFP in clones (Figure 3C). We then assessed the potential time delay between ERK modulation and miniCic relocalization. Inhibition of ERK through the MEK inhibitor trametinib [27] led to rapid disappearance of dpERK in S2 cells (Figure 3D; 5 min after adding drug at 10 μM) and a rapid nuclear accumulation of miniCic in haemocytes (Figure 3E; Video S2A; significant increase 10 min after adding drug). Moreover, the fast induction of dpERK in the lateral ectoderm of cellularizing embryos [28] was also captured by miniCic, except for a short developmental time window of 10–15 min (Figure S3A). Altogether, this showed that miniCic could be used to assess the variations of endogenous ERK activity with a maximum lag time of 10–15 min.
(A) Primary culture of haemocytes expressing miniCic::mCherry upon treatment with 10 μM Trametinib (MEK inhibitor, left) or DMSO (right) at the onset of the movie. Scale bar, 10 μm. (B) Local z-projections of a live pupal notum expressing endo-Ecad::GFP (green) and miniCic (magenta and right). Time is indicated in hours after pupal formation. Scale bar, 10 μm. (C) Local projection of two nuclei of the midline in a living pupal notum expressing miniCic (left) and Scat3 (FRET signal, right, red-orange = low caspase activity, blue-purple = high caspase activity). Scale bar, 10 μm.
We then used this new sensor to visualize the dynamics of ERK in the pupal notum and check whether it could reflect the pattern of ERK activity we previously described by immunostaining (Figures 2A and S1F). As expected, we observed an accumulation of miniCic (low ERK activity) mostly in the midline and in an orthogonal domain in the posterior part of the notum (Figure 3F; Video S2B), both corresponding to zones of high cell death rate (Figure 3F) [9, 15]. The local dynamics of miniCic correlated with the temporal and spatial distribution of cell elimination, which occurred first mostly in the midline and then in the posterior region (Figures S3B–S3D; Video S2B). This suggested again that ERK dynamics and levels correlated with cell death distribution in the notum.
Reduction of ERK Activity Precedes Caspase Activation
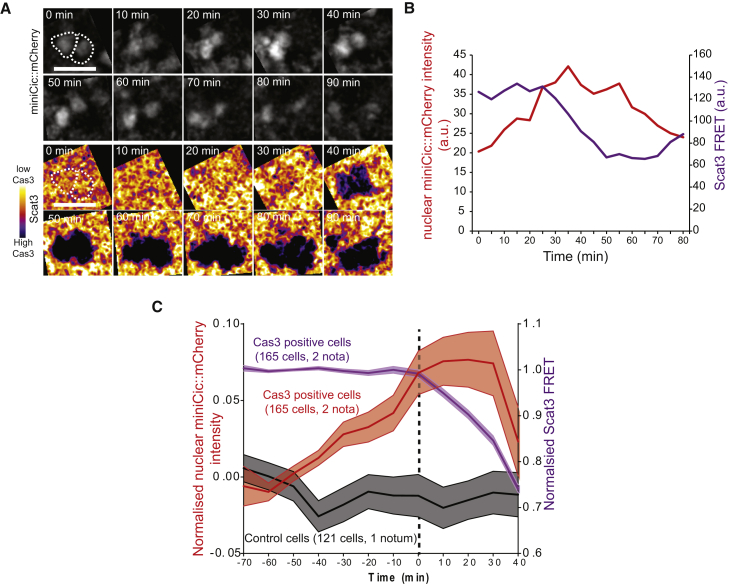
We then checked whether ERK downregulation preceded the activation of caspases at the single-cell level in the midline. Using the fluorescence resonance energy transfer (FRET) caspase sensor Scat3 [29] combined with miniCic, we systematically detected the onset of caspase activation in single cells and measured nuclear miniCic before and after caspase activation. A significant downregulation of ERK signaling preceded the onset of caspase activation for up to 1 hr (Figures 4A–4C; Video S2C), and ERK was slightly upregulated in cells that did not activate caspases (Figure 4C, gray curve; coherent with the global activation observed in Video S2B). Accordingly, 61% of the caspase-activating cells showed an unambiguous increase of miniCic during the last hour preceding caspase activation (n = 165 cells, 2 pupae). Those data confirm at the single-cell level the instructive role of ERK downregulation on caspase activation. However, it also suggests that unknown factors may also trigger caspase activation in the notum.
Figure 4.
ERK Downregulation Precedes Caspase Activation
(A) Snapshots of two nuclei in the midline of the pupal notum showing miniCic signal (top) and Scat3 FRET signal (bottom; dark blue signal, caspase activation). Scale bars represent 10 μm.
(B) miniCic nuclear signal of the left nucleus shown in (A) (red) and FRET signal (purple).
(C) Averaged normalized profiles of miniCic nuclear intensity (red) in the midline aligned at t0 = onset of caspase activation (maximum inflexion of the FRET signal). Light colored areas are ± SEM. The averaged Scat3 FRET signal is shown in purple. Grey curve: normalized averaged miniCic signal of midline cells that do no activate caspase.
See also Video S2.
ERK Can Be Ectopically Activated or Downregulated by Tissue Stretching or Compaction
Cell death in the pupal notum can be downregulated by tissue stretching and ectopically induced by tissue compaction [9]. Because EGFR/ERK is a central regulator of notum cell death, we tested whether ERK activity could be modulated by tissue deformations. Using particle image velocimetry (PIV) to measure local tissue deformations [9], we first compared the local rate of deformation in unperturbed nota with local variations of miniCic signal. We found a good correlation between local tissue stretching (Figure 5A, bottom left) and local activation of ERK activity, especially in the posterior region of the notum, where we observed a global stretch orthogonal to the antero-posterior (AP) axis around 20 hr after pupal formation (APF) (Figures 5A and 5B; Videos S2B and S3). This was reflected by the significant positive cross-correlation between miniCic signal derivative and the compaction rate (Figures 5B and 5C; no significant time delay).
Figure 5.
Tissue Stretching or Compaction Can Increase or Decrease ERK Activity Ectopically
(A, D, and E) Pupal notum expressing endo-Ecad::GFP and miniCic::mCherry during a spontaneous phase of tissue stretching orthogonal to the antero-posterior axis (A; blue arrows) or during stretching induced by laser wounding (E) schematic of the experiment in (D); orange arrows show tissue displacement; A, anterior right; P, Posterior left. From left to right and top to bottom: (1) miniCic::mCherry fluorescent signal; (2) inverted endo-Ecad::GFP; (3) overlay of compaction rate and PIV images averaged over 50 min (PIV biggest arrow: 5 μm/hr in A and 10 μm/hr in E; compaction rate scale bar −0.35 to 0.7 hr−1; blue, stretching; red, compaction); and (4) pseudo-image of the local cross-correlation value between compaction rate and miniCic signal derivative for a lag time of 0 min. The peak of correlation appears in the posterior central region (white region) in (A) and anterior to the wound in (E). Scale bars represent 20 μm.
(B and F) Representative plots of the compaction rate (blue) and miniCic intensity derivative (black) in a single PIV square (small red rectangles, A and E) during the spontaneous stretch (B) and during wounding (F). Insets, normalized cross-correlation of compaction rate versus the derivative of miniCic for those curves is shown.
(C and G) Mean normalized cross-correlation of compaction rate versus miniCic derivative during spontaneous stretching (C) or after wounding (G; red curves) from red square regions in (A) and (E) and control areas (in black) for several nota (n = 3) and subregions. Error bars indicate SEM.
(H) Snapshots of a cell in the stretched region anterior to the wound. Cell contour in blue shows corresponding nucleus with miniCic signal with white dotted circles. Scale bar represents 10 μm.
(I) Averaged normalized cell apical area and miniCic nuclear signal in the stretched region (n = 20 cells). Light colors: SEM. Inset: single curve for the cell shown in (H) is shown.
(J) Local projection of a live pupal notum expressing endo-Ecad::GFP (green) and miniCic::mCherry (magenta, bottom) at different times after laser sectioning of a square (blue) of 110 × 110 μm. Insets: miniCic signal from representative nuclei of the isolated piece of tissue is shown. Scale bar represents 10 μm.
(K) Segmentation of the isolated tissue prior and post-sectioning (apical area in pseudo-colors). Bottom right graph: average variation of cell apical area is shown (one dot per notum; [A final − A initial]/A initial).
(L) Averaged normalized miniCic intensity in the isolated tissue square (t0 = sectioning; 4 nota). Error bars indicate SEM.
Top, middle: Left, miniCic mcherry fluorescent signal (local projection), middle, inverted endo-Ecad::GFP signal (local projection), right, overlay of compaction rate (blue = stretching, red = compaction) and PIV maps all performed on a 16-18h APF notum in normal conditions (top) and upon laser wounding (middle, the wound was done 10 min before the first image of this movie, low intensity rectangle on the left). Black large rectangles on the PIV panels indicate the 30 frames used to analyze correlation between miniCic derivative and compaction rate. Red rectangles underline the zones of stretching used for the analysis. Anterior on the right, posterior on the left. Scale bar, 20 μm. Color map and arrows scales are the same as indicated in Figures 5A and 5C. Bottom: Local z-projections of a live pupal notum (30h APF) expressing endo-Ecad::GFP (green, middle) and miniCic (magenta, right), upon severing of a 400x400px square of tissue with a pulsed UV laser (blue rectangle, ablation between 6 and 8 min). miniCic accumulates transiently in the nuclei of the isolated tissue. Anterior on the right, posterior on the left. Scale bar, 10 μm.
This correlation could be explained by an effect of ERK on tissue mechanics [30, 31, 32] or an effect of tissue deformation or density on ERK [31, 33, 34, 35, 36]. To test those hypotheses, we induced local mechanical perturbations using two laser perturbation approaches. Local laser wounding of the notum induces cell stretching and reduction of cell elimination anterior to the wound because of the opposite movements of the global tissue drift (anterior side) and the wound closure [9] (Figure 5D). Ectopic stretching of the tissue induced by laser wounding was sufficient to transiently upregulate ERK signaling anterior to the wound (Figure 5E, main stretch along AP axis; Video S3). Using PIV, we also observed a significant correlation between zones of stretching and upregulation of ERK activity (Figures 5F and 5G), especially anterior to the wound (bottom right, Figure 5E). This was confirmed by the significant increase of the cell apical area and the concomitant decrease of miniCic nuclear intensity at the single-cell level in the region anterior to the wound (Figures 5H and 5I). The modulation of ERK cannot be driven solely by diffusive factors from the wound, as the upregulation of ERK and the correlation were restricted to the anterior side of the wound, where stretching is stronger (Figures 5D and 5E, bottom right; Video S3). Altogether, we conclude that tissue and/or cell stretching can ectopically upregulate ERK activity.
