Abstract
Seasonality in marine microorganisms has been classically observed in phytoplankton blooms, and more recently studied at the community level in prokaryotes, but rarely investigated at the scale of individual microbial taxa. Here we test if specific marine eukaryotic phytoplankton, bacterial and archaeal taxa display yearly rhythms at a coastal site impacted by irregular environmental perturbations. Our seven-year study in the Bay of Banyuls (North Western Mediterranean Sea) shows that despite some fluctuating environmental conditions, many microbial taxa displayed significant yearly rhythms. The robust rhythmicity was found in both autotrophs (picoeukaryotes and cyanobacteria) and heterotrophic prokaryotes. Sporadic meteorological events and irregular nutrient supplies did, however, trigger the appearance of less common non-rhythmic taxa. Among the environmental parameters that were measured, the main drivers of rhythmicity were temperature and day length. Seasonal autotrophs may thus be setting the pace for rhythmic heterotrophs. Similar environmental niches may be driving seasonality as well. The observed strong association between Micromonas and SAR11, which both need thiamine precursors for growth, could be a first indication that shared nutritional niches may explain some rhythmic patterns of co-occurrence.
Subject terms: Water microbiology, Microbial biooceanography, Next-generation sequencing
Introduction
Regular and predictable fluctuations of environmental parameters have a great impact on life. Seasonality sets the pace for many reoccurring life events, such as mating or migrations in animals, flowering in plants and blooms in plankton communities [1–3]. Phytoplanktonic blooms in temperate oceanic areas are a typical example of seasonal events. Several classical theories, from Sverdrup’s “Critical Depths Hypothesis” [4] to Behrenfeld’s “Dilution-Recoupling Hypothesis” [5], have attempted to explain the mechanisms triggering bloom formation. However, these theories do not aim to explain the reoccurrence and seasonality of specific microbial taxa. In macroscopic organisms, seasonality results from a fine interplay between external environmental factors and the internal circadian clock, which is an endogenous timekeeper [6]. In marine microorganisms, circadian rhythms are less well known and they have been reported only in cyanobacteria and in some eukaryotic microalgae [7–10]. However, the effect of environmental forcing on the seasonality of entire bacterial communities has been studied more extensively and reoccurring microbial communities are often observed responding to environmental changes [11–15].
Oceans are fluctuating habitats that are often marked by a strong seasonality. These regular environmental changes allow for an overall high microbial community diversity, since the environment can accommodate different species in the same space, but at different times of the year [16]. Within a year, diversity also varies locally with peaks observed in winter at high latitudes [15, 17] and community composition changes with seasons. Seasonal cycles in abiotic and/or biotic factors drive these community changes [18, 19]. To understand the seasonality of marine microbial communities, several long term sampling sites have been established within the last couple decades leading to some important findings on the seasonality of major microbial groups in the surface of the ocean [14, 20–23] and the reoccurring patterns of microbial community composition [12, 24].
Most of earlier studies focused on bacteria and there are only few reports on the seasonality of the other domains of life. For marine archaea, it has been shown that both rare and abundant members of the community were re-occurring seasonally and that different ecotypes of archaea had different seasonal patterns [20, 25]. For phytoplankton, evidence for global patterns of temporal dynamics were obtained by compiling seasonal data of chlorophyll a concentrations [26]. Molecular techniques also revealed that microbial eukaryote assemblages displayed seasonality patterns in surface marine waters [27, 28], but interestingly not always in the deeper ocean [28]. Reports on the seasonality of archaea and eukaryotes are scarce, but there are even fewer time series studies covering simultaneously the three domains of life. Steele et al. [29] identified the microorganisms that co-occurred during a 3-year study at the SPOT station (Southern California, USA). At the same site, a 21-day study of the dynamics of phytoplankton, archaea and bacteria revealed a rapid succession of microbial species during a bloom [30], which highlighted the importance of taking into account microbial interactions when studying the seasonality of marine microbial communities. However, long-term surveys of the annual dynamics and succession of photosynthetic picoeukaryotes, bacteria and archaea are currently lacking. Moreover, most time series have covered open ocean sampling sites and there are very few studies dealing with the long term monitoring of microbial communities at coastal sites. In the Mediterranean Sea, coastal environments are characterized by quite variable conditions caused by land to sea transfer of nutrients, organic matter and pollutants through seasonal river discharge during periods of strong precipitations. In such fluctuating environments, predictable patterns of reoccurring microbial communities would be less likely.
The main objective of this study was to test if the eukaryotic phytoplankton, bacteria and archaea communities demonstrated significant patterns of rhythmicity at a coastal site. We conducted a 7-year survey of the taxonomic diversity of microbial plankton community at the Banyuls Bay microbial observatory (SOLA) in the North Western Mediterranean Sea, and investigated the environmental factors that could contribute to microbial seasonality. We also used statistical tools to quantify the rhythmicity of the picoplankton and to detect patterns of co-occurrence between eukaryotic picophytoplankton (less than 3 µm), bacteria and archaea.
Materials and methods
Environmental sampling
Surface seawater (3 m depth) was collected roughly every 2 weeks from October 2007 to January 2015 at the Service d’Observation du Laboratoire Arago (SOLA) sampling station (42°31′N, 03°11′E) in the Bay of Banyuls-sur-Mer, North Western Mediterranean Sea, France. Seawater was collected in 10 l Niskin bottles and then kept in 10 l carboys until arrival to the laboratory within one hour. A subsample of 5 l was prefiltered through 3 μm pore-size polycarbonate filters (Merck-Millipore, Darmstadt, Germany), and the microbial biomass was collected on 0.22 μm pore-size GV Sterivex cartridges (Merck-Millipore) and stored at –80 °C until nucleic acid extraction.
For cytometry, unfiltered seawater samples were fixed at a final concentration of 1% glutaraldehyde, incubated for 15 min at ambient temperature in the dark, frozen in liquid nitrogen and stored at −80 °C. Cytometry analyses were performed on a Becton Dickinson FacsCalibur. Cells were excited at 488 nm and discriminated by SSC and red fluorescence (measured at 670 nm; chlorophyll content). Orange fluorescence (measured at 585 ± 21 nm), produced by phycoerythrin, was used to discriminate Synechococcus from Prochloroccocus populations [15].
The physicochemical (temperature, salinity, nitrite, nitrate, ammonium, phosphate and silicate) and biological (chlorophyll a) parameters were provided by the Service d’Observation en Milieu Littoral (SOMLIT).
DNA extraction, amplification and sequencing
The nucleic acid extraction followed protocols published earlier [25]. Briefly, the Sterivex filters were thawed on ice, followed by addition of lysis buffer (40 nM EDTA, 50 nM Tris, 0.75 M sucrose) and 25 µl of lysozyme (20 mg ml−1). The filters were then incubated on a rotary mixer at 37 °C for 45 min. The 8 µl of Proteinase K (20 mg ml−1) and 26 µl of sodium dodecyl sulfate (20% v/v) were added before incubating at 55 °C for 1 h. Total DNA was extracted and purified with the Qiagen AllPrep kit (Qiagen, Hilden, Germany) following the kit’s protocol.
Specific primer pairs were used to target different domains of life. We used primers 515 F (5’-GTGYCAGCMGCCGCGGTA) [31] and NSR951 (5’-TTGGYRAATGCTTTCGC) [32] to amplify the V4 region of 18S rRNA eukaryote gene. Primers 27 F (5’-AGRGTTYGATYMTGGCTCAG) [33] and 519 R (5’-GTNTTACNGCGGCKGCTG) [34] were used for regions V1-V3 of the bacterial 16S rRNA gene, and finally primers 519 F (5’-CAGCMGCCGCGGTAA) [35] and 1041 R (5’-GGCCATGCACCWCCTCTC) [36] to amplify regions V4-V6 of the archaeal 16S rRNA gene.
