Abstract
Chloroplast (cp) genome sequences provide a valuable source for DNA barcoding. Molecular phylogenetic studies have concentrated on DNA sequencing of conserved gene loci. However, this approach is time consuming and more difficult to implement when gene organization differs among species. Here we report the complete re-sequencing of the cp genome of Capsicum pepper (Capsicum annuum var. glabriusculum) using the Illumina platform. The total length of the cp genome is 156,817 bp with a 37.7% overall GC content. A pair of inverted repeats (IRs) of 50,284 bp were separated by a small single copy (SSC; 18,948 bp) and a large single copy (LSC; 87,446 bp). The number of cp genes in C. annuum var. glabriusculum is the same as that in other Capsicum species. Variations in the lengths of LSC; SSC and IR regions were the main contributors to the size variation in the cp genome of this species. A total of 125 simple sequence repeat (SSR) and 48 insertions or deletions variants were found by sequence alignment of Capsicum cp genome. These findings provide a foundation for further investigation of cp genome evolution in Capsicum and other higher plants.
Keywords: Capsicum, bird pepper, chloroplast, DNA sequencing
1. Introduction
Chloroplasts (cp) are membrane-bound organelles, mainly involved in the photosynthetic conversion of atmospheric CO2 into carbohydrates in which light energy is stored as chemical energy. Cp possess their own genome that encodes a range of genes, involved mainly in photosynthesis and some essential metabolic pathways [1,2]. The first reports on complete cp genome sequences from tobacco and liverwort were reported in 1986 [3,4]. Since then, with emerging rapid and cost-effective NGS sequencing approaches, 342 cp genome sequences from different lineages have been reported [5]. Analyses of the cp genome among land plants show that their genome structure and organization are highly conserved with a quadripartite structure [6,7,8].
Capsicum L. (pepper) is a genus of the highly diverse Solanaceae family and comprises approximately 32 recognized species [9]. Capsicum originated in the New World and is cultivated in temperate and tropical regions [10,11], but knowledge of its domestication is incomplete. Capsicum annuum var. glabriusculum, is a unique Capsicum species commonly known as the American bird pepper well-known for its rich variation in flavor and aroma. Peppers play important roles in various aspects of the economy, food and pharmaceutics [12]. Therefore, knowledge regarding the genetic diversity among the germplasm is vital for strategic germplasm collection, maintenance, conservation and utilization.
DNA barcoding is a taxonomic method that aims to provide rapid and accurate species identification using a standard DNA region. The highly conserved structure of cp genome organization is a potential source of information for phylogenetic reconstruction of species relationships among plants. The cp genome has a simple and stable genetic structure, and universal primers can be used to amplify target sequences. In land plants, the highly variable cp gene sequences, such as matK, rbcL and psbA-trnH, are considered efficient DNA barcodes [13,14,15]. The advent of DNA barcoding to identify plant species appears to be promising, but most of the individual plastid candidate barcodes lack species level resolution [16,17].
For finding multi-locus DNA barcodes of high resolution at species level, it is essential to determine the distribution and location of highly arranged sequences information present in the cp genome. Until now, only three complete cp genome sequences from Capsicum species, American bird pepper (Capsicum annuum var. glabriusculum) [18], Korean landrace “subicho” pepper (Capsicum annuum var. annuum) [19] and a cultivated pepper (Capsicum annuum L.) [20], have been reported. The complete cp genome sequence of Capsicum pepper, C. annuum var. glabriusculum, reported here augments the genetic information for Capsicum species which will facilitate multi-locus choice for plant barcoding, population, phylogenetic and cp genetic engineering studies of this species.