Conversely, we then tested whether ectopic tissue compaction and/or relaxation could downregulate ERK activity. The pupal notum is globally stretched and laser severing of a piece of epithelium leads to a rapid relaxation and reduction of the isolated cells apical area [37]. Using a pulsed UV laser, we isolated a 110 × 110 μm tissue square (∼300 cells), which rapidly relaxed (Figures 5J and 5K; Video S3). Tissue relaxation and/or densification was followed by a transient nuclear accumulation of miniCic peaking ∼30 min after sectioning (Figures 5J and 5L; Video S3). Thus, stress release and/or tissue densification can transiently downregulate ERK activity. In this study, we used two sets of laser perturbation experiments (line wound, Figures 5E–5I; square severing, Figures 5J–5L) that led to opposite effects on ERK activity (activation or inhibition, respectively) and opposite tissue deformations (stretching or compaction). This excludes again that ERK variations were driven by diffusive factors from the laser-induced wounds. Finally, induction of tissue crowding and folding by the expression of a constitutively active form of Yorkie (UAS-YkiS111A,S168A,S250A [38], a strong regulator of growth and survival) [39, 40] was sufficient to downregulate ERK everywhere in the notum (Figure S4). Altogether, this shows that tissue stretching or compaction can transiently upregulate or downregulate ERK activity in the pupal notum.
Compaction-Driven ERK Downregulation Is Required for Cell Elimination Near RasV12 Clones and Accelerates Clone Expansion
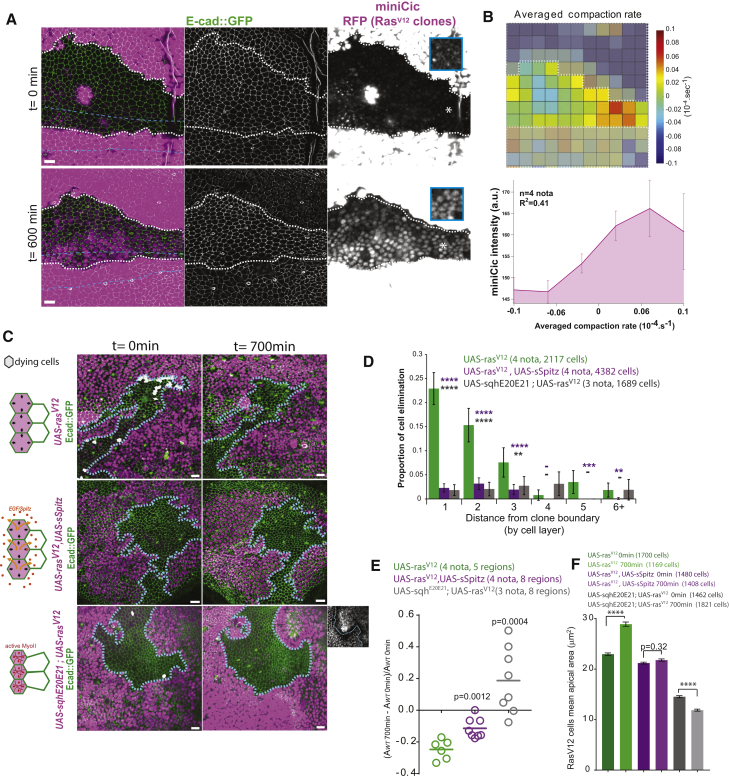
We next tested whether ERK activity was modulated in compacted zones in between RasV12 clones. Induction of ectopic tissue compaction through growth upregulation in RasV12 clones led to a progressive downregulation of ERK signaling in the neighboring WT cells (Figure 6A; Video S4; region where we normally do not observe ERK downregulation; Figure 3F). The pattern of ERK inhibition correlated with the zones of high compaction rate measured by PIV (Figure 6B; R2 = 0.41). Moreover, we observed a clear enrichment of apoptotic cells in zones of low ERK activity in WT cells surrounded by Ras clones (Figure S5A). We thus concluded that ERK is downregulated in compacted cells in between RasV12 clones.
Figure 6.
Mechanical Competition Eliminates Cells through Compaction-Driven ERK Downregulation
(A) Snapshots (0 and 600 min; local z-projections) of a live pupal notum; endo-Ecad::GFP (green) miniCic (magenta); induction of RasV12 in clones (RFP, strong magenta, white dashed lines); midline: blue dotted lines; blue rectangles: single plane miniCic signal from asterisk regions. Scale bars represent 10 μm.
(B) Top: map of the averaged compaction rate (PIV, white dashed lines: clone boundaries). Red: compaction; blue: stretching. Bottom: averaged local miniCic intensity at 600 min for a given local compaction rate is shown (4 nota). R2: Pearson correlation coefficient. Error bars indicate SEM.
(C) z-projections (0 min and after 700 min) of live pupal nota expressing endo-Ecad::GFP (green) upon conditional induction of RasV12, RasV12 and sSpitz (Drosophila EGF), or RasV12 and sqhE20E21 (constitutively active Myosin II regulatory light chain) in clones (RFP, magenta, blue dashed lines: contours). Dying cells are shown in white. Left schemes: expected deformations of the cells are shown (clone, purple). Bottom right inset: E-cad::GFP in Ras and active MyoII clones are shown. Scale bars represent 10 μm.
(D) Probability of cell death for a given distance to clone boundaries (in cell rows). ∗∗∗∗p < 10−4; ∗∗∗p < 10−3; ∗∗p < 10−2; -p > 0.05; Fisher exact test with UAS-rasV12 (green bars). Error bars indicate 95% confidence interval.
(E) Evolution of the area covered by WT cells (one circle = one region surrounded by several clones). Bars indicate averages; t test with UAS-rasV12 (green).
(F) Average rasV12 cell apical area (three first cells layers of the clones) at 0 and 700 min. Mann-Whitney tests between time points; ∗∗∗∗p < 10−4 from 4, 4, and 3 nota. Error bars indicate SEM. Upon activation of MyoII, cells are already smaller at t0 because transcription was activated 8 hr before.
Top: Local z-projection of a pupal notum expressing endo-Ecad::GFP (green) and miniCic (magenta, right panel) upon induction of RasV12 in clones (UAS-RFP, strong magenta). Note that the blurry rapid fluctuating signals are wandering macrophages below the tissue. Anterior on the left, posterior on the right. Scale bar, 10 μm. Bottom: Local z-projections of pupal nota expressing endo-Ecad::GFP (green) upon induction of RasV12 in clones (UAS-nlsRFP, magenta, top left panel), upon induction of RasV12 and secreted Spitz (active Drosophila EGF, magenta top right panel), upon induction of RasV12 and active MyoII (UAS-sqhE20E21, magenta bottom left panel), and upon induction of RasV12 and downregulation of Argos (UAS-argos dsRNA, magenta bottom right panel),. Dying cells are marked in white prior to their extrusion. Anterior on the left, posterior on the right. Scale bars, 10 μm.
Next, we tested whether ERK downregulation was necessary for neighboring cell elimination. Overexpression of a secreted form of Spitz (sSptiz; Drosophila EGF) by RasV12 clones could activate ERK in the neighboring cells without impacting ERK activation in RasV12 clones (Figure S5B). This was sufficient to prevent neighboring cell elimination (Figures 6C and 6D; Video S4). It also slowed down the shrinkage of the area covered by WT cells (Figure 6E) and the expansion of the apical area of RasV12 cells close to the clone boundaries (Figure 6F; 79% increase over 700 min for RasV12 cells; 24% for RasV12, sSpitz cells), which are the cells that should compensate for most of the area left by the WT dying cells [41]. This suggests that cell elimination through ERK downregulation contributes to clone expansion and global replacement of the WT cells.
Finally, we wanted to make sure that WT cell elimination was indeed driven by mechanical cues rather than diffusive factors from the clones. Increasing contractility in RasV12 cells through the overexpression of an active form of MyoII (UAS-sqhE20E21) [42] was sufficient to prevent RasV12 cell expansion (Figure 6F; average cell area decrease of 18%) and strongly downregulated WT cell elimination (Figures 6C–6E; Video S4), suggesting that RasV12 cell expansion is indeed necessary for WT cell elimination. WT cell elimination could also be driven by the secretion of Argos from RasV12 clones and the consequent sequestration of Spitz/EGF [43]. However, preventing Argos accumulation in RasV12 clones by dsRNA did not abolish WT cell elimination near active Ras clones (Figures S5C–S5E; Video S4). Those experiments confirmed that cell elimination was driven by mechanical cues rather than purely diffusive factors from the Ras clones.
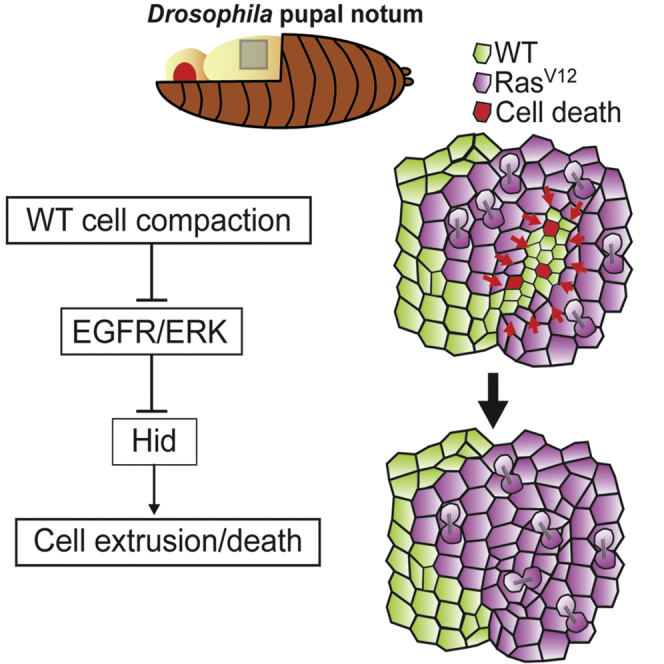
Altogether, we conclude that tissue compaction and/or a reduction of cell tension can rapidly downregulate ERK activity in vivo, which triggers cell elimination through the upregulation of Hid. This modulation of ERK is responsible for cell elimination near RasV12 clones, which contributes significantly to clone expansion and may eventually lead to replacement of the entire WT cell population.