As with all primers, there can biases introduced during the amplification steps, either because some taxa can be preferentially amplified, or because of the uneven number of rRNA gene copies between taxa. A known example is the absence of haptophytes when classical 18S rRNA V4 primers are used [37]. Our eukaryote primers do amplify haptophytes, but no primers are perfect, we hope to have reduced primer biases in this study.
Sequencing was carried out with Illumina MiSeq 2 × 300 bp kits by Research and Testing Laboratory (Lubbock, Texas). We noticed that the R2 reads were of lower quality and therefore chose to conduct our analysis with R1 reads only (300 bp). Having a good quality R2 reads would have been more informative. It could have improved taxa differentiation, taxonomic assignation and overall sequence quality. However, we remain confident, considering the length of the R1, that our data are robust. All the sequences were deposited in NCBI under accession number SRP139203.
Sequence analysis
The analysis of the raw sequences was done by following the standard pipeline of the DADA2 package (https://benjjneb.github.io/dada2/index.html, version 1.6) in “R” (https://cran.r-project.org) with the following parameters: trimLeft = 21, maxN = 0, maxEE = c(5,5), truncQ = 2. Briefly, the package includes the following steps: filtering, dereplication, sample inference, chimera identification, and merging of paired-end reads [38]. DADA2 infers exact amplicon sequence variants (ASVs) from sequencing data, instead of building operational taxonomic units from sequence similarity. In total, we had 159, 160 and 158 samples for the eukaryotic phytoplankton, bacteria and archaea datasets respectively, and an average of ca. 27,000, 29,000 and 16,000 reads per sample respectively. The sequence data were normalized by dividing counts by sample size. This could influence our seasonality analyses, but considering our raw data, we found that the most appropriate transformation was to use proportional abundances [39]. The taxonomy assignments were done with the SILVA v.128 database (https://www.arb-silva.de/documentation/release-128/) and the “assignTaxonomy” function in DADA2 that implements the RDP naive Bayesian classifier method described in Wang et al. [40]. For some ASVs, in order to obtain a finer taxonomical resolution, we did an additional BLAST [41] search (blastn, 95% minimum similarity), which results can be found in the column “Blast” of the supplementary table 1. We also did a PR2 [42] assignation for the rhythmic eukaryotic phytoplankton (supplementary table 1). In this study, we aimed to focus more specifically on autotrophic picoeukaryotes in order to highlight the co-occurrence patterns and rhythmicity of phototrophs versus heterotrophs. We have therefore selected a subset of the eukaryotic datasets by retaining sequences belonging to the divisions: Chlorophyta, Dinoflagellata (without including Syndiniales, which are parasitic), Ochrophyta and Haptophyta. Here we considered all non-parasitic Dinoflagellata to be photosynthetic, but it should be noted that organisms from this group display a range of metabolisms: phototrophic, mixotrophic and heterotrophic [43].
Statistics
The Lomb Scargle periodogram (LSP) was used to determine if periodic patterns were present in microbial ASVs. The LSP, based on the Fourier transform, was originally adapted by astrophysicists to detect periodic signals in time series that were unevenly sampled due to limited access to telescopes and varying weather conditions [44, 45]. The LSP was then successfully used in biological studies to determine the periodicity of an unevenly sampled signal [46]. Owing to the robustness of the method and the fact that the sampling effort at SOLA was unevenly spaced, the LSP appeared as the best tool for our study. Computing the peak normalized power (PNmax) of the LSP was accomplished via the “Lomb” package (https://cran.r-project.org/web/packages/lomb/) in the “R” software. ASVs were considered rhythmic when they had a PNmax > 10. The threshold for PNmax is automatically calculated by the package. In summary, the LSP gives both the significance of the rhythmicity and the period of the rhythm. The LSP looks for all possible rhythmic patterns in a signal, regardless of their period. To estimate the time of the year of maximal abundance, we determined for each year and each rhythmic ASV the week of the year with the highest number of sequences. Then we selected, over the entire time series, the week that most often showed highest number of sequences.
The Shannon index, to estimate community diversity, was calculated for each sample and for eukaryotic phytoplankton, bacteria and archaea, respectively, with the function “diversity” from the “Vegan” package in “R” (https://cran.r-project.org/web/packages/vegan/).
Distances between samples were calculated for eukaryotic phytoplankton, bacteria and archaea based on community composition with a canonical correspondence analyses (CCA). Contribution of environmental factors were added as arrows, and their significance was tested with an analysis of variance (ANOVA) from the “Vegan” package in "R".
Patterns of co-occurrences between taxa were measured with the sparse partial least squares (sPLS) regression [47]. The sPLS was used to relate the abundance matrices of eukaryotic phytoplankton against bacteria and archaea with these parameters: ncomp = 3, mode = ‘regression’, in the “mixOmics” package (https://cran.r-project.org/web/packages/mixOmics/) in "R". Relationships between taxa were then visualized by a heatmap with the “CIM” function, from the same package.
Eukaryotic phytoplankton, bacteria and archaea ASV tables containing reference sequences, taxonomy and proportional abundance in the different samples are available as supplementary table 1.
Results
Environmental conditions
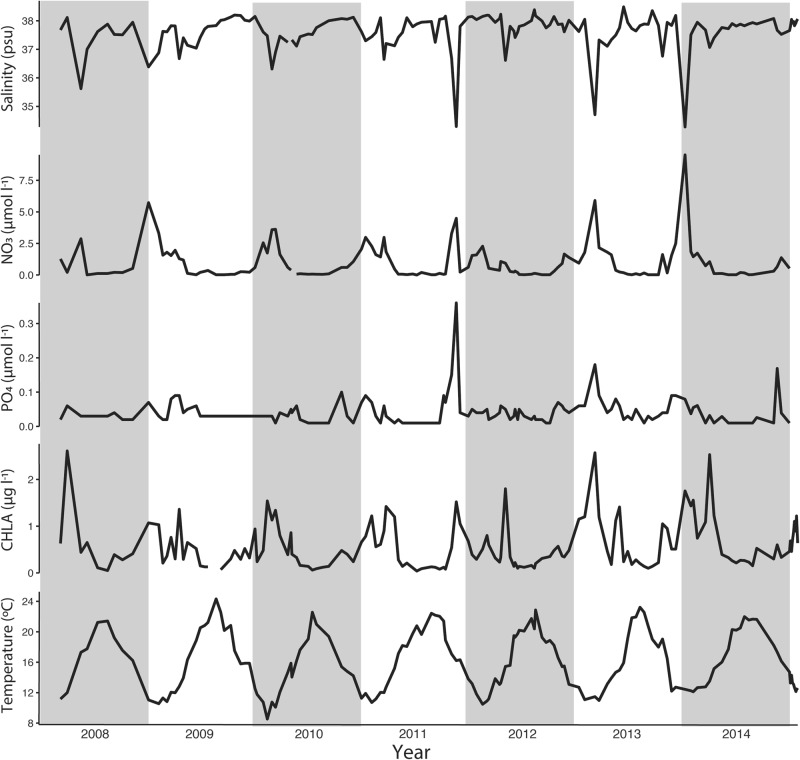
Chlorophyll a concentrations showed yearly reoccurring patterns with maxima reaching up to 2.50 µg l−1 during the winter to spring transitions, and minima at 0.04 µg l−1 during summer months (Fig. 1). Similarly, temperature levels showed yearly patterns but with much less pronounced inter-annual variations. Water temperature at SOLA were warmest during the months of August and September usually, reaching 22 °C, and coldest between February and March, with values as low as 10 °C. Salinity fluctuated from 38.49 to 34.27 psu, with an average of 37.63 psu. Nitrate levels extended from undetectable to 9.52 µmol l−1 with an average of 0.90 µmol l −1. Phosphate concentrations varied from 0.01 µmol l−1 to 0.36 µmol l −1 with an average of 0.04 µmol l–1. Nitrate, phosphate and chlorophyll a concentrations had highest values at the winter/spring transition and lowest in summer. However, salinity, nitrate and phosphate concentrations varied more than average in November 2011, March 2013 and January 2014 when decreases in salinity levels co-occurred with increases in nitrate and phosphate levels (Fig. 1).
