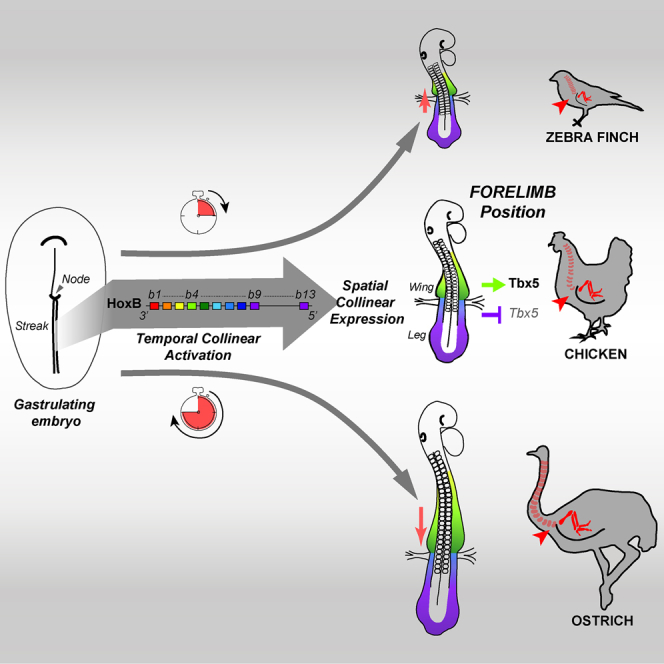
Summary
Limb position along the body is highly consistent within one species but very variable among vertebrates. Despite major advances in our understanding of limb patterning in three dimensions, how limbs reproducibly form along the antero-posterior axis remains largely unknown. Hox genes have long been suspected to control limb position; however, supporting evidences are mostly correlative and their role in this process is unclear. Here, we show that limb position is determined early in development through the action of Hox genes. Dynamic lineage analysis revealed that, during gastrulation, the forelimb, interlimb, and hindlimb fields are progressively generated and concomitantly patterned by the collinear activation of Hox genes in a two-step process. First, the sequential activation of Hoxb genes controls the relative position of their own collinear domains of expression in the forming lateral plate mesoderm, as demonstrated by functional perturbations during gastrulation. Then, within these collinear domains, we show that Hoxb4 anteriorly and Hox9 genes posteriorly, respectively, activate and repress the expression of the forelimb initiation gene Tbx5 and instruct the definitive position of the forelimb. Furthermore, by comparing the dynamics of Hoxb genes activation during zebra finch, chicken, and ostrich gastrulation, we provide evidences that changes in the timing of collinear Hox gene activation might underlie natural variation in forelimb position between different birds. Altogether, our results that characterize the cellular and molecular mechanisms underlying the regulation and natural variation of forelimb positioning in avians show a direct and early role for Hox genes in this process.
Keywords: limb, lateral plate mesoderm, gastrulation, Hox genes, Tbx5, patterning, chicken, birds, collinearity
Graphical Abstract
Highlights
-
•
Forelimb, interlimb, and hindlimb domains are sequentially laid during gastrulation
-
•
Temporal collinear activation controls Hox sequential expression in these domains
-
•
Hox domains differentially instruct (Hox4) or repress (Hox9) limb formation
-
•
Changes in collinear activation underlie bird natural variation in limb position
How limbs reproducibly form along the vertebrate body remains largely unknown. Whereas Hox genes have long been suspected to regulate limb position, their role in this process is unclear. Here, Moreau et al. show a direct and early role for Hox genes in the regulation and natural variation of the forelimb position in birds.
Introduction
In tetrapods, limbs are always positioned at the level of the cervico-thoracic (forelimb) or lumbo-sacral (hindlimb) vertebral transitions; however, the position of these frontiers varies greatly among species. Almost all mammals form forelimbs at the level of the 8th vertebrae; birds display tremendous variation, with sparrow, chicken, and swans forming forelimbs at the level of the 10th, 15th, and 25th vertebrae, respectively. Frogs, in turn, exhibit forelimbs at the level of the 2nd vertebrae. Hox genes have long been proposed to regulate limb position during development (for review, see [1, 2, 3]). Organized in four different clusters, they display a chromosomal organization that reflects their sequential timing of expression and their successive domains of expression along the antero-posterior (A-P) axis, named temporal and spatial collinearity, respectively [4, 5, 6]. The observation that anterior boundaries of expression of specific Hox genes match forelimb, interlimb, and hindlimb borders across tetrapod species [7, 8, 9] and more recently that Hox genes can bind an enhancer of Tbx5, a transcription factor essential for forelimb initiation [10, 11], led to the proposition that Hox genes might regulate limb position. However, whereas gain- and loss-of-function of single or multiple Hox genes result in the transformation of vertebral identity [12], aside from Hoxb5 mutants, which display an anterior shift of the limb with incomplete penetrance [13], functional studies in support of a role for Hox genes in limb positioning have been lacking. Therefore, whether Hox genes control limb initiation and position has not been established.
Limbs originate from the somatopleural lateral plate mesoderm (LPM), an epithelial layer of tissue that flanks axial embryonic structures. Surprisingly, whereas the cellular events underlying the formation of embryonic compartments adjacent to the LPM are well described (i.e., neural tube, notochord, somites, and intermediate mesoderm) [14, 15, 16, 17], little is known about how the LPM is generated during gastrulation. Lineage-tracing experiments in the chick have revealed that LPM precursor cells arise from the central third of the primitive streak (PS) [16, 17, 18, 19, 20]. Only one lineage analysis, using colored chalk dust [21], traced back the origin of forelimb, interlimb, and hindlimb fields up to stage 7 (i.e., after LPM cells have initiated their ingression through the PS). A series of grafting studies demonstrated that the forelimb identity becomes determined by at least stage 9 [22] and that the forelimb field carries the morphogenetic potential to induce a limb in the flank by stage 11 [23]. However, such studies were all designed an interpreted with regards to the determination of limb field identity (forelimb versus hindlimb) [22], polarity (antero-posterior or dorso-ventral) [21, 24], or morphogenetic inductive potential [23, 25, 26] and did not address the question of limb field positioning within the LPM. Thus, how and even when the LPM becomes determined into limb- and non-limb-forming domains remains to be investigated.
Here, we examine how the forelimbs are positioned along the A-P axis during avian development. We find that the LPM is progressively formed and concomitantly patterned by Hox genes into limb- and non-limb-forming domains during the process of gastrulation. Moreover, we provide data suggesting that relative changes in the timing of Hox collinear activation might underlie natural variation in the forelimb position of different bird species.
Results
The Forelimb Position Is Already Determined by the End of Gastrulation
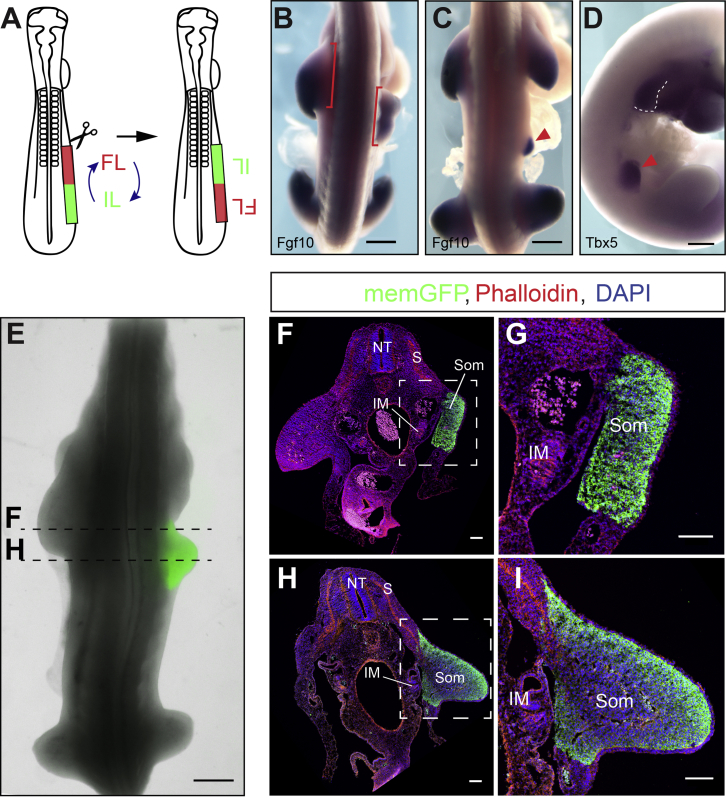
We first sought to elucidate when the position of the future forelimb is determined. The LPM that is generated during gastrulation epithelializes into the somatopleure by stage 11 (i.e., 2 days of development). To test whether limb position is already determined in the freshly generated LPM, we micro-dissected, rotated, and grafted back the right somatopleure encompassing both forelimb and interlimb prospective domains of stage 11 chicken embryos (Figure 1A). Operated embryos were re-incubated for 48 hr until limbs have clearly formed. 65% of operated embryos exhibited either a total or partial posterior shift of the forelimb, as revealed by expression of the limb marker Fgf10 (Figures 1B and 1C; n = 15/23). Shifted limb buds also expressed Tbx5, demonstrating that the position of the forelimb field has been displaced by the surgical procedure (Figure 1D; n = 2/2). To rule out that the shift of the limb bud territory resulted from artificial induction of donor surrounding tissues, we performed quail-chick grafting experiments. To directly visualize the grafted tissue, we generated a transgenic quail line expressing membrane-bound EGFP under the control of the ubiquitous human ubiquitin C (hUbC) promoter (hUbC:memGFP) (Figure 1E). Transverse sections of grafted embryos clearly show that only the somatopleural LPM (GFP positive) was transplanted at the level of forelimb or interlimb (Figures 1F–1I). Altogether, these experiments show that the position of the forelimb domain relative to the interlimb domain is established autonomously within the somatopleural LPM and as early as stage 11.
Figure 1.
The Forelimb Position Is Already Determined by the End of Gastrulation
(A) LPM rotation procedure in a stage 11 chicken embryo. FL, Forelimb; IL, Interlimb.
(B–D) Fgf10 (B and C) and Tbx5 (D) expression 48 hr after LPM rotation, showing complete (B; n = 3/23 embryos, red brackets) or partial (C and D; n = 12/23 embryos, red arrowhead) displacement of the forelimb bud.
(E) Chicken embryo grafted with memGFP transgenic quail somatopleure (green) showing a posterior displacement of the forelimb upon rotation (n = 3/3 embryos).
(F–I) Transverse sections of quail-chick chimera at the forelimb (F and G) and interlimb (H and I) levels, stained with phalloidin (red), GFP antibody (green), and DAPI (blue). (G) and (I) are higher magnification of (F) and (H), respectively. NT, neural tube; S, somite; IM, intermediate mesoderm; Som, somatopleure.
Scale bars are 500 μm in (B)–(E) and 100 μm in (F)–(I).