2. Results and Discussion
2.1. Chloroplast Genome Assembly
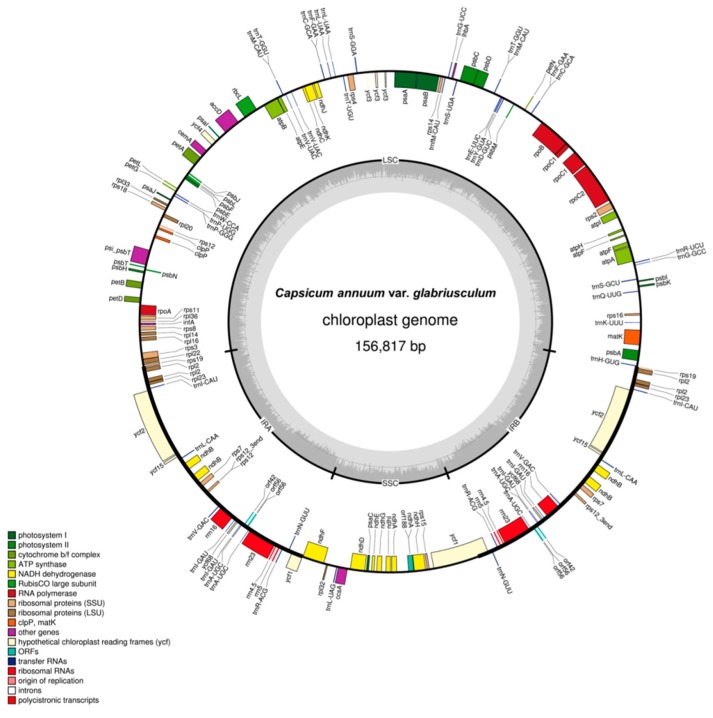
We sequenced the cp genome of C. annuum var. glabriusculum using the Illumina genome analyzer platform. Illumina paired-end (2 × 300 bp) sequencing produced a total of 7,716,442 paired-end reads, with an average fragment length of 277 bp, which were then analyzed to generate 1,964,163,823 bp of sequence. Low quality reads (Q20) were filtered out, and the remaining high quality reads were mapped to the reference cp genome of Capsicum, which contains 29,609,440 mapped nucleotides with an average coverage of 188× on the cp genome. The cp reads extracted from the Illumina dataset were assembled into a total of four contigs. Contig alignment and scaffolding based on paired-end data resulted in a complete circular C. annuum var. glabriusculum cp genome sequence (Figure 1). The genome sequence was deposited into GenBank under the accession number KR078311.
Figure 1.
Complete chloroplast genome map of C. annuum var. glabriusculum. Genes drawn inside the circle are transcribed clockwise, and those outside are transcribed counterclockwise and marked by two arrows. Differential functional gene groups are color-coded. The GC content variation is shown in the middle circle.
2.2. Features of the C. annuum var. glabriusculum Chloroplast Genome
The C. annuum var. glabriusculum cp genome is 156,817 bp in length. The GC content of the cp genome was 37.7%. The inverted repeats (IRs) had higher GC contents (43.05%) than those of the large single copy (LSC) (35.74%) or small single copy (SSC) (32.01%) regions due to the presence of GC-rich rRNA genes. The C. annuum var. glabriusculum cp genome is circular with quadripartite organization (Figure 1). The quadripartite structure includes two single copy DNA fragments, a LSC of 87,380 bp and a SSC of 17,853 bp, separated by a pair of IRs of 25,792 bp on a single circular molecule. The cp genome contains a total of 132 predicted genes (Table 1), including 87 protein-coding genes, 8 ribosomal RNA (rRNA) genes and 37 transfer RNA (tRNA) genes. Seven of these genes are duplicated in the IR regions, nine genes (Rps16, atpF, rpoC1, petB, petD, rpl16, rpl2 (IR), ndhB (IR), ndhA) and six tRNA genes contain one intron, and two genes (clpP, rps12) and one ycf (ycf3) contain two introns.
Table 1.
