Abstract
Plenty of studies have investigated the effect of methionine synthase (MTR) A2756G polymorphism on risk of developing pediatric acute lymphoblastic leukemia (ALL), but the available results were inconsistent. Therefore, a meta-analysis was conducted to derive a more precise estimation of the association between MTR A2756G polymorphism and genetic susceptibility to pediatric ALL. The PubMed, Embase, Google Scholar, Web of Science, ScienceDirect, Wanfang Databases and China National Knowledge Infrastructure were systematically searched to identify all the previous published studies exploring the relationship between MTR A2756G polymorphism and pediatric ALL risk. Odds ratios (ORs) and 95% confidence intervals (CIs) were applied to evaluate the strength of association. Sensitivity analysis and publication bias were also systematically assessed. This meta-analysis finally included ten available studies with 3224 ALL cases and 4077 matched controls. The results showed that there was significant association between MTR A2756G polymorphism and risk of pediatric ALL in overall population (AG vs. AA: OR = 1.13, 95%CI = 1.02–1.26, P = 0.02; AG+GG vs. AA: OR = 1.13, 95%CI = 1.02–1.25, P = 0.01; G allele vs. A allele: OR = 1.10, 95%CI = 1.01–1.20, P = 0.03). In the stratification analyses by ethnicity, quality score and control source, significant association was found in Caucasians, population-based designed studies and studies assigned as high quality. In conclusion, this meta-analysis suggests that MTR A2756G polymorphism may influence the development risk of pediatric ALL in Caucasians. Future large scale and well-designed studies are required to validate our findings.
Keywords: leukaemia, methionine synthase, meta analysis, single nucleotide polymorphisms, susceptibility
Introduction
Acute lymphoblastic leukemia (ALL) is the most common childhood cancer, which accounts for 30% of all malignancy diagnosed in children and 80% of pediatric leukemia [1]. However, the etiology and biological mechanisms underlying ALL development have yet to be elucidated [2–4]. As for many cancers, the interactions between susceptibility genes and environmental factors are likely to implicate in the development of ALL. Epidemiological studies suggest that the imbalance of folate metabolism may be involved in predisposition to carcinogenesis, which is based on its involvement in both DNA biosynthesis and DNA methylation [5]. The low availability of folate causes uracil misincorporation into DNA replication, which leads to double-strand breakage and chromosomal deletion [6,7]. Moreover, gene-specific hypermethylation and global DNA hypomethylation are two of the most frequently observed altered DNA methylation patterns in tumors [8,9]. Accumulating studies have reported that polymorphisms in genes encoding folate-metabolizing enzymes disturb the balance of folate metabolism and have been associated with an altered predisposition to cancer [10–12].
The methionine synthase (MTR) plays a crucial role in the folate metabolic network. It is a vitamin B12-dependent enzyme, which remethylates homocysteine to methionine and simultaneously generates tetrahydrofolate by removing methyl group from 5-methyltetrahydrofolate. MTR helps to maintain the levels of adequate intracellular folate and normal homocysteine and methionine concentrations, which are used for proper DNA methylation or other methylation processes [13]. The MTR gene is mapped on 1q43, and the extensively investigated A2756G polymorphism (rs1805087) leads to a change from aspartate to glycine at codon 919 (D919G), resulting in reduced enzyme activity [14]. It has been reported that this polymorphism can increase homocysteine levels through suppressing methionine metabolism and consequentially can lead to DNA hypomethylation and promote tumorigenesis [15,16]. Plenty of studies have found that MTR A2756G polymorphism has been linked to various cancer, such as prostate cancer, retinoblastoma and lymphoma [17–19]. A number of studies have attempted to explore the effect of MTR A2756G polymorphism on pediatric ALL risk, yet the reported results are inconsistent. The inconsistencies of results might be attributed to some variables in study of population like genetic backgrounds difference and relatively small size of sampling in single study. Therefore, a meta-analysis was conducted to derive a more precise estimation of the association between MTR A2756G polymorphism and genetic susceptibility to pediatric ALL.
Methods
Identification and eligibility of relevant studies
The present study was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines. The PubMed, Embase, Google Scholar, Web of Science, ScienceDirect, Wanfang Databases and China National Knowledge Infrastructure were systematically searched to identify the published case–control studies on the relationship between MTR A2756G polymorphism and pediatric ALL risk with the following subject terms or keywords: ‘methionine synthase’ or ‘MTR’ or ‘MS’ or ‘5-methyltetrahydrofolate-homocysteine methyltransferase’, ‘polymorphism’ or ‘variation’ or ‘variant’ or ‘mutation’, ‘acute lymphoblastic leukemia’ or ‘leukemia’ or ‘ALL’, and ‘pediatric’ or ‘children’ or ‘childhood’. The latest web-based literature search was conducted on May 20, 2018 and no language restriction was applied. In addition, the reference lists in the primary studies and review articles were also examined manually to identify additional potentially relevant studies.
