Abstract
Shexiang Tongxin dropping pill (STP) is a traditional Chinese medicine formula that consists of total saponins of ginseng, synthetic Calculus bovis, bear gall, Venenum bufonis, borneol and Salvia miltiorrhiza. STP has been widely used in China and Southeast Asia for the treatment of cardiovascular diseases. In this study, a qualitative analytical method using high performance liquid chromatography coupled with quadrupole time-of-flight tandem mass spectrometry was developed for identification of the major constituents in STP. Based on the retention time and MS spectra, 41 components were identified by comparison with reference compounds and literature data. Moreover, using ultra-performance liquid chromatography coupled with triple-quadrupole tandem mass spectrometry in multiple-reaction monitoring mode, we quantified 13 of the identified constituents (ginsenoside Rg1, ginsenoside Rk3, cinobufagin, arenobufagin, bufalin, resibufogenin, tanshinone IIA, taurine, tauroursodeoxycholic acid, taurocholic acid, cholic acid, deoxycholic acid, and chenodeoxycholic acid). These results suggest that this new approach is applicable for the routine analysis and quality control of STP products and provides fundamental data for further in vivo pharmacokinetical studies.
Keywords: liquid chromatography, quadrupole time-of-flight tandem mass spectrometry, triple-quadrupole tandem, Shexiang Tongxin dropping pill
1. Introduction
Traditional Chinese Medicine (TCM) typically involves the prescription of natural products in a composite formula, and has been widely used due to its specificity, effectiveness and low toxicity. There are a growing number of people who are using TCMs to achieve optimal health and prevent diseases [1,2,3,4,5]. Any prescribed formulas in TCM involve a complex system containing hundreds or even thousands of different chemical constituents. It is well accepted that the efficacy of TCM formulas is based on the synergistic effects of these components. Extensive efforts have been made to establish specific analytical methods in order to distinguish the chemical components in TCM prescriptions [6,7,8,9,10,11,12]. Shexiang Tongxin dropping pill (STP) is a Chinese FDA-approved formula that includes a combination of seven medicinal ingredients, including moschus, Radix rhizoma ginseng, Calculus bovis, bear gall, Venenum bufonis, borneol and Salvia miltiorrhiza. STP has been widely used in China and Southeast Asia for the clinical treatment of cardiovascular diseases. Previous pharmacological studies have proposed that STP can protect endothelial cells from atherosclerotic lesions by decreasing levels of endothelin-1 (ET-1), C reactive protein (CRP), tumor necrosis factor-α (TNF-α) and increasing levels of nitrogen oxide in the blood [13]. Moreover, STP can decrease levels of harmful lipids and improve abnormal hemorheology indices [14]. These therapeutic properties have been attributed to the major components in the prescription formula of STP [15,16,17,18,19,20,21].
Although the beneficial effects of STP have been well documented, the actual bioactive components of STP are still poorly understood. The constituents of each single herbal component in STP have been reported, however the chemical analysis of the final STP formula as a whole has not yet been elucidated [22,23,24,25,26,27,28]. This is necessary because the effects of TCMs do not simply involve the accumulated effects of individual components, but rather encompass the interactions between various components as a whole. In addition, changes and losses of certain constituents often occur during the production process of STP extraction, concentration and infusion, hence the contents of each component may also be changed. Therefore, qualitative and quantitative analysis of the total constituents in STP as a whole provides a more meaningful and specific outcome compared to the individual analysis of each component. In order to provide valuable information for quality control, as well as screening of the critical bioactive components, it is necessary to develop a specific and sensitive analytical method to identify and characterize the compounds in STP.
High performance liquid chromatography coupled with quadrupole time-of-flight tandem mass spectrometry (HPLC-Q-TOF-MS/MS) has become one of the most powerful analytical tools due to its high resolution and accurate mass measurements [29,30,31,32,33,34,35,36,37,38,39]. By comparing multiple-stage mass spectra with those of reference compounds and literatures, the unknown constituents can be identified. In addition, simultaneous quantitation of multi-components by ultra-performance liquid chromatography coupled with triple-quadrupole tandem mass spectrometry (UPLC-QqQ-MS/MS) has also been widely used in the analysis of TCM [40,41,42,43].
In this study, HPLC-ESI-Q-TOF method was established for separating and identifying the components in STP. All 41 components were identified based on their retention times and MS spectra data. Subsequently, UPLC-QqQ-MS/MS method was developed for the simultaneous quantitative analysis of 13 of the compounds in STP. This study represents the first detailed investigation into the components of STP and provides a valid method for its quality control and evaluation.
2. Results and Discussion
2.1. Optimization of HPLC Conditions
Chromatographic parameters were optimized to achieve a higher separation quality of the fingerprint chromatogram and a reduced analysis time. In order to obtain optimal chromatograms with good peaks and high sensitivity, different mobile phases including water/methanol and water/acetonitrile were examined. We determined that optimal separation of the analytes was achieved using water/acetonitrile as the mobile phase (Figure S1). The retention behavior of the compounds on reversed-phase HPLC columns was significantly affected by the pH of the mobile phase. Therefore we compared the effect of different acids, including formic acid, trifluoroacetic acid and phosphoric acid, as well as the amount of acid added into the mobile phase. We determined that the use of formic acid provided the greatest improvement during chromatographic separation, and also enhanced the formation of ions (Figure S2). The optimal conditions for a given factor was selected based on the observed peak capacity. The combination of the optimal factors and parameters, including chromatographic column, column temperature, mobile phase, flow rate and gradient elution program were analyzed in order to achieve the highest degree of separation for each peak (Figure S3 and Figure S4).
2.2. HPLC-Q-TOF-MS/MS Qualitative Analysis of Chemical Constituents in STP
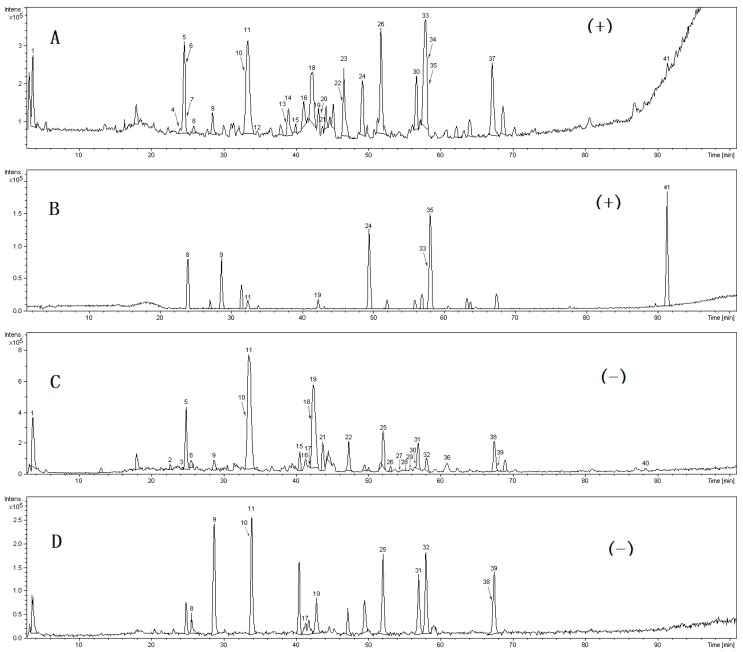
To characterize the chemical constituents in STP, HPLC-Q-TOF-MS/MS method was established. As shown in Table 1, a total of 41 components were initially identified or characterized, and 15 of those were identified unambiguously by comparing their retention times, mass accuracy, and fragmentation behaviors with data from the corresponding reference standards [37,39,44]. The remaining compounds were tentatively characterized based on MS data, and reference to available literature data. The mass error for molecular ions of all identified compounds was within ±9 ppm. The total ion chromatograms in positive and negative ion modes were displayed in Figure 1. All compounds were grouped into six types according to their structural characteristics, which included bufadienolides, triterpene saponins, tanshinones, salvianolic acids, bile acids and other types. The structures of all compounds are shown in Figure 2.
Table 1.
