Abstract
Glucosinolates are anti-carcinogenic, anti-oxidative biochemical compounds that defend plants from insect and microbial attack. Glucosinolates are abundant in all cruciferous crops, including all vegetable and oilseed Brassica species. Here, we studied the expression of glucosinolate biosynthesis genes and determined glucosinolate contents in the edible organs of a total of 12 genotypes of Brassica oleracea: three genotypes each from cabbage, kale, kohlrabi and cauliflower subspecies. Among the 81 genes analyzed by RT-PCR, 19 are transcription factor-related, two different sets of 25 genes are involved in aliphatic and indolic biosynthesis pathways and the rest are breakdown-related. The expression of glucosinolate-related genes in the stems of kohlrabi was remarkably different compared to leaves of cabbage and kale and florets of cauliflower as only eight genes out of 81 were expressed in the stem tissues of kohlrabi. In the stem tissue of kohlrabi, only one aliphatic transcription factor-related gene, Bol036286 (MYB28) and one indolic transcription factor-related gene, Bol030761 (MYB51), were expressed. The results indicated the expression of all genes is not essential for glucosinolate biosynthesis. Using HPLC analysis, a total of 16 different types of glucosinolates were identified in four subspecies, nine of them were aliphatic, four of them were indolic and one was aromatic. Cauliflower florets measured the highest number of 14 glucosinolates. Among the aliphatic glucosinolates, only gluconapin was found in the florets of cauliflower. Glucoiberverin and glucobrassicanapin contents were the highest in the stems of kohlrabi. The indolic methoxyglucobrassicin and aromatic gluconasturtiin accounted for the highest content in the florets of cauliflower. A further detailed investigation and analyses is required to discern the precise roles of each of the genes for aliphatic and indolic glucosinolate biosynthesis in the edible organs.
Keywords: glucosinolates, biosynthetic genes, expression analysis, Brassica oleracea, subspecies, edible organs
1. Introduction
Glucosinolates, β-thioglucoside-N-hydroxysulfates (cis-N-hydroximinosulfate esters), are sulfur-enriched, anionic secondary metabolites of plants synthesized from amino acids and sugars. They are synthesized in all vegetables and oilseed plants of the order Brassicales [1]. Upon hydrolysis, these metabolites not only confer characteristic flavors to Brassica vegetables [2,3] but also serve to prevent carcinogenesis in animals by regulating the cell cycle and stimulating apoptosis [4]. Hydrolysis by the myrosinase enzyme degrades glucosinolates into different bioactive products, mostly isothiocyanates [5,6,7]. Isothiocyanates such as sulforaphane [8,9] and indole-3-carbinol [10] are strongly anti-carcinogenic, whereas phenethyl isothiocyanate inhibits the transformation of carcinogens from one form to another [11,12]. In addition to their anti-carcinogenic properties in the animals that consume them, glucosinolates are anti-oxidative [13] and help defend against herbivores and microbes [14,15]. Apart from the various benefits of glucosinolates, a few of them, for example progoitrin, are also reported to have adverse effects in animals, with goitrogenic effects (i.e., enlargement of the thyroid) [16], although no evidence of any such effect has been reported in humans from Brassica consumption [17]. It is important to understand the genetics of biosynthesis and accumulation of health-promoting glucosinolates in order to increase their content for human and animal health and plant protection.
Plants contain over 200 structurally different glucosinolates, which are generally classified as aliphatic, indolic or aromatic based on their primary precursor amino acids [13,18]. The basic precursors of aliphatic, indolic and aromatic glucosinolates are methionine (or alanine, leucine, isoleucine and valine), tryptophan and phenylalanine (or tyrosine), respectively [13,19]. All three types of glucosinolates are generated by a characteristic biosynthetic pathway that involves elongation of the amino acid side chain by the addition of methylene groups, formation of core structure and subsequent secondary modification of amino acid side chains by oxidation, hydroxylation, methoxylation, sulfation, and glycosylation, etc. [20,21,22]. In Brassica species, most of the glucosinolates are biosynthesized from methionine. Elongation of the methionine side chain involves methylthioalkylmalate synthase (MAM), bile acid:sodium symporter family protein 5 (BASS5) and branched-chain aminotransferase (BCAT) [23,24,25,26]. Formation of the core structure is a five-step process that includes formation of aldoxime by cytochromes P450 of the CYP79 and CYP83 families, oxidation of aldoxime by members of the CYP83 family, formation of thiohydroximic acid followed by C–S cleavage, and formation of desulfoglucosinolate by S-glucosyltransferase and glucosinolates by sulfotransferase [27,28]. Subsequent secondary modification involves several gene loci, for example those encoding GS-OX, GS-AOP, GS-OH, BZO1 and CYP81F2. R2R3-Myb transcript factors and other nucleus-localized regulators participate in glucosinolate biosynthesis [18,29,30,31,32,33,34]. Moreover, the sulfate assimilatory pathway, which provides glutathione and 3′-phosphoadenosine 5′-phosphosulfate co-substrates during glucosinolate biosynthesis on desulfo precursor, also involves several other genes [20].
Brassica oleracea is an important diversified vegetable species in which it has become clear that glucosinolate biosynthetic and catabolism pathways are different compared to those in Arabidopsis and B. rapa. B. oleracea also shows greater glucosinolate profile diversity than B. rapa and B. napus [35]. B. oleracea and B. rapa respectively have 105 and 101 glucosinolate metabolism-related genes, among which 22 genes are related to catabolism [35]. The coding DNA sequences of 84 B. oleracea genes related to glucosinolate biosynthesis [35] are available in two independent databases: Bolbase and EnesmblPlants, but expression analysis has been carried out for none of those genes to date. Therefore, a comparative validation of the coding sequences deposited in the two databases is necessary prior to functional analysis. The glucosinolate biosynthesis and catabolism among Arabidopsis, B. rapa and B. oleracea is likely related but also shows variation in the proportion of tandem genes, and the number and functions of genes for MAM and 2-oxoglutarate-dependent dioxygenase (AOP) [35]. Functions of MAM family members for condensation, side chain elongation and chain length production during glucosinolate biosynthesis differs in Arabidopsis compared to B. rapa and B. oleracea. MYB76 transcription factor is present in Arabidopsis but B. oleracea and B. rapa lack in that factor [36]. In addition to 4C glucosinolate, biosynthesis of sinigrin, a 3C glucosinolate, in B. oleracea is assumed to be related to high expression of the Bol017070 gene, while its ortholog, Bra013007, remain silenced in B. rapa [35]. By contrast, B. rapa biosynthesizes more of the 5C glucosinolate glucobrassicanapin due to higher expression of MAM3 compared to that in B. oleracea [35]. B. oleracea has only one functional AOP gene (AOP2) whereas Arabidopsis and B. rapa have four and three functional AOP genes, respectively [35].
Under different environmental conditions a complex network of transcription factors from the R2R3-MYB family regulate the glucosinolate biosynthetic pathways [18,29,30,31,32,33,34,37,38,39]. MYB28 and MYB29 are related to biosynthesis of aliphatic glucosinolates [18,33,34,38] while MYB51, MYB122 and MYB34 are related to indolic glucosinolate biosynthesis (Figure 1) [29,31,32,39]. Transcription factors and/or their stimulators might vary depending on herbivory [12] and sulfur metabolism [40]. Glucosinolate biosynthesis and catabolism differ not only among species [35] but also across developmental stages of tissues and organs during plant development [18,38,39,40,41,42,43,44]. B. oleracea is a vegetable-producing species that includes cabbage, broccoli, cauliflower, kale, brussels sprouts and kohlrabi. The edible organs are different in different subspecies; leaf is the edible organ for cabbage (B. oleracea capitata) and kale (B. oleracea acephala), whereas stem and floret are the edible organs of kohlrabi (B. oleracea italica) and cauliflower (B. oleracea botrytis), respectively. Currently, little is known about the expression of genes related to glucosinolate biosynthesis at in the edible organs of B. oleracea cultivars from qualitative and quantitative perspectives. In this study, we explored the genes related to glucosinolate biosynthesis and studied their expression in the edible organs of B. oleracea cultivars. We also measured glucosinolate contents in different edible organs of cabbage, kale, kohlrabi and cauliflower.
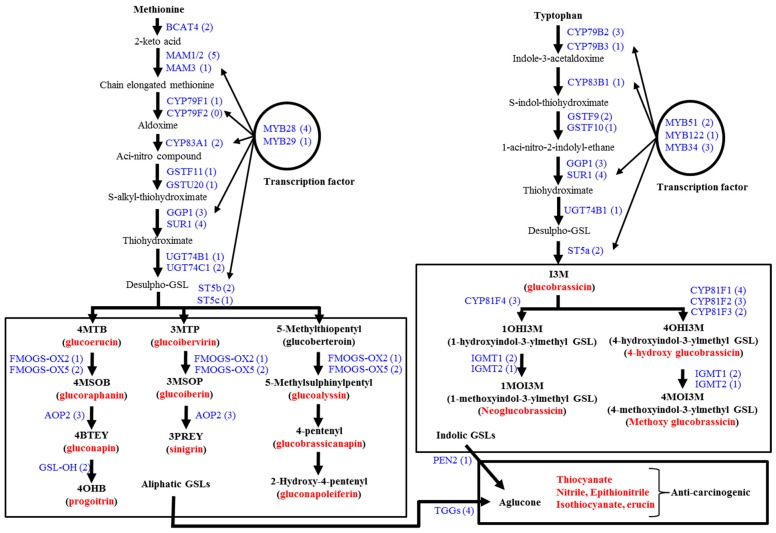
Figure 1.
Aliphatic and indolic glucosinolate metabolic pathways along with glucosinolate (GSL)-related genes in B. oleracea, after Liu et al. [35]. Red bold denotes the GSLs measured in this work by HPLC. Blue bold indicates enzymatic activities for which gene expression was monitored via RT-PCR. Numbers in parentheses are the numbers of genes identified. 4MTB, 4-methylthiobutyl GSL; 4MSOB, 4-methylsulfinylbutyl GSL; 4BTEY, 3-butenyl GSL; 4OHB, 4-hydroxybutyl GSL; 3MTP, 3-methylthiopropyl GSL; 3MSOP: 3-methylsulfinylpropyl GSL; 3PREY: 2-Propenyl GSL; I3M: indolyl-3-methyl GSL.
