Abstract
Ostreococcus tauri, a picoeukaryotic alga that contributes significantly to primary production in oligotrophic waters, has a highly streamlined genome, lacking the genetic capacity to grow without the vitamins thiamine (B1) and cobalamin (B12). Here we demonstrate that the B12 and B1 auxotrophy of O. tauri can be alleviated by co-culturing with a heterotrophic bacterial partner Dinoroseobacter shibae, a member of the Rhodobacteraceae family of alpha-proteobacteria, genera of which are frequently found associated with marine algae. D. shibae lacks the complete pathway to synthesise three other B-vitamins: niacin (B3), biotin (B7), and p-aminobenzoic acid (a precursor for folate, B9), and the alga is in turn able to satisfy the reciprocal vitamin requirements of its bacterial partner in a stable long-term co-culture. Bioinformatics searches of 197 representative marine bacteria with sequenced genomes identified just nine species that had a similar combination of traits (ability to make vitamin B12, but missing one or more genes for niacin and biotin biosynthesis enzymes), all of which were from the Rhodobacteraceae. Further analysis of 70 species from this family revealed the majority encoded the B12 pathway, but only half were able to make niacin, and fewer than 13% biotin. These characteristics may have either contributed to or resulted from the tendency of members of this lineage to adopt lifestyles in close association with algae. This study provides a nuanced view of bacterial–phytoplankton interactions, emphasising the complexity of the sources, sinks and dynamic cycling between marine microbes of these important organic micronutrients.
Subject terms: Metabolism, Microbial communities
Introduction
Photosynthetic picoeukaryotic (PPE) algae are the main primary producers in many marine and freshwater aquatic ecosystems, playing a significant role in biogeochemical processes and food-web dynamics [1–3]. Prasinophytes, a paraphyletic group of green algae, are a major group of PPE [4]. Members of the Ostreococcus genus in this class have cells typically < 1 µm in diameter, making them the smallest free-living eukaryotes described to date. Ostreococcus are found globally in a range of conditions and habitats including the nutrient poor oligotrophic oceans, some of the largest and most challenging biomes on Earth [5]. This underscores their important ecological role and raises interesting questions about how the metabolism of these species equips them for survival.
Analysis of several Ostreococcus species reveals highly reduced genomes, although they retain the majority of metabolic attributes essential for autotrophic growth, including a complete set of genes for transport and assimilation of ammonium, nitrate and urea [6]. However, like many other algae their growth is dependent on the presence of certain organic micronutrients in the environment, specifically thiamine (vitamin B1) [7] and cobalamin (vitamin B12) [8], both of which are required as cofactors for enzymes involved in central metabolism. Genomic evidence revealed that Ostreococcus tauri has only a partial biosynthetic pathway for thiamine, suggesting it may have lost its ability to synthesise this co-factor de novo [9, 10]. The B12-biosynthetic pathway, comprising over 20 enzyme-catalysed steps, is found only in certain prokaryotes [11]. The requirement for this vitamin by B12-dependent algae is as a co-factor for the essential B12-dependent enzyme, methionine synthase (METH), rather than the inability to synthesise the vitamin. Those species that can grow independently of an external supply of B12 encode an alternative methionine synthase enzyme (METE) that does not use B12 as a co-factor [8, 12]. O. tauri encodes METH but not METE, and thus requires vitamin B12 for growth [8].
Algae may acquire vitamins in their natural environment via different routes. Seasonal upwelling of deep nutrient-rich water may provide a source for some coastal strains [13]. However, some vitamins are photolabile and would not persist for long in the water column. Moreover, using a sensitive analytical method Sanudo-Wilhelmy et al. [14] found that ambient concentrations along the coast of California were significantly lower than needed for the growth of vitamin-dependent algae such as O. tauri, and were below the detection threshold in some regions [14]. Addition of vitamin B12 (together with Fe and N) enhanced chlorophyll a levels threefold (beyond stimulating effects of adding just Fe + N) in the eastern boundary of the South Atlantic Gyre [15]. This highlights the crucial role of this compound in controlling phytoplankton growth in natural marine microbial communities, and demonstrates that the vitamin auxotrophy of O. tauri is representative of the natural world, rather than a particularity of this species.
Although small populations may persist under limiting micronutrient conditions, many PPEs including Ostreococcus periodically form characteristic blooms, implying that levels of vitamins must increase. Prokaryotes are the ultimate and only source of vitamin B12 [11]. However, structural diversity in the forms of B12 produced by different prokaryote taxa has important implications in terms of their bioavailability to eukaryote auxotrophs. In particular, evidence indicates that B12-synthesising cyanobacteria produce a variant, pseudocobalamin, which is considerably less bioavailable to eukaryotic algae [16]. This suggests that cobalamin-producing heterotrophic bacteria and archaea are likely to produce the majority of the co-factor that is bioavailable for algae [12, 17]. Further, certain bacterial phyla are more likely to be found associated with algae. In an analysis of over 40 different species of both macro and microalgae, six phyla (Bacteroidetes, Proteobacteria, Firmicutes, Actinobacteria, Verrucomicrobia and Planctomycetes) accounted for the majority of bacteria [18], and both alpha- and gamma-Proteobacteria are almost invariably found in studies of algal-associated microbiomes [19]. In particular, species from the family Rhodobacteraceae [20] have been shown to deliver vitamins and other metabolites to marine algae. For example, Dinoroseobacter shibae, a cosmopolitan obligately aerobic bacterium, originally isolated as a symbiont to cultured marine dinoflagellates [21], could supply thiamine and B12 to its hosts in exchange for a source of fixed carbon [22]. Similarly, Ruegeria pomeroyi supported the B12 requirements of the diatom Thalassiosira pseudonana in exchange for organosulphur compounds [23]. Analysis of environmental isolates of another diatom, Pseudo-nitzschia multiseries, showed association with a number of bacteria one of which, a Sulfitobacter species, promoted algal cell division by providing indole acetic acid (IAA) [24].
The hypothesis of mutualistic exchange of nutrients between microorganisms is challenging to test in the field. Most rely on correlations between co-existing species [25, 26], because in the absence of tight physical associations, which are not necessary for mutualism, nutrient flux is difficult to deduce even using the most advanced methodologies [27]. In light of this, simplified laboratory-based systems are invaluable for the study of nutrient cycling between microbes [28], and may provide important clues for our understanding of biogeochemical cycling of nutrients in natural systems. In this study, we established a stable co-culture between O. tauri and D. shibae, organisms that are reported to co-exist in marine microbial assemblages, such as in the North Pacific subtropical gyre [29]. Using this system in the laboratory, we examined the nature of the metabolic exchange, probing the dynamics and stability of the interaction over successive generations. Our findings demonstrate that there is a complex two-way exchange of B-vitamins that underpins this stable mutualistic relationship.
Materials and methods
Algal and bacterial strains and culture conditions
Ostreococcus tauri (OTH95) was a gift from Herve Moreau at the Oceanological Observatory of Banyuls-sur-mer, France. This culture is a non-axenic environmental isolate from the Thau lagoon of the Mediterranean sea, just off the south coast of France [6]. The strain was made axenic by fluorescence activated cell sorting (FACS), which provided starter cultures growing from single cells (Figure S2). The FACS protocol was specifically adapted and optimised to allow sorting, in collaboration with Nigel Miller (Department of Pathology, University of Cambridge, UK) for a MoFlo MLS high-speed cell sorter (Becton Dickinson). Chlorophyll fluorescence was measured through a 670/40 nm band pass filter (FL3) after excitation using a 488-nm argon laser (dot plots shown in Figure S2). Cells positive for chlorophyll fluorescence were sorted into single wells of a 24-well plate containing 2 ml of L1 medium, monitored for growth, and treated with ampicillin (1 mg/ml), neomycin (0.25 mg/ml), kanamycin (1 mg/ml) and streptomycin (50 µg/ml) for 14 days. Cultures were then tested for the presence of bacteria by replica plating onto Marine Broth (MB) agar. For this, 10 μl of liquid medium from stationary phase algal cultures was streaked onto MB plates, and incubated at room temperature for 14 days, which yielded no bacterial colonies. Its purity was further verified by staining with the nucleic acid-specific stain 4′,6- diamidino-2-phenylindole. Cells were stained using 1 ng/ml of the dye, incubated for 5 min at 20C, placed in a microscopy dish and viewed under epifluorescent illumination (excitation 330–380 nm, emission above 420 nm). Bacteria were clearly visible in non-treated control cultures but not in the FAC-sorted, antibiotic treated O. tauri cultures. Cultures, which tested negative for bacteria in this way, were used for subsequent experiments.