Characterization of chemical constituents of STP by HPLC-Q-TOF-MS/MS.
| No. | Source | Identification | Rt (min) | Formula | Negative Ion (m/z) | |||
|---|---|---|---|---|---|---|---|---|
| [M − H]− | Quasi-Molecular Ion | Error (ppm) | MSMS (m/z) | |||||
| 1 | C & B | Taurine | 3.7 | C2H7NO3S | 124.0063 | 124.0065 | 2 | |
| 2 | R | Salvianolic acid A | 23.8 | C26H22O10 | 493.1129 | 493.1133 | 1 | |
| 3 | C & B | Bilirubin | 24.2 | C33H35N4O6 | 583.2551 | 583.2518 | 6 | 583.2518, 469.3944,356.4968, 243.8168 |
| 4 | G | Ginsenoside Rk3 | NA | C36H60O8 | 619.4204 | NA | ||
| 5 | G | Ginsenoside Rd | 24.9 | C48H82O18 | 945.5417 | 945.5405 | 1 | |
| 6 | R | Tanshinaldehyde | NA | C19H16O4 | 307.0965 | NA | ||
| 7 | G | Ginsenoside Rg6 | NA | C42H70O12 | 765.4784 | NA | ||
| 8 a | R | Salvianolic acid B | 25.5 | C36H30O16 | 717.1450 | 717.1454 | 0 | 717.1454, 537.0995, 527.9935, 493.1069, 339.0521, 321.0405, 295.0633 |
| 9 a | V | Arenobufagin | 28.7 | C24H32O6 | 415.2115 | 415.2115 | 0 | 415.2115, 397.2063, 371.2165, 353.2112, 277.1492, 196.5435, 151.0702 |
| 10 a | C & B | TDCANa | 33.8 | C26H44NNaO6S | 498.2884 | 498.2885 | 0 | 498.2907, 475.3216, 458.8943, 391.5463 |
| 11 a | C & B | TCA | 33.8 | C26H44NO7S | 514.2833 | 514.2856 | 4 | 514.2856, 496.2486, 482.3441, 124.1263 |
| 12 | G | Ginsenoside Rf | NA | C35H76O19 | 799.4897 | NA | ||
| 13 | V | Gamabufotalin | NA | C24H34O5 | 401.2323 | NA | ||
| 14 | G | Notoginsenoside R2 | NA | C41H70O13 | 769.4733 | NA | ||
| 15 | G | Ginsenoside Rb1 | 40.5 | C54H92O23 | 1107.5946 | 1107.5927 | 2 | 1107.5927, 945.5389, 783.4901, 765.4580, 621.4343, 553.2887, 472.2575, 459.3803, 323.0972, 263.0742, 221.0678, 179.0538, 179.0538, 161.0395, 143.0272, 131.0323, 125.0221, 119.0351, 113.0260, 101.0208 |
| 16 | V | Bufotalin | 41.1 | C26H36O6 | 443.2428 | 443.2452 | 5 | 443.2452, 407.2732, 380.9754, 375.2916, 256.8133, 203.8251, 167.9242, 138.0325 |
| 17 a | G | Ginsenoside Rh1 | 41.8 | C37H64O11 | 683.4403 | 683.4384 | 3 | 683.4384, 637.4330, 475.3704, 179.0470, 161.0398 |
| 18 | C & B | GCA | 42.6 | C26H43NO6 | 464.3007 | 464.3015 | 2 | |
| 19 a | C & B | TDCA | 42.8 | C26H45NO6S | 498.2884 | 498.2907 | 5 | 498.2907, 464.2945, 451.3111, 391.1261, 321.1686, 201.2573 |
| 20 | V | Resibufagin | NA | C24H30O5 | 397.2010 | NA | ||
| 21 | G | Ginsenoside Rb2 | 43.7 | C53H90O22 | 1077.5840 | 1077.5795 | 4 | 1077.5795, 945.5318, 915.1144, 783.0026, 765.1212, 621.3652, 311.9821, 293.6514, 191.8123, 149.1853 |
| 22 | G | Ginsenoside Re | 47.2 | C48H82O18 | 945.5417 | 945.5433 | 2 | 945.5433, 880.9436, 799.2672, 765.1661, 637.2542, 475.2767, 218.9817 |
| 23 | G | Ginsenoside Rk1 | NA | C42H70O12 | 765.4784 | NA | ||
| 24 a | V | Bufalin | NA | C24H34O4 | 385.2373 | NA | ||
| 25 a | C & B | CA | 52.1 | C24H40O5 | 407.2792 | 407.2811 | 5 | 407.2811, 389.2677, 363.2526, 325.6244, 289.2744, 233.8977, 215.3690, 205.9169 |
| 26 | V | Argentinogenin-3-lutarate-arginine | 53.6 | C38H58N408 | 697.4171 | 697.4121 | 7 | 697.4121, 651.4153, 535.5406, 489.3649, 179.0546 |
| 27 | C & B | TLCA | 55.0 | C22H44NO10 | 482.2960 | 482.2918 | 9 | |
| 28 | G | Ginsenoside Rg1 | 55.3 | C43H72O14 | 811.4838 | 811.4813 | 3 | 811.4813, 775.8988, 765.4577, 619.3911, 421.2024, 391.2774 |
| 29 | C & B | DCA | 55.8 | C24H40O4 | 391.2843 | 391.2854 | 3 | |
| 30 | C & B | GCDGA | 56.2 | C26H43NO5 | 448.3058 | 448.3065 | 2 | 448.3065, 409.2741, 391.2853, 389.2658, 365.3004 |
| 31 a | C & B | UDCA | 56.8 | C24H40O4 | 391.2843 | 391.2848 | 1 | 409.2741, 391.2739, 373.8139, 354.4757, 152.9965 |
| 32 a | C & B | HDCA | 57.4 | C24H40O4 | 391.2843 | 391.2851 | 2 | 409.2741, 391.2853, 389.2658, 118.1248 |
| 33 a | V | Resibufogenin | NA | C24H32O4 | 383.2217 | NA | ||
| 34 | G | Ginsenoside Rs3 | NA | C44H74O14 | 825.4995 | NA | ||
| 35 a | V | Cinobufagin | NA | C26H34O6 | 441.2272 | NA | ||
| 36 | G | Chikusetsusaponin Iva | 61.6 | C42H66O14 | 793.4374 | 793.4330 | 6 | 793.4330, 598.6542, 481.7612, 524.4589 |
| 37 | G | Ginsenoside 20S-Rg3 | NA | C42H72O13 | 783.4889 | NA | ||
| 38 a | G | ginsenoside Rg2 | 68.9 | C42H72O13 | 783.4889 | 783.4873 | 2 | 783.4873, 765.1686, 617.0004, 409.2952, 391.2852, 313.1097 |
| 39 a | C & B | CDCA | 68.9 | C24H40O4 | 391.2843 | 391.2853 | 3 | 391.2739, 345.4757, 329.1213, 97.0589 |
| 40 | C & B | LCA | 89.4 | C24H40O3 | 375.2894 | 375.2888 | 2 | |
| 41 a | R | Tanshinone IIA | NA | C19H18O3 | 293.1172 | NA | ||
| No. | Source | Identification | Rt (min) | Formula | Positive Ion (m/z) | |||
| [M + H]+ | Quasi-Molecular Ion | Error (ppm) | MSMS (m/z) | |||||
| 1 | C & B | Taurine | 3.7 | C2H7NO3S | 126.0219 | 126.0222 | 2 | 126.0222, 115.3256, 108.8434, 97.2221, 91.7216 |
| 2 | R | Salvianolic acid A | NA | C26H22O10 | 495.1286 | NA | ||
| 3 | C & B | Bilirubin | NA | C33H35N4O6 | 584.2629 | NA | ||
| 4 | G | Ginsenoside Rk3 | 24.5 | C36H60O8 | 621.4361 | 621.4362 | 0 | |
| 5 | G | Ginsenoside Rd | 24.6 | C48H82O18 | 947.5574 | 947.5501 | 8 | |
| 6 | R | Tanshinaldehyde | 24.6 | C19H16O4 | 309.1121 | 309.1147 | 8 | |
| 7 | G | Ginsenoside Rg6 | 24.6 | C42H70O12 | 767.4940 | 767.4931 | 1 | |
| 8 a | R | Salvianolic acid B | 25.8 | C36H30O16 | 719.1607 | 719.1613 | 1 | 719.1613, 323.0496, 295.0572, 181.0460 |
| 9 a | V | Arenobufagin | 28.4 | C24H32O6 | 417.2272 | 417.2273 | 0 | 417.2273, 399.2188, 362.7495, 223.1056 |