Discussion
EGFR/ERK/Hid pathway has been previously involved in tissue homeostasis and cell number regulation [44, 45]. For instance, modulation of segment size in Drosophila embryo can be adjusted by cell death regulated by EGFR/Hid and the limited source of Spitz/EGF [45, 46]. A similar mechanism is involved in the regulation of the number of interommatidial cells in the fly retina [19, 47, 48, 49, 50] or the number of glia cells in the embryo [51]. Those studies are based on the modulation of EGFR/ERK and death triggered by limiting extracellular ligands. Here, we show that ERK activity can also be modified by tissue mechanics in vivo, which changes the probability of cell survival. Modulation of ERK activity by mechanical stress and/or tissue density has been previously described in cell culture [31, 33, 34, 36, 52] and in vivo [35]. Here, we provide the first evidence of a mechanical modulation of ERK playing an instructive role for cell survival and death during competitive interactions between two cell types in vivo. So far, most of the studies of mechanotransduction in vivo focused on the regulation of Hippo/Yap-Taz pathway [11], whose transcriptional outputs should act on hours timescale. Our results suggest that ERK modulation could act on cell survival in a few tens of minutes (see Figure 4) through the phosphorylation [18] and/or transcriptional regulation [19] of Hid.
Interestingly, global EGFR depletion increased the rate of cell elimination everywhere in the notum (Figures 2B–2D), irrespective of the deformation status of the cells. Accordingly, we found that cells are not any more sensitive to stretching upon EGFR depletion (Figures S6A–S6C; Video S5). This suggests that a ubiquitous basal activity of EGFR is required for cell survival everywhere in the notum, which could be then modulated by tissue deformation. We found that tissue stress and/or compaction can modulate ERK activity and that part of the ERK dynamics correlated with tissue deformations. However, it is very likely that the complex spatiotemporal pattern of ERK activity in the pupal notum (e.g., global downregulation 16 or 17 hr APF and global upregulation 20 hr APF; Video S2) is also controlled by currently unknown patterning genes. EGFR/ERK modulation by deformation may be required to fine tune its activity, to coordinate in time and space cell elimination, and to regulate the number of cells that will be eliminated. A high rate of cell elimination could lead to higher cell spacing and/or an increase of tissue tension, which would feedback negatively on cell elimination through ERK activation. Moreover, the mutual regulation of ERK and mechanics [30, 31, 32] could generate complex temporal dynamics and self-organized properties [31].
z projection of a pupal notum expressing ubi-Ecad::GFP (green) and Gal4 UAS-RFP clones (magenta) where EGFR is depleted (UAS-egfr dsRNA) after laser wounding (white rectangle). The midline is encompassed by the blue lines and the extruding cells in the clone are marked in yellow at the beginning of the movie. Anterior is on the left, posterior on the right. Scale bar, 10 μm.
Although we do not know which molecular effectors of ERK pathway are sensitive to mechanical stress, epistasis experiments suggest that modulation occurs upstream and/or at the level of EGFR (see Figures S6A–S6C; Video S5). Accordingly, a large pool of EGFR is located at adherens junctions (Figure S6D) compatible with a modulation of EGFR activity by apical cell geometry and/or mechanical stress. So far, we did not observe obvious variations of the location and concentration of EGFR in the midline compared to the rest of the notum (Figure S6D), arguing for a mechanism based on a modulation of EGFR activity rather than a direct modulation of its concentration/localization.
The correlation between tissue stretching or compaction and ERK activity could be explained by different parameters, including membrane tension, cell apical surface, and/or changes in cell volume. The transient ERK downregulation observed after tissue severing and the correlation between compaction rate and ERK activity suggest that ERK is sensitive to strain rate rather than absolute cell size and/or tissue density. Similarly, compressive forces rather than absolute tissue density are responsible for spontaneous MDCK cell elimination [6]. Further exploration of the single cell parameters correlating with ERK fluctuations will help to identify the relevant factors modulating ERK.
Although our data support a central role for ERK in modulating cell survival in the pupal notum, we cannot predict fully accurately which cell will engage in apoptosis based on miniCic signal. For instance, we do not know whether caspase activation is triggered by the temporal dynamics or ERK or/and by its absolute levels. Moreover, the probability of cell elimination is still higher in the midline compared to the rest of the tissue upon EGFR depletion (Figures 2C and 2D, 0.75 and 0.42, respectively, versus 0.29 and 0.03 for control clones; Figures 1C and 1D), suggesting that other currently unknown factors also modulate the susceptibility to cell death in the midline. This is in agreement with the absence of ERK downregulation preceding caspase activation in 25% of the midline dying cells (Figure 4). Further work will be required to identify all the factors modulating cell survival in the notum.
Finally, the contribution of compaction-induced ERK inhibition to cell elimination and RasV12 clone expansion may be relevant for other competition scenarios and in pathological conditions. Yki activation in clones was also shown to trigger WT cell deformation and their death in the pupal notum [41]. Cells mutant for the apico-basal polarity protein Scribble are also eliminated through the downregulation of EGFR/ERK [53], although this is driven in that case by the ligand Sas and the tyrosine phosphatase PTP10D. Finally, compaction-driven ERK downregulation could be relevant for the elimination of misspecified cells in Drosophila wing imaginal disc, which has been associated with an increase of contractility at the clone boundary, leading to cell compaction within the clone [54]. Constitutively active mutant forms of Ras are present in one third of human cancers [55]. Our study suggests that blocking mechanical-induced cell elimination by ERK activation can significantly slow down the expansion of tumoral cells. This opposite role of ERK in tumors (promotion of tumoral cell growth and survival) and the surrounding cells (resistance to mechanical stress) may explain the limited success of Ras/Raf/ERK-targeted cancer therapies [55].
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rat anti E-cad | DSHB | DCAD2 concentrate; RRID:AB_528120 |
| guinea pig anti Hid | gift of Don Hyong Ryu | N/A |
| mouse anti EcR | DSHB | Ag10.2 concentrate; RRID:AB_528208 |
| rabbit anti dpERK | Cell Signaling | Phospho-p44/42 MAPK #4370; RRID:AB_2315112 |
| chicken anti GFP | abCam | #13970; RRID:AB_300798 |
| chicken anti mCherry | abCam | #205402; RRID:AB_2722769 |
| rabbit anti-cleaved Dcp-1 | Cell Signaling | #9578; RRID:AB_2721060 |
| mouse anti-Argos | DSHB | Argos 85/2/16 concentrate; RRID:AB_528088 |
| monoclonal mouse anti EGFR | Sigma | E29006 clone C-273; RRID:AB_609900 |
| rabbit anti ERK | Cell Signaling | P44/42 MAPK #4695; RRID:AB_390779 |
| Mouse anti alpha-tubulin | DSHB | 12G10 concentrate; RRID:AB_1157911 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Trametinib | Santa Cruz | CAS 871700-17-3 |
| Experimental Models: Cell Lines | ||
| Drosophila S2R+ cells | François Schweisguth lab | FBrf0024118 |
| Experimental Models: Drosophila melanogaster lines | ||
| pnr-gal4 (III) | Bloomington | BDSC_3039 |
| w1118 | Bloomington | BDSC_3605 |
| UAS-lacZ (II) | Bloomington | BDSC_8529 |
| UAS-diap1 (III) | Bloomington | BDSC_6657 |
| UAS-hid ds RNA (III) | VDRC | GD 8269 |
| UAS-rpr dsRNA (II) | VDRC | KK 101234 |
| UAS-grim dsRNA (II) | VDRC | GD 21830 |
| hs-flp22;ubi-ECad::GFP,UAS-mRFP; act < y+ < Gal4 | L. Legoff (Marseille) | N/A |
| UAS-ras1V12 (III) | From [56] | N/A |
| cic-Cic::GFP (Bac clone) (II) | Bloomington | BDSC_42267 |
| UAS-egfr dsRNA (II) | VDRC | KK 107130 |
| UAS-egfr dsRNA TRiPJF01084 (III) | Bloomington | BDSC_31526 |
| UAS-hEC::EGFR (II) | Bloomington | BDSC_58415 |
| endo-Ecad::GFP | Bloomington | BDSC_60584 |
| tub-GFPBantamsensor/Cyo (II) | From [17] | N/A |
| UAS-dsRed Bantam sponge (III) | From [57] | N/A |
| ubi-Ecad::RFP (II) | From [58] | N/A |
| UAS-pvr dsRNA (II) | VDRC | KK 105353 |
| UAS-fak dsRNA (III) | VDRC | GD 108608 |
| UAS-src42A dsRNA (II) | VDRC | KK 100708 |
| UAS-src64B dsRNA (I) | VDRC | GD 35252 |
| UAS-pi3kCAAX (I) | Bloomington | BDSC_8294 |
| UAS-rafGOF(III) | Bloomington | BDSC_2033 |
| tub-miniCic::mCherry (II) | This study | N/A |
| hs-flp22 ; endo-Ecad::GFP, tub-miniCic::mCherry ; act < cd2 < G4, UAS-eGFP | Bloomington and this study | BDSC_60584 |
| endo-Ecad::GFP,tub-miniCic::mCherry | Bloomington and this study | BDSC_60584 |
| UAS-scat3::myc (II) | From [29] | N/A |
| tub-miniCic::mcherry; UAS-ykiS111A S168A S250A | Bloomington combined with miniCic | BDSC_28817 |
| hs-flp22; endo-Ecad::GFP, tub-gal80ts ; act < y+ < gal4 UAS-nlsRFP/TM6b | From [59] | N/A |
| hs-flp22; endo-Ecad::GFP, tub-gal80ts ; act < cd2 < gal4 UAS-eGFP/TM6b | This study | N/A |
| UAS-sSpitzCS, UAS-rasV12 (III) | Bloomington recombined with UAS-rasV12 | BDSC_63134 |
| UAS-sqhE20E21 (II) ; UAS-rasV12 (III) | From [60] combined with UAS-rasV12 | N/A |
| UAS-argos dsRNA (II) ; UAS-rasV12 (III) | VDRC combined with UAS-rasV12 | GD 47181 |
| Oligonucleotides | ||
| miniCicF: ATCGCTGCGTGCGCGCTTAGGCGGCC GCAACATGCCAAAAAAGAAGAGAAAGGTATCCG CCTCCGGAGGGGGCGTGGTC |
This study | N/A |
| miniCicR: AGAACCACCACCAGAACCACCACCGTA ATATTGAAAAACATCTGCC |
This study | N/A |
| mCherryF: GGTGGTGGTTCTGGTGGTGGTTCTGT GTCCAAGGGCGAAGAGGAC |
This study | N/A |
| mCherryR: GCGCGATGCCGACTGAGTAGGTCTAG ATTATTTATACAGCTCGTCCAT |
This study | N/A |
| Recombinant DNA | ||
| pCaSperR4-tubp-Gal80 plasmid | Addgene | #17748 |
| Software and Algorithms | ||
| MATLAB with Image processing toolbox and Statistics and Machine learning toolbox | Mathworks | https://fr.mathworks.com/ |
| MatPIV | Department of Mathematics, University of Oslo | https://www.mn.uio.no/math/english/people/aca/jks/matpiv/ |
| Fiji (ImageJ) | Fiji | https://fiji.sc/ |
| Mypic Zen autofocus macro | Jan Ellenberg | https://git.embl.de/grp-ellenberg/mypic |
| Adaptive local z-projection macro (MATLAB) | This study | Supplementary information (Methods S1) |
| PIV algorithm (MATLAB) | [61] | N/A |
| Graphpad Prism8 | Graphpad | https://www.graphpad.com/ |
| Packing analyzer (now tissue miner in Fiji) | [62], Benoit Aigouy | https://idisk-srv1.mpi-cbg.de/∼eaton/ |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Romain Levayer (romain.levayer@pasteur.fr).