Dinoroseobacter shibae DF-12 was a gift from Michael Cunliffe (Marine Biological Association, UK). It was maintained on 1.5% MB plates (Difco™) at 24°C. Its purity was verified by polymerase chain reaction (PCR) amplification of the V3–V4 variable region of the 16 S rRNA gene using a universal primer set [30], as described in [12, 31]. The sequenced PCR product matched the correct bacterial ribotype by searching against the NCBI 16 S rRNA genomic database (http://blast.ncbi.nlm.nih.gov/).
All axenic and co-cultures containing O. tauri were grown at 18°C with a 16:8 h light:dark cycle (irradiance of 150 μmol photons m−2 s−1) in unmodified L1 medium [32] unless otherwise stated. For experiments testing exchange of B-vitamins, mono-cultures of D. shibae and O. tauri were pre-washed three times in L1 medium and then starved of vitamins for 7 days prior to the start of the experiment. B-vitamins were added to cultures at the following concentrations: 0.40 nM cobalamin, 2.05 nM biotin, 296 nM thiamine, 812 nM niacin and 291 nM p-aminobenzoic acid. These are concentrations known to be sufficient for unhindered algal cell growth in L1 medium. However, it is likely that these amounts are in excess of requirements, and the effect on cellular metabolism of providing a different ratio of these metabolites has not been demonstrated in dedicated experimental analyses.
Measuring cell densities of O. tauri and D. shibae
Optical densities of O. tauri cultures were measured using excitation at 750 nm (UV1, Thermo Spectronic). For cultures with OD values between 0.1 and 0.45, the measured scatter was proportional to cell concentrations (Figure S1). For lower cell concentrations, cell density was determined using FACS with a FacsScan500 flow cytometer (Becton-Dickinson). O. tauri cell cultures at stationary phase were diluted 2–40 times using L1 medium and spiked with CountBright™ absolute counting beads (Life Technologies). Cells were discriminated by forward scatter and red (chlorophyll) fluorescence and CountBright™ beads discriminated by forward scatter and orange fluorescence. Samples were analysed until 100–300 beads events had been noted, with three technical replicates per biological sample. Cyflogic software (version 1.2.1, developed by CyFlo Ltd) was used to determine cell density from the collected data.
For bacteria, colony-forming units were used as a proxy for population density, determined using a variation of the replica plating method as described by [33]. Harvested bacterial cells were diluted serially with L1 medium, and 3 × 10 µl of the diluted solution placed as drops on a single agar plate. The plate was then angled so the liquid formed three vertical lanes, and left to dry. After 10 days at room temperature colonies were counted from each individual lane, and counts from one plate averaged together as technical replicates per biological sample.
Bioinformatics analysis
Sequence similarity searches were performed to assess the presence of vitamin biosynthesis genes in different organisms. Enzymes required for the biosynthesis of each vitamin were identified from a literature search and reference pathways from the Kyoto Encyclopaedia of Genes and Genomes (KEGG) [34]. Validated reference protein sequences for enzymes of vitamin biosynthesis were identified from the Universal Protein Resource (UniProt) archive. BLASTP [35] was used to search for each of these enzymes in the O. tauri (GenBank ID: GCA_000214015.1) and D. shibae (GenBank ID: GCA_000018145.1) genome assembly hosted at the National Centre for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/), using the online BLAST server at the European Bioinformatics Institute (EMBL-EBI; https://www.ebi.ac.uk/Tools/sss/ncbiblast/). Default parameters including a word size of three and the BLOSUM62 scoring matrix were used for all searches. Low complexity regions were also filtered out. Only hits with an expectation coefficient of less than 1 × e−15 were taken through for extended analysis. The protein sequences of the putative biosynthesis enzymes were aligned using the ClustalW algorithm [36] with the query protein sequences using the AlignX module of the vector NTI software (Invitrogen) to confirm sequence similarities. Conserved functional domains of putative proteins were also confirmed using the NCBI’s conserved domain searchable database. These database and literature searches were also used to ascertain conserved active sites of enzymes if available. For whole genome searches to identify vitamin pathways in sequenced bacterial genomes, the automated approach as described in [16] was carried out, using the same query sequences.
Results
Complementary B-vitamin synthesis capabilities of O. tauri and D. shibae revealed by in silico pathway analyses
Figure 1a shows the growth requirements of O. tauri and D. shibae in minimal medium with different supplements. O. tauri requires both vitamins B12 and B1 [7, 8], as cofactors for cobalamin-dependent methionine synthase (B12) and several enzymes of intermediary carbon metabolism including transketolase (B1). D. shibae DFL-12 on the other hand encodes the genes for the biosynthesis of B12 and B1 [22], and can grow without either supplement. However, D. shibae is known to need exogenous sources of three other B-vitamins: biotin (vitamin B7), niacin (B3) and p-aminobenzoic acid a precursor for folate (B9) biosynthesis, whereas O. tauri does not require these molecules for growth. All three of these compounds have vital roles in central metabolism: biotin is a co-factor for enzymes necessary for essential carboxylation, transcarboxylation and decarboxylation reactions, folate is required for enzymes of 1-carbon metabolism, and niacin is a precursor to the hydrogen carriers NAD+ & NADP+, redox carriers used ubiquitously within the cell (reviewed in ref. [37]). Despite these fundamental roles, the exact genetic basis underlying the auxotrophy of D. shibae for these vitamins is currently unknown. We therefore carried out a detailed assessment of the genomes of the two organisms to investigate this, and whether they may be able to support one another’s growth in co-culture.
Fig. 1.
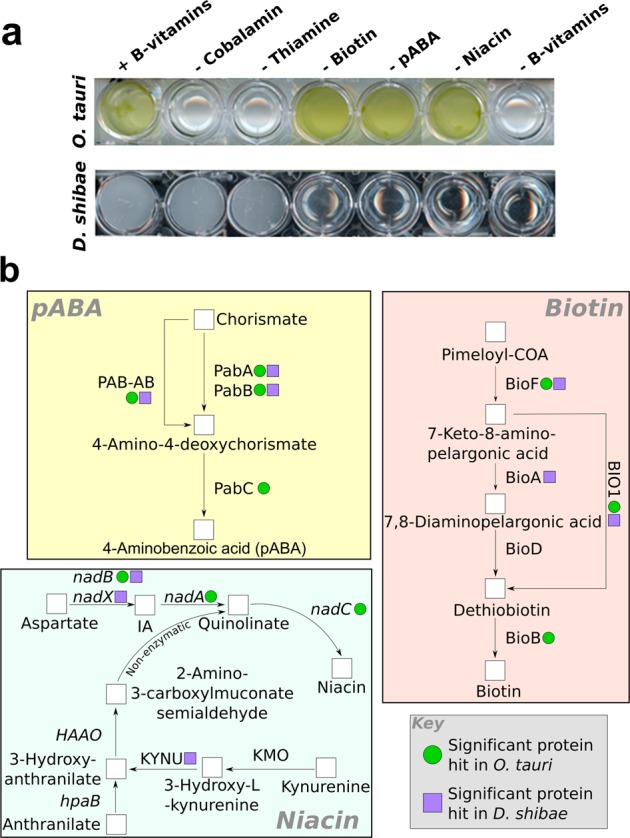
Determining the genetic basis for vitamin dependency. a Growth of axenic O. tauri and D. shibae (with 1% glucose) with and without various B-vitamins at stationary phase after two sub-cultures. b Schematic of biosynthetic pathways for p-aminobenzoic acid, niacin and biotin. Results of similarity sequence searches of D. shibae and O. tauri genomes for genes encoding biosynthetic enzymes for these vitamins are indicated. O. tauri encodes the gene set necessary for synthesis for p-aminobenzoic acid, niacin and biotin (Supplementary Information), whereas D. shibae lacks a complete biosynthetic pathway for all three of these vitamins
The possible biosynthetic pathways of biotin, niacin and pABA are shown in Fig. 1b. In bacteria, biotin is synthesised from its precursor molecule pimeloyl-CoA by four enzymes: 8-amino-7-oxononanoate synthase (BioF) [38], 7,8-diamino-pelargonic acid aminotransferase (BioA) [39], dethiobiotin synthase (BioD) [40] and finally biotin synthase (BioB) [41], which converts dethiobiotin into biotin. In plants and some algae, the reactions of BioA and BioD can be performed by a bi-functional enzyme, known as BIO1 [42]. A BLASTP search for biotin synthesis enzymes in O. tauri identified a significant hit for this bi-functional enzyme, alongside the biosynthesis proteins BIOF and BIOB (green dots, Fig. 1a; Table S1) supporting the physiological data. In contrast, the genome for D. shibae contains putative genes for only two of the four required enzymes: BioF and BioA (two putative hits were found for BioA) (blue dots, Fig. 1b; Table S2). No orthologues of genes for BioD or BioB were identified, explaining the inability of D. shibae to grow without an exogenous source of biotin.