Forelimb, Interlimb, and Hindlimb Fields Are Sequentially Formed during Gastrulation
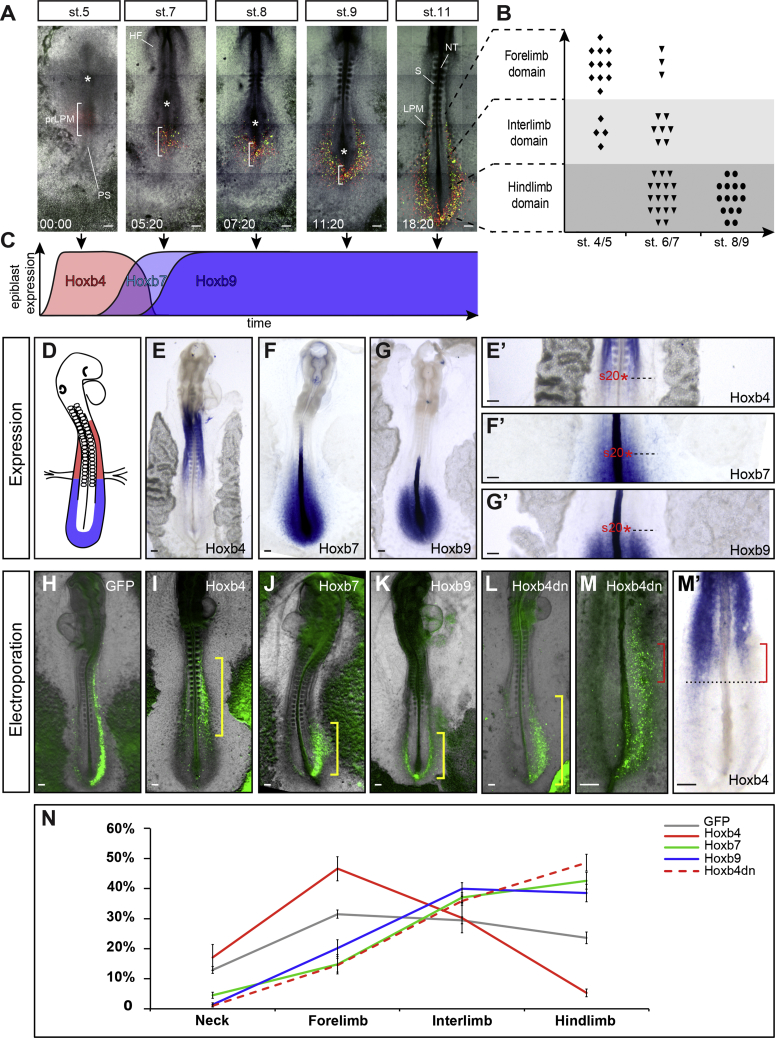
The finding that the LPM is patterned into forelimb and interlimb domains by stage 11 raises the possibility that it might be patterned even earlier. The origin of the LPM in the epiblast has been traced back to the middle third of the PS in chicken embryos [16, 17, 20]. However, how forelimb, interlimb, and hindlimb cells are specifically generated has not been characterized. We performed a dynamic lineage analysis of prospective limb precursor cells by electroporating fluorescent reporters (GFP and nuclear H2b-RFP) into the presumptive LPM territory of stage 4 chicken embryos. Electroporated embryos were then cultured ex vivo [27] and imaged using 2-photon video microscopy for approximately 24 hr (Figure 2A; Video S1). Retrospective tracking of LPM precursors identified the epiblast origin of forelimb, interlimb, and hindlimb fields (Video S2). First, we observed that the formation of the LPM takes place between stages 4 and 10 (i.e., spanning 24 hr of development). Second, we could determine that most forelimb precursor cells are generated between stages 4 and 5, whereas interlimb and hindlimb precursor cells are gradually generated at later stages, mostly at stages 6–7 and 8–9, respectively (Figure 2B). To confirm these results using a different lineage-tracing technique, we generated a transgenic quail line expressing the green-to-red photoconvertible fluorescent protein mEOS2 under the control of the ubiquitous hUbC promoter (hUbC:mEOS2FP). In hUbC:mEOS2FP transgenic quail embryos, regions could be very precisely photoconverted and readily tracked for 24 hr. We thus photoconverted the prospective LPM cells in the PS of stages 6, 7, 9, and 10 mEOS2 transgenic embryos (Figures S1A–S1D) and followed their fate for 24 hr. As expected, the later the cells were photoconverted in the PS, the more posterior they localized in the LPM (Figures S1A’–S1D’ and S1A’’–S1D’’; Video S3). Notably, cells photoconverted past stage 10 did not contribute to the LPM, further confirming that, by this stage, the contribution of the PS to the LPM has ended.
Figure 2.
Progressive Formation of the LPM and Concomitant Patterning by Hox Genes
(A) Time series showing LPM formation from stage 5 to stage 11. LPM precursors are electroporated with H2b-RFP (red) and GFP (green). White brackets outline the presumptive LPM in the PS; white asterisks, Hensen’s node; PS, primitive streak; prLPM, presumptive LPM; HF, head folds. See also Video S1.
(B) Position of electroporated cells in the LPM (y axis) as a function of their timing of ingression (x axis); n = 57 tracked cells, 8 embryos (see Video S2).
(C) Timeline of Hoxb4 (red), Hoxb7 (light blue), and Hoxb9 (blue) activation in the presumptive LPM with regard to LPM formation (black arrows). For detailed expression data, see Figure S2.
(D) Schematic summarizing anterior (red) and posterior (blue) Hox genes expression in the LPM.
(E–G) Expression of Hoxb4 (E), Hoxb7 (F), and Hoxb9 (G) at stage 13. (E’)–(G’) are higher magnifications of (E)–(G), showing Hox genes posterior (Hoxb4; E’) or anterior (Hoxb7 and Hoxb9; F’ and G’, respectively) border of expression in the LPM (dashed black line). Red asterisks mark the somite 20.
(H–L) Stage 13 embryos, electroporated at stage 4 with GFP (H), Hoxb4/GFP (I), Hoxb7/GFP (J), Hoxb9/GFP (K), or Hoxb4 dn-GFP (L). Yellow brackets highlight different distribution of electroporated cells in the LPM.
(M and M’) Hoxb4 endogenous expression (M’) in a Hoxb4 dn-GFP electroporated embryo (M). Note the decrease in Hoxb4 expression on the electroporated side (red brackets, n = 13/21 embryos) compared to the control side (black dashed line).
(N) Distribution of electroporated cells along the A-P axis for GFP-only (gray, 16 embryos, 6,994 cells), Hoxb4/GFP (red, 12 embryos, 3,203 cells), Hoxb7/GFP (green, 11 embryos, 2,359 cells), Hoxb9/GFP (blue, 15 embryos, 3,922 cells), and Hoxb4 dn-GFP (dashed red, 22 embryos, 20,528 cells) electroporated embryos. Distribution is presented as mean ± SEM.
Scale bars are 100 μm. See also Figures S1, S2, and S3A and Videos S1, S2, and S3.
17h time-lapse video of a chicken embryo electroporated with cytoplasmic GFP and nuclear H2b-RFP fluorescent reporters, showing the dynamic process of LPM formation (from stage 5 to stage 11). Images were acquired on an inverted 2-photon microscope, using a long distance 10X objective. White bracket in the 1st frame outlines the presumptive LPM (prLPM); white asterisk represents the Hensen’s node; white dashed line outlines the PS; white brackets in the last frame outline the forelimb (FL), interlimb (IL) and hindlimb (HL) domains.
Example of retrospective cell tracking of forelimb, interlimb and hindlimb LPM cells to identify their epiblast origin on a 24h time-lapse video of a GFP and H2b-RFP-electroporated embryo. Images were acquired on an inverted 2-photon microscope, using a long distance 10X objective. Left panel, bright field image of the embryo (from stage 11 back to stage 4). Middle panel, GFP electroporated cells. Right panel, tracking of 9 LPM cells back to their epiblast origin. Note that the video plays backward.
hUbC:mEOS2FP transgenic quail embryos in which the presumptive LPM in the PS has been photoconverted. Embryos were further imaged every 2h to follow the fate of the photoconverted region. Photoconversion and acquisition were performed on an inverted confocal microscope with a long distance 10X objective. Left, middle-left, middle-right and right panels show embryos where the presumptive LPM was photoconverted at stage 6, 7, 9 and 10, respectively. Black arrowheads outline the anterior limit of photoconverted cells.
Hox Genes Progressively Pattern the LPM during Gastrulation
During paraxial mesoderm formation, the collinear activation of Hoxb genes controls the establishment of their own expression domains by regulating the ingression timing of epiblast cells [28]. Hoxb genes also display collinear activation in the prospective LPM territory of the PS, with Hoxb4 expression starting at stage 4, Hoxb7 at stage 5, and Hoxb9 at stage 6-7 (Figure S2). Importantly, activation of Hoxb4 correlates with the timing of ingression of forelimb precursor cells in the PS, whereas activation of Hoxb7 and Hoxb9 genes correlates with the ingression of interlimb precursor cells (summarized in Figure 2C). By stage 11, Hox genes could be classified into 2 groups: a group of genes expressed anteriorly (anterior to the 20th somite level, e.g., Hoxb4, but also Hoxb3 and Hoxb5) in a domain encompassing the prospective forelimb domain and another group of genes expressed posteriorly (posterior to the 20th somite level, e.g., Hoxb7 and Hoxb9 but also Hoxb6 and Hoxb8) in a domain encompassing the prospective interlimb-hindlimb domain (Figures 2D–2G and 2E’–2G’; data not shown). Therefore, the dynamics of expression of Hoxb4 and Hoxb7/9 correlate with the generation of forelimb and interlimb mesoderm, respectively: first temporally (activation in the PS) and then spatially (domains in the LPM).
To test the role of Hox genes in the patterning of the LPM during gastrulation, we electroporated the presumptive LPM territory of the PS in stage 4 embryos, either with GFP alone or in combination with Hoxb4, Hoxb7, or Hoxb9. Although control GFP-expressing cells were distributed uniformly along the A-P axis (Figures 2H and 2N; n = 16/16), Hoxb4-expressing cells were predominantly found in the anterior part of the embryo (within the forelimb domain; Figures 2I and 2N; n = 12/14). In contrast, Hoxb7- and Hoxb9-expressing cells concentrated in the posterior-most part of the embryo (within the interlimb-hindlimb domain; Figures 2J, 2K, and 2N; n = 11/12 and n = 15/18, respectively). Notably, cells overexpressing a given Hox gene always distributed in the corresponding endogenous expression domain of this gene (compare Figures 2I–2K, yellow brackets, with Figures 2E–2G). In a converse experiment, we interfered with Hoxb4 function using a dominant-negative (dn) form of Hoxb4. Such dn form was constructed by deleting the C-terminal portion of the homeodomain (required for DNA binding; for review, see [29]) while conserving its paralog-specific motives (Figure S3A). The resulting truncated HOXB4 protein retains the ability to bind paralog-specific co-factors but lacks the ability to bind target DNA, presumably acting by competing for co-factors. The action of such truncated HOX protein has been proposed to be dominant over paralogs [30, 31, 32]. When Hoxb4 dn-GFP was electroporated in the PS of stage 4 embryos, cells subsequently distributed in the posterior-most LPM, very much alike Hoxb7 and Hoxb9 electroporated cells, in a pattern almost complementary to Hoxb4 overexpressing cells and endogenous expression (Figures 2L and 2N; n = 18/22). We next checked for an effect on Hoxb4 endogenous expression, which was detected using a C-terminal-specific probe to avoid detection of Hoxb4 dn construct. Strikingly, Hoxb4 expression was strongly reduced at its posterior border, specifically where electroporated cells could be observed, resulting in an anterior-ward shortening of Hoxb4 expression domain on the electroporated side (Figures 2M and 2M’, red brackets; n = 13/21). Altogether, these results show that, as for paraxial mesodermal cells, activation of different Hox genes differentially regulates cell ingression of LPM precursors in the PS and, as a consequence, the relative position of their own expression domains in the LPM.