Experimental Model and Subject Details
Drosophila melanogaster husbandry
All the experiments were performed with Drosophila melanogaster fly lines with regular husbandry technics. The fly food used contains agar agar (7.6 g/l), saccharose (53 g/l) dry yeast (48 g/l), maize flour (38.4 g/l), propionic acid (3.8 ml/l), Nipagin 10% (23.9 ml/l) all mixed in one liter of distilled water. Flies were raised at 25°C in plastic vials with a 12h/12h dark light cycle at 60% of moisture unless specified in the legends and in the table below (alternatively raised at 18°C or 29°C). Females and males were used without distinction for all the experiments. We did not determine the health/immune status of pupae, adults, embryos and larvae, they were not involved in previous procedures, and they were all drug and test naive.
Drosophila melanogaster strains
We used the following fly lines: w1118 on I (Bloomington BDSC_3605), pnr-gal4 on III (Bloomington, BDSC_3039), UAS-lacZ on II (Bloomington, BDSC_8529), UAS-diap1 on III (Bloomington, BDSC_6657), UAS-hid dsRNA on III (VDRC, GD 8269), UAS-rpr dsRNA on II (VDRC, KK 101234), UAS-grim dsRNA on II (VDRC, GD 21830), hs-flp22;ubi-ECad::GFP,UAS-mRFP; act < y+ < Gal4 (L. Legoff, Marseille), UAS-ras1V12 on III (From [56]), cic-Cic::GFP (Bac clone) on II (Bloomington, BDSC_42267), UAS-egfr dsRNA on II (VDRC, KK 107130), UAS-egfr dsRNA on III (Bloomington, TRiPJF01084, BDSC_31526), UAS-hEC::EGFR on II (Bloomington, BDSC_58415), endo-Ecad::GFP on II (Bloomington BDSC_60584), tub-GFPBantamsensor/Cyo on II (from [17]), UAS-dsRed Bantam sponge on III (from [57]), ubi-Ecad::RFP on II (from [58]), UAS-pvr dsRNA on II (VDRC, KK 105353), UAS-fak dsRNA on III (VDRC, GD 108608), UAS-src42A dsRNA on II (VDRC, KK 100708), UAS-src64B dsRNA on I (VDRC, GD 35252), UAS-pi3kCAAX on I (Bloomington, BDSC_8294), UAS-rafGOF on III (Bloomington, BDSC_2033), tub-miniCic::mCherry on II (this study), hs-flp22 ; endo-Ecad::GFP, tub-miniCic::mCherry ; act < cd2 < G4, UAS-eGFP (Bloomington BDSC_60584 and this study), endo-Ecad::GFP,tub-miniCic::mCherry (Bloomington BDSC_60584 and this study), UAS-scat3::myc on II (from [29]), tub-miniCic::mcherry; UAS-yki S111A S168A S250A (Bloomington BDSC_28817 combined with miniCic), hs-flp22; endo-Ecad::GFP, tub-gal80ts ; act < y+ < gal4 UAS-nlsRFP/TM6b (from [59]), hs-flp22; endo-Ecad::GFP, tub-gal80ts ; act < cd2 < gal4 UAS-eGFP/TM6b (this study), UAS-sSpitzCS, UAS-rasV12 on III (Bloomington BDSC_63134 recombined with UAS-rasV12), UAS-sqhE20E21 (on II) ; UAS-rasV12 (on III) (from [60] combined with UAS-rasV12), UAS-argos dsRNA (on II) ; UAS-rasV12 (on III) (VDRC GD 47181 combined with UAS-rasV12).
Genotypes of the experimental model
We list here the fly genotypes used for each figure as well as the time of heat shock when relevant and the days after clone induction (ACI). h APF stands for hours After Pupal Formation. The number of samples (n), the stage (pupal stage, larval stage and embryos) are indicated in the table and the figure legends.
| Figure | Genotype | Heat shock 37°C | Stage | n |
|---|---|---|---|---|
| 1B | UAS-lacZ/+ ; pnr-gal4/+ | N/A | adult | 35 |
| 1B | pnr-gal4/UAS-diap1 | N/A | adult | 19 |
| 1B | pnr-gal4/UAS-hid dsRNA | N/A | adult | 26 |
| 1B | UAS-rpr dsRNA/+ ; pnr-gal4/+ | N/A | adult | 25 |
| 1C | hs-flp22/+; ubi-Ecad::GFP,UASmRFP/+; act < y+ < G4/+ | 12 min, 3 days ACI | Pupal notum (16h APF) | 4 |
| 1C | hs-flp22/+ ; ubi-Ecad::GFP, UASmRFP/+; act < y+ < G4/UAS-hid dsRNA | 12 min, 3 days ACI | Pupal notum (16h APF) | 5 |
| 1C | hs-flp22/+ ; ubi-Ecad::GFP, UASmRFP/UAS-rpr dsRNA; act < y+ < G4/+ | 12 min, 3 days ACI | Pupal notum (16h APF) | 2 |
| 1E | w1118 | N/A | Pupal notum (30h APF) | 7 |
| 1F | hs-flp22/+; endo-Ecad::GFP, tub-gal80ts/+ ; act < y+ < gal4, UAS-nlsRFP/UAS-rasV12 | 30 min, 1 week 18°C, 24h 29°C | Pupal notum (30h APF) | 4 |
| 2A | cic-Cic::GFP (Bac clone) (II) | N/A | Pupal notum (30h APF) | 7 |
| 2B | UAS-lacZ/+ ; pnr-gal4/+ | N/A | adult | 35 |
| 2B | UAS-egfr dsRNA/+ ; pnr-gal4/+ | N/A | adult | 23 |
| 2B | pnr-gal4/UAS-hid dsRNA | N/A | adult | 26 |
| 2B | UAS-egfr dsRNA/+ ; UAS-hid dsRNA/pnr-gal4 | N/A | adult | 20 |
| 2C | hs-flp22/+ ; ubi-Ecad::GFP, UASmRFP/UAS-egfr dsRNA; act < y+ < G4/+ | 12 min, 3 days ACI | Pupal notum (16h APF) | 3 |
| 2C | hs-flp22/+ ; ubi-Ecad::GFP, UASmRFP/UAS-hegfr::GFP; act < y+ < G4/+ | 12 min, 3 days ACI | Pupal notum (16h APF) | 3 |
| S1A | endo-Ecad::GFP | N/A | Pupal notum (30h APF) | 3 |
| S1B | tub-GFPBantamsensor/ubi-Ecad::RFP | N/A | Pupal notum (16h APF) | 3 |
| S1C | pnr-gal4/UAS-dsRed Bantam sponge | N/A | Pupal notum (30h APF) | 4 |
| S1D,E | hs-flp22/+ ; ubi-Ecad::GFP, UASmRFP/+; act < y+ < G4/UAS-dsRed Bantam sponge | 12 min, 3 days ACI | Pupal notum (16h APF) | 2 |
| S1F | w1118 | N/A | Pupal notum (30h APF) | 5 |
| S2A | UAS-pvr dsRNA/+ ; pnr-gal4/+ | N/A | adult | 10 |
| S2A | UAS-fak dsRNA/pnr-gal4 | N/A | adult | 10 |
| S2A | UAS-src42A dsRNA/+ ; pnr-gal4/+ | N/A | adult | 13 |
| S2A | UAS-src64B dsRNA/+ ; ;pnr-gal4/+ | N/A | adult | 13 |
| S2B | hs-flp22/UAS-pi3kCAAX; ubi-Ecad::GFP, UASmRFP/+; act < y+ < G4/+ | 12 min, 3 days ACI | Pupal notum (16h APF) | 2 |
| S2C | hs-flp22/+;ubi-Ecad::GFP,UASmRFP/+; act < y+ < G4/UAS-rafGOF | 12 min, 2 days ACI | Pupal notum (16h APF) | 2 |
| 3B | tub-miniCic::mCherry (II) | N/A | Wing imaginal disc and eye imaginal disc from L3 wandering stage | 10 |
| 3C | hs-flp22/+ ; endo-Ecad::GFP, tub-miniCic::mCherry/UAS-egfr dsRNA ; act < cd2 < G4, UAS-eGFP/+ | 12 min, 2 days ACI | Pupal notum (20h APF) | 6 |
| 3C | hs-flp22/+ ; endo-Ecad::GFP, tub-miniCic::mCherry/UAS-hegfr::GFP; act < cd2 < G4, UAS-eGFP/+ | 12 min, 2 days ACI | Pupal notum (20h APF) | 4 |
| 3E | tub-miniCic::mCherry (II) | N/A | Haemocytes from L3 wandering stage larvae | 2 cultures |
| 3F | endo-Ecad::GFP, tub-miniCic::mCherry | N/A | Pupal notum (16h APF) | 3 |
| S3A | tub-miniCic::mCherry | N/A | Embryos stage 5 to 7 | 56 |
| S3B | endo-Ecad::GFP, tub-miniCic::mCherry | N/A | Pupal notum (16h APF) | 1 |
| 4A-C | tub-miniCic::mCherry/UAS-Scat3 ; pnr-gal4/+ | N/A | Pupal notum (16h APF) | 2 |
| 5A-C | endo-Ecad::GFP, tub-miniCic::mCherry | N/A | Pupal notum (20h APF) | 3 |
| 5E-I | endo-Ecad::GFP, tub-miniCic::mCherry | N/A | Pupal notum (20h APF) | 3 |
| 5J-L | endo-Ecad::GFP, tub-miniCic::mCherry | N/A | Pupal notum (30h APF) | 4 |
| S4A-C | hs-flp22/+ ; endo-Ecad::GFP, tub-gal80ts/tub-miniCic::mCherry; act < cd2 < G4, UAS-eGFP/ UAS-ykiS111A S168A S250A | 40 min, 1 week 18°C, 24h 29°C | Pupal notum (30h APF) | 10 |
| 6A,B | hs-flp22/+; endo-Ecad::GFP, tub-gal80ts/tub-miniCic::mCherry ; act < y+ < gal4, UAS-nlsRFP/UAS-rasV12 | 30 min, 1 week 18°C, 24h 29°C | Pupal notum (20h APF) | 4 |
| 6C-F | hs-flp22/+; endo-Ecad::GFP, tub-gal80ts/+ ; act < y+ < gal4, UAS-nlsRFP/ UAS-rasV12 | 30 min, 1 week 18°C, 24h 29°C | Pupal notum (20h APF) | 4 |
| 6C-F | hs-flp22/+; endo-Ecad::GFP, tub-gal80ts/+ ; act < y+ < gal4, UAS-nlsRFP/ UAS-rasV12, UAS-sSpitzCS | 30 min, 1 week 18°C, 24h 29°C | Pupal notum (20h APF) | 4 |
