Abstract
In recent years, heavy metal pollution has become a more serious global problem, and all countries are actively engaged in finding methods to remediate heavy metal-contaminated soil. We conducted transcriptome sequencing of the roots of cotton grown under three different cadmium concentrations, and analysed the potential strategies for coping with cadmium stress. Through Gene Ontology analysis, we found that most of the genes differentially regulated under cadmium stress were associated with catalytic activity and binding action, especially metal iron binding, and specific metabolic and cellular processes. The genes responsive to cadmium stress were mainly related to membrane and response to stimulus. The KEGG pathways enriched differentially expressed genes were associated with secondary metabolite production, Starch and sucrose metabolism, flavonoid biosynthesis, phenylalanina metalism and biosynthesis, in order to improve the activity of antioxidant system, repair systems and transport system and reduction of cadmium toxicity. There are three main mechanisms by which cotton responds to cadmium stress: thickening of physical barriers, oxidation resistance and detoxification complexation. Meanwhile, identified a potential cotton-specific stress response pathway involving brassinolide, and ethylene signaling pathways. Further investigation is needed to define the specific molecular mechanisms underlying cotton tolerance to cadmium stress. In this study potential coping strategies of cotton root under cadmium stress were revealed. Our findings can guide the selection of cotton breeds that absorb high levels of cadmium.
Introduction
Cadmium pollution is a global environmental issue, and the development of modern industry and agriculture has led to more and more water and soil being polluted by cadmium every year1–3. Cadmium can accumulate for long periods of time inside animals and plants, affecting growth and development and posing a great danger to human health. The high tolerance of cotton to cadmium stands out from all extensively planted crops. Cotton is capable of absorbing a considerable amount of cadmium, and lower concentrations of cadmium has even been shown to promote development, productivity and fiber quality, and the effects of cadmium on a variety of physiological and biochemical characteristics as well as agronomic characteristics are very limited when the cadmium concentration is below 0.2 mmol. In addition, the cadmium content in cotton fibers, the main product of cotton, remains relatively low4–6. Therefore, cotton is a promising crop for treating cadmium-polluted soil.
Cadmium mainly causes damage to the plant by disrupting the balance of oxidation-reduction in cells. Specifically, cadmium accumulation leads to the production of a large quantity of reactive oxygen species (ROS) that oxidize membrane lipids, causing damage to the membrane system, affecting normal cellular functions and even leading to the collapse and death of the endomembrane system7–9. Previous studies have shown that there are three main cadmium resistance strategies in plants. The first strategy is the absorption of cadmium or isolation of cadmium inside plant. The second is alleviation of cadmium (Cd) toxicity and cadmium (Cd) removal through a series of chelating mechanisms. The third is the removal of the oxygen species (ROS) that accumulate during cadmium (Cd) stressPlants store cadmium at specific locations inside the plant and reduce the cadmium concentration in plant tissues that are metabolically active10,11. For example, hyper-accumulators like Brassica juncea store absorbed cadmium in the epidermis and epidermic hair cells, which are physiologically less active in order to alleviate the toxicity of cadmium (Cd) in other tissues. Cadmium absorbed via the symplast pathway in Oryza sativa accumulates in the vacuoles of root cortex cells, so that the number of cadmium ions entering the microtubules cells is reduced. When tobacco is exposed to cadmium stress, a large quantity of cadmium accumulates in the root cytoderm, which prevents cadmium ions from entering microtubules cell via the apoplast pathway. In addition, some cadmium ions bind to the active groups of cellulose and lignin, such as carboxyl and hydroxyl groups, reducing the quantity of cadmium that enters the protoplasm, and alleviating damage to the plant12–14.
Strategies for removal of cadmium that has accumulated inside cells mainly involve the induction of phytochelatin, metallothionein and relevant transport proteins that ultimately remove chelates out of the cell. The Arabidopsis thaliana mutants, cad1, cad2 and vtc1, which lack a phytochelatin (PC) synthesis system, have increased sensitivity to cadmium15–17. Overexpression of CAD1 in Brassica juncea increases its cadmium resistance, and overexpressing wheat phytochelatin (PC) synthase gene in tobacco also show increased cadmium resistance18,19. metallothionein (MT) genes such as CeMT2b, SaMT2 and TaMT3 overexpressing in Nicotiana tabacum L show even higher cadmium resistance20–22. The anti-oxidative mechanism of plants against the accumulation of oxygen species (ROS) is now clear, and includes the up-regulation of antioxidant enzymes such as superoxide superoxide dismutase, catalase, peroxidase, ascorbate peroxidase, and glutathione S-transferase, the improvement of the activity of anti-oxidative system, and the synthesis of a large quantity of anti-oxidative substances that accelerate the removal of oxygen species (ROS)23.
Even though the mechanism of cadmium resistance is known for some plants, the specific factors that affect the growth of cotton under cadmium stress still remain unclear. In this study, we investigated the cadmium resistance mechanism of cotton by analyzing the transcriptome data of cotton roots under cadmium stress and using existing studies as references. Our findings will provide insight that guide further studies of the cadmium resistance signaling pathways in cotton, as well as breeding of cadmium-resistant cotton species.
Results and Discussion
It is known that heavy metal ions enter plants via the symplast pathway and the apoplast pathway, and the root is the primary site of heavy metal absorption. Because the root is the first tissue to be poisoned by cadmium, it is prudent to investigate the mechanisms for heavy metal tolerance by analyzing plant root. Hence, we have conducted transcriptomic analysis of roots treated with different concentrations of cadmium.
Illumina Solexa paired-end RNA sequencing and read mapping
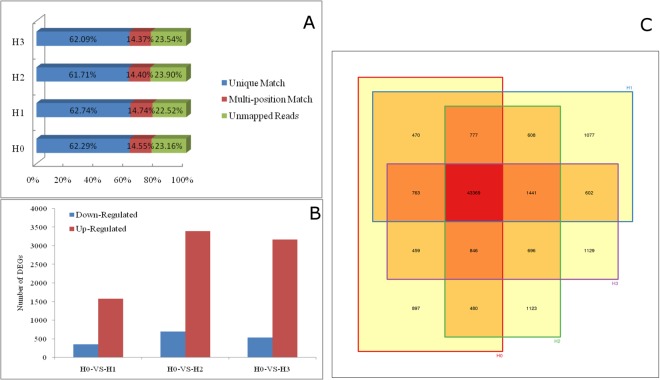
Seedlings of the upland cotton cultivar C184 were exposed to three different concentrations of cadmium using a hydroponics system. RNA from the root tissues of the control group (H0) and cadmium-treated groups (H1, H2 and H3) was sequenced on the Illumina HiSeqTM 2000 platform. The clean reads from the four samples accounted for nearly 93% of the raw data. The clean reads were mapped to the reference genome with Gossypium hirsutum TM-1 (http://mascotton.njau.du.cn), and in all four samples nearly 76% matched reference sequences. of these mapped reads, about 62% were unique matches, and 14%~15% mapped to multiple locations (Fig. 1A). A total of 48061, 49107, 49340 and 49305 transcripts were detected in H0, H1, H2 and H3, respectively, and the number transcripts iidentified in all four samples was 43,369 (Fig. 1B). The number of differentially expressed gene in all cadmium-treated groups compared with the control group was 1441, and the number of genes with log2(Relative value) change in expression equal to or greater than 1 or equal or less than −1 was 1151 (Fig. 1C). The number of differentially expressed genes identified by comparing H1 and H0 was 1908, among which the expression of 1565 genes were up-regulated in H1. The number of differentially expressed genes between H2 and H0 was 4075, of which 3165 were up-regulated in H3. These differentially expressed genes in H1, H2 and H3 were mapped to 115, 119, and 120 KEGG categories respectively.
Figure 1.
RNA-sequencing results and differentially expressed genes. (A) The percentage of RNA-sequencing reads that mapped to the reference. (B) The number of significantly up-regulated (red) and down-regulated (blue) genes between cotton roots treated with three cadmium concentrations (H1, H2 and H3) and the untreated control (H0). (C) Venn diagram illustrating the overlap in transcripts between samples.
