Abstract
Red skin colour is an important target trait in various pear breeding programmes. In this study, the genetic control of red skin colour was investigated in an interspecific population derived using the descendants of the red sport European pear cultivar ‘Max Red Bartlett’ (MRB) and the red-blushed Chinese pear cultivar ‘Huobali’. Approximately 550 seedlings from nine families were phenotyped for red skin over-colour coverage (Ocolcov) and the intensity of red over-colour (Ocolint) on a 0–9 scale, and genotyped using genotyping-by-sequencing. Genome-wide association analyses were conducted using 7500 high-quality single nucleotide polymorphisms (SNPs). Genomic regions on linkage groups (LG) 4 and 5 were found to be associated, and the best SNP (S578_25116) on LG4 accounted for ~15% of phenotypic variation in Ocolcov and Ocolint. The association of S578_25116 with Ocolcov and Ocolint was successfully validated in a sample of ~200 European and Asian pear accessions. The association with red skin at locus S578_25116 was not present in Asian pear accessions, suggesting its close proximity to the MRB’s Cardinal gene. Several putative candidate genes, including MYB transcription factors (PCP027962 and PCP027967), were identified in the quantitative trait locus region on LG4 and await functional validation.
Subject terms: Genetics, Genomics
Crop genetics: What makes a pear red?
Researchers in New Zealand have produced a map of genetic variants linked with red skin color in pears, opening the door to identifying the genes responsible. Satish Kumar and others at the New Zealand Institute for Plant and Food Research Limited measured the skin color of 550 hybrid pear seedlings and sequenced their genomes. Combining these data produced a map of 7,500 variants throughout the genome and identified those associated with red skin color. The most significant variant accounted for about 15% of the color variation. Further analysis of that genomic region revealed several genes which might be related to red skin color. The genomic map produced by this study will improve breeding efficiency by making it possible to screen seedlings for fruit color, but further research is necessary to characterize the candidate genes.
Introduction
Pear (Pyrus sp.) has been cultivated for at least 2000–3000 years, and is currently grown commercially in more than 50 countries in Europe, Northern Africa, Asia, Australasia and North America1. Red skin colour, which enhances the appearance of pears and can make them more attractive to potential customers, is considered an important trait to target in breeding programmes around the world, including South Africa, New Zealand, Italy, Spain, China and Australia. Interspecific hybrids are sometimes used in pear breeding programmes to produce new cultivars with novel combinations of colour and texture2. Red skin expression in pear results mainly from the concentration and composition of anthocyanin; environmental factors including temperature and UV light can affect anthocyanin concentrations and perceived intensity of colour3.
The underlying genetics of anthocyanin biosynthesis and the metabolic pathway have been investigated in various studies and this topic has been extensively reviewed recently4–6. Regulation of the anthocyanin pathway in pear is most likely primarily controlled by the MYB-bHLH-WD40 (MBW) transcription factor complex, as it has been demonstrated to be conserved across a broad range of species7. For example, in apple, MYB10 and its allelic counterparts have been shown to control anthocyanin expression4. In pear, two MYB transcription factors have been associated with anthocyanin production, MYB10 and MYB114, and these appear to act synergistically to enhance pigment concentrations8. However, as has been demonstrated across a range of species, anthocyanin biosynthesis may not be regulated by the MBW complex alone9. Other transcription factors, such as MADS-box and ethylene response factors (ERF), have also been implicated in pear anthocyanin production8,10.
Genetic mechanism behind red skin or blush colour of various European and Asian pear cultivars have been investigated in segregating families. The red skin colour derived from ‘Max Red Bartlett’ (MRB) appears to be controlled by a single dominant gene, whereas the blush colour for ‘Huobali’ may be controlled by complimentary actions of at least two dominant genes11. Dondini et al.12 reported that the red-skinned phenotype of MRB was controlled by a single dominant gene located on linkage group (LG) 4. Genome-wide dense genotyping, facilitated by the publication of draft genome of Asian13 and European14 pears, has contributed to the fine mapping of quantitative trait loci (QTL) for colour and fruit quality traits15. Using bulked-segregant analysis next-generation sequencing of pooled DNA from red- and green-skinned pear seedlings derived from a cross between red-skinned Asian pear cultivars, Xue et al.16 mapped the red/green (R/G) locus to the bottom of LG5.
