Abstract
Staining with Congo Red (CR) is a qualitative method used for the identification of amyloids in vitro and in tissue sections. However, the drawbacks and artefacts obtained when using this dye can be found both in vitro and in vivo. Analysis of scientific data from previous studies shows that CR staining alone is not sufficient for confirmation of the amyloid nature of protein aggregates in vitro or for diagnosis of amyloidosis in tissue sections. In the present paper, we describe the characteristics and limitations of other methods used for amyloid studies. Our historical review on the use of CR staining for amyloid studies may provide insight into the pitfalls and caveats related to this technique for researchers considering using this dye.
Keywords: amyloid dye, amyloid staining, amyloids, amyloid detection, amyloidosis, Congo red
Introduction
In amyloidosis diseases, amyloids can be detected in the kidney, liver, brain and other human and animal organs. The development of neurodegenerative human diseases, such as Alzheimer’s disease and Parkinson’s disease, is thought to be the consequence of accumulation of insoluble amyloid plaques in the nerve tissue [1]. However, amyloids have been observed serving biological functions beyond the development of pathological processes. Since the beginning of the 21st century, researchers have reported evidence that amyloids form in the body to perform specific functional roles [2]. These so-called ‘functional amyloids’ have been observed in both prokaryotes [3–5] and eukaryotes [2,6–8]. For instance, in Escherichia coli, amyloids contribute to the formation of biofilms [5]. Amyloids are also involved in melanin formation in the melanosomes of human skin cells [2].
The study of the amyloid formation process is currently one of the most important tasks in this field. Research in this area involves not only understanding how amyloidosis develops and how to prevent amyloid plaque formation but also investigating functional amyloids.
Staining with Congo Red dye (CR) is one of the major methods used to detect the amyloid structure of protein aggregates. However, a series of experiments have shown that CR staining is insufficient for confirmation of the amyloid nature of protein aggregates. This review details the history of amyloid research using CR dye. Some drawbacks and artefacts obtained when using this dye are described.
Discovery and study of amyloids
In 1814, Colin and Gaultier de Claubry identified the blue staining reaction of starch with iodine and sulphuric acid [9]. One of the founders of the cell theory, Matthias Schleiden (1804–1881), who was interested in studying the chemical composition and anatomical structure of plants, proposed application of the iodine-sulphuric acid test for the detection of starch in plants (1838) (cited in [10]). In his later works, Schleiden wrote that starch formation occurred in plants, and the term ‘amyloid’ was introduced for the description of ‘a normal amylaceous constituent of plants’ [9].
Rudolph Virchow employed the term ‘amyloid’ in 1854 when he described the peculiar reaction of the corpora amylacea in the nervous system with iodine [9]. Interestingly, it is now known that these corpora are not amyloids [9]. Virchow suggested that cerebral corpora amylacea are similar to carbohydrates in nature and form deposits during ‘lardaceous’ or ‘waxy’ changes in the liver [9]. In 1859, Schmidt [11] denied this opinion, reporting a high proportion of nitrogen in organs infiltrated by amyloids. In the same year, Friedreich and Kekule [12] showed that waxy spleen tissue contained no material that corresponded chemically to amylon (that is, starch) or cellulose. These were the first steps toward investigation of the nature of amyloid deposits, which had been observed in organs since the 17th century.
The first mention of amyloidosis was most likely made by Fonteyn in 1639 [9,13]. He described an enlarged human spleen filled with sizable white inclusions. It is currently assumed that these inclusions were amyloid in nature [9,13]. In 1789, Antoine Portal was the first to describe liver amyloidosis [9]. Consequently, the term ‘amyloidosis’ encompasses a group of diseases associated with amyloid deposits in tissues and organs.
In 1842, Carl Rokitansky reported that the livers of patients with tuberculosis or syphilis became enlarged after infiltration by a grey, albuminous, gelatinous substance [9]. It appears that he was the first person to state that lardaceous/amyloid deposits occurred in cases of tuberculosis, syphilis and mercury poisoning [9].
In 1856, Samuel Wilks studied lardaceous viscera in elderly patients and concluded that the observed changes in organ tissue were not related to syphilis or tuberculosis. In 1865, Wilks found similar changes in the liver, kidney and adrenal glands in patients without diseases such as syphilis, tuberculosis or bony disease [9]. In 1867, Weber detected amyloidosis in a patient with multiple myeloma. In this patient, amyloids were found in the left ventricle of the hypertrophied heart, the kidneys and the spleen [9].
Further investigation of amyloidoses continued with the application of histopathological dyes, such as CR (1922) [14] and thioflavin (1959) [15], which were used instead of iodine. Until 1959, a histochemical approach was the common method for detecting amyloid deposits.
Structural investigations of amyloids began in the 1930s with the use of X-ray diffraction [16]. In 1935, Astbury and Dickinson, in the paper ‘The X-ray interpretation of denaturation and the structure of the seed globulins’ based on the process of egg albumin denaturation, concluded that ‘heat-denaturation of the albumins, for instance, merely makes the X-ray photograph more like that of a random arrangement of fibres of β-keratin’ [16]. Hence, Astbury and Dickinson [16] first noted the distinctive X-ray fibre diffraction pattern later called ‘cross-β’. With active use of the X-ray diffraction method, it was concluded that amyloids are composed of polypeptide chains extended in the so-called cross-β conformation [17,18]. In the cross-β structure, the individual strands of each β-sheet run perpendicular to the fibril axis (4.7 Å spacing) whereas the β-sheet (∼10 Å spacing) are parallel to the fibril axis [16,19].
In 1959, Cohen and Calkins [20] undertook electron microscopic studies of amyloid tissues in rat kidneys and human kidneys and skin. They observed amyloid deposits with a fibrillar structure ∼75–140 Å wide and ∼1000–16000 Å long [20]. The authors supposed that a challenge in protein detection within the deposits is the insolubility of amyloids in organic solvents [20].
In 1997, atomic force microscopy, cryoelectron microscopy [21,22] and solid-state NMR spectroscopy [23,24] were employed for investigations of amyloid morphology. Researchers began to elucidate the morphology of amyloid protofilaments, protofibrils and fibrils [26–28]. It was shown that the same protein can form amyloid fibrils, which can have different morphologies, such as coiled and ribbon-like fibrils of Аβ-peptide [25]. It was later found that amyloids may form not only structured fibrils but also amorphous aggregates [28–31].
Research focused on amyloids includes the study of prions, which were discovered by Prusiner in the 1980s [32]. Prions are a unique class of infectious agents where proteins are the basis of infectivity. In mammals, prions cause fatal neurodegenerative diseases, such as Creutzfeldt–Jakob disease in humans, sheep scrapie and bovine spongiform encephalopathy [33,34]. All of these diseases are caused by the PrP protein, which has a conformationally altered form (PrPSc) that can convert the normal host-encoded protein (PrPC) into this altered form [35]. The resulting altered form is the amyloid cross-β conformation.
In addition to the discovery of amyloids and prions that are responsible for the development of diseases, functional amyloids have also been discovered [2–8]. Since the beginning of the 21st century, data on protein aggregates with a cross-β structure that do not cause development of a pathological process but play a specific role in the organism have been reported. Functional amyloids have been observed in both prokaryotes [3–5] and eukaryotes [2,6–8]. It has been shown that in humans, functional amyloid fibrils are formed from proteolytic fragments of the Pmel17 protein in melanosomes [2]. These fibrils are involved in the polymerisation of melanin precursor into melanin and play a cytoprotective role by sequestering toxic intermediates produced during melanin synthesis and/or by templating and accelerating melanin production [2]. It was also suggested that amyloids play an essential role in the formation of long-term memory in animals. This idea is based on a study showing that the translational regulator CPEB of Aplysia californica, with prion-like properties, plays a key role in long-term synaptic changes [36].
In recent studies, it has been reported that many proteins, under certain in vitro conditions, are able to form amyloid-like aggregates or fibrils [37]. However, it is known that not all of these proteins form amyloids in humans or animals [38]. The term ‘amylome’ has been introduced to describe the universe of proteins that can potentially generate amyloid-like fibrils [38].
Today, there are multiple methods available for the study of amyloids (Table 1). With these methods, it is possible to detect amyloids in vitro and in vivo, to determine their structure and morphology in tissue sections and in vitro, and to explore the aggregation kinetics of amyloidogenic proteins (Table 1).
Table 1. Methods used to study amyloids in vitro and in vivo.
| Method | Characteristics and peculiarities | Applicability | Limitations | |||
|---|---|---|---|---|---|---|
| Detection of amyloids | Study of amyloid structure | Study of amyloid morphology | Study of aggregation kinetics | |||
| Immunohistochemistry and Immunochemistry | Immunohistochemistry is applied in pathology to visualise and localise protein aggregates and inclusions found in tissue sections of individuals with amyloidosis [39]. This technique is widely available and easily applicable in most pathology laboratories [40]. Immunohistochemistry uses antibodies for detection of amyloids. The availability of novel monoclonal antibodies targeting amyloidogenic precursors [41,42] and antibodies is directed against the amyloid fibrils [41,43]. Conformation-specific antibodies recognise soluble oligomers [44,45] or fibrils from many types of amyloid proteins [46–49], regardless of sequence. A small, bispecific antibody-based radioligand capable of crossing the blood–brain barrier can bind to intracerebral Aβ, allowing for in vivo visualisation [50]. This technique also uses luminescent–conjugated oligothiophenes as a unique class of amyloid dyes [51,52,53]. |
Yes | No | No | No | Commercially available antibodies for various proteins are often suboptimal for the identification of the same proteins in an amyloid fibril conformation [54]. This is caused by the proteins adopting different conformations and a variety of modifications during fibril formation, such as N- or C-terminal truncation [54]. To overcome this limitation, novel conformation-specific antibodies were designed [44–47]. At present, they are not yet widely used and require additional testing. Novel techniques that use luminescent–conjugated oligothiophenes [51–53] and the antibody-based radioligand [50] also need to be tested further. These novel developments are primarily used to detect Aβ fibrils and occasionally α-synuclein fibrils [46]. |
| Staining with CR | The binding of CR to amyloids in vitro induces a characteristic increase in CR absorption leading to a red shift of its absorbance peak from 490 to 512 nm and the presence of a unique shoulder peak at approximately 540 nm [55,56]. Amyloid is detected by the increased optical anisotropy after CR binding [57], which is called the ‘apple-green birefringence’ (under crossed polarisers) [58]. ‘Apple-green birefringence’ is used to detect amyloid deposits in tissues and in in vitro studies of amyloids [15,57,58]. Today, this method is commonly used in histopathology laboratories because it is simple and cost-effective. There are different modifications of the method, including CR fluorescence (CRF) [59,60]. |
Yes | No | No | No | There exists some limitations (additional information is given in the main text). |
| Staining with Thioflavin Т/S | Thioflavin fluorescence is a classical method for detecting and analysing amyloids in tissue samples [61]. Thioflavin T is selectively localised to amyloid deposits, thereupon exhibiting a dramatic increase in fluorescent brightness [61]. This staining is also used in in vitro studies [62,63]. Upon binding to amyloid fibrils, Thioflavin T gives a strong fluorescence signal at approximately 482 nm when excited at 450 nm [64]. The fluorescence intensity scales linearly with amyloid fibril mass (e.g., with the number of available binding sites). Based on increasing fibril mass, the concentration of amyloid is calculated [65]. Reductions in Thioflavin T emission intensity are often interpreted as an indicator of fibril growth inhibition [66]. | Yes | No | No | Yes | Staining with Thioflavin T/S is easy to perform, but the requirement of fluorescence microscopy limits the usefulness of this staining method [61]. Other tissues, such as cartilage, elastic fibres and mucopolysaccharides, can also be stained with ThT [67–69]. Sometimes, even at substoichiometric dye/monomer ratios, ThT undergoes substantial self-quenching that results in a non-linear relation between its binding and emission properties [63]. In this regard, without preliminary experiments, the use of this method for the study of fibril formation kinetics is problematic. ThT fluorescence is not generally a suitable tool for the detection of oligomeric intermediates during amyloid fibril growth [62]. Nevertheless, this dye binds to some oligomers [70–72]. |
| Circular dichroism | This method is used to determine the secondary structures of proteins and peptides in amyloid aggregates [73,74]. This technique operates by the differential absorption of the left- and right-handed components of circularly polarised light by chiral molecules in solution [75]. |
No | Yes | No | No | This technique can be used only for determination of changes in secondary structure and is sensitive to aromatic compounds present in the sample. It can be employed only in in vitro studies. |
| Fourier transform IR spectroscopy | Fourier transform IR spectroscopy is an absorption spectroscopy in which the transitions detected are those arising from vibrational modes of bonds involving heteroatoms [77,78]. The presence and relative abundance of β-sheet structure in peptides and proteins can be assessed by this technique [76] also for amyloid studies [79,80]. Methodologies exist to acquire spectra from proteins in any physical state, including crystals, powders, thin films, and aqueous solutions, as well as from membrane-bound proteins [76]. | No | Yes | No | No | There is water interference [76]. High protein concentration is necessary [76]. This technique can be employed only in in vitro studies. |
| NMR | Molecular and supramolecular structures of amyloid fibrils can be probed by various solid-state NMR techniques [24,82–89]. | No | Yes | Yes | Yes | This technique is expensive and difficult [81]. Analysis is limited to small proteins [81]. It can be employed only in in vitro studies. |
| X-ray diffraction | The method is used to examine the structures of insoluble amyloid fibres [90,91]. With this method, it is possible to reveal the presence of the cross β-sheet structure by detection of the 4–5 Å equatorial and 10–12 Å axial reflections that correspond to the inter-chain distance and to the face-to-face separation of β-sheets, respectively [92,93]. |
No | Yes | No | No | This method can only be used when the sample is in the crystallised form. It is difficult to employ this method to study intermediates at the initial aggregation stages. This technique can be employed only in in vitro studies. |
| Small-angle X-ray scattering | This technique can be used to investigate the structure, folding and conformational dynamics of globular proteins, including multidomain and multisubunit proteins [94]. Small-angle X-ray scattering is a powerful and flexible technique for the characterisation of structural variations in amyloid fibrils [95–98]. |
No | Yes | Yes | No | This technique is expensive and difficult. This technique can be employed only in in vitro studies. |
| Cryoelectron microscopy | Cryoelectron microscopy technology allows for high resolution (less than 5 Å) determination of the atomic structures of amyloid fibrils in vitro [99–101]. Also in this technique, it is possible to explore amyloids extracted from tissues (ex vivo) [102,103]. | No | Yes | Yes | No | It is difficult or impossible to study amorphous amyloid aggregates using this technique. The process of sample preparation is very complicated. |
| TEM | Since amyloid fibrils have unique electron microscopy characteristics [104,105], electron microscopy is routinely used in the analysis of kidney biopsies in the United States and other countries [104]. All types of amyloid deposits seen in different tissues, are mainly composed of bundled, not branched, straight fibrils, ranging from 6 to 13 nm in diameter (average 7.5–10 nm) and 100–1600 nm in length [106]. However, other morphological characteristics can also exist [107]. To increase the diagnostic significance of the method, immunogold electron microscopy, a technique that combines immunohistochemistry with electron microscopy, can be used [108]. With this technique, it is possible to explore both amyloids extracted from tissues and amyloids generated in vitro [106]. | Yes | No | Yes | No | In some cases, this technique is not suitable to diagnose amyloidosis. For instance, immunotactoid glomerulopathy and fibrillary glomerulonephritis can be misdiagnosed as immunoglobulin light and heavy chain amyloidosis [109]. |
| Atomic force microscopy | This technique can be used to investigate the size and morphology of protein aggregates in solution [110]. This method is also used to probe intrinsic properties of amyloid fibrils, such as mechanical strength and Young’s modulus [111]. | No | No | Yes | Yes | When contact mode is used, high shear forces cause damage to the fibrils and may require immobilisation strategies [112]. The tapping-mode in air requires a ‘dried’ sample, so it cannot probe fibril processes directly; as a result, potential artefacts from dehydration (e.g., salt crystals, fibril damage) may be observed, and the degree of hydration is not known and cannot be controlled [112]. For the tapping-mode in liquid, current scan speeds are too slow to image rapid processes, and often, samples (fibrils) need to be well-adhered to a surface. Other atomic force microscopic techniques, including the use of support surfaces (e.g., lipid bilayers) are often difficult to use [112]. |
| Dynamic light scattering | Dynamic light scattering is a laser scattering technique capable of unbiased analysis of size distributions for diffusing particles in the nanometre to micrometre size range [113]. The ability to resolve multimodal size distributions and make absolute size measurements makes dynamic light scattering a powerful technique for systems with heterogeneous species. It has been used to quantitatively study fibril formation in a range of systems from different proteins [114–117]. |
No | No | No | Yes | A hydrodynamic radius of the particle is measured, but this can differ from the true radius. As a result, determination of the real size and shape of macromolecules is difficult. |
| Fluorescence correlation spectroscopy | This highly sensitive analytical technique is used to measure dynamic molecular parameters, such as diffusion time (from which particle size can be calculated), conformation and concentration of fluorescent molecules [118,119]. It has been particularly powerful for characterising size distributions in molecular associations (e.g., dimer/multimer and fibril formation) both in well-behaved thermodynamically equilibrated systems in vitro and in more complex environments in vivo. Fluorescence correlation spectroscopy could therefore be used as a highly sensitive and specific competition assay to identify potential inhibitors of fibril formation [118]. | No | No | No | Yes | This technique requires the use of fluorescent tags to label amyloid samples. This makes it difficult and rarely used. |
| Analytical size-exclusion chromatography | The method is used to separate a diverse range of differently sized particles by passing a solution containing the particles through a partially permeable gel medium. It is used to analyse intermediates (mainly, oligomers) and identify soluble aggregates in tissue [120,121]. | No | No | No | Yes | It is impossible to study aggregates with high molecular weight. This method is inefficient for scale-up because size-exclusion chromatography performs poorly on large liquid volumes. |
| Analytical ultracentrifugation | This technique is based on the sedimentation velocity analysis used to determine the size, shape, and hydrodynamic behaviour of soluble macromolecules, including amyloid fibrils, as well as to study the process of amyloid aggregation [122–125]. | No | No | No | Yes | This technique is used only for specific tasks. |
A considerable number of scientific studies on the structure and function of amyloids have been conducted since the beginning of the second half of the 19th century. During this time, staining with CR dye remained a conventional technique for determining the amyloid nature of protein aggregates.