Characterization of differentially expressed genes
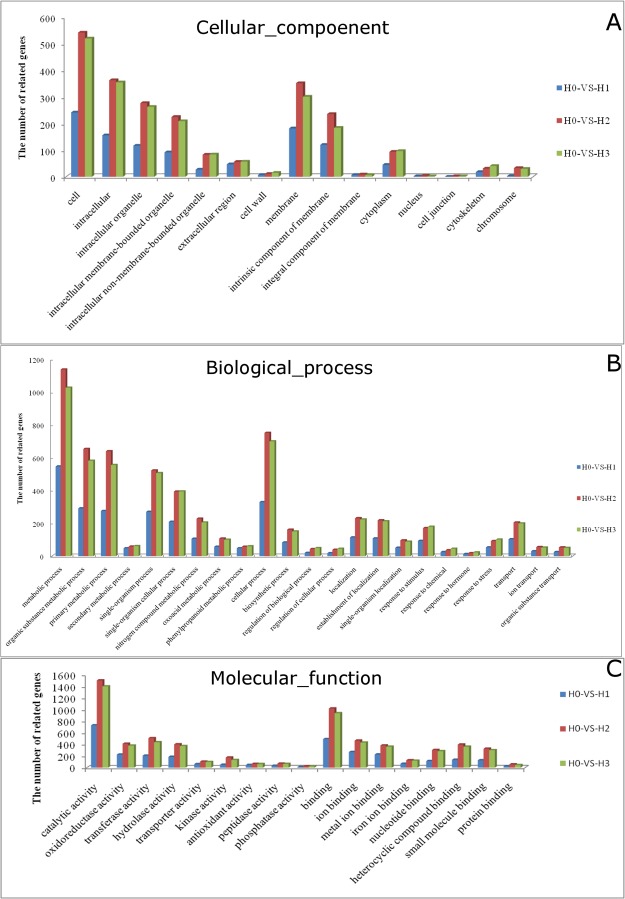
We used gene ontology annotations to classify the differentially expressed genes based on function and found that for each of the three cadmium-treated samples (H1, H2 and H3), the proportions of differentially expressed genes compared with H0 in gene ontology (GO) functional cluster were basically consistent, but there were differences in specific clusters. gene ontology (GO) annotations can be grouped into three broad categories: molecular function, cellular composition and biological process. With respect to cellular composition, the proportion of differentially expressed genes that function in intracellular regions is relatively larger, and the ratio of intracellular to extracellular differentially expressed genes increases as cadmium (Cd) concentration increases. The proportion of genes related to intracellular membranous organelles accounts for a considerable portion(Fig. 2A). In addition, the number of differentially expressed genes related to cytomembrane is far greater than the number related to other cellular components, indicating that cadmium (Cd) stress may affects the membrane system of root cells, especially the combination of membrane system. The effect of cadmium (Cd) stress on extracellular structures is limited, for example the cell junctions.
Figure 2.
Gene ontology classification of cotton root genes differentially expressed under different Cd concentrations. (A) cellular component (B) biological process and (C) molecular function.
With respect to biological process (Fig. 2B), the genes differentially expressed in response to cadmium (Cd) stress are mainly related to primary metabolism, with a smaller related to secondary metabolism. The metabolism of nitrogen-containing compounds, the synthesis of phenylpropanoids, and the synthesis of ketonic acid are three major biological processes that have a major impact on cadmium stress24,25. From the perspective of the cellular processes, the effects of cadmium stress are mainly taking place on the localization, response to stimulus, transport, while biological synthesis is a process that accounts for a considerable proportion. There is also a large proportion of differentially expressed genes that are related to the localization of substances and organelles, suggesting that cadmium stress increases the frequency of synthesis and transport of substances in root cells. And a severe change occur both in intracellular and extracellular environment while various response mechanism of cells start activating. Stress signaling under cadmium mainly include response to chemical and response to hormone process. Meanwhile, similarities of the stress processes when being treated with biological threat such as the invasion of pathogen are also demonstrated. With respect to substance transport, differentially expressed genes are mainly related to the transport of ions and organic substances, indicating that cadmium stress may disrupt ionic balance, and a dramatic increase in transport of substances, such as secondary metabolites. A number of studies have shown that ions, especially metal ion transporters do in fact play a key role in the absorption and accumulation of heavy metals in plants.
With respect to molecular functions, a considerable proportion of differentially expressed genes are related to catalytic activity, oxidoreductase activity and transferase activity (Fig. 2C), which is consistent with previous analysis. In addition, we found that a large number of differentially expressed genes are related to antioxidant enzyme activities and kinase activity suggesting that kinases function in cadmium stress signaling pathways. In the process of cell junction, the junction of metal ions, heterocyclic compounds and small molecular substances are of higher number of differentially expressed genes, indicating that metal ions, especially iron ions have more signal pathways of engagement in the reaction of cadmium stress, which is consistent with existing studies, wu study showed that maintaining high iron content in shoot under cadmium (Cd) exposure could alleviate the cadmium (Cd) toxicity. Meanwhile, the hybrid compounds and small molecular substances produced during metabolism are maybe play a important role in cadmium stress signaling pathways.
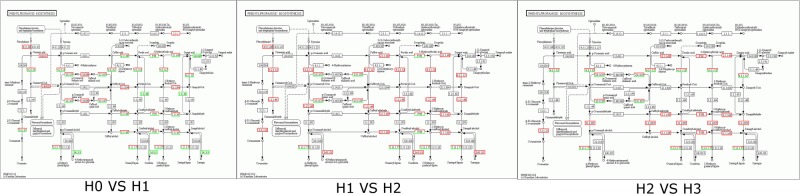
Analysis of the KEGG metabolic pathways enriched in genes differentially expressed in response to cadmium stress indicates that nearly 30% are involved in the synthesis of secondary metabolite pathways, including phenylpropanoid synthesis, phenylalanine metabolism, and flavonoid synthesis. Phenylpropanoid synthesis (https://www.kegg.jp/dbget-bin/www_bget?map00940) and PHE metabolism are closely related to the synthesis of lignin as well as plant disease responses (Fig. 3). Flavonoid metabolites product mainly include pigments, antioxidants, and small signaling molecules, which are closely related to plant stress resistance, indicating that in cotton the mechanisms for responding to cadmium and biological stress are similar. Cadmium stress leads to dramatic increase in metabolic activities on a cellular level, and also the increase of metabolites with hydrolytic activities.
Figure 3.
Impact of cadmium stress on phenylpropanoid biosynthesis (https://www.kegg.jp/ dbget-bin/www_bget?map00940)under different Cd concentrations.
Coping with cadmium stress by forming a physical barrier
Heavy metal tolerance of plants includes two aspects: one is the removal of heavy metals absorbed by the plant or impeding their transport withinin the plant; the other is detoxification by a variety of mechanisms, such as the binding of heavy metal to the cytoderm, which is then removed from the cell through the formation of organic acid and protein complexes. To further understand the mechanism for cadmium tolerance in cotton, we conducted a comprehensive analysis of 1151 genes differentially expressed in all three cadmium stress samples.
Cellulose is the main component of the plant cytoderm. Of the 1151 differentially expressed genes, the expression levels of 19 genes related to the synthesis of cellulose increased significantly; all of these genes are homologs of cellulose synthase (K10999) (Fig. 2). cellulose synthase A7 (CotAD_38396), cellulose synthase A8 (CotAD_23453), CotAD_69280), and cellulose synthase A4 (CotAD_57824, CotAD_54812) are correlated with the synthesis of the cellulose of primary cytoderm26, and cellulose synthase A9 (CotAD_62834, CotAD_51434), cellulose synthase A10 (CotAD_72572), CSLC (CotAD_58043, CotAD_13312) participates in the synthesis of the cellulose of secondary cytoderm27–29 (Table 1). In a previous study by Chen et al. on the effects of cadmium (Cd) on different components of willow cytoderm components, cellulose was found to strengthen by the absorption of cadmium (Cd) by cytoderm30. The increase in cellulose will in turn increase the absorption of cadmium (Cd) ions by cytoderm, reducing the number of cadmium (Cd) ions between cells, and consequently reducing the introduction of cadmium (Cd) into cells via the apoplast pathway, and thus alleviating cadmium (Cd) toxicity. We found that a large number of pectin methylesterase inhibitor genes and polygalacturonase-inhibiting protein genes were also up-regulated, and the hydrolysis process of pectin that is another cell wall component was inhibited. Previous studies of wheat, chilies, and kiwis have shown that overexpression of methylesterase inhibitor genes (PMEI) may lead to increased methyl esterification of pectin in the cytoderm, while suppressing the activities of pectin methylesterase and polygalacturonase, which are related to pectin hydrolysis31–33. Thus, the cytoderm is thickened, enhancing plant resistance to biological and abiotic stress. In addition, decreased methylesterase activity leads to the reduction of the carboxy l produced during pectin de-esterification in the cytoderm of peripheral cells, consequently decreasing the of absorption of positive heavy metal ions, such as cadmium (Cd) by root cells, as well as the amount of heavy metals absorbed by plants34.