Hox Genes Control the Definitive Forelimb Position
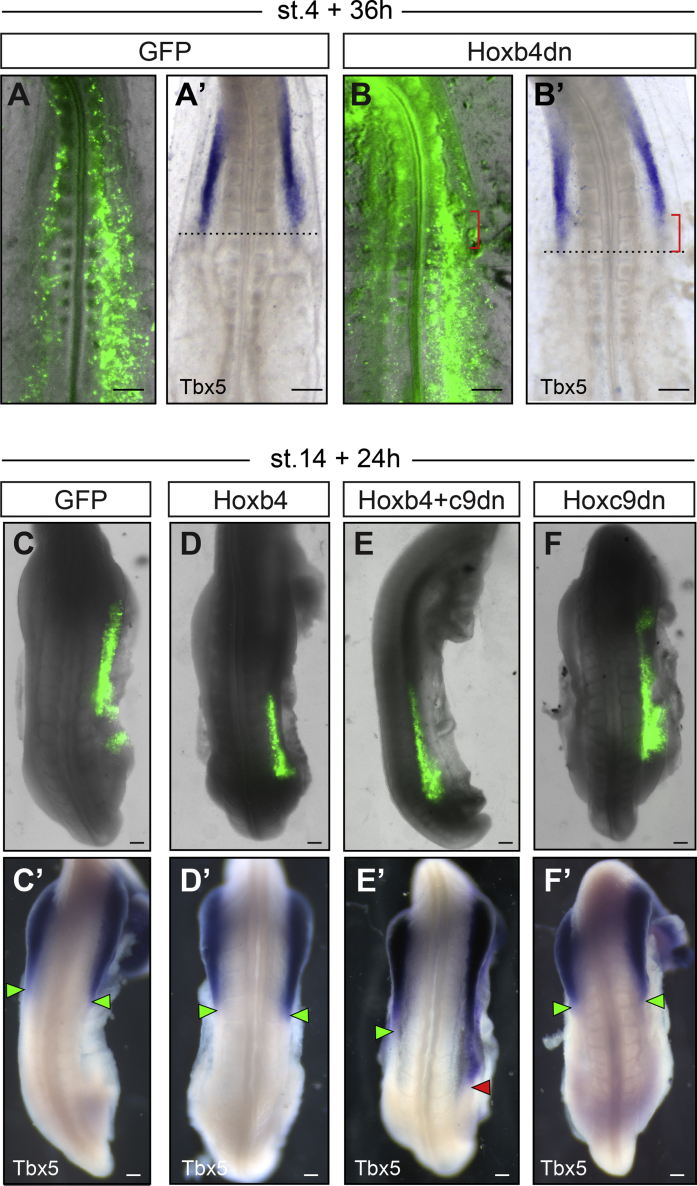
We next decided to test the role of the successive Hox domains in instructing the forelimb position in the LPM. Genes from the Hox4 and Hox5 groups have recently been proposed to control Tbx5 expression [10, 11]. However, whether these Hox genes can drive endogenous or ectopic Tbx5 expression in vivo and whether they can modulate forelimb position has not been tested. We therefore checked for Tbx5 expression in Hoxb4 dn electroporated embryos. Whereas GFP electroporation had no effect on Tbx5 expression (Figures 3A and 3A’; n = 0/4), Hoxb4 dn-GFP electroporated embryos exhibited a decrease in Tbx5 posterior expression, specifically where electroporated cells could be found, which resulted in a unilateral, anterior-ward shortening of the Tbx5 domain of expression (Figures 3B and 3B’; n = 7/9). These results show that Hoxb4 (and presumably other Hox4 genes) acts during gastrulation to establish its own domain of expression, which then establishes the position of the Tbx5-positive forelimb field.
Figure 3.
Hox Genes Determine the Position of the Early Tbx5-Positive Forelimb Field
(A and B) Stage 15 embryos electroporated at stage 4 with GFP (A; n = 4 embryos) or Hoxb4 dn-GFP (B; n = 9 embryos). Note that intense signal in the anterior somite and neural tube in (B) is non-specific auto-fluorescence. (A’ and B’) Tbx5 expression in corresponding embryos is shown. Note the decrease in Tbx5 expression on the electroporated side of Hoxb4 dn-GFP electroporated embryo (B’; red brackets; n = 7/9 embryos).
(C–F) Embryos electroporated with GFP alone (C; n = 20 embryos) or in combination with Hoxb4 (D; n = 16 embryos), Hoxb4+Hoxc9 dn (E; n = 19 embryos), or Hoxc9 dn (F; n = 15 embryos) in the interlimb domain at stage 14 and re-incubated for 24 hr. Note that variation in the position of the electroporated domain is due to technical variation. (C’–F’) Tbx5 expression in corresponding embryos is shown. Ectopic Tbx5 expression in the interlimb region is denoted by red arrowheads compared to normal endogenous expression denoted by green arrowheads (E’; n = 9/19 embryos).
Scale bars are 100 μm. See also Figure S3.
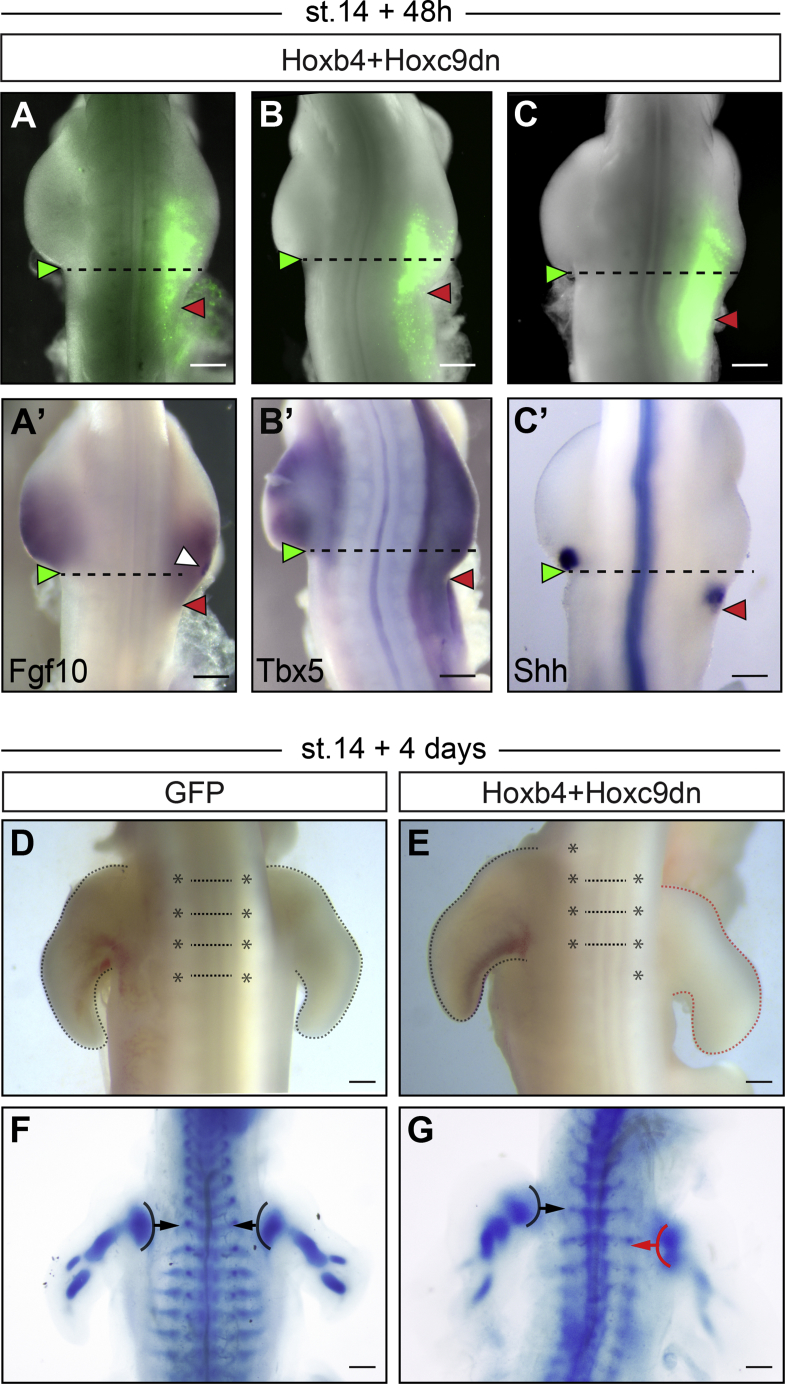
In a converse experiment, we tested whether Hoxb4, when electroporated ectopically in the interlimb region of stage 14 embryos, is able to drive Tbx5 expression and displace limb position posteriorly. To our surprise, as with GFP electroporation, we could not detect ectopic expression of Tbx5 in the interlimb region of embryos overexpressing Hoxb4, 24 hr after electroporation (Figures 3C, 3C’, 3D, and 3D’; n = 0/20 and n = 0/16, respectively). This could be due to expression of Hoxc9 in the interlimb region, as this gene can repress Tbx5 expression [11]. To test this hypothesis, we generated a Hoxc9 dn form using the same strategy described above for Hoxb4 (Figure S3A). The repressive domains of Hoxc9 have been precisely identified and characterized (i.e., hexapeptide motif and N-terminal residues of the homeodomain) [11]. The truncated HOXC9 protein therefore retains the ability to bind HOX9-paralog-specific and repressive co-factors but lacks the ability to bind target DNA. In mouse, deletion of all Hox9 paralogs has been shown to prevent Sonic Hedgehog expression in the developing forelimb buds [33]. We validated the dominant-negative effect of the Hoxc9 dn construct by electroporation in the limb mesenchyme of stage 15 chicken embryos. This led to a partial (very likely because of the mosaic efficiency of electroporation, which only targets a fraction of cells) but significant decrease in Sonic Hedgehog expression, reminiscent of Hox9-null mutant mouse embryos and suggestive of a pan Hox9 inhibition (Figures S3B–S3J). When Hoxb4 was electroporated in combination with Hoxc9 dn and GFP, ectopic expression of Tbx5 was observed in the interlimb region (Figures 3E and 3E’; n = 9/19). Notably, electroporation of Hoxc9 dn alone did not promote ectopic expression of Tbx5 in the interlimb region (Figures 3F and 3F’; n = 0/15), suggesting that in Hoxb4-Hoxc9 dn electroporations, Tbx5 ectopic expression does not arise from the sole release of a repression imposed by Hox9 genes. To observe the effects on limb bud formation, we then allowed electroporated embryos to develop for 48 hr, after limb buds have clearly formed. In embryos electroporated with a combination of Hoxb4, Hoxc9 dn, and GFP, a posterior extension of the limb bud by 1 to 2 somites could be observed in 59% of the cases (Figures 4A–4C; n = 13/22). As expected, neither electroporation of Hoxc9 dn, Hoxb4, nor GFP alone induced a posterior extension of the limb bud (n = 0/14, n = 0/18, and n = 0/15, respectively; Figure S4). Posteriorly extended limb buds expressed the pan-limb and forelimb-specific markers, Fgf10 and Tbx5, respectively (Figures 4A’ and 4B’). High levels of Fgf10 expression were observed in the electroporated region (Figure 4A’, white arrowhead), indicative of the ectopic activation of the limb initiation-outgrowth regulation feedback loop. Sonic Hedgehog expression, which labels the zone of polarizing activity (ZPA), was also posteriorly displaced, indicating that the A-P patterning of the early limb bud is also affected (Figure 4C’). Finally, we tested the effect of such early perturbation of Hox genes function on the forelimb definitive position by allowing embryos to develop for four days after electroporation. In these embryos, a posterior shift of the forelimb, specifically on the right electroporated side, could be readily observed morphologically (Figure 4E, n = 11/22; compare to GFP control Figure 4D, n = 0/7). Alcian blue staining, which marks differentiated cartilage, definitively confirmed a posterior, unilateral shift of limb skeletal elements by about one complete vertebrae (Figures 4F and 4G; n = 0/5 and n = 5/9, respectively). Together with previous data demonstrating Hox binding at the Tbx5 genomic locus [10, 11], these results show that Hox genes regulate the definitive forelimb position through combinatorial activation and repression activities on Tbx5 expression.
Figure 4.
Hox Genes Regulate the Definitive Forelimb Position
(A–C) Embryos electroporated with GFP in combination with Hoxb4+Hoxc9 dn in the interlimb domain at stage 14 and re-incubated for 48 hr. The unilateral expansion of the forelimb is denoted by red arrowheads compared to contralateral side denoted by green arrowhead (n = 13/22 embryos). (A’)–(C’) shows Fgf10 (A’), Tbx5 (B’), and Shh (C’) expression in corresponding embryos. White arrowhead in (A’) points at ectopic Fgf10 expression within the electroporated region.