Inclusion criteria
The following inclusion criteria were applied for selecting literature: (1) confirmed diagnosis for the pediatric ALL cases; (2) case–control study; (3) available genotypes distribution data for both patients and control populations; (4) genotypes distribution of the control group must be in consistent with Hardy–Weinberg equilibrium (HWE). The case reports, letters, commentary and review articles were excluded. If the same or overlapping patient population was reported by several articles, only the most recent or largest sample size was chose in this meta-analysis.
Quality assessment
The quality assessment of included studies was preformed independently by two authors according to the Newcastle-Ottawa Scale (NOS). Discrepancies were adjudicated by the third investigator until consensus was achieved. The NOS is a tool used for assessing the quality of non-randomized studies included in a systematic review and meta-analysis [20]. Using the tool, each study is judged on eight items, categorized into three groups: the study group selection, the comparability of the groups, and the ascertainment of exposure. Stars are awarded such that the highest quality studies are awarded up to nine stars. In this meta-analysis, studies with more than six stars were identified as high quality.
Data collection
From each of the included articles, the following data were collected independently by two authors: the name of first author, year of publication, country and ethnicity of participants, source of controls, total number of cases and controls, genotyping methods, genotyping data of the MTR A2756G polymorphism in cases and controls. Any disagreement was resolved by re-evaluation of the originally included studies.
Statistical analysis
For the controls of each study, the χ2-test was adopted to check HWE of genotypes distribution frequencies, with P<0.05 indicating deviation from HWE. The strength of association between MTR A2756G polymorphism and pediatric ALL risk was assessed by calculating pooled ORs and corresponding 95% CIs under the allele model (G allele vs. A allele), heterozygote model (AG vs. AA), homozygote model (GG vs. AA), recessive model (GG vs. AA+AG) and dominant model (AG+GG vs. AA), respectively. The significance of the overall ORs was determined by the Z-test. The χ2-test based Q-test was performed to estimate the heterogeneity across the eligible studies, and the heterogeneity was further quantified with I2-test. When P>0.05, showing that the effects were assumed to be homogeneous, the fixed-effects model (Mantel–Haenszel method) was selected to calculate the ORs, alternatively, the random-effects model (DerSimonian–Laird method) was used [21]. Stratification analyses were performed by ethnicity (Asian and Caucasian), control source (hospital-based and population-based) and NOS score (low quality and high quality). Sensitivity analysis was conducted by excluding one study each time and recalculating the ORs with corresponding 95%CIs to assess the stability of combined results. The qualitative funnel plot was employed to assess publication bias by calculating the standard error of log(OR) of each study plotted against its log(OR), and the funnel plot asymmetry was further assessed using quantitative Egger’s test [22]. All the statistical tests were done with RevMan v5.3 (The Cochrane Collaboration, Oxford, U.K.) and STATA v12.0 (Stata Corporation, College Station, TX). All P values were two-sided, and P<0.05 was considered statistically significant.
Results
Characteristics of included studies
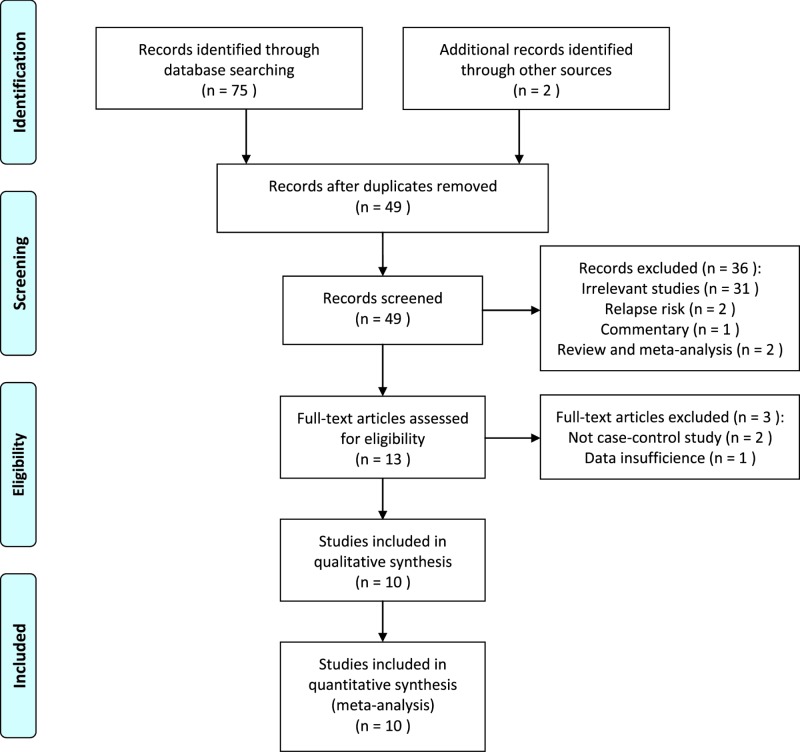
The flow diagram of literature selection was presented in Figure 1. After duplicates removed, 49 relevant articles were identified based on an extensive search. After glancing the titles and abstracts, 36 irrelevant studies and reviews were excluded and three full-text articles were excluded during the further assessment. Finally, a total of ten case–control studies met our inclusion criteria, including 3224 ALL cases and 4077 matched controls [23–32]. Table 1 presented the main characteristics of eligible studies. Of these included studies, there were nine studies carried out among Caucasians [23–31], and one study among Asian descents [32]. When classified by the source of controls, one study was hospital-based [26] and nine were population-based designed [23–25,27–32]. Three studies were divided into low quality and seven were assigned as high quality. The genotypes distribution frequencies among the controls were in agreement with HWE for all included studies. The genotyping data of MTR A2756G polymorphism in cases and controls from the individual studies were shown in Table 2.