There are two biosynthetic routes for synthesis of niacin: (i) from anthranilate produced from the degradation of tryptophan (as in animals), or (ii) de novo synthesis from aspartate (characteristic of plants). Bacteria have been shown to use both pathways [43]. O. tauri does not encode biosynthesis genes from the tryptophan degradation pathway, but has all the genes required to produce nicotinamide mononucleotide through the aspartate pathway, using l-aspartate oxidase (NADB), quinolinate synthase (NADA) and quinolinic acid phosphoribosyl transferase (NADC) (Fig. 1b and supporting material in Table S3). Whereas D. shibae has a significant hit for proteins related to aspartate dehydrogenase (NadX, which is functionally equivalent to NADB [44]) and kynureninase (KYNU) (Table S4), we found no convincing orthologues for any other enzymes necessary for niacin biosynthesis. The lack of biosynthesis genes for the full aspartate or tryptophan pathway in D. shibae corroborates previous physiological evidence that D. shibae cannot make niacin itself [22], and thus requires an exogenous supply of this essential molecule (Fig. 1b).
The compound p-aminobenzoic acid (pABA), an intermediate in the synthesis of folate and its bioactive forms, is produced from chorismate [45]. In bacteria, two enzymes are required for this process. The first, aminodeoxychorismate synthase (ADCS) is made up of two protein sub-units, PabA and PabB [46]. Sub-unit II of the ADCS complex (PabA) acts as a glutamine amidotransferase, transferring the amino group from glutamine to sub-unit I (PabB), which is then used to aminate the chorismate molecule [46]. Arabidopsis thaliana, some yeasts and certain algae have been shown to encode a bi-functional enzyme (PAB-AB) that performs both of these steps [47]. In both bacteria and plants, the final enzyme in the pathway is 4-amino-4-deoxychorismate lyase (ADCL, PabC) that catalyses the reversible conversion of aminodeoxychorismate into p-aminobenzoic acid and pyruvate [48]. O. tauri has significant hits for PAB-AB and PABC (Fig. 1b and supporting material in Table S5), suggesting it is able to synthesise p-aminobenzoic acid. Incidentally, separate PABA and PABB homologues were also identified. D. shibae also encodes pabA and pabB genes (Table S6). In addition, a possible pabC gene was identified, but this was annotated as a d-alanine aminotransferase (D-AAT). ADCL and D-AAT are both class IV pyridoxal 5’-phosphate-dependent enzymes and share similar domain architecture. Using the NCBI conserved domain search function [49], we looked at the affiliation of our hit to reference ADCL and D-AAT sequences (Figure S3). The D. shibae gene (YP_001534484.1, highlighted in red) clusters together with other D-AAT enzymes (green) including that from another member of the Rhodobacteraceae family, Rhodobacter sphaeroides, rather than with ADCL-like enzymes (blue). These results indicate this gene is thus more likely to encode a D-AAT. The incomplete biosynthesis pathway of D. shibae for p-aminobenzoic acid thus corroborates the observation that this bacterium requires an exogenous source of this molecule for growth.
In the present study, we did not investigate the secretory pathways, which are associated with the uptake and potential release of these vitamins into the environment. For algal species, vitamin transporters have not been identified conclusively so bioinformatics characterisation is not possible. This is with the exception of CBA1, a protein important for cobalamin uptake [50]. However, this does not appear to be encoded by O. tauri. Our work therefore only addresses the question of whether the species are capable of metabolic synthesis and modes of exchange—whether passive (such as for example by viral lysis) or active (through transport proteins), remains unknown.
In summary, using curated sequences as queries on the O. tauri genome, complete biosynthetic pathways for p-aminobenzoic acid, niacin and biotin were identified, demonstrating potential biosynthetic capacity for each of these metabolites. In contrast, D. shibae lacks a complete biosynthetic pathway for all three of these vitamins.
Reciprocal complementation O. tauri/D.shibae B-vitamin requirements by mutualism
To explore further the complementary vitamin synthesis capabilities of O. tauri and D. shibae, we investigated their ability to grow together in the absence of exogenous B-vitamins. This required an axenic culture of both organisms. All cultures of O. tauri available from culture collections are maintained as uni-algal, with one or more contaminating bacteria present. Treatment of O. tauri OTH95 with a cocktail of antibiotics over several sub-cultures reduced the load considerably, but one bacterium, Zeaxanthibacterium sp. (Flavobacteriaceae), was always present. Although supplementation with B1 & B12 was still required, indicating that it was not providing the vitamins to O. tauri, we nonetheless wanted to separate the algal and bacterial cells to avoid confounding factors. As these are essentially the same dimensions (~1 µm in diameter), we investigated the possibility of using FACS. Initially, cells were gated on their size and shape, and then gated further based on their pulse width/pulse area, as described in Methods. This process could distinguish between single cells and doublets or triplets, which could be indicative of bacteria attached to algae. Once settings were established that allowed only single cells to flow through the channel, chlorophyll fluorescence was used to separate algal cells from bacteria (Figure S2). After sorting, cells were treated again with antibiotics for 14 days, following which no bacteria were detected by light microscopy or growth on MB plates.
Given the non-overlapping B-vitamin dependencies of O. tauri and D. shibae, we then investigated whether a stable co-culture of the two organisms could be established in medium containing none of the five B-vitamins, to instigate the transfer of complementary micronutrients. Cultures were maintained for three sub-cultures (each of 21–25 day duration) to ensure that any rescue effect was long-term and stable. At stationary phase, total cell numbers for both O. tauri and D. shibae were taken and compared with control mono-cultures grown in parallel (Fig. 2). Maximal growth was evident when O. tauri and D. shibae were grown in mono-culture with all the necessary B-vitamins. However, a significant increase in O. tauri cell density was observed when the alga was grown in co-culture with D. shibae (Fig. 2a, white bars) in medium lacking thiamine, B12 or all B-vitamins compared with the respective axenic mono-cultures (grey bars) (Student’s t test p < 0.05), although the rescue effect varied in magnitude. We found that whereas D. shibae was able to satisfy the requirements of O. tauri for B12, the rescue effect was weaker when thiamine was omitted, with the maximum carrying capacity achieved only ~ 30% of that when the vitamin was included in the growth medium. In turn, co-culturing with O. tauri satisfied the requirements of D. shibae for each of niacin, biotin and p-aminobenzoic acid (Fig. 2b), with statistically significant differences observed compared to growth in mono-culture under the equivalent deficiency profiles (Student’s t test P < 0.05 for each comparison). Interestingly, for axenic cultures of D. shibae (Fig. 2b, grey bars), omission of niacin had the greatest inhibitory effect on the final carrying capacity, and was equivalent to omitting all B-vitamins from the medium.
Fig. 2.