Table 1.
Differential expression genes in cellulose synthesis process under cadmium stress in cotton root.
| Gene and Blast | log2Ratio(H0-VS-H1) | log2Ratio(H0-VS-H2) | log2Ratio(H0-VS-H3) |
|---|---|---|---|
| The cellulose synthesis genes of primary cell wall | |||
| CotAD_38396 CESA7 | 1.557482 | 1.713838 | 1.983378 |
| CotAD_23453 CESA8 | 1.591243 | 2.027124 | 1.774615 |
| CotAD_69280 CESA8 | 1.058526 | 1.281549 | 1.351638 |
| CotAD_57824 Cellulose synthase A4 | 2.456065 | 2.864345 | 2.462791 |
| CotAD_54812 Cellulose synthase A4 | 3.571157 | 3.405992 | 3.560715 |
| CotAD_00575 Cellulose synthase A1 | 1.837912 | 2.082523 | 2.219553 |
| CotAD_10480 Cellulose-synthase-like C5 | 1.98907 | 1.144908 | 1.882366 |
| CotAD_56775 Cellulose-synthase-like C5 | 2.391528 | 1.295149 | 1.885731 |
| CotAD_05132 Cellulose synthase-like B3 | −3.263034 | −1.607683 | −2.29956 |
| CotAD_46937 Cellulose synthase-like E1 | −0.293683 | −1.034949 | −0.138555 |
| CotAD_53925 Cellulose synthase like G3 | −0.722466 | −1.552541 | −2.956931 |
| CotAD_19832 Cellulose synthase like G3 | −11.049849 | −0.88243 | −0.568049 |
| The cellulose synthesis genes of Secondary cell wall | |||
| CotAD_62834 CESA9 | 1.69329 | 1.822638 | 2.359511 |
| CotAD_51434 CESA9 | 1.938548 | 2.234141 | 2.487153 |
| CotAD_72572 CESA10 | 1.501478 | 2.286739 | 2.522806 |
| CotAD_58043 Cellulose-synthase-like C12 | 1.460226 | 1.528496 | 2.233591 |
| CotAD_13312 Cellulose-synthase-like C4 | 1.2935 | 2.003514 | 2.295651 |
| Other related cellulose synthesis genes | |||
| CotAD_48408 Cellulose synthase | 1.419384 | 1.639206 | 2.051312 |
| CotAD_51651 Cellulose synthase | 1.864864 | 2.215985 | 2.176928 |
| CotAD_61788 Cellulose synthase 2-Dt | 1.661831 | 2.121881 | 2.292562 |
| CotAD_63213 Cellulose synthase family protein | 1.113962 | 1.30248 | 1.651 |
| CotAD_10636 Cellulose synthase like G2 | 1.379658 | 2.02403 | 1.714423 |
| CotAD_10635 Cellulose synthase like G2 | 1.23078 | 2.061952 | 1.665133 |
| CotAD_59099 Beta tubulin | 3.04459 | 3.585502 | 5.046353 |
| CotAD_21797 ATP binding microtubule motor family protein | 1.429138 | 1.463174 | 1.78061 |
| Pectin hydrolysis related genes | |||
| CotAD_11961 Pectin methylesterase inhibitor superfamily | 4.137504 | 2.918386 | 3.996389 |
| CotAD_01805 Pectin methylesterase inhibitor superfamily | 2.632695 | 3.375199 | 3.27894 |
| CotAD_22628 Pectin methylesterase inhibitor superfamily | 2.650687 | 3.059871 | 3.184791 |
| CotAD_45983 Pectin methylesterase inhibitor superfamily | 3.063326 | 4.725126 | 3.411565 |
| CotAD_59548 Pectin methylesterase inhibitor superfamily | 2.881072 | 3.27894 | 3.441269 |
| CotAD_71768 Pectin methylesterase inhibitor superfamily | 2.849094 | 3.333737 | 3.500173 |
| CotAD_50138 Pectin methylesterase inhibitor superfamily | 1.357719 | 2.728178 | 1.879046 |
| CotAD_70731 Pectin methylesterase inhibitor superfamily | 1.765261 | 3.091386 | 1.794206 |
| CotAD_32591 Pectin methylesterase inhibitor superfamily | 2.189825 | 2.369234 | 2.254241 |
| CotAD_76273 Pectin methylesterase inhibitor superfamily | 1.72259 | 2.10728 | 1.578135 |
| CotAD_45985 Pectin methylesterase inhibitor superfamily | −1.515442 | −2.166455 | −1.438093 |
| CotAD_28069 Pectin methylesterase inhibitor superfamily | −0.630113 | −0.462516 | −1.613652 |
| CotAD_63272 Pectin methylesterase inhibitor superfamily | −0.734779 | −0.81509 | −1.655598 |
| CotAD_06244 Polygalacturonase-inhibiting protein | 1.22272 | 2.802103 | 1.630121 |
| CotAD_07327 Polygalacturonase-inhibiting protein | 1.540627 | 2.810026 | 1.674442 |
| CotAD_17205 Polygalacturonase-inhibiting protein | −1.179787 | −0.96138 | −1.171961 |
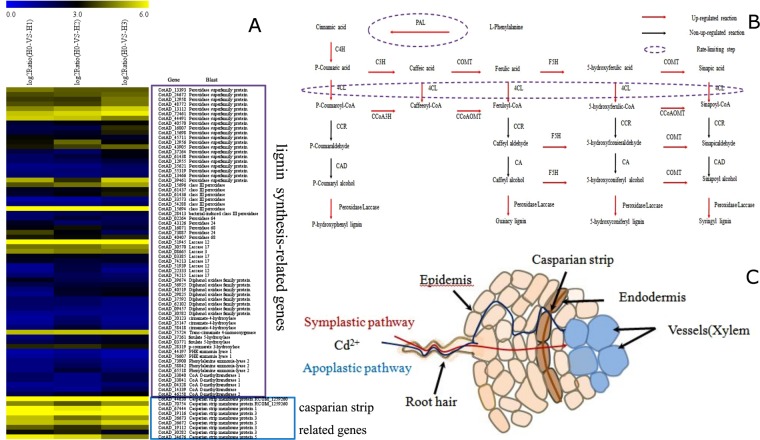
Cadmium stress led to a significant increase in the expression of genes involved in the synthesis of another cytoderm component, lignin35. Enzymes involved in the general lignin synthesis pathways were all up-regulated, and the expression levels of genes encoding the rate-limiting enzyme of lignin synthesis, Phenylalanine ammonia lyase, including CotAD_73900, CotAD_58842, CotAD_65518, doubled36. The expression levels of genes encoding another rate-limiting enzyme, 4-coumarate: CoA ligase, including CotAD_20123, CotAD_35147, CotAD_58418, were also significantly increased37. The expression levels of other genes, such as cinnamate-4-hydroxylase, ferulate 5-hydroxylase, p-coumarate 3-hydroxylase and CoA O-methyltransferase, which are involved in other lignin synthesis processes38, also increased to different extents (Fig. 4A,B). Lignin is the main component of secondary cytoderm, and cadmium stress increases lignin synthesis in cotton roots, the thickening of secondary cytoderm, and hence the reduction of the absorption of cadmium (Cd) by the root system, and the improvement of cadmium (Cd) tolerance in cotton. The cotton similar Matricaria chamomilla under Cu and cadmium (Cd) stress, lignin is found to accumulate in the cytoderm, forming a barrier against the absorption of heavy metal, and the cadmium (Cd) ions bind to acidic functional groups of lectin on the cell surface and to phenolic aldehyde led reduce cadmium (Cd) poisoning39. Therefore, some genes related to lignin synthesis are upregulated suggest that lignin plays an important role in cadmium tolerance in cotton, and its absorption of cadmium may reduce the cadmium (Cd) that enters root cells, consequently alleviating effects of cadmium (Cd) on the growth and development of the cotton root system.
Figure 4.
The physical barrier strategy for cadmium tolerance in cotton. (A) A heat map illustrating the differential expression of genes related to the formation of a physical barrier under Cd stress. x axis is log2x value of relative expression level. (B) Lignin biosynthetic pathway genes differentially expressed under cadmium stress. (C) A schematic diagram of the obstruction of cadmium ion transport by the Casparian strip in cotton roots.