Fig. 1.
Salinity, nitrates (NO3), phosphates (PO4), chlorophyll a (CHLA) and temperature from 2008 to 2015 at the SOLA station in the Banyuls Bay
Eukaryotic phytoplankton, bacteria and archaea community composition
Overall, the datasets yielded 6398, 6242 and 918 ASVs for the eukaryotic phytoplankton, bacterial and archaeal communities respectively. Within the eukaryotes, 1801 ASVs corresponded to autotrophs (eukaryotic phytoplankton). The Shannon index showed similar patterns of diversity for autotrophic eukaryotes, bacteria and archaea, with higher values at the beginning and the end of winter, and lower values during late summer (Supplementary Fig. 1). Bacterial communities had, on average, the highest diversity, followed by eukaryotic phytoplankton and then archaeal communities.
Canonical correspondence analyses (CCA) were performed on the eukaryotic phytoplankton, bacteria and archaea datasets to investigate the relationships between community composition and measured environmental variables (Fig. 2a–c). The communities showed a strong seasonal pattern but the environmental parameters that we measured explained only 7, 12 and 14% of the variance for the eukaryotic phytoplankton, bacteria and archaea communities respectively (Supplementary Table 2). The main explaining factors were temperature (T), day length (DL) for the three datasets, and also Nitrate (NO3) and Salinity (S) for bacteria (ANOVA, p = 0.001). Temperature and day length explained close to half of the total variance for eukaryotic phytoplankton, bacteria and archaea (Supplementary Table 2). The eukaryotic phytoplankton communities grouped together according to the month of sampling. The communities showed more divergence on the CCA plots during the months of April and May, whereas they were grouped during the other months (Fig. 2a). Bacterial communities showed a similar seasonal structure with higher separation between samples from March to June (Fig. 2b). Finally, the archaea had a comparable structure of monthly successions, but with highest variability between samples from July to October (Fig. 2c).
Fig. 2.
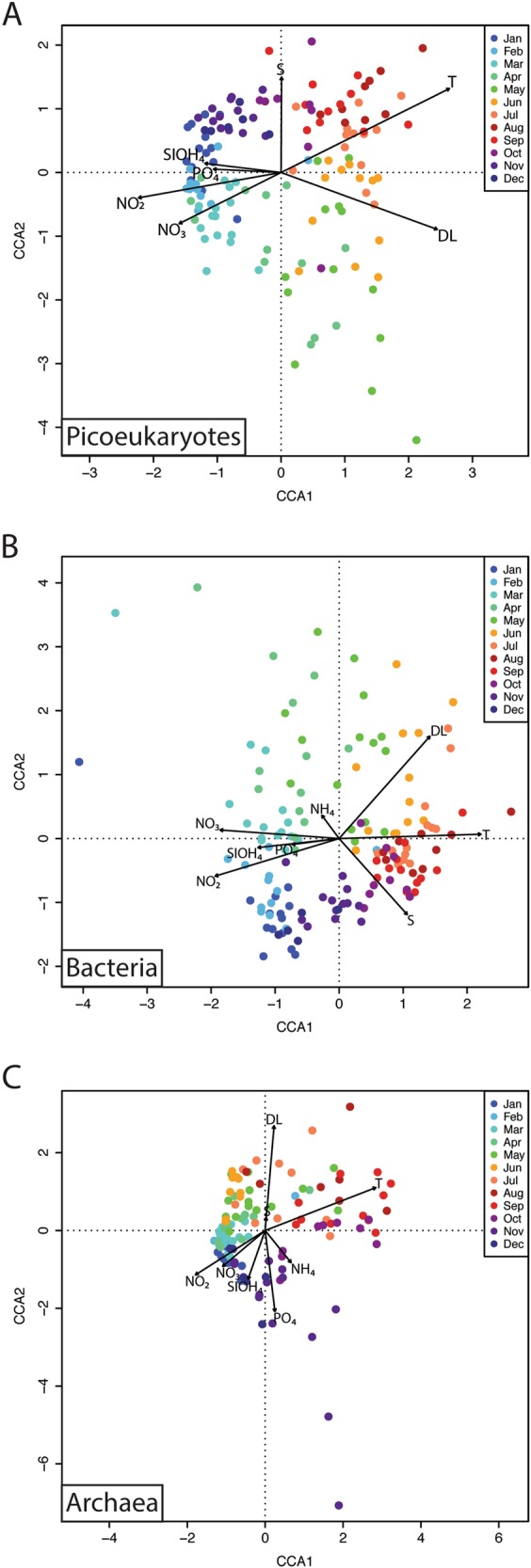
Canonical correspondence analyses (CCA) of the eukaryotic phytoplankton (a), bacteria (b), and archaea (c) community composition in relation to environmental factors. The communities are color coded according to the month of sampling. The arrows represent the different environmental factors (T: temperature, DL: day length, NH4: ammonium, NO3: nitrates, NO2: nitrites, PO4: phosphates, SIOH4: silicates, S: salinity)
From 2007 to 2015, at the division level, the photosynthetic picoeukaryote community was composed of Dinoflagellata, Chlorophyta, Ochrophyta and Haptophyta (44.01% of the sequences, 29.45, 13.23, 13.31% respectively) (Supplementary Fig. 2). Dinoflagellata were dominated by Dinophyceae (99.19% of the sequences) and Chlorophyta by Mamiellophyceae (94.36%). Within Mamiellophyceae, three main genera were found, Micromonas, Bathycoccus and Ostreococcus (64.59, 31.89 and 3.49%, respectively) (Supplementary Fig. 2). Bacteria (Supplementary Fig. 3) were dominated by the phyla Proteobacteria (76.74%) and Cyanobacteria (12.12%). The main contributors of the Proteobacteria were Alphaproteobacteria (89.79%, mainly SAR11) and Gammaproteobacteria (9.93%). Synechococcus ASVs represented 95.8% of Cyanobacteria sequences. Finally, archaea were divided between the Thaumarchaeota (64.36%) and the Euryarchaeota (35.07%) (Supplementary Fig. 4).
Rhythmicity of the environmental and biological compartments
In order to test if environmental factors and microbial taxa had significant rhythmic patterns during the 7-year time series, the Lomb Scargle periodogram (LSP) algorithm was applied to the eukaryotic phytoplankton, bacteria, archaea and environmental datasets. The most rhythmic environmental parameters were day length and temperature with a PNmax score of 60.00 and 55.67 respectively. Other rhythmic factors were NO2, NO3, chlorophyll a and NH4 but with lower PNmax scores of 37.17, 24.27, 21.37 and 13.44, respectively. SIOH4, PO4 and salinity had PNmax scores that did not cross the statistical threshold to be considered rhythmic (PNmax scores of 9.95, 7.03 and 5.45, respectively). A total of 15 picoeukaryote, 89 bacteria and 31 archaea ASVs had significant patterns of rhythmicity. The rhythmic ASVs and environmental factors all had a period of one year. Theses rhythmic microbial ASVs were selected for further detailed analysis.