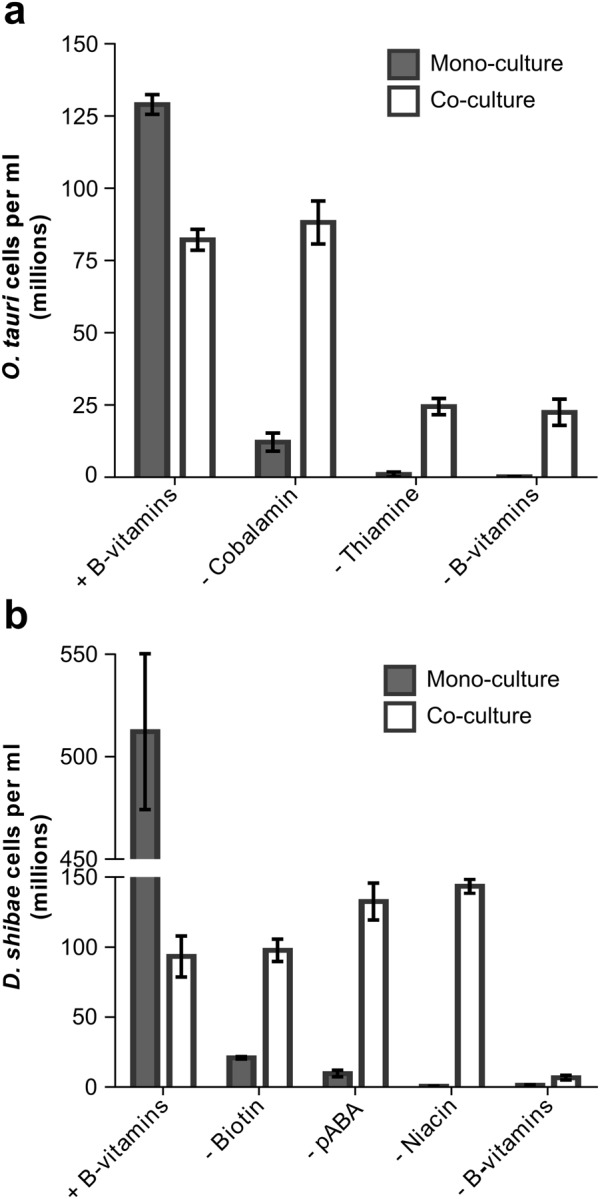
Growth of O. tauri and D. shibae under different B-vitamin combinations. a Growth assay to confirm B-vitamin auxotrophy in axenic O. tauri and D. shibae. Bacterial cultures were supplemented with 1% glucose. b Growth of O. tauri in mono-culture (grey) or in co-culture with D. shibae (white) under different B-vitamin profiles. Cell density was determined at stationary phase. c Growth of D. shibae in mono-culture (grey) or in co-culture with O. tauri (white) under different B-vitamin profiles. Error bars show standard deviation for three biological replicates
O. tauri can satisfy D. shibae requirements for niacin and p-aminobenzoic acid in stable long-term co-culture
The previous experiment indicated that growth yields of both O. tauri (OTH95) and D. shibae DFL-12 were lower in co-culture compared with mono-cultures. The observation that carrying capacities of organisms engaged in mutualisms are different to mono-culture is common, and has been termed regulation [51]. This is particularly well documented for plants in association with rhizobial symbionts, where the balance of host to mutualist is carefully maintained, and allows a carefully maintained exchange of resources (e.g., ref. [52]). For algal and bacterial systems in mutualism regulation has been observed [53] but often the signalling mechanism remains unclear (reviewed in [54]).
To determine whether O. tauri and D. shibae actively regulate each other’s growth, we inoculated the two organisms at different relative proportions into minimal medium without niacin, p-aminobenzoic acid or B12; both thiamine and biotin were included to enable reasonable growth rates and minimise the time needed for each experiment (omission of these vitamins had the greatest inhibitory effect on O. tauri and D. shibae cell density in co-culture, respectively (Fig. 2)). For those cultures with higher initial algal numbers, the starting inoculation density for the algae was set to ~ 2 × 106 cells per ml, with bacterial numbers adjusted accordingly to obtain the different ratios. When bacteria were initially in excess, the starting inoculation density of bacterial cells was ~ 1 × 106 cells per ml. Growth of the cultures were monitored over time by cell counting as described in the Methods. We found that regardless of starting numbers of either algal or bacterial cells, after 21–25 days in co-culture a ~ 1:1 ratio of the two organisms was reached (Fig. 3a). D. shibae cells have a faster growth rate than O. tauri, and so the 1:1 equilibrium was reached within 8 days when starting cultures had a higher ratio of algae to bacteria, whereas 25 days were required for O. tauri cells to reach parity from an initial inoculum with more bacterial cells.
Fig. 3.
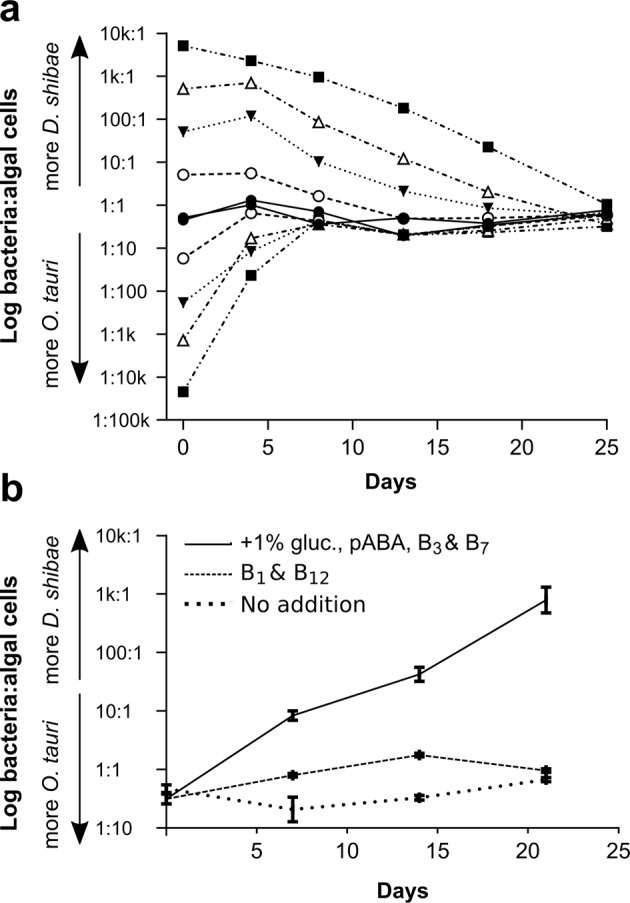
Growth dynamics of O. tauri in co-culture with D. shibae. a The ratio of D. shibae to O. tauri cells during growth in co-cultures initiated at different starting ratios. Cell densities of algae and bacteria were monitored until O. tauri reached stationary phase at 25 days. b The effect of adding-back nutrients on the established ratio. Ratio of O. tauri to D. shibae cells over the course of one culture when a stable co-culture is inoculated into medium favouring the bacteria (supplemented with 1% glucose and vitamins p-aminobenzoic acid (pABA), B3 and B7), medium favouring the alga (with B1 and B12) or no nutrient addback
Previously, we had shown that a stable co-culture could be established between the B12-dependent freshwater green alga Lobomonas rostrata with the soil bacterium Mesorhizobium loti [53]. An exogenous supply of either vitamin B12 or a fixed carbon source disrupted the nature of the co-culture. We therefore attempted to disrupt the O. tauri/D. shibae interaction through the addition of different B-vitamins and a carbon source that favour bacterial growth. When the suite of vitamins required by the bacteria were provided exogenously into the algal-bacterial co-culture together with a source of fixed carbon, the ratio of bacteria:algae altered significantly in favour of D. shibae, releasing the co-cultures from the established 1:1 ratio (Fig. 3b). Addition of just thiamine and B12 to the co-cultures also encouraged the growth of bacteria slightly relative to controls, even though these conditions satisfy the vitamin requirements of O. tauri and would be expected to favour the growth of the alga. This suggests that O. tauri is unable to regulate the growth of D. shibae when not engaged in mutualism with the bacteria. Nevertheless, when conditions for mutualism were maintained in minimal medium over successive sub-cultures (i.e., without supplementation with fixed carbon or B-vitamins), O. tauri and D. shibae continued to grow in a stable self-sustaining mutualistic relationship over four consecutive sub-cultures regardless of starting inoculum, resulting in a ratio of around 1:1 for the two organisms (Fig. 4).
Fig. 4.