| 6C-F | hs-flp22/+; endo-Ecad::GFP, tub-gal80ts/UAS-sqhE20E21 ; act < y+ < gal4, UAS-nlsRFP/ UAS-rasV12 | 30 min, 1 week 18°C, 24h 29°C | Pupal notum (20h APF) | 3 |
| S5A | hs-flp22/+;endo-Ecad::GFP,tub-gal80ts/ tub-miniCic::mCherry ; act < y+ < gal4, UAS-nlsRFP/UAS-rasV12 | 30 min, 1 week 18°C, 24h 29°C | Pupal notum (30h APF) | 4 |
| S5B | hs-flp22/+;endo-Ecad::GFP,tub-gal80ts/ tub-miniCic::mCherry ; act < y+ < gal4, UAS-nlsRFP/UAS-rasV12 | 30 min, 1 week 18°C, 24h 29°C | Pupal notum (30h APF) | 4 |
| S5B | hs-flp22/+;endo-Ecad::GFP,tub-gal80ts/ tub-miniCic::mCherry; act < y+ < gal4, UAS-nlsRFP/ UAS-rasV12, UAS-sSpitzCS | 30 min, 1 week 18°C, 24h 29°C | Pupal notum (30h APF) | 6 |
| S5C | hs-flp22/+;endo-Ecad::GFP,tub-gal80ts/+; act < y+ < gal4, UAS-nlsRFP/ UAS-rasV12 | 30 min, 1 week 18°C, 24h 29°C | Pupal notum (30h APF) | 3 |
| S5C | hs-flp22/+;endo-Ecad::GFP,tub-gal80ts/UAS-argos dsRNA; act < y+ < gal4,UAS-nlsRFP/UAS-rasV12 | 30 min, 1 week 18°C, 24h 29°C | Pupal notum (30h APF) | 3 |
| S5D,E | hs-flp22/+;endo-Ecad::GFP,tub-gal80ts/UAS-argos dsRNA; act < y+ < gal4,UAS-nlsRFP/UAS-rasV12 | 30 min, 1 week 18°C, 24h 29°C | Pupal notum (20h APF) | 2 |
| S6A-C | hs-flp22/+ ; ubi-Ecad::GFP, UASmRFP/UAS-egfr dsRNA; act < y+ < G4/+ | 12 min, 3 days ACI | Pupal notum (16h APF) | 3 |
| S6D | endo-Ecad::GFP | N/A | Pupal notum (25h APF) | 5 |
S2R+ cells
S2R+ cells were cultured in Schneider’s Drosophila medium at 25°C with 10% fetal bovine serum, penicillin and streptomycin. The line was provided by the group of François Schweisguth who originally obtained the line from the DGRC.
Method Details
Replication, sample size and strategy for randomization
Data were not analyzed blindly. At least two independent replicates were performed for each experiment (the exact number is given in Figure legends and in the Experimental Model and Subject Details section). No specific method was used to predetermine the number of samples. We did not exclude any data/subject.
Immunostaining
Dissection and immunostainings of nota were performed as indicated in [63] with standard formaldehyde fixation and permeabilisation/washes in PBT 0.4% Triton. Briefly, the nota where pinched on a small plates containing silicone, dissected in PBS, fixed in 4% formaldehyde solution, washed in PBT 0.4% triton, stained with primary antibody solution washed and then stained with secondary antibody solution. Immunostaining of wing imaginal discs and eye imaginal discs were performed on L3 wandering stage larvae and fixed in 4% formaldehyde and permeabilised/washed in PBT. Immunostaining of embryos were performed using standard formaldehyde and heptane/methanol protocol, with permeabilisation in PBS Tween (0.05%). The following antibodies/markers were used: rat anti E-cad (DCAD2 concentrate, DSHB, 1/50), guinea pig anti Hid (1/50, strepatavidin amplified, gift of Don Hyong Ryu), mouse anti EcR (DSHB Ag10.2 concentrated, 1/100), rabbit anti dpERK (Cell signaling, #4370, 1/100), chicken anti GFP (abCam, #13970, 1/500), chicken anti mCherry (abCam, #205402, 1/200), rabbit anti-cleaved Dcp-1 (Cell Signaling, #9578, 1/50), mouse anti-Argos (DSHB concentrated, 1/50), monoclonal mouse anti EGFR (Sigma, E29006 clone C-273, 1/50). Secondary antibodies were all Invitrogen secondary antibodies produced in goat with Alexa 488,555 or 633. Dissected nota were mounted in Vectashield with DAPI (Vectorlab) and stained embryos were mounted in Aquapolymount media. They were imaged on a Leica confocal SP8 or a Zeiss lsm880 using a 63X oil immersion objective N.A. 1.3. Unless specified, images shown are maximal z-projection containing adherens junction plane.
Design of the miniCic sensor
The C-ter part of Capicua containing the C2 and C1 domains but lacking the DNA binding domain (HMG box) was PCR amplified from a cDNA clone (DGRC clone FI04109, starting at base number 2687, sequence: TCCGCCTCCGGAGGGGGCGTGGTC) adding a NLS site in N-ter right after the ATG. The following primers were used:
miniCicF
ATCGCTGCGTGCGCGCTTAGGCGGCCGCAACATGCCAAAAAAGAAGAGAAAGGTATCCGCCTCCGGAGGGGGCGTGGTC
miniCicR
AGAACCACCACCAGAACCACCACCGTAATATTGAAAAACATCTGCC
The reverse primer contains 23 overlapping nucleotides for the fusion with mCherry. mCherry was PCR amplified with the following primers containing the flexible linker GGGSGGGS in Nter:
mCherryF
GGTGGTGGTTCTGGTGGTGGTTCTGTGTCCAAGGGCGAAGAGGAC
mCherryR
GCGCGATGCCGACTGAGTAGGTCTAGATTATTTATACAGCTCGTCCAT
Fusion was performed by performing a PCR on the two first PCR products using the miniCicF and the mCherryR primers alone. The fusion product was inserted in pC4-tub-gal80 using the NotI and XbaI sites after removal of the Gal80 through digestion, purification and ligation. The construct was validated by sequencing and injected by Bestgene (P-element insertion).
Western blot
S2R+ cells were cultured in Schneider’s Drosophila Medium with 10% fetal bovine serum, penicillin and streptomycin. Cells were plated at a density of 2x106 cells into twelve well plates and incubated at 25°C. 24h after plating, they were treated with Trametinib 10 μM (initial stock 10mM in DMSO) or with DMSO alone (1/1000). Cellular extracts were prepared at indicated time post treatment and analyzed by immunoblotting. The following primary antibodies were used: phospho-Erk(Thr202/Tyr204) (1/1000, Cell signaling, #4370), total-Erk (1/1000, Cell signaling, #4695) and alpha-tubulin(1/5000, DSHB, 12G10 concentrated).
Analysis of adult thorax defects
Adult thorax defects were analyzed on adult female thorax raised at 25°C. For Figure 1B we measured the width of the midline divided by the averaged width of the two adjacent SOP rows along a line connecting the two aDC macrochaetaes (log2 scale). For Figure 2B we measured the width of the pnr domain (distance between the two aDC macrochaetaes) divided by the total width of the thorax (distance between the two pSA macrochaetae) and plotted the log2 of the ratio.
Imaging of haemocytes and drug treatment
Primary culture of haemocytes was performed by bleeding ten larvae expressing tub-miniCic::mCherry that were first washed in ethanol, dried on a piece of paper and resuspended in 200 μL of S2 medium on a square of parafilm. After bleeding with forceps each larvae, the 200 μL were deposed in a MaTek Petri dish (P35G-1.5-10-C) and we let the cells sediment and adhere for 30min. The media was then supplemented by 2mL of S2 medium. The dish was placed on a LSM800 point scanning confocal (63X oil N.A. 1.3) and acquisition was launched with a transmitted light channel and mCherry channel (one z stack/min). Trametinib (Santa Cruz, 10mM stock in DMSO, final concentration 10 μM) or DSMO (1/1000) were added in the medium at the onset of the movie. Nuclear miniCic signal was measured by tracking manually the nucleus using the transmission light channel on Fiji, measuring mCherry intensity and normalizing intensity by intensity at t0.