CR dye
Dyes are usually aromatic, heterocyclic compounds, some of which are toxic and possibly carcinogenic [126]. CR is a direct diazo dye that is intended primarily for the colouration of paper products. It is toxic and possibly carcinogenic and mutagenic [126–128]. CR is the sodium salt of benzidinediazo-bis-1-naphtylamine-4-sulphonic acid (formula: C32H22N6Na2O6S2; molecular weight: 696.66 g/mol; see the chemical structure in Table 2) [126]. The Colour Index Number of CR is 22120.
Table 2. Relation between different dyes and selective staining of amyloid (by [129] with our modifications).
| Dye* | Amyloid | Collagen | Elastic fibres** | Cytoplasm |
|---|---|---|---|---|
CR (C32H22N6Na2O6S2) 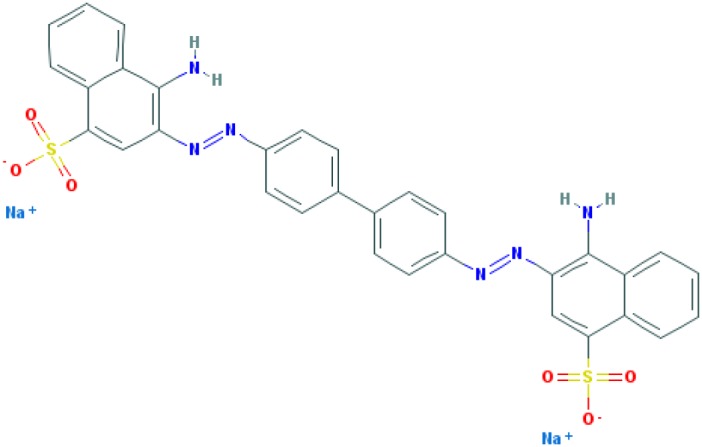
|
+++++ | ± | +++ | ± |
Congo Corinth (C32H23N5NaO7S2+) 
|
++++ | ± | +++ | ± |
Benzopurpurin 4B (C34H26N6Na2O6S2) 
|
++++ | 0 | ++ | 0 |
Vital Red (C34H25N6Na3O9S3) 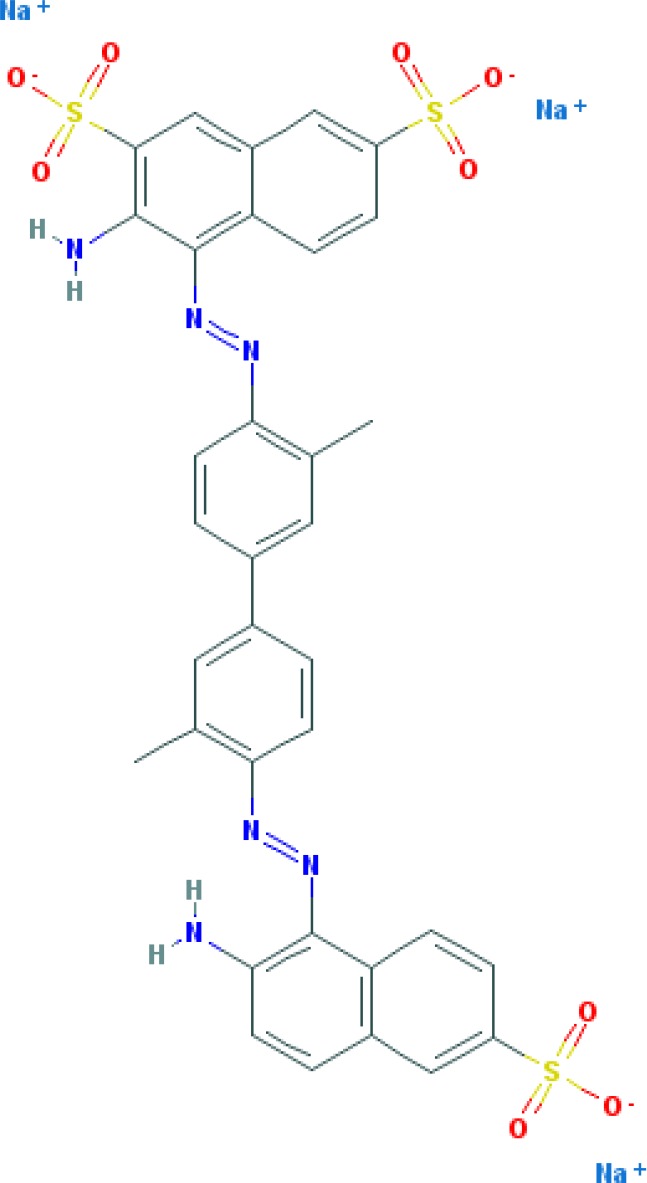
|
+++ | 0 | + | 0 |
Trypan Blue (C34H24N6Na4O14S4) 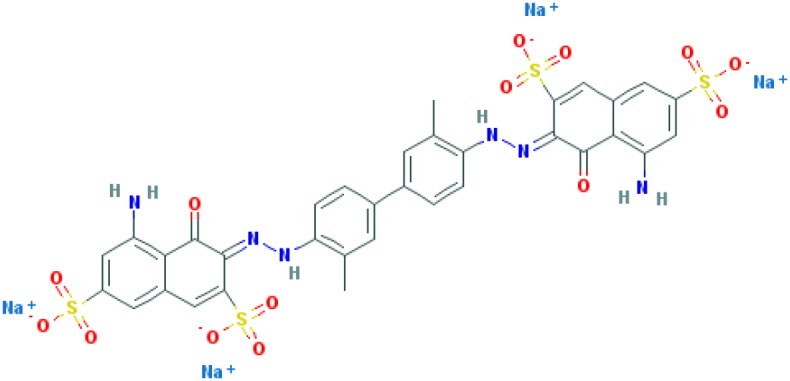
|
++ | 0 | ± | 0 |
Amidoblack 10B (C22H14N6Na2O9S2) 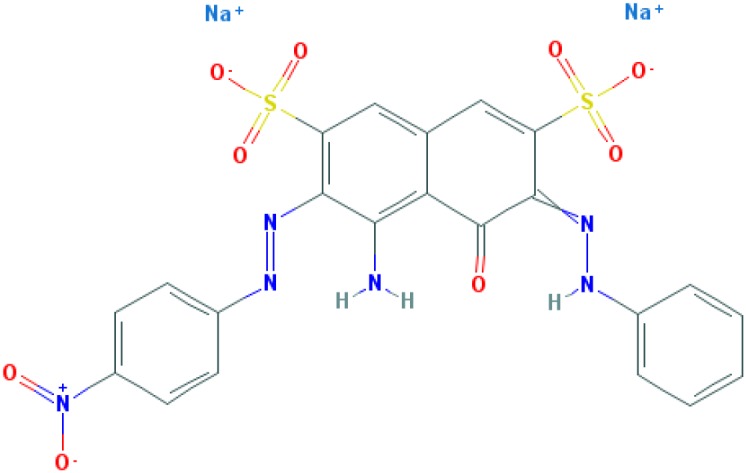
|
+ | + | + | + |
Biebrich Searlet WS (C22H14N4Na2O7S2) 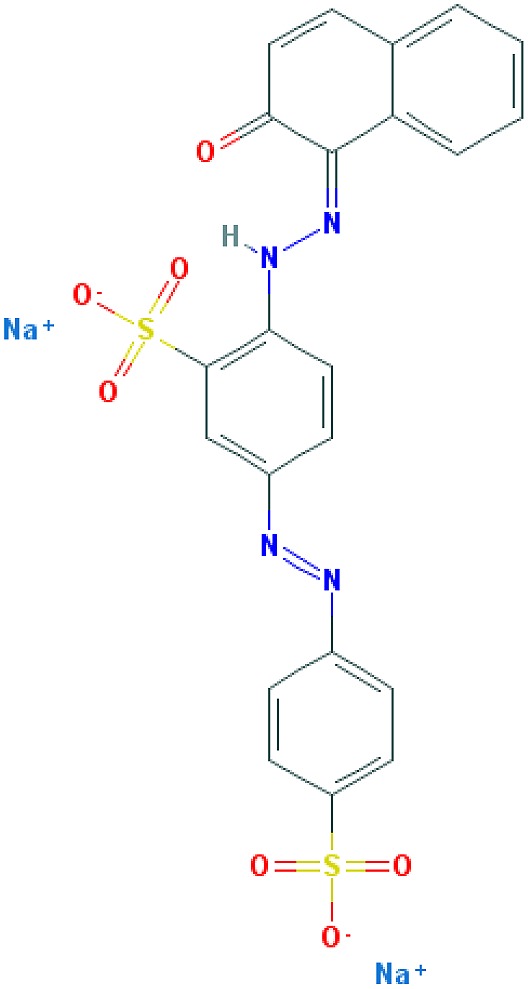
|
0 | 0 | 0 | 0 |
Aniline Blue WS (C32H25N3Na2O9S3)
|
± | ± | ± | ± |
Acid Fuchsin (C20H17N3Na2O9S3)
|
+++++ | +++++ | +++++ | +++++ |
Tissue colouration degree: 0 denotes no colour, ± → + → ++ → +++ → ++++ → +++++ is from poor colour to rich colour.
Formulas and pictures are taken from the Open Chemistry Database, PubChem.
Membrana elastic interna of arteriae interlobares in kidney.
CR was discovered in 1883 by chemist Paul Böttiger when he tried to synthesise a substance that could be used as a pH indicator [130]. In 1885, this dye was named as CR [130]. When CR is present as a disodium salt in alkaline or weakly acidic solutions, a red colour is produced. In strongly acidic conditions, as the free acid, the maximum absorption moves to longer wavelengths in the yellow and orange regions, giving a violet or blue colour [129].
This dye became the first of many so-called ‘Congo’ direct dyes produced at the dyestuff chemical laboratory of the Friedrich Bayer Company in Elberfeld, Germany, where Böttiger worked [130]. After the discovery of CR, many textile dyes received the Congo name: Congo Rubine, Congo Corinth, Brilliant Congo, Congo Orange, Congo Brown and Congo Blue. The word ‘Congo’ in the dye name was used for marketing purposes [130]. Many aniline dyes were tested as possible histological dyes [131,132], which included testing for staining of amyloids (Table 2).
In 1886, CR was employed as a pH indicator to detect acid in the intestinal tracts of animals [133]. In 1922, Bennhold was the first to discover that CR can be used for identification of amyloids in organisms after injecting the dye into blood [14,134,135]. Since then, CR has been used for identification of amyloids both in vivo and in vitro.