Previous studies that mapped QTL for pear red skin used a single bi-parental mapping population of small size (~100), and hence lacked statistical power. To address this deficiency, association mapping in multiple family populations, where large numbers of progeny derived from controlled crosses in various mating schemes, has been used for genome-wide association (GWA) studies17–19. In this study, we applied GWA analysis in combination with genotyping-by-sequencing (GBS) on a population of genetically related interspecific hybrid families to fine-map the genetic control of red skin colour. Genotype-phenotype associations identified from this GWA approach were validated in an independent population. The identified candidate genes within the QTL region lay a solid base for further functional genomics studies. Furthermore, it is expected that the single nucleotide polymorphic (SNP) markers identified in this study could improve efficiency of pear breeding programmes via marker-assisted selection.
Material and methods
Plant material and phenotypes
First-generation families were created at the New Zealand Institute for Plant and Food Research Limited (PFR) in 1986 and 1989 using commercial cultivars of European, Chinese and Japanese pear as parents. MRB, ‘Huobali’ and ‘Worden Seckel’ were the primary sources of red skin/blush. Second-generation families were created in 1996 using the best selections (high proportion of red skin and superior eating quality) produced from first-generation families as parents. Nine third-generation families were created in 2007–2008 using 12 second-generation selections as parents. All 12 parents had ‘PremP45’ (Maxie), a selection from a first-generation family (‘Nijisseiki’ × MRB), in their pedigree. In addition to varying degrees of genetic relatedness among the 12 parents, some of these parents were represented in more than one family (Table 1), which resulted in partial sharing of genomes among hybrid progenies.
Table 1.
Parentage (each of the 12 colours represents a parent) of the nine interspecific hybrid families, and the parental phenotypes (0–9 scale) for over-colour coverage (Ocolcov) and over-colour intensity (Ocolint)

|
The segregation ratio of genotypes at the best SNP locus S578_25116 (C/T), and the observed frequency of allele ‘T’ are also shown.
aTT genotype was not observed in this family despite both parents a being heterozygous
A random selection of seedlings that reached a minimum height after growing in a field nursery for 1 year, were propagated onto Quince C rootstock, inter-stocked with ‘Beurré Hardy’ and planted in PFR’s orchard in Motueka during 2011. Six fruit harvested from each seedling over two consecutive seasons (2015 and 2016) were stored for 4 weeks at 3 °C, then a further 1 day at 20 °C before assessment. Red skin over-colour coverage (Ocolcov) was scored on a scale from 0 (no red colour) to 9 (completely red). The intensity of red over-colour (Ocolint) was scored on a scale from 0 (none) to 9 (highest).
DNA extraction and variants discovery from GBS
Protocols for DNA extraction, GBS library preparation and variant discovery were the same as those reported earlier by Kumar et al.20. Briefly, GBS libraries were sequenced on the Illumina HiSeq2000 platform and the sequence data were analysed using TASSEL/GBS21 pipeline. The fastq files were mapped separately to the P. × bretschneideri (cultivar ‘Suli’)13 and P. communis (cultivar ‘Bartlett’)14 reference genomes using Bowtie2 (version 2.2.1)22 to maximise the number of SNP calls in interspecific hybrid progenies. The various data quality filters used for SNP calling were described earlier by Kumar et al.20.
Linkage map construction
Resulting SNPs from the previous step were used to construct a consensus linkage map. Genotypes of the parents of one family (P449) were not available, hence data from eight families were used for constructing a consensus linkage map. Paternal and maternal maps (16 maps in total) were constructed using Joinmap v4.1 software. Then, common markers shared by at least three different paternal (or maternal) maps were selected as a bridge to merge the maps using R package MergeMap23. Linkage group (LG) identification was determined based on those SNPs common with the previously published map of ‘Old Home’ × ‘Louise Bon of Jersey’24.
Genome-wide association (GWA) analysis
Genome-wide tests for marker-trait associations were conducted using a mixed-linear model approach. A genomic relationship matrix25 and principal components (PCs) derived from the genotypic data matrix were used to account for cryptic relatedness and population stratification, respectively. Studies have shown that using genomic (or realised) relationship matrix is more efficient than the pedigree-based relationship matrix to account for Mendelian sampling and segregation distortion26,27. Estimate of narrow-sense heritability (h2) of each trait and genetic correlation between Ocolcov and Ocolint were also obtained. GAPIT28 software was used to conduct the above described genetic analyses.