Figure 1. Flow diagram of study selection process.
Table 1. Main characteristics of studies included in the meta-analysis.
| Reference | Year | Country | Ethnicity | Control source | Genotyping methods | Quality score |
|---|---|---|---|---|---|---|
| de Jonge et al. [23] | 2009 | Netherlands | Caucasian | PB | PCR-RFLP | 5 |
| Gast et al. [24] | 2007 | Germany | Caucasian | PB | Allelic discrimination | 8 |
| Kamel et al. [25] | 2007 | Egypt | Caucasian | PB | PCR-RFLP | 6 |
| Lautner-Csorba et al. [26] | 2013 | Hungary | Caucasian | HB | MassARRAY | 7 |
| Lightfoot et al. [27] | 2010 | U.K. | Caucasian | PB | TaqMan Assay | 9 |
| Metayer et al. [28] | 2011 | U.S.A. | Caucasian | PB | GoldenGate Assay | 8 |
| Milne et al. [29] | 2015 | Australia | Caucasian | PB | PCR-RFLP | 9 |
| Petra et al. [30] | 2007 | Slovenia | Caucasian | PB | PCR-RFLP | 5 |
| Rahimi et al. [31] | 2012 | Iran | Caucasian | PB | PCR-RFLP | 7 |
| Nikbakht et al. [32] | 2012 | India | Asian | PB | PCR-RFLP | 9 |
Abbreviations: HB, hospital-based; PB, population-based; PCR, polymerase chain reaction; RFLP, restriction fragment length polymorphism.
Table 2. Genotypes distribution of MTR A2756G polymorphism in cases and controls.
| Reference | Sample size | Case group | Control group | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | AA | AG | GG | A | G | AA | AG | GG | A | G | PHWE | |
| de Jonge et al. [23] | 245 | 489 | 162 | 74 | 9 | 398 | 92 | 340 | 137 | 12 | 817 | 161 | 0.68 |
| Gast et al. [24] | 446 | 547 | 280 | 153 | 13 | 713 | 179 | 375 | 151 | 21 | 901 | 193 | 0.24 |
| Kamel et al. [25] | 87 | 306 | 55 | 29 | 3 | 139 | 35 | 194 | 97 | 15 | 485 | 127 | 0.53 |
| Lautner-Csorba et al. [26] | 543 | 529 | 344 | 175 | 24 | 863 | 223 | 341 | 163 | 25 | 845 | 213 | 0.34 |
| Lightfoot et al. [27] | 870 | 759 | 531 | 288 | 51 | 1350 | 390 | 510 | 223 | 26 | 1243 | 275 | 0.79 |
| Metayer et al. [28] | 376 | 447 | 237 | 123 | 16 | 597 | 155 | 292 | 137 | 18 | 721 | 173 | 0.70 |
| Milne et al. [29] | 391 | 514 | 251 | 130 | 10 | 632 | 150 | 337 | 158 | 19 | 832 | 196 | 0.93 |
| Petra et al. [30] | 68 | 258 | 51 | 16 | 1 | 118 | 18 | 161 | 82 | 15 | 404 | 112 | 0.30 |
| Rahimi et al. [31] | 73 | 128 | 42 | 26 | 5 | 110 | 36 | 75 | 47 | 6 | 197 | 59 | 0.69 |
| Nikbakht et al. [32] | 125 | 100 | 74 | 44 | 7 | 192 | 58 | 58 | 35 | 7 | 151 | 49 | 0.59 |
Abbreviations: HWE, Hardy–Weinberg equilibrium; MTR, methionine synthase.