2. Experimental Section
2.1. Plant Materials and Growth Conditions
Seeds of 12 different genotypes from four different groups, three genotypes from each group of B. oleracea L., were purchased from Asia Seed Co., Ltd. (Seoul, Korea). The groups were B. oleracea capitata (cabbage), B. oleracea acephala (curly kale), B. oleracea italica (kohlrabi) and B. oleracea botrytis (cauliflower) (Table 1). The seedlings were raised in garden soil composed of peat moss, coco peat, perlite, zeolite and vermiculite in a growth chamber. Four-week-old seedlings were transferred to a glasshouse. Plants were grown for four months in the glasshouse before samples were destructively excised from several plants. Sampling sites were the edible organ of the plants (Table 1). The collected samples were snap frozen in liquid nitrogen and freeze-dried and stored at −80 °C for RNA isolation and/or high performance liquid chromatography (HPLC) analysis.
Table 1.
List of B. oleracea subspecies, genotypes and edible organs used to study the expression of glucosinolate-related genes and to estimate content of different glucosinolates.
| No. | Common Name | Genotype | Sampling Site/Edible Organ | Scientific Name of the Genotype |
|---|---|---|---|---|
| 1 | Cabbage | White cabbage | Leaf | B. oleracea L. convar capitata (L) Alef. var. alba DC |
| 2 | Cabbage | Cabbage | Leaf | B. oleracea var. capitata alba |
| 3 | Cabbage | Sprouting red cabbage | Leaf | B. oleracea capitata rubra |
| 4 | Kale | Curly kale Halftall | Leaf | B. oleracea acephala |
| 5 | Kale | Curly kale | Leaf | B. oleracea L. convar. acephala |
| 6 | Kale | Curly kale | Leaf | B. oleracea L. convar. acephala |
| 7 | Kohlrabi | Kohlrabi | Stem | B. oleracea var. italica Plenck |
| 8 | Kohlrabi | Kohlrabi | Stem | B. oleracea var. italica Plenck |
| 9 | Kohlrabi | Kohlrabi | Stem | B. oleracea L. convar. acephala (DC) Alef. var. gongyodes |
| 10 | Cauliflower | Cauliflower | Floret | B. oleracea L. convar. botrytis |
| 11 | Cauliflower | Cauliflower | Floret | B. oleracea L. convar. botrytis |
| 12 | Cauliflower | Cauliflower | Floret | B. oleracea L. convar. botrytis |
2.2. In Silico Analysis
For in silico analysis, the following databases were utilized: B. rapa genome database (http://brassicadb.org/brad/glucoGene.php), Bolbase, a comprehensive genomics database for B. oleracea, (http://www.ocri-genomics.org/bolbase/index.html) and EnsemblPlants (http://plants.ensembl.org/index.html). Gene symbols and annotated names for glucosinolate-related genes such those for transcription factors and enzymes related to side-chain elongation, core structure formation, secondary modification and co-substrate pathways of B. rapa were obtained from the two B. rapa databases [42]. B. oleracea orthologs along with full-length coding sequence (CDS) and % matching sequence between Bolbase and EnsemblPlants databases were then obtained (Table 2). In cases where Bolbase data were not available, EnsemblPlants data were used.
Table 2.
Glucosinolate (GSL) biosynthesis-related genes in B. oleracea as compared between Bolbase and EnsemblPlants.
| Gene Name | Biosynthesis Pathway | B. rapa | B. oleracea (Bolbase) | Total CDS of Bolbase | B. oleracea (Ensemblplants) | Total CDS of EnsemblPlants | Matching Sequence (%) *** |
|---|---|---|---|---|---|---|---|
| Transcription factors | Aliphatic and indolic GSLs | Bra012961 | Bol007795 (Cun) | 558 | Bo2g161590.1 | 1059 | 554(98.0) |
| MYB28 | Bra035929 | Bol036286 (C09) | 615 | Bo9g014610.1 | 1083 | 476(93.0) | |
| Bra029311 | Bol017019 (C06) | 426 | Bo7g098590.1 ** | 1068 | 426(99.0) | ||
| Bol036743 (C05) | 426 | Bo7g098590.1 ** | 1068 | 426(98.0) | |||
| MYB29 | Bra009245 | Bol008849 (C03) | 513 | Bo3g004500.1 | 993 | 513(94.0) | |
| Bra005949 | |||||||
| MYB34 | Bra013000 | Bol017062 (C06) | 951 | Bo7g098110.1 | 948 | 951(97.0) | |
| Bra035954 | Bol007760 (Cun) | 843 | Bo2g161180.1 | 843 | 843(97.0) | ||
| Bra029349 | Bol036262 (C09) | 294 | Bo9g014380.1 | 882 | 294(100.0) | ||
| Bra029350 | |||||||
| MYB51 | Bra016553 | Bol013207 (C08) | 1002 | Bo8g067910.1 | 1002 | 1002(100.0) | |
| Bra031035 | Bol030761 (C05) | 990 | Bo5g025570.1 | 990 | 990(99.0) | ||
| Bra025666 | |||||||
| MYB122 | Bra015939 | Bol026204 (C07) | 981 | Bo6g118350.1 | 1113 | 981(100.0) | |
| Bra008131 | |||||||
| Dof1.1 | Bra030696 | Bol023400 (C08) | 1005 | Bo8g010700.1 | 1350 | 915(99.0) | |
| Bra031588 | Bol041144 (C05) | 1011 | Bo5g008360.1 | 1011 | 1011(100.0) | ||
| Bol006511 (C08) | 936 | Bo8g112940.1 | 936 | 936(100.0) | |||
| IQD1 | Bra034081 | Bol023096 (C01) | 1656 | Bo1g144340.1 | 1368 | 1260(95.0) | |
| Bra001299 | Bol033935 (Cun) | 1437 | Bo3g061890.1 | 1437 | 1437(99.0) | ||
| TFL2 | Bra023629 | Bol021358 (C02) | 1146 | Bo2g013840.1 | 1152 | 1146(94.0) | |
| Bra013958 | Bol000201 (Cun) ** | 1236 | Bo9g159960.1 ** | 1372 | 1236(99.0) | ||
| Bra006417 | Bol019784 (C09) ** | 1236 | Bo9g159960.1 ** | 1372 | 1236(99.0) | ||
| Bol034455 (C03) | 999 | Bo3g012730.1 | 1248 | 906(95.0) | |||
| BCAT-4 | Aliphatic GSLs | Bra022448 | Bol018130 (C05) | 1083 | Bo5g113720.1 | 1083 | 1083(99.0) |
| Bra001761 | Bol026690 (C03) | 1083 | Bo3g073430.1 | 825 | 825(100.0) | ||
| MAM1/2 | Bra029355 | Bol017071 (C06) | 1236 | Bo7g098000.1 | 1527 | 1243(81.0) | |
| Bra013009 | Bol020647 (C03) | 1518 | Bo2g161100.1 ** | 1518 | 1518(99.0) | ||
| Bra029356 | Bol020646 (C03) | 804 | Bo2g161100.1 ** | 1518 | 760(82.0) | ||
| Bra021947 | Bol037823 (C04) | 1302 | Bo2g102060.1 | 1494 | 880(78.0) | ||
| Bra013011 | Bol017070 (C06) | 1527 | Bo7g098000.1 | 1527 | 1527(99.0) | ||
| MAM3 | Bra018524 | Bol016496 (C02) | 759 | Bo2g102060.1 | 1494 | 759(100.0) | |
| CYP79F1 | Bra026058 | Bol038222 (C05) | 1551 | Bo5g021810.1 | 1623 | 1551(99.0) | |
| CYP83A1 | Bra032734 | Bol040365 (C04) | 1506 | Bo4g130780.1 | 1506 | 1505(99.0) | |
| Bra016908 | Bol005188 (C04) | 570 | Bo4g191120.1 | 1509 | 570(100.0) | ||
| GSTF11 | Bra032010 | Bol000843 (Cun) | 633 | Bo5g150180.1 | 648 | 633(98.0) | |
| GSTU20 | Bra003645 | Bol021558 (C07) | 654 | Bo6g081630.1 | 654 | 654(99.0) | |
| UGT74C1 | Bra021743 | Bol006450 (Cun) | 1371 | Bo4g177530.1 | 657 | 647(99.0) | |
| Bra005641 | Bol014127 (C04) | 1371 | Bo4g049480.1 | 1372 | 1371(98.0) | ||
| STb | Bra015938 | Bol026202 (C07) | 1035 | Bo6g118360.1 | 1035 | 1035(100.0) | |