The most significant SNP markers associated with Ocolcov and Ocolint were validated in a germplasm set of about 200 accessions comprising Asian and European pears and their hybrids. More details about this germplasm can be found in Kumar et al.20. Searches for potential candidate genes in the vicinity of significant SNPs were also conducted using the Asian13 and European pear (double haploid ‘Bartlett’; unpublished) genome assemblies. Candidate gene sequences were then compared to Arabidopsis genome with the Basic Local Alignment Search Tool (BLAST) from The Arabidopsis Information Resource (TAIR)29 to determine sequence homologs.
Results
Genetic analysis of colour
A total of 550 seedlings with phenotypic and genotypic information were available for genetic analysis (Table 1). The number of seedlings from each family varied between 12 (P484) and 163 (P449). ‘PremP45’, a carrier of a dominant red skin gene of MRB, was represented as a parent or a grandparent in all nine families of this study. Phenotypes of the 12 parents varied considerably; for example, on a 0–9 scale, four parents had Ocolcov and Ocolint scores less than 4, and one parent had a score of >8 for both traits (Table 1). The distribution of trait values, after adjusting for fixed effects, demonstrated the quantitative nature of the red phenotype (Fig. 1). Narrow-sense heritability of Ocolcov and Ocolint was 0.68 and 0.70, respectively, and the estimated genetic correlation between these two traits was about 0.90.
Fig. 1.
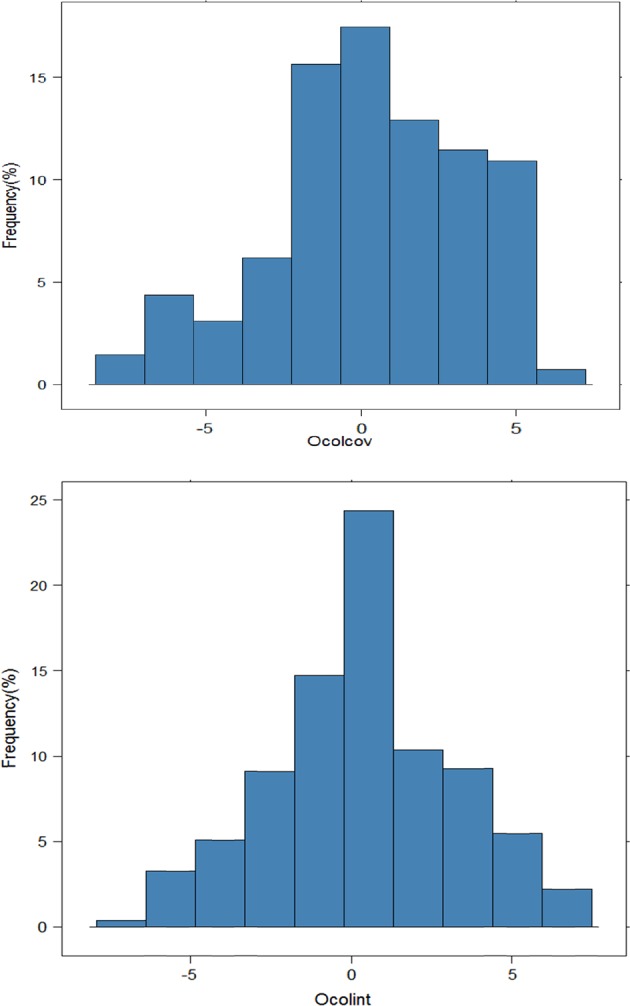
Distribution of over-colour coverage (Ocolcov) and over-colour intensity (Ocolint), after adjusting for fixed effects (e.g. year and assessor), in 550 interspecific pear hybrid seedlings
Genetic map and GWA analysis
Nearly 50,000 SNPs were identified, across nine families, following the earlier described SNP calling criteria. After some additional quality controls (i.e. missing data frequency <10%, minor allele frequency >0.05), over 10,000 SNPs were retained for map construction. A total of about 7500 SNPs could reliably be mapped onto a consensus linkage map spanning about 4100 cM in total. The length of the 17 LGs showed a wide range, from 54 (LG16) to 362 cM (LG13). The average number of markers per LG and the average marker interval were 441 and 0.60 cM, respectively (Supplementary Table S1; Supplementary Figure S1). Overall, there were about 4434 unique genetic positions on this GBS-based consensus map, which means that on average there were 1.7 markers per unique genetic position.