The use of CR in the study of amyloidosis
As mentioned above, the use of CR for amyloidosis identification began with experiments performed by Bennhold [14]. From the moment this new class of diseases, ‘amyloidoses’, was discovered, clinicians and scientists have searched for ways to diagnose these diseases and develop treatment methods. Bennhold described, for the first time, an approach for laboratory diagnosis of amyloidosis in 1923 [14]. He intravenously injected 10 ccs of a 1% solution of CR into 21 healthy subjects and 21 patients with different diseases [14]. In patients with nephrotic syndrome, a disappearance of CR in the blood occurred faster than in healthy individuals. In ten patients with amyloidosis secondary to pulmonary tuberculosis, clearance was apparently even more rapid, owing to an erroneous interpretation of the absence of CR in the urine. One of the patients with amyloidosis died 20 h after injection of the dye, and autopsy tissues were then obtained. The liver and spleen appeared to be stained by the dye. Microscopic studies of unstained frozen sections of the organs showed red areas [14]. This was the first microscopic demonstration of CR binding to amyloid [136]. Bennhold regarded the disappearance of 60% of the CR from circulating blood 1 h after injection as presumptive evidence of amyloid disease [134,135].
Subsequent studies concluded that the use of CR staining of amyloids also gives many false-positive results [141]. As a result, Bennhold’s method for diagnosis of amyloidosis was modified by many investigators including Friedman and Auerbach [137], Taran and Eckstein [138], Lipstein [139], Stemmerman and Auerbach [140] and others. The proposed modifications involved reducing the time between patient blood sampling and analysis and changing the criteria for clinically relevant dye content.
In 1948, Unger et al. [141], who criticised Bennhold’s CR test and its different modifications, pointed three significant mistakes made during the application of this method: the first lies in the assumption that the injected dye is completely mixed at the time of analysis; the second source of error lies in the assumption that little or no absorption of the injected CR takes place in amyloidosis at the reduced time (before 2- or 4-min specimens are obtained [138]); and the third source of error is the personal equation which enters into the colour matchings whenever optical colorimeters are used]. It should be noted that Unger et al. [141] also modified the CR test, calculating the theoretical initial concentration rather than using 2- or 4-min specimens for comparison and using 30 min rather than an hour as the end point [141].
In 1964, Stemmerman and Auerbach [140] reviewed the results of 649 cases of CR tests for amyloidosis performed on 446 patients, reported that the cases included 24.3% false negatives and 4.2% false positives and concluded that the chief cause of false-negative results was the minimal presence of amyloids and the principal reason for false-positive results was probably due to technical errors.
Notably, CR has been used for applications beyond detection of amyloidosis. Two years after Bennhold’s experiments (1925), Alder and Reimann [142] introduced the CR test as a reticuloendothelial system function test.
In 1976, Ouchi et al. [143] stated that there were no appreciable data on Bennhold’s test for the diagnosis of amyloidosis in scientific Japanese literature. This probably points to the fact that amyloidosis is an extremely rare disease in Japan [143]. Hence, it is clear why Japanese investigators explored other properties of CR. Japanese researchers presented data showing that intravenously injected CR was selectively taken up by Kupffer’s cells [144,145]. CR passes mainly through Kupffer’s cells and is later excreted into bile via hepatocytes; it also partially passes through other cells in the reticuloendothelial system [143–145].
Ouchi et al., while evaluating the CR test for its ability to detect amyloidosis of the liver, observed that the ‘CR test is not the best test for the diagnosis of amyloidosis’ [58]. No pathological features specifically influencing the CR index (CRI) were found by Japanese researchers during histological studies. At the same time, it was shown that the CRI is usually higher in liver diseases than in various other diseases [143]. Ouchi et al. [143] showed that an increase in the CRI is most visible in cases of liver cirrhosis and can also be explained by an obstructive change in liver blood flow followed by hepatic tissue damage. It should be noted that the authors did not provide any explanation for what CRI represented. At times, the authors used symbols (−/+/++/+++) as subjective estimates for the degrees of tissue colouration [146], and the results were occasionally given as percentages [143].
Although the CR test was recognised as suboptimal for the diagnosis of amyloidosis [143] and has been criticised [141], the development of CR-staining methods for the diagnosis of amyloidosis has continued to this day. Intravenous injection of CR, used in clinical practice for approximately 40 years, was replaced by staining of histological sections and biopsies with the dye [147].
It is commonly thought that CR-stained amyloid has an orange-red appearance under light microscopy and apple-green birefringence under polarised light during in vitro investigations and in vivo histological studies using tissue sections [9,147]. Several papers reported that this phenomenon was discovered by Divry (1927) and/or Divry and Florkin (1927) [148,149]. These authors described an increase in the intensity of the birefringence in CR-stained amyloid, but not a change in the colour of the specimen ([147]). Colours in CR-stained amyloid were first described by Ladewig (1945) [150] in his work ‘Double-refringence of the amyloid-Congo-red-complex in histological sections’. Green was not the only colour observed in his experiments; he stated that the colour ‘changes during rotation through 360° of the optical axes of the preparation twice from yellow to green, if conditions are optimal’ [150]. In this work, Ladewig also wrote about limitations when using CR: ‘its specificity is compromised by certain hyaline, mucous, fibrinoid or elastic tissue components also ‘taking’ CR’. The author suggested that this phenomenon is seen because other CR-positive, non-amyloid tissue components, such as certain hyalines, etc. ‘may be strongly anisotropic by themselves, a fact which is argument enough against their being regarded as amyloid’ [150].
Because of the geopolitical situation in Istanbul at the time Ladewig was conducting this research, he had no access to scientific literature on similar topics, and Howie and Brewer [147] noted that Ladewig’s investigations can be considered accurate and independent observations.
Ladewig was not the only investigator who described different colours under polarised light while studying CR-stained amyloids. Cooper [151] and Taylor et al. (1974) [152], who explored tissue sections with amyloid damage using CR staining, described that counter clockwise rotation of the analyser by 1-2 changes the colour to yellow-orange; a similar clockwise rotation produces a blue-green colour. Howie and Brewer stated in 2009 [147] that confusion regarding the origin of the term ‘apple-green birefringence’, which describes the change in colour when CR binds to amyloids in birefringence, began with a report by Hans-Peter Missmahl who was a physician in Tübingen under the direction of Bennhold (1920–2008). In 1957, Missmahl claimed that ‘Divry first noticed the increase in the strength of the birefringence of CR-stained amyloid. He was also struck by the appearance of a green polarisation colour’ (translated version from the German language [147]). Later, other researchers wrote that ‘Divry and Florkin were the first to observe that amyloid structures stained with CR, when observed in polarised light, as well as the increase in birefringence, lit up green’ (Diezel and Pfleiderer (1959) [153]). Wolman and Bubis (1965) [154], Benditt et al. (1970) [155] and others repeated similar incorrect explanations of the initial observation of apple-green birefringence in CR bound to amyloids (by [147]).
The phrase ‘apple-green birefringence’, often mentioned in articles on amyloid investigations, has apparently been used since 1972 [156]. This terminology probably originates from the word combination ‘apple-green immunofluorescence’, which was used in the 1950s. The use of ‘apple-green birefringence’ and ‘apple-green dichroism’ in textbooks and articles apparently promoted their subsequent citation [145]. Until now, observation of a green colour under polarised light is thought to indicate the presence of amyloids in CR-stained tissues. When this phenomenon was not observed, possible causes were considered, including inconsistencies in sample preparation. Specifically, it was shown that tissue sections <5 μm in thickness can produce false-positive results [15,61]. Conversely, other experiments showed that tissue thickness is not a significant factor in false-positive results and that the same area of a section or smear can appear green or yellow depending on the setup of the microscope [147].
Analysis of experimental data available before 2012 on CR apple-green birefringence has shown inconsistencies between the results of different studies [157]. In 160 scientific papers on staining of amyloids in tissue sections with CR, researchers reported only green birefringence or apple-green birefringence, even though only 31% of the illustrations showed a pure green colour [58,157]. In 66% of the papers, there were discrepancies between the descriptions of the colours reported in the text and the visual images shown in the figures [157]. These works mainly described discrepancies between reports of green-only and green-with-another-colour in the figures, or even between reports of green colour and total lack of green in the figures. Consequently, regardless of the colour observed, for example yellow/green and blue/green, the researchers described these colours as yellow or green. Combinations of yellow and blue colours are seen in practice more often than a pure green colour [147]. The various other colours, apart from the red that is seen when the polariser or analyser is progressively rotated from the crossed position, are also so-called ‘anomalous’ colours and are explained by the combination of absorption and changes in birefringence due to anomalous dispersion of the refractive index, usually with additional effects of strain birefringence in the optical system [147].
Howie and Owen-Casey [157] suggested that a regular use of the word ‘green’ in texts, papers, meetings and lectures provided the basis for the scientists to believe that observation of ‘green’ (including apple-green birefringence) is a reliable tool for diagnosis of amyloidosis [158].
Since 1965, CR fluorescence (CRF) has been used in research [59]. It is thought that this technique increases the recognition of tissue-bound CR by its property as a fluorochrome [159].
Nevertheless, until recently, CR has been used as a specific dye for amyloids. The question arises of how CR binds to substrate.
The binding of CR to amyloids
CR was the first direct dye for cotton, which did not require a mordant or chemical to fix the colour to the material [130]. Cotton is composed of cellulose, the substance that constitutes most of the plant cell wall. It is known that the CR molecules align themselves along the linear molecules in cellulose and that hydrogen bonds form between these two substances [160].
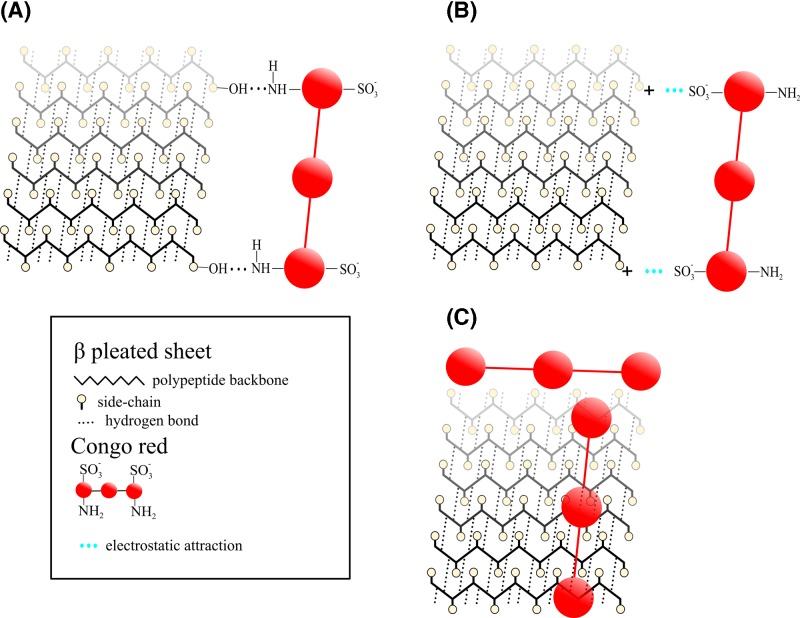
Figure 1 presents the main concepts of the binding of CR to amyloids. Amyloids were discovered during histological studies of human tissues with the iodine-sulphuric acid test for starch [9]. Virchow, who visualised amyloids using this test, began to use the term ‘amyloid’, and he thought that their structure resembled cellulose rather than starch [9]. Considering this fact, it is not difficult to understand why the researchers assumed that the binding of CR to amyloids occurred in a similar manner as its binding to cellulose [132]. Puchtler et al. (1962) [132] believed that dye binding was mediated by hydrogen bonding between primary hydroxyl groups of the polysaccharide chain and the amino groups of CR (Figure 1A). They also showed that not all direct cotton dyes bind amyloid selectively [129]. Therefore, the mechanism of the binding of CR to amyloids may be different from the mechanism of cellulose staining mentioned above.
Figure 1. The main hypothetical models of the binding of CR to amyloids.
(A) Dye binding mediated by hydrogen bonding between primary hydroxyl groups of the peptide chain (similar to the polysaccharide chain) and the amino groups of CR (Puchtler et al., 1962 [132]). (B) CR molecule could bind to positively charged amino acid residues along the peptide chains (Klunk, W.E. et al., 1989 [161]). (C) Polar contacts drive CR binding (Reinke and Gestwicki, 2011 [166]).
Cooper (1974) [151] suggested that CR may bind to amyloid by being partially entrapped and bound in ‘channels’ of the β-pleated sheet amyloid by non-specific close-range forces [161]. Still, this mechanism did not explain binding on the biochemical level.
In a paper by Klunk et al. in 1989 [161], it was shown that CR binding correlates well with the number of positively charged amino acids in a sample of amyloid fibrils (Figure 1B) [161].
The modern notion of CR binding to amyloids lies on the assumption that linear ligands can exploit the regular patterns (e.g., grooves) on the amyloid fibril surface in primary binding modes [162].
Molecular docking simulation results suggested that CR binds to sites parallel to the fibril axis (antiparallel to the β-sheets) [163] on amyloid fibrils [162–165] and protofibrils [162]. With the use of molecular docking simulations, a second site at the ‘end’ of the protofibril in an orientation antiparallel to the fibril axis and parallel to the β-sheets was also described (Figure 1C) [166]. Notably, this latter binding mode is represented only in approximately 11% of the total CR binding clusters, while the antiparallel orientation is clearly preferred (78% of the total binding) [166].
There is evidence demonstrating the binding of CR to proteins with different secondary structures [167]. This fact indicates that a specific amyloid structure is not mandatory for the binding of CR [167]. It has also been found that the binding of CR to protein molecules leads to protein oligomerisation [167]. Based on these data, the following conclusion on the possible mechanism of CR binding to proteins was made: ‘It is most likely that both the hydrophobic and the electrostatic components of the structure of CR are critical for its binding to proteins’ (2001) [167].
It is thought that the orientation of CR molecules on amyloid fibrils determines the dichroic and birefringent effects seen in tissue sections [168]. However, the physical basis of colours seen in CR-stained amyloid in polarised light was thoroughly described only in 2008 [168]. Birefringence indicates that a material has two refractive indices, depending on its orientation in polarised light. Usually, in studies on CR-stained amyloids, researchers report that experiments were carried out ‘under polarised light’. In most cases, this finding implies that examination of the sample was performed when the polariser and analyser were crossed [168]. In this case, the word combination ‘apple-green dichroism’ is used incorrectly [168]. Dichroism means that either a material has different amounts of absorption of light polarised in different planes, or the material varies from coloured to colourless, depending on the plane of polarisation. To explore this property, it is necessary to examine the material on a microscope fitted with either a polariser or an analyser, but not both, and when either the stage or the polarising filter is rotated, the appearance of the material should change from its deepest colour to colourless [169].