Quantitative data synthesis
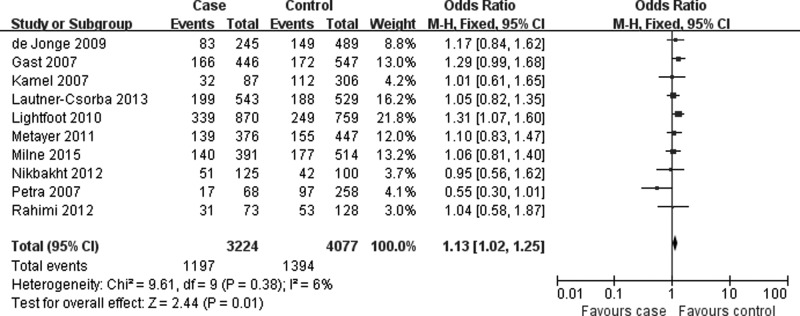
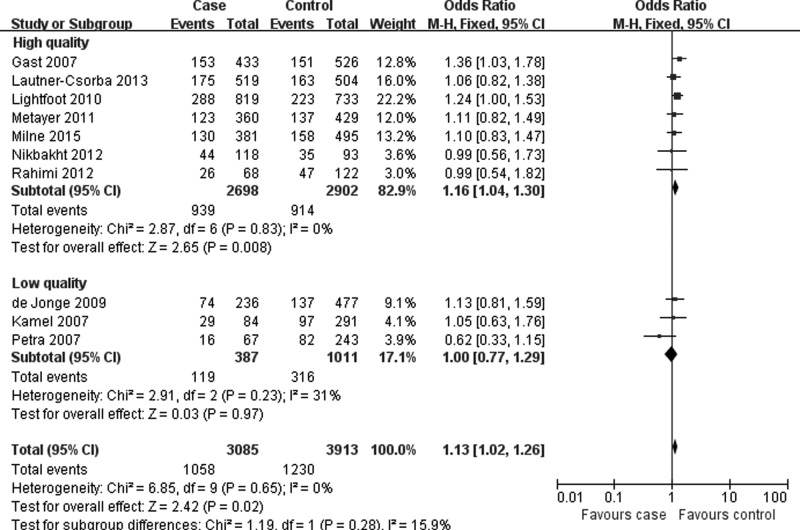
Table 3 listed the main results of quantitative synthesis. When all eligible studies were pooled together, the results found that there was statistically significant association between MTR A2756G polymorphism and risk of pediatric ALL under three genetic models (AG vs. AA: OR = 1.13, 95%CI = 1.02–1.26, P = 0.02; AG+GG vs. AA: OR = 1.13, 95%CI = 1.02–1.25, P = 0.01; G allele vs. A allele: OR = 1.10, 95%CI = 1.01–1.20, P = 0.03) (Figure 2 and Table 3). In the stratification analyses according to ethnicity, control source and quality score, significant association was also found in Caucasians, population-based designed studies, and studies assigned as high quality (Figures 3 and 4, Table 3).
Table 3. Results of meta-analysis for MTR A2756G polymorphism with pediatric ALL risk.
| Variables | No. | Sample size | AG vs. AA | GG vs. AA | GG vs. AA+AG | AG+GG vs. AA | G Allele vs. A Allele | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | OR (95% CI) | P | Ph* | OR (95% CI) | P | Ph* | OR (95% CI) | P | Ph* | OR (95% CI) | P | Ph* | OR (95% CI) | P | Ph* | ||
| Overall | 10 | 3224 | 4077 | 1.13 (1.02–1.26) | 0.02 | 0.65 | 1.10 (0.86–1.39) | 0.45 | 0.28 | 1.05 (0.83–1.34) | 0.66 | 0.33 | 1.13 (1.02–1.25) | 0.01 | 0.38 | 1.10 (1.01–1.20) | 0.03 | 0.18 |
| Ethnicity | ||||||||||||||||||
| Caucasian | 9 | 3099 | 3977 | 1.14 (1.03–1.27) | 0.01 | 0.58 | 1.11 (0.87–1.42) | 0.39 | 0.23 | 1.07 (0.84–1.37) | 0.58 | 0.27 | 1.14 (1.03–1.26) | 0.01 | 0.33 | 1.11 (1.02–1.21) | 0.02 | 0.15 |
| Asian | 1 | 125 | 100 | 0.99 (0.56–1.73) | 0.96 | 0.78 (0.26–2.36) | 0.66 | 0.79 (0.27–2.33) | 0.67 | 0.95 (0.56–1.62) | 0.86 | 0.93 (0.60–1.44) | 0.75 | |||||
| Control source | ||||||||||||||||||
| PB | 9 | 2681 | 3548 | 1.15 (1.03–1.28) | 0.02 | 0.58 | 1.13 (0.87–1.47) | 0.37 | 0.23 | 1.08 (0.83–1.41) | 0.55 | 0.27 | 1.15 (1.03–1.28) | 0.01 | 0.33 | 1.11 (1.02–1.22) | 0.02 | 0.15 |
| HB | 1 | 543 | 529 | 1.06 (0.82–1.38) | 0.64 | 0.95 (0.53–1.70) | 0.87 | 0.93 (0.53–1.65) | 0.81 | 1.05 (0.82–1.35) | 0.71 | 1.03 (0.83–1.27) | 0.82 | |||||
| Quality | ||||||||||||||||||
| High | 7 | 2824 | 3024 | 1.16 (1.04–1.30) | 0.008 | 0.83 | 1.14 (0.88–1.48) | 0.32 | 0.29 | 1.09 (0.84–1.41) | 0.52 | 0.31 | 1.16 (1.04–1.29) | 0.006 | 0.71 | 1.12 (1.03–1.23) | 0.01 | 0.49 |
| Low | 3 | 400 | 1053 | 1.00 (0.77–1.29) | 0.97 | 0.23 | 0.86 (0.45–1.64) | 0.65 | 0.16 | 0.87 (0.45–1.66) | 0.67 | 0.20 | 0.98 (0.77–1.26) | 0.89 | 0.10 | 0.90 (0.60–1.35) | 0.60 | 0.05 |
Ph value used to evaluate the heterogeneity between included studies. ALL, acute lymphoblastic leukemia; CI, confidence interval; HB, hospital-based; MTR, methionine synthase; OR, odds ratio; PB, population-based.