The profiles of FDR-adjusted p-values (in terms of –log10(p)) for all tested SNPs for both traits are illustrated in Fig. 2. A genome-wide p-value of <0.01, which roughly equates to a comparison-wise p-values of p < 1 × 10−6 as we tested over 7500 SNPs, was used as a significance threshold for individual SNP testing. The ten most significant SNPs were common to both Ocolcov and Ocolint, and were clustered within a genomic region between 196 and 256 cM on LG4 (Fig. 2). This clustering of significant SNPs suggests the presence of a large-effect QTL on LG4. The most significant SNP (S578_25116), mapped at 196.6 cM on LG4, explained about 15% of phenotypic variation in Ocolcov and Ocolint. For both traits, the Q-Q plots showed a close adherence of the observed and expected –log10(p) values over most of the range (results not shown). Although not statistically significant, a couple of SNPs (S210_30053 and S210_30038) mapped at the bottom of LG5 and accounted for about 5% of phenotypic variation in Ocolint and Ocolcov, respectively (Fig. 2).
Fig. 2.
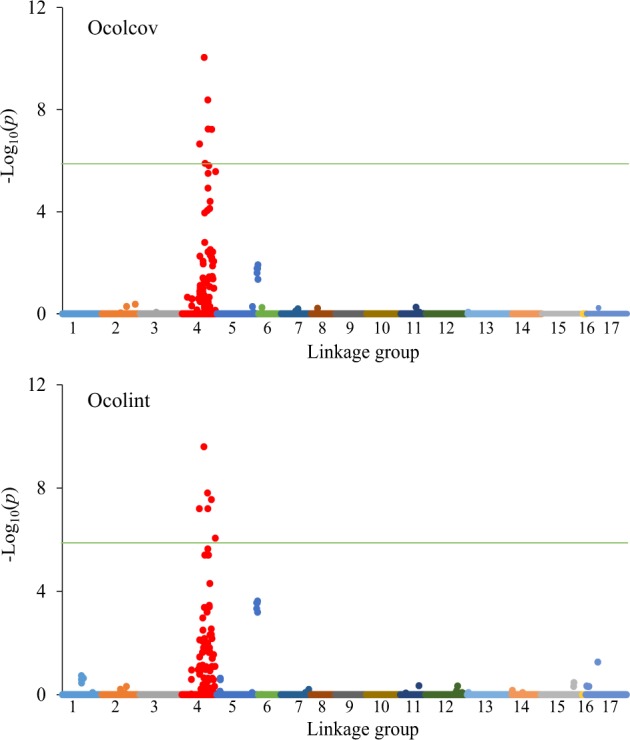
Manhattan plots of the –log10(p) values for over-colour coverage (Ocolcov) and over-colour intensity (Ocolint) from a genome-wide scan plotted against position on each of 17 Pyrus linkage groups (represented by different colours). Green horizontal line indicates the genome-wide significance threshold
Unexpectedly, the ‘TT’ genotype was not observed in families where both parents were heterozygous (CT) at SNP locus S578_25116 (Table 1). Observed frequencies of the allele ‘T’ at SNP locus S578_25116 varied considerably between families (Table 1), and a high correlation (~0.92) was observed between family performance for Ocolcov and Ocolint and allele frequency (Fig. 3). The average Ocolcov of families P507 and P508 with zero frequency of allele ‘T’ was 4.2 compared with 6.2 for families P493 and P491 with the highest frequency (0.32). Similarly, the average Ocolint varied between 3.0 and 5.3 for families with the lowest and the highest allele frequencies, respectively (Fig. 3).
Fig. 3.
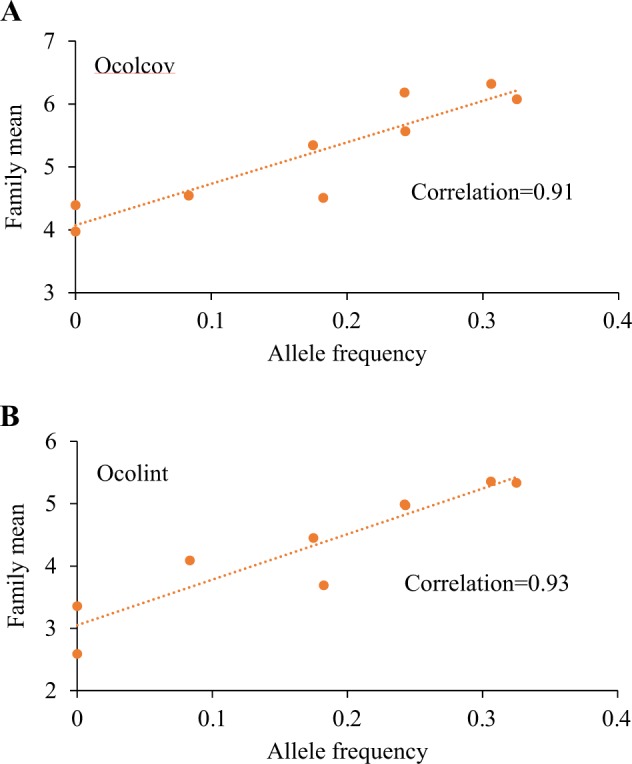
Relationship between frequency of allele ‘T’ at SNP locus S578_25116 (C/T) in a family and family mean for over-colour coverage (Ocolcov) and over-colour intensity (Ocolint) in interspecific pear hybrids
Separate analyses of various subsets of families identified S39_462595 and S161_15221, which are in close proximity of the overall best SNP S578_25116, as the most significant markers. Joint analysis of families P507 and P508, which did not segregate for S578_25116 (see Table 1), did not show any genomic region significantly associated with skin colour.