It has been shown that CR is dichroic [160,170] and, as a result, the change in intensity of red can be easy to see in smears of CR. Since amyloid fibrils are often haphazardly arranged, it is rather difficult to identify such a change in samples of CR-stained amyloid [168]. It has been shown that since the most typical property of CR-stained amyloid is the development of anomalous colours (yellow/green and blue/green) when using a crossed polariser and analyser setup, ‘apple-green birefringence’ should not be considered as the main feature to describe the properties of CR-stained amyloid [168].
When spectral methods were employed, characteristic pH-dependent changes in the CR absorption spectrum were defined. In strongly acidic conditions, CR has a maximum absorption that moves to longer wavelengths in the yellow and orange regions, giving a violet or blue colour [129]. This explains how CR can be used as a pH indicator, changing colour in the pH range 3–5 [172,173]. The peak absorption may also move to longer wavelengths when there is increased binding to a substrate, which is called the bathochromic shift [174]. The effects of pH upon binding may explain some reported differences in the optical properties of CR-stained amyloid and orientated CR, for instance in the wavelengths at which a property is at its maximum [147]. In addition, it has been found that the change in pH value of the medium leads to a change not only in colour but also in the solubility of CR [172].
In the 1970–80s, it was shown in stained tissue sections that observed acid-induced colour changes may be affected by the type of dye–substrate binding [156,177–179]. New modifications of CR staining methods were made to improve staining of tissue sections, however none of these procedures abolished concomitant staining of elastic fibres [171]. The researchers believed that staining of elastic fibres and amyloids was a result of non-ionic dye-binding forces [175,176]. However, studies that used a variety of different solutions and dye bath modifications designed to inhibit ionic, hydrogen or hydrophobic bonding did not show abolished staining of these two tissue substrates [177].
The authors of these studies concluded that the colour distinction between different stained substrates may reflect the dye indicator response to the acidic or basic nature of the substrate [171]. It appears that acidic and neutral substrates stained by CR display the colour induced by the acidic buffer, whereas basic substrates apparently accept protons from the cationic dye and, therefore, retain red colouration. In the case of amyloids, the red colouration observed in acidic conditions suggests the presence of basic groups [171]. According to these findings, the red colour of amyloids that develops after staining with CR might be a consequence of the presence of a comparatively large proportion of basic amino acids. However, this assumption does not explain the specific blue staining of collagen by CR, although this protein also has a large proportion of basic amino acids [178]. One explanation for this difference may be the variation in the positions of the basic amino acids within the protein and their availability in relationship to the dye molecules [171]. Nonetheless, removal of the ionisable basic groups in tissue by nitrous acid deamination resulted in minimal changes in the colour intensity of the red stain [171].
We next consider the known exceptions to the rules.
Exceptions to the rules
There are facts that indicate histochemical staining with CR is non-specific. In particular, it has been shown that CR is not specific for amyloid and can also stain elastotic dermis, as well as hyaline deposits in colloid milium and in lipid proteinosis [179]. It was also found that CR can stain native proteins such as elastin [174–176] and collagen [61].
In medical practice, Fernandez-Flores [180] discovered that a heat artefact due to cautery in surgical specimens can cause false-positive results in the diagnosis of amyloidosis when staining biopsies with this dye.
False-positive results with CR also depend on salt concentration [137,181]. It was shown that the use of sodium chloride at 50% saturation led to strong staining of collagen and eosinophilic granules [181]. Sodium chloride saturation has been reported to restrict staining to ‘true’ amyloid, although collagen and elastic fibres can also show red fluorescence [136,181].
There are several parameters, such as choice of biopsy site, as well as staining and analysis of the tissue, that can affect the sensitivity and specificity of histochemical analysis [182]. Notably, fat pad biopsies are less invasive and reasonably have 73% sensitivity, but fat pad needle biopsies have sensitivities ranging from 58 to 85% [182]. The overall staining specificity lies within the range 75–100% for biopsies [182,183]. However, some papers report that fat pad aspirates stained with CR exhibited lower sensitivity [184]. Devata et al. [188] wrote: “this may be due to multiple variables including type of patient population, severity of disease with scant versus abundant amyloid, experience level of the interpreters, microscope type, polariser quality, room darkness, and time spent to detect amyloid” [189].
The inter-observer variability in interpretation of fat pad aspirates stained with CR was also reported [185–187]. Devata et al. [188] demonstrated that when detecting amyloid in abdominal fat pad aspirates in early amyloidosis, the polarised light microscopy method for CR-stained sections resulted in frequent false-negative results (9 out of 9, 100%). In addition, the investigators estimated the effects of inter-observer variability among four pathologists: sensitivity was 25–75% and specificity was 50–100% [188].
Interestingly, Klatskin [189] showed as early as 1969 that it is possible to reveal foci of green birefringence in connective liver tissue and several other organs in both humans and rats under appropriate processing conditions. Regrettably, early publications describing cases of amyloidosis with CR staining, lack a ‘materials and methods’ section. This makes it difficult to determine CR staining procedures, taking into consideration that staining conditions influence the result. Typically, assays are performed under extreme conditions with 50–80% ethanol, high salt and alkaline pH conditions for successful binding to amyloid [132]. Despite these extreme conditions, false-positive results associated with the binding of CR to collagen fibres and cytoskeletal proteins were obtained [61].
By 2012, it was shown that the sensitivity of CR staining for amyloidosis is related to the tissue source, specifically bone marrow (63%), kidney, liver and cardiac tissues (87–98%) and rectal (69–97%) tissues [182]. The difficulty is that sampling of biological material is invasive and a highly qualified and skilled surgeon is necessary to perform this task. Linke [60] also wrote: “CR-stained sections should be evaluated by an experienced observer and always using a positive control. An appropriate microscope, with a powerful light source, used in a dimmed room, is an essential requisite”.
The aforementioned sensitivity values for CR staining of amyloidosis in tissue sections are astonishing compared with the results of in vitro studies of amyloid aggregates/fibrils. It should be considered that amyloid plaques also contain non-proteinaceous components, including nucleic acids [190], lipids [191], metal ions [192] and glycosaminoglycans [193].
Despite all aforementioned challenges, CR stain apple-green birefringence under polarised light is the most popular method for detecting amyloid in tissue sections; however, it has limitations. It was shown, that CR stain by fluorescent microscopy significantly enhances the diagnostic yield [194]. CRF has been used since 1965 [59]. For instance, when slides of amyloidosis stained with CR were examined under UV light, the following results were obtained: ‘amyloid was coloured bright red, and other tissue structures were pale greenish grey’ [59]. In 2003, CRF was employed to determine cutaneous amyloidosis. The results obtained were as follows: ‘while the amyloid stained either weakly or very weakly with CR, while CRF was strong or intensive’ [159]. In 2010, while studying cutaneous amyloidosis, Fernandez-Flores reported that the results of his study [195] were not in agreement with those previously published [159]. For instance, colouration after the use of CRF was intense only when CR staining was pronounced [195]. In this paper, the author concluded that immunohistochemistry offers advantages over CR staining and CRF [195]. Some investigators suggest that sensitivity can be enhanced by combining CR staining with fluorescence and antibodies, since the classical CR method by itself is insensitive [60].
Results from in vitro studies on non-specificity of amyloid staining with CR are known [155,161,167]. In 1989, comparative studies on the binding of this dye to proteinaceous substances with various internal structures, including β-pleated sheets and α-helical conformations, were performed [161]. The researchers found that CR binds to both the β-pleated sheet conformation and the α-helical conformation of poly-l-lysine [161]. These authors also found that CR did not bind as well to either poly-l-serine or polyglycine as it did to insulin fibrils or β-poly-l-lysine, even though all of these homopolymers exist in the β-pleated sheet conformation [196]. It was also shown that CR can bind to native α-proteins, such as citrate synthase and interleukin-2 [168] and to recombinant α-proteins, such as recombinant human growth hormone and recombinant human interferon-α2b [197]. These findings indicate that the β-pleated sheet peptide backbone alone is insufficient to form the optimal structural substrate for CR binding. Considering the two negatively charged sulphonic acid groups in the dye molecule, it was suggested that CR binding depends on the content of charged amino acid groups in protein molecules [161]. In this regard, it became clear why peptides with no positively charged amino acid residues (other than at the N-terminus), such as poly-l-serine and polyglycine, possess very few CR binding sites and fail to induce a spectral shift, even though they have a β-pleated sheet conformation [161], and vice versa; an α-helical peptide with an abundance of positively charged residues, such as α-poly-l-lysine, binds well to CR [198,199]. Nevertheless, it should be noted that the presence of positively charged residues alone are not enough to promote CR binding, as evidenced by the binding of CR to native insulin [161]. The β-pleated sheet conformation is apparently necessary to orient cations into the optimal conformation for CR binding [161]. The authors pointed out that the binding of CR to β-poly-l-lysine was also pH dependent [161].
It is known that binding of CR to β-amyloid fibrils formed in vitro is dependent on рН and on the histidine residue content in the peptide molecules [200]. The Cegelski group, while exploring the Escherichia coli amyloid fibre curli using surface plasmon resonance, showed that CR binding is dependent on pH but is independent of the mutant curli fibres that lack histidine residues [201].
The difficulty of using CR to study amyloids lies in the fact that the dye can interfere with the processes of protein misfolding and aggregation [202,203]. In 1994, it was shown that CR and certain sulphated glycans are potent inhibitors of protease-resistant PrP accumulation in scrapie-infected cells [202]. Inhibition of huntingtin fibrillogenesis by CR was shown in vitro in 2000 [203]. Undoubtedly, these properties of CR may affect interpretation of the results.
In support of the fact that CR binds preferentially to β-sheets containing amyloid fibrils and can specifically inhibit oligomerisation and disrupt preformed oligomers, evidence for inhibition of the effect of CR on polyglutamine oligomerisation exists [204]. It has also been reported that, in vitro, CR inhibits the fibrillisation of β-amyloid [205], insulin [206] and amylin [207]. As a result of these findings, some researchers recommended employing CR, not for diagnosing, but for treating amyloidosis [208]. However, the anti-aggregation effect of CR is not observed for all amyloids. Evidence that this dye did not inhibit amyloid production in vivo, for instance are available in microbial systems such as E. coli and Salmonella [201].
Notably, by 2001, the question ‘Is CR an amyloid-specific dye?’ had already been raised in scientific literature [167]. Researchers studied the structural specificity of CR binding to amyloid fibrils using an induced CD assay [167]. The authors found that the native conformations of insulin and Ig light chain induced CR circular dichroism, but with spectral shapes that differed from those of fibrils of the same proteins [167]. Khurana et al. [167] also showed that a wide variety of native proteins exhibited induced CR circular dichroism, indicating that CR bound to representative proteins from different secondary structure classes, such as α (citrate synthase), α+β (lysozyme), β (concavalin A) and parallel β-helical proteins (pectate lyase) [167]. These scientists also demonstrated that CR induced oligomerisation of native proteins using small angle X-ray scattering and cross-linking analysis [167].
Researchers have also tested the feasibility of using CR to detect amyloids in tissue sections [167]. The results of this study showed that CR binds to many native proteins and is not specific to secondary structure. These findings explain the false-positive results obtained on cytoskeletal proteins that are stable under the conditions used for CR staining in tissue sections [167].
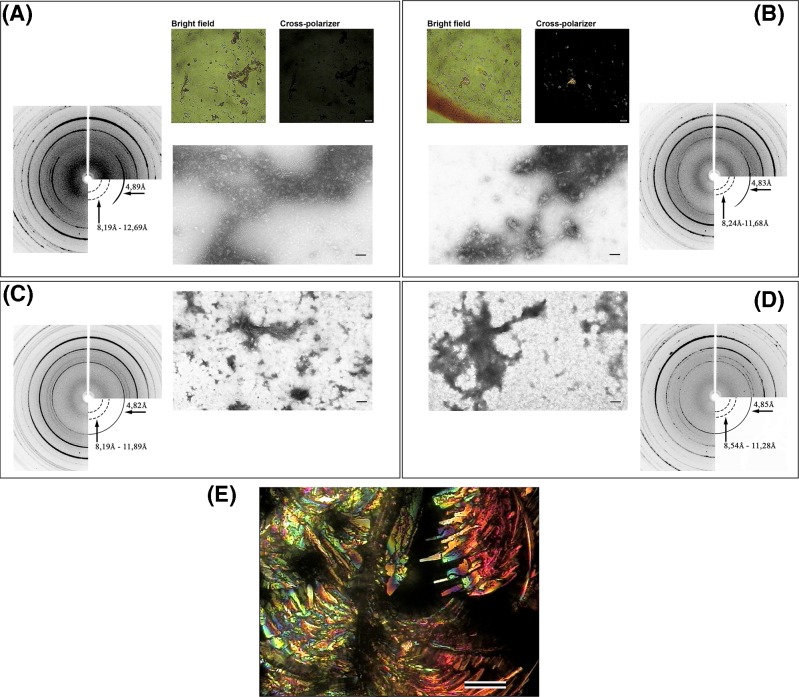
The non-specificity of CR as an amyloid dye was also shown in our in vitro studies [31]. We found that smooth muscle titin (SMT) from chicken gizzards can form amorphous aggregates in two different solutions containing: (i) 0.2 M KCl, 10 mM imidazole, pH 7.0 (SMT(KCl) aggregates) (Figure 2A) and (ii) 0.15 M glycine-KOH, pH 7.2–7.4 (SMT(Gly) aggregates) (Figure 2B). The amyloid natures of SMT(KCl) and SMT(Gly) aggregates were confirmed by X-ray diffraction [31]. CR-stained SMT(Gly) aggregates had ‘yellow to apple-green birefringence under polarised light’, while no such birefringence for SMT(KCl) aggregates was observed (Figure 2A,B). Interestingly, SMT(KCl) and SMT(Gly) aggregates, dialysed against a solution containing 0.6 M KCl, 30 mM KH2PO4, 1 mM DTT and 0.1 M NaN3 at pH 7.0, also had amyloid X-ray patterns (Figure 2C,D). In both cases, ‘yellow to apple-green birefringence under polarised light’ was absent [31]. In our in vitro study, we did not observe a characteristic shift in the CR maximum optical absorbance from 490 to 540 nm in any sample, indicating the binding of the dye to titin amyloids [31].