| 10 a | C & B | TDCANa | 33.2 | C26H44NNaO6S | 500.4245 | NA | ||
| 11 a | C & B | TCA | 33.5 | C26H44NO7S | 516.2990 | 516.2984 | 1 | |
| 12 | G | Ginsenoside Rf | 34.0 | C35H76O19 | 801.5054 | 801.5075 | 3 | |
| 13 | V | Gamabufotalin | 38.9 | C24H34O5 | 403.2251 | 403.2267 | 4 | 403.2267, 385.9531, 367.1296, 349.4632, 331.8123 |
| 14 | G | Notoginsenoside R2 | 39.0 | C41H70O13 | 771.4889 | 771.4861 | 4 | |
| 15 | G | Ginsenoside Rb1 | 39.9 | C54H92O23 | 1109.6102 | 1109.6030 | 6 | 1109.6030, 935.5613, 878.7854, 646.6751, 443.3798, 407.3652, 325.1114, 217.1888, 163.0566, 145.0507, 127.0384, 85.0294 |
| 16 | V | Bufotalin | 41.1 | C26H36O6 | 445.2585 | 445.2585 | 0 | |
| 17 a | G | Ginsenoside Rh1 | NA | C37H64O11 | 685.4521 | NA | ||
| 18 | C & B | GCA | 42.3 | C26H43NO6 | 466.3163 | 466.3132 | 7 | 466.3132, 337.2523, 319.2353, 295.2026, 288.1602, 213.1621, 209.1339, 201.1658 |
| 19 a | C & B | TDCA | 42.8 | C26H45NO6S | 500.3040 | 500.3042 | 0 | |
| 20 | V | Resibufagin | 42.8 | C24H30O5 | 399.2166 | 399.2158 | 2 | 399.2158, 387.2519, 297.7655, 223.1439, 211.1011, 145.0999, 131.0982, 179.0841, 105.0709, 91.0525 |
| 21 | G | Ginsenoside Rb2 | 43.1 | C53H90O22 | 1079.5997 | 1079.5942 | 5 | |
| 22 | G | Ginsenoside Re | 46.5 | C48H82O18 | 947.5574 | 947.5510 | 7 | |
| 23 | G | Ginsenoside Rk1 | 46.6 | C42H70O12 | 767.4940 | 767.4931 | 1 | |
| 24 a | V | Bufalin | 49.1 | C24H34O4 | 387.253 | 387.2527 | 1 | 387.2527, 370.9842, 352.9666, 340.2608, 255.2028 |
| 25 a | C & B | CA | NA | C24H40O5 | 409.2949 | NA | ||
| 26 | V | Argentinogenin-3-lutarate-arginine | 53.2 | C38H58N408 | 699.4327 | 699.4299 | 4 | 699.4299, 681.4053, 598.7017, 582.3791, 331.2027, 278.1446, 250.1590, 175.1200, 157.1072 |
| 27 | C & B | TLCA | NA | C22H44NO10 | 483.3038 | NA | ||
| 28 | G | Ginsenoside Rg1 | NA | C43H72O14 | 813.4995 | NA | ||
| 29 | C & B | DCA | NA | C24H40O4 | 393.3000 | NA | ||
| 30 | C & B | GCDGA | 56.1 | C26H43NO5 | 450.3214 | 450.3204 | 2 | 450.3204, 415.3174, 321.2607, 278.7283, 215.1679, 175.1452, 161.1349, 147.1140, 107.0885 |
| 31 a | C & B | UDCA | NA | C24H40O4 | 393.3000 | NA | ||
| 32 a | C & B | HDCA | NA | C24H40O4 | 393.3000 | NA | ||
| 33 a | V | Resibufogenin | 57.7 | C24H32O4 | 385.2373 | 385.2369 | 1 | 385.2369, 367.2214, 350.2243, 341.2244, 332.2025 |
| 34 | G | Ginsenoside Rs3 | 57.8 | C44H74O14 | 827.5151 | 827.5165 | 2 | |
| 35 a | V | Cinobufagin | 57.9 | C26H34O6 | 443.2428 | 443.2425 | 1 | 443.2425, 401.2288, 385.2322, 367.2210, 349.2129, 187.1473, 151.0382 |
| 36 | G | Chikusetsusaponin Iva | NA | C42H66O14 | 795.4525 | NA | ||
| 37 | G | Ginsenoside 20S-Rg3 | 68.6 | C42H72O13 | 785.5046 | 785.5063 | 2 | 785.5063, 621.6871, 357.2785, 339.2647, 321.2458, 275.2027, 221.1526, 161.1345 |
| 38 a | G | ginsenoside Rg2 | NA | C42H72O13 | 785.5046 | NA | ||
| 39 a | C & B | CDCA | NA | C24H40O4 | 393.3000 | NA | ||
| 40 | C & B | LCA | NA | C24H40O3 | 377.3050 | NA | ||
| 41 a | R | Tanshinone IIA | 92.3 | C19H18O3 | 295.1269 | 295.1266 | 0 | 295.1266, 280.4501, 277.1173, 266.0878, 262.0933, 249.1212, 235.0725, 225.1178, 221.1251, 207.0771 |
G: Ginsenoside; V: Venenum bufonis; R: Red-rooted salvia; C: Calculus bovis; B: Bear gall; a Compared with a reference standard.
Figure 1.
HPLC-Q-TOF-MS/MS total ion chromatogram (A) TIC of STP drug in positive mode (B) TIC of reference substances in positive mode (C) TIC of STP drug in negative mode (D) TIC of reference substances in negative mode. Standards are as follows: salvianolic acid B, arenobufagin, TDCA, TCA, Rg2, Rh1, bufalin, CA, UDCA, HDCA, resibufogenin, cinobufagin, CDCA, tanshinone IIA, and TDCANa.
Figure 2.
Chemical structures of the compounds identified in STP.
2.2.1. Compounds from Ginseng
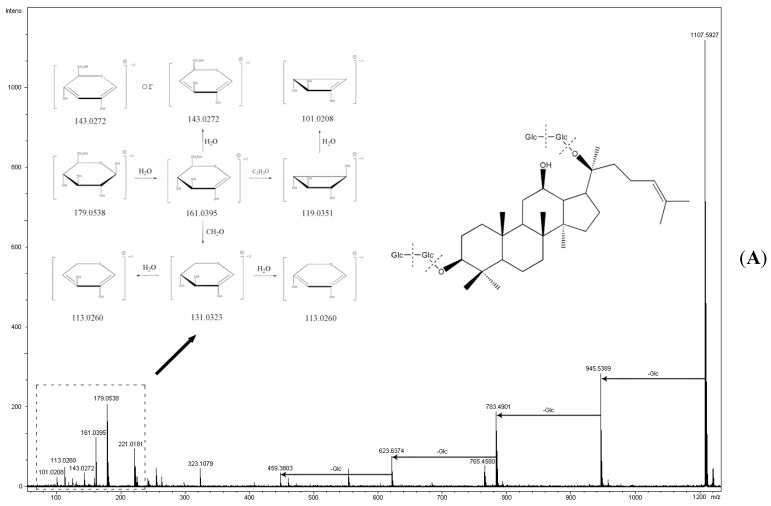
A total of 13 ginseng saponins were identified in this study, and these can be divided into those belonging to the protopanaxadiol (PPD) group, including ginsenoside Rd (Peak 5), ginsenoside Rf (Peak 12), ginsenoside Rg2 (Peak 38), ginsenoside Rk1 (Peak 23), ginsenoside Rs3 (Peak 34), and ginsenoside Rg3 (Peak 37); and the protopanaxatriol (PPT) group, including ginsenoside Rk3 (Peak 4), ginsenoside Re (Peak 22), ginsenoside Rb1 (Peak 15), ginsenoside Rb2 (Peak 21), ginsenoside Rh1 (Peak 17), ginsenoside Rg6 (Peak 7), and ginsenoside Rg1 (Peak 28). For saponin compounds, successive or simultaneous losses of sugar moieties are commonly produced in the negative MS/MS ion mode. In this study, ginsenoside Rb1 was used as an example to illustrate the fragmentation pathway of saponins as shown in Figure 3A. The precise molecular weight of ginsenoside Rb1 is 1107.5946 g/mol, and the main fragment ions including m/z 1107.5927 [M − H]−, 945.5389 [M − H-Glc]−, 783.4901 [M − H-2Glc]−, 765.4580 [M − H-(Glc-H2O)]−, and 459.3803 [C32H56O3]− were observed via MS/MS screening. In addition, highly abundant ions at m/z 179.0538, 161.0395, 143.0272, 131.0323, 119.0351, 113.0260, and 101.0208 produced via cross fragmentation were also found.
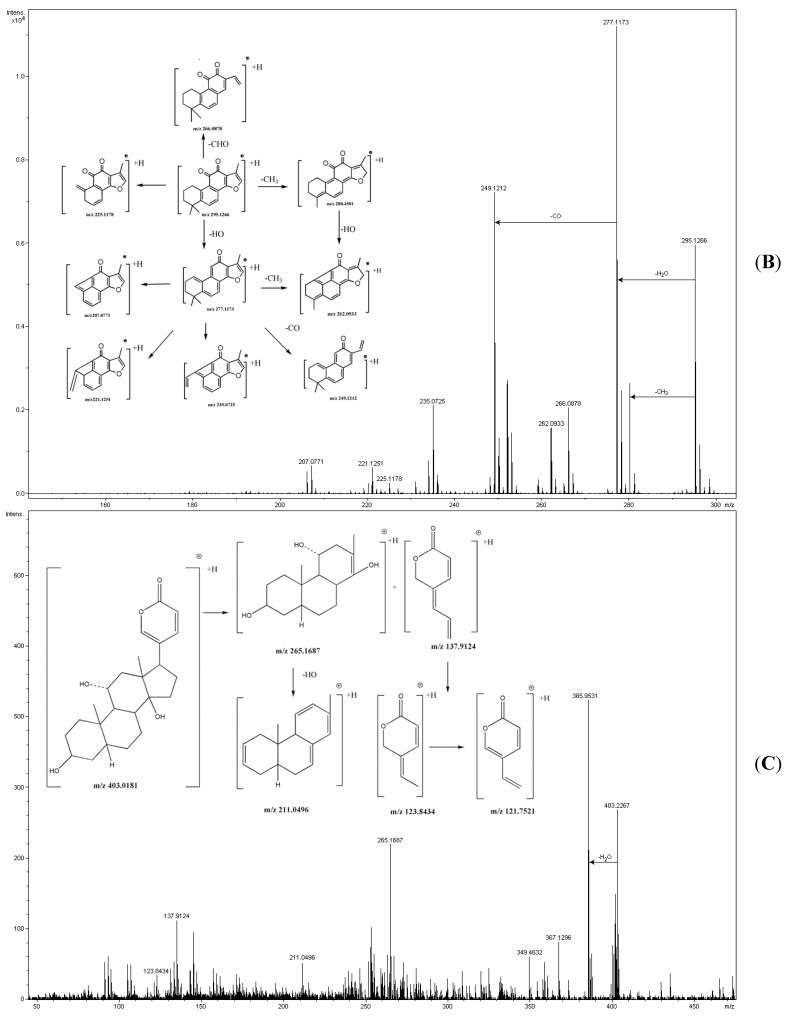
Figure 3.
(A) The ESI-QTOF-MS spectra and the proposed fragmentation pathway of ginsenoside Rb1; (B) The ESI-QTOF-MS spectra and the proposed fragmentation pathway of tanshinone IIA; (C) The ESI-QTOF-MS spectra and the proposed fragmentation pathway of gamabufotalin; (D) The ESI-QTOF-MS spectra and the proposed fragmentation pathway of TDCA.