Timing of yearly reoccurrences and relative abundance of rhythmic ASVs
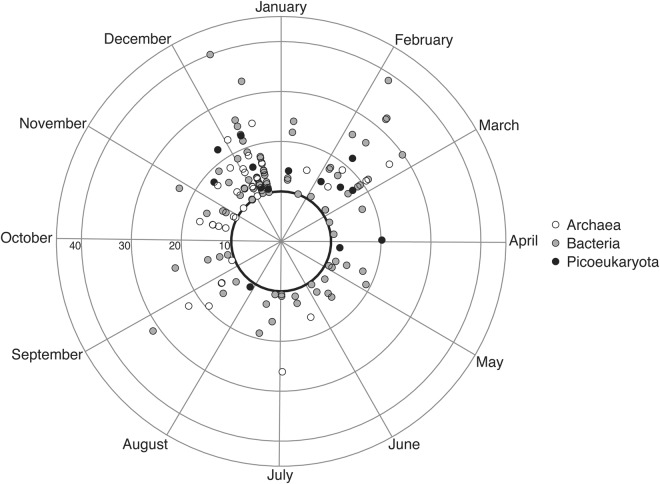
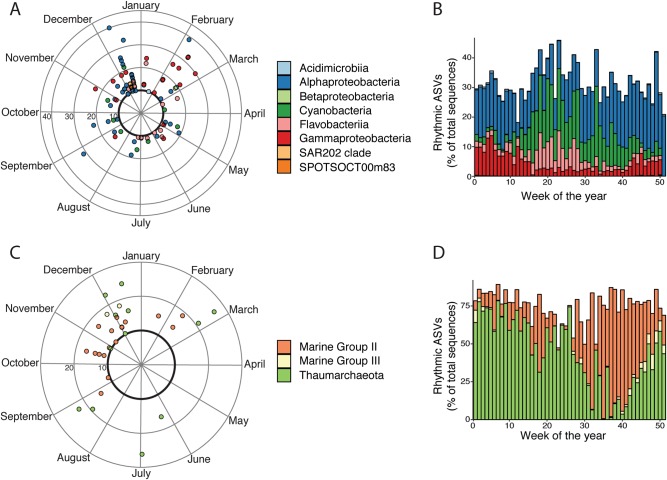
Among the 135 ASVs (Fig. 3, Supplementary Table 3) that showed significant reoccurrences throughout the year, different domains displayed different patterns. Bacterial rhythmic ASVs showed phases of maximal abundance that spread throughout the year, whereas eukaryotic phytoplankton and archaeal rhythmic ASVs phases were confined to certain moments of the year. Eukaryotic phytoplankton rhythmic ASVs had maximal abundance from November to April, while archaeal rhythmic ASVs had maximal abundance from September to March.
Fig. 3.
Polar plot showing when during the year the rhythmic ASVs reoccur and the strength of reoccurrence (PNmax, calculated via the LSP). The black circle shows the statistical threshold for significant rhythmicity (PNmax = 10). The ASVs are color coded according to which domain of life they belong to
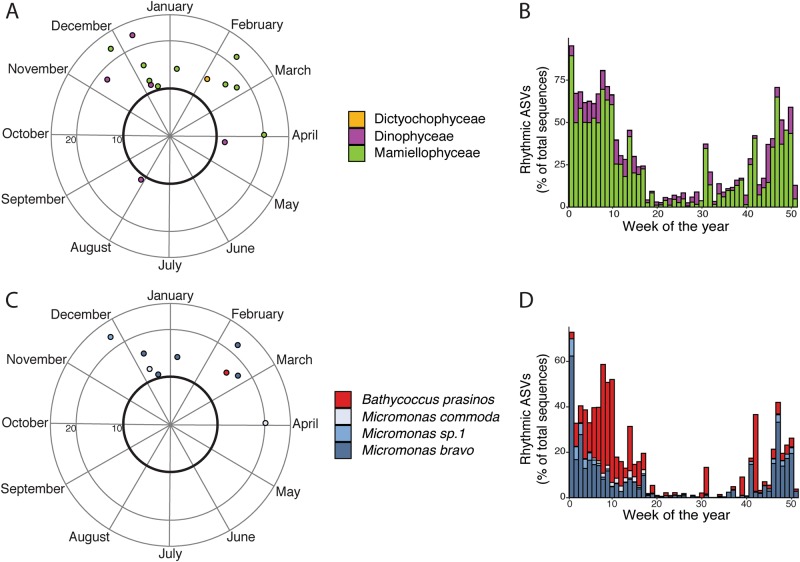
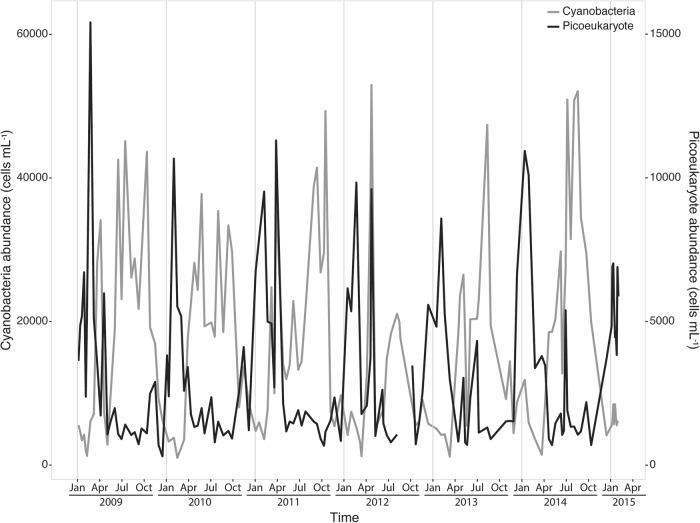
On average 30.5% of the eukaryotic phytoplankton sequences were rhythmic but the proportion varied throughout the year. Rhythmic ASVs represented up to 96% of the sequences in January and as low as 2.5% of the sequences in July (Fig. 4b). All classes followed a similar pattern with high levels (50 to 60% of total sequences) from mid-Autumn to mid-Spring (October to April) and lower levels (less than 15% of total sequences) during the rest of the year. The lowest number of rhythmic sequences were seen during the summer months (Fig. 4b). Flow cytometry showed that picoeukaryotes had low abundances during the summer months and high abundances during winter months (Fig. 5).
Fig. 4.
Polar plots representing the rhythmic eukaryotic phytoplankton classes (a) and the rhythmic Mamiellophyceae ASVs (c). The bar plots show the proportion of sequences belonging to rhythmic ASVs averaged per week of the year for eukaryotic phytoplankton classes (b) and Mamiellophyceae ASVs (d)
Fig. 5.
Photosynthetic picoeukaryote and cyanobacteria abundance determined by flow cytometry from 2009 to 2015 at the SOLA station in the Banyuls Bay
At the eukaryotic class level, (Fig. 4a), the Mamiellophyceae rhythmic ASVs were found mostly from the end of November to the end of March. The Dinophyceae rhythmic ASVs had peaks of abundances year-round. The Dictyochophyceae rhythmic ASV was only abundant at the beginning of February. Within Mamiellophyceae, the Bathycoccus prasinos ASV peaked around the middle of February (7th week of the year) (Fig. 4c) with a distribution going from January to April (Fig. 4d). Micromonas commoda was recurrent from December to the end of March (Fig. 4c) and distributed from February to April (Fig. 4d). Micromonas sp.1 ASV was more present at the end of November (Fig. 4c) with a distribution from November to February (Fig. 4d). Micromonas bravo, however, had ASVs peaks from December to February (Fig. 4c) and was present from October to April (Fig. 4d).
Rhythmic bacterial ASVs were present throughout the year (Fig. 6a), and represented in average 31.3% of the sequences, with variations from 18 to 45.7% of the sequences (Fig. 6b). The contributors to the rhythmic ASVs were Acidimicrobiia, Alphaproteobacteria, Betaproteobacteria, Cyanobacteria, Flavobacteria, Gammaproteobacteria, SAR202 and candidate Proteobacteria SPOTSOCT00m83 (Fig. 6a, b).
Fig. 6.