| Bra015936 | Bol026201 (C07) | 1035 | Bo6g118370.1 | 1035 | 1035(100.0) | ||
| STc | Bra025668 | Bol030757 (C05) | 1014 | Bo5g025610.1 | 1014 | 1014(99.0) | |
| FMOGS-OX2 | Bra027035 | Bol010993 (Cun) | 1386 | Bo9g037180.1 | 1386 | 1386(100.0) | |
| FMOGS-OX5 | Bra016787 | Bol029100 (C08) | 1347 | Bo8g062610.1 | 1347 | 1347(99.0) | |
| Bra026988 | Bol031350 (C08) | 1380 | Bo8g108390.1 | 1380 | 1380(100.0) | ||
| AOP2 | Bra034180 | Bol045938 | ? | Bo9g006240.1 | 1032 | ||
| Bra018521 | Bol045939 | ? | Bo2g102190.1 | 1104 | 1105 **** | ||
| Bra000848 | Bol045940 | ? | Bo3g052110.1 | 948 | 497 **** | ||
| GS-OH | Bra022920 | Bol033373 (C04) ** | 243 | Bo4g173530.1 ** | 1077 | 219(99.0) | |
| Bra021670 | Bol033374 (C04) ** | 243 | Bo4g173530.1 ** | 1077 | 219(99.0) | ||
| CYP79B2 | Indolic GSLs | Bra010644 | Bol032767 (C03) | 1557 | Bo3g152800.1 | 1557 | 1557(100.0) |
| Bra011821 | Bol028852 (C01) | 1623 | Bo1g002970.1 | 1623 | 1623(97.0) | ||
| Bra017871 | Bol018585 (C06) | 1626 | Bo7g118840.1 | 1626 | 1626(100.0) | ||
| CYP79B3 | Bra030246 | Bol031784 (Cun) | 1632 | Bo4g149550.1 | 1632 | 1632(99.0) | |
| CYP83B1 | Bra034941 | Bol033477 (C08) | 1473 | Bo8g024390.1 | 1500 | 1473(100.0) | |
| GSTF9 | Bra021673 | Bol033376 (C04) | 648 | Bo4g173610.1 | 648 | 648(99.0) | |
| Bra022815 | Bol004624 (C03) | 648 | Bo3g024840.1 | 648 | 648(99.0) | ||
| GSTF10 | Bra022816 | Bol004625 (C03) | 648 | Bo3g024850.1 | 648 | 648(99.0) | |
| STa | Bra008132 | Bol039395 (C02) | 1014 | Bo2g080910.1 | 1014 | 1014(100.0) | |
| Bra015935 | Bol026200 (C07) | 1017 | Bo6g118380.1 | 1017 | 1017(100.0) | ||
| CYP81F1 | Bra011762 | Bol028913 (C01) | 1500 | Bo1g003680.1 | 1437 | 882(97.0) | |
| Bra011761 | Bol028914 (C01) | 1497 | Bo1g003710.1 | 1497 | 1497(98.0) | ||
| Bol017375 (C07) | 369 | Bo6g095040.1 ** | 942 | 324(99.0) | |||
| Bol017376 (C07) | 246 | Bo6g095040.1 ** | 942 | 232(97.0) | |||
| CYP81F2 | Bra002747 | Bol012237 (C09) | 933 | Bo9g131960.1 | 1581 | 933(99.0) | |
| Bra020459 | Bol014239 (C02) | 1482 | Bo2g032590.1 | 1482 | 1482(99.0) | ||
| Bra006830 | Bol026044 (C03) | 1482 | Bo3g019420.1 | 1482 | 1482(100.0) | ||
| CYP81F3 | Bra010597 | Bol032711 (C03) | 1491 | Bo3g153480.1 | 1491 | 1491(99.0) | |
| Bra011758 | Bol028919 (C01) | 1500 | Bo1g004740.1 | 1500 | 1500(99.0) | ||
| CYP81F4 | Bra010598 | Bol032712 (C03) | 1506 | Bo01007s020.1 ** | 1506 | 1506(99.0) | |
| Bra011759 | Bol032714 (C03) | 960 | Bo01007s020.1 ** | 1506 | 807(98.0) | ||
| Bol028918 (C01) | 1503 | Bo1g004730.1 | 1506 | 1503(97.0) | |||
| IGMT1 | Bra012270 | Bol007029 (C08) | 1119 | Bo8g070650.1 | 1119 | 1119(100.0) | |
| Bra012269 | Bol020663 (C05) | 342 | Bo09472s010.1 | 312 | 342(100.0) | ||
| IGMT2 | Bra016432 | Bol007030 (C08) | 1125 | Bo8g070660.1 | 1350 | 1120(99.0) | |
| GGP1 * | Both aliphatic and indolic GSLs | Bra024068 | Bol033672 (C06) | 753 | Bo7g114570.1 | 753 | 753(99.0) |
| Bra011201 | Bol018073 (C01) | 753 | Bo1g012070.1 | 753 | 753(100.0) | ||
| Bra010283 | Bol012989 (C03) | 753 | Bo3g175530.1 | 753 | 753(100.0) | ||
| SUR1 * | Bra036490 | Bol038767 (C09) | 1209 | Bo7g003330.1 | 1371 | 1209(99.0) | |
| Bra036703 | Bol038764 (C09) ** | 459 | Bo28705s010.1 ** | 207 | 207(100.0) | ||
| Bol038765 (C09) ** | 459 | Bo28705s010.1 ** | 207 | 207(100.0) | |||
| Bol029775 (Cun) | 1008 | Bo7g003330.1 | 1371 | 991(89.0) | |||
| UGT74B1 * | Bra024634 | Bol005786 (C05) | 1311 | Bo5g041080.1 | 1401 | 1311(99.0) | |
| TGG1 | Breakdown aliphatic and indolic GSLs | Bra039825 | Bol017328 (C07) | 822 | Bo6g095780.1 | 411 | 373(99.0) |
| Bra039824 | |||||||
| Bra039823 | |||||||
| Bra016676 | |||||||
| Bra004012 | |||||||
| TGG2 | Bra036914 | Bol028319 (C08) | 1179 | Bo8g039420.1 | 1638 | 1163(99.0) | |
| Bol025706 (C03) | 663 | Bo2g155820.1 | 714 | 187(98.0) | |||
| TGG4 | |||||||
| TGG5 | Bol031599 (C07) | 1326 | Bo09266s010.1 | 163 | 163(100.0) | ||
| PEN2 | Bra004840 | Bol030092 (C04) | 1299 | Bo4g023800.1 | 3350 | 937(99.0) | |
| Bra004839 |
* Participates in biosynthesis of both aliphatic and indolic GSLs; ** These genes have the same CDS sequence in both databases; *** the number of matching base pairs between the sequence in Bol sequence (http://www.ocri-genomics.org/bolbase/) and that in EnsemblPlants (http://plants.ensembl.org/Brassica_oleracea/Info/Index?db=core); **** comparison between B. rapa and B. oleracea EnsemblPlants sequence.
2.3. cDNA Synthesis and Reverse-Transcriptase PCR Analysis
Total RNA of the samples harvested from 12 genotypes of four B. oleracea subspecies was extracted using the RNeasy mini kit (Catalogue No. 74106, Qiagen, Valencia, CA, USA). For cDNA synthesis, 5 μg total RNA, 1 μL gene-specific primer, 1 μL annealing buffer and 8 μL RNase were combined in a 0.2 mL thin-walled PCR tube on ice. Gene specific primers were designed using Primer3 website, http://primer3.ut.ee/ (Table 3). PrimeScript-based kit (Takara Bio, Inc., Shiga, Japan) was used for cDNA synthesis. There were two biological replicates for each genotype and gene combination. The RT-PCR experiment was repeated twice for each gene and genotype combination.
Table 3.
Primers used to amplify 81 glucosinolate biosynthetic genes in B. oleracea through RT-PCR.
| Gene Name | Acc. Number | cDNA Size | Forward | Reverse | Product Size (bp) |
|---|---|---|---|---|---|
| Aliphatic GSL Pathway | |||||
| BCAT-4 | Bol018130 | 1083 | TACGCGAATGTGAAGTGGGA | CCCCTTCTTATCCTCGACCC | 987 |
| Bol026690 | 1083 | TACGCGAATGTGAAGTGGGA | CACCGTCCACCCCTTCTTAT | 996 | |
| MAM1/2 | Bol017071 | 1236 | TGTTGCCCAGTGTGGAAAGG | TGAATGATACAGTTGGCTCCA | 970 |
| Bol020647 | 1518 | GTGACGGCGAACAATCTCC | AGCTTTCCAAGAACAATGCCT | 1000 | |
| Bol020646 | 804 | AATGATCCCTACCACCGGTTCAAACA | CTTGCGGCATGTTGATCTCC | 650 | |
| Bol037823 | 1302 | CAAGCTTCCCGACACGAATT | CGTCCGCTAAGCTTTCCAAG | 975 | |
| Bol017070 | 1527 | GCTCTTACTCCACCGCAGAA | CAACCCCAACATCTTCTGGC | 937 | |
| MAM3 | Bol016496 | 759 | CTACCGCCAACACAATCTC | CGTCCGTAATCCTCTTTTTCT | 504 |
| CYP79F1 | Bol038222 | 1551 | GAACCATCGGAGGCAATCAC | AGGTGACGCTCTGGTTTGTA | 970 |
| CYP83A1 | Bol040365 | 1506 | ATGTCAACTTCACGAACCGG | GTTAGGGCCCCACTCTTTCT | 953 |
| Bol005188 | 570 | GGGGTTAACGTTCGTCACTG | GAGCCCAGTCATGACATCCA | 442 | |
| GSTF11 | Bol000843 | 633 | TTGGGCAGATAAAAGCAGGT | GCAGCCATTTCCATAAGTTGC | 610 |