The casparian strip, which consists of proteins, lignin, suberinite and cellulose, is another important barrier that prevents positive metal ions such as cadmium (Cd) from entering the cortex via the apoplast pathway (Fig. 4C). Among them, casparianstrip membrane domain proteins (CASPs) are the key signals for the initiation and localization of the casparian strip (CASP), which as casparian strip (CASP) polymer composites are firmly stuck to plasma membranes. And the casparian strip (CASP) formed by the accumulation of substances such as lignin and suberinite, impedes the transport of positive ions. For example, previous studies have found that calcium and lanthanum ions are incapable of penetrating casparian strip (CASP) of corn roots40,41. Seregin et al. found that the concentration of cadmium (Cd) ions within the root cortex of corn seedlings was significantly lower than that outside casparian strip, indicating that casparian strip (CASP) did impede the transport of cadmium ions42. As a result, corn seedlings are tolerate certain cadmium concentrations. Here we found that the expression level of casparian strip (CASP) genes, including one CASP1, five CASP3 and one CASP5 homologs, significantly increased upon cadmium treatment, indicating that the casparian strip (CASP) in the cotton root endodermis becomes broader, wider and deeper in response to cadmium stress. The expression levels of genes encoding specific enzymes involved in the synthesis of fatty acids and extension of fatty acid chains also increased. These results together with the increased expression levels of lignin and cellulose synthesis genes, are consistent with thickening of casparian strip of roots and the impediment of ion transport.
In conclusion, in cotton root cells cadmium stress leads to the thickening of the cytoderm, the thickening and broadening of casparian strip, and blocking of the transport of cadmium ions from both apoplast and symplast pathways. This excludes cadmium ions from the cortex and alleviates the effects of cadmium toxicity.
Oxidation resistance and complexation detoxification
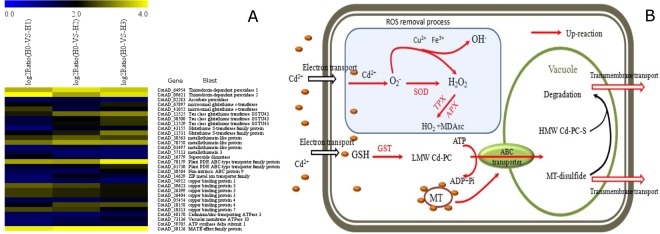
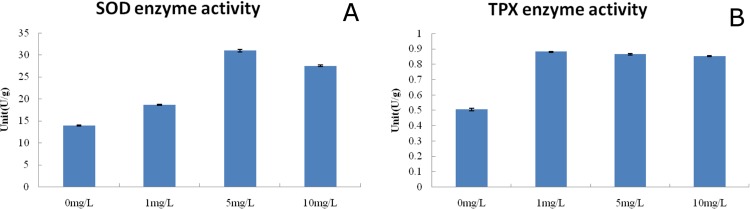
Studies have shown that as soon as cadmium (Cd) ions enter plant cells, oxygen species (ROS) begin to accumulate, oxygen species (ROS) which results in a series of physiological and metabolic disorders. Analysis of the cadmium stress transcriptome data revealed that the accumulation of cadmium (Cd) ions and oxygen radicals in cotton root tissue may induce stress responses, including oxidation resistance and heavy metal complexation (Fig. 5A,B). The expression level of the key enzyme involved in oxidation resistance, superoxide (SOD), was upregulated. Cu2+ transport proteins were also up-regulated, indicating a potential increase in Cu2+ concentration in cells, and hence the potential increase in the activities of several antioxidative metalloenzymes, such as superoxide (SOD), laccase and polyphenol oxidase43,44. Thioredoxin peroxidase and ascorbate peroxidase (APx), which are involved in the decomposition of H2O2, a secondary product of oxygen species (ROS), were also significantly up-regulated. The activity of superoxide (SOD), Thioredoxin peroxidase (TPx) and ascorbate peroxidase (APx) remove oxygen species (ROS) and prevents cell metabolic disorders. The increased activity of superoxide (SOD) and Thioredoxin peroxidase (TPx) in the cotton root under cadmium stress was verified by enzyme assays (Fig. 6).
Figure 5.
The oxidation resistance and complexation detoxification strategies for cadmium tolerance in cotton. (A) A heat map illustrating the differential expression of genes related to oxidation resistance and complexation detoxification under Cd stress. x axis is log2x value of relative expression level. (B) A diagram illustrating the functions of differentially expressed genes in oxidation resistance and complexation detoxification
Figure 6.
The determination of Superoxide Dismutase (SOD) and thioredoxin peroxidase (TPx) enzyme activity under cadmium stress in cotton roots.
cadmium (Cd) ions already located inside cells are detoxified by heavy metal chelating agents such as phytochelatin (PC), glutathione and metallothionein (MT). While the expression quantity of PDR-type ABC transport protein (CotAD_70179, CotAD_61730), MATE transport protein gene (CotAD_30126) and ATP synthetase involved in the transport of heavy metal chelates also increased for cadmium (Cd) detoxification. When cotton root tissues are treated with cadmium, glutathione (GSH) transferase (GST) catalyzes conjugation of glutathione (GSH) to heavy metals. Cotton GST as well multiple metallothionein (MT) genes were up-regulated by cadmium stress. The expression level of genes encoding PDR-type ABC transport protein (CotAD_70179, CotAD_61730) and MATE transport protein (CotAD_30126), which are involved in the transport of heavy metal chelates, and ATP synthetase, which supplies engery needed for transport, were also upregulated. Previous studies by Kim et al. and Ogawa et al. showed that the expression of Oryza sativa and A. thaliana PDR genes and MATE transport protein genes were induced by metal ions; among them, PRD8 was shown to be involved in the transport of Cd2+ or cadmium (Cd) chelates in arabidopsis45–47.
Effect of cadmium on the development of the cotton root
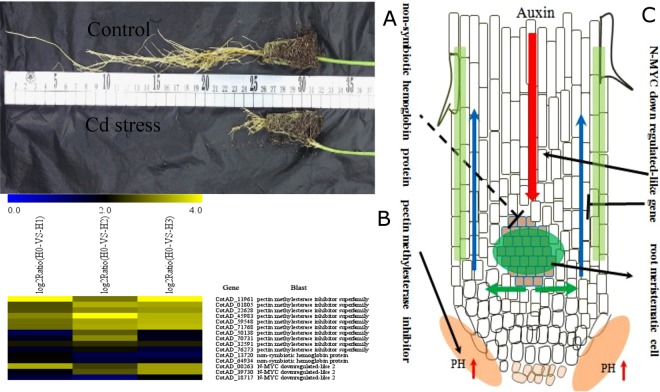
Exposure of cotton grown in solution to cadmium stress significantly suppressed root development, especially the development of primary roots (Fig. 7A). Similar phenomena have been found by Liu in cotton cadmium stress experiment48. We found that three types of genes related to root development, methylesterase inhibitor (PMEI), nonsymbiotic hemoglobin, and N-MYC downregulated-like 2, were differentially expressed upon cadmium treatment (Fig. 7B). The up-regulation of methylesterase inhibitor (PMEI) genes suppresses the de-esterification of pectin, which also reduces the amount of oxygen ions produced during the de-esterification, leading to an increase in the pH in the root. According to the acid curvature theory, this increase in pH will inevitably affect the extension of root tip cells, consequently suppressing root growth49 and potentially explains why cadmium suppresses cotton root development. Expression of nonsymbiotic hemoglobin (nsHb) is an important strategy for stress resistance. Previous studies by Igamberdiev et al. and Yang et al. showed that overexpression of nonsymbiotic hemoglobin (nsHb) genes increase plant antioxidase activity and strengthens resistance against oxidative stress50,51. Moreover, Parent et al. discovered that the nonsymbiotic hemoglobin (nsHb) gene was highly expressed specifically in the protoderm and xylem of root tissues52. Therefore, the up-regulation of cotton nonsymbiotic hemoglobin (nsHb) by cadmium stress may be another factor suppressing root development. Studies have shown that the downregulated-like 2 (NDL2) gene functions with heterotrimeric G-protein to regulate auxin transport within plants, and Yashwanti et al. verified that NDL increases the transport of basipetal auxin, while suppressing the transport of auxin towards the top of the plant. Hence, the up-regulation of downregulated-like 2 (NDL2) by cadmium stress may lead to accumulation of auxin in root meristems, leading to impeded development of primary roots, such as observed under cadmium stress53. Taken together, our finding that methylesterase inhibitor (PMEI), nonsymbiotic hemoglobin (nsHb) and downregulated-like 2 (NDL2) genes are upregulated by cadmium stress, is consistent with the suppression of root development (Fig. 7C).
Figure 7.