(D–G) Embryos electroporated with GFP alone (D and F; n = 7 embryos) or in combination with Hoxb4+Hoxc9 dn (E and G; n = 22 embryos) in the interlimb domain at stage 14 and re-incubated for 4 days. (F and G) Alcian Blue staining in GFP (F; n = 5 embryos) or Hoxb4-Hoxc9 dn-GFP (G; n = 9 embryos) electroporated embryos is shown. Note the posterior shift of the forelimb on the right electroporated side of Hoxb4-Hoxc9 dn-GFP electroporated embryos (E and G; n = 11/22 embryos) highlighted with red dashed line (E) and red arrow (G). Stars, somites; dashed lines, forelimb outline; arrows, vertebral level of the forelimb.
Scale bars are 200 μm in (A)–(C) and (A’)–(C’) and 500 μm in (D)–(G). See also Figure S4.
Relative Changes in Hox Collinear Activation Timing during Gastrulation Prefigure Bird Natural Variation in Limb Position
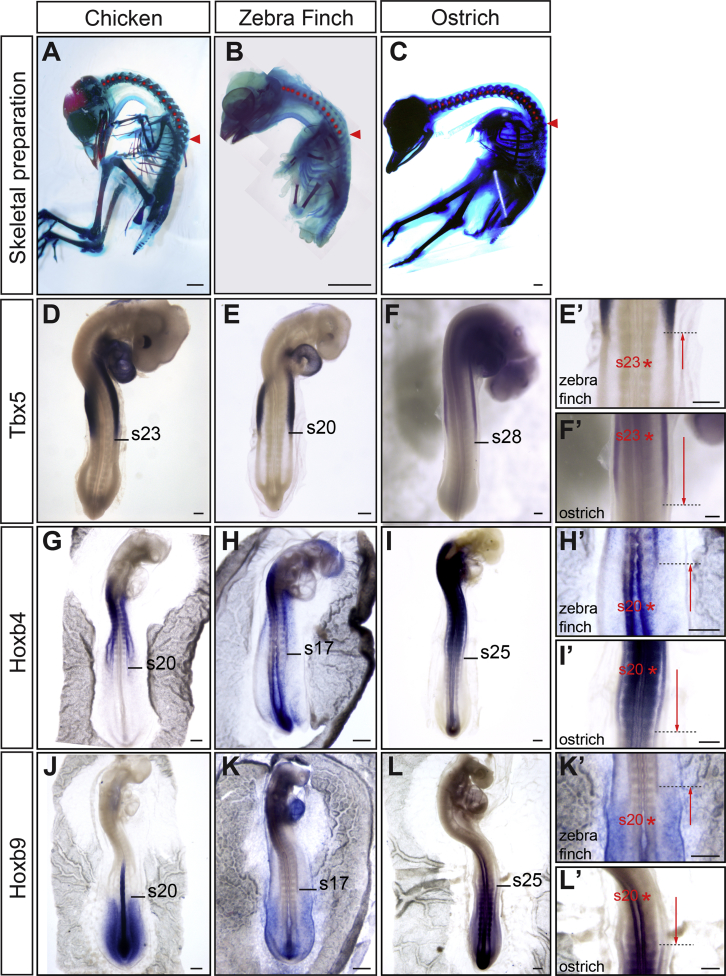
Altogether, these results argue that the timing of Hox activation during gastrulation defines the future positioning of their expression domain, which then determines the position of forelimb and interlimb fields. We next reasoned that natural variation in forelimb position between bird species, which exhibit neck of variable vertebrae number, should therefore be traced back to changes in the timing of Hox activation during gastrulation. In order to test this hypothesis, we compared the early development of three bird species, zebra finch (Guttata taeniopygia), chicken, and ostrich (Struthio camelus), whose forelimbs begin at the level of the 13th, 15th, and 18th vertebrae, respectively (Figures 5A–5C). Surprisingly, when compared to chicken embryo, the Tbx5-positive forelimb field was not simply translated anteriorly in zebra finch or posteriorly in ostrich (Figures 5D–5F). Instead, only the posterior border of the Tbx5 domain displayed an anterior shift (of about 3 somites) in zebra finch (Figures 5E and 5E’) and a posterior shift (of about 5 somites) in ostrich embryos (Figures 5F and 5F’). As a result, it is the extent of the forelimb field that varies between embryos of these species. A concomitant shift of the Hoxb4/Hoxb9 border by about 3–5 somites could be observed anteriorly in zebra finch (Figures 5H, 5H’, 5K, and 5K’) and posteriorly in ostrich embryos (Figures 5I, 5I’, 5L, and 5L’), when compared to chicken embryos (Figures 5G and 5J). Therefore, a shift in the Hoxb4/Hoxb9 border and in the posterior border of Tbx5 foreshadows the differences in limb position.
Figure 5.
Variations in Wing Position Correlate with Changes in Tbx5, Hoxb4, and Hoxb9 Expression Domains in the LPM
(A–C) Alcian blue-Alizarin red staining of chicken (A; E20), zebra finch (B; E13), and ostrich (C; E37). Red arrowheads point at the wing position level (15th, 13th, and 18th vertebrae in chicken, zebra finch, and ostrich embryos, respectively); red dots mark each cervical vertebra.
(D–F) Tbx5 expression in stage 18 chicken (D), zebra finch (E), and ostrich embryos (F). The position of Tbx5 posterior border of expression is indicated in somite number. (E’) and (F’) are higher magnification of (E) and (F), respectively.
(G–I) Hoxb4 expression in chicken (G; 20-somite stage), zebra finch (H; 20-somite stage), and ostrich embryos (I; 34-somite stage). The position of Hoxb4 posterior border of expression is indicated in somite number. (H’) and (I’) are higher magnification of (H) and (I), respectively.
(J–L) Hoxb9 expression in chicken (J; 20-somite stage), zebra finch (K; 20-somite stage), and ostrich embryos (L; 36-somite stage). The position of Hoxb9 anterior border of expression is indicated in somite number. (K’) and (L’) are higher magnification of (K) and (L), respectively.
Dashed black lines show variation in posterior-anterior border of expression in zebra finch (E’, H’, and K’) and ostrich embryos (F’, I’, and L’) compared to chicken embryo (represented by red asterisks).
Scale bars are 3 mm in (A)–(C) and 100 μm in (D)–(L), (E’), (F’), (H’), (I’), (K’), and (L’).
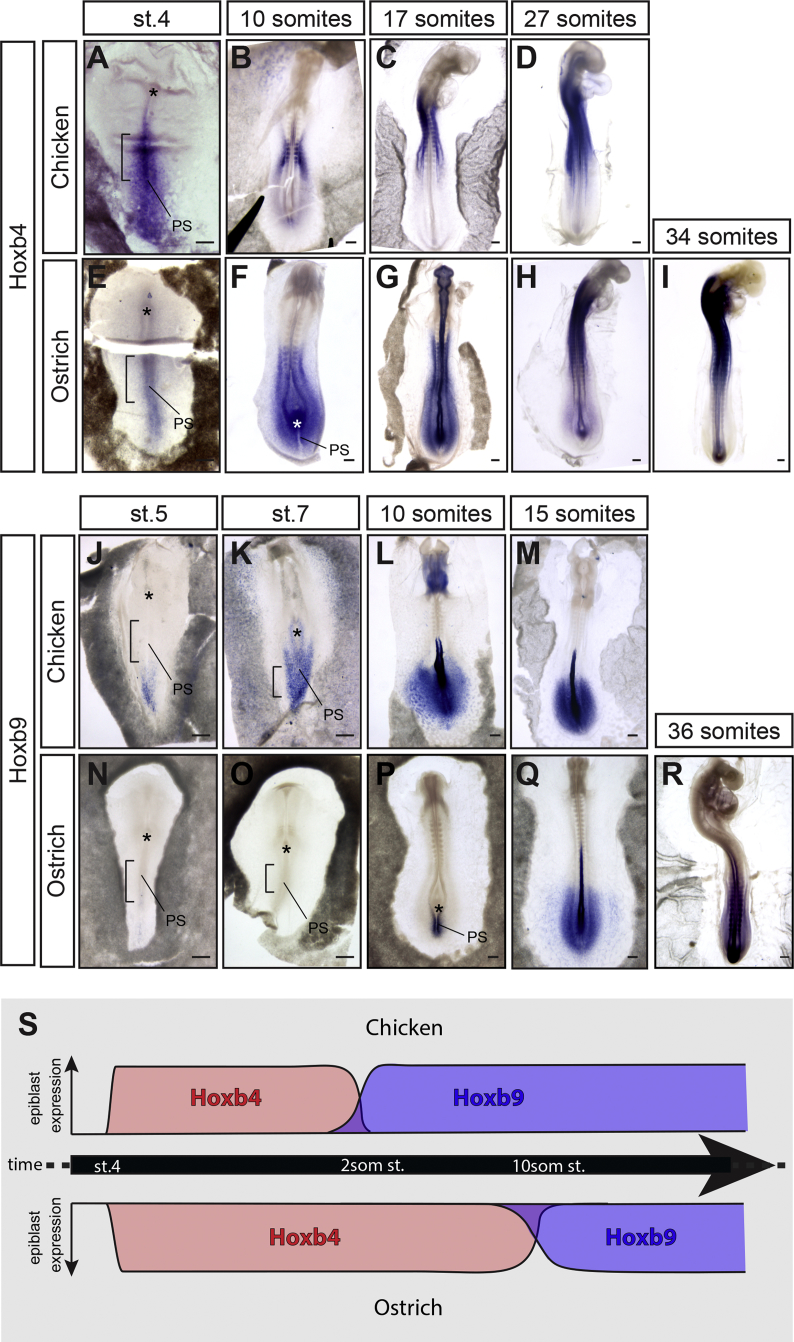
We then investigated whether temporal differences in the collinear activation of Hoxb4 and Hoxb9 genes during gastrulation could be linked to the spatial variation of the Hoxb4/Hoxb9 border in chicken and ostrich embryos. We found that Hoxb4 is activated at the same stage (stage 4) in both chicken and ostrich embryos (Figures 6A and 6E). However, Hoxb4 remained expressed in the epiblast for much longer in ostrich (10-somite stage) than in chicken embryos (2-somite stage; Figures 6B, 6F, and S2C). Concomitant to this delay in Hoxb4 arrest of expression in the epiblast, Hoxb9 activation was delayed in ostrich (10-somite stage; Figures 6N–6P) compared to chicken embryos (2-somite stage; Figures 6J and 6K). Ultimately, the Hoxb4/Hoxb9 border became posteriorly shifted in ostrich embryos when compared to chicken embryos, although at a later stage (Figures 6C, 6D, 6G–6I, 6L, 6M, 6Q, and 6R). Altogether, these results show that relative changes in the activation timing of Hoxb genes, in particular in the transition between Hoxb4 and Hoxb9, prefigure variation in the relative position of Hox domains and in the definitive limb position (summarized in Figure 6S).
Figure 6.
Relative Changes in Hox Collinear Activation Timing Underlie Variation in Limb Position between Chicken and Ostrich
(A–I) Hoxb4 expression in chicken (A–D) and ostrich (E–I) embryos.
(J–R) Hoxb9 expression in chicken (J–M) and ostrich (N–R) embryos.
(S) Timeline of Hoxb4 (red) and Hoxb9 (blue) activation in chicken (top diagram) and ostrich (bottom diagram).
Scale bars are 100 μm. Asterisks represent the Hensen’s node; black brackets outline the presumptive LPM.