Figure 2. Investigation of SMT aggregates.
(A) SMT(KCl) aggregates: X-ray diffraction (left); CR polarisation microscopy of the aggregates, scale: 1 μm (top right); electron microscopy of negatively stained aggregates, scale: 100 nm (bottom right). (B) SMT(Gly) aggregates: CR polarisation microscopy of the aggregates, scale: 1 μm (top left); electron microscopy of negatively stained aggregates, scale: 100 nm (bottom left); X-ray diffraction (right). (C) SMT(KCl) aggregates after partial disaggregation: X-ray diffraction (left), electron microscopy of negatively stained titin aggregates, scale: 100 nm (right). (D) SMT(Gly) aggregates after partial disaggregation: electron microscopy of negatively stained aggregates, scale: 100 nm (left); X-ray diffraction (right). (E) Microscopy under polarised light of a dried drop of buffer containing 0.15 M glycine-KOH, pH 7.2–7.4 and CR, scale: 100 μm. For electron microscopy, 2% aqueous uranyl acetate staining was used.
Note that a dried buffer (0.15 M glycine-KOH, pH 7.2–7.4) containing CR without protein had yellow to apple-green birefringence under polarised light along with other colours (Figure 2E). In our experiments, it was apparent that differences found in titin aggregates stained with CR were associated with the buffer (despite performing a water wash) and not with the amyloid nature of the protein aggregates.
Conclusion
CR, now a classic dye, has played a role in the history of amyloid research. However, there is currently ample evidence of nonspecific binding of CR in studies on the identification of amyloids. The dye has been shown to bind non-amyloid substances, in addition to not binding amyloids in in vitro experiments and in in vivo histochemical studies. There is evidence that a specific amyloid cross-β structure is not necessary for binding to the dye, and the mechanism of CR binding to amyloids depends on a number of conditions, including the type of solvent, the composition of the solution, pH etc.
Due to modern scientific and methodological developments, researchers have a variety of possible methods available to study amyloids, according to their interest (Table 1) and can take into account the characteristic features and limitations of different methods. In this review, we discussed in detail the disadvantages of the CR staining method. Care should be taken when using the CR staining method to avoid misinterpretation of data.
Nevertheless, this technique has some merits. It is quick and easy to stain amyloids in vitro. CR can be employed for amyloid research in studies that utilise appropriate control samples (e.g., buffer alone and a sample of amyloid precursor) and other additional techniques to detect or characterise amyloid aggregates/fibrils.
Notwithstanding the imperfect specificity of CR binding to amyloids, the dye can be used in histochemical studies. It should be considered that positively stained tissue elements, such as elastin, collagen, eosinophilic granules and others, can usually be identified by their appearance or location. Moreover, when amyloidosis is suspected, in the presence of negative results obtained after CR staining, it is necessary to use immunohistochemistry for confirmation. Researchers must also remember that green is not the only colour that can be observed in the CR birefringence assay. Lastly, the use of CR alone is insufficient for diagnosing amyloidosis.
In conclusion, this historical analysis shows that the relationship between CR and amyloids is more complex than it appears initially. We anticipate that this review will provide insight into the expected outcomes following the use of CR.
Abbreviations
- CR
Congo red
- CRI
CR index
- CRF
CR fluorescence
- SMT
smooth muscle titin
- SMT(KCl) aggregate
aggregate of SMT in solution containing 0.2 M KCl, 10 mM imidazole, pH 7.0
- SMT(Gly) aggregate
aggregate of SMT in solution containing 0.15 M glycine-KOH, pH 7.2–7.4
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the Russian Foundation for Basic Research [grant number 18-315-00012]; and the Russian Science Foundation [grant number 14-14-00879].
References
- 1.Chiti F. and Dobson C.M. (2006) Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366 10.1146/annurev.biochem.75.101304.123901 [DOI] [PubMed] [Google Scholar]
- 2.Fowler D.M., Koulov A.V., Alory-Jost C., Marks M.S., Balch W.E. and Kelly J.W. (2006) Functional amyloid formation within mammalian tissue. PLoS Biol. 4, e6 10.1371/journal.pbio.0040006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Romling U., Bian Z., Hammar M., Sierralta W.D. and Normark S. (1998) Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 180, 722–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claessen D., Rink R., de Jong W., Siebring J., de Vreugd P., Boersma F.G.. et al. (2003) A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev. 17, 1714–1726 10.1101/gad.264303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Otzen D. and Nielsen P.H. (2008) We find them here, we find them there: functional bacterial amyloid. Cell. Mol. Life Sci. 65, 910–927 10.1007/s00018-007-7404-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wöesten H.A. and de Vocht M.L (2000) Hydrophobins, the fungal coat unraveled. Biochim. Biophys. Acta 1469, 79–86 10.1016/S0304-4157(00)00002-2 [DOI] [PubMed] [Google Scholar]
- 7.Iconomidou V.A., Chryssikos G.D., Gionis V., Galanis A.S., Cordopatis P., Hoenger A.. et al. (2006) Amyloid fibril formation propensity is inherent into the hexapeptidetandemly repeating sequence of the central domain of silk moth chorion proteins of the A-family. J. Struct. Biol. 156, 480–488 10.1016/j.jsb.2006.08.011 [DOI] [PubMed] [Google Scholar]
- 8.Slotta U., Hess S., Spiess K., Stromer T., Serpell L. and Scheibel T. (2007) Spider silk and amyloid fibrils: a structural comparison. Macromol. Biosci. 7, 183–188 10.1002/mabi.200600201 [DOI] [PubMed] [Google Scholar]
- 9.Kyle R.A. (2001) Amyloidosis: a convoluted story. Br. J. Haematol. 114, 529–538 10.1046/j.1365-2141.2001.02999.x [DOI] [PubMed] [Google Scholar]
- 10.Scleiden M.J., Schwann T. and Schulze M.. Klassische Schriften zur Zellenlehre. Ostwalds klassiker der exakten Wissenschaften, band 275, Verlag Harri Deutsch, http//books.google/fi/books/about/Klassische_Schriften_zur_Zellenlehre.html?=h9_WFhLD8uYC&redir_esc=y [Google Scholar]
- 11.Schmidt C. (1859) Ueber das sogenannte «thierische Amyloid» (Substanzder corpuscula Amylacea). Ann. Chern. Pharm. 110, 250–254 10.1002/jlac.18591100220 [DOI] [Google Scholar]
- 12.Friedreich N. and Kekule A. (1859) Zur amyloidfrage. Virch. Arch. Path. Anat. 16, 50–65 10.1007/BF01945246 [DOI] [Google Scholar]
- 13.Fonteyn N. (1639) Responsionum et Curationum Medicinalium, Amstelodami, Amsterdam [Google Scholar]
- 14.Bennhold H. (1922) Specific staining of amyloid by Congo red (in German). MuEnchener Medizinische Wochenschrift 69, 1537–1538 [Google Scholar]
- 15.Elghetany M.T. and Saleem A. (1988) Methods for staining amyloid in tissues: a review. Stain Technol. 63, 201–212 10.3109/10520298809107185 [DOI] [PubMed] [Google Scholar]
- 16.Astbury W.T. and Dickinson S. (1935) The X-ray interpretation of denaturation and the structure of the seed globulins. Biochem. J. 29, 2351–2360.1 10.1042/bj0292351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eanes E.D. and Glenner G.G. (1968) X-ray diffraction studies on amyloid filaments. J. Histochem. Cytochem. 16, 673–677 10.1177/16.11.673 [DOI] [PubMed] [Google Scholar]
- 18.Bonar L., Cohen A.S. and Skinner M.M. (1969) Characterization of the amyloid fibril as a cross-beta protein. Proc. Soc. Exp. Biol. Med. 131, 1373–1375 10.3181/00379727-131-34110 [DOI] [PubMed] [Google Scholar]
- 19.Maji S.K., Wang L., Greenwald J. and Riek R. (2009) Structure-activity relationship of amyloid fibrils. FEBS Lett. 583, 2610–2617 10.1016/j.febslet.2009.07.003 [DOI] [PubMed] [Google Scholar]
- 20.Cohen A.S. and Calkins E. (1959) Electron microscopic observation on a fibrous component in amyloid of diverse origins. Nature 183, 1202–1203 10.1038/1831202a0 [DOI] [PubMed] [Google Scholar]
- 21.Jimenez J.L., Guijarro J.I., Orlova E., Zurdo J., Dobson C.M., Sunde M.. et al. (1999) Cryo-electron microscopy structure of an SH3 amyloid fibril and model of the molecular packing. EMBO J. 18, 815–821 10.1093/emboj/18.4.815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jimenez J.L., Nettleton E.J., Bouchard M., Robinson C.V., Dobson C.M. and Saibil H.R. (2002) The protofilament structure of insulin amyloid fibrils. Proc. Natl. Acad. Sci. U.S.A. 99, 9196–9201 10.1073/pnas.142459399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petkova A.T., Ishii Y., Balbach J.J., Antzutkin O.N., Leapman R.D., Delaglio F.. et al. (2002) A structural model for Alzheimer’s beta -amyloid fibrils based on experimental constraints from solid state NMR. Proc. Natl. Acad. Sci. U.S.A. 99, 16742–16747 10.1073/pnas.262663499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaroniec C., MacPhee C.E., Bajaj V.S., McMahon M.T., Dobson C.M. and Griffin R.G. (2004) High-resolution molecular structure of a peptide in an amyloid fibril determined by magic angle spinning NMR spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 101, 711–716 10.1073/pnas.0304849101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldsbury C.S., Wirtz S., Müller S.A., Sunderji S., Wicki P., Aebi U.. et al. (2000) Studies on the in vitro assembly of a beta 1-40: implications for the search for a beta fibril formation inhibitors. J. Struct. Biol. 130, 217–231 10.1006/jsbi.2000.4259 [DOI] [PubMed] [Google Scholar]
- 26.Harper J.D., Wong S.S., Lieber C.M. and Lansbury P.T. (1997) Observation of metastable Abeta amyloid protofibrils by atomic force microscopy. Chem. Biol. 4, 119–125 10.1016/S1074-5521(97)90255-6 [DOI] [PubMed] [Google Scholar]
- 27.Walsh D.M., Hartley D.M., Kusumoto Y., Fezoui Y., Condron M.M., Lomakin A.. et al. (1999) Amyloid b-protein fibrillogenesis–structure and biological activity of protofibrillar intermediates. J. Biol. Chem. 274, 25945–25952 10.1074/jbc.274.36.25945 [DOI] [PubMed] [Google Scholar]
- 28.Huang T.H., Yang D.S., Fraser P.E. and Chakrabartty A. (2000) Alternate aggregation pathways of the Alzheimer beta-amyloid peptide. An in vitro model of preamyloid. J. Biol. Chem. 275, 36436–36440 10.1074/jbc.M005698200 [DOI] [PubMed] [Google Scholar]
- 29.Kumar S., Sharma P., Arora K., Raje M. and Guptasarma P. (2014) Calcium binding to beta-2-microglobulin at physiological pH drives the occurrence of conformational changes which cause the protein to precipitate into amorphous forms that subsequently transform into amyloid aggregates. PLoS ONE 9, e95725 10.1371/journal.pone.0095725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berthelot K., Lecomte S., Estevez Y., Coulary-Salin B., Bentaleb A., Cullin C.. et al. (2012) Rubber elongation factor (REF), a major allergen component in Hevea brasiliensis latex has amyloid properties. PLoS ONE 7, e48065 10.1371/journal.pone.0048065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yakupova E.I., Vikhlyantsev I.M., Bobyleva L.G., Penkov N.V., Timchenko A.A., Timchenko M.A.. et al. (2018) Different amyloid aggregation of smooth muscles titin in vitro. J. Biomol. Struct. Dyn. 36, 2237–2248 10.1080/07391102.2017.1348988 [DOI] [PubMed] [Google Scholar]
- 32.Prusiner S.B. (1982) Novel proteinaceous infectious particles cause scrapie. Science 216, 136–144 10.1126/science.6801762 [DOI] [PubMed] [Google Scholar]
- 33.Horwich A.L. and Weissman J.S. (1997) Deadly conformations–protein misfolding in prion disease. Cell 89, 499–510 10.1016/S0092-8674(00)80232-9 [DOI] [PubMed] [Google Scholar]
- 34.Prusiner S.B., Scott M.R., DeArmond S.J. and Cohen F.E. (1998) Prion protein biology. Cell 93, 337–348 10.1016/S0092-8674(00)81163-0 [DOI] [PubMed] [Google Scholar]
- 35.Kushnirov V.V., Vishnevskaya A.B., Alexandrov I.M. and Ter-Avanesyan M.D. (2007) Prion and nonprion amyloids. Prion 1, 179–184 10.4161/pri.1.3.4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Si K., Lindquist S. and Kandel E.R. (2003) A neuronal isoform of the Aplysia CPEB has prion-like properties. Cell 115, 879–891 10.1016/S0092-8674(03)01020-1 [DOI] [PubMed] [Google Scholar]
- 37.Dorta-Estremera S.M., Li J. and Cao W. (2013) Rapid generation of amyloid from native proteins in vitro. J. Vis. Exp. 82, 50869 10.3791/50869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldschmidt L., Teng P.K., Riek R. and Eisenberg D. (2010) Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc. Natl. Acad. Sci. U.S.A. 107, 3487–3492 10.1073/pnas.0915166107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Galvin J.E. (2003) Detection of aggregates and protein inclusions by staining of tissues. Methods Mol. Biol. 232, 149–164 [DOI] [PubMed] [Google Scholar]