| GSTU20 | Bol021558 | 654 | CTGGCCAAGCATGTTCTGTA | TTCCTAAACTCAGCGGCGTA | 612 |
| UGT74C1 | Bol006450 | 1371 | CACACGAACACCCTCAAACC | AGCCTTCACTCTAACCCCAA | 989 |
| Bol014127 | 1371 | CCCTCACGCCAAGATCAAAG | CGGTCTTCACTCTAACCCCA | 980 | |
| ST5b | Bol026202 | 1035 | CGTACCGAACCAAGACAAGA | ACCATGTTCAAGCAAACCTGT | 1000 |
| Bol026201 | 1035 | GGACCAAACCAGGACGAGA | ACCATGTTCAAGCAAACCTGT | 999 | |
| ST5c | Bol030757 | 1014 | TCCAAATCCGAAAACGACGT | GCAAGAAAGCCAGTTCCTCT | 995 |
| FMOGS-OX2 | Bol010993 | 1386 | AGTCTCTCCGAACCAACCTC | AACCACATTCTTCTGCGACC | 978 |
| FMOGS-OX5 | Bol029100 | 1347 | CATAGTCCACTCCAGCGTCT | ACTTGCTCGGCTATCCAGTT | 993 |
| Bol031350 | 1380 | AACCGTAGTCCACTCTAGCG | CTGACCGACGACACCAAGA | 970 | |
| GSL-OH | Bol033373 | 243 | GATTGTGCAAAAGGCTTGT | AGAGCATTAGGATTAGGAGGA | 188 |
| Bol033374 | same as Bol033374 CDS | ||||
| AOP2 | Bo9g006240.1 | CCAGGAAGTGAGAAGTGGGT | TAGCACCATCACCAGCATCA | 517 | |
| Bo2g102190.1 | GGAACGTGTCTCCAAAACCC | TAGCACCATCACCAGCATCA | 354 | ||
| Bo3g052110.1 | CCAGGAAGTGAGAAGTGGGT | ACCAACATCCGCACCAGTAT | 552 | ||
| Indolic GSL pathway | |||||
| CYP79B2 | Bol032767 | 1557 | GTCAAGTCCTCCTTAGCCGT | CTTGAAGAAGTCTCGCGAGC | 219 |
| Bol028852 | 1623 | ATCACCGTCACATGCCCTAA | CCAGCCCATATCGACTGAGA | 991 | |
| Bol018585 | 1626 | CACTCTTACCTCAAACTCTTC | TTTCTTCCGCTCTCTTCT | 530 | |
| Indolic GSL pathway | |||||
| CYP79B3 | Bol031784 | 1632 | CGTCATTCCAGTCACATGTCC | ACGACCAAGTCCGTAACG | 1000 |
| CYP83B1 | Bol033477 | 1473 | GGACCTCAATTTCACCGCTC | TCCACTCCTTTCTGCTCGTT | 999 |
| GSTF9 | Bol033376 | 648 | TGTACGGACCTCACTTTGCT | TCAAGAGTCTCCTTCCAAGCA | 613 |
| Bol004624 | 648 | TGTACGGACCTCACTTTGCT | AAGAGTCTCCTTCCACGCAG | 611 | |
| GSTF10 | Bol004625 | 648 | TTGGTGAAGTGCTTGACGTG | CCGGCAATGCGTATTTCTCA | 228 |
| ST5a | Bol039395 | 1014 | CGGTCTTCACTCTAACCCCA | TCATGTTCAAGCAAGCCAGT | 983 |
| Bol026200 | 1017 | GATCCCAACTCGAGCTCTCA | TCATGTTGAAGCAAGCCAGT | 985 | |
| CYP81F1 | Bol028913 | 1500 | GAGACCTCCGCAGTAACCTT | GTCCTCCGTCGGTCTTCTAG | 222 |
| Bol028914 | 1497 | ACTTGATCCTCATCCCTCTCC | CATCGGAGTGAGTTGTGTCAC | 483 | |
| Bol017375 | 369 | AAGCAGAGCGGTTCAAGAAG | GCGTGACCATTGTGTTACCA | 204 | |
| Bol017376 | 246 | CCGTCTCCTTCAACGGTTCT | CGACGTATTTACCGGTGAGC | 170 | |
| CYP81F2 | Bol012237 | 933 | CTACGGAGACCAGGTTCACA | GTCATAATGGGACGCTGATGG | 897 |
| Bol014239 | 1482 | CGTGATCTCTTCTTTGCCCC | TCATCCCATAGCTTCGGGTC | 978 | |
| Bol026044 | 1482 | CCAACTCCCTTTCCCATC | TTGCTTTCCCCATCTCTTC | 689 | |
| CYP81F3 | Bol032711 | 1491 | TCTCACCCAAAAACCAAC | CTCCCTCCCAATACTTTC | 637 |
| Bol028919 | 1500 | CCTTACTCTCTCCCCATC | CTCCATAATATCTCTTCCCC | 478 | |
| CYP81F4 | Bol032712 | 1506 | CGTAGTTGTATCGAACCGCC | TTTCTCCTTCTCCTCCACCG | 980 |
| Bol032714 | 960 | GTTTCAACCTCCCTCCCTCT | CTCTGGGTCTTGTTGTTGCA | 818 | |
| Bol028918 | 1503 | CACTCTCTCCCCATTATT | ACCATCTTTATTTCCCCTAC | 687 | |
| IGMT1 | Bol007029 | 1119 | GTGTTCCTCTCACCTTCCGA | GTGTTGAGGAAGACGCTGTC | 260 |
| Bol020663 | 342 | AGATGCCATGATCTTGAAACGT | CCAGCAATGATAAGCCTGACA | 298 | |
| IGMT2 | Bol007030 | 1125 | TAGGTTTGATGGCCGTGAGA | CGAATTTGCAATGGGTGAAGC | 999 |
| Both aliphatic and indolic GSL pathway | |||||
| GGP1 | Bol033672 | 753 | TGTTTCTAGCAACTCCTGATTCA | AGTTCTTGCAAATCGTCTCCA | 699 |
| Bol018073 | 753 | TGTTTCTTGCAACTCCTGATTCA | AAATTCTTGCAAATCGTCTCCAA | 700 | |
| Bol012989 | 753 | TGTTTCTAGCGACTCTTGATTCA | AAATTCTTGCAAATCGTCTCCAA | 700 | |
| SUR1 | Bol038767 | 1209 | TCCGCACCTGTATCGAGG | CTCATCCAGTTCTTCACCCC | 1000 |
| Bol038764 | 459 | CTTCCGTCTATCCTTGCTT | CTGTATCTGTCTTCTTGGT | 360 | |
| Bol038765 | same as Bol038764 CDS | ||||
| Both aliphatic and indolic GSL pathway | |||||
| UGT74B1 | Bol029775 | 1008 | TGGTTCCCGCGTTTAAAACT | AGTGGAAGGGTCAGGAGTTA | 923 |
| Bol005786 | 1311 | AATCCTTCAAGCTCAACGGC | TCAAACACCTCACCACCTCA | 993 | |
| Breakdown of aliphatic and indolic GSLs | |||||
| TGG1 | Bol017328 | 822 | TCTTAACGTGTGGGATGGCT | CCTCCTTTGTTCACTCCCCT | 210 |
| TGG2 | Bol028319 | 1179 | AGATGTGCTGGACGAACTCA | CGGCGTAACAGGTAGGATCA | 401 |
| Bol025706 | 663 | CGTTTGGGATGGCTTCAGTC | TTCCTCGGTGAAGTTGGGAA | 421 | |
| TGG5 | Bol031599 | 1326 | TGCAGCACATAGAGCACTT | CGGTTCCAGAATCTCCTCC | 402 |
| PEN2 | Bol030092 | 1299 | GCATCATCATCCAACAGCGT | ACGCCTTGATCAGTTCTCCA | 207 |
| Transcription factors for aliphatic and indolic GSL pathways | |||||
| MYB28 | Bol007795 | 558 | GAGAGGTTCCTTGAGTTGCAAC | GAGAAATCGTAACCCTGATCCA | 238 |
| Bol036286 | 615 | GAAGGTAGCTTGAATGCTAATAC | TATGAGATGCTTTCCGAGGG | 414 | |
| Bol017019 | 426 | GTTGCGGCTAAGGTCACTTCT | CAGAAGTAGCGTTGATCTCATGC | 223 | |
| Bol036743 | 426 | GGTTTCTTGGGCGCTGCTAC | CCTCGATCATCAACGCTTGTT | 328 | |
| MYB29 | Bol008849 | 513 | CGCCCAAGACTTCTGAGTT | TGATATTGCCCATGGAAGCTG | 234 |
| MYB34 | Bol017062 | 951 | TGAGAACACCATGCTGCAAA | ACGAGCTTACAAACTTCTCCA | 918 |
| Bol007760 | 843 | ACCATGTTGCAAAGAAGAAGGA | CCAAACCATCTTCTTCGTTCCA | 812 | |
| Bol036262 | 294 | GGTTTCTCCGACAACTGTTCT | ACGAACTCACAAACTTCTCCA | 250 | |
| MYB51 | Bol013207 | 1002 | GCTTGTCTCCTACGTCAACC | GTCCTCCTCAAGAAACCTCGA | 850 |
| Bol030761 | 990 | GGACTCCCGAGGAAGATCAG | CCTCGACGTCATTGTTCACA | 537 | |
| MYB122 | Bol026204 | 981 | CCTTAGGGCCATCATCAGCT | ACCAGTTGTCAATCCCTTCAA | 510 |
| Dof1.1 | Bol023400 | 1005 | GACGAAACATAGCAGCTCCG | ACCGGGTTGTTCTTCCATCT | 227 |
| Bol041144 | 1011 | TTGGTCACAGCCTACGAACT | TCGTTAGAAGAAGGACCCGA | 975 | |
| Bol006511 | 936 | CCAATTGGTCACAGCCTACG | AGAAGGACCCGAGAAATCCG | 896 | |
| IQD1 | Bol023096 | 1656 | GGGGTAATTGGAACGACAGC | CTTTCCAACCAGCTCCAACC | 202 |
| Bol033935 | 1437 | CCACAAAACCAGCCGATGAA | AGGATCGTCTTGGTTTGGGT | 273 | |
| TFL2 | Bol021358 | 1146 | ACGATGCTGCTGAGAAGTCT | CCTGGTCCCCTTAACTCGTT | 199 |
| Bol000201 | 1236 | CTCTGCGGTTCAGGAGATGGG | CTCCAACACACCAGGATACTC | 381 | |
| Bol019784 | same as Bol000201 CDS | ||||
| Bol034455 | 999 | CTATCCGTCATAAGCGAGTTC | CGATGTCCAAGTTTGGTGTC | 531 | |
EmeraldAmp GT PCR Master Mix Cat, No/ID RR310A (Takara Bio, Inc., Shiga, Japan) was used for PCR mixture preparation. A typical PCR reaction included denaturation at 94 °C for 5 min, followed by 34 cycles at 94 °C for 30 s, 62 °C or 55 °C for 30 s, 72 °C for 1 min, and a final extension at 72 °C for 7 min. The actin gene (GenBank accession No. FJ969844) was used as a reference gene as it is expressed consistently in different organs of different species [45,46,47].