Polar plots representing the rhythmic bacteria ASVs (a) and the rhythmic archaea ASVs (c). The bar plots show the proportion of sequences belonging to rhythmic ASVs averaged per week of the year, for bacteria (b) and archaea (d)
The different rhythmic bacterial classes showed different types of patterns. The Acidimicrobiia, Gammaproteobacteria, SAR202 and candidate Proteobacteria SPOTSOCT00m83 showed high numbers from October to April and were almost absent during the summer months (Fig. 6b). They displayed similar reoccurrence patterns as well, mainly from December to February (Fig. 6a). Cyanobacteria rhythmic ASVs demonstrated an opposite pattern, with high levels during the warm summer and autumn months (March to October) and low levels the rest of the year (Fig. 6b). Cytometry data showed the same seasonal pattern in terms of cell abundance (Fig. 5). Flavobacteria had similar patterns as cyanobacteria. However, their reoccurrence patterns were different. Rhythmic Cyanobacteria ASVs reoccurred from the end of March to October, whereas Flavobacteria ASVs had two periods of maximal reoccurrence, one from March to July and another during December (Fig. 6a). Betaproteobacteria ASVs were more abundant from January to May and were absent the rest of the year (Fig. 6b), and were only recurrent at the end of February (Fig. 6a). Alphaproteobacteria rhythmic ASVs displayed similar sequence numbers throughout the year, accounting for half of the rhythmic ASVs sequence numbers (15%) (Fig. 6b). Similarly, the Alphaproteobacteria ASVs reoccurrences covered the whole year except for March (Fig. 6a).
Amongst the rhythmic Alphaproteobacteria, a majority of ASVs belonged to SAR11. All sub-groups of SAR11 (SAR11Ia, SAR11Ib, SAR11Ic, SAR11IIa, SAR11IIIa and SAR11IV) had high numbers of rhythmic ASVs from September to the end of February (Supplementary Fig. 5A). These groups also showed higher number of sequences during winter months (Supplementary Fig. 5B), except for SAR11IIIa which had higher sequence abundance from June to November (Supplementary Fig. 5B).
Finally, archaeal rhythmic ASVs had maximum occurrences from the end of August to March, both for Euryarchaeota and Thaumarchaeota (Fig. 6c). Rhythmic ASVs dominated the dataset as they represented an average of 74.6% of total sequence numbers, ranging from 47.3 to 89.2% (Fig. 6d). Within the Euryarchaeota phylum, rhythmic ASVs of Marine group II (MGII) and Marine group III (MGIII) were found. Rhythmic MGII ASVs showed reoccurrence patterns from September to March (Fig. 6c) and highest relative sequence numbers from July to October (Fig. 6d). MGIII rhythmic ASVs had a more restrained occurrence, from end of November to beginning of December (Fig. 6c) and were less present in relative abundance (Fig. 6d). The Thaumarchaeota rhythmic ASVs displayed high levels of presence throughout the year with the exception of the months of September. The months preceding and succeeding September showed a steady decrease and increase of relative sequence number, respectively (Fig. 6d). Thaumarchaeota had high occurrences all year, except from March to May (Fig. 6c).
We also observed a large number of ASVs that were not rhythmic and thus had peaks of abundance at different moments from year to year. Non-rhythmic ASVs had different patterns of seasonal dynamics. Some ASVs, like the Gymnodiniphycidae ASV00020, were absent from most of the samples but shows sudden and irregular peaks of abundance (Supplementary Fig. 6). Other, like the Gymnodiniphycidae ASV00036, were more frequent and had irregular peaks of sequence abundance that co-occurred with irregular environmental events such as freshening sea surface waters and increased nitrate concentrations (Supplementary Fig. 6).
Co-occurrence at the ASV level
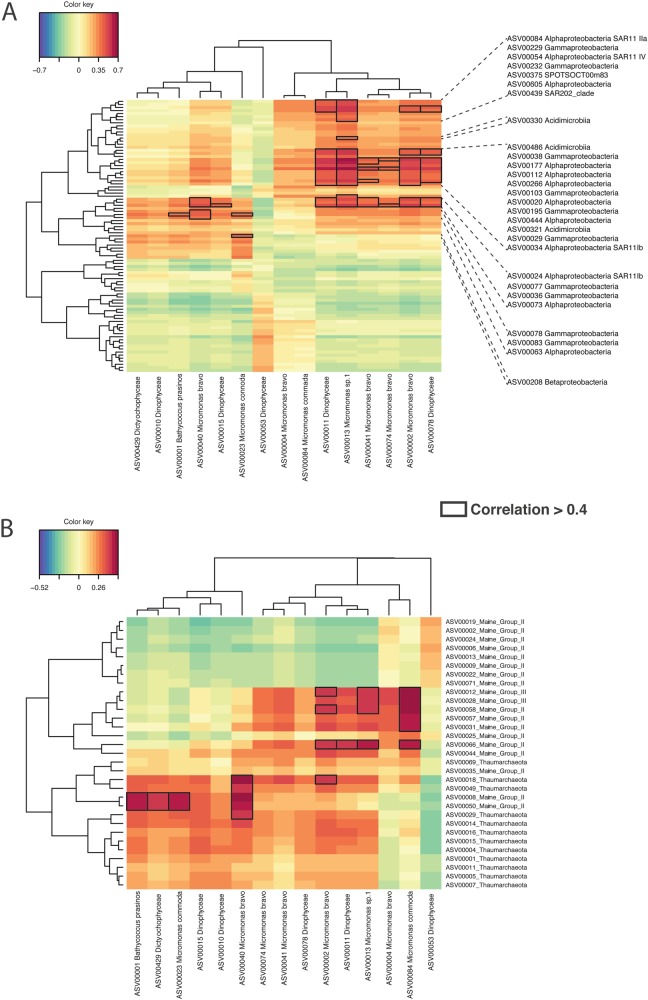
To determine co-occurrences, heatmaps were created with the rhythmic ASVs after calculating Sparse Partial Least Squares (sPLS) regressions for one dataset against the other (bacteria vs. picoeukaryote, bacteria vs. archaea and archaea vs. picoeukaryote). For bacteria vs. picoeukaryotes (Fig. 7a), the highest correlation scores (>0.6) were between Micromonas sp.1 (ASV 00013) and a SAR11 sequence (ASV 00054) as well as 3 Rhodospirillaceae (ASV 00020, ASV 00112 and ASV 00266). A Dinophyceae (ASV 00011) also had a high correlation (0.55) with the same Alphaproteobacteria ASVs. Other high correlations were found between Bathycoccus prasinos and Alpha- and Gammaproteobacteria. Micromonas bravo (ASV 00002) also had high correlations with an Alphaproetobacteria (ASV 00112). A Dinophyceae ASV (ASV00053) displayed a specific high correlation with a group of bacteria that were not correlated to other eukaryotic phytoplankton. This is probably due to the fact that Dinophyceae is the only rhythmic picoeukaryote to peak in summer (Fig. 4a).
Fig. 7.
Heatmap based on a sPLS regression showing co-occurrences between rhythmic eukaryotic phytoplankton ASVs and bacteria ASVs (a) and between rhythmic eukaryotic phytoplankton ASVs and archaea ASVs (b). Correlations > 0.4 are highlighted
The archaea vs. picoeukaryote heatmap revealed high correlation ( > 0.5) between Bathycoccus prasinos and MGII ASVs (ASV 00050 and ASV 00008). Micromonas bravo (ASV 00040) showed a similar trend. On the other hand, Micromonas commada (ASV 00084) had high correlations ( > 0.5) with MGIII ASVs (ASV 00012 and ASV 00028). As with the bacteria dataset, the Dinophyceae, ASV 00053, displayed high correlations when all other eukaryotic phytoplankton ASVs had low correlations (Fig. 7b).
In the bacteria vs. archaea heatmap the main co-occurrences were observed between a Thaumarchaeota ASV and a Gammaproteobacteria ASV, as well as between a MGII and Alphaproteobacteria ASV (Supplementary Fig. 7).