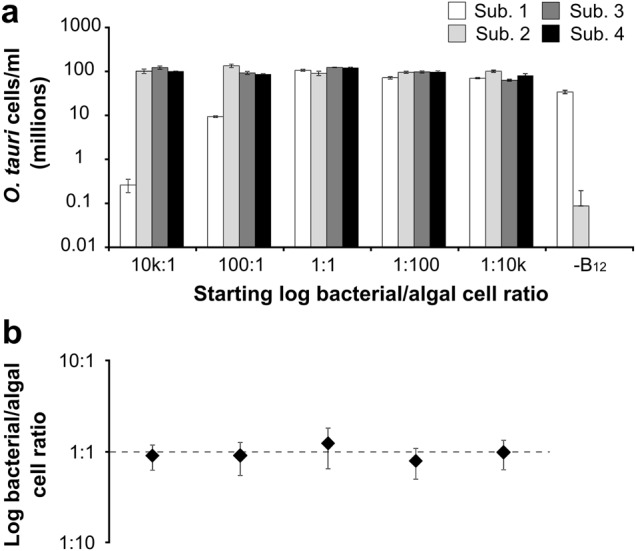
Rescue effect of O. tauri and D. shibae for required B-vitamins is stable and long-term. a Log cell density of O. tauri cells at stationary phase over four sub-cultures that were inoculated at differing initial ratios to D. shibae. b The average ratio of D. shibae: O. tauri at each different starting bacterial: algal ratio at stationary phase. Error bars indicate standard error for mean of ratios at stationary phase across all sub-cultures (n = 4)
Niacin and biotin auxotrophy is enriched in B12-biosynthesising bacteria belonging to the Rhodobacteraceae
The co-culture between O. tauri (OTH95) and D. shibae DFL-12 demonstrates a novel role for the vitamin cofactors niacin, biotin and p-aminobenzoic acid in nutrient exchange. To assess the broader relevance of this observation, we investigated whether the potential to form mutualistic interactions with eukaryotic marine algae based on reciprocal B-vitamin exchanges is a common characteristic of algae-associated bacteria. A survey of reports in the literature of bacteria found associated with marine algae from seven different locations identified a total of 28 genera (Table S8). Strikingly, in each location species of the Rhodobacteraceae family (order Rhodobacterales) were found (asterisked in Table S8). We chose 70 species from this family that had sequenced genomes, and analysed the gene complement for the presence of the 63 genes involved in the biosynthesis of cobalamin, niacin and biotin, using our previously established bioinformatics approach that allows rapid searching of complete bacterial genomes for large numbers of genes [16]. We were not able to use this method to search for genes encoding pABA biosynthesis enzymes because the close similarity between sequences for other enzymes requires manual inspection of the hits to verify their identity. The results revealed that that the majority (87%) of the species possess the genes required for vitamin B12 synthesis, whereas those for niacin were found in half, and only 9 (13%) appeared to encode the entire biotin biosynthetic pathway (Supplementary data set S1). In total, 29 species (41%) shared the genotype of D. shibae, namely the ability to synthesise vitamin B12 but not biotin or niacin.
To assess whether the Rhodobacteraceae exhibit a disproportionately high frequency of this trait compared with other bacteria, we extended the search more generally, using the GOLD database [55], which characterises species from known locations. A total of 197 verified marine eubacteria from 39 different orders, including 19 members of the Rhodobacteraceae family, were searched for the presence or absence of these 63 genes (Supplementary data set S2). Only nine species capable of producing B12 and at the same time lacking the capacity to synthesise both biotin and niacin were identified, all of which were Rhodobacteraceae. For the remaining 178 species from non-Rhodobacteraceae families, 149 (84%) encoded the complete niacin pathway, and 123 (69%) the entire biotin pathway, much higher percentages than for the Rhodobacteraceae (37 and 16%). Thus, using two independent datasets combined with information from previous studies, we find that, in contrast to marine bacteria generally, bacteria of the Rhodobacteraceae family, which includes D. shibae, are characterised by the ability to synthesise vitamin B12, but not B3 or B7 (Fig. 5). Members of this family are also commonly found together with eukaryotic marine algae [19, 54]. We therefore find it plausible that they could supply algae with their required cobalamin, while receiving niacin and biotin in return.
Fig. 5.
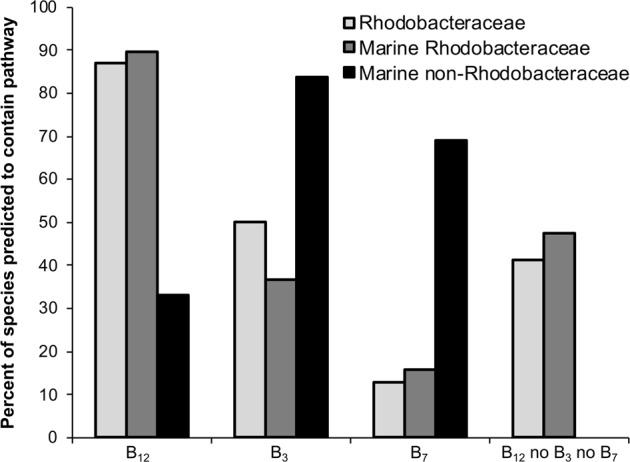
Distribution of pathways for the synthesis of vitamins B3, B7 and B12 in selected bacterial species. A range of bacterial species with sequenced genomes were subjected to a bioinformatics pipeline to predict their vitamin biosynthesis capabilities [16]. A set of 197 verified marine bacteria was obtained from the GOLD database [51], and a selection of 70 members of the Rhodobacteraceae family were sampled from NCBI. Their taxonomic positions were determined using ETE 3 [66], and species that were unclassified at the family level were discarded from the final analysis. The data were summarised at the family level for each of the different data sets—marine non-Rhodobacteraceae n = 178, marine Rhodobacteraceae n = 19, Rhodobacteraceae n = 70
Discussion
The notion that photosynthetic microorganisms are at the bottom of the food web in aquatic communities, supplying metabolites and energy to heterotrophic bacteria and macro-organisms in progressive trophic levels is out-dated [56, 57]. Instead, studies on aquatic microbial interactions are unveiling a suite of different lifestyles and cross-feeding interactions at the heart of these complex communities. Vitamins have emerged as important players in associations between aquatic microbes (e.g., ref. 23, 24, 56, 57), as by definition they are required obligately by auxotrophs for survival, so the demands for them must be met by other species capable of their biosynthesis, even if not directly [58]. The findings presented here are the first physiological demonstration of a bilateral B-vitamin exchange between aquatic microorganisms. Perhaps particularly important is our finding that O. tauri was able to provide D. shibae with niacin, the vitamin for which D. shibae exhibited the highest dependency. Very few studies have attempted to determine the concentration of niacin in oceanic environments: at the time of writing the only available publication is from half a century ago [59] and states niacin was undetectable in the Alaskan surface waters sampled. Our in silico survey found 20% of marine bacteria sampled do not have the genetic capacity to synthesise it de novo (Supplementary data set S2), but niacin auxotrophy is significantly enriched in the Rhodobacteracaeae (35/70 species sampled), whereas in contrast these are more likely to synthesise vitamin B12 than marine bacteria generally. Two Roseobacter species, R. litoralis and R. denitrificans, have previously been shown experimentally to require an exogenous source of niacin and biotin for growth [22]. These bacteria were isolated from the surface of green seaweeds, and were not found in open waters. More recently, another member of the Rhodobacteraceae [20], a Sulfitobacter species named SA11, was identified as a key symbiont with the toxic bloom forming alga Pseudo-nitzschia [24]. Previous studies have also identified niacin auxotrophy in host-associated bacteria, including in pathogenic and enteric bacteria [60]. For example, Shigella flexneri, the pathogen responsible for shigellosis in primates, is a niacin auxotroph, having lost the nadA and nadB genes responsible for synthesis of quinolinate from aspartate, a precursor to niacin [61]. The central role played by the bioactive forms of niacin as redox carriers and signalling molecules [62] makes it a crucial micronutrient for cell growth and survival in all organisms. The inability of some microbes to synthesise it is therefore perhaps surprising, especially considering that it requires just three enzymatic steps from central metabolites [37]. It is tempting to conclude that niacin auxotrophy in enteric bacteria, commensalists and pathogens, as well as in those found associated with algae, may be a common type of genetic streamlining in bacteria that live closely with other organisms.