2.4. Extraction of Desulfo-Glucosinolates and HPLC Analysis
Desulfoglucosinolates from the selected samples were isolated using the HPLC protocol previously used by Choi et al. [47] with modifications. Fresh leaf tissue (100 mg) was sampled and snap-frozen in liquid nitrogen and stored at −80 °C freezer. The frozen samples were ground and treated with 1 mL 70% alcohol followed by incubation at 70 °C in a water bath for 10 min and at room temperature for 1 h. The tissue and proteins were precipitated by centrifugation for 8 min at 10,000 g at 4 °C and the supernatant was collected for anion-exchange chromatography. The extraction process was repeated twice and the combined supernatant was collected in a 5 mL tube. The combined supernatants represented the crude glucosinolate extracts. The supernatant was mixed with 0.5 mL each 50 mM barium acetate and 50 mM lead acetate. The crude glucosinolates were centrifuged again at 2000 g for 10 min and loaded onto a pre-equilibrated column and rinsed three times with 1 mL distilled water and 250 μL aryl sulfatase was added for desulfation. The desulfation process was allowed to continue for 16 h and then the desulfated glucosinolates were eluted with 1 mL distilled water. The eluted glucosinolates were centrifuged at 20,000 g for 4 min at 4 °C and passed through a filter to remove any impurities (PTFE, 13 mm, 0.2 μm; Advantec, Pleasanton, CA, USA).
The samples were then used for analysis with an HPLC system (Waters 2695, Waters, Milford, MA, USA) equipped with a C18 column (Zorbax Eclipse XBD C18, 4.6 mm × 150 mm, Agilent Technologies, Palo Alto, CA, USA). Water and acetonitrile were used as mobile phase solvents. A flow rate of 0.4 mL·min−1 was set at 30 °C. The desulfoglucosinolates were detected at 229 nm using a UV-visible detector (PDA 996, Waters) with commercially available sinigrin as a glucosinolate standard for quantification. A sinigrin standard curve was used to quantify the amount of glucosinolates in the samples. Individual glucosinolates were identified after mass spectrometry analysis (HPLC/MS, Agilent 1200 series, Agilent Technologies) using an Electrospray ionization interface operated in positive ion mode.
2.5. Statistical Analysis
The HPLC measurements of glucosinolate contents were analyzed via one-way analysis of variance using MINITAB 17 statistical software (Minitab Inc., State College, PA, USA). Pairwise comparisons of means were conducted following Tukey’s procedure for the significant statistical difference.
3. Results and Discussion
3.1. Genes Related to Glucosinolate Biosynthesis and Breakdown
A total of 84 B. oleracea genes orthologous to B. rapa genes related to glucosinolate biosynthesis, transcriptional regulation and breakdown were identified (Table 2).These 84 genes were distributed across all nine chromosomes of B. oleracea (Supplementary Table S1). Aliphatic biosynthesis, indolic biosynthesis and transcription factor-related genes identified in this study were not clustered in any particular chromosome, but rather were distributed across all nine chromosomes (Supplementary Table S1). The highest numbers of aliphatic and indolic biosynthesis genes were located on chromosomes C4 and C3, respectively (Supplementary Table S1). Nine genes were not able to be assigned to a chromosome and three AOP2 genes, related to aliphatic glucosinolate biosynthesis, were absent in Bolbase although they were present in EnsemblPlants (Table 2, Supplementary Table S1). Per Bolbase, the highest and the lowest number of glucosinolate genes per chromosome were 14 and four in chromosomes C3 and C2, respectively. There was more than 93% identity in coding sequence (CDS) for 77 genes between Bolbase and EnsemblPlants (Table 2). Three MAM1/2 genes and one SUR1 gene, showed 78%–89% identity in the two databases. CDS regions differed in number of nucleotides between the two databases for several genes (Table 2). The aliphatic and indolic glucosinolate pathways involved two sets of 32 genes and seven shared genes including three genes for GGP1, four genes for SUR1 (Bol038764 and Bol038765 have identical CDS) and one gene for UGT74B1 (Table 2, Figure 1). Twenty genes were transcription related and five genes were related to aglucone biosynthesis through the breakdown of glucosinolates (Figure 1). MYB28 and MYB29 are aliphatic transcription factor-related and MYB51, MYB122 and MYB34 are indolic transcription factor-related genes in B. oleracea (Figure 1). The pairs of genes Bol000201 and Bol019784 for TFL2, Bol033373 and Bol033374 for GS-OH, and Bol038764 and Bol038765 for SUR1 each shared the same CDS (Table 2).
3.2. Glucosinolate-Related Gene Expression in B. oleracea Subspecies
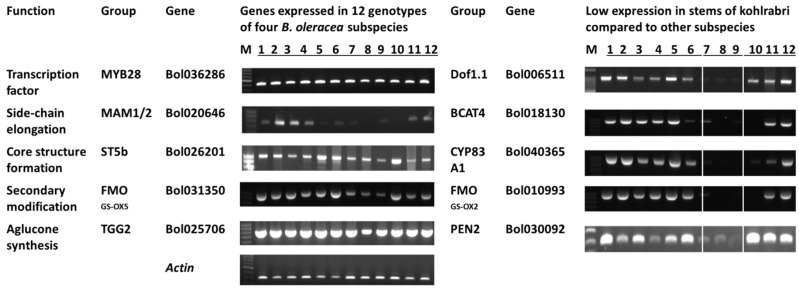
Compared to the edible organs of the other three subspecies, the stem samples of kohlrabi had much lower expression of many glucosinolate-related genes (Figure 2, Supplementary Figure S1). However, at least one gene from each of the following categories was expressed in the stems of kohlrabi: transcription factor, core structure formation, secondary modification and aglucone biosynthesis (Figure 2). Expression of side chain elongation-related genes was low in all four subspecies (Figure 2). The majority of other genes were highly expressed only in the leaves of cabbage and kale and in the florets of cauliflower (Figure 2). Only eight genes out of 84 were expressed similarly in the stems of kohlrabi as in leaves of cabbage and kale and floret of cauliflower (Supplementary Figure S1). These genes were MYB28, MYB51, IQD1 and TFL2 of the transcription-factor related set, AOP2 and ST5b aliphatic genes; the CSTF9 indolic gene and the TGG2 aglucone biosynthesis-related gene (Supplementary Figure S1). Genotypic differences in gene expression within subspecies for particular genes were also remarkable (Supplementary Figure S1). Some genes yielded notably different product size compared to the expected, for example: SUR1 (Bol038764, Bol038765), TGG2 (Bol025706), TGG5 (Bol031599). The observed product size of Bol038764, Bol025706 and Bol031599 were approximately 850, 800 and 900 bp whereas the expected product sizes from the primers designed based on Bolbase data were 360, 421 and 402 respectively (Supplementary Figure S1). Eight aliphatic pathway genes, three indolic pathway genes, five transcription factor-related genes and one breakdown-related gene were expressed in almost all genotypes of four cultivars (Supplementary Figure S1). These included aliphatic pathway CYP83A1 (Bol005188), GSTF11 (Bol000843), ST5b (Bol026201, Bol026202), ST5c (Bol030757), FMOGS-OX5 (Bol031350) and GSL-OH (Bol033373, Bol033374); indolic pathway GSTF9 (Bol033376, Bol004624) and GSTF10 (Bol004625); transcription factor-related MYB28 (Bol036286), Dof1.1 (Bol041144), IQD1 (Bol023096) and TFL2 (Bol000201, Bol019784) and breakdown-related TGG2 (Bol025706). Notably, expression levels of ST5b (Bol026202) and GSTF9 (Bol004624) were quite similar across all 12 genotypes (Supplementary Figure S1). Ten genes were expressed only in cabbage, kale and cauliflower but not in the stem of kohlrabi (Supplementary Figure S1). These genes were CYP83A1 (Bol040365), ST5a (Bol026200), SUR1 (Bol038764, Bol038765, Bol029775), UGT74B1 (Bol005786), Dof1.1 (Bol023400, Bol006511), TFL2 (Bol021358), and PEN2 (Bol030092).
Figure 2.
RT-PCR analysis of selected glucosinolate biosynthesis genes reveals differences in expression between stems of kohlrabi and edible organs of three other subspecies of B. oleracea. Genotypes 1–3, cabbage; 4–6, kale; 7–9, kohlrabi; 10–12, cauliflower.
3.3. Glucosinolate Analysis in B. oleracea Subspecies
HPLC analysis revealed the presence of 16 different types of glucosinolates in three different edible organs of four different subspecies of B. oleracea (Supplementary Table S2, Table 4). Cabbage leaves contained 12 glucosinolates, kale leaves contained 10, kohlrabi stems contained 11 and the cauliflower florets contained 14. Gluconapin, glucoalyssin, gluconapoleiferin and 4-hydroxy glucobrassicin were identified only the florets of cauliflower (Table 4). Glucoerucin was only found in the cabbage leaves, and glucoiberverin was identified in the cabbage leaves and kohlrabi stems (Table 4). The absolute amount of the three aliphatic glucosinolates gluconapin, glucoiberverin and glucobrassicanapin differed significantly or marginally among the edible organs (Table 4). Stems of kohlrabi contained the most glucoiberverin and glucobrassicanapin while the florets of cauliflower contained the highest gluconapin content compared to other edible organs (Table 4). Cauliflower florets recorded the highest content of methoxyglucobrassicin and gluconasturtiin, which are respectively indolic and aromatic glucosinolates (Table 4). Out of 11 aliphatic glucosinolates identified in three types of edible organs in 12 genotypes of four B. oleracea subspecies, only gluconapin, glucoalyssin and gluconapoleiferin were expressed in the florets of cauliflower (Table 4, Supplementary Table S3).
Table 4.