Discussion
Our 7-year survey in the NW Mediterranean Sea showed that within all domains of life some taxa showed significant patterns of rhythmicity with a one year period. The number of rhythmic taxa differed between domains. Phototrophic picoeukaryotes had 1% of rhythmic ASVs, bacteria 3.1 % and archaea 3.4%, but these ASVs represented a large proportion of the total number of sequences (31.3, 31.6 and 75.5%, respectively). The large proportion of rhythmic sequences supports the idea of microbial communities that come back year after year at the same season. The concept of re-occurring communities has been demonstrated in several long term studies [14, 20, 24] but coastal observations are quite scarce [48]. The Banyuls Bay is a coastal site with seasonal characteristics specific to the NW Mediterranean. It has a marked seasonality but interestingly it is also characterized by strong and ephemeral inputs of nutrients brought from sediment mixing during episodic winter storms and during flash floods from incoming rivers [49]. Nutrients are known to strongly structure communities by promoting planktonic blooms and by stimulating the growth of certain microbes [12, 50]. However, despite irregular nutrient supply from year to year, as illustrated by salinity and phosphate variations during winter and spring (Fig. 1), we could still observe a large number of rhythmic eukaryotic phytoplankton, bacteria and archaea sequences. The CCA analysis (Fig. 2) confirmed that day length and temperature were major factors structuring the communities and we can suppose that they directly or indirectly control the dynamics of the rhythmic taxa.
Day length has been shown to be a strong driver of community structure in temperate and polar marine environments such as the English Channel [14], or a high-Arctic fjord [50]. Temperature is another strong driver as it can affect gene expression and subsequently the structure and the function of the microbial communities [51]. The availability of nutrients has also been shown to be an important factor in community composition as demonstrated in the BATS time series in the Atlantic Ocean [3]. Our data from the Banyuls Bay shows that in a coastal ecosystem, environmental parameters like temperature and day length have such a structuring effect that sporadic meteorological events do not appear to impact the overall microbial rhythms of re-occurring dominant groups of eukaryotic phytoplankton, bacteria and archaea. However, even though rhythmic ASVs could be predominately influenced by day length and temperature, we observed non-rhythmic ASVs, which were influenced by irregular environmental factors. For example, the dynamics of the Gymnodiniphycidae ASV00036 was associated to the irregular peaks of nitrate concentration (Supplementary Fig. 6).
The importance of day length in driving the rhythm of individual microorganisms brings the question whether seasonality is driven by circadian clocks in marine microbes. The presence of a functional circadian clock governing day/night biological processes bas been demonstrated in the mamiellophyceae Ostreococcus [8, 52], however, the existence of a photoperiod dependent regulation of blooms remains to be established formally in this order. In diatoms from northern Norwegian coastal waters, it has been reported that the timing of the spring bloom varies little from year to year whether water stratification had occurred or not [53]. The authors hypothesized that the photoperiod was the major factor that relieved diatoms resting spores from dormancy, leading to seasonal blooms. However, the internal mechanisms triggering these rhythms remain unknown since the presence of circadian clocks remain to be shown in diatoms. Amongst the prokaryotes, cyanobacteria are the only known group to have a genuine circadian clock [7] and the occurrence of circadian clock remain to be established in heterotrophic bacteria and archaea. The rhythmicity of some heterotrophic microorganisms could thus be governed directly by day length or indirectly through interactions with the rhythmic autotrophs. Interestingly, altogether, eukaryotic and prokaryotic autotrophs were present during the entire year, but they showed clear differences in their seasonal dynamics. Picoeukaryotes had highest abundance from autumn to spring, and cyanobacteria during the summer.
We observed a large number of rhythmic ASV sequences, which were mainly seen within abundant members of the communities. The eukaryotic phytoplankton were represented primarily by Mamiellophyceae and Dinophyceae, which have important ecological roles as primary producers and as links in the predator/prey food chain [54]. Among prokaryotic rhythmic ASVs, there were many representative of the SAR11, known for being the most common group of marine bacteria [23]. Seasonality has been observed for SAR11 and Flavobacteria groups earlier [15, 55]. Rhythmic archaea were found in MGII, MGIII and Thaumarchaeota, which have already been shown to have reoccurring yearly patterns [20, 25]. The dominance of abundant groups within rhythmic ASVs raises the question as whether our analysis underestimated the rhythmicity of less abundant ASVs. It should be noted, however, that some rare ASVs, with occurrences of 0.034, 0.009 and 0.115% respectively for the eukaryotic phytoplankton, bacteria and archaea dataset, were also found to be rhythmic. In agreement with our observations, Alonso-Sáez et al. [55] recently showed that, also in a coastal system, both rare and abundant bacterial species had patterns of rhythmicity in the Atlantic Ocean [55] and that many species that remained rare all year long also showed significant patterns of rhythmicity. Rhythmicity of marine microbes, at the ASV level, remains to be verified in other sites as there have been only few studies conducted at this level of resolution. While the re-occurrence of entire communities is now well documented [13, 14, 24], the long-term monitoring of individual taxa is not common [12, 21] and the use of statistics to test patterns of ASV rhythmicity is even less frequent.
There have been very few studies looking at the temporal dynamics across the three domains of life in marine microbial ecosystems. One of the first long term study covering the three domains did not focus on the rhythm of the individual taxa but rather looked at co-occurrence networks [29]. They showed that correlations between microbes were more prevalent than correlations between microbes and environmental factors. This is probably due to the stability of the deep chlorophyll maximum at their study site [29]. More recently, Needham and Fuhrman looked at the succession of phytoplankton, archaea and bacteria, but only during 6 months [30]. Another study in the same ecosystem, relying on automated sampling, showed daily and highly dynamic population variations in the three domains of life, and extensively described the biological interactions that took place during the sampling period [56]. A study looking at bacterioplankton diversity and phytoplankton microscopy counts, has shown that despite inter-annual variations in phytoplankton blooms, bacterioplankton microdiversity patterns seem stable in both bloom and non-bloom conditions [57]. The present dataset showed high co-occurrence between some eukaryotic phytoplankton and prokaryotes ASVs. The most significant correlations were found between Mamiellophyceae and the alphaproteobacteria SAR11. This co-occurrence could be explained by the fact that Micromonas and SAR11 might interact by exchanging compounds such as vitamins, growth factors and organic carbon [58, 59]. However, SAR11 was recently shown to be auxotrophic to the thiamine precursor 4-amino-5-hydroxymethyl-2-methyl pyrimidine [60], thus resulting in similar needs as Micromonas for thiamin precursors [61]. The co-occurrence of these two microbes therefore may be explained by their requirement for similar nutritional niches rather than by a relationship of interdependency depending on environmental factors.
In conclusion, through the analysis of our time series we demonstrated that a large proportion of members of eukaryotic phytoplankton, bacteria and archaea datasets, showed rhythmicity with a one year period of reoccurrence over then entire time series. The main drivers of seasonality were photoperiod and temperature. Sporadic meteorological events and irregular nutrient supply characteristic of our coastal site did not affect significantly the seasonality, indicating that the yearly rhythms were robust. Rhythmicity was found in both autotrophs (picoeukaryotes and cyanobacteria) and heterotrophic prokaryotes. Seasonal autotrophs, which respond to light, may be setting the pace for rhythmic heterotrophs but similar environmental niches may be driving seasonality as well.
Electronic supplementary material
Acknowledgements
We are grateful to the captain and the crew of the RV ‘Nereis II’ for their help in acquiring the samples. We thank the “Service d’Observation”, particularly Eric Maria and Paul Labatut, for their help in obtaining and processing of the samples. MT was supported by a PhD fellowship from the Sorbonne Université and the Région Bretagne. We would like to thank the ABIMS platform in Roscoff for access to bioinformatics resources. This work was supported by the French Agence Nationale de la Recherche through the projects Photo-Phyto (ANR-14-CE02-0018) to FYB, and EUREKA (ANR-14-CE02-0004-01) to PEG.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
François-Yves Bouget, Phone: (+33) 468887350, Email: francois-yves.bouget@obs-banyuls.fr.