2.2.2. Compounds from Salvia miltiorrhiza
The main ingredients of Salvia miltiorrhiza were detected in this study, which included salvianolic acid A (Peak 2), salvianolic acid B (Peak 8), and tanshinone IIA (Peak 41). The main fragment ions of tanshinone IIA as analyzed by MS/MS screening were observed at m/z 295.1266 [M + H]+, 280.4501 [M + H-CH3]+, 225.1178 [M + H-CHO]+, 277.1173 [M + H-H2O]+, 262.0933[M + H-H2O-CH3]+, and 249.1212 [M + H-H2O-CO]+ in the positive ion spectrum (Figure 3B).
2.2.3. Compounds from Toad Skin Secretion (Venenum bufonis)
Eight toad skin secretion compounds from Venenum bufonis were identified in this study. These compounds can be categorized into the Venenum bufonis I (VB I) group, which included arenobufagin (Peak 9), gamabufotalin (Peak 13), bufotalin (Peak 16), bufalin (Peak 24), and argentinogenin-3-adipate-arginine (Peak 26); and the Venenum bufonis II (VB II) group, including resibufagin (Peak 20), resibufogenin (Peak 33), and cinobufagin (Peak 35).
The mass fragmentation patterns were analyzed to verify the identification of these compounds. The results showed that the unconjugated bufosteroids can produce a series of neutral ions or positive ions containing rings A, B and C following the continuous loss of neutral H2O (18 Da) ion in the remaining steroidal rings. The main fragment ions of gamabufotalin include m/z 403.2267 as [M + H]+, 385.9531 [M + H-H2O]+, 367.1296 [M − H2O-CO]+, 349.4632 [M + H-C2OH3]+, and 265.1687 [M + H-C8H9O2]+ were observed in this study. The cleavage method of the bufosteroids without 16-hydroxyl substation showed obvious ion peaks of the lactonic ring E at m/z 121.7521 [C7H5O2]+ and m/z 137.9124 [C8H8O2 + H]+ in positive ion mode (Figure 3C).
2.2.4. Compounds from Calculus bovis and Bear Gall
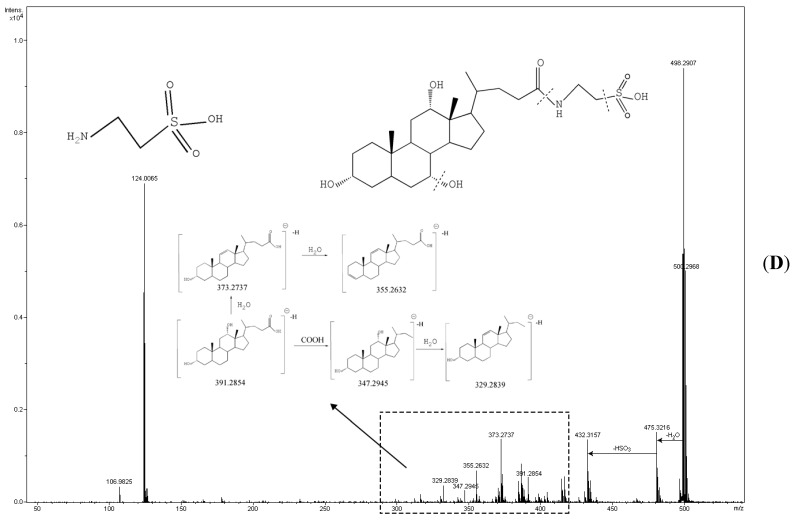
The major constituents of Calculus bovis and bear gall all have similar bile acid composition, and therefore we performed simultaneous analysis of these compounds. Eleven bile acids were identified, including taurine (Peak 1), bilirubin (Peak 3), TDCA (Peak 19), TCA (Peak 11), GCA (Peak 18), CA (Peak 25), UDCA (Peak 31), TLCA (Peak 27), GCDCA (Peak 30), HDCA (Peak 32), CDCA (Peak 39), DCA (Peak 29), and lithocholic acid (Peak 40).
In this study, four peaks (compounds 31, 32, 39, and 29) showed identical ions at m/z 391.2850 ± 4 ppm [M* − H]− and m/z 783.4830 ± 3ppm [2M* − H]−. By comparison with reference standards, these four compounds were identified as UDCA (391.2848 [M* − H]−), HDCA (391.2851[M* − H]−), CDCA (391.2853 [M* − H]−), and DCA (391.2854[M* − H]−). The main fragment ions of compound 19 were observed at m/z 498.2907 [M − H]−, 475.3216 [M − H-H2O]−, 432.3157 [M − H-HSO3]−, and 391.2854 [M + H-H2O-C2H7N]− in the negative ion spectrum. Furthermore, compound 19 showed the same fragmentation pathway to DCA at m/z 373.2737 [M* − H-H2O]−, m/z 355.2632 [M* − H-2H2O]−, m/z 347.2945 [M* − H-HCOO]− and m/z 329.2839 [M* − H-H2O-HCOO]− in the MS/MS spectrum, and therefore we deduced that compound 19 was TDCA (Figure 3D).
2.3. UPLC-QqQ-MS/MS Quantitative Analysis of Chemical Constituents from STP
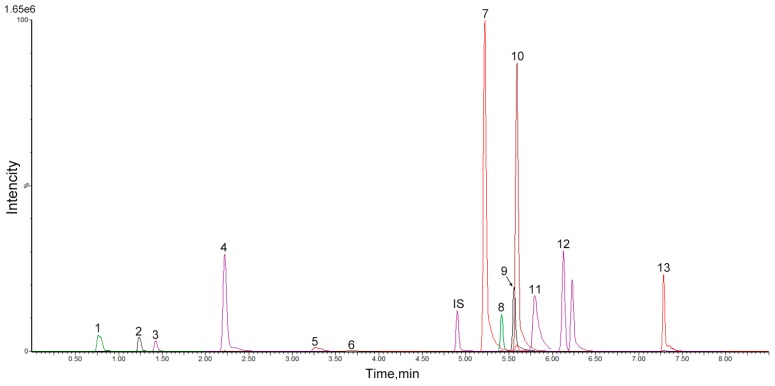
The qualitative results indicated that ginsenoside Rg1, ginsenoside Rg3, cinobufagin, arenobufagin, bufalin, resibufogenin, tanshinone IIA, taurine, astragaloside, tauroursodeoxycholic acid, taurocholic acid, cholic acid, deoxycholic acid, and chenodeoxycholic acid were the major constituents in STP (Figure 4).
Figure 4.
The MRM chromatograms of 13 markers and internal standard: (1) taurine, (2) ginsenoside Rg1, (3) arenobufagin, (4) TDCA, (5) TCA, (6) bufalin, (7) CA, (8) cinobufagin, (9) resibufogenin, (10) ginsenoside Rg3, (11) CDCA, (12) DCA, (13) tanshinone IIA, (IS) astragaloside.
Furthermore, because of the similarity between their structures, retention time and ionization response in ESI-MS, astragaloside was chosen as the internal standard for the saponins, bile acids and tanshinones. In order to develop a sensitive and accurate quantitative method, the MS/MS fragmentation for each compound was investigated by direct infusion of the single standard solution into the mass spectrometer, and the resulting mass spectra of product ions were recorded. All chemical constituents were characterized according to their mass spectra in order to ascertain their precursor ions and to select proper product ions for MRM analysis. Finally, the most sensitive transition in MRM was selected.
2.4. Method Validation
2.4.1. Linearity and Detection Limit
The calibration curves plotted against the standard solution were constructed from the peak area ratios of each standard vs. concentrations of each constituent. Therefore we can determine that this method is sensitive for the quantitative determination of the major components in STP samples. The linearity of the calibration curves were verified by correlation study, where the correlation coefficients were better than 0.9990 within the test ranges. The LODs and LOQs for the 13 compounds were less than 0.51 and 1.70 μg/L, respectively (Table 2).
Table 2.
Calibration curves, linear ranges, LOQ and LOD of 13 detected compounds (n = 3).
| Compound | Linear | Linear Range (μg/mL) | r | LOQ (μg/mL) | LOD (μg/mL) |
|---|---|---|---|---|---|
| Taurine | y = 0.0707x − 0.0005 | 10.03–0.40 | 0.9999 | 0.1300 | 0.0390 |
| Ginsenoside Rg1 | y = 0.0574x − 0.0102 | 7.51–0.75 | 0.9998 | 0.6000 | 0.1800 |
| Arenobufagin | y = 0.3438x − 0.0003 | 5.02–0.40 | 0.9996 | 0.0300 | 0.0090 |
| TDCA | y = 0.0125x − 0.0061 | 25.28–3.75. | 0.9992 | 1.7000 | 0.5100 |
| TCA | y = 0.0525x − 0.0094 | 10.53–0.75 | 0.9996 | 0.6000 | 0.1800 |
| Bufalin | y = 1.1663x + 0.0624 | 4.01–0.40 | 0.9999 | 0.0032 | 0.0010 |
| CA | y = 0.0909x − 0.0211 | 10.07–1.01 | 0.9998 | 0.7800 | 0.2340 |
| Cinobufagin | y = 0.1767x + 0.0523 | 5.02–0.75 | 0.9998 | 0.0003 | 0.0010 |
| Resibufogenin | y = 0.9504x − 0.0596 | 4.01–0.50 | 0.9993 | 0.2100 | 0.0630 |
| Ginsenoside Rg3 | y = 0.048x + 0.0256 | 2.01–0.01 | 0.9994 | 0.0041 | 0.0012 |
| CDCA | y = 0.2824x − 0.0241 | 5.01–0.75 | 0.9993 | 0.2900 | 0.0870 |
| DCA | y = 0.2049x + 0.0145 | 4.01–0.40 | 0.9990 | 0.0050 | 0.0015 |
| Tanshinone IIA | y = 34.5600x − 0.0030 | 0.05–0.01 | 0.9995 | 0.0042 | 0.0013 |
2.4.2. Precision, Repeatability, Stability and Recovery
Precision, repeatability and stability of the method were also validated for each analyte. The RSDs were less than 4.88%, respectively. In conclusion, our experimental method provided high precision, repeatability, and stability. The accuracy of the method was determined by a recovery test (Table 3). As shown in Table 4, the recovery rate of the 13 standards varied from 98.24%–101.02% (RSD ≤ 4.48%). These results verified the high recovery and accuracy of this method.