General features of the C. annuum var. glabriusculum chloroplast genome.
| Features | Chloroplast |
|---|---|
| Genome size (bp) | 156,817 |
| GC content (%) | 37.7 |
| Total number of genes | 132 |
| Protein coding genes | 87 |
| No. of rRNA genes | 8 |
| No. of tRNA genes | 37 |
| No. of gene duplicated in IR regions | 7 |
| Total introns | 12 |
| Single intron (gene) | 9 |
| Double introns (gene) | 3 |
| Single intron (tRNA) | 6 |
2.3. Discovery of SSRs and SNPs
A total of 125 potential SSRs motifs were identified which are located mostly in the non-coding regions (Table S1), and the majority belonged to tetra-nucleotide (50%) and tri-nucleotide (26%) repeats. All other types of SSRs such as di and penta-nucleotide motifs were relatively low (25%), and the majority of tetra-nucleotide SSRs had the AAAT/AATA/ATAA motif, followed by those with the ATAA/TAAA/AAAT motif, and the remaining those with the TTTG/TTGT/TGTT, TCTT/CTTT/TTTC, and AATT/ATTA/TTAA motifs were found with similar proportion (7.2%). Two different repeats those with the TTTTA/TTTAT/TTATT, and TTATT/TATTT/ATTTT motifs were identified among penta-nucleotide SSRs. The TTC/TCT/CTT and TTA/TAT/ATT motifs were identified among the tri-nucleotide SSRs. Only, the TA/AT motif was identified as the dinucleotide SSRs (Table S1). Comparison of C. annuum var. glabriusculum cp genome sequence with the reference cp sequence of C. annuum revealed a total of 48 mutations (15 SNPs and 33 InDels) and 32 of these variants involving more than one nucleotide (Tables S2 and S3). Amongst the detected variants, 5 SNPs and 3 InDels were observed in the coding region of the cp genome. Amongst these SNPs and InDels, there were 43 and 5 mutations located in LSC and SSC region, respectively.
2.4. Discussion
Here we report the re-sequencing and assembly of a cp genome using the Illumina sequencing platform in which we recovered four contigs comprising 156,817 bp covering the entire C. annuum var. glabriusculum cp genome. Reported Capsicum cp genomes range in size from 156,612 to 156,781 bp, and the size of the C. annuum var. glabriusculum cp genome identified here is consistent with those reported previously in plants of the same species [18,20]. The entire cp genome of C. annuum var. glabriusculum was 36 bp longer than the reported C. annuum L. cp genome (GenBank accession NC_018552) and 205 bp longer than another C. annuum var. glabriusculum cp genome (GenBank accession KJ619462). Also, the SSC and IR regions of C. annuum var. glabriusculum were 3 and 9 bp longer, respectively, and the LSC region was 14 bp shorter and 167 bp longer, respectively, than those of the previously reported cp genomes. The average GC content in the C. annuum cp genome is 37.7%, similar to other Capsicum species. The data generated using the Illumina platform covered a greater depth (188×) of the cp genome whereas, in the previous studies cp genome sequence coverage was not reported and were able to resolve the ambiguities present in the GS-FLX pyrosequencing. Thus, the data from the cp assembly reported here supports previous findings that Illumina can produce high quality sequence assemblies covering a greater genome depth [21].
The organization and gene order of the Capsicum cp genome exhibited the general cp genome structure of angiosperms [22]. The Capsicum cp genome contained 132 genes (Table 2), of which there were 8 rRNA genes, 37 tRNA genes, 21 ribosomal subunit genes (12 small subunit and 9 large subunit) and 4 DNA-directed RNA polymerase genes. Forty-six genes were involved in photosynthesis, of which 11 encoded subunits of the NADH-oxidoreductase, 7 for photosystem I, 15 for photosystem II, 6 for the cytochrome b6/f complex, 6 for different subunits of ATP synthase and 1 for the large chain of ribulose bisphosphate carboxylase. Five genes were involved in different functions, and three genes were of unknown function. As shown in Figure 1 and Table 2, genome organization appeared to be more conserved with unique gene sequences, as discovered previously in Capsicum species [18,19,20]. However, in this newly determined cp genome, we found 132 predicted genes and size variations were observed in the IR and LSC regions. A total of 125 cpSSRs markers were identified in 156.8 kb sequence of the Capsicum chloroplast genome. The observed frequency of SSRs was approximately 1/1.25 kb of chloroplast genome. More interestingly, the cpSSRs were only observed in the non-coding region of the cp genome. Similarly, most of the SNPs and InDels in the cp genome present in intergenic region, and only 8 variants were located in genic region (Tables S2 and S3).
Table 2.