Glucosinolate content (μmol·g−1 DW) in edible organs of cabbage, kale, kohlrabi and cauliflower.
| Common Name (Edible Organ) | Aliphatic | Indolic | Aromatic | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GER | GRA | GNA | PRO | GIV | GIB | SIN | GAL | GBN | GNL | GRE | GBS | 4HGBS | MGBS | NGBS | GST | |
| Cabbage (Leaf) | 0.090 | 1.024 | 0.000b | 0.126 | 0.004b | 1.176 | 0.065 | 0.000 | 0.020ab | 0.000 | 0.851 | 1.906 | 0.000 | 0.078b | 0.105 | 0.141ab |
| Kale (Leaf) | 0.000 | 0.204 | 0.000b | 0.148 | 0.000b | 0.427 | 0.201 | 0.000 | 0.013ab | 0.000 | 0.074 | 1.146 | 0.000 | 0.126b | 0.637 | 0.119ab |
| Kohlrabi (Stem) | 0.000 | 0.303 | 0.000b | 0.333 | 2.115a | 1.247 | 0.601 | 0.000 | 0.071a | 0.000 | 0.079 | 0.434 | 0.000 | 0.046b | 0.120 | 0.006b |
| Cauliflower (Floret) | 0.000 | 0.060 | 0.053a | 0.080 | 0.000b | 0.406 | 0.073 | 0.115 | 0.006b | 0.117 | 0.524 | 0.252 | 0.044 | 1.093a | 0.621 | 1.185a |
| SE | 0.255 | 0.009 | 0.040 | 0.323 | 0.318 | 0.133 | 0.029 | 0.010 | 0.029 | 0.211 | 0.493 | 0.008 | 0.165 | 0.171 | 0.187 | 0.187 |
| p (subspecies) | 0.44 | 0.61 | 0.052 | 0.10 | 0.011 | 0.73 | 0.50 | 0.44 | 0.06 | 0.44 | 0.55 | 0.69 | 0.07 | 0.033 | 0.57 | 0.056 |
Different letters indicate significant difference. GER, glucoerucin; GRA, glucoraphanin; GNA, gluconapin; PRO, progoitrin; GIV, glucoiberverin; GIB, glucoiberin; SIN, sinigrin; GAL, glucoalyssin; GBN, glucobrassicanapin; GNL, gluconapoleiferin; GRE, glucoraphenin; GBS, glucobrassicin; 4HGBS; 4-hydroxy glucobrassicin; MGBS, methoxyglucobrassicin; NGBS, neoglucobrassicin; GST, gluconasturtiin.
3.4. Discussion
In this study a total of 84 genes related to glucosinolate biosynthesis in B. oleracea were compared. Furthermore, the expression of those genes and biosynthesis of glucosinolates in the edible organs were monitored in four B. oleracea subspecies. The study revealed a disparity in chromosome position for glucosinolate biosynthesis genes between Bolbase and EnsemblPlants databases (Supplementary Table S1). Moreover, the number of nucleotides in the CDS for several glucosinolate-related genes differs between Bolbase and EnsemblPlants (Table 2). These observations suggest that further investigation and validation of those two databases are required. In a future experimentation cloning and sequencing of the mismatched CDS would be targeted. In the present study, the 84 genes identified and expressed are expected to have high similarity with Arabidopsis thaliana and Brassica rapa, which have high ancestral synteny [35]. Other than those 84 genes, a recent study revealed that bHLH04, bHLH05, and bHLH06/MYC2 factors as novel regulators of glucosinolate biosynthesis in Arabidopsis, which belong to basic helix-loop-helix transcription factors and are essential for basal glucosinolate levels and response to jasmonic acid signal pathway; GTR1 and GTR2, which are involved in glucosinolate translocation [48]. Therefore in future investigation these three genes should be also included along with 84 genes reported in Liu et al. [35]. A previous study compared 52 glucosinolate biosynthetic genes between A. thaliana GLS (AtGS) and the draft B. rapa genome using nucleotide BLAST analysis [42]; high nucleotide sequence identity of about 72%–92% for the transcription factor-related genes was noted.
Kim et al. [44] studied a total of 17 transcription factor-related genes in B. rapa ssp. pekinensis involved in glucosinolate biosynthesis through aliphatic and indolic pathways in leaves, flower, stem and root. Similar to our study, expression of transcription factor-related genes was strikingly different in stem samples compared to leaves and florets [44]. Their relative expression level, compared to the reference gene, in young leaves and flowers was much higher compared to in stem [44], similar to the results of the present study. In B. rapa, the highest glucosinolate content was measured in seeds and the lowest in roots and old leaves [44]. The gene Bra035929 (encoding MYB28) in B. rapa exhibited 16- to 552-fold higher transcript levels in stems compared to seeds, young leaves and roots. Notably, the only B. oleracea orthologue of Bra035929, namely Bol036286, was expressed in all three genotypes of stem samples of kohlrabi, along with other edible organs (Figure 2). A MYB29 gene, Bol08849, an orthologue of Bra005949, which has 11- to 92-fold higher gene expression in stems of B. rapa [44], was expressed only in two genotypes of kale and one genotype of kohlrabi (Figure 2). These results are subject of further investigation as those genes were differentially expressed among genotypes within subspecies.
Both transcription factor-related genes and glucosinolate biosynthesis genes showed differences in expression in different plant organs such as seeds, stems, leaves and flowers in previous studies [25,44]. In A. thaliana, some important glucosinolate biosynthetic genes, such as CYP79B2, UGT74B1, CYP79F1, CYP79F2, IQD1, and Dof1.1, are expressed only in vascular tissues [19,30,31,49,50,51,52,53]. Desulfoglucosinolate sulfotransferases (BrST) isoforms, involved in core glucosinolate biosynthesis in B. rapa, were found to be expressed in mature leaf and root highly compared to other tissues, displaying functional redundancy for differential expression [53]. In our study, the edible organs of kohlrabi (stems) and those of cauliflower (florets) have much different types of structural and vascular tissues compared to the leaves of the other two subspecies analyzed, cabbage and kale, and hence the variation in expression of glucosinolate biosynthesis genes is expected. The fact that only one gene, namely Bol036286, out of five aliphatic transcription factor-related genes was expressed in all 12 genotypes including the stems of kohlrabi suggests that expression of this particular gene is essential in B. oleracea to induce desulfo-glucosinolates as a precursor of different aliphatic glucosinolates (Figure 2). Similarly, only one indolic transcription factor-related gene, Bol030761, was expressed in all 12 genotypes, suggesting that the presence of that gene is needed for continuation of the glucosinolate biosynthetic pathway (Supplementary Figure S1). The genes Bol025706 and Bol030092 should be essential for aglucone biosynthesis from the aliphatic and indolic glucosinolate pathways, respectively (Figure 1 and Figure 2). Expression analysis further suggests that in the aliphatic biosynthetic pathway two genes Bol031350 (FMOGS-OX5) and Bo9g006240 (AOP2) successively carry out glucosinolate transformation in the stems of kohlrabi from the primary glucosinolates glucoerucin and glucoibervirin derived from desulfo-glucosinolates produced by three ST5 genes (Figure 1, Supplementary Figure S1). Our results thus indicate that: (i) expression of all genes simultaneously is not required for glucosinolate biosynthesis in a particular organ; and (ii) the expression of a single gene or a few genes from each step is required to complete the glucosinolate biosynthesis. In addition, as in the stems of kohlrabi, expression of MYB28 and contents of aliphatic glucosinolate were detected, but expression of genes related to side-chain elongation were extremely low compared to that in other subspecies, suggesting the involvement of other transcription factors recently reported [48], or there is possibility that glucosinolates were transported.
Glucosinolate concentrations are commonly estimated on a tissue dry weight basis. The variation in glucosinolate concentrations we found in the different edible parts might be related to the fact that leaves, stems and florets have differences in water content. Accordingly, glucosinolate concentration on a tissue fresh weight basis could be different from that on a tissue dry weight basis. Thus, the variation observed in glucosinolate content in our study comparing tissues on a dry weight basis might be explained as a methodological variation.
Velasco et al. [54] found that glucosinolate concentration in the floral parts of B. oleracea acephala subspecies greatly increases from 300 days of age, but that it decreases rapidly in the leaf samples of the same plants. In this study, we measured glucosinolate concentration only at one time point. Similar to our study, the presence of glucoiberin, sinigrin and glucobrassicin was previously reported in all different subspecies of B. oleracea [55,56,57]. Likewise, in this study, other glucosinolates such as glucoraphanin, progoitrin, glucobrassicin, methoxyglucobrassicin, neoglucobrassicin and gluconasturtiin were also expressed in all three types of edible organs, such as in the leaves, stems and florets (Table 4). In B. oleracea var. italica, the patterns of glucosinolates were found to be mainly controlled genetically and less affected by environmental factors but several agronomic and environmental factors strongly influence the absolute content of various glucosinolates [2,58]. In particular, biosynthesis of aliphatic glucosinolates was found strongly genetically controlled in broccoli whereas that of indolic glucosinolates was controlled by genetic and environmental factors and by their interactions [59,60]. For example, high nitrogen and high sulphur content were found to increase the content of indolic glucobrassicin in cabbage cultivars [61,62].
Glucoraphanin and glucoiberin are the two most desirable glucosinolates from a nutritional perspective, whereas 2-hydroxy-3-butenyl (progoitrin) glucosinolate is undesirable as upon hydrolysis it produces oxazolidine-2-thione, which causes goiters in mammals and other harmful effects [56,63]. Glucoraphanin and glucoiberin were found in all four subspecies (Table 4). In this study, one of the cauliflower genotype measured no progoitrin (Supplementary Table S3). Wang et al. [56] found comparatively higher progoitrin in commercial broccoli genotypes compared to inbred lines, 1.77–6.07 μmol·g−1 and reported that it contributed around 20% of the total glucosinolates measured in that subspecies. Generally, B. rapa is abundant in that undesired glucosinolate [63]. The glucosinolates gluconapin, sinigrin, progoitrin, glucobrassicin and neoglucobrassicin show chemoprotective activity, but produce bitter and pungent isothiocyanates [60], so an excessive content might decrease consumer preference [64]. Cauliflower florets contained all five of these glucosinolates, whereas all but gluconapin were identified in all four subspecies under study (Table 4).
Among the four subspecies, the florets of cauliflower contained the highest number of glucosinolates with the lowest absolute content of progoitrin. This study thus identified that natural variation in glucosinolates and their absolute content exist among the edible organs of different B. oleracea subspecies, the results of which might be useful in breeding for glucosinolate contents or in transformation studies.