The rescue effect of the mutualistic symbiosis that we describe here between O. tauri and D. shibae is stable over multiple generations for both individual B-vitamins and combinations thereof (Figs. 2 and 4). D. shibae was found to support the bulk of the B12 requirement for growth of O. tauri in co-culture but only a small proportion of its thiamine requirement (Fig. 2a). This matches the known Ks values of thiamine for several algal species, which have been reported to be up to four orders of magnitude higher than those for B12 [63]. Our findings indicate that thiamine is the most important regulator of O. tauri growth. For D. shibae, biotin limitation had the greatest effect on bacterial cell density in co-culture. We also investigated whether the interaction between the algae and bacteria exhibited regulation of population size, a phenomenon observed both in terrestrial systems and aquatic symbioses (e.g., ref. [54, 64]). Our findings suggest that regulation between D. shibae and O. tauri is likely, as the ratio of bacterial to algal cells reproducibly stabilises at ~ 1:1 during the exponential phase of growth, regardless of initial starting cell concentrations (Fig. 3a). This is a higher proportion of bacterial cells than the 1:30 measured in another co-culture, between L. rostrata and M. loti [53], and may reflect the fact that L. rostrata is much larger cell than M. loti, whereas O. tauri and D. shibae are essentially the same size. However, as for the L. rostrata/M. loti system the 1:1 equilibrium in cell numbers was only maintained when the medium was minimal, without any B-vitamins or an exogenous source of fixed carbon. Addition of these micronutrients favoured the growth of both algae and bacteria, disrupting their dependency on one another and destabilising the 1:1 ratio observed (Fig. 3b). This indicates vitamin (and carbon) availability are partially responsible for the observed regulatory effect. Thiamine and biotin may be particularly important in this regard, given that omission of these vitamins had the greatest inhibitory effect on O. tauri and D. shibae cell density in co-culture, respectively (Fig. 2). However, since bacterial and algal cell density in co-culture is reduced even in the presence of all B-vitamins (Fig. 2) other factors must be important too. For instance, it remains possible that the equilibrium observed is reached due to competition of the cells for other nutrients in the medium. This can be excluded by further studies under chemostat or semi-continuous batch culturing, which replenish the demand for macronutrients. However, factors beyond nutrient limitation may control algal-bacterial cell densities too. Other work [65] observed 'Jekyll and Hyde' dynamics regulated algal cell density in co-cultures of the marine alga Emiliania hyxleyi and bacterium Phaeobacter gallaeciensis. In this instance, population regulation occurs via production of the algicidal compound p-coumaric acid. Further experiments into possible signalling between the cells and quorum sensing is required to confirm the presence of regulation in this interaction.
Recent evidence has highlighted further complex exchange between algae and bacteria. Several marine bacteria have been demonstrated to synthesise auxins such as IAA, which have growth-promoting roles for microalgae [24, 65]. Moreover, algal-derived tryptophan may provide the precursor for IAA synthesis. Together with our results, these studies identify a remarkably complex and important aspect of marine microbial ecology that we are only beginning to understand. By providing the genetic basis underlying the physiological observations, a portion of the 'meta-metabolome' of two organisms in symbiosis representing the combined metabolic capabilities of the interacting species) can be visualised with respect to B-vitamins, highlighting the nodes of exchange (Fig. 6). Without considering co-limitation of macronutrients as a factor, when both these organisms are present in oligotrophic waters containing little or no B-vitamins free in solution they could support each other’s growth. Our experiments add to accumulating evidence of generalised 'niche-fitting' [66] such as observed for example between microbial species Chlamydomonas reinhardtii and the budding yeast Saccharomyces cerevisiae, which were shown to form complex physiological structures in the laboratory, providing each other with limiting nutrients [67]. Similarly, here, O. tauri could provide and receive B-vitamins from D. shibae. It should be mentioned that the formation of non-specific mutualisms between organisms in co-limiting conditions is not a given; for example, complementary auxotrophs of S. cerevisiae deficient in various aspects of amino acid and nucleotide metabolism regularly fail to rescue each other upon co-culturing [68] even though they are the same species. Consequently, our study makes an important contribution to the exploration of microbial interactions by providing an example where metabolic complementarity is achieved between partners from different branches of life. The fact that this is mediated by exchange of B-vitamins highlights the significance of these compounds, and expands the repertoire of nutrients involved in interactions in natural ecosystems.
Fig. 6.
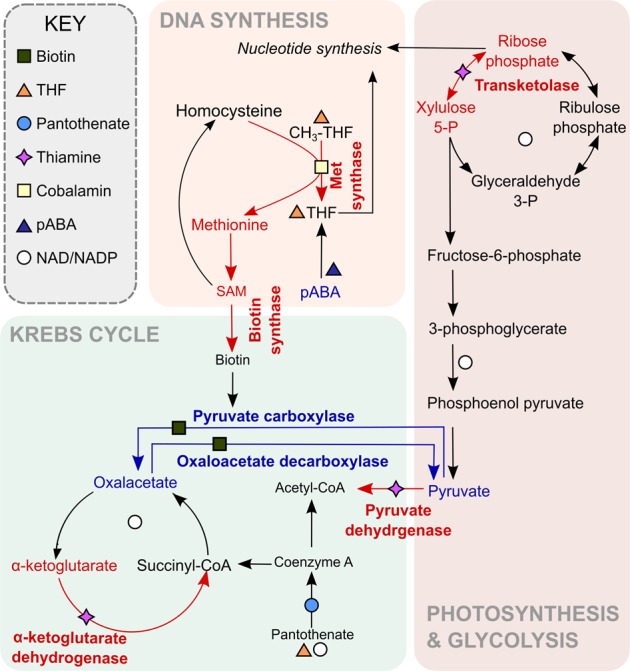
The role of cofactors in the meta-metabolome of the O. tauri-D. shibae partnership. Many essential metabolic processes depend on enzyme cofactors derived from B-vitamins, including biosynthesis of some of the cofactors themselves. Red arrows indicate reactions dependent on vitamins synthesised by D. shibae (cobalamin, thiamine), blue arrows indicate reactions dependent on vitamins synthesised by O. tauri: biotin, pABA (leading to folate) and niacin (leading to NAD+/NADP+). Reactions shown are not exhaustive, and in particular there are many more NAD+/NADP+ dependent enzymes
Evidence is accumulating to demonstrate a range of complex ecological interactions within microbial communities, so it is important to consider how such interactions may arise in an environment such as the ocean, where species assemblages are likely to be transient due to the fluid and dynamic medium. It remains an open question whether these symbiotic interactions are the product of evolutionary history, which presumes a number of generations of co-occurrence with metabolic exchange as the outcome rather than the driver for co-existence, or the opposite [69]. Two hypotheses for the evolution of mutualism within microbes have been proposed, which explore these contrasting scenarios: the Black Queen Hypothesis [70] and the Forager-to-Farmer model [57]. The identification and characterisation of microbial interactions at the genetic and physiological levels, as described here, will provide data to enable the hypotheses to be tested and refined.
Electronic supplementary material
Acknowledgements
We thank Herve Moreau at Banyuls-sur-mer Oceanological Observatory in France for the gift of O. tauri OTH95. We are grateful to Nigel Miller (Department of Pathology, University of Cambridge) for help with FACS. We acknowledge funding from BBSRC grant (BB/I013164/1) and EU FP7 DEMA (Project no. 309086). MBC was in recipient of a CASE studentship with the UK Biotechnology and Biological Sciences Research Council (BBSRC) and Plymouth Marine Laboratory Applications Ltd, UJK was supported by EU FP7 Marie Curie ITN Photo.Comm (Project No 317184), and AS by the Cambridge BBSRC Doctoral Training Partnership.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0274-y) contains supplementary material, which is available to authorised users.