Notum live imaging, image processing and cell death analysis
Notum live imaging was performed as described previously [9]. Briefly, the pupae were collected at the white stage (0 hour after pupal formation), aged at 29°, glued on double sided tape on a slide and surrounded by two spacers composed of stacks of 4 #1 20x20mm coverslips glued with nail polisher. The pupal case was opened up to the abdomen using forceps and mounted with a 20x40mm #1.5 coverslip where we buttered halocarbon oil 10S. The coverslip was then glued on the spacers using fast drying nail polisher. Pupae were collected 48 or 72h after clone induction and dissected 16-18h after pupae formation (APF). Pupae were dissected and imaged on a confocal spinning disc microscope (Till photonics) with a 40X oil objective (N.A. 1.35) using tile imaging (6 to 12 tiled positions) or a point scanning confocal microscope Leica SP8 with a 63X objective (N.A. 1.3), or a Zeiss LSM800 or a LSM880 equipped with a fast Airyscan using an oil 40X objective (N.A. 1.3), Z stacks (1 μm/slice), every 5min using autofocus at 25°C. The autofocus was performed using E-cad::GFP plane as a reference (Tillphotonics and Leica LAS provided option) or the autofluorescence of the cuticle in far red (using a Zen Macro developed by Jan Ellenberg laboratory, MyPic). Movies were performed in the nota close to the scutellum region containing the midline and the aDC and pDC macrochaetae. Movies shown are maximum projections or adaptive local z-projection (see below for details). Total duration was always 700min (except for Figure 6A, 600min, Figures 3F, 4, and 5). For imaging of UAS-rasV12 clones, the cross and the progeny were kept at 18°C, and the pupae were switched to 29°C 8 hours prior to the movie for conditional activation (clones induced by the following lines: hs-flp22; endo-Ecad::GFP, tub-gal80ts ; act < y+ < gal4 UAS-nlsRFP/TM6b or hs-flp22; endo-Ecad::GFP, tub-gal80ts ; act < cd2 < gal4 UAS-GFP) and imaged at 29°C. The midline covers all the cells surrounded by the two most central lines of sensory organ precursors (SOP, located at the end of the movies). Every cell extrusion event was localized manually and marked using Fiji. The probability of extrusion was obtained by dividing the total number of extruding cells in the clones divided by the initial number of cells in the clones or by doing the same for each cell layer next to Ras clones. Values from the control clones (ayGal4 alone) are coming from [9] as part of the clonal experiments were performed during the same period. Note that we did the same experiment with UAS-grim dsRNA (GD21830) and observed a significant increase of the rate of cell death in and outside the midline (not shown here, all Grim dsRNA lines have multiple off targets). In the figures, cells were marked as dying cells if they died or if at least one of their daughter cells died before the end of the movie.
For the quantification of the area covered by WT cells during competition with Ras, regions of WT cells surrounded by at least two Ras clones were defined on Fiji and their area was measured at the onset of the movie and after 700min. Ras cell area was obtained after segmentation of the images at the onset of the movie and after 700min using packing analyzer Fiji plugin [62]. We measured apical area of the three first layers of cells near clone boundaries.
Adaptive local z-projection and analysis of miniCic::mCherry
All the image of miniCic::mCherry in the notum were obtained through a custom made adaptive local z-projection procedure wrote on MATLAB (to avoid projection of the autofluorescence signal of the cuticle and projection of more basal planes). The procedure can be found in the Supplemental information (Methods S1). For every time point, a reference channel was used (here E:cad::GFP), the image was subdivided in 40x40px (1px = 0.18 μm) square and the plane with the highest standard deviation was selected. The search window was moved in x and y 10px by 10px. The obtained z-profile was then smoothened in x and y using a Gaussian blur (2px width). miniCic signal was then obtained by calculating the median of the intensity over seven planes centered on a reference plane located 6 μm basally to the local E-cad reference plane. Note that similar results could be obtained by using maximum projection instead of the median. For every movie, we checked on the raw files that no nuclear signal of miniCic was lost because of projection errors.
Nuclear miniCic quantification and Scat3 imaging
Live pupal nota expressing UAS-Scat3 with pnr-gal4 driver and miniCic were imaged on a LSM880 point scanning confocal (one stack every 5min or 10min). Images were pretreated with local z-projection (see above, YFP used as a reference channel, average of 7 planes around the plane of reference). Image ratios of YFP over CFP fluorescent channels were then calculated (FRET signal). Cells showing a clear caspase activation (low YFP/CFP ratio) were selected manually. The miniCic channel was also cut according to those positions and smaller stacks of 15 time frames were created (1 stack/10 min). Positions of the nuclei were selected manually over the 15 frames for all the selected cells and both YFP/CFP ratio and miniCic mean intensity values were quantified in boxes of 2 μm squares centered on those positions. The average profiles were obtained by aligning the curves on the point of maximum inflexion of the FRET signal (local minimum of the 2nd derivative). Each single curve was then normalized by the values at the three first time points (70 to 50 min before caspase activation). For miniCic, the average intensity value at the three first time points was also subtracted. All the curves were then averaged. Non-ambiguous ERK downregulation (as mentioned in the text) corresponds to curves with at least a 5% increase of miniCic signal over one hour prior to caspase activation. The same procedure was performed for the control curve on randomly selected cells that do not activate caspase in same region and time developmental window as used for caspase activating cells. For those curves, alignment was made at T0 (onset of the tracking).
Particle Image Velocimetry, cross-correlation and tissue wounding
For Figure 5, we used a PIV algorithm based on the MATLAB routine described in [61]. Parameters for this PIV were analysis boxes of 64 pixels (11.5x11.5 μm ∼4-5 cells), 50% overlap boxes. PIV displacements at time t were determined based on images t-5 and t+5min. Displacement maps were spatially filtered to remove noise coming from outliers.
Quantification of miniCic mCherry signal (Figures 5A–5G) are median values of mCherry signal over the analysis boxes of the PIV after performing local projection. For each notum, the region of interest (zones of maximum deformation) were subdivided in 25 squares of analysis (50% overlap). Compaction rate maps were calculated as minus the “divergence” function of MATLAB on the PIV vector fields. miniCic signals were smoothen over ten time frames before calculating the derivative, compaction rate were smoothen the same way. The average value was subtracted from each function to obtain variations around 0 value. Maps of cross-correlation and normalized cross-correlation between compaction rate and derivative of miniCic levels were then calculated with the “xcorr” MATLAB function either without or with the ‘coef’ option (non-normalized and normalized cross-correlation). Quantification of cross-correlations were established in stretching areas for time periods of 2 hours. Number of nota rather than number of square of analysis was used to calculate the s.e.m.. To obtain control cross-correlation curves, we computed the normalized cross-correlation of miniCic and compaction rates in regions with no obvious deformations. We also checked that there were no correlation between compaction rate of the stretched area and miniCic signals from regions outside the stretch area to make sure that the miniCic variations were specific of the zones of deformation (data not shown). Single cell tracking and segmentation (Figures 5H and 5I) was performed using packing analyzer Fiji plugin [62] and by manually tracking the corresponding miniCic nuclei for each cell. For the cross-correlation between miniCic signal and cell elimination (Figure S3D), we measured mean minCic intensity from local projections in 100x100px squares (50% overlap) and quantified the number of cell elimination occurring in those squares (disappearance of cell apical area). We then computed the normalized cross-correlation on MATLAB between miniCic intensity and the smooth of the cell elimination function (smooth window 29) and averaged cross-correlation curves for every square of analysis.
Notum wounding was performed as described previously [9], using exposure at full power with an Argon laser (488 and 458nm) and a 405nm diode in the wounded region over 3min. For experiment in Figures 5J and 5K, tissue square dissection was performed using a pulsed 355nm laser (Teem photonics, 20kHz, peak power 0.7kW) controlled by the ILas pulse system (Gataca systems) on an inverted Nikon eclipse Ti2 microscope (plan fluor 40X 1.3 N.A, oil) equipped with a Yokogawa CSU-W1 spinning disc system and a prime95 sCMOS camera (Photometrics). The region of ablation was a 400x400px square (110x110 μm) with a band of 10px width and 10 repetitions (AOTF 35%). Relaxation was imaged using z-acquisitions in green and red channels every 2min combined with an autofocus between each time point. Images shown are local z-projections. Quantification of cell area was performed in Tissue analyzer [62], intensity of miniCic was quantified after tissue drift correction (Fiji plugin StackReg), bleaching correction (on the full movie) and removal of the background (rolling ball radius 50px).
For Figure 6, PIV analysis was performed as described previously [9], using MatPIV toolbox on MATLAB with 64px windows with 50% overlap and a rolling window of time averaging of 9 frames. Compaction rate is defined as “- Divergence” of the vector field (calculated on MATLAB). Averaged compaction rates were calculated by averaging on one PIV window the compaction rate over the full movie (700 min). The same squares were used to quantify miniCic signal at the last time point of the movie excluding any zone that partially overlapped with the Ras clones. The mean miniCic signal was then calculated for each bin of compaction rate values and the full dataset was used to calculate a Pearson correlation coefficient.
Quantification and Statistical Analysis
Data were not analyzed blindly. No specific method was used to predetermine the number of samples. The definition of n and the number of samples is given in each figure legend and in the table of the Experimental Model and Subject Details section. Error bars are standard error of the mean (s.e.m.). p values are calculated through t test if the data passed normality test (Shapiro-Wilk test), or Mann-Whitney test if the distribution were not normal. For proportion (death probability), the error bars are 95% confidence interval and p value are calculated through a Fisher exact test. Statistical tests were performed on Graphpad Prism 8.
Data and Software Availability
The custom made adaptive local z-projection procedure was written on MATLAB. Basically it uses a reference channel (here E-cad::GFP) to find locally the plane of focus and perform a local projection around this plane of reference for a second channel. The code can be found in the Supplemental information (Methods S1). The principle is explained in the Method Details section.
Acknowledgments
We thank members of R.L. lab and F. Schweisguth for critical reading of the manuscript. We are also grateful to F. Janody, M. Miura, C. Bökel, H.D. Ryoo, G. Jiménez, the Bloomington Drosophila Stock Center, the Drosophila Genetic Resource Center, the Vienna Drosophila Resource Center, and the Developmental Studies Hybridoma Bank for sharing stocks and reagents. We also thank B. Aigouy for the Packing Analyzer software and J. Ellenberg group for MyPic autofocus macro. L.V. is supported by a post-doctoral grant “Aide au Retour en France” from the FRM (Fondation pour la Recherche Médicale) (ARF20170938651); work in R.L. lab is supported by the Institut Pasteur (G5 starting package) and the ERC starting grant CoSpaDD (Competition for Space in Development and Disease) (grant number 758457). Work in E.M. lab is supported by the European Research Council, The Swiss National Foundation, and the Champalimaud Foundation.