Figure 2. Forest plot describing the association of MTR A2756G polymorphism and pediatric ALL risk (AG+GG vs. AA).
Figure 3. Forest plot describing the association of MTR A2756G polymorphism and pediatric ALL risk (G allele vs. A allele; stratified by ethnicity).
Figure 4. Forest plot describing the association of MTR A2756G polymorphism and pediatric ALL risk (AG vs. AA; stratified by quality score).
Heterogeneity and sensitivity analysis
No significant heterogeneity was detected across the eligible studies under all five genetic models for MTR A2756G polymorphism, so the fixed-effects model based Mantel–Haenszel method was selected for the combined analysis (Table 3). Sensitivity analysis, in which the pooled ORs were recalculated after sequential omission of individual studies, revealed that the combined results remained virtually unchanged, suggesting the robustness of quantitative synthesis results.
Publication bias
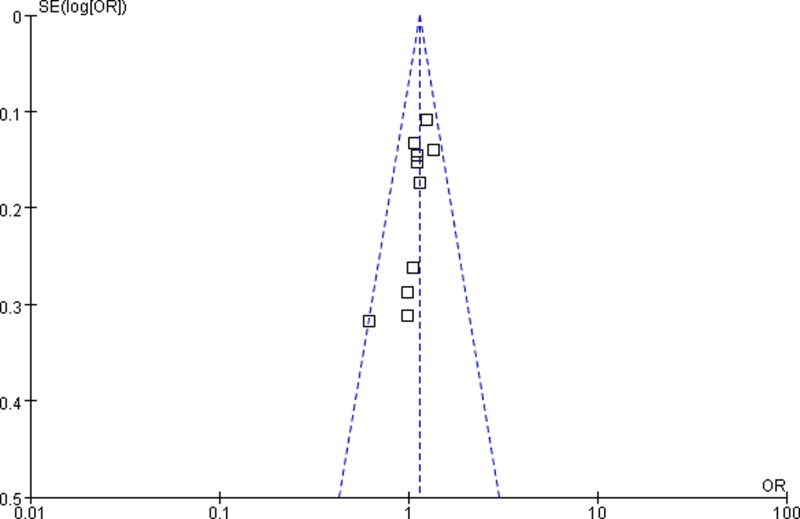
The shapes of funnel plots appeared symmetrical, suggesting that there was no obvious publication bias (Figures 5 and 6). In addition, the results of Egger’s test also indicated a lack of publication bias of the current meta-analysis.
Figure 5. Funnel plot assessing publication bias in heterozygote model (AG vs. AA).
Figure 6. Funnel plot assessing publication bias in dominant model (AG+GG vs. AA).
Discussion
MTR gene encodes a vitamin B12-dependent enzyme, which catalyzes the remethylation of homocysteine to methionine, the precursor to S-adenosylmethionine, which acts as the universal methyl group donor [33]. The MTR reaction also releases tetrahydrofolate, which is remethylated to 5,10-methylene tetrahydrofolate for further participating in nucleotide synthesis. It is reported that MTR A2756G polymorphism can convert the codon for aspartate to glycine, resulting in a lower enzyme activity followed by homocysteine elevation and DNA hypomethylation [14,15]. In addition, the G-variant could enhance the flux of one-carbon moieties available for DNA methylation [13]. Therefore, MTR A2756G polymorphism might lead to alterations in DNA biosynthesis and methylation pattern, and contribute to the genetic susceptibility to cancer including leukemia, as hypermethylation is important in acute leukemia [34,35].
Numerous investigations have examined the association of MTR A2756G polymorphism with pediatric ALL susceptibility, yet have generated conflicting results. Petra et al. [30] found that the presence of at least one polymorphic MTR 2756 G allele showed some, but insignificant, tendency to reduce the risk for pediatric ALL. However, a dose–response relationship between the number of copies of the MTR 2756 G allele and increased risk of pediatric ALL was observed in the study by Lightfoot et al. [27]. Specifically, heterozygosity for the variant allele (AG) was associated with a 1.24-fold increased risk of ALL (95%CI = 1.00–1.53, P = 0.05), and homozygosity (GG) with a 1.88-fold increased risk of ALL (95%CI = 1.16–3.07, P = 0.01). de Jonge et al. [23] found no statistical differences in genotype distribution for MTR A2756G polymorphism between children ALL and the controls. To elucidate this inconsistency, a meta-analysis was conducted to derive a more precise estimation of the association.
In the present study, the combined results found that there was significant association between MTR A2756G polymorphism and risk of pediatric ALL in overall comparison. Individuals with the MTR 2756 G allele had increased risk of developing pediatric ALL compared to those with the A allele. Moreover, individuals with the AG genotype or the AG+GG genotype had raised risk of pediatric ALL compared to those with the AA genotype. Significant association was also found in Caucasians, population-based designed studies, and studies assigned as high quality. Our results were in accordance with the conclusion reported by Xia et al. [36], which showed MTR 2756 A allele was associated with a decreased risk of pediatric ALL compared with the G allele. In present meta-analysis, more web-based databases including English and non-English databases were systematically searched to minimize the selection bias and the potential risk of missing eligible literature [37]. Since our analysis included several new studies and included 3224 cases and 4077 controls, allowing for sufficient statistical power and more precise estimation, our conclusion is more credible.