The influence of cadmium stress on cotton root development. (A) The root systems of 30 day old control and Cd stress (5 mg/L) cotton treated seedlings. x axis is log2x value of relative expression level. (B) Genes A heat map illustrating the differential expression of genes related to root development under cadmium stress. (C) Diagram illustrating the regulation of root growth under cadmium stress in cotton.
Differentially expressed transcription factors under cadmium stress in cotton roots
We found that 28 transcription factors were differentially expressed in all three cadmium-treated cotton samples compared with the control group. Of these, 20 were upregulated including, MYB, zinc finger (GATA-type, CCCH-type, and C2H2-type), leucine zipper and NAC transcription factors (Table 2). Based on studies in A. thaliana the homologs of these transcription factors with known functions are mainly involved in the transduction of light signals, the synthesis of brassinolide (BR), brassinolide (BR) signal transduction, the synthesis of the secondary cytoderm, and ethylene signal transduction.
Table 2.
Differential expression transcription factors under cadmium stress in cotton root.
| Gene ID | log2Ratio(H0-VS-H1) | log2Ratio(H0-VS-H2) | log2Ratio(H0-VS-H3) | Blast and description | The function of homologous gene in other species |
|---|---|---|---|---|---|
| Up-regulated transcripts | |||||
| CotAD_02717 | 5.229531 | 4.714754 | 5.49896 | Myb domain protein 18[Theobroma cacao] | Positive regulator of the phyA photoresponse |
| CotAD_23906 | 3.424715 | 3.49136 | 4.690159 | Myb domain protein 55 [Theobroma cacao] | involved in Brassinosteroid homeostasis and growth responses |
| CotAD_19868 | 2.710493 | 2.890771 | 3.460425 | Myb domain protein 40 [Theobroma cacao] | function unknown |
| CotAD_76285 | 2.297773 | 2.479582 | 2.641895 | Myb domain protein 40 [Theobroma cacao] | function unknown |
| CotAD_50930 | 1.830316 | 1.64293 | 1.60539 | Myb-like HTH transcriptional regulator protein [Theobroma cacao] | function unknown |
| CotAD_62038 | 1.674672 | 2.007526 | 2.324341 | MYb transcription factor [Gossypium hirsutum] | involved in the response to UV-B |
| CotAD_39114 | 1.063645 | 1.621674 | 2.095315 | Myb domain protein 55 [Theobroma cacao] | involved in Brassinosteroid homeostasis and growth responses |
| CotAD_48495 | 2.645083 | 2.98263 | 3.303392 | GATA transcription factor 9 [Theobroma cacao] | function unknown |
| CotAD_23031 | 2.341828 | 2.160992 | 2.989946 | GATA transcription factor 9 [Theobroma cacao] | function unknown |
| CotAD_75922 | 1.556025 | 1.49199 | 2.442026 | GATA transcription factor 9 [Theobroma cacao] | function unknown |
| CotAD_59298 | 1.369646 | 1.944693 | 2.293563 | GATA transcription factor 2 [Theobroma cacao] | positive regulator of photomorphogenesis |
| CotAD_36519 | 1.357823 | 2.199698 | 1.54739 | GATA zinc finger protein regulating nitrogen assimilation [Theobroma cacao] | function unknown |
| CotAD_55882 | 1 | 1.053503 | 1.195975 | GATA zinc finger protein regulating nitrogen assimilation [Theobroma cacao] | function unknown |
| CotAD_38727 | 2.49114 | 3.372333 | 2.093109 | Zinc finger protein [Theobroma cacao] | involved in secondary wall biosynthesis |
| CotAD_30228 | 1.973305 | 2.385891 | 2.909453 | C2H2-type zinc finger family protein[Theobroma cacao] | function unknown |
| CotAD_53425 | 3.342392 | 3.1836 | 3.336963 | Basic-leucine zipper transcription factor family protein [Theobroma cacao] | involved in lipid metabolism or cellular transpor |
| CotAD_16854 | 1.905141 | 2.242857 | 2.20543 | Basic-leucine zipper transcription factor family protein [Theobroma cacao] | involved in lipid metabolism or cellular transpor |
| CotAD_21422 | 1.999432 | 1.737874 | 1.377638 | Integrase-type DNA-binding superfamily protein[Theobroma cacao] | involved in ethylene signaling pathways |
| CotAD_11710 | 1.654185 | 1.162601 | 2.048023 | ethylene-responsive element-binding protein 5 [Gossypium barbadense] | involved in ethylene signaling pathways |
| CotAD_61187 | 1.843845 | 2.563242 | 1.513578 | LOB domain-containing protein 4 | function unknown |
| Dn-regulated transcripts | |||||
| CotAD_13693 | −2.066401 | −2.447005 | −3.066401 | Basic helix-loop-helix (bHLH) DNA-binding superfamily protein [Theobroma cacao] | involved in iron homeostasis |
| CotAD_55579 | −2.024977 | −1.152438 | −3.169925 | Basic helix-loop-helix (bHLH) DNA-binding superfamily protein [Theobroma cacao] | involved in iron homeostasis |
| CotAD_12483 | −1.683411 | −1.834735 | −2.361483 | Basic helix-loop-helix (bHLH) DNA-binding superfamily protein [Theobroma cacao] | involved in Interactions between Ethylene and Auxin |
| CotAD_54397 | −1.374919 | −1.533229 | −1.290792 | Basic helix-loop-helix (bHLH) DNA-binding superfamily protein [Theobroma cacao] | involved in Interactions between Ethylene and Auxin |
| CotAD_08994 | −1.201244 | −1.241635 | −1.704707 | Basic helix-loop-helix (bHLH) DNA-binding superfamily protein [Theobroma cacao] | involved in salicylic-dependent defense signaling response |
| CotAD_50063 | −1.613672 | −2.009292 | −1.449406 | NAC domain protein 9 [Gossypium hirsutum] | function unknown |
| CotAD_73593 | −1.210922 | −1.524039 | −1.079658 | NAC domain protein 9 [Gossypium hirsutum] | function unknown |
| CotAD_70812 | −1.214829 | −2.034157 | −1.538036 | Myb-like transcription factor family protein [Theobroma cacao] | function unknown |
The transduction of light signals, synthesis of brassinolide (BR) and brassinolide (BR) signal transduction pathways are involved in plant photomorphogenesis. Processes such as brassinolide (BR) synthesis, synthesis of secondary cytoderm and ethylene signal response are closely related to the resistance of plants against abiotic stress. The A. thaliana homolog of CotAD_02717, ATMYB18, was shown to be a transcriptional activator involved in regulating the response to phytochrome signals, especially under low red light and far red light54. AtMYB4 was shown to be involved in the response to drought, high salt and ultraviolet radiation. In addition, Silvia et al. discovered that AtMYB4 is involved in the synthesis of flavonol, and the up-regulation of AtMYB4 leads to increased stress resistance55–57. So, the up-regulation of the expression of a large number of genes involved in phenylpropanoid and flavonoid synthesis pathways is consistent with increased resistance against cadmium stress in cotton. Interestingly, two types of genes that are involved in ethylene response are also related to stress resistance and tissue development. First, the A. thaliana homolog of CotAD_21422 and CotAD_11710, which are ethylene response factors, functions in oxidative stress and osmotic pressure signal transduction pathways58,59. The up-regulated expression of ethylene response genes suggests that the improved resistance to cadmium (Cd) in cotton is mediated via ethylene response pathways. Second, GATA2 and MYB55 have been reported to be key genes connecting the responses to brassinolide (BR) and developmental signals, and the expression levels of both genes were repressed by brassinolide (BR) though the BR-activated transcription factor BZR160,61. Therefore, the increased expression of GATA2 and MYB55 suggests that the accumulation of brassinolide (BR) is decreased under cadmium (Cd) stress in cotton, and less brassinolide (BR) in root cells would impede root growth, leading to a shorter taproot. Other differentially expression transcription factors are predicted to be involved in cell wall synthesis, the metabolism and cellular transport of ester compounds and iron ion balance in the cell; all of these functions are associated with plant stress resistance. Thus, it indicates that the brassinolide (BR) signaling pathway activated in response to cadmium (Cd) stress in A. thaliana and cotton. The functions of other differentially expressed transcription factors still remain unclear and need to be further studied.