References
- 1.Li WKW. Primary production of prochlorophytes, cyanobacteria, and eukaryotic ultraphytoplankton: measurements from flow cytometric sorting. Limnol Oceanogr. 1992;39:169–75. doi: 10.4319/lo.1994.39.1.0169. [DOI] [Google Scholar]
- 2.Countway PD, Caron DA. Abundance and distribution of Ostreococcus sp. in the San Pedro Channel, California, as revealed by quantitative PCR. Appl Environ Microbiol. 2006;72:2496–506. doi: 10.1128/AEM.72.4.2496-2506.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Worden AZ, Nolan JK, Palenik B. Assessing the dynamics and ecology of marine picophytoplankton: The importance of the eukaryotic component. Limnol Oceanogr. 2004;49:168–79. doi: 10.4319/lo.2004.49.1.0168. [DOI] [Google Scholar]
- 4.Massana R. Eukaryotic picoplankton in surface oceans. Annu Rev Microbiol. 2011;65:91–110. doi: 10.1146/annurev-micro-090110-102903. [DOI] [PubMed] [Google Scholar]
- 5.Demir-Hilton E, Sudek S, Cuvelier ML, Gentemann CL, Zehr JP, Worden AZ. Global distribution patterns of distinct clades of the photosynthetic picoeukaryote Ostreococcus. ISME J. 2011;5:1095–107. doi: 10.1038/ismej.2010.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derelle E, Ferraz C, Rombauts S, Rouzé P, Worden AZ, Robbens S, et al. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci USA. 2006;103:11647–52. doi: 10.1073/pnas.0604795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paerl RW, Bertrand EM, Allen AE, Palenik B, Azam F. Vitamin B1 ecophysiology of marine picoeukaryotic algae: Strain-specific differences and a new role for bacteria in vitamin cycling. Limnol Oceanogr. 2015;60:215–28. doi: 10.1002/lno.10009. [DOI] [Google Scholar]
- 8.Helliwell KE, Wheeler GL, Leptos KC, Goldstein RE, Smith AG. Insights into the evolution of vitamin B12 auxotrophy from sequenced algal genomes. Mol Biol Evol. 2011;28:2921–33. doi: 10.1093/molbev/msr124. [DOI] [PubMed] [Google Scholar]
- 9.Bertrand EM, Allen AE. Influence of vitamin B auxotrophy on nitrogen metabolism in eukaryotic phytoplankton. Front Microbiol. 2012;3:375. doi: 10.3389/fmicb.2012.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McRose D, Guo J, Monier A, Sudek S, Wilken S, Yan S, et al. Alternatives to vitamin B1 uptake revealed with discovery of riboswitches in multiple marine eukaryotic lineages. ISME J. 2014;8:2517–29. doi: 10.1038/ismej.2014.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warren MJ, Raux E, Schubert HL, Escalante-Semerena JC. The biosynthesis of adenosylcobalamin (vitamin B12) Nat Prod Rep. 2002;19:390–412. doi: 10.1039/b108967f. [DOI] [PubMed] [Google Scholar]
- 12.Croft MT, Lawrence AD, Raux-deery E, Warren MJ, Smith AG. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature. 2005;438:90–93. doi: 10.1038/nature04056. [DOI] [PubMed] [Google Scholar]
- 13.Karl DM. Nutrient dynamics in the deep blue sea. Trends Microbiol. 2002;10:410–9. doi: 10.1016/S0966-842X(02)02430-7. [DOI] [PubMed] [Google Scholar]
- 14.Sañudo-Wilhelmy S, Cutter LS, Durazo R, Smail E, Gómez-Consarnau L, Webb E, et al. Multiple B-vitamin depletion in large areas of the coastal ocean. Proc Natl Acad Sci USA. 2012;109:14041–5. doi: 10.1073/pnas.1208755109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Browning TJ, Achterberg EP, Rapp I, Engel A, Bertrand EM, Tagliabue A, et al. Nutrient co-limitation at the boundary of an oceanic gyre. Nature. 2017;551:242. doi: 10.1038/nature24063. [DOI] [PubMed] [Google Scholar]
- 16.Helliwell KE, Lawrence AD, Holzer A, Kudahl UJ, Sasso S, Kräutler B, et al. Cyanobacteria and eukaryotic algae use different chemical variants of vitamin B12. Curr Biol. 2016;26:999–1008. doi: 10.1016/j.cub.2016.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doxey AC, Kurtz DA, Lynch MD, Sauder LA, Neufeld JD. Aquatic metagenomes implicate Thaumarchaeota in global cobalamin production. ISME J. 2015;9:461–71. doi: 10.1038/ismej.2014.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goecke F, Labes A, Wiese J, Imhoff JF. Phylogenetic analysis and antibiotic activity of bacteria isolated from the surface of two co-occurring macroalgae from the Baltic Sea. Eur J Phycol. 2013;48:47–60. doi: 10.1080/09670262.2013.767944. [DOI] [Google Scholar]
- 19.Cooper MB, Smith AG. Exploring mutualistic interactions between microalgae and bacteria in the omics age. Curr Opin Plant Biol. 2015;26:147–53. doi: 10.1016/j.pbi.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 20.Simon M, Scheuner C, Meier-Kolthoff JP, Brinkhoff T, Wagner-Döbler I, Ulbrich M, et al. Phylogenomics of Rhodobacteraceae reveals evolutionary adaptation to marine and non-marine habitats. ISME J. 2017;11:1483–99. doi: 10.1038/ismej.2016.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biebl H, Allgaier M, Tindall BJ, Koblizek M, Lünsdorf H, Pukall R, et al. Dinoroseobacter shibae gen. nov., sp. nov., a new aerobic phototrophic bacterium isolated from dinoflagellates. Int J Syst Evol Microbiol. 2005;55:1089–96. doi: 10.1099/ijs.0.63511-0. [DOI] [PubMed] [Google Scholar]
- 22.Wagner-Döbler I, Ballhausen B, Berger M, Brinkhoff T, Buchholz I, Bunk B, et al. The complete genome sequence of the algal symbiont Dinoroseobacter shibae: a hitchhiker’s guide to life in the sea. ISME J. 2010;4:61–77. doi: 10.1038/ismej.2009.94. [DOI] [PubMed] [Google Scholar]
- 23.Durham BP, Sharma S, Luo H, Smith CB, Amin SA, Bender SJ, et al. Cryptic carbon and sulfur cycling between surface ocean plankton. Proc Natl Acad Sci USA. 2015;112:453–7. doi: 10.1073/pnas.1413137112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amin SA, Hmelo LR, van Tol HM, Durham BP, Carlson LT, Heal KR, et al. Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature. 2015;522:98–101. doi: 10.1038/nature14488. [DOI] [PubMed] [Google Scholar]
- 25.Bertrand EM, McCrow JP, Moustafa A, Zheng H, McQuaid JB, Delmont TO, et al. Phytoplankton-bacterial interactions mediate micronutrient colimitation at the coastal Antarctic sea ice edge. Proc Natl Acad Sci USA. 2015;112:9938–43. doi: 10.1073/pnas.1501615112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lima-Mendez G, Faust K, Henry N, Decelle J, Colin S, Carcillo F, et al. Determinants of community structure in the global plankton interactome. Science. 2015;348:1262073–3. doi: 10.1126/science.1262073. [DOI] [PubMed] [Google Scholar]
- 27.Giovannoni SJ. Vitamins in the sea. Proc Natl Acad Sci USA. 2012;109:13888–9. doi: 10.1073/pnas.1211722109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helliwell KE, Pandhal J, Cooper MB, Longworth J, Kudahl UJ, Russo DA, et al. Quantitative proteomics of a B12 -dependent alga grown in coculture with bacteria reveals metabolic tradeoffs required for mutualism. New Phytol. 2018;217:599–612. doi: 10.1111/nph.14832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poretsky RS, Hewson I, Sun S, Allen AE, Zehr JP, Moran MA. Comparative day/night metatranscriptomic analysis of microbial communities in the North Pacific subtropical gyre. Environ Microbiol. 2009;11:1358–75. doi: 10.1111/j.1462-2920.2008.01863.x. [DOI] [PubMed] [Google Scholar]
- 30.Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic Acid Techniques in Bacterial Systematic. New York, NY: John Wiley and Sons; 1991. pp. 116–75. [Google Scholar]
- 31.Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:740–3 [DOI] [PubMed]
- 32.Guillard RRL, Hargreaves PE. Stichochrysis immobilis is a diatom, not a crysophyte. Phycologia. 1993;32:234–6. doi: 10.2216/i0031-8884-32-3-234.1. [DOI] [Google Scholar]
- 33.Jett BD, Hatter KL, Huycke MM, Gilmore MS. Simplified agar plate method for quantifying viable bacteria. Biotechniques. 1997;23:648–50. doi: 10.2144/97234bm22. [DOI] [PubMed] [Google Scholar]