Author Contributions
E.M. and R.L. initiated the project. L.V. performed the wounding experiments, the cross-correlation analysis, the miniCic and caspase correlation, and the local projection algorithm. F.L. performed the dpERK western blot. R.L. wrote the manuscript and performed all the other experiments and analysis. Every author has commented and edited the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: December 13, 2018
Footnotes
Supplemental Information includes six figures, one Methods file, and five videos and can be found with this article online at https://doi.org/10.1016/j.cub.2018.11.007.
Contributor Information
Eduardo Moreno, Email: eduardo.moreno@research.fchampalimaud.org.
Romain Levayer, Email: romain.levayer@pasteur.fr.
Supplemental Information
References
- 1.Merino M.M., Levayer R., Moreno E. Survival of the fittest: essential roles of cell competition in development, aging, and cancer. Trends Cell Biol. 2016;26:776–788. doi: 10.1016/j.tcb.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Petridou N.I., Spiró Z., Heisenberg C.P. Multiscale force sensing in development. Nat. Cell Biol. 2017;19:581–588. doi: 10.1038/ncb3524. [DOI] [PubMed] [Google Scholar]
- 3.Guillot C., Lecuit T. Mechanics of epithelial tissue homeostasis and morphogenesis. Science. 2013;340:1185–1189. doi: 10.1126/science.1235249. [DOI] [PubMed] [Google Scholar]
- 4.Irvine K.D., Shraiman B.I. Mechanical control of growth: ideas, facts and challenges. Development. 2017;144:4238–4248. doi: 10.1242/dev.151902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eisenhoffer G.T., Loftus P.D., Yoshigi M., Otsuna H., Chien C.B., Morcos P.A., Rosenblatt J. Crowding induces live cell extrusion to maintain homeostatic cell numbers in epithelia. Nature. 2012;484:546–549. doi: 10.1038/nature10999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saw T.B., Doostmohammadi A., Nier V., Kocgozlu L., Thampi S., Toyama Y., Marcq P., Lim C.T., Yeomans J.M., Ladoux B. Topological defects in epithelia govern cell death and extrusion. Nature. 2017;544:212–216. doi: 10.1038/nature21718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagstaff L., Goschorska M., Kozyrska K., Duclos G., Kucinski I., Chessel A., Hampton-O’Neil L., Bradshaw C.R., Allen G.E., Rawlins E.L. Mechanical cell competition kills cells via induction of lethal p53 levels. Nat. Commun. 2016;7:11373. doi: 10.1038/ncomms11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marinari E., Mehonic A., Curran S., Gale J., Duke T., Baum B. Live-cell delamination counterbalances epithelial growth to limit tissue overcrowding. Nature. 2012;484:542–545. doi: 10.1038/nature10984. [DOI] [PubMed] [Google Scholar]
- 9.Levayer R., Dupont C., Moreno E. Tissue crowding induces caspase-dependent competition for space. Curr. Biol. 2016;26:670–677. doi: 10.1016/j.cub.2015.12.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pereira A.M., Tudor C., Kanger J.S., Subramaniam V., Martin-Blanco E. Integrin-dependent activation of the JNK signaling pathway by mechanical stress. PLoS ONE. 2011;6:e26182. doi: 10.1371/journal.pone.0026182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Panciera T., Azzolin L., Cordenonsi M., Piccolo S. Mechanobiology of YAP and TAZ in physiology and disease. Nat. Rev. Mol. Cell Biol. 2017;18:758–770. doi: 10.1038/nrm.2017.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Norman M., Wisniewska K.A., Lawrenson K., Garcia-Miranda P., Tada M., Kajita M., Mano H., Ishikawa S., Ikegawa M., Shimada T., Fujita Y. Loss of Scribble causes cell competition in mammalian cells. J. Cell Sci. 2012;125:59–66. doi: 10.1242/jcs.085803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vincent J.P., Fletcher A.G., Baena-Lopez L.A. Mechanisms and mechanics of cell competition in epithelia. Nat. Rev. Mol. Cell Biol. 2013;14:581–591. doi: 10.1038/nrm3639. [DOI] [PubMed] [Google Scholar]
- 14.Brás-Pereira C., Moreno E. Mechanical cell competition. Curr. Opin. Cell Biol. 2018;51:15–21. doi: 10.1016/j.ceb.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Guirao B., Rigaud S.U., Bosveld F., Bailles A., López-Gay J., Ishihara S., Sugimura K., Graner F., Bellaïche Y. Unified quantitative characterization of epithelial tissue development. eLife. 2015;4:e08519. doi: 10.7554/eLife.08519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang C., Baehrecke E.H., Thummel C.S. Steroid regulated programmed cell death during Drosophila metamorphosis. Development. 1997;124:4673–4683. doi: 10.1242/dev.124.22.4673. [DOI] [PubMed] [Google Scholar]
- 17.Brennecke J., Hipfner D.R., Stark A., Russell R.B., Cohen S.M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 18.Bergmann A., Agapite J., McCall K., Steller H. The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell. 1998;95:331–341. doi: 10.1016/s0092-8674(00)81765-1. [DOI] [PubMed] [Google Scholar]
- 19.Kurada P., White K. Ras promotes cell survival in Drosophila by downregulating hid expression. Cell. 1998;95:319–329. doi: 10.1016/s0092-8674(00)81764-x. [DOI] [PubMed] [Google Scholar]
- 20.Astigarraga S., Grossman R., Díaz-Delfín J., Caelles C., Paroush Z., Jiménez G. A MAPK docking site is critical for downregulation of Capicua by Torso and EGFR RTK signaling. EMBO J. 2007;26:668–677. doi: 10.1038/sj.emboj.7601532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parsons J.T. Focal adhesion kinase: the first ten years. J. Cell Sci. 2003;116:1409–1416. doi: 10.1242/jcs.00373. [DOI] [PubMed] [Google Scholar]
- 22.Kim L.C., Song L., Haura E.B. Src kinases as therapeutic targets for cancer. Nat. Rev. Clin. Oncol. 2009;6:587–595. doi: 10.1038/nrclinonc.2009.129. [DOI] [PubMed] [Google Scholar]
- 23.Shilo B.Z. The regulation and functions of MAPK pathways in Drosophila. Methods. 2014;68:151–159. doi: 10.1016/j.ymeth.2014.01.020. [DOI] [PubMed] [Google Scholar]
- 24.Inaki M., Vishnu S., Cliffe A., Rørth P. Effective guidance of collective migration based on differences in cell states. Proc. Natl. Acad. Sci. USA. 2012;109:2027–2032. doi: 10.1073/pnas.1115260109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prober D.A., Edgar B.A. Interactions between Ras1, dMyc, and dPI3K signaling in the developing Drosophila wing. Genes Dev. 2002;16:2286–2299. doi: 10.1101/gad.991102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar J.P., Hsiung F., Powers M.A., Moses K. Nuclear translocation of activated MAP kinase is developmentally regulated in the developing Drosophila eye. Development. 2003;130:3703–3714. doi: 10.1242/dev.00556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slack C., Alic N., Foley A., Cabecinha M., Hoddinott M.P., Partridge L. The Ras-Erk-ETS-signaling pathway is a drug target for longevity. Cell. 2015;162:72–83. doi: 10.1016/j.cell.2015.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim B., Dsilva C.J., Levario T.J., Lu H., Schüpbach T., Kevrekidis I.G., Shvartsman S.Y. Dynamics of inductive ERK signaling in the Drosophila embryo. Curr. Biol. 2015;25:1784–1790. doi: 10.1016/j.cub.2015.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takemoto K., Nagai T., Miyawaki A., Miura M. Spatio-temporal activation of caspase revealed by indicator that is insensitive to environmental effects. J. Cell Biol. 2003;160:235–243. doi: 10.1083/jcb.200207111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanimura S., Takeda K. ERK signalling as a regulator of cell motility. J. Biochem. 2017;162:145–154. doi: 10.1093/jb/mvx048. [DOI] [PubMed] [Google Scholar]
- 31.Aoki K., Kondo Y., Naoki H., Hiratsuka T., Itoh R.E., Matsuda M. Propagating wave of ERK activation orients collective cell migration. Dev. Cell. 2017;43:305–317.e5. doi: 10.1016/j.devcel.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 32.Ogura Y., Wen F.L., Sami M.M., Shibata T., Hayashi S. A switch-like activation relay of EGFR-ERK signaling regulates a wave of cellular contractility for epithelial invagination. Dev. Cell. 2018;46:162–172.e5. doi: 10.1016/j.devcel.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 33.Aoki K., Kumagai Y., Sakurai A., Komatsu N., Fujita Y., Shionyu C., Matsuda M. Stochastic ERK activation induced by noise and cell-to-cell propagation regulates cell density-dependent proliferation. Mol. Cell. 2013;52:529–540. doi: 10.1016/j.molcel.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 34.Kawabata N., Matsuda M. Cell density-dependent increase in tyrosine-monophosphorylated ERK2 in MDCK cells expressing active Ras or Raf. PLoS ONE. 2016;11:e0167940. doi: 10.1371/journal.pone.0167940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sano T., Kobayashi T., Negoro H., Sengiku A., Hiratsuka T., Kamioka Y., Liou L.S., Ogawa O., Matsuda M. Intravital imaging of mouse urothelium reveals activation of extracellular signal-regulated kinase by stretch-induced intravesical release of ATP. Physiol. Rep. 2016;4:e13033. doi: 10.14814/phy2.13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirata H., Gupta M., Vedula S.R., Lim C.T., Ladoux B., Sokabe M. Actomyosin bundles serve as a tension sensor and a platform for ERK activation. EMBO Rep. 2015;16:250–257. doi: 10.15252/embr.201439140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonnet I., Marcq P., Bosveld F., Fetler L., Bellaïche Y., Graner F. Mechanical state, material properties and continuous description of an epithelial tissue. J. R. Soc. Interface. 2012;9:2614–2623. doi: 10.1098/rsif.2012.0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oh H., Irvine K.D. In vivo analysis of Yorkie phosphorylation sites. Oncogene. 2009;28:1916–1927. doi: 10.1038/onc.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson B.J., Cohen S.M. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell. 2006;126:767–774. doi: 10.1016/j.cell.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 40.Huang J., Wu S., Barrera J., Matthews K., Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 41.Tsuboi A., Ohsawa S., Umetsu D., Sando Y., Kuranaga E., Igaki T., Fujimoto K. Competition for space is controlled by apoptosis-induced change of local epithelial topology. Curr. Biol. 2018;28:2115–2128.e5. doi: 10.1016/j.cub.2018.05.029. [DOI] [PubMed] [Google Scholar]