Quantitative RT-PCR validation
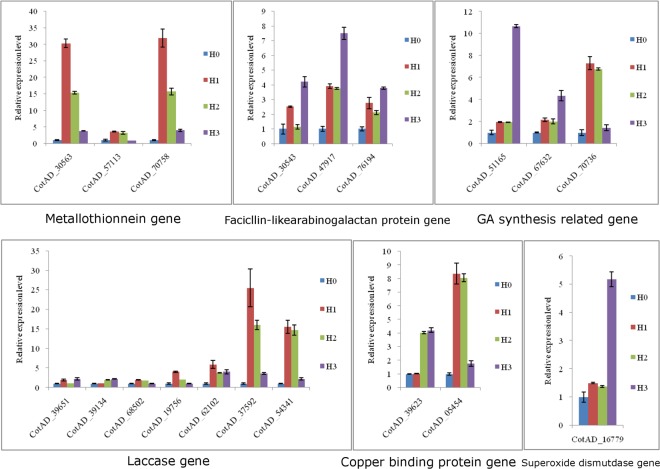
We performed real-time quantitative PCR to validate the expression of selected differentially expressed genes, including genes related to the formation of a physical barrier, anti-oxidization and chelation and root development. The expression levels determined by quantitative real-time PCR were basically consistent with the transcriptome data (Fig. 8). The expression level of FLA, which is a laccase involved in lignin synthesis and may be related to the sedimentation and thickening of the secondary cytoderm, increased under cadmium stress. The key anti-oxidative gene superoxide dismutase (SOD) (CotAD16779) and metallothionein (MT) genes that encode proteins that chelate heavy metal ions (CotAD30563, CotAD57113, CotAD70758) are all up-regulated, indicating an increase in the activity of antioxidant enzymes. Gibberellic acid synthetase, which is involved in root development was dramatically upregulated by cadmium stress. In a previous study, Gou et al. suggested that excessive amounts of gibberellin may suppress the development of the side roots of Populus tremula62. So, the accumulation of gibberellin may be one of the reasons why the development of cotton roots is impeded by cadmium stress.
Figure 8.
Verification of gene expression in untreated cotton seedlings (H0) and cotton seedlings treated with three different concentrations of cadmium (H1, H2 and H3) by real-time RT-PCR. (A) Metallothionein genes. (B) Facicllin-like arabinogalactan genes. (C) GA synthesis-related genes. (D) Laccase genes. (E) Copper binding protein genes. (F) Superoxide dismutase gene.
Conclusion
Transcriptome analysis of cotton roots exposed to cadmium stress revealed that physical barriers, including thichening of the cell wall and the casparian strip (CASP), may play an important role in the response to cadmium stress. In addition, an increase in pH in the root may enhance cadmium tolerance and also affect root development. Resistance to oxidation stress and detoxification via heavy metal complexation likely also play important roles. In addition, analysis of the functions of transcription factors differentially expressed in response to cadmium stress may have revealed a cotton-specific brassinolide (BR) signaling pathway that mediates the response cadmium stress. It was previously found that in L. Chinense response to cadmium stress is mediated by an endogenous salicylic acid signaling pathway, and overexpression of the Lycium chinense glutathione synthetase (LcGSHS) gene increased cadmium tolerance in transgenic A. thaliana63. The salicylic acid signaling pathway is often synergistic with the brassinolide (BR) signal pathway in environmental stress. Meanwhile, we found NPR1 which plays a key cross-talk between SA and brassinolide (BR) which via brassinolide (BR) signal activate the oxygen species (ROS) clearance mechanism was down-regulated signally under cadmium (Cd) stress in cotton64,65. So, it is may have a crosstalk between brassinolide (BR), ethylene signaling and cadmium (Cd) stress that help to improve cadmium (Cd) tolerance in cotton.
Methods
Plant Growth and Treatment
The C184 cotton species used for the experiment was provided by Hunan Cotton Science Institute. The cadmium solution was prepared with CdCl2·2.5H2O. The cotton seedlings were grown in Kimura B nutrient solution containing the following macronutrients66 (mM): (NH4)2SO4 (0.18), MgSO4·7H2O (0.27), KNO3 (0.09), Ca(NO3)2·4H2O (0.18), and KH2PO4 (0.09); and the following micronutrients (µM): MnCl2·4H2O (0.5), H3BO3 (3), (NH4)6Mo7O24·4H2O (1), ZnSO4·7H2O(0.4), Fe-EDTA (20), and 0.2 µM CuSO4·5H2O. The pH of the nutrient solution was adjusted to 5.5. Growth conditions were as follows: 27/24 °C day/night temperatures, 60–80% relative humidity, and a 14/10-h day/night photoperiod. The cultures were placed in an artificial intelligence (AI) climatic chamber for all-weather illumination. Four solutions containing different concentrations of CdCl2 added to the cultures when the seedlings developed two leaves and one core. The concentrations of the solutions were as follows: 0 mmol/L (H0, control), 1 mmol/L (H1), 5 mmol/L (H2) and 10 mmol/L (H3). Seedlings were grown for an additional 10 days (from October 12th to October 21st). All root systems of seedlings from the four different treatments were taken out at the end of treatment, cleaned with tap water before rinsing with double distilled water, and then dried with sterile absorbent paper. The root tissues were then placed in a 5 ml germ-free centrifuge tube and then quickly frozen in liquid nitrogen, and submitted to the beijing genomics Institute (BGI) for sequencing analysis.
Unigene annotation
Unigene annotations include functional annotation and information about expression. The comment information covers cluster of orthologous group (COG) functional annotation, protein functional annotation and gene ontology (GO) (http://geneontology.org/) functional annotation of unigenes based on BLASTX alignments to protein databases such as nr (http://www.ncbi.nlm.nih.gov/sites/entrez?db=protein), Swiss-Prot (http://www.expasy.org), KEGG (www.kegg.jp/kegg/kegg1.html) and COG (E-value < 0.00001) (http://www.ncbi.nlm.nih.gov/COG/). The proteins with the highest sequence similarity to unigenes are retrieved along with their protein and functional annotations. The gene ontology (GO) terms for upland cotton genes were obtained using the software Blast2GO (http://www.blast2go.com). At the same time, pathway analysis done using the KEGG mapping method. Enzyme commission numbers were assigned to unique sequences that had BLASTX scores with cutoff values of E < 1.0e5, as determined based on protein database searches. The Unigene sequences were mapped to the KEGG biochemical pathways according to the EC distribution in the pathway database.
Quantitative RT-PCR
Total RNA was extracted from cotton root tissues using the Aidlab RNA extraction kit and was synthesized into cDNA using the Promega reverse transcription kit. The volumes of RNA from the four samples were adjusted with RNA-ase free H2O to ensure that the concentrations were the same before reverse transcription. Primer-BLAST was used to design the primers for fluorescent quantitation. RT-PCR was conducted on the ABI 7500 fast platform. Each reaction was done in a total volume of 20 µl, including 1 µl primer, 1 µl 10-fold diluted cDNA, 10 µl SYBR Green PCR mix, and brought up to volume with H2O. The cycling conditions were as follows: 95 °C 10 min followed by 95 °C 5 s, 60 °C 30 s, 72 °C 30 s, for a total of 40 cycles. The following conditions were used for melting curve analysis: 95 °C 15 s, 60 °C 1 min, 95 °C 30 s, 60 °C 15 s. Ghactin7 was used as the reference gene, and 2−ΔΔCT was used to calculate the relative expression level67. Each reaction was repeated three times for each gene. The primer of Real-time fluorescent quantitative PCR see (supplementary material- primer).
Enzyme activity assays
Superoxide dismutase enzyme and Thioredoxin peroxidase (TPx) enzyme activity was tested by kit of Sangon Biotech. Weighing 0.2 g root tissue and added to 0.8 ml phosphate buffer to 20% tissue homogenate for enzyme assays. Homogenates were spun at 4000 rpm/min in a centrifuge for 10 min. A volume of 0.1 ml supernatant was removed for superoxide dismutase enzyme activity determination, and double distilled water was used as the control. The samples were incubated in a water bath for 40 min and 37 °C, thermostatic and sufficient mixing, After adding 2 ml color developing reagent, samples were incubated 10 min at room temperature and then the absorbance at 550 nm of each sample was measured. The formula used to calculate superoxide dismutase activity is as follows:
A 0.1 ml aliquot of the same supernatant used for superoxide dismutase enzyme activity determination was used to assay for thioredoxin peroxidase (TPx) activity. thioredoxin peroxidase (TPx) decomposes H2O2 in the presence of a reducing agent, and we detemermined thioredoxin peroxidase (TPx) enzyme activity by measuring the rate of decrease in absorbance at 240 nm and subtracting the catalase (CAT) enzyme activity. The formulas used to calculate thioredoxin peroxidase (TPx) activity are as follows:
Electronic supplementary material
Acknowledgements
This research was supported by the National Key Research and Development Program of China (2016YFD0100203, 2016YFD0101409), the State Key Laboratory of Cotton Biology Open Fund (CB2017A09) and Hunan Natural Science Foundation of Changde Mutual Funds (2016JJ5014). This program was financially sponsored by the National Key Research and Development Program of China (2016YFD0100203, 2016YFD0101409), the State Key Laboratory of Cotton Biology Open Fund (CB2017A09), Hunan Natural Science Foundation of Changde Mutual Funds (2016JJ5014). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
H.C. has designed the experiments,carried out all computational analyses and drafted the manuscript. Y.L. has conceived the experiments, analyzed the results, carried out all computational analyses and drafted the manuscript. X.M. has drafted the manuscript and carried out the subsequent revision of paper. L.G. has carried out all computational analyses and proof read revised the manuscript. Y.H. has carried out all computational analyses and proof read revised the manuscript. Z.R. has carried out the subsequent revision of paper. Z.K. has drafted the manuscript. X.Z. has designed the experiments and revised the manuscript. Z.Z. has designed the experiments and revised the manuscript.