Table 3.
Precision, repeatability, stability of 13 detected components of STP.
| Analyte | Repeatability | Precision RSD% | Stability | ||
|---|---|---|---|---|---|
| Content (mg/g) ± SD | RSD% n = 6 | Intra-Day n = 6 | Inter-Day n = 9 | RSD% n = 6 | |
| Taurine | 35.2978 ± 0.7821 | 2.22 | 2.35 | 2.42 | 2.71 |
| Ginsenoside RG1 | 8.7050 ± 0.2457 | 2.82 | 4.69 | 4.30 | 3.24 |
| Arenobufagin | 10.9163 ± 0.2826 | 2.59 | 3.13 | 3.95 | 4.33 |
| TDCA | 43.8546 ± 1.8145 | 4.14 | 3.96 | 3.08 | 2.47 |
| TCA | 29.7562 ± 1.3109 | 4.41 | 4.17 | 3.34 | 3.55 |
| Bufalin | 6.9432 ± 0.2124 | 3.06 | 3.88 | 4.20 | 3.40 |
| CA | 33.6313 ± 0.8419 | 2.50 | 3.52 | 3.58 | 2.58 |
| Cinobufagin | 19.1693 ± 0.4315 | 2.25 | 2.71 | 2.63 | 3.91 |
| Resibufogenin | 8.1053 ± 0.1211 | 1.49 | 2.88 | 3.00 | 3.00 |
| Ginsenoside RG3 | 8.5907 ± 0.2642 | 3.08 | 2.63 | 2.60 | 2.25 |
| CDCA | 14.7924 ± 0.5840 | 3.95 | 4.79 | 3.46 | 2.12 |
| DCA | 5.7912 ± 0.1679 | 2.90 | 2.01 | 2.37 | 2.24 |
| Tanshinone IIA | 0.0208 ± 0.0007 | 3.54 | 4.00 | 4.55 | 4.88 |
Table 4.
Recoveries of 13 detected components of STP.
| Analyte | Original (μg) | Added (μg) | Detected (μg) (±SD n = 3) | Recovery (%) (±SD n = 9) | RSD (%) |
|---|---|---|---|---|---|
| Taurine | 1764.89 | 2000.00 | 3734.82 ± 73.55 | 99.20 ± 3.00 | 3.02 |
| 1500.00 | 3263.21 ± 55.01 | ||||
| 1000.00 | 2756.97 ± 27.37 | ||||
| Ginsenoside RG1 | 435.25 | 500.00 | 934.19 ± 22.09 | 98.86 ± 3.77 | 3.82 |
| 400.00 | 826.68 ± 12.69 | ||||
| 300.00 | 732.04 ± 14.85 | ||||
| Arenobufagin | 545.82 | 900.00 | 1422.16 ± 4.83 | 99.28 ± 3.49 | 3.52 |
| 600.00 | 1156.98 ± 26.84 | ||||
| 300.00 | 841.64 ± 10.52 | ||||
| TDCA | 2192.73 | 3000.00 | 5115.89 ± 43.01 | 98.64 ± 2.96 | 3.00 |
| 2000.00 | 4195.34 ± 57.22 | ||||
| 1000.00 | 3176.37 ± 43.66 | ||||
| TCA | 1487.81 | 2000.00 | 3435.51 ± 54.39 | 99.26 ± 3.31 | 3.34 |
| 1500.00 | 2994.86 ± 70.89 | ||||
| 1000.00 | 2487.12 ± 24.57 | ||||
| Bufalin | 347.16 | 500.00 | 843.70 ± 24.18 | 99.00 ± 4.18 | 4.23 |
| 400.00 | 744.40 ± 18.15 | ||||
| 300.00 | 642.24 ± 15.03 | ||||
| CA | 1681.57 | 2000.00 | 3682.31 ± 57.38 | 99.20 ± 3.09 | 3.11 |
| 1500.00 | 3149.48 ± 52.01 | ||||
| 1000.00 | 2678.33 ± 37.17 | ||||
| Cinobufagin | 958.47 | 1500.00 | 2428.88 ± 47.17 | 98.64 ± 3.38 | 3.00 |
| 1000.00 | 1963.89 ± 36.52 | ||||
| 500.00 | 1434.10 ± 3.47 | ||||
| Resibufogenin | 405.27 | 500.00 | 896.21 ± 13.03 | 98.24 ± 3.20 | 3.25 |
| 400.00 | 795.04 ± 12.45 | ||||
| 300.00 | 702.54 ± 14.18 | ||||
| Ginsenoside RG3 | 429.54 | 500.00 | 928.54 ± 19.34 | 99.63 ± 3.55 | 3.56 |
| 400.00 | 825.86 ± 17.65 | ||||
| 300.00 | 729.51 ± 11.68 | ||||
| CDCA | 739.62 | 1500.00 | 2222.5 ± 54.47 | 98.67 ± 2.97 | 3.01 |
| 750.00 | 1475.11 ± 17.65 | ||||
| 500.00 | 1234.91 ± 19.78 | ||||
| DCA | 289.56 | 600.00 | 888.35 ± 23.94 | 99.38 ± 3.23 | 3.25 |
| 300.00 | 584.70 ± 11.05 | ||||
| 150.00 | 439.51 ± 4.75 | ||||
| Tanshinone IIA | 1.04 | 2.00 | 3.01 ± 0.09 | 101.02 ± 4.53 | 4.48 |
| 1.00 | 2.05 ± 0.05 | ||||
| 0.50 | 1.56 ± 0.03 |
2.5. Sample Analysis
The established UPLC-QqQ-MS/MS analytical approach was subsequently used to determine the representative constituents in 8 batches of commercial STP products. The contents of the investigated compounds, based on their respective calibration curves are summarized in Table 3. There were significant variations among the contents of constituents between different batches of STP (Table 5). Among these, the main ingredients can be divided into bufadienolides, triterpene saponins, tanshinones, salvianolic acids, and bile acids. For instance, Sample 2 had the lowest contents of taurine, ginsenoside Rg1, and arenobufagin, whereas Sample 6 had the highest contents of CDCA, CA, cinobufagin, and resibufogenin. Moreover, the four bufadienolides, which includes arenobufagin, bufalin, cinobufagin, and resibufogenin, were found to be the major constituents, accounting for approximately 3.23%–14.06%, 2.75%–7.17%, 6.17%–19.17%, and 3.07%–8.11% composition of all batches, respectively. However, the contents of these four compounds varied significantly among the 13 different samples, which could result in the varying quality and efficacy of STP samples. Real sample data also demonstrated that UPLC-QqQ-MS/MS is a suitable method for the analysis of the active components in STP samples. Therefore, further studies of STP quality control should also focus on the composition of raw materials, as well as the optimization of parameters during processing and manufacturing.
Table 5.
Contents of 13 analytes in different STP samples.