Pierre E. Galand, Phone: (+33) 430192451, Email: pierre.galand@obs-banyuls.fr
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0281-z) contains supplementary material, which is available to authorized users.
References
- 1.Antle MC, Silver R. Circadian insights into motivated behavior. In: Simpson EH, Balsam PD, editors. Behavioral neuroscience of motivation. Cham: Springer International Publishing; 2015. pp. 137–69. [Google Scholar]
- 2.MacDonald CC, McMahon KW. The flowers that bloom in the spring: RNA processing and seasonal flowering. Cell. 2003;113:671–72. doi: 10.1016/S0092-8674(03)00426-4. [DOI] [PubMed] [Google Scholar]
- 3.Treusch AH, Demir-Hilton E, Vergin KL, Worden AZ, Carlson CA, Donatz MG, et al. Phytoplankton distribution patterns in the northwestern Sargasso Sea revealed by small subunit rRNA genes from plastids. ISME J. 2012;6:481–92. doi: 10.1038/ismej.2011.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sverdrup HU. On vernal blooming of phytoplankton. J Cons Exp Mer. 1953;18:287–95. doi: 10.1093/icesjms/18.3.287. [DOI] [Google Scholar]
- 5.Behrenfeld MJ. Abandoning Sverdrup’s critical depth hypothesis on phytoplankton blooms. Ecology. 2010;91:977–989. doi: 10.1890/09-1207.1. [DOI] [PubMed] [Google Scholar]
- 6.Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, et al. GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 1999;18:4679–88. doi: 10.1093/emboj/18.17.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen SE, Golden SS. Circadian rhythms in Cyanobacteria. Microbiol Mol Biol Rev. 2015;79:373–85. doi: 10.1128/MMBR.00036-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corellou F, Schwartz C, Motta JP, Djouani-Tahri EB, Sanchez F, Bouget FY. Clocks in the green lineage: comparative functional analysis of the circadian architecture of the picoeukaryote ostreococcus. Plant Cell. 2009;21:3436–49. doi: 10.1105/tpc.109.068825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edmunds L, Adams K. Clocked cell cycle clocks. Science. 1981;211:1002–13. doi: 10.1126/science.7008196. [DOI] [PubMed] [Google Scholar]
- 10.Jacquet S, Partensky F, Lennon JF, Vaulot D. Diel patterns of growth and division in marine picoplankton in culture. J Phycol. 2001;37:357–69. doi: 10.1046/j.1529-8817.2001.037003357.x. [DOI] [Google Scholar]
- 11.Alonso-Sáez L, Balagué V, Sá EL, Sánchez O, González JM, Pinhassi J, et al. Seasonality in bacterial diversity in north-west Mediterranean coastal waters: assessment through clone libraries, fingerprinting and FISH: Seasonality in marine bacterial diversity. FEMS Microbiol Ecol. 2007;60:98–112. doi: 10.1111/j.1574-6941.2006.00276.x. [DOI] [PubMed] [Google Scholar]
- 12.Chow CET, Sachdeva R, Cram JA, Steele JA, Needham DM, Patel A, et al. Temporal variability and coherence of euphotic zone bacterial communities over a decade in the Southern California Bight. ISME J. 2013;7:2259–73. doi: 10.1038/ismej.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cram JA, Chow CET, Sachdeva R, Needham DM, Parada AE, Steele JA, et al. Seasonal and interannual variability of the marine bacterioplankton community throughout the water column over ten years. ISME J. 2015;9:563–80. doi: 10.1038/ismej.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert JA, Steele JA, Caporaso JG, Steinbrück L, Reeder J, Temperton B, et al. Defining seasonal marine microbial community dynamics. ISME J. 2012;6:298–308. doi: 10.1038/ismej.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salter I, Galand PE, Fagervold SK, Lebaron P, Obernosterer I, Oliver MJ, et al. Seasonal dynamics of active SAR11 ecotypes in the oligotrophic Northwest Mediterranean Sea. ISME J. 2015;9:347–60. doi: 10.1038/ismej.2014.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tonkin JD, Bogan MT, Bonada N, Rios-Touma B, Lytle DA. Seasonality and predictability shape temporal species diversity. Ecology. 2017;98:1201–16. doi: 10.1002/ecy.1761. [DOI] [PubMed] [Google Scholar]
- 17.Ladau J, Sharpton TJ, Finucane MM, Jospin G, Kembel SW, O’Dwyer J, et al. Global marine bacterial diversity peaks at high latitudes in winter. Isme J. 2013;7:1669. doi: 10.1038/ismej.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bunse C, Pinhassi J. Marine bacterioplankton seasonal succession dynamics. Trends Microbiol. 2017;25:494–505. doi: 10.1016/j.tim.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 19.Fuhrman JA, Cram JA, Needham DM. Marine microbial community dynamics and their ecological interpretation. Nat Rev Microbiol. 2015;13:133–146. doi: 10.1038/nrmicro3417. [DOI] [PubMed] [Google Scholar]
- 20.Galand PE, Gutiérrez-Provecho C, Massana R, Gasol JM, Casamayor EO. Inter-annual recurrence of archaeal assemblages in the coastal NW Mediterranean Sea (Blanes Bay Microbial Observatory) Limnol Oceanogr. 2010;55:2117–25. doi: 10.4319/lo.2010.55.5.2117. [DOI] [Google Scholar]
- 21.Lindh MV, Sjöstedt J, Andersson AF, Baltar F, Hugerth LW, Lundin D, et al. Disentangling seasonal bacterioplankton population dynamics by high-frequency sampling: high-resolution temporal dynamics of marine bacteria. Environ Microbiol. 2015;17:2459–76. doi: 10.1111/1462-2920.12720. [DOI] [PubMed] [Google Scholar]
- 22.Treusch AH, Vergin KL, Finlay LA, Donatz MG, Burton RM, Carlson CA, et al. Seasonality and vertical structure of microbial communities in an ocean gyre. ISME J. 2009;3:1148–63. doi: 10.1038/ismej.2009.60. [DOI] [PubMed] [Google Scholar]
- 23.Vergin KL, Beszteri B, Monier A, Cameron Thrash J, Temperton B, Treusch AH, et al. High-resolution SAR11 ecotype dynamics at the Bermuda Atlantic Time-series study site by phylogenetic placement of pyrosequences. ISME J. 2013;7:1322–32. doi: 10.1038/ismej.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuhrman JA, Hewson I, Schwalbach MS, Steele JA, Brown MV, Naeem S. Annually reoccurring bacterial communities are predictable from ocean conditions. Proc Natl Acad Sci USA. 2006;103:13104–09. doi: 10.1073/pnas.0602399103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hugoni M, Taib N, Debroas D, Domaizon I, Jouan Dufournel I, Bronner G, et al. Structure of the rare archaeal biosphere and seasonal dynamics of active ecotypes in surface coastal waters. Proc Natl Acad Sci USA. 2013;110:6004–9. doi: 10.1073/pnas.1216863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winder M, Cloern JE. The annual cycles of phytoplankton biomass. Philos Trans R Soc B: Biol Sci. 2010;365:3215–26. doi: 10.1098/rstb.2010.0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brannock PM, Ortmann AC, Moss AG, Halanych KM. Metabarcoding reveals environmental factors influencing spatio‐temporal variation in pelagic micro‐eukaryotes. Mol Ecol. 2016;25:3593–604. doi: 10.1111/mec.13709. [DOI] [PubMed] [Google Scholar]
- 28.Kim DY, Countway PD, Jones AC, Schnetzer A, Yamashita W, Tung C, et al. Monthly to interannual variability of microbial eukaryote assemblages at four depths in the eastern North Pacific. ISME J. 2014;8:515–30. doi: 10.1038/ismej.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steele JA, Countway PD, Xia L, Vigil PD, Beman JM, Kim DY, et al. Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J. 2011;5:1414–25. doi: 10.1038/ismej.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Needham DM, Fuhrman JA. Pronounced daily succession of phytoplankton, archaea and bacteria following a spring bloom. Nat Microbiol. 2016;1:16005. doi: 10.1038/nmicrobiol.2016.5. [DOI] [PubMed] [Google Scholar]
- 31.Parada AE, Needham DM, Fuhrman JA. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol. 2016;18:1403–14. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 32.Mangot JF, Domaizon I, Taib N, Marouni N, Duffaud E, Bronner G, et al. Short-term dynamics of diversity patterns: evidence of continual reassembly within lacustrine small eukaryotes: short-term dynamics of small eukaryotes. Environ Microbiol. 2013;15:1745–58. doi: 10.1111/1462-2920.12065. [DOI] [PubMed] [Google Scholar]
- 33.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, United Kingdom: John Wiley and Sons; 1991. pp. 115–175.