Validation of significant SNPs
Genotypes from 160 validation samples were available for the most significant SNP (S578_25116), and the estimated frequency of the positive allele in these samples was about 0.10. The number of accessions with genotype CC, CT and TT were 130, 17 and 13, respectively. Accessions with only Asian ancestry did not carry allele T irrespective of their Ocolcov and Ocolint scores. Although, the average Ocolcov and Ocolint of accessions carrying allele T (i.e. with genotype CT or TT) was significantly higher compared with CC genotypes, the pattern of additive association between marker and phenotype was possibly diluted by the small number of accessions with CT and TT genotypes (Fig. 4a). The second best SNP (S123069_330) with somewhat larger sample sizes for CT and TT genotypes, clearly displayed the additive association of allele T with phenotypes (Fig. 4b). Genotypes of some pear cultivars at locus S578_25116 are documented in Supplementary Table S2.
Fig. 4.
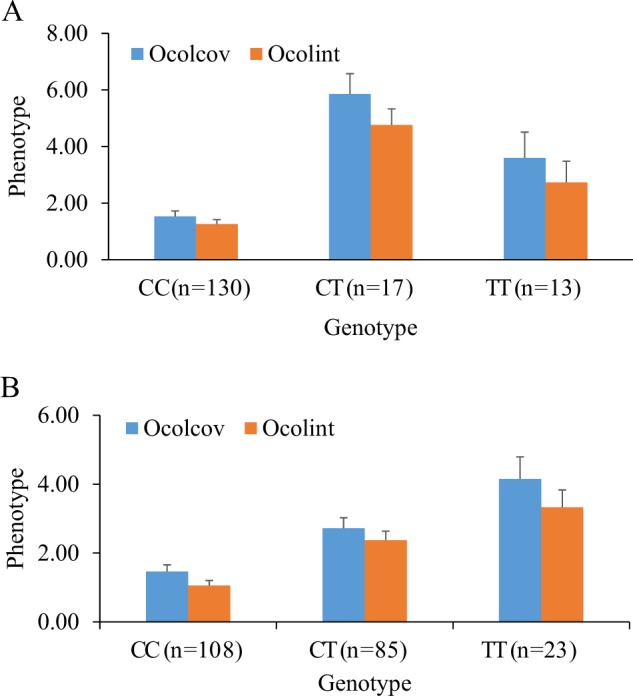
Validation of the effect of best SNPs S578_25116 (a) and S123069_330 (b) on over-colour coverage (Ocolcov) and over-colour intensity (Ocolint) in an independent population of European and Asian pear accessions. Error bar indicates standard error of mean phenotype (y-axis)
Candidate genes for large-effect QTL
To identify candidate genes around the major QTL on LG4, searches were carried out on the genomic scaffolds of the most recent versions of P. communis and P. bretschneideri genomes. A list of potential candidate genes in the vicinity of some of the most significant SNPs on LG4 and LG5 are listed in Supplementary Table S3. The largest-effect SNP on LG4 was located on scaffold S578: this scaffold contained 16 gene models and from these, two MYB-related transcription factors (gene ID: PCP027962 and PCP027967) and three E3 Ubiquitin genes (gene IDs: PCP044630; Pbr024602.1 and Pbr024617.1) were identified as potential candidates (Supplementary Table S3). The MYB transcription factor genes PCP027962 and PCP027967 exhibited sequence similarity to the Arabidopsis transcription factor genes At5G47390 (MYB-like transcription domain) and At5G35550 (TT2 gene, which encodes a R2R3 MYB domain protein), respectively. The potential candidate genes aligned with the three other scaffolds (S39, S161 and S210) of significance included a family of basic helix-loop-helix (bHLH) transcription factors (e.g. Pbr009756.1; Pbr009770.1; Pbr008711.1; PCP028905; PCP031885 and PCP014862) (Supplementary Table S3).