When interpreting the results, some limitations of our meta-analysis should be considered. First, our results were based on unadjusted estimates, which may cause confounding bias. A more precise analysis could be performed if all raw data were available, which would allow for the adjustment by other confounders including sex, age, lifestyles and other potential factors. Second, the quantitative synthesis of some subgroups may have no sufficient testing power to accurately assess the real association, for instance, only one study was conducted among Asians. In addition, the gene–environment interactions which may modify genetic susceptibility to cancer were not taken into account in the present study due to the limited data. Last but not least, we also did not consider other genes in folate metabolic network that might be associated with the risk of pediatric ALL. The etiological mechanism of ALL is very complicated, in which gene–gene, and gene–environment interactions are involved [4,38]. Several case–control studies have reported that MTR 2756AG individuals who were SHMT1 1420CT/TT had a 5.6-fold reduction in ALL risk [39]. In contrast, MTR 2756 G was a risk allele for ALL on itself but also in combination with the MTHFR 677 T allele in adults [38]. The possibility cannot be ruled out that the role of MTR A2756G polymorphism is somewhat diluted or concealed by other gene–gene interactions. Future studies combining other genes in folate metabolism with MTR are encouraged.
Conclusion
In conclusion, this meta-analysis suggests that MTR A2756G polymorphism influences the genetic susceptibility to pediatric ALL, especially in Caucasians. However, large scale and well-designed studies are required to validate our findings, and the biochemical mechanism and function of MTR A2756G polymorphism should also be investigated in the future.
Abbreviations
- ALL
acute lymphoblastic leukemia
- CI
confidence interval
- HB
hospital-based
- HWE
Hardy–Weinberg equilibrium
- MTHFR
methylenetetrahydrofolate reductase
- MTR
methionine synthase
- NOS
Newcastle–Ottawa Scale
- OR
odds ratio
- PB
population-based
- PCR
polymerase chain reaction
- RFLP
restriction fragment length polymorphism
- SHMT1
serine hydroxymethyltransferase 1
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
The authors declare that there are no sources of funding to be acknowledged.
Author Contribution
H.-P.Y. designed and supervised the study. L.-M.M. drafted the manuscript, analyzed and interpreted the data. L.-M.M. and X.-W.Y. carried out the literature search and quality assessment, and extracted the data from the eligible studies. H.-P.Y. and L.-H.R. critically reviewed and all authors approved the final manuscript.
References
- 1.Pui C.-H. and Evans W.E (2006) Treatment of acute lymphoblastic leukemia. N. Engl. J. Med. 354, 166–178 10.1056/NEJMra052603 [DOI] [PubMed] [Google Scholar]
- 2.Bhojwani D., Yang J.J. and Pui C.-H. (2015) Biology of childhood acute lymphoblastic leukemia. Pediatr. Clin. North Am. 62, 47–60 10.1016/j.pcl.2014.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greaves M. (2006) Infection, immune responses and the aetiology of childhood leukaemia. Nat. Rev. Cancer 6, 193–203 10.1038/nrc1816 [DOI] [PubMed] [Google Scholar]
- 4.Bonaventure A., Goujon-Bellec S., Rudant J., Orsi L., Leverger G., Baruchel A.. et al. (2012) Maternal smoking during pregnancy, genetic polymorphisms of metabolic enzymes, and childhood acute leukemia: the ESCALE study (SFCE). Cancer Causes Control. 23, 329–345 10.1007/s10552-011-9882-9 [DOI] [PubMed] [Google Scholar]
- 5.Stover P.J. (2004) Physiology of folate and vitamin B12 in health and disease. Nutr. Rev. 62, S3–S12, discussion S13 10.1111/j.1753-4887.2004.tb00070.x [DOI] [PubMed] [Google Scholar]
- 6.Blount B.C., Mack M.M., Wehr C.M., MacGregor J.T., Hiatt R.A., Wang G.. et al. (1997) Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc. Natl. Acad. Sci. U.S.A. 94, 3290–3295 10.1073/pnas.94.7.3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beetstra S., Thomas P., Salisbury C., Turner J. and Fenech M. (2005) Folic acid deficiency increases chromosomal instability, chromosome 21 aneuploidy and sensitivity to radiation-induced micronuclei. Mutat. Res. 578, 317–326 10.1016/j.mrfmmm.2005.05.012 [DOI] [PubMed] [Google Scholar]