Retinoic Acid Signaling Modulation during Gastrulation Changes the Extent of the Forelimb Field
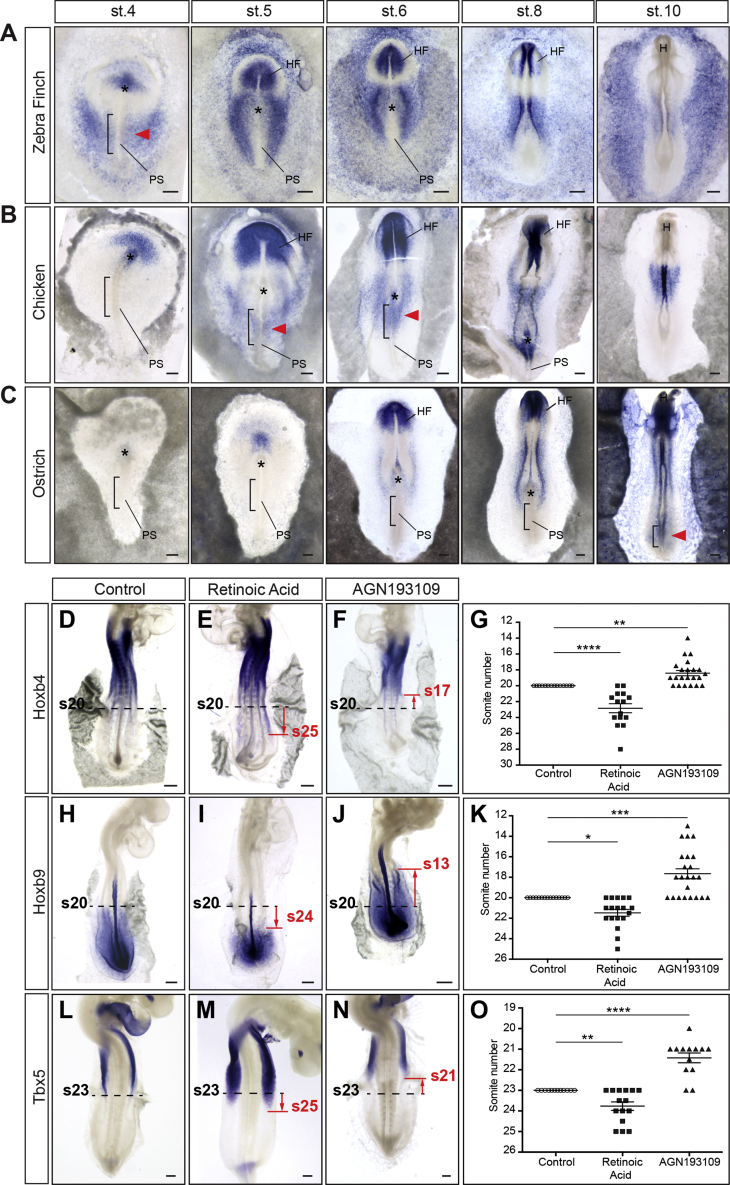
We next sought to understand how variation in Hox activation timing is controlled between zebra finch, chicken, and ostrich. Differences in the timing of expression of Gdf11 have recently been proposed to account for variation in hindlimb position between tetrapods [34]. Because Gdf11 acts through modulation of retinoic acid (RA) signaling by inducing Cyp26a1 expression (a RA catabolizing enzyme) [35] and because RA was shown to activate anterior Hox genes in the neural tube [36] and repress posterior Hox genes in the tail bud [37], Cyp26a1 stood as an excellent candidate to regulate variation in Hox activation timing. We therefore compared Cyp26a1 expression between the different species and found that it is first detected in the prospective LPM territory of the PS at stage 4 in zebra finch, stages 5–6 in chicken, and stage 10 in ostrich (Figures 7A–7C, red arrowheads). Therefore, Cyp26a1 is prematurely expressed in zebra finch and delayed in ostrich, compared to chicken embryo. Notably, the onset of Cyp26a1 expression in the PS precedes by a few hours the transition between Hoxb4 and Hoxb9 expression in the epiblast.
Figure 7.
Changes in Retinoic Acid Signaling during Gastrulation Modulate the Forelimb Field Position
(A–C) Cyp26a1 expression in zebra finch (A), chicken (B), and ostrich (C) embryos. Red arrowheads highlight the onset of Cyp26a1 expression in the presumptive LPM (black brackets); black asterisks, Hensen’s node; H, head.
(D–F) Hoxb4 expression in stage 13 control (D), retinoic-acid-treated (E), and AGN193109-treated (F) embryos.
(G) Position of Hoxb4 posterior border of expression in control (n = 14 embryos), retinoic-acid-treated (n = 15 embryos), and AGN193109-treated embryos (n = 22 embryos).
(H–J) Hoxb9 expression in stage 13 control (H), retinoic-acid-treated (I), and AGN193109-treated (J) embryos.
(K) Position of Hoxb9 anterior border of expression in control (n = 15 embryos), retinoic-acid-treated (n = 18 embryos), and AGN193109-treated embryos (n = 23 embryos).
(L–N) Tbx5 expression in stage 18 control (L), retinoic-acid-treated (M), and AGN193109-treated (N) embryos.
(O) Position of Tbx5 posterior border of expression in control (n = 12 embryos), retinoic-acid-treated (n = 15 embryos), and AGN193109-treated embryos (n = 13 embryos).
Red lines show changes in posterior-anterior border of expression in treated embryos compared to control embryos (dashed black lines).
Scale bars are 100 μm. (G, K, and O) Each dot represents one embryo; error bars represent mean ± SEM. Statistical analysis, ANOVA with Fisher least significant difference (LSD) post hoc test; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
Finally, we tested whether modulating RA signaling, specifically during gastrulation, could alter the spatial organization of Hox expression domains and the forelimb field position. When stage 4 chicken embryos were incubated with a 12-hr pulse (between stage 4 and stage 8; see STAR Methods) of RA or AGN193109 (a pan RA receptor antagonist), the posterior border of Hoxb4 became shifted of about 3–5 somites posteriorly and anteriorly, respectively (Figures 7D–7G). A complementary posterior and anterior shift of Hoxb9 anterior border was observed in embryos treated with RA and AGN193109, respectively (Figures 7H–7K). Therefore, the modulation of RA signaling during gastrulation changes the relative position of the Hoxb4/Hoxb9 border in the LPM at later stages. We next checked the effect on Tbx5 expression and observed a similar shift of its posterior border of about 2 or 3 somites posteriorly in embryos treated with RA and anteriorly when these embryos were treated with AGN193109 (Figures 7L–7O). Notably, this led to changes in the extent of Tbx5-positive forelimb field in RA- and AGN193109-treated embryos, which strikingly resemble the extent of forelimb fields in ostrich and zebra finch embryos, respectively (compare Figures 7M and 7N with Figures 5F and 5E, respectively). Altogether, these results show that modulation of RA signaling during gastrulation affects the axial extent of Hox expression domains in the lateral plate mesoderm and provokes modification in the A-P extent of Tbx5 expression, which is linked to variation in limb position.
Discussion
Our work identifies an early role for Hox genes in the regulation and variation of forelimb position in birds. During gastrulation, the lateral plate mesoderm is progressively generated and concomitantly patterned by Hox genes. As observed for the adjacent paraxial mesoderm, collinear activation of Hox genes in the epiblast progressively establishes their own collinear domains of expression in the LPM. It is the position of these expression domains that determines the position of forelimb-forming and interlimb-forming domains through a combination of activation (e.g., Hoxb4) and repression (e.g., Hox9) of limb initiation (i.e., Tbx5 expression) and eventually the definitive limb position. Furthermore, relative changes in the collinear activation of Hox genes during gastrulation prefigure variation in the spatial organization of Hox genes expression domains and natural variation in limb position between birds. We also observe differences in the onset of expression of the RA-catabolizing enzyme Cyp26a1 during gastrulation and show that modulation of RA signaling provokes changes in limb field position. Based on our findings, we therefore propose that timely controlled Hox collinear activation during gastrulation is responsible for the regulation and variation in forelimb position in birds.
Progressive Formation and Patterning of the LPM by Collinear Activation of Hox Genes
Our results show that the LPM is progressively formed during gastrulation. Surprisingly, whereas the prospective territory of the LPM had been traced back to the middle third of the PS [16, 17, 20], how the forelimb, interlimb, and hindlimb arise from this field had not been addressed. Here, we show that these fields sequentially form between stages 4 and 10, (between 24 hr and 48 hr of development). By characterizing the precise timing of forelimb, interlimb, and hindlimb domain formation, we could link the formation of these domains to the concomitant Hox genes collinear activation. The comparison of Hox activation in zebra finch, chicken, and ostrich embryos further revealed that relative differences in the timing of Hox activation prefigure spatial variation of their domain of expression as predicted by experiments performed in chicken. A previous study has shown that, in the context of paraxial mesoderm formation, Hox genes, collinearly activated, establish their own spatial collinear characteristic pattern through the regulation of cell ingression at the PS [28]. Using both overexpression and loss-of-function approaches, we show that a similar mechanism is at work during the generation of LPM, with a direct implication on its patterning into limb- and non-limb-forming domains but also on variation in body plan organization. The finding that paraxial and LPM are similarly generated and patterned during gastrulation provides a simple explanation for the concomitant patterning of the cervico-thoracic frontier in the somites and the associated forelimb position in the LPM.
In birds, the variation of limb position involves meristic variations (characterized by changes in the total number of component parts) [38]. As an example, whereas sparrows have 9 cervical vertebrae, swans exhibit 16 additional vertebrae. These meristic variations imply that, at the embryonic level, a larger number of mesodermal cells have to be produced to form both the somitic and corresponding LPM tissue. A recent study has linked the collinear activation of posterior Hox genes (Hox9 to 13) to the regulation of axis elongation and its termination. The temporal collinear activation of posterior Hox genes was shown to slow down the influx of mesodermal cells through the PS in a collinear trend, thereby controlling the elongation rate [30]. Coupling both the patterning of mesodermal tissues and the rate of axis elongation to the same regulatory mechanism would ensure that addition of anterior mesodermal tissue (somites and LPM) is not made at the expense of the posterior mesoderm. Such coupling would therefore allow meristic variations observed in birds while maintaining the vertebrate body integrity.
Hox Genes Expression Determines Limb- and Non-limb-Forming Domains
Among Hox gene deletions in mouse, the Hoxb5 mutant is the only one reported to exhibit a change in the position of its forelimbs. Whereas these mice were described to exhibit a mildly penetrant anterior shift in the definitive position of the forelimb, the underlying mechanisms remained unclear, because only late stages were analyzed [13]. The anterior-ward shortening of Hoxb4 and Tbx5 domains we obtained upon electroporation of Hoxb4 dn is in full agreement with a role of Hox4/5 genes in the regulation of forelimb domain position and suggests that the shift in limb position in Hoxb5 mutant mice might have arisen from patterning defects at gastrulation stages. Aside from Hoxb5 mutants, none of the single or compound Hox mutants along with Hox overexpression approaches in mice have been reported to provoke major phenotypes regarding limb positioning, putting their role in this process into question [39]. More recently, evidences reinforcing the role of Hox genes in positioning limb fields have accumulated. Hox paralogs 4 and 5 can bind to a Tbx5-forelimb-specific enhancer and activate transcription of a downstream reporter [10, 11]; however, this has not been shown in vivo on the endogenous Tbx5 expression. Intriguingly, whereas Tbx5 expression is greatly conserved among vertebrates, the above-mentioned Tbx5 limb-specific enhancer, located in intron 2 in mouse, could not be located in chicken, raising doubts about the requirement of this particular enhancer for Tbx5 forelimb expression. Moreover, the fact that Tbx5 has recently been shown to be insufficient (although necessary) for forelimb initiation [40] also raised concerns about the sufficiency of displacing forelimb position by solely displacing Tbx5 domain. Hoxc9, in turn, was shown to bind directly Tbx5 limb enhancer and to inhibit its expression. As a consequence, it has been proposed that there is a latent potential in the caudal LPM to express Tbx5, which is normally masked by the presence of Hoxc8–10 genes [11, 41]. Our data, which show that ectopic expression of Hoxb4 and Hoxc9 dn can induce Tbx5 expression and displace the forelimb, are in full support with a role of Hox4/5 genes in regulating Tbx5 expression in vivo. However, the finding that Hoxc9 dn on its own does not promote Tbx5 expression or forelimb displacement but does so only in combination with Hoxb4 has several important implications with respect to the regulation and variation of limb position. First, it suggests that there is no latent potential of forelimb forming activity in the interlimb but “just” a repressive forelimb-forming activity. Second and corollary to this, it suggests that, to shift limb position, both a shift of the forelimb field (e.g., Hoxb4 expression, which is normally not expressed posteriorly) and of the interlimb field (e.g., Hoxb9 anterior border of expression) must be performed. In other words, changes in limb position can be induced only if the overall spatial sequence of Hox expression pattern is changed. This might explain why the vast majority of the single and compound mutants for a variety of Hox genes, including Hox5 and Hox9 groups, do not show a major phenotype on limb position [33, 42]. Indeed, our data argue that, to induce a shift in forelimb position in mouse, a combination of gain and loss of function for forelimb-activator and forelimb-repressor Hox genes, respectively, should be performed. Based on these results, we propose that the sequence limb-forming/non-limb-forming domains is embedded in the collinear organization of Hox genes through their specific activation-repression function on limb initiation; it is in turn the timing of collinear activation that sets the relative position of these domains along the main axis.