Availability of Data and Materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Haodong Chen and Yujun Li contributed equally.
Contributor Information
Xiling Zhang, Email: zhangxiling1962@126.com.
Zhigang Zhang, Email: zhangzhig@126.com.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-36228-z.
References
- 1.Zhang L, Wong MH. Environmental mercury contamination in China: sources and impacts. Environment international. 2007;33:108–121. doi: 10.1016/j.envint.2006.06.022. [DOI] [PubMed] [Google Scholar]
- 2.Huang M, et al. Heavy metals in wheat grain: assessment of potential health risk for inhabitants in Kunshan. China. Science of the Total Environment. 2008;405:54–61. doi: 10.1016/j.scitotenv.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Dai XP, et al. Concentration level of heavy metals in wheat grains and the health risk assessment to local inhabitants from Baiyin, Gansu, China. Advanced Materials Research. 2012;518:951–956. doi: 10.4028/www.scientific.net/AMR.518-523.951. [DOI] [Google Scholar]
- 4.Li L, et al. Characterization of physiological traits, yield and fiber quality in three upland cotton cultivars grown under cadmium stress. Australian Journal of Crop Science. 2012;6:1527. [Google Scholar]
- 5.Daud, M. K. et al. In vitro cadmium-induced alterations in growth and oxidative metabolism of upland Cotton (Gossypium hirsutum L.). The Scientific World Journal. 5, 10.1155; 10.1155/309409 (2014). [DOI] [PMC free article] [PubMed]
- 6.Liu LT, et al. Cotton seedling plants adapted to cadmium stress by enhanced activities of protective enzymes. Plant Soil Environ. 2016;62:80–85. [Google Scholar]
- 7.Cho UH, Seo NH. Oxidative stress in Arabidopsis thaliana exposed to cadmium is due to hydrogen peroxide accumulation. Plant Science. 2005;168:113–120. doi: 10.1016/j.plantsci.2004.07.021. [DOI] [Google Scholar]
- 8.Sun Y, et al. Cadmium tolerance and accumulation characteristics of Bidens pilosa L. as a potential Cd-hyperaccumulator. Journal of Hazardous Materials. 2009;161:808–814. doi: 10.1016/j.jhazmat.2008.04.030. [DOI] [PubMed] [Google Scholar]
- 9.Cuypers A, et al. Cadmium stress: an oxidative challenge. Biometals. 2010;23:927–940. doi: 10.1007/s10534-010-9329-x. [DOI] [PubMed] [Google Scholar]
- 10.Hall JL. Cellular mechanisms for heavy metal detoxification and tolerance. Journal of experimental botany. 2002;53:1–11. doi: 10.1093/jexbot/53.366.1. [DOI] [PubMed] [Google Scholar]
- 11.Viehweger K. How plants cope with heavy metal. Botanical Studies. 2014;55:35. doi: 10.1186/1999-3110-55-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salt DE, Wagner GJ. Cadmium transport across tonoplast of vesicles from oat roots. Evidence for a Cd2+/H+ antiport activity. Journal of Biological Chemistry. 1993;268:12297–12302. [PubMed] [Google Scholar]
- 13.Clemens S, et al. Plant science: the key to preventing slow cadmium poisoning. Trends in plant science. 2013;18:92–99. doi: 10.1016/j.tplants.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 14.Nedelkoska TV, Doran PM. Hyperaccumulation of cadmium by hairy roots of Thlaspi caerulescens. Biotechnology and Bioengineering. 2000;67:607–615. doi: 10.1002/(SICI)1097-0290(20000305)67:5<607::AID-BIT11>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 15.Howden R, Cobbett CS. Cadmium-sensitive mutants of Arabidopsis thaliana. Plant Physiology. 1992;100:100–107. doi: 10.1104/pp.100.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cobbett CS, et al. The glutathione-deficient, cadmium-sensitive mutant, cad2-1, of Arabidopsis thalianais deficient in γ-glutamylcysteine synthetase. The Plant Journal. 1998;16:73–78. doi: 10.1046/j.1365-313x.1998.00262.x. [DOI] [PubMed] [Google Scholar]
- 17.Jozefczak M, et al. Both the concentration and redox state of glutathione and ascorbate influence the sensitivity of arabidopsis to cadmium. Annals of botany. 2015;116:601–612. doi: 10.1093/aob/mcv075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu YL, et al. Overexpression of glutathione synthetase in Indian mustard enhances cadmium accumulation and tolerance. Plant physiology. 1999;119:73–80. doi: 10.1104/pp.119.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim YO, et al. Zn tolerance of novel Colocasia esculenta metallothionein and its domains in Escherichia coli and tobacco. Journal of plant research. 2012;125:793–804. doi: 10.1007/s10265-012-0492-8. [DOI] [PubMed] [Google Scholar]
- 20.Martínez M, et al. An engineered plant that accumulates higher levels of heavy metals than Thlaspi caerulescens, with yields of 100 times more biomass in mine soils. Chemosphere. 2006;64:478–485. doi: 10.1016/j.chemosphere.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 21.Zhou B, et al. The metallothionein gene, TaMT3, from Tamarix androssowii confers Cd2+ tolerance in tobacco. International journal of molecular sciences. 2014;15:10398–10409. doi: 10.3390/ijms150610398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, et al. Metallothionein 2 (SaMT2) from Sedum alfredii Hance confers increased Cd tolerance and accumulation in yeast and tobacco. PloS one. 2014;9:e102750. doi: 10.1371/journal.pone.0102750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sytar O, et al. Heavy metal-induced oxidative damage, defense reactions, and detoxification mechanisms in plants. Acta physiologiae plantarum. 2013;35:985–999. doi: 10.1007/s11738-012-1169-6. [DOI] [Google Scholar]
- 24.Nishizono H, et al. The role of the root cell wall in the heavy metal tolerance ofAthyrium yokoscense. Plant and soil. 1987;101:15–20. doi: 10.1007/BF02371025. [DOI] [Google Scholar]
- 25.Küpper H, et al. Cellular compartmentation of nickel in the hyperaccumulators Alyssum lesbiacum, Alyssum bertolonii and Thlaspi goesingense. Journal of Experimental Botany. 2001;52:2291–2300. doi: 10.1093/jexbot/52.365.2291. [DOI] [PubMed] [Google Scholar]
- 26.Handakumbura PP, et al. Perturbation of Brachypodium distachyon CELLULOSE SYNTHASE A4 or 7 results in abnormal cell walls. BMC plant biology. 2013;13:131. doi: 10.1186/1471-2229-13-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stork J, et al. CELLULOSE SYNTHASE9 serves a nonredundant role in secondary cell wall synthesis in Arabidopsis epidermal testa cells. Plant physiology. 2010;153:580–589. doi: 10.1104/pp.110.154062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cocuron JC, et al. A gene from the cellulose synthase-like C family encodes a β-1, 4 glucan synthase. Proceedings of the National Academy of Sciences. 2007;104:8550–8555. doi: 10.1073/pnas.0703133104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watanabe Y, et al. Visualization of cellulose synthases in Arabidopsis secondary cell walls. Science. 2015;350:198–203. doi: 10.1126/science.aac7446. [DOI] [PubMed] [Google Scholar]
- 30.Chen G, et al. Cadmium adsorption by willow root: the role of cell walls and their subfractions. Environmental Science and Pollution Research. 2013;20:5665–5672. doi: 10.1007/s11356-013-1506-3. [DOI] [PubMed] [Google Scholar]
- 31.Hong MJ, et al. Functional characterization of pectin methylesterase inhibitor (PMEI) in wheat. Genes & genetic systems. 2010;85:97–106. doi: 10.1266/ggs.85.97. [DOI] [PubMed] [Google Scholar]
- 32.An SH, et al. Pepper pectin methylesterase inhibitor protein CaPMEI1 is required for antifungal activity, basal disease resistance and abiotic stress tolerance. Planta. 2008;228:61–78. doi: 10.1007/s00425-008-0719-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Camardella L, et al. Kiwi protein inhibitor of pectin methylesterase. The FEBS Journal. 2000;267:4561–4565. doi: 10.1046/j.1432-1327.2000.01510.x. [DOI] [PubMed] [Google Scholar]