- 34.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 36.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 37.Gerdes S, Lerma-Ortiz C, Frelin O, Seaver SMD, Henry CS, de Crecy-Lagard V, et al. Plant B vitamin pathways and their compartmentation: a guide for the perplexed. J Exp Bot. 2012;63:5379–95. doi: 10.1093/jxb/ers208. [DOI] [PubMed] [Google Scholar]
- 38.Webster SP, Alexeev D, Campopiano DJ, Watt RM, Alexeeva M, Sawyer L, et al. Mechanism of 8-amino-7-oxononanoate synthase: spectroscopic, kinetic, and crystallographic studies. Biochemistry. 2000;39:516–28. doi: 10.1021/bi991620j. [DOI] [PubMed] [Google Scholar]
- 39.Stoner GL, Eisenberg MA. Purification and properties of 7, 8-diaminopelargonic acid aminotransferase. J Biol Chem. 1975;250:4029–36. [PubMed] [Google Scholar]
- 40.Krell K, Eisenberg MA. The purification and properties of dethiobiotin synthetase. J Biol Chem. 1970;245:6558–66. [PubMed] [Google Scholar]
- 41.Berkovitch F, Nicolet Y, Wan JT, Jarrett JT, Drennan CL. Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science. 2004;303:76–9. doi: 10.1126/science.1088493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muralla R, Chen E, Sweeney C, Gray JA, Dickerman A, Nikolau BJ, et al. A bifunctional locus (BIO3-BIO1) required for biotin biosynthesis in arabidopsis. Plant Physiol. 2007;146:60–73. doi: 10.1104/pp.107.107409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roje S. Vitamin B biosynthesis in plants. Phytochemistry. 2007;68:1904–21. doi: 10.1016/j.phytochem.2007.03.038. [DOI] [PubMed] [Google Scholar]
- 44.Yang Z, Savchenko A, Yakunin A, Zhang R, Edwards A, Arrowsmith C, et al. Aspartate dehydrogenase, a novel enzyme identified from structural and functional studies of TM1643. J Biol Chem. 2003;278:8804–8. doi: 10.1074/jbc.M211892200. [DOI] [PubMed] [Google Scholar]
- 45.Green JM, Matthews RG. (2007). FolateBiosynthesis, reduction, and polyglutamylation and the interconversion of folate derivatives. EcoSal Plus. 2007;2. 10.1128/ecosalplus.3.6.3.6 [DOI] [PubMed]
- 46.Viswanathan VK, Green JM, Nichols BP. Kinetic characterization of 4-amino 4-deoxychorismate synthase from Escherichia coli. J Bacteriol. 1995;177:5918–23. doi: 10.1128/jb.177.20.5918-5923.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Camara D, Richefeu-Contesto C, Gambonnet B, Dumas R, Rébeillé F. The synthesis of pABA: Coupling between the glutamine amidotransferase and aminodeoxychorismate synthase domains of the bifunctional aminodeoxychorismate synthase from Arabidopsis thaliana. Arch Biochem Biophys. 2011;505:83–90. doi: 10.1016/j.abb.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 48.Nakai T, Mizutani H, Miyahara I, Hirotsu K, Takeda S, Jhee KH, et al. Three-dimensional structure of 4-amino-4-deoxychorismate lyase from Escherichia coli. J Biochem. 2000;128:29–38. doi: 10.1093/oxfordjournals.jbchem.a022727. [DOI] [PubMed] [Google Scholar]
- 49.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–9. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bertrand EM, Allen AE, Dupont CL, Norden-Krichmar TM, Bai J, Valas RE, et al. Cobalamin acquisition protein from diatoms. PNAS. 2012;109:E1762–71. doi: 10.1073/pnas.1201731109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodriguez RJ, Freeman CD, McArthur DE, Kim YO, Redman RS. Symbiotic regulation of plant growth, development and reproduction. Commun Integr Biol. 2009;2:1–3. doi: 10.4161/cib.7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Horvath B, Bachem CWB, Schell J, Kondorosi A. Host-specific regulation of nodulation genes in Rhizobium is mediated by a plant-signal, interacting with the nodD gene product. EMBO J. 1987;6:841–8. doi: 10.1002/j.1460-2075.1987.tb04829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kazamia E, Czesnick H, Nguyen TTV, Croft MT, Sherwood E, Sasso S, et al. Mutualistic interactions between vitamin B12-dependent algae and heterotrophic bacteria exhibit regulation. Environ Microbiol. 2012;14:1466–76. doi: 10.1111/j.1462-2920.2012.02733.x. [DOI] [PubMed] [Google Scholar]
- 54.Amin SA, Parker MS, Armbrust EV. Interactions between diatoms and bacteria. Microbiol Mol Biol Rev. 2012;76:667–84. doi: 10.1128/MMBR.00007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mukherjee S, Stamatis D, Bertsch J, Ovchinnikova G, Verezemska O, Isbandi M, et al. Genomes OnLine Database (GOLD) v.6: data updates and feature enhancements. Nucleic Acids Res. 2017;45:D446–56. doi: 10.1093/nar/gkw992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Flynn KJ, Stoecker DK, Mitra A, Raven JA, Glibert PM, Hansen PJ, et al. Misuse of the phytoplankton–zooplankton dichotomy: the need to assign organisms as mixotrophs within plankton functional types. J Plankton Res. 2013;35:3–11. doi: 10.1093/plankt/fbs062. [DOI] [Google Scholar]
- 57.Kazamia E, Helliwell KE, Purton S, Smith AG. How mutualisms arise in phytoplankton communities: building eco-evolutionary principles for aquatic microbes Fussmann G (ed) Ecol Lett. 2016;19:810–22. doi: 10.1111/ele.12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Helliwell KE. The roles of B-vitamins in phytoplankton nutrition: new perspectives and prospects. New Phytol. 2017;216:62–68. doi: 10.1111/nph.14669. [DOI] [PubMed] [Google Scholar]
- 59.Natarajan KV. Distribution of thiamine, biotin, and niacin in the sea. Appl Microbiol. 1968;16:366–9. doi: 10.1128/am.16.2.366-369.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ford JE, Perry KD, Briggs CA. Nutrition of lactic acid bacteria isolated from the rumen. J Gen Microbiol. 1958;18:273–84. doi: 10.1099/00221287-18-1-273. [DOI] [PubMed] [Google Scholar]
- 61.Prunier AL, Schuch R, Fernández RE, Mumy KL, Kohler H, McCormick BA, et al. nadA and nadB of Shigella flexneri 5a are antivirulence loci responsible for the synthesis of quinolinate, a small molecule inhibitor of Shigella pathogenicity. Microbiology. 2007;153:2363–72. doi: 10.1099/mic.0.2007/006916-0. [DOI] [PubMed] [Google Scholar]
- 62.Opitz CA, Heiland I. Dynamics of NAD-metabolism: everything but constant. Biochem Soc Trans. 2015;43:1127–32. doi: 10.1042/BST20150133. [DOI] [PubMed] [Google Scholar]
- 63.Tang YZ, Koch F, Gobler CJ. Most harmful algal bloom species are vitamin B-1 and B-12 auxotrophs. Proc Natl Acad Sci USA. 2010;107:20756–61. doi: 10.1073/pnas.1009566107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parniske M. Intracellular accommodation of microbes by plants: a common developmental program for symbiosis and disease? Curr Opin Plant Biol. 2000;3:320–8. doi: 10.1016/S1369-5266(00)00088-1. [DOI] [PubMed] [Google Scholar]
- 65.Seyedsayamdost MR, Case RJ, Kolter R, Clardy J. The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat Chem. 2011;3:331–5. doi: 10.1038/nchem.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Agosta SJ, Klemens JA. Ecological fitting by phenotypically flexible genotypes: implications for species associations, community assembly and evolution. Ecol Lett. 2008;11:1123–34. doi: 10.1111/j.1461-0248.2008.01237.x. [DOI] [PubMed] [Google Scholar]
- 67.Hom EFY, Murray AW. Plant-fungal ecology. Niche engineering demonstrates a latent capacity for fungal-algal mutualism. Science. 2014;345:94–98. doi: 10.1126/science.1253320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Campbell K, Vowinckel J, Mülleder M, Malmsheimer S, Lawrence N, Calvani E, et al. Self-establishing communities enable cooperative metabolite exchange in a eukaryote. Elife. 2015;4:e09943. doi: 10.7554/eLife.09943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Helliwell KE, Wheeler GL, Smith AG. Widespread decay of vitamin-related pathways: coincidence or consequence? Trends Genet. 2013;29:469–78. doi: 10.1016/j.tig.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 70.Morris JJ, Lenski RE, Zinser ER. (2012) The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. mBio. 3;pii: e00036-12. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