Posterior Border of the Early Limb Field and Final Limb Position
Our results point at the posterior border of the forelimb (Hoxb4/Hoxb9 border) as a critical regulator of limb positioning because only the position of the posterior (and not the anterior) border of Hoxb4 and Tbx5 is gradually shifted in zebra finch, chicken, and ostrich. This automatically leads to a gradual extension of the forelimb field rather than a gradual posterior translation; however, the limb becomes eventually posteriorly shifted between these species. How can a variation in forelimb field size lead to variation in limb position? The early limb mesenchyme is pre-patterned by opposing gradients of Gli3 and dHand, which set the position of the ZPA [43]. Modulation of this Gli3-dHand pre-patterning (by modulating Tbx3 activity) subsequently modifies the position of the ZPA and eventually the position of the limb [43]. Our results showing that, upon Hoxb4-Hoxc9 dn electroporation, Shh expression is shifted posteriorly suggest that such pre-pattern is downstream of Hox genes function and that it might well play a role in defining the definitive limb position. Indeed, Hox5 mutant mice show derepression of Shh anteriorly [42], whereas in Hox9 mutants, Shh is not expressed posteriorly due to defects in Gli3-dHand pre-patterning of the early mesenchyme [33]. It is thus tempting to speculate that Hox4/5, Hox9, and presumably other Hox genes not only set forelimb fields of variable size in different species but also pre-pattern the early mesenchyme accordingly, provoking variation in the position of the ZPA, which would, in turn, be responsible for refining the definitive position of the forelimb. However, neither Shh mutant mice nor the oligozeugodactyly (OZD) mutant in chicken have been reported to exhibit variation in limb position [44, 45]. Therefore, in this view, the ZPA would not be a regulator of forelimb positioning per se but rather a posterior anchor, refining the position of forelimb fields of different A-P extents by biasing outgrowth toward the posterior border, although this remains just speculative. Furthermore, the fact that Tbx5 activation upon electroporation of Hoxb4-Hoxc9 dn does not induce an independent, ectopic limb bud but instead expands the pre-existing forelimb domain is in agreement with a concomitant re-patterning into a single larger limb field. It also supports the recent report of Tbx5 insufficiency in promoting (ectopic) limb bud formation [40] as opposed to what was previously published [46].
RA Signaling as Regulator of Both Forelimb and Hindlimb Position?
We show that Cyp26a1 is prematurely expressed in zebra finch and delayed in ostrich when compared to chicken and correlates with the activation of interlimb-specific Hox genes, such as Hoxb7 or Hoxb9, in each of these species. Furthermore, we find that modulation of RA signaling during gastrulation leads to changes in the extent of Hox genes expression patterns while preserving overall collinearity. Consequently, we observe a variation in the posterior border of the forelimb domain, as revealed by Tbx5 expression, which recapitulates natural variation observed between zebra finch, chicken, and ostrich. The possibility that Cyp26a1, which has been implicated in positioning hindlimbs [35], might regulate forelimb position is particularly interesting because a single signaling pathway would therefore be responsible for the regulation of both forelimb and hindlimb position by acting at different developmental timings.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-GFP | Torrey Pines Biolabs | Cat#TP-401; RRID: AB_10013661 |
| Goat anti-Rabbit IgG (H+L) Secondary Antibody, Alexa Fluor Plus 488 | Thermo Fisher Scientific | Cat#A32731; RRID:AB_2633280 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Alexa Fluor 555 Phalloidin | Thermo Fisher Scientific | Cat#A34055 |
| Fluoromount-G, with DAPI | Thermo Fisher Scientific | Cat#00-4959-52 |
| RA | Sigma-Aldrich | Cat#R2625 |
| AGN 193109 | Tocris | Cat#5758 |
| Alcian Blue 8GX | Sigma-Aldrich | Cat#A9186 |
| Alizarin Red S | Sigma-Aldrich | Cat#A5533 |
| Experimental Models: Organisms/Strains | ||
| Fertilized chicken eggs | EARL Morizeau | N/A |
| Fertilized ostrich eggs | SARL Le père Louis | N/A |
| Fertilized zebra finch eggs | Laboratory of Marie Manceau | N/A |
| Transgenic quail: hUbC:memGFP | This paper | N/A |
| Transgenic quail: hUbC:mEOS2FP | This paper | N/A |
| Oligonucleotides | ||
| Primer Hoxb4 dn Forward: AATAAATCTAGAATGGCCATGAGCT CGTTTTTGATCAACTCC |
This paper | N/A |
| Primer Hoxb4 dn Reverse: TTTTGGCGCGCCTCAGATTTTGATCTGGC | This paper | N/A |
| Primer Hoxb4 C-terminal Forward: CCAGAACAGGAGGATGAAATGG | This paper | N/A |
| Primer Hoxb4 C-terminal Reverse: TTATAGGCTGCTGGCTGGTCC | This paper | N/A |
| Primer Hoxc9 dn Forward: TTTGCTAGCATGTCGGCTTCTGGCCCC ATAAGC |
This paper | N/A |
| Primer Hoxc9 dn Reverse: TTTGGCGCGCCTCAGATTTTGACTTGGC | This paper | N/A |
| Primer cCyp26a1 Forward: ATCCTGCTGGGCTTCCAGCCC | This paper | N/A |
| Primer cCyp26a1 Reverse: GGCCGCTGAAACCTATGAATTTGG | This paper | N/A |
| Recombinant DNA | ||
| pCAGGS-H2b-RFP | Laboratory of S. Tajbakhsh | N/A |
| pFlox-pA-EGFP | [47] | N/A |
| pCX-Cre | [47] | N/A |
| pCAG-GFP | [48] | Addgene Cat#11150 |
| pCAGGS-cHoxb4-IRES2-Venus | This paper | N/A |
| pCAGGS-cHoxb4 dn-IRES2-Venus | This paper | N/A |
| pCAGGS-cHoxb7-IRES2-Venus | This paper | N/A |
| pCAGGS-cHoxb9-IRES2-Venus | This paper | N/A |
| pCAGGS-cHoxc9 dn-IRES2-Venus | This paper | N/A |
| pBS-Hoxb4 | [28] | N/A |
| pBSKS-Hoxb7 | [28] | N/A |
| pGEMT easy-Hoxb9 | [28] | N/A |
| pSLAX-cTbx5 | Laboratory of C. Tabin | N/A |
| pBSSK-cFgf10 | Laboratory of C. Tabin | N/A |
| pBSSK-cShh | Laboratory of C. Tabin | N/A |
| pGEMT easy-cCyp26a1 | This paper | N/A |
| pGEMT easy-cHoxb4Cter | This paper | N/A |
| Software and Algorithms | ||
| ImageJ | [49] | RRID:SCR_003070 |
| GraphPad Prism 6 | GraphPad Software | RRID:SCR_002798 |
Contact for Reagents and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jerome Gros (jgros@pasteur.fr).
Experimental Model and Subject Details
Avian Embryos
Fertilized chicken and ostrich eggs were ordered from commercial sources (chicken: EARL Morizeau; ostrich: SARL Le père Louis), fertilized zebra finch eggs were generously provided by Dr Marie Manceau from Collège de France (Paris) and transgenic quail eggs were produced in the lab. Chicken, quail and zebra finch eggs were incubated at 38°C and ostrich eggs at 36°C up to the appropriate developmental stage (details provided in the Methods Details section). All embryos were staged according to the Hamburger and Hamilton classification system [50].
Animals
All experimental methods and animal husbandry procedures to generate transgenic quails were carried out in accordance with the guidelines of the European Union 2010/63/UE, approved by the Institut Pasteur ethics committee, and under the GMO agreement #2432.
Generation of transgenic quail lines
Two transgenic lines were created in this study (hUbC:memGFP and hUbC:mEOS2FP) by following previously published method [51]. Briefly, non-incubated quail eggs (Coturnix japonica) were windowed and a solution of high titer lentivirus was injected into the subgerminal cavity of stage X embryos. Eggs were sealed with a plastic piece and paraffin wax. Injected eggs were incubated at 38°, 56% humidity until hatching. For the hUbC:memGFP line, a total of 42 embryos were injected with the lentivirus solution (titer 1010/ml). Three F0 mosaic founder males successfully hatched and reached sexual maturity (7%). They were bred to WT female and all three produced transgenic offspring (transmission rate: 8.8%). One line was selected on the basis of a single copy of the transgene, checked by Southern Blot, and high intensity of the memGFP signal. For the hUbC:mEOS2FP line, a total of 141 embryos were injected with lentivirus (titer 6,4.1010/ml). Five F0 mosaic founder successfully hatched and reached sexual maturity (3.5%). All five produced transgenic offspring (transmission rate: 6.1%) and one line was selected by Southern Blot analysis for single transgene integration and high intensity of the mEOS2 fluorescent signal.
Method Details
LPM rotation
LPM rotations were performed in stage 11 embryos. The ectoderm was carefully detached in order to only microdissect the right somatopleure encompassing both forelimb and interlimb prospective domains, as previously proposed [21, 52]. This tissue was rotated along its A-P axis and grafted back into the same embryo. Eggs were further sealed with tape and re-incubated at 38°C. After 48h, embryos were harvested and fixed in 4% formaldehyde for 2h at room temperature.
A similar procedure was used to perform quail-chick chimeric grafts. Stage 11 hUbC:memGFP transgenic quail embryos were collected and placed into a Petri dish filled with PBS, the somatopleure was microdissected as described above and grafted to stage 11 host chicken embryos.
Embryo Culture
Embryos were prepared for ex ovo culture using a modified version of the EC culture system [27]. Briefly, embryos were collected at stage 4 using filter paper rings and cultured on a semisolid nutritive medium containing a mix of albumen, agarose (0,2%), glucose and NaCl. Embryos were then incubated at 38°C in a humidified chamber and cultured for up to 48h.
Embryo Electroporation
Ex ovo Electroporation
Stage 4 embryos collected on a filter paper ring were electroporated in a custom-made electroporation chamber using the SuperElectroporator NEPA21 type II® (NEPAGENE) with two 5ms poring pulses of 15V, 50ms delay, and three 50ms transfer pulses of 10V, 500ms delay. Solutions of plasmid DNA were prepared as previously described [53] with a final DNA concentration of 1 μg/μl. Embryos were then cultured ex ovo for up to 48h, harvested and fixed in 4% formaldehyde for 2h at room temperature.
In ovo Electroporation
In ovo electroporation was performed as previously described [53]. Briefly, eggs were incubated up to stage 14 (52h incubation). DNA solution with a final concentration of 5 μg/μl was injected in the coelomic cavity at the level of the forelimb and interlimb domains. All the electroporations were performed on the right side of the embryo. Electroporation was performed using homemade electrodes and the SuperElectroporator NEPA21 type II® (NEPAGENE) with two 1ms poring pulses of 70V, 100ms delay and three 2ms transfer pulses of 40V, 500ms delay. Eggs were sealed with tape and re-incubated for 24h, 48h or 4 days. Embryos were then harvested and fixed in 4% formaldehyde for 2h at room temperature.
Time-Lapse Microscopy
Stage 4 embryos co-electroporated with pCAGGS-H2b-RFP (1 μg/μl), pFlox-pA-EGFP (1 μg/μl) and pCX-Cre (50ng/μl) plasmid DNAs, were placed into medium-containing glass-bottom Petri dishes (MatTek inc.). Embryos were imaged at 38°C using an inverted 2-photon microscope (Zeiss, NLO LSM 7MP) coupled to a Chameleon Ti/Saph femtosecond pulsed laser and OPO system (Coherent inc.) at 840nm (GFP) and 1100nm (RFP) wavelengths using a 10X long-distance objective. Embryos were acquired every 5min for about 24 hours.