Discussion
Pear cultivar breeding is a lengthy process, largely because of how long it takes for seedlings to flower (juvenility) and the subsequent, but necessary, time for reliable tree/fruit performance (phenotyping), testing and selection. Decreasing that time can be achieved through reducing the time to fruit, breeding for increased precocity, or using molecular tools2. Marker-assisted selection would further improve the efficiency of breeding programmes by enabling screening of young seedlings for red skin/blush colour and elimination of those that do not have the desirable alleles.
In our study, a large number of multi-generation hybrid seedlings with varying degrees of genome sharing with the European and Asian pear cultivars MRB and ‘Huobali’, respectively, presented a mosaic of major and minor genes imparting variation in skin colour. Using advanced genotyping and analysis methods, a cluster of SNPs on LG4 displayed significant association with skin colour. The same set of SNPs were associated with Ocolcov and Ocolint, supporting the observed high genetic correlation (about 0.90) between these traits. We independently validated the additive effect of allele T at SNP locus S578_25116 in a large sample (about 200) of European and Asian pear accessions, adding further credibility to the marker-trait association reported in this study. A moderate performance of P507 and P508 families, which did not segregate for allele T at SNP locus S578_25116, highlighted the contribution of minor genes, especially those inherited from ‘Huobali’, but that might also have come from ‘Worden Seckel’ or ‘Sanguinole’.
The maximum observed frequency of allele T was about 32% despite both parents of a family being heterozygous (CT × CT) (Table 1). The absence of TT genotypes suggested a ‘fitness penalty’ for offspring that were homozygous (TT) for the dominant red allele (T). From these results, we hypothesise that the homozygous dominant form of red skin gene could affect survival of seeds or seedlings, leading to non-Mendelian segregation ratios. Dominant genes underpinning skin colour in some animal species such as mouse were reported to segregate in non-Mendelian ratio, because one-quarter of the offspring (homozygous for the dominant allele) from crosses between heterozygous parents died during embryonic development30.
Unlike previous studies, interspecific hybrid seedlings in our study inherited genes for red fruit colour from both European (MRB) and Asian cultivars (‘Huobali’). A previous study12 using 90 F1 seedlings of cross ‘Abbé Fétel’ × MRB, mapped the red fruit colour as a morphological marker on LG4 of MRB, which is consistent with results from this study. This dominant gene in MRB is also associated with the red foliage, and was named C, the Cardinal red colour gene31,32. In ‘Bayuehong’, a red-blushed cultivar derived from the European pear cultivar ‘Clapp’s Favorite’, a major QTL for red phenotype was mapped to LG5, and a gene PyMYB114 was identified within the QTL region8,15. Xue et al.16, using a cross between Asian pear cultivars ‘Mantianhong’ and ‘Hongxiangsu’ (both derived from ‘Huobali’), identified a QTL for red phenotype at the bottom of LG5. A genomic region at the bottom of LG5 identified in our study is consistent with these previous studies8,16, and suggests that LG5 could potentially harbour QTLs underpinning red blush development in Asian and European pears as well as interspecific hybrids.
Similar to the apple MdMYB10 gene, the pear PcMYB10 gene was mapped by Pierantoni et al.33 to the bottom of LG9, and the importance of MYB10 transcription factors in the expression of red skin phenotype has been widely reported in functional genomics studies4,5,33–35. However, there was no significant marker-trait association detected on LG9 in our study, which is consistent with other QTL mapping reports12,15,16,36. The molecular basis for this disconnect between QTL mapping and the major anthocyanin-controlling MYB transcription factor is as yet unknown.
Two MYB transcription factors (PCP027962 and PCP027967) were aligned with the scaffold S578 where the most significant SNP (S578_25116) resides. In particular, PCP027962 appears to be homologous to the Arabidopsis MYB-like domain transcription factor (At5G47390), that is reported to be a positive regulator of anthocyanin biosynthesis and is itself regulated by the anthocyanin-associated elongated hypocotyl (HY5) gene37. The other gene, PCP027967, is similar to Arabidopsis gene (At5G35550) TT2, which encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation38. In addition to these MYB transcription factors, the bHLH, E3 Ubiquitin and ABC transporter classes were among the candidate genes identified within the QTL mapping region on LG4 and LG5. Functions of some of these candidate genes have been investigated in various studies4,5,8,10. For example, Yao et al.8 identified a gene PyMYB114 within the QTL region on LG5, and the transient assays demonstrated its role in biosynthesis of anthocyanins in pear fruits. Taken together with our results, we hypothesise that LG5 also harbours genes underpinning red skin colour development on both Asian and European pears. SNP markers identified in this study will help screening of young seedlings for red fruit colour and enhance the efficiency of pear breeding programmes. More research is required to test the function of candidate genes, especially PCP027962 and PCP027967, identified on LG4 in this study.