- 34.Arancibia RA, Motsenbocker CE. Pectin methylesterase activity in vivo differs from activity in vitro and enhances polygalacturonase-mediated pectin degradation in tabasco pepper. Journal of plant physiology. 2006;163:488–496. doi: 10.1016/j.jplph.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 35.Sewalt VJH, et al. Reduced lignin content and altered lignin composition in transgenic tobacco down-regulated in expression of L-phenylalanine ammonia-lyase or cinnamate 4-hydroxylase. Plant physiology. 1997;115:41–50. doi: 10.1104/pp.115.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kajita S, Katayama Y, Omori S. Alterations in the biosynthesis of lignin in transgenic plants with chimeric genes for 4-coumarate: coenzyme A ligase. plant and cell physiology. 1996;37:957–965. doi: 10.1093/oxfordjournals.pcp.a029045. [DOI] [PubMed] [Google Scholar]
- 37.Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annual review of plant biology. 2003;54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- 38.Kováčik J, Klejdus B. Dynamics of phenolic acids and lignin accumulation in metal-treated Matricaria chamomilla roots. Plant cell reports. 2008;27:605–615. doi: 10.1007/s00299-007-0490-9. [DOI] [PubMed] [Google Scholar]
- 39.Nagahashi G, Thomson WW, Leonard RT. The Casparian strip as a barrier to the movement of lanthanum in corn roots. Science. 1974;183:670–671. doi: 10.1126/science.183.4125.670. [DOI] [PubMed] [Google Scholar]
- 40.Kuhn AJ, Schröder WH, Bauch J. The kinetics of calcium and magnesium entry into mycorrhizal spruce roots. Planta. 2000;210:488–496. doi: 10.1007/PL00008156. [DOI] [PubMed] [Google Scholar]
- 41.Kuhn AJ, Bauch J, Schröder WH. Springer Netherlands. 1995. Monitoring uptake and contents of Mg, Ca and K in Norway spruce as influenced by pH and Al, using microprobe analysis and stable isotope labelling; pp. 135–150. [Google Scholar]
- 42.Seregin IV, Shpigun LK, Ivanov VB. Distribution and toxic effects of cadmium and lead on maize roots. Russian Journal of Plant Physiology. 2004;51:525–533. doi: 10.1023/B:RUPP.0000035747.42399.84. [DOI] [Google Scholar]
- 43.Ma X, Deng D, Chen W. Inhibitors and Activators of SOD, GSH-Px, and CAT. InTech. 2017;9:65936. doi: 10.5772/65936. [DOI] [Google Scholar]
- 44.Zhou C, et al. Effect of Common Metal Ions and Anions on Laccase Catalysis of Guaiacol and Lignocellulosic Fiber. BioResources. 2017;12:5102–5117. [Google Scholar]
- 45.Ogawa I, et al. Time course analysis of gene regulation under cadmium stress in rice. Plant and soil. 2009;325:1–2. doi: 10.1007/s11104-009-0116-9. [DOI] [Google Scholar]
- 46.Kim DY, et al. The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. The Plant Journal. 2007;50:207–218. doi: 10.1111/j.1365-313X.2007.03044.x. [DOI] [PubMed] [Google Scholar]
- 47.Kim DY, et al. AtATM3 is involved in heavy metal resistance in Arabidopsis. Plant Physiology. 2006;140:922–932. doi: 10.1104/pp.105.074146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu LT, et al. Effects of Cadmium Stress on Growth and Cadmium Accumulation in Cotton (Gossypium hirsutum L.) Seedlings. Cotton Science. 2014;5:466–470. [Google Scholar]
- 49.Rayle DL, Cleland RE. The Acid Growth Theory of auxin-induced cell elongation is alive and well. Plant physiology. 1992;99:1271–1274. doi: 10.1104/pp.99.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Igamberdiev AU, Bykova NV, Hill RD. Nitric oxide scavenging by barley hemoglobin is facilitated by a monodehydroascorbate reductase-mediated ascorbate reduction of methemoglobin. Planta. 2006;223:1033–1040. doi: 10.1007/s00425-005-0146-3. [DOI] [PubMed] [Google Scholar]
- 51.Yang L, et al. AtGLB1 enhances the tolerance of Arabidopsis to hydrogen peroxide stress. Plant and cell physiology. 2005;46:1309–1316. doi: 10.1093/pcp/pci140. [DOI] [PubMed] [Google Scholar]
- 52.Parent C, et al. A novel nonsymbiotic hemoglobin from oak: cellular and tissue specificity of gene expression. New Phytologist. 2008;177:142–154. doi: 10.1111/j.1469-8137.2007.02250.x. [DOI] [PubMed] [Google Scholar]
- 53.Mudgil Y, et al. Arabidopsis N-MYC DOWNREGULATED-LIKE1, a positive regulator of auxin transport in a G protein-mediated pathway. The Plant Cell. 2009;21:3591–3609. doi: 10.1105/tpc.109.065557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ballesteros ML, et al. LAF1, a MYB transcription activator for phytochrome A signaling. Genes & development. 2001;15:2613–2625. doi: 10.1101/gad.915001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao J, et al. SAD2, an importin β-like protein, is required for UV-B response in Arabidopsis by mediating MYB4 nuclear trafficking. The Plant Cell. 2007;19:3805–3818. doi: 10.1105/tpc.106.048900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vannini C, et al. Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. The Plant Journal. 2004;37:115–127. doi: 10.1046/j.1365-313X.2003.01938.x. [DOI] [PubMed] [Google Scholar]
- 57.Fornalé S, et al. AtMYB7, a new player in the regulation of UV-sunscreens in Arabidopsis thaliana. Plant and Cell Physiology. 2014;55:507–516. doi: 10.1093/pcp/pct187. [DOI] [PubMed] [Google Scholar]
- 58.Papdi C, et al. The low oxygen, oxidative and osmotic stress responses synergistically act through the ethylene response factor VII genes RAP2. 12, RAP2. 2 and RAP2.3. The Plant Journal. 2015;82:772–784. doi: 10.1111/tpj.12848. [DOI] [PubMed] [Google Scholar]
- 59.Schellingen K, et al. Ethylene signalling is mediating the early cadmium-induced oxidative challenge in Arabidopsis thaliana. Plant Science. 2015;239:137–146. doi: 10.1016/j.plantsci.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 60.Luo XM, et al. Integration of light-and brassinosteroid-signaling pathways by a GATA transcription factor in Arabidopsis. Developmental cell. 2010;19:872–883. doi: 10.1016/j.devcel.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He JX, et al. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gou J, et al. Gibberellins regulate lateral root formation in Populus through interactions with auxin and other hormones. The Plant Cell. 2010;22:623–639. doi: 10.1105/tpc.109.073239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guan C, et al. A GSHS-like gene from Lycium chinense maybe regulated by cadmium-induced endogenous salicylic acid and overexpression of this gene enhances tolerance to cadmium stress in Arabidopsis. Plant cell reports. 2015;34:871–884. doi: 10.1007/s00299-015-1750-8. [DOI] [PubMed] [Google Scholar]
- 64.Divi UK, Rahman T, Krishna P. Brassinosteroid-mediated stress tolerance in Arabidopsis shows interactions with abscisic acid, ethylene and salicylic acid pathways. BMC plant biology. 2010;10:151. doi: 10.1186/1471-2229-10-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sharma I, Kaur N, Pati PK. Brassinosteroids: a promising option in deciphering remedial strategies for abiotic stress tolerance in rice[J] Frontiers in Plant Science. 2017;8:2151. doi: 10.3389/fpls.2017.02151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng L, et al. YSL16 is a phloem-localized transporter of the copper-nicotianamine complex that is responsible for copper distribution in rice. The Plant Cell. 2012;24:3767–3782. doi: 10.1105/tpc.112.103820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.