Photoconversion of hUbC:mEOS2FP transgenic quail embryos was performed using an inverted confocal microscope (ZEISS LSM 880). Before photoconversion, mEOS2 was visualized under standard imaging conditions for GFP using a 488nm Argon laser. A 405nm diode laser (40% power with 1x25 iterations) was used for photocoversion. Photoconverted mEOS2 was subsequently imaged using a 561nm HeNe laser. Embryos were imaged every 2 hours to follow the photoconverted region of the embryo, without bleaching.
Transient Drug Treatments
Chicken embryos were incubated up to stage 4 and RA (Sigma, 400 μM in DMSO) or AGN193109 (Tocris, 100 μM in DMSO) were injected in between the vitelline membrane and the embryo, on top of the PS. Note that lower concentrations did not yield any phenotype. Control embryos were treated with equivalent concentration of DMSO-only. Eggs were sealed with tape and re-incubated for 12h (up to stage 8, before the end of LPM formation). Embryos were then collected on a filter paper ring, washed in PBS solution to stop exposition to RA or AGN193109 and cultured ex ovo in EC culture plates for 24h (up to stage 13, 19-22 somites) or 36h (up to stage 18, 30-36 somites). Embryos were then harvested and fixed in 4% formaldehyde for 2h at room temperature.
DNA constructs
Hoxb4, Hoxb7 and Hoxb9 were subcloned into the pCAGGS-IRES2-Venus vector [28, 30] and were used alone or in combination with pCAGGS-GFP [48]. The constructs pFlox-pA-EGFP and pCX-Cre were kindly provided by X. Morin [47] and used in combination with pCAGGS-H2b-RFP, kindly provided by S. Tajbakhsh. The truncated form of HOXB4 and HOXC9 were generated by inserting a stop codon to replace the highly conserved tryptophan amino acid on the α-helix III of the Homeodomain (amino acid 50 of the Homeodomain) as previously described [30, 31, 32] and further subcloned into the pCAGGS-IRES2-Venus vector.
In situ hybridization
In situ hybridization in chick embryos was performed as previously described [54]. Briefly, formaldehyde fixed embryos were dehydrated in PBS/0,1%Tween with increasing methanol concentrations (25%, 50%, 75% and 100%) and further rehydrated. Embryos were then treated with Proteinase K (10 μg/mL) and refixed with 4% formaldehyde/0,1% glutaraldehye. Hybridization with DIG-labeled RNA probes was performed under stringent conditions (5X SSC pH4,5, 50% formamide, 1% SDS, 50 μg/mL yeast tRNA, 50 μg/mL heparin; at 65°C). Washed embryos were treated with MABT(20mM Maleic Acid pH7,5; 30mM NaCl; 0,02% Tween-20)/2%BBR(Boehringer blocking reagent)/20%Lamb Serum and incubated overnight with AP-coupled anti-DIG antibody. Washed embryos were then stained with NBT/BCIP® liquid substrate (Sigma). Pictures of whole embryos were made using a macroscope (SteREO Discovery.V8, ZEISS) with a 1X objective and a color camera (AxioCam MRc, Zeiss).
DIG-labeled probes were generated from plasmids containing cDNA fragments of cHoxb4, cHoxb7, cHoxb9 (as previously published [28]), cTbx5, cFgf10, cShh (gifts from C. Tabin). A 900bp-fragment of the Cyp26a1 coding sequence (from nucleotide 571 to 1471) was PCR-amplified from chicken cDNA and cloned in pGEM®-T Easy vector (Promega) to be used as a probe. The Hoxb4 C-terminal specific probe was generated by PCR-amplification of the last 145 nucleotides of the chicken Hoxb4 coding sequence (from nucleotide 597 to 741) corresponding to the C-terminal portion truncated in the Hoxb4 dn construct, and further cloned in pGEM®-T Easy vector (Promega).
Immunostaining and Labeling
Fixed embryos were embedded in 7.5% gelatin/15% sucrose and sectioned using a Leica CM3050S cryostat. Sections were degelatinized in PBS at 37°C for 30min, blocked in PBS containing 0,1% Triton and 20% goat serum for 30min at room temperature. Sections were incubated with Alexa Fluor 555 Phalloidin (1:100, Invitrogen) and rabbit anti-GFP primary antibody (1:500, Torrey Pines Biolabs) overnight at 4°C, washed with PBS for 24h, incubated 2h at room temperature with goat anti-rabbit Alexa Fluor 488 secondary antibody (1:1000, Invitrogen) and washed with PBS for 24h. Sections were then mounted with DAPI-containing Fluoromount-G and imaged using an inverted confocal microscope (Zeiss LSM700) with a 20X objective.
Skeleton analysis
Embryos electroporated with GFP alone or together with Hoxb4+Hoxc9 dn were harvested 4 days after electroporation, placed in ice-cold 95% ethanol for 1h, and transferred to fresh 95% ethanol overnight at room temperature. Embryos were then stained in freshly prepared Alcian Blue solution (5% of 0,4%Alcian Blue 8GX in 70% ethanol/5% glacial acetic acid/70% of 95% ethanol/20% water) for 48h at room temperature to label cartilage. Embryos were briefly rinsed in water, transferred to 1% KOH solution for 30min and cleared in successive solutions of glycerol/0,25%KOH with increased glycerol concentration (20%, 33% and 50%) for 1h at room temperature. Embryos were then transferred in fresh 50%glycerol/0,25%KOH for further clearing and storage.
Chicken (ch), zebra finch (zf) and ostrich (os) embryos were collected at late development stage (ch: E20; zf: E13; os: E37) and stained with Alcian Blue and Alizarin Red to label cartilage and bone tissues, respectively, as previously described [55]. Briefly, dissected embryos were fixed in 95% ethanol overnight at 4°C, further stained in Alcian Blue solution (ch: 17 days; zf: overnight; os: 26 days) and washed in 95% ethanol. Embryos were cleared in 1% KOH and stained with Alizarin Red solution (ch: 5 days; zf: 4h; os: 7 days). Embryos were transferred in 1% KOH and and equilibrated in successive solutions of glycerol/1%KOH with increased glycerol concentration (20%, 50% and 90%) for further clearing and storage.
Note that no strategy of randomization and/or stratification, blinding, sample-size estimation or inclusion/exclusion of data or subjects were required to perform these experiments.
Quantification and Statistical Analysis
Time-lapse videos of embryos electroporated with cytoplasmic GFP and nuclear H2b-RFP fluorescent reporters were analyzed using ImageJ software [49] and retrospective cell tracking was performed using the Manual Tracking plugin from Fabrice Cordelières.
Stage 13 embryos electroporated at stage 4 with GFP, GFP/Hoxb4, GFP/Hoxb7, GFP/Hoxb9 or GFP/Hoxb4 dn, were analyzed for electroportated cell distribution. The LPM was segmented along the A-P axis into neck, forelimb, interlimb and hindlimb regions. Using ITCN (Imaged-based Tool for Counting Nuclei) plugin from Thomas Kuo and Jiyun Byun (UC Santa Barbara) in ImageJ software, the number of electroporated cells was quantified in each region and normalized relative to the area of these regions.
For each experiment, n represents the number of embryos analyzed. Statistical analyzes were performed using Prism 6 software (GraphPad software). Results were considered significant when p < 0,05. ∗ p < 0,05; ∗∗ p < 0,01; ∗∗∗ p < 0,001; ∗∗∗∗ p < 0,0001. Error bars represent mean ± SEM. Statistical details of each experiment are provided in the figures and corresponding legends.
Acknowledgments
We thank Francois Schweisguth and Cliff Tabin for critical reading of the manuscript and Marie Manceau for providing zebra finch fertilized eggs. C.M. was supported by a fellowship from the French Ministère de l’Enseignement supérieur de la Recherche et de l’Innovation. The research leading to these results has received funding from the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013)/ERC grant agreement no. 337635 and from the Institut Pasteur, the Centre National de la Recherche Scientifique, and the Fondation Schlumberger pour l’Education et la Recherche.
Author Contributions
Conceptualization, C.M. and J.G.; Methodology, D.R. and J.R.; Investigation, C.M., P.C., and J.G.; Writing – Original Draft, J.G.; Writing – Review and Editing, J.G., C.M., and O.P.; Resources, N.D. and O.P.; Funding Acquisition, J.G.; Supervision, J.G.
Declaration of Interests
The authors declare no competing interests.
Published: December 13, 2018
Footnotes
Supplemental Information includes four figures and three videos and can be found with this article online at https://doi.org/10.1016/j.cub.2018.11.009.