Supplementary information
Acknowledgements
This research was partly funded by the New Zealand Ministry of Business, Innovation and Employment (MBIE). Funding from the National Science Foundation of China (31820103012) is thankfully acknowledged. The PFR Markers & Mapping Team is thanked for help in collecting leaf samples. Marlene Aldworth, Chris Morgan, David Anderson and Angela Shirtcliff helped in fruit harvest and phenotyping. Constructive feedback from Dr Andrew Allan is thankfully acknowledged.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41438-018-0112-4).
References
- 1.Silva GJ, Souza TM, Barbieri RL, Oliveira ACD. Origin, domestication, and dispersing of pear (Pyrus spp.) Adv. Agr. 2014;2014:e541097. [Google Scholar]
- 2.Brewer LR, Palmer JW. Global pear breeding programmes: goals, trends and progress for new cultivars and new rootstocks. Acta Hort. 2011;909:105–119. doi: 10.17660/ActaHortic.2011.909.10. [DOI] [Google Scholar]
- 3.Steyn W, et al. Anthocyanin degradation in detached pome fruit with reference to preharvest red color loss and pigmentation patterns of blushed and fully red pears. J. Am. Soc. Hort. Sci. 2004;129:13–19. [Google Scholar]
- 4.Lin-Wang K, et al. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 2010;10:1. doi: 10.1186/1471-2229-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Y, et al. Expression differences of anthocyanin biosynthesis genes reveal regulation patterns for red pear coloration. Plant Cell Rep. 2014;34:189–198. doi: 10.1007/s00299-014-1698-0. [DOI] [PubMed] [Google Scholar]
- 6.Thomson GE, Turpin S, Goodwin I. A review of pre-harvest anthocyanin development in full red and blush cultivars of European pear. N. Z. J. Crop Hort. Sci. 2018;46:81–100. doi: 10.1080/01140671.2017.1351378. [DOI] [Google Scholar]
- 7.Allan AC, Hellens RP, Laing WA. MYB transcription factors that colour our fruit. Trends Plant Sci. 2008;13:99–102. doi: 10.1016/j.tplants.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 8.Yao G, et al. Map‐based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to co-ordinately regulate fruit anthocyanin biosynthesis. Plant J. 2017;92:437–451. doi: 10.1111/tpj.13666. [DOI] [PubMed] [Google Scholar]
- 9.Jaakola L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013;18:477–483. doi: 10.1016/j.tplants.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 10.Wu J, et al. Identification of differentially expressed genes related to coloration in red/green mutant pear (Pyrus communis L.) Tree Genet. Genome. 2013;9:75–83. doi: 10.1007/s11295-012-0534-3. [DOI] [Google Scholar]
- 11.Volz RK, White AG, Brewer LR. Breeding for red skin colour in interspecific pears. Acta Hortic. 2008;800:469–474. doi: 10.17660/ActaHortic.2008.800.59. [DOI] [Google Scholar]
- 12.Dondini L, et al. The inheritance of the red colour character in European pear (Pyrus communis) and its map position in the mutated cultivar ‘Max Red Bartlett’. Plant Breed. 2008;127:524–526. doi: 10.1111/j.1439-0523.2008.01500.x. [DOI] [Google Scholar]
- 13.Wu J, et al. The genome of the pear (Pyrus bretschneideri Rehd.) Genome Res. 2013;23:396–408. doi: 10.1101/gr.144311.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chagné D, et al. The draft genome sequence of European pear (Pyrus communis L.‘Bartlett’) PLoS One. 2014;9:e92644. doi: 10.1371/journal.pone.0092644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu J, et al. High-density genetic linkage map construction and identification of fruit-related QTLs in pear using SNP and SSR markers. J. Exp. Bot. 2014;65:5771–5781. doi: 10.1093/jxb/eru311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xue H, et al. Interval mapping for red/green skin colour in Asian pears using a modified QTL-seq method. Hortic. Res. 2017;4:17053. doi: 10.1038/hortres.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kover PX, et al. A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 2009;5:e1000551. doi: 10.1371/journal.pgen.1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo B, Sleper D, Beavis WD. Nested association mapping for identification of functional markers. Genetics. 