| Lot No. Compounds (mg/g) | Sample 1 | Sample 2 | Sample 3 | Sample 4 | Sample 5 | Sample 6 | Sample 7 | Sample 8 |
|---|---|---|---|---|---|---|---|---|
| Taurine | 13.8735 | 10.0161 | 15.1052 | 14.4826 | 13.6382 | 35.2978 | 24.4059 | 17.9399 |
| Ginsenoside RG1 | 10.4003 | 4.7917 | 6.0041 | 5.4945 | 8.1709 | 8.7050 | 10.5062 | 9.8985 |
| Arenobufagin | 6.5724 | 3.2283 | 3.9044 | 6.4789 | 4.8138 | 10.9163 | 7.2296 | 14.0551 |
| TDCA | 43.2012 | 73.8723 | 52.7846 | 76.3674 | 96.1933 | 43.8546 | 78.2465 | 32.8261 |
| TCA | 21.2145 | 21.3171 | 33.0645 | 44.2204 | 52.5620 | 29.7562 | 47.4044 | 21.5164 |
| Bufalin | 4.3781 | 5.2646 | 2.7532 | 4.9919 | 3.4709 | 6.9432 | 5.4234 | 7.1698 |
| CA | 13.2086 | 25.3780 | 15.5592 | 15.1331 | 16.1522 | 33.6313 | 23.6953 | 23.8216 |
| Cinobufagin | 10.3166 | 6.1709 | 6.9125 | 11.3476 | 9.3716 | 19.1693 | 13.6489 | 15.7044 |
| Resibufogenin | 5.0181 | 3.0739 | 3.1118 | 5.4689 | 4.1013 | 8.1053 | 6.3188 | 7.4538 |
| Ginsenoside RG3 | 17.1353 | 3.2188 | 8.0643 | 8.0672 | 13.4715 | 8.5907 | 13.3079 | 6.4563 |
| CDCA | 3.7939 | 4.6744 | 10.4556 | 6.6600 | 8.0945 | 14.7924 | 11.2672 | 5.8183 |
| DCA | 5.5619 | 3.6310 | 2.2421 | 5.1843 | 6.1205 | 5.7912 | 10.0939 | 9.4447 |
| Tanshinone IIA | 0.0026 | 0.0028 | 0.0207 | 0.0031 | 0.0134 | 0.0208 | 0.0239 | 0.0154 |
3. Experimental Section
3.1. Materials and Reagents
Reference substances of arenobufagin, bufalin, resibufogenin, cinobufagin, turo-ursodesoxycholic acid (TDCA), taurocholic acid (TCA), cholic acid (CA), chenodeoxycholic acid (CDCA), ursodesoxycholic acid (UDCA), and hyodeoxycholic acid (HDCA) were purchased from Must Bio-Technology Co., Ltd. (Chengdu, China). Tanshinone IIA, salvianolic acid B, gensenoside Rg2, and gensenoside Rh1 were purchased from the National Institute for the Control of Pharmaceutical and Biological Products (Beijing, China). The purity of each compound was determined to be higher than 98% by HPLC. LC-grade acetonitrile and formic acid were purchased from Merck (Darmstadt, Germany). Deionized water for HPLC analysis was purified using a Milli-Q system (Millipore, Billerica, MA, USA). Methanol for sample extraction was from Merck. STP raw materials were provided by the Inner Mongolia Conba Pharmaceutical Co., Ltd. (Inner Mongolia, China).
3.2. Preparation of Standard and Sample Solutions
Stock solutions of 13 standards (ginsenoside Rg1, ginsenoside Rk3, cinobufagin, arenobufagin, bufalin, resibufogenin, tanshinone IIA, taurine, tauroursodeoxycholic acid, taurocholic acid, cholic acid, deoxycholic acid, chenodeoxycholic acid and astragaloside as internal standard) were prepared individually by dissolving an accurately weighed amount of reference compound in methanol at concentrations of 1.397, 1.273, 0.975, 0.635, 1.272, 1.115, 0.167, 1.096, 0.885, 2.491, 1.119, 1.065, and 1.300 mg/mL, respectively. Next, aliquots of each stock solution were mixed, and diluted with methanol to achieve a series of standard working solutions for the construction of calibration curves.
In order to obtain the purest possible chemical constituents from STP extract, eight batches of STP samples were ground into fine powder without the capsule outer casing and thoroughly mixed. Next, 0.5 g powder was accurately weighed and extracted with methanol (50 mL) in an ultrasonic water bath (40 kHz, 500 W) for 30 min at room temperature. The samples were then centrifuged at 16,000× g for 10 min after replenishing methanol. The solution was incubated for 24 h at 4 °C and the resulting supernatant was used as the sample solution for LC/MS analysis. The resulting solution was filtered through a membrane with 0.22 µm pores prior to use. Sample injected volume was 6 µL.
3.3. Qualitative and Quantitative Analysis Conditions
3.3.1. HPLC Method for Qualitative Analysis
HPLC-Q-TOF-MS/MS method for qualitative analysis was performed using a LC20AT HPLC instrument (Shimadzu, Kyoto, Japan) coupled with a micro-TOF-QII mass spectrometer (Bruker, Karlsruhe, Germany) equipped. The HPLC instrument includes a binary pump, an online degasser, and a thermostatically controlled column compartment. Chromatographic separation was carried out at 25 °C on an Inertsil ODS-SP C18 column (4.6 mm × 250 mm, 5 µm, GL Sciences Inc., Kyoto, Japan). The chromatographic conditions were as follows: flow rate of 0.8 mL/min, sample injection volume of 6 µL, mobile phase A (0.1% formic acid-water) and mobile phase B (100% acetonitrile) with a gradient elution program as follows: 0–5 min, 5%–5% B; 5–10 min, 5%–25% B; 10–60 min, 25%–48% B; 60–80 min, 48%–57% B; 80–100 min, 57%–90% B. Re-equilibration duration was 10 min between individual runs.
3.3.2. Q-TOF Method for Qualitative Analysis
The micrOTOF-Q II mass spectrometer was equipped with electrospray ionization (ESI) source and operated in positive and negative mode. For the negative ion mode MS detection, the optimized operating parameters were as follows: capillary, +3500 V; end plate offset, +500 V; transfer time of 80 µs. For the positive ion mode, the optimized operating parameters were as follows: capillary, −4500 V; end plate offset, −500 V; transfer time of 120 μs. For both negative and positive ion mode, the same operation parameters were as follows: drying gas (N2) flow rate, 4.0 L/min; drying gas temperature, 180 °C; nebulizer, 2.0 Bar; hexapole Rf, 100.0 Vpp; quadrupole ion energy, 3.0 eV; collision Rf, 150.0 Vpp; prepulse storage time, 5μs. Argon was applied as the collision gas, The sample collision energy was set at 25–55 eV to obtain the fragment ions data. Acquisition and analysis of data was performed with Data Analysis Version 4.0 SP1 (Bruker Technologies). Each sample was analyzed in both positive and negative modes to provide abundant information for structural identification. Mass spectra were recorded across the range 50–1500 m/z with accurate mass measurement of all mass peaks. Accurate mass measurements of each peak from the total ion chromatogram (TIC) were obtained by means of an automated calibration delivery system using a dual-nebulizer ESI source that introduces a low flow (100 μL/min) of a calibrating solution (sodium formate solution, Bruker Technologies), which was performed daily before sample injection, and contains the internal reference masses at m/z 90.9766 and 1518.7125 in positive ion mode and m/z 112.9856 and 1472.7341 in negative ion mode.
3.3.3. UPLC-MS Method for Quantitative Analysis
UPLC-QqQ-MS/MS method was used for qualitative analysis, and a Waters (Milford, MA, USA) Acquity UPLC H-Class system coupled with Xevo TQD QqQ-MS with ESI was employed. Separations were accomplished on Waters CORTECS C18 (2.1 mm × 100 mm, 1.6 μm) at a flow rate of 0.25 mL/min. The column temperature was maintained at 45 °C. The mobile phase was acetonitrile (phase A) and water (containing 0.1% formic acid, phase B) with a gradient elution program as follows: 0–3 min, 30%–30% A; 3–5 min, 30%–70% A; 5–6 min, 70%–85% A; 6–7 min, 85%–85% A; 7–8 min, 85%–100% A, 8.01–10 min, 30%–30% A. The MS spectra were acquired in multiple reaction monitoring (MRM) mode. The collision gas was argon. The nebulizer gas and the heater gas both were nitrogen. The MS conditions were optimized as follows: capillary voltage 2500 V in positive ion mode; source temperature, 150 °C; dwell time, 20 ms, dry gas flow, 800 L/h, the temperature of dry gas, 200 °C. The most appropriate setting such as precursor ion, daughter ion, cone voltage, collision energy (CE) was adjusted according to each analyte (Table 6).
Table 6.
The transitions and optimized MS parameters of 13 target markers and internal standard in the UPLC-QqQ-MS/MS analysis.
| Compounds | Rt (min) | Precursor Ion (m/z) | Daughter Ion (m/z) | Con Voltage (V) | Collision Energy (eV) |
|---|---|---|---|---|---|
| Taurine | 0.82 | 126.00 | 108.20 | 22 | 14 |
| Ginsenoside Rg1 | 1.25 | 823.30 | 643.50 | 80 | 40 |
| Arenobufagin | 2.23 | 417.20 | 399.10 | 45 | 30 |
| TDCA | 3.39 | 522.28 | 486.61 | 50 | 25 |
| TCA | 3.41 | 538.00 | 538.00 | 60 | 2 |
| Astragaloside | 4.91 | 807.00 | 807.00 | 80 | 30 |
| Bufalin | 5.22 | 387.30 | 351.20 | 35 | 22 |
| CA | 5.41 | 391.30 | 355.30 | 0 | 20 |
| Cinobufagin | 5.56 | 443.50 | 365.20 | 60 | 20 |
| Resibufogenin | 5.59 | 385.30 | 367.20 | 35 | 18 |
| Ginsenoside Rg3 | 5.75 | 807.00 | 365.00 | 90 | 5 |
| CDCA | 6.13 | 357.60 | 161.20 | 40 | 20 |
| DCA | 6.23 | 357.60 | 161.20 | 40 | 20 |
| Tanshinone IIA | 7.28 | 295.10 | 277.00 | 40 | 20 |
3.4. Validation of Quantitative Method
3.4.1. Calibration Curve, LOD, and LOQ
For the calibration curves, at least six concentrations of calibration standard solution were made and analyzed in triplicate. Next, the calibration curve of each analyte was constructed by plotting the peak area vs. the corresponding concentration.
The mixed standard solution with the lowest concentration was further diluted to a certain concentration to evaluate the LODs and LOQs. The LODs and LOQs were determined at an S/N ratio of 3 and 10, respectively.