4. Conclusions
In this study, a total of 84 genes related to aliphatic, indolic and aromatic glucosinolate pathways or transcription/breakdown were subjected to RT-PCR-based analysis of expression in the edible organs of four species of B. oleracea. Only eight genes were expressed in the stem samples of kohlrabi, whereas majority of those genes were expressed in leaves of cabbage or kale and florets of cauliflower. The results are subject of further investigation as genotypic variation within subspecies is also evident along with subspecies difference. Out of 16 different types of identified glucosinolates, only five differed among the edible organs of four subspecies. Stems of kohlrabi contained the most glucoiberverin and glucobrassicanapin, whereas the florets of cauliflower had the highest contents of glucoraphanin, methoxyglucobrassicin and gluconasturtiin in the four-month-old plants. Overall, cauliflower florets had the highest number of glucosinolates and lacked undesirable progoitrin a genotype-dependent manner.
Acknowledgments
We thank the Asia Seed Co., Ltd., Republic of Korea for providing B. oleracea seeds. This study was supported by the Golden Seed Project (Center for Horticultural Seed Development, No. 213003-04-3-SB110) of the Ministry of Agriculture, Food and Rural affairs in the Korea (MAFRA).
Supplementary Information
Supplementary materials can be accessed at: http://www.mdpi.com/1420-3049/20/07/13089/s1.
Author Contributions
I.S.N., T.J.Y., J.G.K. and J.P. conceived and designed the study. K.Y. conducted in silico analysis. G.Y. managed the experimental plants, collected samples, prepared cDNA and conducted HPLC analysis. A.H.K.R. provided technical advice to prepare tables and figures to G.Y. and K.Y., analyzed data and wrote the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Seeds of the genotypes and cDNA are available from the authors.
References
- 1.Rodman J.E., Karol K.G., Price R.A., Sytsma K.J. Molecules, morphology, and Dahlgren’s expanded order Capparales. Syst. Bot. 1996:289–307. doi: 10.2307/2419660. [DOI] [Google Scholar]
- 2.Schonhof I., Krumbein A., Brückner B. Genotypic effects on glucosinolates and sensory properties of broccoli and cauliflower. Food Nahr. 2004;48:25–33. doi: 10.1002/food.200300329. [DOI] [PubMed] [Google Scholar]
- 3.Padilla G., Cartea M.E., Velasco P., de Haro A., Ordás A. Variation of glucosinolates in vegetable crops of Brassica rapa. Phytochemistry. 2007;68:536–545. doi: 10.1016/j.phytochem.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 4.Hayes J.D., Kelleher M.O., Eggleston I.M. The cancer chemopreventive actions of phytochemicals derived from glucosinolates. Eur. J. Nutr. 2008;47:73–88. doi: 10.1007/s00394-008-2009-8. [DOI] [PubMed] [Google Scholar]
- 5.Rask L., Andréasson E., Ekbom B., Eriksson S., Pontoppidan B., Meijer J. Plant Molecular Evolution. Springer; Dordrecht, The Netherlands: 2000. Myrosinase: Gene family evolution and herbivore defense in Brassicaceae; pp. 93–113. [PubMed] [Google Scholar]
- 6.Wittstock U., Halkier B.A. Cytochrome P450 CYP79A2 from Arabidopsis thaliana L. catalyzes the conversion of l-phenylalanine to phenylacetaldoxime in the biosynthesis of benzylglucosinolate. J. Biol. Chem. 2000;275:14659–14666. doi: 10.1074/jbc.275.19.14659. [DOI] [PubMed] [Google Scholar]
- 7.Bones A.M., Rossiter J.T. The enzymic and chemically induced decomposition of glucosinolates. Phytochemistry. 2006;67:1053–1067. doi: 10.1016/j.phytochem.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 8.Keck A.S., Finley J.W. Cruciferous vegetables: Cancer protective mechanisms of glucosinolate hydrolysis products and selenium. Integr. Cancer Ther. 2004;3:5–12. doi: 10.1177/1534735403261831. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y., Kensler T.W., Cho C.G., Posner G.H., Talalay P. Anticarcinogenic activities of sulforaphane and structurally related synthetic norbornyl isothiocyanates. Proc. Natl. Acad. Sci. USA. 1994;91:3147–3150. doi: 10.1073/pnas.91.8.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi H.S., Cho M.C., Lee H.G., Yoon D.Y. Indole-3-carbinol induces apoptosis through p53 and activation of caspase-8 pathway in lung cancer A549 cells. Food Chem. Toxicol. 2010;48:883–890. doi: 10.1016/j.fct.2009.12.028. [DOI] [PubMed] [Google Scholar]
- 11.Hecht S.S. Inhibition of carcinogenesis by isothiocyanates 1 *. Drug Metab. Rev. 2000;32:395–411. doi: 10.1081/DMR-100102342. [DOI] [PubMed] [Google Scholar]
- 12.Nakajima M., Yoshida R., Shimada N., Yamazaki H., Yokoi T. Inhibition and inactivation of human cytochrome P450 isoforms by phenethyl isothiocyanate. Drug Metab. Dispos. 2001;29:1110–1113. [PubMed] [Google Scholar]
- 13.Halkier B.A., Gershenzon J. Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 2006;57:303–333. doi: 10.1146/annurev.arplant.57.032905.105228. [DOI] [PubMed] [Google Scholar]
- 14.Kusznierewicz B., Bartoszek A., Wolska L., Drzewiecki J., Gorinstein S., Namieśnik J. Partial characterization of white cabbages (Brassica oleracea var. capitata f. alba) from different regions by glucosinolates, bioactive compounds, total antioxidant activities and proteins. LWT-Food Sci. Technol. 2008;41:1–9. [Google Scholar]
- 15.Fahey J.W., Haristoy X., Dolan P.M., Kensler T.W., Scholtus I., Stephenson K.K., Lozniewski A. Sulforaphane inhibits extracellular, intracellular, and antibiotic-resistant strains of Helicobacter pylori and prevents benzo [a] pyrene-induced stomach tumors. Proc. Natl. Acad. Sci. USA. 2002;99:7610–7615. doi: 10.1073/pnas.112203099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Z., Hirani A.H., McVetty P.B., Daayf F., Quiros C.F., Li G. Reducing progoitrin and enriching glucoraphanin in Braasica napus seeds through silencing of the GSL-ALK gene family. Plant Mol. Biol. 2012;79:179–189. doi: 10.1007/s11103-012-9905-2. [DOI] [PubMed] [Google Scholar]
- 17.Mithen R.F. Glucosinolates and their degradation products. Adv. Bot. Res. 2001;35:213–262. [Google Scholar]
- 18.Sønderby I.E., Hansen B.G., Bjarnholt N., Ticconi C., Halkier B.A., Kliebenstein D.J. A systems biology approach identifies a R2R3 MYB gene subfamily with distinct and overlapping functions in regulation of aliphatic glucosinolates. PLoS ONE. 2007;2 doi: 10.1371/journal.pone.0001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grubb C.D., Abel S. Glucosinolate metabolism and its control. Trends Plant Sci. 2006;11:89–100. doi: 10.1016/j.tplants.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 20.Sønderby I.E., Geu-Flores F., Halkier B.A. Biosynthesis of glucosinolates-gene discovery and beyond. Trends Plant Sci. 2010;15:283–290. doi: 10.1016/j.tplants.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 21.Sørensen H. Canola and Rapeseed. Springer; New York, NY, USA: 1990. Glucosinolates: Structure-properties-function; pp. 149–172. [Google Scholar]
- 22.Mikkelsen M.D., Petersen B.L., Olsen C.E., Halkier B.A. Biosynthesis and metabolic engineering of glucosinolates. Amino Acids. 2002;22:279–295. doi: 10.1007/s007260200014. [DOI] [PubMed] [Google Scholar]
- 23.Kroymann J., Textor S., Tokuhisa J.G., Falk K.L., Bartram S., Gershenzon J., Mitchell-Olds T. A gene controlling variation in Arabidopsis glucosinolate composition is part of the methionine chain elongation pathway. Plant Physiol. 2001;127:1077–1088. doi: 10.1104/pp.010416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Textor S., de Kraker J.W., Hause B., Gershenzon J., Tokuhisa J.G. MAM3 catalyzes the formation of all aliphatic glucosinolate chain lengths in Arabidopsis. Plant Physiol. 2007;144:60–71. doi: 10.1104/pp.106.091579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gigolashvili T., Yatusevich R., Rollwitz I., Humphry M., Gershenzon J., Flügge U.I. The plastidic bile acid transporter 5 is required for the biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana. Plant Cell. 2009;21:1813–1829. doi: 10.1105/tpc.109.066399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sawada Y., Toyooka K., Kuwahara A., Sakata A., Nagano M., Saito K., Hirai M.Y. Arabidopsis bile acid: Sodium symporter family protein 5 is involved in methionine-derived glucosinolate biosynthesis. Plant Cell Physiol. 2009;50:1579–1586. doi: 10.1093/pcp/pcp110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wittstock U., Halkier B.A. Glucosinolate research in the Arabidopsis era. Trends Plant Sci. 2002;7:263–270. doi: 10.1016/S1360-1385(02)02273-2. [DOI] [PubMed] [Google Scholar]
- 28.Brader G., Mikkelsen M.D., Halkier B.A., Tapio Palva E. Altering glucosinolate profiles modulates disease resistance in plants. Plant J. 2006;46:758–767. doi: 10.1111/j.1365-313X.2006.02743.x. [DOI] [PubMed] [Google Scholar]