- 42.Jordan P., Karess R. Myosin light chain-activating phosphorylation sites are required for oogenesis in Drosophila. J. Cell Biol. 1997;139:1805–1819. doi: 10.1083/jcb.139.7.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klein D.E., Nappi V.M., Reeves G.T., Shvartsman S.Y., Lemmon M.A. Argos inhibits epidermal growth factor receptor signalling by ligand sequestration. Nature. 2004;430:1040–1044. doi: 10.1038/nature02840. [DOI] [PubMed] [Google Scholar]
- 44.Liang J., Balachandra S., Ngo S., O’Brien L.E. Feedback regulation of steady-state epithelial turnover and organ size. Nature. 2017;548:588–591. doi: 10.1038/nature23678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parker J. Control of compartment size by an EGF ligand from neighboring cells. Curr. Biol. 2006;16:2058–2065. doi: 10.1016/j.cub.2006.08.092. [DOI] [PubMed] [Google Scholar]
- 46.Crossman S.H., Streichan S.J., Vincent J.P. EGFR signaling coordinates patterning with cell survival during Drosophila epidermal development. PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.3000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baker N.E., Yu S.Y. The EGF receptor defines domains of cell cycle progression and survival to regulate cell number in the developing Drosophila eye. Cell. 2001;104:699–708. doi: 10.1016/s0092-8674(01)00266-5. [DOI] [PubMed] [Google Scholar]
- 48.Yang L., Baker N.E. Cell cycle withdrawal, progression, and cell survival regulation by EGFR and its effectors in the differentiating Drosophila eye. Dev. Cell. 2003;4:359–369. doi: 10.1016/s1534-5807(03)00059-5. [DOI] [PubMed] [Google Scholar]
- 49.Yu S.Y., Yoo S.J., Yang L., Zapata C., Srinivasan A., Hay B.A., Baker N.E. A pathway of signals regulating effector and initiator caspases in the developing Drosophila eye. Development. 2002;129:3269–3278. doi: 10.1242/dev.129.13.3269. [DOI] [PubMed] [Google Scholar]
- 50.Miller D.T., Cagan R.L. Local induction of patterning and programmed cell death in the developing Drosophila retina. Development. 1998;125:2327–2335. doi: 10.1242/dev.125.12.2327. [DOI] [PubMed] [Google Scholar]
- 51.Bergmann A., Tugentman M., Shilo B.Z., Steller H. Regulation of cell number by MAPK-dependent control of apoptosis: a mechanism for trophic survival signaling. Dev. Cell. 2002;2:159–170. doi: 10.1016/s1534-5807(02)00116-8. [DOI] [PubMed] [Google Scholar]
- 52.Tschumperlin D.J., Dai G., Maly I.V., Kikuchi T., Laiho L.H., McVittie A.K., Haley K.J., Lilly C.M., So P.T., Lauffenburger D.A. Mechanotransduction through growth-factor shedding into the extracellular space. Nature. 2004;429:83–86. doi: 10.1038/nature02543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamamoto M., Ohsawa S., Kunimasa K., Igaki T. The ligand Sas and its receptor PTP10D drive tumour-suppressive cell competition. Nature. 2017;542:246–250. doi: 10.1038/nature21033. [DOI] [PubMed] [Google Scholar]
- 54.Bielmeier C., Alt S., Weichselberger V., La Fortezza M., Harz H., Jülicher F., Salbreux G., Classen A.K. Interface contractility between differently fated cells drives cell elimination and cyst formation. Curr. Biol. 2016;26:563–574. doi: 10.1016/j.cub.2015.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Samatar A.A., Poulikakos P.I. Targeting RAS-ERK signalling in cancer: promises and challenges. Nat. Rev. Drug Discov. 2014;13:928–942. doi: 10.1038/nrd4281. [DOI] [PubMed] [Google Scholar]
- 56.Karim F.D., Rubin G.M. Ectopic expression of activated Ras1 induces hyperplastic growth and increased cell death in Drosophila imaginal tissues. Development. 1998;125:1–9. doi: 10.1242/dev.125.1.1. [DOI] [PubMed] [Google Scholar]
- 57.Herranz H., Hong X., Cohen S.M. Mutual repression by bantam miRNA and Capicua links the EGFR/MAPK and Hippo pathways in growth control. Curr. Biol. 2012;22:651–657. doi: 10.1016/j.cub.2012.02.050. [DOI] [PubMed] [Google Scholar]
- 58.Michel M., Raabe I., Kupinski A.P., Pérez-Palencia R., Bökel C. Local BMP receptor activation at adherens junctions in the Drosophila germline stem cell niche. Nat. Commun. 2011;2:415. doi: 10.1038/ncomms1426. [DOI] [PubMed] [Google Scholar]
- 59.Levayer R., Hauert B., Moreno E. Cell mixing induced by myc is required for competitive tissue invasion and destruction. Nature. 2015;524:476–480. doi: 10.1038/nature14684. [DOI] [PubMed] [Google Scholar]
- 60.Bertet C., Rauzi M., Lecuit T. Repression of Wasp by JAK/STAT signalling inhibits medial actomyosin network assembly and apical cell constriction in intercalating epithelial cells. Development. 2009;136:4199–4212. doi: 10.1242/dev.040402. [DOI] [PubMed] [Google Scholar]
- 61.Valon L., Marín-Llauradó A., Wyatt T., Charras G., Trepat X. Optogenetic control of cellular forces and mechanotransduction. Nat. Commun. 2017;8:14396. doi: 10.1038/ncomms14396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aigouy B., Farhadifar R., Staple D.B., Sagner A., Röper J.C., Jülicher F., Eaton S. Cell flow reorients the axis of planar polarity in the wing epithelium of Drosophila. Cell. 2010;142:773–786. doi: 10.1016/j.cell.2010.07.042. [DOI] [PubMed] [Google Scholar]
- 63.Jauffred B., Bellaiche Y. Analyzing frizzled signaling using fixed and live imaging of the asymmetric cell division of the Drosophila sensory organ precursor cell. Methods Mol. Biol. 2012;839:19–25. doi: 10.1007/978-1-61779-510-7_2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
z projections of 16–18 hr APF pupal nota expressing ubi-Ecad::GFP (green) and Gal4 UAS-RFP clones (magenta) in control (UAS-lacZ, left), upon Hid downregulation in clones (UAS-hid dsRNA, middle), Rpr downregulation in clones (UAS-rpr dsRNA, right), overexpressing Bantam sponge (UAS-bantam sponge), an active from of PI3K (UAS-pi3kCA, middle), an active form of Raf (UAS-rafCA, right), EGFR downregulation in clones (UAS-egfr dsRNA) and upon overexpression of hEGFR::GFP (UAS-EGFR::GFP, clones are marked by the strong membrane GFP signal, note that the movie was filtered to obtain similar intensity ranges for E-cad and EGFR::GFP in the clones). The midline is encompassed by the white lines and the extruding cells in the clone are marked in yellow at the beginning of the movie. Anterior on the left, posterior on the right. Scale bars, 10 μm.
(A) Primary culture of haemocytes expressing miniCic::mCherry upon treatment with 10 μM Trametinib (MEK inhibitor, left) or DMSO (right) at the onset of the movie. Scale bar, 10 μm. (B) Local z-projections of a live pupal notum expressing endo-Ecad::GFP (green) and miniCic (magenta and right). Time is indicated in hours after pupal formation. Scale bar, 10 μm. (C) Local projection of two nuclei of the midline in a living pupal notum expressing miniCic (left) and Scat3 (FRET signal, right, red-orange = low caspase activity, blue-purple = high caspase activity). Scale bar, 10 μm.
Top, middle: Left, miniCic mcherry fluorescent signal (local projection), middle, inverted endo-Ecad::GFP signal (local projection), right, overlay of compaction rate (blue = stretching, red = compaction) and PIV maps all performed on a 16-18h APF notum in normal conditions (top) and upon laser wounding (middle, the wound was done 10 min before the first image of this movie, low intensity rectangle on the left). Black large rectangles on the PIV panels indicate the 30 frames used to analyze correlation between miniCic derivative and compaction rate. Red rectangles underline the zones of stretching used for the analysis. Anterior on the right, posterior on the left. Scale bar, 20 μm. Color map and arrows scales are the same as indicated in Figures 5A and 5C. Bottom: Local z-projections of a live pupal notum (30h APF) expressing endo-Ecad::GFP (green, middle) and miniCic (magenta, right), upon severing of a 400x400px square of tissue with a pulsed UV laser (blue rectangle, ablation between 6 and 8 min). miniCic accumulates transiently in the nuclei of the isolated tissue. Anterior on the right, posterior on the left. Scale bar, 10 μm.
Top: Local z-projection of a pupal notum expressing endo-Ecad::GFP (green) and miniCic (magenta, right panel) upon induction of RasV12 in clones (UAS-RFP, strong magenta). Note that the blurry rapid fluctuating signals are wandering macrophages below the tissue. Anterior on the left, posterior on the right. Scale bar, 10 μm. Bottom: Local z-projections of pupal nota expressing endo-Ecad::GFP (green) upon induction of RasV12 in clones (UAS-nlsRFP, magenta, top left panel), upon induction of RasV12 and secreted Spitz (active Drosophila EGF, magenta top right panel), upon induction of RasV12 and active MyoII (UAS-sqhE20E21, magenta bottom left panel), and upon induction of RasV12 and downregulation of Argos (UAS-argos dsRNA, magenta bottom right panel),. Dying cells are marked in white prior to their extrusion. Anterior on the left, posterior on the right. Scale bars, 10 μm.
z projection of a pupal notum expressing ubi-Ecad::GFP (green) and Gal4 UAS-RFP clones (magenta) where EGFR is depleted (UAS-egfr dsRNA) after laser wounding (white rectangle). The midline is encompassed by the blue lines and the extruding cells in the clone are marked in yellow at the beginning of the movie. Anterior is on the left, posterior on the right. Scale bar, 10 μm.