- 8.Pufulete M., Al-Ghnaniem R., Leather A. J.M., Appleby P., Gout S., Terry C.. et al. (2003) Folate status, genomic DNA hypomethylation, and risk of colorectal adenoma and cancer: a case control study. Gastroenterology 124, 1240–1248 10.1016/S0016-5085(03)00279-8 [DOI] [PubMed] [Google Scholar]
- 9.Song M.-A., Brasky T.M., Marian C., Weng D.Y., Taslim C., Llanos A.A.. et al. (2016) Genetic variation in one-carbon metabolism in relation to genome-wide DNA methylation in breast tissue from heathy women. Carcinogenesis 37, 471–480 10.1093/carcin/bgw030 [DOI] [PubMed] [Google Scholar]
- 10.Ebrahimi A., Hosseinzadeh Colagar A. and Karimian M (2017) Association of human methionine synthase-A2756G transition with prostate cancer: a case–control study and in silico analysis. Acta Med. Iran. 55, 297–303 [PubMed] [Google Scholar]
- 11.Nakao H., Wakai K., Ishii N., Kobayashi Y., Ito K., Yoneda M.. et al. (2016) Associations between polymorphisms in folate-metabolizing genes and pancreatic cancer risk in Japanese subjects. BMC Gastroenterol. 16, 83 10.1186/s12876-016-0503-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nazki F.H., Sameer A.S. and Ganaie B.A. (2014) Folate: metabolism, genes, polymorphisms and the associated diseases. Gene 533, 11–20 10.1016/j.gene.2013.09.063 [DOI] [PubMed] [Google Scholar]
- 13.Harmon D.L., Shields D.C., Woodside J.V., McMaster D., Yarnell J.W., Young I.S.. et al. (1999) Methionine synthase D919G polymorphism is a significant but modest determinant of circulating homocysteine concentrations. Genet. Epidemiol. 17, 298–309 10.1002/(SICI)1098-2272(199911)17:4%3c298::AID-GEPI5%3e3.0.CO;2-V [DOI] [PubMed] [Google Scholar]
- 14.Chen J., Stampfer M.J., Ma J., Selhub J., Malinow M.R., Hennekens C.H.. et al. (2001) Influence of a methionine synthase (D919G) polymorphism on plasma homocysteine and folate levels and relation to risk of myocardial infarction. Atherosclerosis 154, 667–672 10.1016/S0021-9150(00)00469-X [DOI] [PubMed] [Google Scholar]
- 15.Paz M.F., Avila S., Fraga M.F., Pollan M., Capella G., Peinado M.A.. et al. (2002) Germ-line variants in methyl-group metabolism genes and susceptibility to DNA methylation in normal tissues and human primary tumors. Cancer Res. 62, 4519–4524 [PubMed] [Google Scholar]
- 16.Bleich S., Semmler A., Frieling H., Thumfart L., Muschler M., Hillemacher T.. et al. (2014) Genetic variants of methionine metabolism and DNA methylation. Epigenomics 6, 585–591 10.2217/epi.14.54 [DOI] [PubMed] [Google Scholar]
- 17.Shao H.-B., Ren K., Gao S.-L., Zou J.-G., Mi Y.-Y., Zhang L.-F.. et al. (2018) Human methionine synthase A2756G polymorphism increases susceptibility to prostate cancer. Aging (Albany NY) 10, 1776–1788 10.18632/aging.101509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akbari M.T., Naderi A., Saremi L., Sayad A., Irani S. and Ahani A (2015) Methionine synthase A2756G variation is associated with the risk of retinoblastoma in Iranian children. Cancer Epidemiol 39, 1023–1025 10.1016/j.canep.2015.11.002 [DOI] [PubMed] [Google Scholar]
- 19.Guo S.-J., Luo S.-C., Liu W.-Y., Zuo Q.-N. and Li X.-H. (2017) Methionine synthase A2756G polymorphism and lymphoma risk: a meta-analysis. Eur. Rev. Med. Pharmacol. Sci. 21, 3075–3082 [PubMed] [Google Scholar]
- 20.Stang A. (2010) Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 25, 603–605 10.1007/s10654-010-9491-z [DOI] [PubMed] [Google Scholar]
- 21.Barili F., Parolari A., Kappetein P.A. and Freemantle N. (2018) Statistical primer: heterogeneity, random- or fixed-effects model analyses? Interact. Cardiovasc. Thorac. Surg. 27, 317–321 10.1093/icvts/ivy163 [DOI] [PubMed] [Google Scholar]
- 22.Egger M., Davey Smith G., Schneider M. and Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315, 629–634 10.1136/bmj.315.7109.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Jonge R., Tissing W. J.E., Hooijberg J.H., Jansen G., Kaspers G. J.L., Lindemans J.. et al. (2009) Polymorphisms in folate-related genes and risk of pediatric acute lymphoblastic leukemia. Blood 113, 2284–2289 10.1182/blood-2008-07-165928 [DOI] [PubMed] [Google Scholar]
- 24.Gast A., Bermejo J.L., Flohr T., Stanulla M., Burwinkel B., Schrappe M.. et al. (2007) Folate metabolic gene polymorphisms and childhood acute lymphoblastic leukemia: a case-control study. Leukemia 21, 320–325 10.1038/sj.leu.2404474 [DOI] [PubMed] [Google Scholar]