3.4.2. Precision, Stability, Repeatability, and Recovery
The analysis of intra- and inter-day precisions was carried out by six repetitive injection of a mixed standard solution, in the same day and once a day for three consecutive days, respectively. Both assays were determined by performing three different concentration levels of the standards. Six samples were prepared independently to verify the repeatability of the experiment. To investigate the stability of the samples, each sample solution was analyzed every 4 h within 24 h, and stored at room temperature. The recovery was used to evaluate the accuracy of the method, and was determined by adding the mixed standard solutions at three different concentration levels (low, medium, and high) to 0.10 g of the known amounts of STP sample (Sample 6). The mixture was then extracted and analyzed. Three replicates were performed at each level. The percentages of recovery were calculated according to the following equation: (found amount − original amount) × 100%/spiked amount. The RSD was used to evaluate the results.
4. Conclusions
Our study demonstrates for the first time, the chemical profile of STP through a thorough and systematic investigation using HPLC-Q-TOF-MS/MS. In this study, a rapid and sensitive method based on HPLC-Q-TOF-MS/MS and UPLC-QqQ-MS/MS was established for the separation, identification and determination of the chemical constituents of STP. We identified a total of 41 components, including triterpene saponins, bufadienolides, bile acids, phenylally compounds and other compounds, which were successfully separated and identified by HPLC-Q-TOF-MS/MS. We also established the optimized UPLC-QqQ-MS/MS method for simultaneous determination of 13 compounds in STP under optimized UPLC conditions. The MRM mode of QqQ-MS/MS using our method enabled identification of target compounds with high sensitivity even at low concentrations, through comparison with standards. Rapid analysis performed within 10 min also facilitated the efficient quantitation of the target compounds in STP. This analytical method also verified its efficiency through method validation (linearity, precision and recovery test). Consequently, our method provided an accurate and efficient tool for the quality control of STP. The results of this study will aid in the discovery and analysis of biologically active compounds in STP, as well as facilitating improvements in the quality control standard of this commonly used multi-component TCM formula.
Acknowledgments
This study was sponsored by the Foundation of Fujian University of Traditional Chinese Medicine (X2013026) and the Developmental Fund of Chen Keji Integrative Medicine (CKJ2013016 & CKJ2013017 & CKJ2015003) and the Education Department of Fujian Province (JA14163). We also thank Inner Mongolia Conba Pharmaceutical Co., Ltd for their support.
Supplementary Materials
Supplementary materials can be accessed at: http://www.mdpi.com/1420-3049/20/10/18597/s1.
Author Contributions
D.C. and J.P. conceived and designed the experiments; D.C. and S.L. performed the experiments; W.X., J.L. and F.X. analyzed the data; J.C. contributed reagents/materials/analysis tools; D.C. and M.H. wrote the paper.
Conflicts of Interest
The authors declare no conflicts of interest.
Footnotes
Sample Availability: Samples of the compounds (1) taurine, (2) ginsenoside Rg1, (3) arenobufagin, (4) TDCA, (5) TCA, (6) bufalin, (7) CA, (8) cinobufagin, (9) resibufogenin, (10) ginsenoside Rg3, (11) CDCA, (12) DCA, (13) tanshinone IIA are available from the authors.
References
- 1.Chernetsova E.S., Crawford E.A., Shikov A.N., Pozharitskaya O.N., Makarov V.G., Morlock G.E. ID-CUBE direct analysis in real time high-resolution mass spectrometry and its capabilities in the identification of phenolic components from the green leaves of Bergenia crassifolia L. Rapid Commun. Mass Spectrom. 2012;26:1329–1337. doi: 10.1002/rcm.6226. [DOI] [PubMed] [Google Scholar]
- 2.Wang B., Zhang Y.-B., Li G.-Q., Guan H.-S., Tong G.-Z. Fingerprint chromatogram analysis of Radix Glehniae by LC coupled with hierarchical clustering analysis. Chromatographia. 2009;70:811–816. doi: 10.1365/s10337-009-1230-9. [DOI] [Google Scholar]
- 3.Wang R., Luo J., Kong L. Screening of radical scavengers in Scutellaria baicalensis using HPLC with diode array and chemiluminescence detection. J. Sep. Sci. 2012;35:2223–2227. doi: 10.1002/jssc.201200156. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Q., Hong B., Zheng L., Wang X., Cai D. Matrix solid-phase dispersion extraction followed by HPLC-diode array detection method for the determination of major constituents in a traditional Chinese medicine Folium isatidis (Da-qing-ye) J. Sep. Sci. 2012;35:2453–2459. doi: 10.1002/jssc.201200422. [DOI] [PubMed] [Google Scholar]
- 5.Gautam K., Kumar P., Poonia S. Larvicidal activity and GC-MS analysis of flavonoids of Vitex negundo and Andrographis paniculata against two vector mosquitoes Anopheles stephensi and Aedes aegypti. J. Vector Borne Dis. 2013;50:171–178. [PubMed] [Google Scholar]
- 6.Liakopoulou-Kyriakides M., Kyriakidis D. Croscus sativus-biological active constitutents. Stud. Nat. Prod. Chem. 2002;26:293–312. [Google Scholar]
- 7.Shen L., Cong W.-J., Lin X., Hong Y.-L., Hu R.-W., Feng Y., Xu D.-S., Ruan K.-F. Characterization using LC/MS of the absorption compounds and metabolites in rat plasma after oral administration of a single or mixed decoction of Shaoyao and Gancao. Chem. Pharm. Bull. 2012;60:712–721. doi: 10.1248/cpb.60.712. [DOI] [PubMed] [Google Scholar]
- 8.Wang D., Liang J., Yang W., Hou J., Yang M., Da J., Wang Y., Jiang B., Liu X., Wu W. HPLC/qTOF-MS-oriented characteristic components data set and chemometric analysis for the holistic quality control of complex TCM preparations: Niuhuang Shangqing pill as an example. J. Pharm. Biomed. Anal. 2014;89:130–141. doi: 10.1016/j.jpba.2013.10.042. [DOI] [PubMed] [Google Scholar]
- 9.Pfister S., Meyer P., Steck A., Pfander H. Isolation and structure elucidation of carotenoid-glycosyl esters in gardenia fruits (Gardenia jasminoides Ellis) and saffron (Crocus sativus Linne) J. Agric. Food Chem. 1996;44:2612–2615. doi: 10.1021/jf950713e. [DOI] [Google Scholar]
- 10.Wang Y., He S., Cheng X., Lu Y., Zou Y., Zhang Q. UPLC-Q-TOF-MS/MS fingerprinting of Traditional Chinese Formula SiJunZiTang. J. Pharm. Biomed. Anal. 2013;80:24–33. doi: 10.1016/j.jpba.2013.02.021. [DOI] [PubMed] [Google Scholar]
- 11.Yin Q., Wang P., Zhang A., Sun H., Wu X., Wang X. Ultra-performance LC-ESI/quadrupole-TOF MS for rapid analysis of chemical constituents of Shaoyao-Gancao decoction. J. Sep. Sci. 2013;36:1238–1246. doi: 10.1002/jssc.201201198. [DOI] [PubMed] [Google Scholar]
- 12.Zhong X., Guo J., Wang L., Luo D., Bei W., Chen Y., Yan K., Peng J. Analysis of the constituents in rat serum after oral administration of Fufang Zhenzhu Tiaozhi capsule by UPLC-Q-TOF-MS/MS. Chromatographia. 2012;75:111–129. doi: 10.1007/s10337-011-2164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H., Xu D., Yang L. The Protective Function of Shexiang on Endothelial Injury Induced by Angiotensin II Osmotic Pump in the Rat. Chin. J. Clin. Med. 2009;16:669–672. [Google Scholar]
- 14.Ning H. The analysis of Shexiang Tongxin dropping pill on treating coronary heart disease angina pectoris curative effect. Chin. J. Geriatr. Care. 2012;10:46. [Google Scholar]
- 15.Wu H., Høiby N., Yang L., Givskov M., Song Z. Effects of Radix Ginseng on microbial infections: A narrative review. J. Tradit. Chin. Med. 2014;34:227–233. doi: 10.1016/S0254-6272(14)60083-2. [DOI] [PubMed] [Google Scholar]
- 16.Lai C., Li S., Yu H., Wan J., Kan K., Wang Y. A rapid HPLC–ESI-MS/MS for qualitative and quantitative analysis of saponins in “XUESETONG” injection. J. Pharm. Biomed. Anal. 2006;40:669–678. doi: 10.1016/j.jpba.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Sui W. Clinical observation of Shexiang Tongxin dropping pill in treating senile unstable agina pectoris. Chin. Community Dr. 2011;9:177–178. [Google Scholar]
- 18.Wang X., Sun W., Sun H., Lv H., Wu Z., Wang P., Liu L., Cao H. Analysis of the constituents in the rat plasma after oral administration of Yin Chen Hao Tang by UPLC/Q-TOF-MS/MS. J. Pharm. Biomed. Anal. 2008;46:477–490. doi: 10.1016/j.jpba.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J., Xu X., Wang W. Shexiang Tongxin dropping pill Evaluate the Efficacy of the Treatment of Unstable Angina. Chin. J. Integr. Med. Cardio-/Cerebrovasc. Dis. 2011;9:917–918. [Google Scholar]