Genes present in the C. annuum var. glabriusculum chloroplast genome.
| Gene Products of Capsicum annuum var. glabriusculum | |
|---|---|
| Photosystem I | psaA, B, C, I, J, ycf3 2, ycf4 |
| Photosystem II | psbA, B, C, D, E, F, H, I, J, K, L, M, N, T, Z |
| Cytochrome b6/f | petA, B 1, D 1, G, L, N |
| ATP synthase | atpA, B, E, F 1, H, I |
| Rubisco | rbcL |
| NADH oxidoreductase | ndhA 1, B 1,3, C, D, E, F, G, H, I, J, K |
| Large subunit ribosomal proteins | rpl2 1,3, 14, 16 1, 20, 22, 23 3, 32, 33, 36 |
| Small subunit ribosomal proteins | rps2, 3, 4, 7 3, 8, 11, 12 2,3,4, 14, 15, 16 1, 18, 19 |
| RNA polymerase | rpoA, B, C1 1, C2 |
| Unknown function protein coding gene | ycf1 3, 2 3, 15 3 |
| Other genes | accD, ccsA, cemA, clpP 2, matK |
| rRNAs | rrn16 3, 23 3, 4.5 3, 5 3 |
| tRNAs | trnA-UGC 1,3, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-UCC 1, trnG-GCC, trnH-GUG, trnI-CAU 3, trnI-GAU 1,3, trnK-UUU 1, trnL-UAA 1, trnL-UAG, trnL-CAA 3, trnfM-CAU, trnM-CAU, trnN-GUU 3, trnP-UGG, trnQ-UUG, trnR-ACG 3, trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-UAC 1, trnV-GAC 3, trnW-CCA, trnY-GUA |
1 Genes containing a single intron; 2 Genes containing two introns; 3 Two gene copies in IR; 4 Trans-splicing gene.
3. Experimental Section
3.1. Sampling and DNA Extraction
Sample (accession No. IT158289) was obtained from the National Agrobiodiversity Center, Rural Development Administration, Korea. Fresh leaves were collected from 40-day-old seedlings, and DNA was extracted to construct cp DNA libraries.
3.2. Library Preparation and Sequencing
An Illumina paired-end cp DNA library (average insert size of 500 bp) was constructed using the Illumina TruSeq library preparation kit following the manufacturer’s instructions. The libraries were sequenced with 2 × 300 bp on the MiSeq instrument at LabGenomics (http://www.labgenomics.co.kr/).
3.3. Chloroplast Genome Assembly
Prior to cp de novo assembly, low quality sequences (quality score < 20; Q20) were filtered out, and the remaining high quality reads were assembled using the CLC Genome Assembler (version beta 4.6, CLC Inc. Aarhus, Denmark) with a 200-600-bp overlap size. Cp contigs were selected from the initial assembly by performing a BLAST search against known cp sequences (GenBank accession NC_018552). The selected contigs were oriented to construct the complete cp genome structure. Ambiguous nucleotides or gaps were corrected manually to build the complete cp genome.
3.4. Gene Annotation
The web-based program Dual OrganellarGenoMe Annotator (DOGMA, http://dogma.ccbb.utexas.edu/) was used to annotate the assembled genome using default parameters to predict protein coding, tRNA and rRNA genes. Subsequently, BLASTN was used to further identify intron-containing gene positions by searching a published cp genome database. A cp gene map was constructed using the OrganellarGenomeDRAW software (OGDRAW, http://ogdraw.mpimp-golm.mpg.de).
3.5. Discovery of SNPs and SSRs
Sputnik (http://espressosoftware.com/pages/sputnik.jsp) software was used to find the SSR markers present in the cp genome of C. annuum var. glabriusculum. It uses a recursive algorithm to search for repeats with length between 2 and 5, and finds perfect, compound and imperfect repeats. Sputnik has been applied for SSR identification in many species including Arabidopsis and barley [23]. To identify SNP and INDEL variants in C. annuum var. glabriusculum cp genome, we used BWA [24] and Samtools [25] software. More detailed method and algorithm are descripted in Li (2012) [26].