- 25.Kamel A.M., Moussa H.S., Ebid G.T., Bu R.R. and Bhatia K.G (2007) Synergistic effect of methyltetrahydrofolate reductase (MTHFR) C677T and A1298C polymorphism as risk modifiers of pediatric acute lymphoblastic leukemia. J. Egypt. Natl. Cancer Inst. 19, 96–105 [PubMed] [Google Scholar]
- 26.Lautner-Csorba O., Gézsi A., Erdélyi D.J., Hullám G., Antal P., Semsei Á.F.. et al. (2013) Roles of genetic polymorphisms in the folate pathway in childhood acute lymphoblastic leukemia evaluated by Bayesian relevance and effect size analysis. PLoS One 8, e69843 10.1371/journal.pone.0069843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lightfoot T.J., Johnston W.T., Painter D., Simpson J., Roman E., Skibola C.F.. et al. (2010) United Kingdom Childhood Cancer Study Genetic variation in the folate metabolic pathway and risk of childhood leukemia. Blood 115, 3923–3929 10.1182/blood-2009-10-249722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metayer C., Scélo G., Chokkalingam A.P., Barcellos L.F., Aldrich M.C., Chang J.S.. et al. (2011) Genetic variants in the folate pathway and risk of childhood acute lymphoblastic leukemia. Cancer Causes Control 22, 1243–1258 10.1007/s10552-011-9795-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milne E., Greenop K.R., Scott R.J., Haber M., Norris M.D., Attia J.. et al. (2015) Folate pathway gene polymorphisms, maternal folic acid use, and risk of childhood acute lymphoblastic leukemia. Cancer Epidemiol. Biomarkers Prev. 24, 48–56 10.1158/1055-9965.EPI-14-0680 [DOI] [PubMed] [Google Scholar]
- 30.Petra B.G., Janez J. and Vita D (2007) Gene-gene interactions in the folate metabolic pathway influence the risk for acute lymphoblastic leukemia in children. Leuk. Lymphoma 48, 786–792 10.1080/10428190601187711 [DOI] [PubMed] [Google Scholar]
- 31.Rahimi Z., Ahmadian Z., Akramipour R., Vaisi-Raygani A., Rahimi Z. and Parsian A (2012) Thymidylate synthase and methionine synthase polymorphisms are not associated with susceptibility to childhood acute lymphoblastic leukemia in Kurdish population from Western Iran. Mol. Biol. Rep. 39, 2195–2200 10.1007/s11033-011-0968-y [DOI] [PubMed] [Google Scholar]
- 32.Nikbakht M., MalekZadeh K., Jha A.K., Askari M., Marwaha R.K., Kaul D.. et al. (2012) Polymorphisms of MTHFR and MTR genes are not related to susceptibility to childhood ALL in North India. Exp. Oncol. 34, 43–48 [PubMed] [Google Scholar]
- 33.Matsuo K., Suzuki R., Hamajima N., Ogura M., Kagami Y., Taji H.. et al. (2001) Association between polymorphisms of folate- and methionine-metabolizing enzymes and susceptibility to malignant lymphoma. Blood 97, 3205–3209 10.1182/blood.V97.10.3205 [DOI] [PubMed] [Google Scholar]
- 34.Davidsson J., Lilljebjörn H., Andersson A., Veerla S., Heldrup J., Behrendtz M.. et al. (2009) The DNA methylome of pediatric acute lymphoblastic leukemia. Hum. Mol. Genet. 18, 4054–4065 10.1093/hmg/ddp354 [DOI] [PubMed] [Google Scholar]
- 35.Mullighan C.G. (2013) Genomic characterization of childhood acute lymphoblastic leukemia. Semin. Hematol. 50, 314–324 10.1053/j.seminhematol.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia J., Wang Y., Zhang H. and Hu Y (2014) Association between MTR A2756G polymorphism and childhood acute lymphoblastic leukemia: a meta-analysis. Leuk. Lymphoma 55, 1388–1393 10.3109/10428194.2013.830304 [DOI] [PubMed] [Google Scholar]
- 37.Li S.-Y., Ye J.-Y., Liang E.-Y. and Yang M (2014) The protective role of MTR A2756G polymorphisms in childhood acute lymphoblastic leukemia remains inconclusive. Leuk. Lymphoma 55, 2217–2218 10.3109/10428194.2013.867490 [DOI] [PubMed] [Google Scholar]
- 38.Gemmati D., Ongaro A., Scapoli G.L., Della Porta M., Tognazzo S., Serino M.L.. et al. (2004) Common gene polymorphisms in the metabolic folate and methylation pathway and the risk of acute lymphoblastic leukemia and non-Hodgkin’s lymphoma in adults. Cancer Epidemiol. Biomarkers Prev. 13, 787–794 [PubMed] [Google Scholar]
- 39.Skibola C.F., Smith M.T., Hubbard A., Shane B., Roberts A.C., Law G.R.. et al. (2002) Polymorphisms in the thymidylate synthase and serine hydroxymethyltransferase genes and risk of adult acute lymphocytic leukemia. Blood 99, 3786–3791 10.1182/blood.V99.10.3786 [DOI] [PubMed] [Google Scholar]