- 40.Flodrova P., Flodr P., Pika T., Vymetal J., Holub D., Dzubak P.. et al. (2018) Cardiac amyloidosis: from clinical suspicion to morphological diagnosis. Pathology 50, 261–268 10.1016/j.pathol.2017.10.012 [DOI] [PubMed] [Google Scholar]
- 41.Strege R.J., Saeger W. and Linke R. (1998) Diagnosis and immunohistochemical classification of systemic amyloidoses. Report of 43 cases in an unselected autopsy series. Virch. Arch. 433, 19–27 10.1007/s004280050211 [DOI] [PubMed] [Google Scholar]
- 42.De Genst E., Messer A. and Dobson C.M. (2014) Antibodies and protein misfolding: from structural research tools to therapeutic strategies. Biochim. Biophys. Acta 1844, 1907–1919 [DOI] [PubMed] [Google Scholar]
- 43.Higaki J.N., Chakrabartty A., Galant N.J., Hadley K.C., Hammerson B., Nijjar T.. et al. (2016) Novel conformation-specific monoclonal antibodies against amyloidogenic forms of transthyretin. Amyloid 23, 86–97 10.3109/13506129.2016.1148025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kayed R. and Glabe C.G. (2006) Conformation-dependent anti-amyloid oligomer antibodies. Methods Enzymol. 413, 326–344 10.1016/S0076-6879(06)13017-7 [DOI] [PubMed] [Google Scholar]
- 45.Murakami K., Tokuda M., Suzuki T., Irie Y., Hanaki M., Izuo N.. et al. (2016) Monoclonal antibody with conformational specificity for a toxic conformer of amyloid β42 and its application toward the Alzheimer’s disease diagnosis. Sci. Rep. 6, 29038 10.1038/srep29038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kayed R., Head E., Sarsoza F., Saing T., Cotman C.W., Necula M.. et al. (2007) Fibril specific, conformation dependent antibodies recognize a generic epitope common to amyloid fibrils and fibrillar oligomers that is absent in prefibrillar oligomers. Mol. Neurodegener. 2, 18 10.1186/1750-1326-2-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarsoza F., Saing T., Kayed R., Dahlin R., Dick M., Broadwater-Hollifield C.. et al. (2009) A fibril-specific, conformation-dependent antibody recognizes a subset of Abeta plaques in Alzheimer disease, Down syndrome and Tg2576 transgenic mouse brain. Acta Neuropathol. 118, 505–517 10.1007/s00401-009-0530-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dumoulin M. and Dobson C.M. (2004) Probing the origins, diagnosis and treatment of amyloid diseases using antibodies. Biochimie 86, 589–600 10.1016/j.biochi.2004.09.012 [DOI] [PubMed] [Google Scholar]
- 49.De Genst E. and Dobson C.M. (2012) Nanobodies as structural probes of protein misfolding and fibril formation. Methods Mol. Biol. 911, 533–558 10.1007/978-1-61779-968-6_34 [DOI] [PubMed] [Google Scholar]
- 50.Fang X.T., Hultqvist G., Meier S.R., Antoni G., Sehlin D. and Syvänen S. (2018) High detection sensitivity with antibody-based PET radioligand for amyloid beta in brain. Neuroimage 184, 881–888 10.1016/j.neuroimage.2018.10.011 [DOI] [PubMed] [Google Scholar]
- 51.Rasmussen J., Mahler J., Beschorner N., Kaeser S.A., Häsler L.M., Baumann F.. et al. (2017) Amyloid polymorphisms constitute distinct clouds of conformational variants in different etiological subtypes of Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 114, 13018–13023 10.1073/pnas.1713215114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aslund A., Sigurdson C.J., Klingstedt T., Grathwohl S., Bolmont T., Dickstein D.L.. et al. (2009) Novel pentameric thiophene derivatives for in vitro and in vivo optical imaging of a plethora of protein aggregates in cerebral amyloidosis. ACS Chem. Biol. 4, 673–684 10.1021/cb900112v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Psonka-Antonczyk K.M., Hammarström P., Johansson L.B., Lindgren M., Stokke B.T., Nilsson K.P.. et al. (2016) Nanoscale structure and spectroscopic probing of Aβ1-40 fibril bundle formation. Front. Chem. 4, 44 10.3389/fchem.2016.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Westermark G.T., Ihse E. and Westermark P. (2018) Development of mouse monoclonal antibodies against human amyloid fibril proteins for diagnostic and research purposes. Methods Mol. Biol. 1779, 401–414 10.1007/978-1-4939-7816-8_24 [DOI] [PubMed] [Google Scholar]
- 55.Frid P., Anisimov S.V. and Popovic N. (2007) Congo red and protein aggregation in neurodegenerative diseases. Brain Res. Rev. 53, 135–160 10.1016/j.brainresrev.2006.08.001 [DOI] [PubMed] [Google Scholar]
- 56.Meehan S., Berry Y., Luisi B., Dobson C.M., Carver J.A. and MacPhee C.E. (2004) Amyloid fibril formation by lens crystallin proteins and its implications for cataract formation. J. Biol. Chem. 279, 3413–3419 10.1074/jbc.M308203200 [DOI] [PubMed] [Google Scholar]
- 57.Linke R.P. (2006) Congo Red Staining of amyloid: improvements and practical guide for a more precise diagnosis of amyloid and the different amyloidoses. In Protein Misfolding, Aggregation, and Conformational Diseases. Protein Reviews (Uversky V.N. and Fink A.L., eds), Springer, Boston, MA [Google Scholar]
- 58.Howie A.J. and Owen-Casey M.P. (2010) Discrepancies between descriptions and illustrations of colours in Congo red-stained amyloid, and explanation of discrepant colours. Amyloid 17, 109–117 10.3109/13506129.2010.527448 [DOI] [PubMed] [Google Scholar]
- 59.Puchtler H. and Sweat F. (1965) Congo red as a stain for fluorescence microscopy of amyloid (letter). J. Histochem. Cytochem. 13, 693–694 10.1177/13.8.693 [DOI] [PubMed] [Google Scholar]
- 60.Linke R.P. (2012) Diagnosis of minimal amyloid deposits using the congo red fluorescence method: a review. 175–185 10.1007/978-3-319-19294-9_14 Picken M., Herrera G. and Dogan A. (2015) Amyloid and Related Disorders. Human Press, Heidelberg [DOI] [Google Scholar]
- 61.Westermark G.T., Johnson K.H. and Westermark P. (1999) Staining methods for identification of amyloid in tissue. Methods Enzymol. 309, 3–25 10.1016/S0076-6879(99)09003-5 [DOI] [PubMed] [Google Scholar]
- 62.Persichilli C., Hill S.E., Mast J. and Muschol M. (2011) Does thioflavin-T detect oligomers formed during amyloid fibril assembly. Biophys. J. 100, 538a 10.1016/j.bpj.2010.12.3140 [DOI] [Google Scholar]
- 63.Lindberg D.J., Wenger A., Sundin E., Wesén E., Westerlund F. and Esbjörner E.K. (2017) Binding of thioflavin-T to amyloid fibrils leads to fluorescence self-quenching and fibril compaction. Biochemistry 56, 2170–2174 10.1021/acs.biochem.7b00035 [DOI] [PubMed] [Google Scholar]
- 64.Naiki H., Higuchi K., Hosokawa M. and Takeda T. (1989) Fluorometric determination of amyloid fibrils in vitro using the fluorescent dye, thioflavin T1. Anal. Biochem. 177, 244–249 10.1016/0003-2697(89)90046-8 [DOI] [PubMed] [Google Scholar]
- 65.Cohen S.I.A., Linse S., Luheshi L.M., Hellstrand E., White D.A., Rajah L.. et al. (2013) Proliferation of amyloid-β42 aggregates occurs through a secondary nucleation mechanism. Proc. Natl. Acad. Sci. U.S.A. 110, 9758–9763 10.1073/pnas.1218402110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Groenning M., Norrman M., Flink J.M., van de Weert M., Bukrinsky J.T., Schluckebier G.. et al. (2007) Binding mode of thioflavine T in insulin amyloid fibrils. J. Struct. Biol. 159, 483–497 10.1016/j.jsb.2007.06.004 [DOI] [PubMed] [Google Scholar]
- 67.Vassar P.S. and Culling C.F.A. (1959) Fluorescent stains with special reference to amyloid and connective tissues. Arch. Pathol. 68, 487–494 [PubMed] [Google Scholar]
- 68.LeVine H., III (1995) Thioflavin T interaction with amyloid β-sheet structures Amyloid. Int. J. Exp. Clin. Invest. 2, 1–6 [Google Scholar]
- 69.Kelényi G. (1967) On the histochemistry of azo group-free thiazole dyes. J. Histochem. Cytochem. 15, 172–180 10.1177/15.3.172 [DOI] [PubMed] [Google Scholar]
- 70.Lindgren M., Sörgjerd K. and Hammarström P. (2005) Detection and characterization of aggregates, prefibrillar amyloidogenic oligomers, and protofibrils using fluorescence spectroscopy. Biophys. J. 88, 4200–4212 10.1529/biophysj.104.049700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maezawa I., Hong H.S., Liu R., Wu C.Y., Cheng R.H., Kung M.P.. et al. (2008) Congo red and thioflavin-T analogs detect Abeta oligomers. J. Neurochem. 104, 457–468 [DOI] [PubMed] [Google Scholar]
- 72.Grudzielanek S., Smirnovas V. and Winter R. (2006) Solvation-assisted pressure tuning of insulin fibrillation: from novel aggregation pathways to biotechnological applications. J. Mol. Biol. 356, 497–509 10.1016/j.jmb.2005.11.075 [DOI] [PubMed] [Google Scholar]
- 73.McCubbin W.D., Kay C.M., Narindrasorasak S. and Kisilevsky R. (1988) Circular-dichroism studies on two murine serum amyloid A proteins. Biochem. J. 256, 775–783 10.1042/bj2560775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Abe H. and Nakanishi H. (2003) Novel observation of a circular dichroism band originating from amyloid fibril. Anal. Sci. 19, 171–173 10.2116/analsci.19.171 [DOI] [PubMed] [Google Scholar]
- 75.Scarlett G., Siligardi G. and Kneale G.G. (2015) Circular dichroism for the analysis of protein-DNA interactions. Methods Mol. Biol. 1334, 299–312 10.1007/978-1-4939-2877-4_19 [DOI] [PubMed] [Google Scholar]
- 76.Calero M. and Gasset M. (2005) Fourier transform infrared and circular dichroism spectroscopies for amyloid studies. In Amyloid Proteins. Methods in Molecular Biology™ (Sigurdsson E.M., ed.), Humana Press; [DOI] [PubMed] [Google Scholar]
- 77.Smith B.C. (1995) Fundamentals of Fourier Transform Infrared Spectroscopy, Smith B.C. ed. CRC Press, Boca Raton, Florida [Google Scholar]
- 78.Stuard B. (1997) Biological applications of infrared spectroscopy. In ACOL series, John Wiley & Sons, New York [Google Scholar]
- 79.Bouchard M., Zurdo J., Nettleton E.J., Dobson C.M. and Robinson C.V. (2000) Formation of insulin amyloid fibrils followed by FTIR simultaneously with CD and electron microscopy. Protein Sci. 9, 1960–1967 10.1110/ps.9.10.1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zou Y., Li Y., Hao W., Hu X. and Ma. G. (2013) Parallel β-sheet fibril and antiparallel β-sheet oligomer: new insights into amyloid formation of hen egg white lysozyme under heat and acidic condition from FTIR spectroscopy. J. Phys. Chem. B 117, 4003–4013 10.1021/jp4003559 [DOI] [PubMed] [Google Scholar]
- 81.Karamanos T.K., Kalverda A.P., Thompson G.S. and Radford S.E. (2015) Mechanisms of amyloid formation revealed by solution NMR. Prog. Nucl. Magn. Reson. Spectrosc. 86–104 10.1016/j.pnmrs.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tycko R. (2011) Solid-state NMR studies of amyloid fibril structure. Annu. Rev. Phys. Chem. 62, 279–299 10.1146/annurev-physchem-032210-103539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Scheidt H.A., Morgado I., Rothemund S., Huster D. and Fändrich M. (2011) Solid-state NMR spectroscopic investigation of Aβ protofibrils: implication of a β-sheet remodeling upon maturation into terminal amyloid fibrils. Angew. Chem. Int. Ed. (Engl.) 50, 2837–2840 10.1002/anie.201007265 [DOI] [PubMed] [Google Scholar]
- 84.Aimoto S., Elliott J.I., Van Nostrand W.E. and Smith S.O. (2010) Structural conversion of neurotoxic amyloid β(1-42) oligomers to fibrils. Nat. Struct. Mol. Biol. 17, 561–567 10.1038/nsmb.1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tycko R. (2004) Progress towards a molecular-level structural understanding of amyloid fibrils. Curr. Opin. Struct. Biol. 14, 96–103 10.1016/j.sbi.2003.12.002 [DOI] [PubMed] [Google Scholar]
- 86.Lansbury P.T. Jr, Costa P.R., Griffiths J.M., Simon E.J., Auger M., Halverson K.J.. et al. (1995) Structural model for the beta-amyloid fibril based on interstrand alignment of an antiparallel-sheet comprising a C-terminal peptide. Nat. Struct. Biol. 2, 990–998 10.1038/nsb1195-990 [DOI] [PubMed] [Google Scholar]
- 87.Burkoth T.S., Burkoth T.S., Benzinger T.L.S., Urban V., Morgan D.M., Gregory D.M.. et al. (2000) Structure of the β-Amyloid(10-35) Fibril. J. Am. Chem. Soc. 122, 7883–7889 10.1021/ja000645z [DOI] [Google Scholar]
- 88.Balbach J.J., Petkova A.T., Oyler N.A., Antzutkin O.N., Gordon D.J., Meredith S.C.. et al. (2002) Supramolecular structure in full-length Alzheimer’s beta-amyloid fibrils: evidence for a parallel beta-sheet organization from solid-state nuclear magnetic resonance. Biophys. J. 83, 1205–1216 10.1016/S0006-3495(02)75244-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Antzutkin O.N., Leapman R.D., Balbach J.J. and Tycko R. (2002) Supramolecular structural constraints on Alzheimer’s beta-amyloid fibrils from electron microscopy and solid-state nuclear magnetic resonance. Biochemistry 41, 15436–15450 10.1021/bi0204185 [DOI] [PubMed] [Google Scholar]
- 90.Makin O.S. and Serpell L.C. (2005) X-ray diffraction studies of amyloid structure. Methods Mol. Biol. 299, 67–80 [DOI] [PubMed] [Google Scholar]
- 91.Morris K.L. and Serpell L.C. (2012) X-ray fibre diffraction studies of amyloid fibrils. Methods Mol. Biol. 849, 121–135 10.1007/978-1-61779-551-0_9 [DOI] [PubMed] [Google Scholar]