4. Conclusions
The cp genome sequences of Capsicum species, such as C. annuum var. glabriusculum, C. annuum var. annuum and C. annuum L., have been reported previously; however, information on cp gene content is limited. The complete cp genome sequence of Capsicum pepper (C. annuum var. glabriusculum) reported here enhances the genomic information for C. annuum and contributes to the study of germplasm diversity. These data represent a valuable source of markers for future studies on Capsicum populations. Moreover, the complete cp genome sequence also provides data on functional protein variability in the cp.
Acknowledgments
This study was carried out with the support of the “Research Program for Agricultural Science and Technology Development (Project No. PJ008623)” and was supported by the 2014 Postdoctoral Fellowship Program of the National Academy of Agricultural Science, Rural Development Administration, Korea.
Supplementary Materials
Supplementary materials can be accessed at: http://www.mdpi.com/1420-3049/20/07/13080/s1.
Author Contributions
S.R. and Y.-W.N. analyzed the data and wrote the paper, J.R.L. and D.S. performed the experiments, K.-H.M. and S.-Y.L. contributed the materials, and J.-W.C. conceived and designed the experiments. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Footnotes
Sample Availability: Samples are not available.
References
- 1.Rodriguez-Ezpeleta N., Brinkmann H., Burey S.C., Roure B., Burger G., Loffelhardt W., Bohnert H.J., Philippe H., Lang B.F. Monophyly of primary photosynthetic eukaryotes: Green plants, red algae, and glaucophytes. Curr. Biol. 2005;15:1325–1330. doi: 10.1016/j.cub.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 2.Tetlow I.J., Rawsthorne S., Raines C., Emes M.J. Plastid metabolic pathways. In: Moller S.G., editor. Annual Plant Reviews, Plastids. Volume 13. Blackwell Publishing; Hoboken, NJ, USA: 2009. pp. 60–125. [Google Scholar]
- 3.Ohyama K., Fukuzawa H., Kohchi T., Shirai H., Sano T., Sano S., Umesono K., Shiki Y., Takeuchi M., Chang Z., et al. Chloroplast gene organization deduced from complete sequence of liverwort marchantia polymorpha chloroplast DNA. Nature. 1986;322:572–574. doi: 10.1038/322572a0. [DOI] [Google Scholar]
- 4.Shinozaki K., Ohme M., Tanaka M., Wakasugi T., Hayashida N., Matsubayashi T., Zaita N., Chunwongse J., Obokata J., Yamaguchi-Shinozaki K., et al. The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. EMBO J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. [(accessed on 6 July 2015)]. Available online: http://chloroplast.ocean.washington.edu/cpbase.
- 6.Sugiura M., Hirose T., Sugita M. Evolution and mechanism of translation in chloroplasts. Ann. Rev. Genet. 1998;32:437–459. doi: 10.1146/annurev.genet.32.1.437. [DOI] [PubMed] [Google Scholar]
- 7.Daniell H., Lee S.B., Grevich J., Saski C., Quesada-Vargas T., Guda C., Tomkins J., Jansen R.K. Complete chloroplast genome sequences of solanum bulbocastanum, solanum lycopersicum and comparative analyses with other solanaceae genomes. Theor. Appl. Genet. 2006;112:1503–1518. doi: 10.1007/s00122-006-0254-x. [DOI] [PubMed] [Google Scholar]
- 8.Leseberg C.H., Duvall M.R. The complete chloroplast genome of coix lacryma-jobi and a comparative molecular evolutionary analysis of plastomes in cereals. J. Mol. Evol. 2009;69:311–318. doi: 10.1007/s00239-009-9275-9. [DOI] [PubMed] [Google Scholar]
- 9.Moscone E.A., Scaldaferro M.A., Grabiele M., Cecchini N.M., Sanchez Garcia Y., Jarret R., Davina J.R., Ducasse D.A., Barboza G.E., Ehrendorfer F. The evolution of chili peppers (Capsicum-Solanaceae): A cytogenetic perspective. Acta Hortic. 2007;745:137–170. [Google Scholar]