- 20.Xu S., Yang L., Zeng X., Zhang M., Wang Z. Characterization of compounds in the Chinese herbal drug Mu-Dan-Pi by liquid chromatography coupled to electrospray ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2006;20:3275–3288. doi: 10.1002/rcm.2717. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H.-Y., Hu P., Luo G.-A., Liang Q.-L., Wang Y.-L., Yan S.-K., Wang Y.-M. Screening and identification of multi-component in Qingkailing injection using combination of liquid chromatography/time-of-flight mass spectrometry and liquid chromatography/ion trap mass spectrometry. Anal. Chim. Acta. 2006;577:190–200. doi: 10.1016/j.aca.2006.06.053. [DOI] [PubMed] [Google Scholar]
- 22.Lee H.-J., Koung F.-P., Kwon K.-R., Kang D.-I., Cohen L., Yang P.-Y., Yoo H.-S. Comparative analysis of the bufonis venenum by using TLC, HPLC, and LC-MS for different extraction methods. J. Pharmacopunct. 2012;15:52–65. doi: 10.3831/KPI.2012.15.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu Y., Yu Z., Yang Z.J., Zhu G., Fong W. Comprehensive chemical analysis of Venenum bufonis by using liquid chromatography/electrospray ionization tandem mass spectrometry. J. Pharm. Biomed. Anal. 2011;56:210–220. doi: 10.1016/j.jpba.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 24.Shellie R.A., Marriott P.J., Huie C.W. Comprehensive two-dimensional gas chromatography (GC× GC) and GC× GC-quadrupole MS analysis of Asian and American ginseng. J. Sep. Sci. 2003;26:1185–1192. doi: 10.1002/jssc.200301404. [DOI] [Google Scholar]
- 25.Wan J.-Y., Liu P., Wang H.-Y., Qi L.-W., Wang C.-Z., Li P., Yuan C.-S. Biotransformation and metabolic profile of American ginseng saponins with human intestinal microflora by liquid chromatography quadrupole time-of-flight mass spectrometry. J. Chromatogr. A. 2013;1286:83–92. doi: 10.1016/j.chroma.2013.02.053. [DOI] [PubMed] [Google Scholar]
- 26.Wang C.-Z., Kim K.E., Du G.-J., Qi L.-W., Wen X.-D., Li P., Bauer B.A., Bissonnette M.B., Musch M.W., Chang E.B. Ultra-performance liquid chromatography and time-of-flight mass spectrometry analysis of ginsenoside metabolites in human plasma. Am. J. Chin. Med. 2011;39:1161–1171. doi: 10.1142/S0192415X11009470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wen X.-D., Yang J., Ma R.-H., Gao W., Qi L.-W., Li P., Bauer B.A., Du G.-J., Zhang Z., Somogyi J. Analysis of Panax notoginseng metabolites in rat bile by liquid chromatography–quadrupole time-of-flight mass spectrometry with microdialysis sampling. J. Chromatogr. B. 2012;895:162–168. doi: 10.1016/j.jchromb.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng C., Tian J., Lv M., Huang Y., Tian Y., Zhang Z. Development and Validation of a Sensitive LC-MS-MS Method for the Simultaneous Determination of Multicomponent Contents in Artificial Calculus bovis. J. Chromatogr. Sci. 2014;52:128–136. doi: 10.1093/chromsci/bms256. [DOI] [PubMed] [Google Scholar]
- 29.García-Reyes J.F., Hernando M.D., Molina-Díaz A., Fernández-Alba A.R. Comprehensive screening of target, non-target and unknown pesticides in food by LC-TOF-MS. TrAC Trends Anal. Chem. 2007;26:828–841. doi: 10.1016/j.trac.2007.06.006. [DOI] [Google Scholar]
- 30.Jiang P., Dou S., Liu L., Zhang W., Chen Z., Xu R., Ding J., Liu R. Identification of multiple constituents in the TCM-formula Shexiang Baoxin pill by LC coupled with DAD-ESI-MS-MS. Chromatographia. 2009;70:133–142. doi: 10.1365/s10337-009-1142-8. [DOI] [Google Scholar]
- 31.Díaz R., Ibáñez M., Sancho J.V., Hernández F. Target and non-target screening strategies for organic contaminants, residues and illicit substances in food, environmental and human biological samples by UHPLC-QTOF-MS. Anal. Methods. 2012;4:196–209. doi: 10.1039/C1AY05385J. [DOI] [Google Scholar]
- 32.Hernández F., Sancho J., Ibáñez M., Grimalt S. Investigation of pesticide metabolites in food and water by LC-TOF-MS. TrAC Trends Anal. Chem. 2008;27:862–872. doi: 10.1016/j.trac.2008.08.011. [DOI] [Google Scholar]
- 33.Yan Y., Chai C.-Z., Wang D.-W., Yue X.-Y., Zhu D.-N., Yu B.-Y. HPLC-DAD-Q-TOF-MS/MS analysis and HPLC quantitation of chemical constituents in traditional Chinese medicinal formula Ge-Gen Decoction. J. Pharm. Biomed. Anal. 2013;80:192–202. doi: 10.1016/j.jpba.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez-Aller M., Gurny R., Veuthey J.-L., Guillarme D. Coupling ultra high-pressure liquid chromatography with mass spectrometry: Constraints and possible applications. J. Chromatogr. A. 2013;1292:2–18. doi: 10.1016/j.chroma.2012.09.061. [DOI] [PubMed] [Google Scholar]
- 35.Huang M., Zhang Y., Xu S., Xu W., Chu K., Xu W., Zhao H., Lu J. Identification and quantification of phenolic compounds in Vitex negundo L. var. cannabifolia (Siebold et Zucc.) Hand.-Mazz. using liquid chromatography combined with quadrupole time-of-flight and triple quadrupole mass spectrometers. J. Pharm. Biomed. Anal. 2015;108:11–20. doi: 10.1016/j.jpba.2015.01.049. [DOI] [PubMed] [Google Scholar]
- 36.Schymanski E.L., Singer H.P., Slobodnik J., Ipolyi I.M., Oswald P., Krauss M., Schulze T., Haglund P., Letzel T., Grosse S., et al. Non-target screening with high-resolution mass spectrometry: Critical review using a collaborative trial on water analysis. Anal. Bioanal. Chem. 2015;407:6237–6255. doi: 10.1007/s00216-015-8681-7. [DOI] [PubMed] [Google Scholar]
- 37.Wu Z.-F., Wang Y.-Q., Wan N., Ke G., Yue P.-F., Chen H., Zhan J.-J., Yang M. Structural Stabilities and Transformation Mechanism of Rhynchophylline and Isorhynchophylline by Ultra Performance Liquid Chromatography/Time-of-Flight Mass Spectrometry (UPLC/Q-TOF-MS) Molecules. 2015;20:14849–14859. doi: 10.3390/molecules200814849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fraser K., Harrison S.J., Lane G.A., Otter D.E., Hemar Y., Quek S.-Y., Rasmussen S. Non-targeted analysis of tea by hydrophilic interaction liquid chromatography and high resolution mass spectrometry. Food Chem. 2012;134:1616–1623. doi: 10.1016/j.foodchem.2012.03.045. [DOI] [PubMed] [Google Scholar]
- 39.Guibal R., Lissalde S., Charriau A., Poulier G., Mazzella N., Guibaud G. Coupling passive sampling and time of flight mass spectrometry for a better estimation of polar pesticide freshwater contamination: Simultaneous target quantification and screening analysis. J. Chromatogr. A. 2015;1387:75–85. doi: 10.1016/j.chroma.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 40.Yang Y., Wang H.-J., Yang J., Brantner A.H., Lower-Nedza A.D., Si N., Song J.-F., Bai B., Zhao H.-Y., Bian B.-L. Chemical profiling and quantification of Chinese medicinal formula Huang-Lian-Jie-Du decoction, a systematic quality control strategy using ultra high performance liquid chromatography combined with hybrid quadrupole-orbitrap and triple quadrupole mass spectrometers. J. Chromatogr. A. 2013;1321:88–99. doi: 10.1016/j.chroma.2013.10.072. [DOI] [PubMed] [Google Scholar]
- 41.Xu W., Huang M., Li H., Chen X., Zhang Y., Liu J., Xu W., Chu K., Chen L. Chemical profiling and quantification of Gua-Lou-Gui-Zhi decoction by high performance liquid chromatography/quadrupole-time-of-flight mass spectrometry and ultra-performance liquid chromatography/triple quadrupole mass spectrometry. J. Chromatogr. B. 2015;986:69–84. doi: 10.1016/j.jchromb.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 42.Tomczyk M., Gudej J., Sochacki M. Flavonoids from Ficaria verna Huds. Z. Naturforschung C. 2002;57:440–444. doi: 10.1515/znc-2002-5-606. [DOI] [PubMed] [Google Scholar]
- 43.Huang M., Xu S., Xu W., Chen D., Chu K., Xu W., Peng J., Lu J. Qualitative and quantitative analysis of the major constituents in Jin-Mu-Gan-Mao tablet by high-performance liquid chromatography with diode-array detection and quadrupole time-of-flight tandem mass spectrometry. J. Sep. Sci. 2014;37:3497–3508. doi: 10.1002/jssc.201400631. [DOI] [PubMed] [Google Scholar]
- 44.Zedda M., Zwiener C. Is nontarget screening of emerging contaminants by LC-HRMS successful? A plea for compound libraries and computer tools. Anal. Bioanal. Chem. 2012;403:2493–2502. doi: 10.1007/s00216-012-5893-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.