- 92.Inouye H., Fraser P.E. and Kirschner D.A. (1993) Structure of beta-crystallite assemblies formed by Alzheimer beta-amyloid protein analogues: analysis by x-ray diffraction. Biophys. J. 64, 502–519 10.1016/S0006-3495(93)81393-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sunde M., Serpell L., Bartlam M., Fraser P.E., Pepys M. and Blake C.C. (1997) Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J. Mol. Biol. 273, 729–739 10.1006/jmbi.1997.1348 [DOI] [PubMed] [Google Scholar]
- 94.Jacques D.A. and Trewhella J. (2010) Small-angle scattering for structural biology expanding the frontier while avoiding the pitfalls. Protein Sci. 19, 657 10.1002/pro.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dahal E., Choi M., Alam N., Bhirde A.A., Beaucage S.L. and Badano A. (2017) Structural evaluation of an amyloid fibril model using small-angle x-ray scattering. Phys. Biol. 14, 046001 10.1088/1478-3975/aa776a [DOI] [PubMed] [Google Scholar]
- 96.Vestergaard B., Groenning M., Roessle M., Kastrup J.S., Van De Weert M., Flink J.M.. et al. (2007) A helical structural nucleus is the primary elongating unit of insulin amyloid fibrils. PLoS Biol. 5, e134 10.1371/journal.pbio.0050134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Oliveira C.L., Behrens M.A., Pedersen J.S., Erlacher K., Otzen D. and Pedersen J.S. (2009) A SAXS study of glucagon fibrillation. J. Mol. Biol. 387, 147–161 10.1016/j.jmb.2009.01.020 [DOI] [PubMed] [Google Scholar]
- 98.Chatani E., Inoue R., Imamura H., Sugiyama M., Kato M., Yamamoto M.. et al. (2015) Early aggregation preceding the nucleation of insulin amyloid fibrils as monitored by small angle X-ray scattering. Sci. Rep. 5, 15485 10.1038/srep15485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gremer L., Schölzel D., Schenk C., Reinartz E., Labahn J., Ravelli R.B.G.. et al. (2017) Fibril structure of amyloid-β(1-42) by cryo-electron microscopy. Science 358, 116–119 10.1126/science.aao2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fitzpatrick A.W.P., Falcon B., He S., Murzin A.G., Murshudov G., Garringer H.J.. et al. (2017) Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 547, 185–190 10.1038/nature23002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li Y., Zhao C., Luo F., Liu Z., Gui X., Luo Z.. et al. (2018) Amyloid fibril structure of α-synuclein determined by cryo-electron microscopy. Cell Res. 28, 897–903 10.1038/s41422-018-0075-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jiménez J.L., Tennent G., Pepys M. and Saibil H.R. (2001) Structural diversity of ex vivo amyloid fibrils studied by cryo-electron microscopy. J. Mol. Biol. 311, 241–247 10.1006/jmbi.2001.4863 [DOI] [PubMed] [Google Scholar]
- 103.Fitzpatrick A.W.P., Falcon B., He S., Murzin A.G., Murshudov G., Garringer H.J.. et al. (2017) Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 547, 185–190 10.1038/nature23002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Leung N., Nasr S.H. and Sethi S. (2012) How I treat amyloidosis: the importance of accurate diagnosis and amyloid typing. Blood 120, 3206–3213 10.1182/blood-2012-03-413682 [DOI] [PubMed] [Google Scholar]
- 105.Merlini G. and Bellotti V. (2003) Molecular mechanisms of amyloidosis. N. Engl. J. Med. 349, 583–596 10.1056/NEJMra023144 [DOI] [PubMed] [Google Scholar]
- 106.Sipe J.D. and Cohen A.S. (2000) Review: history of the amyloid fibril. J. Struct. Biol. 130, 88–98 10.1006/jsbi.2000.4221 [DOI] [PubMed] [Google Scholar]
- 107.Markowitz G.S. (2004) Dysproteinemia and the kidney. Adv. Anat. Pathol. 11, 49–63 10.1097/00125480-200401000-00005 [DOI] [PubMed] [Google Scholar]
- 108.Herrera G.A., Paul R., Turbat-Herrera E.A., Work J., Viale G., dell’Orto P.. et al. (1986) Ultrastructural immunolabeling in the diagnosis of light-chain-related renal disease. Pathol. Immunopathol. Res. 5, 170–187 10.1159/000157010 [DOI] [PubMed] [Google Scholar]
- 109.Sethi S., Theis J.D., Leung N., Dispenzieri A., Nasr S.H., Fidler M.E.. et al. (2010) Mass spectrometry-based proteomic diagnosis of renal immunoglobulin heavy chain amyloidosis. Clin. J. Am. Soc. Nephrol. 5, 2180–2187 10.2215/CJN.02890310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Crespo R., Villar-Alvarez E., Taboada P., Rocha F.A., Damas A.M. and Martins P.M. (2016) What can the kinetics of amyloid fibril formation tell about off-pathway aggregation? J. Biol. Chem. 291, 2018–2032 10.1074/jbc.M115.699348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Adamcik J. and Mezzenga R. (2012) Study of amyloid fibrils via atomic force microscopy. Curr. Opin. Colloid Interface Sci. 17, 369–376 10.1016/j.cocis.2012.08.001 [DOI] [Google Scholar]
- 112.Gosal W.S., Myers S.L., Radford S.E. and Thomson N.H. (2006) Amyloid under the atomic force microscope. Prot. Pept. Lett. 13, 261–270 10.2174/092986606775338498 [DOI] [PubMed] [Google Scholar]
- 113.Berne B.J. and Pecora R. (2000) Dynamic Light Scattering: With Applications to Chemistry, Biology, and Physics, Courier Dover Publications, Mineola [Google Scholar]
- 114.Streets A.M., Sourigues Y., Kopito R.R., Melki R. and Quake S.R. (2013) Simultaneous measurement of amyloid fibril formation by dynamic light scattering and fluorescence reveals complex aggregation kinetic. PLoS ONE 8, e54541 10.1371/journal.pone.0054541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hill S.E., Robinson J., Matthews G. and Muschol M. (2009) Amyloid protofibrils of lysozyme nucleate and grow via oligomer fusion. Biophys. J. 96, 3781–3790 10.1016/j.bpj.2009.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Georgalis Y., Starikov E.B., Hollenbach B., Lurz R., Scherzinger E., Saenger W.. et al. (1998) Huntingtin aggregation monitored by dynamic light scattering. Proc. Natl. Acad. Sci. U.S.A. 95, 6118–6121 10.1073/pnas.95.11.6118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lomakin A., Chung D.S., Benedek G.B., Kirschner D.A. and Teplow D.B. (1996) On the nucleation and growth of amyloid beta-protein fibrils: detection of nuclei and quantitation of rate constants. Proc. Natl. Acad. Sci. U.S.A. 93, 1125–1129 10.1073/pnas.93.3.1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tjernberg L.O., Pramanik A., Björling S., Thyberg P., Thyberg J., Nordstedt C.. et al. (1999) Amyloid beta-peptide polymerization studied using fluorescence correlation spectroscopy. Chem. Biol. 6, 53–62 10.1016/S1074-5521(99)80020-9 [DOI] [PubMed] [Google Scholar]
- 119.Sahoo B., Drombosky K.W. and Wetzel R. (2016) Fluorescence correlation spectroscopy: a tool to study protein oligomerization and aggregation in vitro and in vivo. Methods Mol. Biol. 1345, 67–87 10.1007/978-1-4939-2978-8_5 [DOI] [PubMed] [Google Scholar]
- 120.Erlich P., Dumestre-Pérard C., Ling W.L., Lemaire-Vieille C., Schoehn G., Arlaud G.J.. et al. (2010) Complement protein C1q forms a complex with cytotoxic prion protein oligomers. J. Biol. Chem. 285, 19267–19276 10.1074/jbc.M109.071860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Esparza T.J., Wildburger N.C., Jiang H., Gangolli M., Cairns N.J., Bateman R.J.. et al. (2016) Soluble amyloid-beta aggregates from human Alzheimer’s disease brains. Sci. Rep. 6, 38187 10.1038/srep38187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78, 1606–1619 10.1016/S0006-3495(00)76713-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mok Y.F. and Howlett G.J. (2006) Sedimentation velocity analysis of amyloid oligomers and fibrils. Methods Enzymol. 413, 199–217 10.1016/S0076-6879(06)13011-6 [DOI] [PubMed] [Google Scholar]
- 124.Pham C.L., Mok Y.F. and Howlett G.J. (2011) Sedimentation velocity analysis of amyloid fibrils. Methods Mol. Biol. 752, 179–196 10.1007/978-1-60327-223-0_12 [DOI] [PubMed] [Google Scholar]
- 125.Mok Y.F., Howlett G.J. and Griffin M.D. (2015) Sedimentation velocity analysis of the size distribution of amyloid oligomers and fibrils. Methods Enzymol. 562, 241–256 10.1016/bs.mie.2015.06.024 [DOI] [PubMed] [Google Scholar]
- 126.Jalandoni-Buan A.C., Decena-Soliven A.L.A., Cao E.P., Barraquio V.L. and Barraquio W.L. (2010) Characterization and identification of congo red decolorizing bacteria from monocultures and consortia. Philippine J. Sci. 139, 71–78 [Google Scholar]
- 127.Cooksey C.J. (2014) Quirks of dye nomenclature. 2. Congo red. Biotech. Histochem. 89, 384–387 10.3109/10520295.2014.880513 [DOI] [PubMed] [Google Scholar]
- 128.Reid T.M., Morton K.C., Wang C.Y. and King C.M. (1983) Conversion of Congo red and 2-azoxyfluorene to mutagens following in vitro reduction by whole-cell rat cecal bacteria. Mutat. Res. 117, 105–112 10.1016/0165-1218(83)90157-X [DOI] [PubMed] [Google Scholar]
- 129.Horobin R.W. and Kiernan J.A. (2002) Conn’s Biological Stains: A Handbook of Dyes, Stains and Fluorochromes for Use in Biology and Medicine, 10th edition, pp. 132–134, BIOS [Google Scholar]
- 130.Steensma D.P. (2001) “Congo” red: out of Africa? Arch. Pathol. Lab. Med. 125, 250–252 [DOI] [PubMed] [Google Scholar]
- 131.Alturkistani H.A., Tashkandi F.M. and Mohammedsaleh Z.M. (2016) Histological stains: a literature review and case study. Glob. J. Health. Sci. 8, 72–79 10.5539/gjhs.v8n3p72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Puchtler H., Sweat F. and Levine M. (1962) On the binding of Congo red by amyloid. J. Histochem. Cytochem. 10, 355–364 10.1177/10.3.355 [DOI] [Google Scholar]
- 133.Schulz H. (1886) Über das Congorot als reagens auf freie säure [Concerning Congo red as a reagent for free acids]. Centralblatt für die Medicinischen Wissenschaften, 29–30 [Google Scholar]
- 134.Bennhold H. (1922) Eine spezifische amyloidfärbung mit Kongorot [Specific staining of amyloid with Congo red]. Münchener Medizinische Wochenschrifte 69, 1537–1538 [Google Scholar]
- 135.Bennhold H. (1923) Excretion of intravenously injected Congo red in different diseases, especially amyloidosis [in German]. Deutsches Archiv für Klinische Medizin 142, 32–46 [Google Scholar]
- 136.Fernandez-Flores A. (2011) A review of amyloid staining: methods and artifacts. Biotech. Histochem. 86, 293–301 10.3109/10520291003784493 [DOI] [PubMed] [Google Scholar]
- 137.Friedman M.M. and Auerbach O. (1935) An improved Congo Red test for amyloidosis. J. Lab. Clin. Med. 21, 93 [Google Scholar]
- 138.Taran A. and Eckstein A. (1942) The standardization of the Congo Red test for amyloidosis. Am. J. M. Sc. 203, 246 10.1097/00000441-194202000-00011 [DOI] [Google Scholar]
- 139.Lipstein S. (1938) An evaluation of the Congo Red test for amyloidosis. Am. J. M. Sc. 195, 205 10.1097/00000441-193802000-00009 [DOI] [Google Scholar]
- 140.Stemmerman M. and Auerbach O. (1944) The value and limitations of the Congo Red test for amyloidosis. Am. J. M. Sc. 208, 305 10.1097/00000441-194409000-00004 [DOI] [Google Scholar]
- 141.Unger P.N., Zuckerbrod M., Beck G.J., Steele J.M. and Porosowska Y. (1948) Study of the disappearance of Congo red from the blood of non-amyloid subjects and patients with amyloidosis. J. Clin. Invest. 27, 111–118 10.1172/JCI101914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Alder H. and Reimann F. (1925) Beitrag zur Funktionsprufung des retikuloendothelialen Apparates. Z. Exp. Med. 47, 617–633 10.1007/BF02608745 [DOI] [Google Scholar]
- 143.Ouchi E., Nomura N., Watabe S., Seiji K. and Sato J. (1976) Clinical significance of Congo red test. Tohoku J. Exp. Med. 118, 191–198 10.1620/tjem.118.Suppl_191 [DOI] [PubMed] [Google Scholar]
- 144.Ouchi E., Miura K. and Yamagata S. (1973) Reevaluation of Congo red test as reticuloendothelial system function test using tritiated Congo red autoradiography. Tohoku J. Exp. Med. 109, 251–259 10.1620/tjem.109.251 [DOI] [PubMed] [Google Scholar]
- 145.Miura K. and Ouchi E. (1974) Congo red test. Recent Adv. Res. 13, 123–133 [Google Scholar]
- 146.Ouchi E., Miura K. and Yamagata S. (1975) Reticuloendothelial system function test using fall-labeled aggregated albumin. Tohoku J. exp. Med. 116, 141–147 10.1620/tjem.116.141 [DOI] [PubMed] [Google Scholar]
- 147.Howie A.J. and Brewer D.B. (2009) Optical properties of amyloid stained by Congo red: History and mechanisms. Micron 40, 285–301 10.1016/j.micron.2008.10.002 [DOI] [PubMed] [Google Scholar]
- 148.Divry P. (1927) Etude histochimique des plaques. J. Belge de Neurologie et de Psychiatrie 27, 643–657 [Google Scholar]
- 149.Divry P. and Florkin M. (1927) Sur les propriétés optiques de l’amyloïde. Comptes Rendus des Sèances de la Sociètè de Biologie et de ses Filiales 97, 1808–1810 [Google Scholar]
- 150.Ladewig P. (1945) Double-refringence of the amyloid-Congo-red-complex in histological sections. Nature 156, 81–82 10.1038/156081a0 [DOI] [Google Scholar]
- 151.Cooper J.H. (1974) Selective amyloid staining as a function of amyloid composition and structure. Lab. Invest. 31, 232 [PubMed] [Google Scholar]