- 29.Celenza J.L., Quiel J.A., Smolen G.A., Merrikh H., Silvestro A.R., Normanly J., Bender J. The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol. 2005;137:253–262. doi: 10.1104/pp.104.054395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levy M., Wang Q., Kaspi R., Parrella M.P., Abel S. Arabidopsis IQD1, a novel calmodulin-binding nuclear protein, stimulates glucosinolate accumulation and plant defense. Plant J. 2005;43:79–96. doi: 10.1111/j.1365-313X.2005.02435.x. [DOI] [PubMed] [Google Scholar]
- 31.Skirycz A., Reichelt M., Burow M., Birkemeyer C., Rolcik J., Kopka J., Witt I. DOF transcription factor AtDof1. 1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J. 2006;47:10–24. doi: 10.1111/j.1365-313X.2006.02767.x. [DOI] [PubMed] [Google Scholar]
- 32.Gigolashvili T., Berger B., Mock H.P., Müller C., Weisshaar B., Flügge U.I. The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 2007;50:886–901. doi: 10.1111/j.1365-313X.2007.03099.x. [DOI] [PubMed] [Google Scholar]
- 33.Gigolashvili T., Yatusevich R., Berger B., Müller C., Flügge U.I. The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 2007;51:247–261. doi: 10.1111/j.1365-313X.2007.03133.x. [DOI] [PubMed] [Google Scholar]
- 34.Gigolashvili T., Engqvist M., Yatusevich R., Müller C., Flügge U.I. HAG2/MYB76 and HAG3/MYB29 exert a specific and coordinated control on the regulation of aliphatic glucosinolate biosynthesis in Arabidopsis thaliana. New Phytol. 2008;177:627–642. doi: 10.1111/j.1469-8137.2007.02295.x. [DOI] [PubMed] [Google Scholar]
- 35.Liu S., Liu Y., Yang X., Tong C., Edwards D., Parkin I.A., Waminal N.E. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014;5 doi: 10.1038/ncomms4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Araki R., Hasumi A., Nishizawa O.I., Sasaki K., Kuwahara A., Sawada Y., Totoki Y., Toyoda A., Sakaki Y., Li Y., et al. Novel bioresources for studies of Brassica oleracea: Identification of a kale MYB transcription factor responsible for glucosinolate production. Plant Biotechnol. J. 2013;11:1017–1027. doi: 10.1111/pbi.12095. [DOI] [PubMed] [Google Scholar]
- 37.Maruyama-Nakashita A., Nakamura Y., Tohge T., Saito K., Takahashi H. Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell. 2006;18:3235–3251. doi: 10.1105/tpc.106.046458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirai M.Y., Sugiyama K., Sawada Y., Tohge T., Obayashi T., Suzuki A., Saito K. Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. USA. 2007;104:6478–6483. doi: 10.1073/pnas.0611629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malitsky S., Blum E., Less H., Venger I., Elbaz M., Morin S., Aharoni A. The transcript and metabolite networks affected by the two clades of Arabidopsis glucosinolate biosynthesis regulators. Plant Physiol. 2008;148:2021–2049. doi: 10.1104/pp.108.124784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Halkier B.A., Du L. The biosynthesis of glucosinolates. Trends Plant Sci. 1997;2:425–431. doi: 10.1016/S1360-1385(97)90026-1. [DOI] [PubMed] [Google Scholar]
- 41.Yatusevich R., Mugford S.G., Matthewman C., Gigolashvili T., Frerigmann H., Delaney S., Kopriva S. Genes of primary sulfate assimilation are part of the glucosinolate biosynthetic network in Arabidopsis thaliana. Plant J. 2010;62:1–11. doi: 10.1111/j.1365-313X.2009.04118.x. [DOI] [PubMed] [Google Scholar]
- 42.Wang H., Wu J., Sun S., Liu B., Cheng F., Sun R., Wang X. Glucosinolate biosynthetic genes in Brassica rapa. Gene. 2011;487:135–142. doi: 10.1016/j.gene.2011.07.021. [DOI] [PubMed] [Google Scholar]
- 43.Augustine R., Majee M., Gershenzon J., Bisht N.C. Four genes encoding MYB28, a major transcriptional regulator of the aliphatic glucosinolate pathway, are differentially expressed in the allopolyploid Brassica juncea. J. Exp. Bot. 2013;64:4907–4921. doi: 10.1093/jxb/ert280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim Y.B., Li X., Kim S.J., Kim H.H., Lee J., Kim H., Park S.U. MYB transcription factors regulate glucosinolate biosynthesis in different organs of Chinese cabbage (Brassica rapa ssp. pekinensis) Molecules. 2013;18:8682–8695. doi: 10.3390/molecules18078682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manoli A., Sturaro A., Trevisan S., Quaggiotti S., Nonis A. Evaluation of candidate reference genes for qPCR in maize. J. Plant Physiol. 2012;169:807–815. doi: 10.1016/j.jplph.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 46.Han X., Lu M., Chen Y., Zhan Z., Cui Q., Wang Y. Selection of reliable reference genes for gene expression studies using real-time PCR in tung tree during seed development. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0043084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi S.H., Park S., Lim Y.P., Kim S.J., Park J.T., An G. Metabolite profiles of glucosinolates in cabbage varieties (Brassica oleracea var. capitata) by season, color, and tissue position. Hortic. Environ. Biotechnol. 2014;55:237–247. [Google Scholar]
- 48.Frerigmann H., Berger B., Gigolashvili T. bHLH05 is an interaction partner of MYB51 and a novel regulator of glucosinolate biosynthesis in Arabidopsis. Plant Physiol. 2014;166:349–369. doi: 10.1104/pp.114.240887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reintanz B., Lehnen M., Reichelt M., Gershenzon J., Kowalczyk M., Sandberg G., Palme K. Bus, a bushy Arabidopsis CYP79F1 knockout mutant with abolished synthesis of short-chain aliphatic glucosinolates. Plant Cell. 2001;13:351–367. doi: 10.1105/tpc.13.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Douglas Grubb C., Zipp B.J., Ludwig-Müller J., Masuno M.N., Molinski T.F., Abel S. Arabidopsis glucosyltransferase UGT74B1 functions in glucosinolate biosynthesis and auxin homeostasis. Plant J. 2004;40:893–908. doi: 10.1111/j.1365-313X.2004.02261.x. [DOI] [PubMed] [Google Scholar]
- 51.Mikkelsen M.D., Hansen C.H., Wittstock U., Halkier B.A. Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J. Biol. Chem. 2000;275:33712–33717. doi: 10.1074/jbc.M001667200. [DOI] [PubMed] [Google Scholar]
- 52.Zang Y.X., Kim H.U., Kim J.A., Lim M.H., Jin M., Lee S.C., Park B.S. Genome-wide identification of glucosinolate synthesis genes in Brassica rapa. FEBS J. 2009;276:3559–3574. doi: 10.1111/j.1742-4658.2009.07076.x. [DOI] [PubMed] [Google Scholar]
- 53.Kabouw P., Biere A., van der Putten W.H., van Dam N.M. Intra-specific differences in root and shoot glucosinolate profiles among white cabbage (Brassica oleracea var. capitata) cultivars. J. Agric. Food Chem. 2009;58:411–417. doi: 10.1021/jf902835k. [DOI] [PubMed] [Google Scholar]
- 54.Velasco P., Cartea M.E., González C., Vilar M., Ordás A. Factors affecting the glucosinolate content of kale (Brassica oleracea acephala group) J. Agric. Food Chem. 2007;55:955–962. doi: 10.1021/jf0624897. [DOI] [PubMed] [Google Scholar]
- 55.Cartea M.E., Velasco P. Glucosinolates in Brassica foods: Bioavailability in food and significance for human health. Phytochem. Rev. 2008;7:213–229. doi: 10.1007/s11101-007-9072-2. [DOI] [Google Scholar]
- 56.Wang J., Gu H., Yu H., Zhao Z., Sheng X., Zhang X. Genotypic variation of glucosinolates in broccoli (Brassica oleracea var. italica) florets from China. Food Chem. 2012;133:735–741. [Google Scholar]
- 57.Cartea M.E., Velasco P., Obregón S., Padilla G., de Haro A. Seasonal variation in glucosinolate content in Brassica oleracea crops grown in northwestern Spain. Phytochemistry. 2008;69:403–410. doi: 10.1016/j.phytochem.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 58.Vallejo F., Tomás-Barberán F.A., Benavente-García A.G., García-Viguera C. Total and individual glucosinolate contents in inflorescences of eight broccoli cultivars grown under various climatic and fertilisation conditions. J. Sci. Food Agric. 2003;83:307–313. doi: 10.1002/jsfa.1320. [DOI] [Google Scholar]
- 59.Brown A.F., Yousef G.G., Jeffery E.H., Klein B.P., Wallig M.A., Kushad M.M., Juvik J.A. Glucosinolate profiles in broccoli: Variation in levels and implications in breeding for cancer chemoprotection. J. Am. Soc. Hortic. Sci. 2002;127:807–813. [Google Scholar]
- 60.Farnham M.W., Wilson P.E., Stephenson K.K., Fahey J.W. Genetic and environmental effects on glucosinolate content and chemoprotective potency of broccoli. Plant Breed. 2004;123:60–65. doi: 10.1046/j.0179-9541.2003.00912.x. [DOI] [Google Scholar]
- 61.Kim S.J., Matsuo T., Watanabe M., Watanabe Y. Effect of nitrogen and sulphur application on the glucosinolate content in vegetable turnip rape (Brassica rapa L.) Soil Sci. Plant Nutr. 2002;48:43–49. doi: 10.1080/00380768.2002.10409169. [DOI] [Google Scholar]
- 62.Rosen C.J., Fritz V.A., Gardner G.M., Hecht S.S., Carmella S.G., Kenney P.M. Cabbage yield and glucosinolate concentrations as affected by nitrogen and sulfur fertility. HortScience. 2005;40:1493–1498. [Google Scholar]
- 63.Tripathi M.K., Mishra A.S. Glucosinolates in animal nutrition: A review. Anim. Feed Sci. Technol. 2007;132:1–27. doi: 10.1016/j.anifeedsci.2006.03.003. [DOI] [Google Scholar]
- 64.Traka M., Mithen R. Glucosinolates, isothiocyanates and human health. Phytochem. Rev. 2009;8:269–282. doi: 10.1007/s11101-008-9103-7. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.