- 10.Eshbaugh W.H. History and exploitation of a serendipitous new crop discovery. In: Janick J., Simon J.E., editors. New Crops. Wiley; New York, NY, USA: 1993. [Google Scholar]
- 11.Pozzobon M.T., Schifino-Wittmann M.T., Bianchetti L.D.B. Chromosome numbers in wild and semidomesticated Brazilian Capsicum L. (Solanaceae) species: Do x = 12 and x = 13 represent two evolutionary lines? Bot. J. Linn. Soc. 2005;151:259–269. doi: 10.1111/j.1095-8339.2006.00503.x. [DOI] [Google Scholar]
- 12.Kumar S., Kumar R., Singh J. Cayenne/American pepper (Capsicum species) In: Peter K.V., editor. Handbook of Herbs and Spices. Woodhead; Cambridge, UK: 2006. pp. 299–312. [Google Scholar]
- 13.Dong W., Liu J., Yu J., Wang L., Zhou S. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0035071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hollingsworth P.M., Graham S.W., Little D.P. Choosing and using a plant DNA barcode. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0019254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Group C.P.W., Hollingsworth P.M., Forrest L.L., Spouge J.L., Hajibabaei M., Ratnasingham S., van der Bank M., Chase M.W., Cowan R.S., Erickson D.L., et al. A DNA barcode for land plants. Pro. Natl. Acad. Sci. USA. 2009;106:12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Selvaraj D., Sarma R., Shanmughanandhan D., Srinivasan R., Ramalingam S. Evaluation of DNA barcode candidates for the discrimination of the large plant family Apocynaceae. Plant Syst. Evol. 2015;301:1263–1273. doi: 10.1007/s00606-014-1149-y. [DOI] [Google Scholar]
- 17.Spouge J.L., Mariño-Ramírez L. The practical evaluation of DNA barcode efficacy. Methods Mol. Biol. 2012;858:365–377. doi: 10.1007/978-1-61779-591-6_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng F.C., Gao C.W., Gao L.Z. The complete chloroplast genome sequence of American bird pepper (Capsicum annuum var. glabriusculum) Mitochondrial DNA. 2014 doi: 10.3109/19401736.2014.913160. [DOI] [PubMed] [Google Scholar]
- 19.Sebastin R., Young-Ah J., Jung-Ro L., Gi-An L., Kyung Jun L., Gyu-Taek C., Kyung-Ho M., Sok-Young L., Jong-Wook C. The complete chloroplast genome sequence of Korean landrace “subicho” pepper (Capsicum annuum var. annuum) Plant Breed. Biotechnol. 2015;3:88–94. [Google Scholar]
- 20.Jo Y.D., Park J., Kim J., Song W., Hur C.G., Lee Y.H., Kang B.C. Complete sequencing and comparative analyses of the pepper (Capsicum annuum L.) plastome revealed high frequency of tandem repeats and large insertion/deletions on pepper plastome. Plant Cell Rep. 2011;30:217–229. doi: 10.1007/s00299-010-0929-2. [DOI] [PubMed] [Google Scholar]
- 21.Wu Z.H., Gui S.T., Quan Z.W., Pan L., Wang S.Z., Ke W.D., Liang D.Q., Ding Y. A precise chloroplast genome of Nelumbo nucifera (Nelumbonaceae) evaluated with Sanger, Illumina Miseq, and Pacbio RS II sequencing platforms: Insight into the plastid evolution of basal eudicots. BMC Plant Biol. 2014;14 doi: 10.1186/s12870-014-0289-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sugiura M. The chloroplast genome. Plant Mol. Biol. 1992;19:149–168. doi: 10.1007/BF00015612. [DOI] [PubMed] [Google Scholar]
- 23.Cardle L., Ramsay L., Milbourne D., Macaulay M., Marshall D., Waugh R. Computational and experimental characterization of physically clustered simple sequence repeats in plants. Genetics. 2000;156:847–854. doi: 10.1093/genetics/156.2.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H., Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Proc G.P.D. The sequence alignment/map format and samtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H. Exploring single-sample SNP and INDEL calling with whole-genome de novo assembly. Bioinformatics. 2012;28:1838–1844. doi: 10.1093/bioinformatics/bts280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.