- 34.Turner S, Pryer KM, Miao VPW, Palmer JD. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J Eukaryot Microbiol. 1999;46:327–38. doi: 10.1111/j.1550-7408.1999.tb04612.x. [DOI] [PubMed] [Google Scholar]
- 35.Ovreås L, Forney L, Daae FL, Torsvik V. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol. 1997;63:3367–73. doi: 10.1128/aem.63.9.3367-3373.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolganova TV, Kuznetsov BB, Tourova TP. Designing and testing oligonucleotide primers for amplification and sequencing of archaeal 16S rRNA. Genes. 2002;71:4. [PubMed] [Google Scholar]
- 37.Liu H, Probert I, Uitz J, Claustre H, Aris-Brosou S, Frada M, et al. Extreme diversity in noncalcifying haptophytes explains a major pigment paradox in open oceans. Proc Natl Acad Sci USA. 2009;106:12803–808. doi: 10.1073/pnas.0905841106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiss S, Xu ZZ, Peddada S, Amir A, Bittinger K, Gonzalez A, et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 2017;5:27. [DOI] [PMC free article] [PubMed]
- 40.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–67. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 42.Guillou L, Bachar D, Audic S, Bass D, Berney C, Bittner L, et al. The protist ribosomal reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 2013;41:D597–604. doi: 10.1093/nar/gks1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanchez-Puerta MV, Lippmeier JC, Apt KE, Delwiche CF. Plastid genes in a non-photosynthetic dinoflagellate. Protist. 2007;158:105–17. doi: 10.1016/j.protis.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 44.Lomb NR. Least-squares frequency analysis of unequally spaced data. Astrophys Space Sci. 1976;39:447–62. doi: 10.1007/BF00648343. [DOI] [Google Scholar]
- 45.Scargle JD. Studies in astronomical time series analysis. II-Statistical aspects of spectral analysis of unevenly spaced data. Astrophys J. 1982;263:835–53. doi: 10.1086/160554. [DOI] [Google Scholar]
- 46.Ruf T. The lomb-scargle periodogram in biological rhythm research: analysis of incomplete and unequally spaced time-series. Biol Rhythm Res. 1999;30:178–201. doi: 10.1076/brhm.30.2.178.1422. [DOI] [Google Scholar]
- 47.Lê Cao K-A, Rossow D, Robert-Granié C, Besse P. Sparse PLS: Variable selection when integrating omics data. 2008. [DOI] [PubMed]
- 48.Nelson JD, Boehme SE, Reimers CE, Sherrell RM, Kerkhof LJ. Temporal patterns of microbial community structure in the mid-atlantic bight: spatio-temporal variability of coastal marine bacteria. FEMS Microbiol Ecol. 2008;65:484–93. doi: 10.1111/j.1574-6941.2008.00553.x. [DOI] [PubMed] [Google Scholar]
- 49.Charles F, Lantoine F, Brugel S, Chrétiennot-Dinet MJ, Quiroga I, Rivière B. Seasonal survey of the phytoplankton biomass, composition and production in a littoral NW mediterranean site, with special emphasis on the picoplanktonic contribution. Estuar, Coast Shelf Sci. 2005;65:199–212. doi: 10.1016/j.ecss.2005.06.006. [DOI] [Google Scholar]
- 50.Marquardt M, Vader A, Stübner EI, Reigstad M, Gabrielsen TM. Strong seasonality of marine microbial eukaryotes in a high-arctic fjord (Isfjorden, in West Spitsbergen, Norway) Appl Environ Microbiol. 2016;82:1868–80. doi: 10.1128/AEM.03208-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ward CS, Yung CM, Davis KM, Blinebry SK, Williams TC, Johnson ZI, et al. Annual community patterns are driven by seasonal switching between closely related marine bacteria. ISME J. 2017;11:1412–22. doi: 10.1038/ismej.2017.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Monnier A, Liverani S, Bouvet R, Jesson B, Smith JQ, Mosser J, et al. Orchestrated transcription of biological processes in the marine picoeukaryote ostreococcus exposed to light/dark cycles. BMC Genom. 2010;11:192. doi: 10.1186/1471-2164-11-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eilertsen HC, Sandberg S, Tøllefsen H. Photoperiodic control of diatom spore growth: a theory to explain the onset of phytoplankton blooms. Mar Ecol Progress Ser Oldendorf. 1995;116:303–7. doi: 10.3354/meps116303. [DOI] [Google Scholar]
- 54.Massana R. Eukaryotic picoplankton in surface o. Annu Rev Microbiol. 2011;65:91–110. doi: 10.1146/annurev-micro-090110-102903. [DOI] [PubMed] [Google Scholar]
- 55.Alonso-Sáez L, Díaz-Pérez L, Morán XAG. The hidden seasonality of the rare biosphere in coastal marine bacterioplankton: Seasonality of the rare biosphere. Environ Microbiol. 2015;17:3766–80. doi: 10.1111/1462-2920.12801. [DOI] [PubMed] [Google Scholar]
- 56.Needham DM, Fichot EB, Wang E, Berdjeb L, Cram JA, Fichot CG, et al. Dynamics and interactions of highly resolved marine plankton via automated high-frequency sampling. The ISME J. 2018. 10.1038/s41396-018-0169-y. [DOI] [PMC free article] [PubMed]
- 57.Chafee M, Fernàndez-Guerra A, Buttigieg PL, Gerdts G, Eren AM, Teeling H, et al. Recurrent patterns of microdiversity in a temperate coastal marine environment. ISME J. 2018;12:237–52. doi: 10.1038/ismej.2017.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alonso-Saez L, Gasol JM. Seasonal variations in the contributions of different bacterial groups to the uptake of low-molecular-weight compounds in northwestern mediterranean coastal waters. Appl Environ Microbiol. 2007;73:3528–35. doi: 10.1128/AEM.02627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paerl RW, Bouget FY, Lozano JC, Vergé V, Schatt P, Allen EE, et al. Use of plankton-derived vitamin B1 precursors, especially thiazole-related precursor, by key marine picoeukaryotic phytoplankton. ISME J. 2017;11:753–65. doi: 10.1038/ismej.2016.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carini P, Campbell EO, Morré J, Sañudo-Wilhelmy SA, Cameron Thrash J, Bennett SE, et al. Discovery of a SAR11 growth requirement for thiamin’s pyrimidine precursor and its distribution in the Sargasso Sea. ISME J. 2014;8:1727–38. doi: 10.1038/ismej.2014.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Paerl RW, Bertrand EM, Allen AE, Palenik B, Azam F. Vitamin B1 ecophysiology of marine picoeukaryotic algae: strain‐specific differences and a new role for bacteria in vitamin cycling. Limnol Oceanogr. 2015;60:215–28. doi: 10.1002/lno.10009. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.