Supplemental Information
References
- 1.Tanaka M. Molecular and evolutionary basis of limb field specification and limb initiation. Dev. Growth Differ. 2013;55:149–163. doi: 10.1111/dgd.12017. [DOI] [PubMed] [Google Scholar]
- 2.Tanaka M. Developmental mechanism of limb field specification along the anterior-posterior axis during vertebrate evolution. J. Dev. Biol. 2016;4:18. doi: 10.3390/jdb4020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tickle C. How the embryo makes a limb: determination, polarity and identity. J. Anat. 2015;227:418–430. doi: 10.1111/joa.12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dollé P., Izpisúa-Belmonte J.-C., Falkenstein H., Renucci A., Duboule D. Coordinate expression of the murine Hox-5 complex homoeobox-containing genes during limb pattern formation. Nature. 1989;342:767–772. doi: 10.1038/342767a0. [DOI] [PubMed] [Google Scholar]
- 5.Gaunt S.J., Sharpe P.T., Duboule D. Spatially restricted domains of homeo-gene transcripts in mouse embryos: relation to a segmented body plan. Development. 1988;104:169–179. [Google Scholar]
- 6.Izpisúa-Belmonte J.C., Falkenstein H., Dollé P., Renucci A., Duboule D. Murine genes related to the Drosophila AbdB homeotic genes are sequentially expressed during development of the posterior part of the body. EMBO J. 1991;10:2279–2289. doi: 10.1002/j.1460-2075.1991.tb07764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burke A.C., Nelson C.E., Morgan B.A., Tabin C. Hox genes and the evolution of vertebrate axial morphology. Development. 1995;121:333–346. doi: 10.1242/dev.121.2.333. [DOI] [PubMed] [Google Scholar]
- 8.Cohn M.J., Patel K., Krumlauf R., Wilkinson D.G., Clarke J.D., Tickle C. Hox9 genes and vertebrate limb specification. Nature. 1997;387:97–101. doi: 10.1038/387097a0. [DOI] [PubMed] [Google Scholar]
- 9.Hostikka S.L., Gong J., Carpenter E.M. Axial and appendicular skeletal transformations, ligament alterations, and motor neuron loss in Hoxc10 mutants. Int. J. Biol. Sci. 2009;5:397–410. doi: 10.7150/ijbs.5.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minguillon C., Nishimoto S., Wood S., Vendrell E., Gibson-Brown J.J., Logan M.P.O. Hox genes regulate the onset of Tbx5 expression in the forelimb. Development. 2012;139:3180–3188. doi: 10.1242/dev.084814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishimoto S., Minguillon C., Wood S., Logan M.P.O. A combination of activation and repression by a colinear Hox code controls forelimb-restricted expression of Tbx5 and reveals Hox protein specificity. PLoS Genet. 2014;10:e1004245. doi: 10.1371/journal.pgen.1004245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mallo M., Wellik D.M., Deschamps J. Hox genes and regional patterning of the vertebrate body plan. Dev. Biol. 2010;344:7–15. doi: 10.1016/j.ydbio.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rancourt D.E., Tsuzuki T., Capecchi M.R. Genetic interaction between hoxb-5 and hoxb-6 is revealed by nonallelic noncomplementation. Genes Dev. 1995;9:108–122. doi: 10.1101/gad.9.1.108. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Martinez V., Schoenwolf G.C. Primitive-streak origin of the cardiovascular system in avian embryos. Dev. Biol. 1993;159:706–719. doi: 10.1006/dbio.1993.1276. [DOI] [PubMed] [Google Scholar]
- 15.James R.G., Schultheiss T.M. Patterning of the avian intermediate mesoderm by lateral plate and axial tissues. Dev. Biol. 2003;253:109–124. doi: 10.1006/dbio.2002.0863. [DOI] [PubMed] [Google Scholar]
- 16.Psychoyos D., Stern C.D. Fates and migratory routes of primitive streak cells in the chick embryo. Development. 1996;122:1523–1534. doi: 10.1242/dev.122.5.1523. [DOI] [PubMed] [Google Scholar]
- 17.Sawada K., Aoyama H. Fate maps of the primitive streak in chick and quail embryo: ingression timing of progenitor cells of each rostro-caudal axial level of somites. Int. J. Dev. Biol. 1999;43:809–815. [PubMed] [Google Scholar]
- 18.Garcia-Martinez V., Schoenwolf G.C. Positional control of mesoderm movement and fate during avian gastrulation and neurulation. Dev. Dyn. 1992;193:249–256. doi: 10.1002/aja.1001930305. [DOI] [PubMed] [Google Scholar]
- 19.Schoenwolf G.C., Garcia-Martinez V., Dias M.S. Mesoderm movement and fate during avian gastrulation and neurulation. Dev. Dyn. 1992;193:235–248. doi: 10.1002/aja.1001930304. [DOI] [PubMed] [Google Scholar]
- 20.Sweetman D., Wagstaff L., Cooper O., Weijer C., Münsterberg A. The migration of paraxial and lateral plate mesoderm cells emerging from the late primitive streak is controlled by different Wnt signals. BMC Dev. Biol. 2008;8:63. doi: 10.1186/1471-213X-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaube S. On axiation and symmetry in transplanted wing of the chick. J. Exp. Zool. 1959;140:29–77. doi: 10.1002/jez.1401400104. [DOI] [PubMed] [Google Scholar]
- 22.Saito D., Yonei-Tamura S., Kano K., Ide H., Tamura K. Specification and determination of limb identity: evidence for inhibitory regulation of Tbx gene expression. Development. 2002;129:211–220. doi: 10.1242/dev.129.1.211. [DOI] [PubMed] [Google Scholar]
- 23.Kieny M. Les phases d’activité morphogène du mésoderme somatopleural pendant le développement précoce du membre chez l’embryon de poulet. Ann. Embryol. Morphog. 1971;4:281–298. [Google Scholar]
- 24.Hamburger V. Morphogenetic and axial self-differentiation of transplanted limb primordia of 2-day chick embryos. J. Exp. Zool. 1938;77:379–399. [Google Scholar]
- 25.Pinot P.M. Le rôle du mésoderme somitique dans la morphogenèse précoce des membres de l’embryon de Poulet. Development. 1970;23:109–151. [PubMed] [Google Scholar]
- 26.Rudnick D. Limb-forming potencies of the chick blastoderm: including notes on associated trunk structures. Trans. Conn. Acad. Arts Sci. 1945;36:353–377. [Google Scholar]
- 27.Chapman S.C., Collignon J., Schoenwolf G.C., Lumsden A. Improved method for chick whole-embryo culture using a filter paper carrier. Dev. Dyn. 2001;220:284–289. doi: 10.1002/1097-0177(20010301)220:3<284::AID-DVDY1102>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 28.Iimura T., Pourquié O. Collinear activation of Hoxb genes during gastrulation is linked to mesoderm cell ingression. Nature. 2006;442:568–571. doi: 10.1038/nature04838. [DOI] [PubMed] [Google Scholar]
- 29.Gehring W.J., Müller M., Affolter M., Percival-Smith A., Billeter M., Qian Y.Q., Otting G., Wüthrich K. The structure of the homeodomain and its functional implications. Trends Genet. 1990;6:323–329. doi: 10.1016/0168-9525(90)90253-3. [DOI] [PubMed] [Google Scholar]
- 30.Denans N., Iimura T., Pourquié O. Hox genes control vertebrate body elongation by collinear Wnt repression. eLife. 2015;4:e04379. doi: 10.7554/eLife.04379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mortlock D.P., Innis J.W. Mutation of HOXA13 in hand-foot-genital syndrome. Nat. Genet. 1997;15:179–180. doi: 10.1038/ng0297-179. [DOI] [PubMed] [Google Scholar]
- 32.de Santa Barbara P., Roberts D.J. Tail gut endoderm and gut/genitourinary/tail development: a new tissue-specific role for Hoxa13. Development. 2002;129:551–561. doi: 10.1242/dev.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu B., Wellik D.M. Axial Hox9 activity establishes the posterior field in the developing forelimb. Proc. Natl. Acad. Sci. USA. 2011;108:4888–4891. doi: 10.1073/pnas.1018161108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsubara Y., Hirasawa T., Egawa S., Hattori A., Suganuma T., Kohara Y., Nagai T., Tamura K., Kuratani S., Kuroiwa A., Suzuki T. Anatomical integration of the sacral-hindlimb unit coordinated by GDF11 underlies variation in hindlimb positioning in tetrapods. Nat. Ecol. Evol. 2017;1:1392–1399. doi: 10.1038/s41559-017-0247-y. [DOI] [PubMed] [Google Scholar]
- 35.Lee Y.J., McPherron A., Choe S., Sakai Y., Chandraratna R.A., Lee S.-J., Oh S.P. Growth differentiation factor 11 signaling controls retinoic acid activity for axial vertebral development. Dev. Biol. 2010;347:195–203. doi: 10.1016/j.ydbio.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bel-Vialar S., Itasaki N., Krumlauf R. Initiating Hox gene expression: in the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes in two distinct groups. Development. 2002;129:5103–5115. doi: 10.1242/dev.129.22.5103. [DOI] [PubMed] [Google Scholar]
- 37.Abu-Abed S., Dollé P., Metzger D., Wood C., MacLean G., Chambon P., Petkovich M. Developing with lethal RA levels: genetic ablation of Rarg can restore the viability of mice lacking Cyp26a1. Development. 2003;130:1449–1459. doi: 10.1242/dev.00357. [DOI] [PubMed] [Google Scholar]
- 38.Sawin P.B. Preliminary studies of hereditary variation in the axial skeleton of the rabbit. Anat. Rec. 1937;69:407–428. [Google Scholar]
- 39.Jurberg A.D., Aires R., Varela-Lasheras I., Nóvoa A., Mallo M. Switching axial progenitors from producing trunk to tail tissues in vertebrate embryos. Dev. Cell. 2013;25:451–462. doi: 10.1016/j.devcel.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 40.Nishimoto S., Wilde S.M., Wood S., Logan M.P.O. RA acts in a coherent feed-forward mechanism with Tbx5 to control limb bud induction and initiation. Cell Rep. 2015;12:879–891. doi: 10.1016/j.celrep.2015.06.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishimoto S., Logan M.P.O. Subdivision of the lateral plate mesoderm and specification of the forelimb and hindlimb forming domains. Semin. Cell Dev. Biol. 2016;49:102–108. doi: 10.1016/j.semcdb.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 42.Xu B., Hrycaj S.M., McIntyre D.C., Baker N.C., Takeuchi J.K., Jeannotte L., Gaber Z.B., Novitch B.G., Wellik D.M. Hox5 interacts with Plzf to restrict Shh expression in the developing forelimb. Proc. Natl. Acad. Sci. USA. 2013;110:19438–19443. doi: 10.1073/pnas.1315075110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rallis C., Del Buono J., Logan M.P.O. Tbx3 can alter limb position along the rostrocaudal axis of the developing embryo. Development. 2005;132:1961–1970. doi: 10.1242/dev.01787. [DOI] [PubMed] [Google Scholar]
- 44.Ros M.A., Dahn R.D., Fernandez-Teran M., Rashka K., Caruccio N.C., Hasso S.M., Bitgood J.J., Lancman J.J., Fallon J.F. The chick oligozeugodactyly (ozd) mutant lacks sonic hedgehog function in the limb. Development. 2003;130:527–537. doi: 10.1242/dev.00245. [DOI] [PubMed] [Google Scholar]
- 45.Chiang C., Litingtung Y., Lee E., Young K.E., Corden J.L., Westphal H., Beachy P.A. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 46.Takeuchi J.K., Koshiba-Takeuchi K., Suzuki T., Kamimura M., Ogura K., Ogura T. Tbx5 and Tbx4 trigger limb initiation through activation of the Wnt/Fgf signaling cascade. Development. 2003;130:2729–2739. doi: 10.1242/dev.00474. [DOI] [PubMed] [Google Scholar]
- 47.Morin X., Jaouen F., Durbec P. Control of planar divisions by the G-protein regulator LGN maintains progenitors in the chick neuroepithelium. Nat. Neurosci. 2007;10:1440–1448. doi: 10.1038/nn1984. [DOI] [PubMed] [Google Scholar]
- 48.Matsuda T., Cepko C.L. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc. Natl. Acad. Sci. USA. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hamburger V., Hamilton H.L. A series of normal stages in the development of the chick embryo. 1951. Dev. Dyn. 1992;195:231–272. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- 51.Sato Y., Poynter G., Huss D., Filla M.B., Czirok A., Rongish B.J., Little C.D., Fraser S.E., Lansford R. Dynamic analysis of vascular morphogenesis using transgenic quail embryos. PLoS ONE. 2010;5:e12674. doi: 10.1371/journal.pone.0012674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Michaud J.L., Lapointe F., Le Douarin N.M. The dorsoventral polarity of the presumptive limb is determined by signals produced by the somites and by the lateral somatopleure. Development. 1997;124:1453–1463. doi: 10.1242/dev.124.8.1453. [DOI] [PubMed] [Google Scholar]
- 53.Scaal M., Gros J., Lesbros C., Marcelle C. In ovo electroporation of avian somites. Dev. Dyn. 2004;229:643–650. doi: 10.1002/dvdy.10433. [DOI] [PubMed] [Google Scholar]
- 54.Henrique D., Adam J., Myat A., Chitnis A., Lewis J., Ish-Horowicz D. Expression of a Delta homologue in prospective neurons in the chick. Nature. 1995;375:787–790. doi: 10.1038/375787a0. [DOI] [PubMed] [Google Scholar]
- 55.Rigueur D., Lyons K.M. Whole-mount skeletal staining. In: Hilton M., editor. Skeletal Development and Repair Methods in Molecular Biology. Humana Press; Totowa, NJ: 2014. pp. 113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
17h time-lapse video of a chicken embryo electroporated with cytoplasmic GFP and nuclear H2b-RFP fluorescent reporters, showing the dynamic process of LPM formation (from stage 5 to stage 11). Images were acquired on an inverted 2-photon microscope, using a long distance 10X objective. White bracket in the 1st frame outlines the presumptive LPM (prLPM); white asterisk represents the Hensen’s node; white dashed line outlines the PS; white brackets in the last frame outline the forelimb (FL), interlimb (IL) and hindlimb (HL) domains.
Example of retrospective cell tracking of forelimb, interlimb and hindlimb LPM cells to identify their epiblast origin on a 24h time-lapse video of a GFP and H2b-RFP-electroporated embryo. Images were acquired on an inverted 2-photon microscope, using a long distance 10X objective. Left panel, bright field image of the embryo (from stage 11 back to stage 4). Middle panel, GFP electroporated cells. Right panel, tracking of 9 LPM cells back to their epiblast origin. Note that the video plays backward.
hUbC:mEOS2FP transgenic quail embryos in which the presumptive LPM in the PS has been photoconverted. Embryos were further imaged every 2h to follow the fate of the photoconverted region. Photoconversion and acquisition were performed on an inverted confocal microscope with a long distance 10X objective. Left, middle-left, middle-right and right panels show embryos where the presumptive LPM was photoconverted at stage 6, 7, 9 and 10, respectively. Black arrowheads outline the anterior limit of photoconverted cells.