2010;186:373–383. doi: 10.1534/genetics.110.115782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar S, et al. Novel genomic approaches unravel genetic architecture of complex traits in apple. BMC Genom. 2013;14:93. doi: 10.1186/1471-2164-14-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar S, et al. Genotyping-by-sequencing of pear (Pyrus spp.) accessions unravels novel patterns of genetic diversity and selection footprints. Hortic. Res. 2017;4:17015. doi: 10.1038/hortres.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bradbury PJ, et al. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- 22.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu, Y. H., Close, T. J. & Lonardi, S. Accurate construction of consensus genetic maps via integer linear programming, IEEE/ACM Trans. Comput. Biol.8, 381–394 (2011). [DOI] [PubMed]
- 24.Li L, et al. Integrated high-density consensus genetic map of Pyrus and anchoring of the ‘Bartlett’v1. 0 (Pyrus communis) genome. DNA Res. 2017;24:289–301. doi: 10.1093/dnares/dsw063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Raden PM. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008;91:4414–4423. doi: 10.3168/jds.2007-0980. [DOI] [PubMed] [Google Scholar]
- 26.Nejati-Javaremi A, Smith C, Gibson J. Effect of total allelic relationship on accuracy of evaluation and response to selection. J. Anim. Sci. 1997;75:1738–1745. doi: 10.2527/1997.7571738x. [DOI] [PubMed] [Google Scholar]
- 27.Hayes BJ, Visscher PM, Goddard ME. Increased accuracy of artificial selection by using the realized relationship matrix. Genet. Res. 2009;91:47–60. doi: 10.1017/S0016672308009981. [DOI] [PubMed] [Google Scholar]
- 28.Lipka AE, et al. GAPIT: genome association and prediction integrated tool. Bioinformatics. 2012;28:2397–2399. doi: 10.1093/bioinformatics/bts444. [DOI] [PubMed] [Google Scholar]
- 29.Berardini TZ, et al. The Arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome. Genesis. 2015;53:474–485. doi: 10.1002/dvg.22877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paigen K. One hundred years of mouse genetics: an intellectual history. I. The classical period (1902-1980) Genetics. 2003;163:1–7. doi: 10.1093/genetics/163.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zielinski QB. Precocious flowering of pear seedlings carrying the Cardinal red color gene. J. Hered. 1963;54:75–76. doi: 10.1093/oxfordjournals.jhered.a107228. [DOI] [Google Scholar]
- 32.Lespinasse Y, Guérif P. Inheritance of red leaf colour from pear red sports of ‘Doyenne du Comice’, ‘Bartlett’ and ‘Beurré Hardy’. Acta Hortic. 2011;909:97–102. doi: 10.17660/ActaHortic.2011.909.9. [DOI] [Google Scholar]
- 33.Pierantoni L, et al. Mapping of an anthocyanin regulating MYB transcription factor and its expression in red and green pear Pyrus communis. Plant Physiol. Biochem. 2010;48:1020–1026. doi: 10.1016/j.plaphy.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 34.Wang Z, et al. The methylation of the PcMYB10 promoter is associated with green-skinned sport in Max Red Bartlett Pear. Plant Physiol. 2013;162:885–896. doi: 10.1104/pp.113.214700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qian M, et al. Isolation and expression analysis of anthocyanin biosynthesis genes from the red Chinese sand pear, Pyrus pyrifolia Nakai cv. Mantianhong, in response to methyl jasmonate treatment and UV-B/VIS conditions. Plant Mol. Biol. Rep. 2014;32:428–437. doi: 10.1007/s11105-013-0652-6. [DOI] [Google Scholar]
- 36.Yamamoto T, et al. Identification of QTLs controlling harvest time and fruit skin color in Japanese pear (Pyrus pyrifolia Nakai) Breed. Sci. 2014;64:351–361. doi: 10.1270/jsbbs.64.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen NH, et al. MYBD employed by HY5 increases anthocyanin accumulation via repression of MYBL2 in Arabidopsis. Plant J. 2015;84:1192–1205. doi: 10.1111/tpj.13077. [DOI] [PubMed] [Google Scholar]
- 38.Nesi N, et al. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell. 2001;13:2099–2114. doi: 10.1105/TPC.010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