- 152.Taylor D.L., Allen R.D. and Benditt E.P. (1974) Determination of the polarization optical properties of the amyloid-Congo red complex by phase modulation microspectrophotometry. J. Histochem. Cytochem. 22, 1105–1112 10.1177/22.12.1105 [DOI] [PubMed] [Google Scholar]
- 153.Diezel P.B. and Pfleiderer A. (1959) Histochemische und polarisationsoptische Untersuchungen am Amyloid (histochemical and polarising optical investigations of amyloid). Virch. Arch. Pathol. Anat. 332, 552–567 10.1007/BF02438761 [DOI] [PubMed] [Google Scholar]
- 154.Wolman M. and Bubis J.J. (1965) The cause of the green polarization color of amyloid stained with Congo red. Histochemistry 4, 351–356 10.1007/BF00306246 [DOI] [PubMed] [Google Scholar]
- 155.Benditt E.P., Eriksen N. and Berglund C. (1970) Congo red dichroism with dispersed amyloid fibrils, an extrinsic Cotton effect. Proc. Natl. Acad. Sci. U.S.A. 66, 1044–1051 10.1073/pnas.66.4.1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Attwood H.D., Price C.G. and Riddell R.J. (1972) Primary diffuse tracheobronchial amyloidosis. Thorax 27, 620–624 10.1136/thx.27.5.620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Howie A.J. and Owen-Casey M.P. (2012) Apple-green birefringence’ of amyloid stained by Congo red. Kidney Int. 82, 114 10.1038/ki.2012.89 [DOI] [PubMed] [Google Scholar]
- 158.Howie A.J. (2015) “Green (or apple-green) birefringence” of Congo red-stained amyloid. Amyloid 22, 205–206 10.3109/13506129.2015.1054026 [DOI] [PubMed] [Google Scholar]
- 159.Linke R.P. (2000) Highly sensitive diagnosis of amyloid and various amyloid syndromes using Congo red fluorescence. Virch. Arch. 436, 439–448 10.1007/s004280050471 [DOI] [PubMed] [Google Scholar]
- 160.Wälchli O. (1945) Beitrag zur Kenntnis des Aufbaues der Zellulose. Schweiz. Arch. Wiss. Technik. 11, 181–189 [Google Scholar]
- 161.Klunk W.E., Pettegrew J.W. and Abraham D.J. (1989) Quantitative evaluation of congo red binding to amyloid-like proteins with a betapleated sheet conformation. J. Histochem. Cytochem. 37, 1273–1281 10.1177/37.8.2666510 [DOI] [PubMed] [Google Scholar]
- 162.Wu C., Wang Z., Lei H., Zhang W. and Duan Y. (2007) Dual binding modes of Congo red to amyloid protofibril surface observed inmolecular dynamics simulations. J. Am. Chem. Soc. 129, 1225–1232 10.1021/ja0662772 [DOI] [PubMed] [Google Scholar]
- 163.Klunk W.E., Debnath M.L. and Pettegrew J.W. (1994) Developmentof small molecule probes for the beta-amyloid protein of Alz-heimer’s disease. Neurobiol. Aging 15, 691–698 10.1016/0197-4580(94)90050-7 [DOI] [PubMed] [Google Scholar]
- 164.Ma B. and Nussinov R. (2002) Stabilities and conformations of Alz-heimer’s beta-amyloid peptide oligomers (Abeta 16-22, Abeta16-35, and Abeta 10-35): sequence effects. Proc. Natl. Acad. Sci. U.S.A. 99, 14126–14131 10.1073/pnas.212206899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Childers W.S., Mehta A.K., Lu K. and Lynn D.G. (2009) Templating molecular arrays in amyloid’s cross-beta grooves. J. Am. Chem. Soc. 131, 10165–10172 10.1021/ja902332s [DOI] [PubMed] [Google Scholar]
- 166.Reinke A.A. and Gestwick J.E. (2011) Insight into amyloid structure using chemical probes. Chem. Biol. Drug Des. 77, 399–411 10.1111/j.1747-0285.2011.01110.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Khurana R., Uversky V.N., Nielsen L. and Fink A.L. (2001) Is Congo red an amyloid-specific dye? J. Biol. Chem. 276, 22715–22721 10.1074/jbc.M011499200 [DOI] [PubMed] [Google Scholar]
- 168.Howie A.J., Brewer D.B., Howell D. and Jones A.P. (2008) Physical basis of colors seen in Congo red-stained amyloid in polarized light. Lab. Invest. 88, 232–242 10.1038/labinvest.3700714 [DOI] [PubMed] [Google Scholar]
- 169.Born M. and Wolf E. (1999) Principles of Optics. Electromagnetic Theory of Propagation, Interference and Diffraction of Light 49, 7th edn, pp. 95–103, Cambridge University Press, Cambridge, U.K. [Google Scholar]
- 170.Neubert H. (1925) Über Doppelbrechung und Dichroismus gefärbter Gele. Kolloidchem. Beiheft Supp. 20, 244–272 10.1007/BF02558506 [DOI] [Google Scholar]
- 171.Mera S.L. and Davies J.D. (1984) Differential Congo red staining: the effects of pH, non-aqueous solvents and the substrate. Histochem. J. 16, 195–210 10.1007/BF01003549 [DOI] [PubMed] [Google Scholar]
- 172.Horobin R.W. and Kiernan J.A. (2002) Conn’s Biological Stains, 10th edn, pp. 132–134, BIOS Scientific, Oxford [Google Scholar]
- 173.Amelin V.G. and Tret’yakov A.V. (2003) Adsorption-bonded azo reagents in chemical tests based on the principles of precipitation paper chromatography. J. Anal. Chem. 58, 740–747 10.1023/A:1025031510057 [DOI] [Google Scholar]
- 174.Horobin R.W. and James N.T. (1970) The staining of elastic fibres with Direct Blue 152. A general hypotheses for the staining of elastic fibres. Histochemie 22, 324–336 [DOI] [PubMed] [Google Scholar]
- 175.Horobin R.W. and Flemming L. (1980) Structure-staining relationships in histochemistry and biological staining. II. Mechanistic and practical aspects of the staining of elastic fibres. J. Microsc. 119, 357–372 10.1111/j.1365-2818.1980.tb04107.x [DOI] [PubMed] [Google Scholar]
- 176.Lendrum A.C., Slidders W. and Fraser D.S. (1972) Renal hyalin. A study of amyloidosis and diabetic fibrinoid vasculosis with new staining methods. J. Din. Path. 25, 373–396 10.1136/jcp.25.5.373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Davies I.D. and Mera S.L. (1983) Congo “blue” staining: a histochemical problem. Bristol Med. Chi. J. 98, 90 [Google Scholar]
- 178.Kulonen E. and Pikkarainen J. (1970) Comparative studies of the chemistry and chain structure of collagen. In Chemistry and Molecular Biology of the Intercellular Matrix (Balazs E., ed.), pp. 81–97, Academic Press, London [Google Scholar]
- 179.Bayer-Garner I.B. and Smoller B.R. (2002) AL amyloidosis is not present as an incidental finding in cutaneous biopsies of patients with multiple myeloma. Clin. Exp. Dermatol. 27, 240–242 10.1046/j.1365-2230.2002.01022.x [DOI] [PubMed] [Google Scholar]
- 180.Fernandez-Flores A. (2009) Positive staining with Congo red in tissues with heat artifact due to cautery. Rom. J. Morphol. Embryol. 50, 203–206 [PubMed] [Google Scholar]
- 181.Elghetany M.T., Saleem A. and Barr K. (1989) The Congo red stain revisited. Ann. Clin. Lab. Sci. 19, 190–195 [PubMed] [Google Scholar]
- 182.Bowen K., Shah N. and Lewin M. (2012) AL-amyloidosis presenting with negative Congo red staining in the setting of high clinical suspicion: a case report. Case Rep. Nephrol. 2012, 593460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Bardarov S., Michael C.W., Pu R.T. and Pang Y. (2009) Computer-assisted image analysis of amyloid deposits in abdominal fat pad aspiration biopsies. Diagn. Cytopathol. 37, 30–35 10.1002/dc.20948 [DOI] [PubMed] [Google Scholar]
- 184.Halloush R., Lavrovskaya E., Mody D., Lager D. and Truong L.D. (2009) Diagnosis and typing of systemic amyloidosis: The role of abdominal fat pad fine needle aspiration. Cyto J. 6, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Guy C.D. and Jones C.K. (2001) Abdominal fat pad aspiration biopsy for tissue confirmation of systemic amyloidosis: Specificity, positive predictive value, and diagnostic pitfalls. Diagn. Cytopathol. 24, 181–185 10.1002/1097-0339(200103)24:3%3c181::AID-DC1037%3e3.0.CO;2-D [DOI] [PubMed] [Google Scholar]
- 186.Westermark P. and Stenkvist B. (1973) A new method for the diagnosis of systemic amyloidosis. Arch. Intern. Med. 132, 522–523 10.1001/archinte.1973.03650100040007 [DOI] [PubMed] [Google Scholar]
- 187.Westermark P. (1995) Diagnosing amyloidosis. Scand J. Rheumatol. 24, 327–329 10.3109/03009749509095175 [DOI] [PubMed] [Google Scholar]
- 188.Devata S., Hari P., Markelova N., Li R., Komorowski R. and Shidham V.B. (2011) Detection of amyloid in abdominal fat pad aspirates in early amyloidosis: role of electron microscopy and Congo red stained cell block sections. Cyto J. 8, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Klatskin G. (1969) Nonspecific green birefringence in Congo red-stained tissues. Am. J. Pathol. 56, 1–13 [PMC free article] [PubMed] [Google Scholar]
- 190.Bradley-Whitman M.A., Timmons M.D., Beckett T.L., Murphy M.P., Lynn B.C. and Lovell M.A. (2014) Nucleic acid oxidation: an early feature of Alzheimer’s disease. J. Neurochem. 128, 294–304 10.1111/jnc.12444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Han S., Kollmer M., Markx D., Claus S., Walther P. and Fandrich M. (2017) Amyloid plaque structure and cell surface interactions of beta-amyloid fibrils revealed by electron tomography. Sci. Rep. 7, 43577 10.1038/srep43577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Maynard C.J., Bush A.I., Masters C.L., Cappai R. and Li Q.X. (2005) Metals and amyloid-beta in Alzheimer’s disease. Int. J. Exp. Pathol. 86, 147–159 10.1111/j.0959-9673.2005.00434.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Alexandrescu A.T. (2005) Amyloid accomplices and enforcers. Protein Sci. 14, 1–12 10.1110/ps.04887005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Clement C.G. and Truong L.D. (2014) An evaluation of Congo red fluorescence for the diagnosis of amyloidosis. Hum. Pathol. 45, 1766–1772 10.1016/j.humpath.2014.04.016 [DOI] [PubMed] [Google Scholar]
- 195.Fernandez-Flores A. (2010) Comparative study of Congo red fluorescence and immunohistochemistry in cutaneous amyloidosis. Rom. J. Morphol. Embryol. 51, 683–686 [PubMed] [Google Scholar]
- 196.Davidson B., Tooney N. and Fasman G.D. (1966) The optical rotatory dispersion of the beta-structure of poly-L-lysine and poly-L-serine. Biochem. Biophys. Res. Commun. 23, 156–162 10.1016/0006-291X(66)90521-3 [DOI] [PubMed] [Google Scholar]
- 197.Small E.W., Fanconi B. and Peticolas W.L. (1970) Raman spectra and photon dispersion of polyglycine. J. Chem. Phys. 52, 4369–4379 10.1063/1.1673659 [DOI] [PubMed] [Google Scholar]
- 198.Glenner G.G., Eanes E.D. and Page D.L. (1972) The relation of the properties of Congo red-stained amyloid fibrils to the beta-conformation. Histochem. Cytochem. 20, 821–826 10.1177/20.10.821 [DOI] [PubMed] [Google Scholar]
- 199.Bcnditt E.P., Eriksen N. and Bergtund C. (1970) Congo red dichroism with dispersed amyloid fibrits, an extrinsic Cotton effect. Proc. Natl. Acad. Sci. U.S.A. 66, 1044–1051 10.1073/pnas.66.4.1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Inouye H., Nguyen J., Fraser P., Shinchuk L., Packard A. and Kirschner D. (2000) Histidine residues underlie Congo red binding to A beta analogs. Amyloid 7, 179–188 10.3109/13506120009146832 [DOI] [PubMed] [Google Scholar]
- 201.Reichhardt C., Jacobson A.N., Maher M.C., Uang J., McCrate O.A., Eckart M.. et al. (2015) Congo red interactions with curli-producing E. coli and native curli amyloid fibers. PLoS ONE 10, e0140388 10.1371/journal.pone.0140388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Caughey B., Brown K., Raymond G.J., Katzenstein G.E. and Thresher W. (1994) Binding of the protease-sensitive form of PrP (prion protein) to sulfated glycosaminoglycan and congo red [corrected]. J. Virol. 68, 2135–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Heiser V., Scherzinger E., Boeddrich A., Nordhoff E., Lurz R., Schugardt N.. et al. (2000) Inhibition of huntingtin fibrillogenesis by specific antibodies and small molecules: implications for Huntington’s disease therapy. Proc. Natl. Acad. Sci. U.S.A. 97, 6739–6744 10.1073/pnas.110138997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Sánchez I., Mahlke C. and Yuan J. (2003) Pivotal role of oligomerization in expanded polyglutamine neurodegenerative disorders. Nature 421, 373–379 10.1038/nature01301 [DOI] [PubMed] [Google Scholar]
- 205.Podlisny M.B., Walsh D.M., Amarante P., Ostaszewski B.L., Stimson E.R., Maggio J.E.. et al. (1998) Oligomerization of endogeneous and synthetic amyloid beta-protein at nanomolar levels in cell culture and stabilization of monomer by congo red. Biochemistry 37, 3602–3611 10.1021/bi972029u [DOI] [PubMed] [Google Scholar]
- 206.Turnell W.G. and Finch J.T. (1992) Binding of the dye congo red to the amyloid protein pig insulin reveals a novel homology amongst amyloid-forming peptide sequences. J. Mol. Biol. 227, 1205–1223 10.1016/0022-2836(92)90532-O [DOI] [PubMed] [Google Scholar]
- 207.Lorenzo A. and Yankner B.A. (1994) Beta-amyloid neurotoxicity requires fibril formation and is inhibited by congo red. Proc. Natl. Acad. Sci. U.S.A. 91, 12243–12247 10.1073/pnas.91.25.12243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Frid P., Anisimov S.V. and Popovic N. (2007) Congo red and protein aggregation in neurodegenerative diseases. Brain Res. Rev. 53, 135–160 10.1016/j.brainresrev.2006.08.001 [DOI] [PubMed] [Google Scholar]