Abstract
Wuzi-Yanzong-Wan (WZYZW), a classical traditional Chinese medicine (TCM) prescription containing Fructus Lych, Semen Cuscutae (fried), Fructus Rubi, Fructus Schisandrae chinensis (steamed) and Semen Plantaginis (fried with salt), is widely used to treat impotence, sterility, spermatorrhea, premature ejaculation, lumbago and post-micturation dribble. However, the chemical profile of WZYZW has not been established yet. In this work, a rapid and sensitive method for systematically screening and identifying the chemical constituents of WZYZW in both positive and negative ion modes using Ultra-Performance LC coupled with ESI-linear ion trap-Orbitrap tandem mass spectrometry (UPLC-ESI-LTQ-Orbitrap-MS) has been developed. Based on the chromatographic and spectrometric data, and referring to the literature, we could tentatively identify 106 compounds, including organic acids, flavonoids, phenylpropanoids, alkaloids and terpenoids. Fourteen ingredients from Fructus Lych were identified, while 10 ingredients were from Semen Cuscutae (fried), 33 ingredients were from Fructus Rubi, 37 ingredients were from Fructus Schisandrae chinensis (steamed), and 20 ingredients were from Semen Plantaginis (fried with salt). The results may provide essential data for further quality control, pharmacological research and clinical evaluation of WZYZW. Furthermore, this study indicates the developed approach based on UPLC-ESI-LTQ-Orbitrap-MS is suitable for characterizing the chemical profiles of TCM prescriptions. This is the first report to provide a comprehensive analysis of the chemical constituents of WZYZW.
Keywords: Wuzi-Yanzong-Wan, UPLC-ESI-LTQ-Orbitrap-MS, constituents, identification
1. Introduction
Chinese herbal prescriptions, the basic form of clinical application of TCM for thousands of years, have been proven by clinical practice to play a positive role in human health. The multi-chemical constituents of Chinese herbal prescriptions are the key to achieving their prevention and treatment effects, so comprehensive analysis of their chemical constituents is the premise and foundation to promote development and innovation in TCM [1,2,3]. Therefore, it is necessary to develop rapid and sensitive methods for identifying the chemical constituents of TCM prescriptions, to facilitate their quality control, safe and effective use in clinic.
Wuzi-Yanzong-Wan (WZYZW), the most classical Chinese herb prescription for nourishing the kidney and strengthening the essence, is called “ancient and modern seed first choice prescription”, being firstly detailed recorded in She-sheng-zhong-miao-fang treatise of the Ming Dynasty and widely used to treat syndrome of kidney deficiency and damage of essence, including impotence, sterility, spermatorrhea, premature ejaculation, lumbago and post-micturation dribble [4,5,6]. WZYZW is composed of Fructus Lych, Semen Cuscutae (fried), Fructus Rubi, Fructus Schi-sandrae chinensis (steamed), and Semen Plantaginis (fried with salt).
Over the years, the chemical work on WZYZW has mainly focused on either qualitative or quantitative analyses of its major components using different techniques [7,8,9,10], such as HPLC, LC-MS, etc. These methods have contributed to the study of WZYZW to some extent, but there has been no systematic characterization of the chemical constituents in this prescription, which could contribute to interpret the material basis for its therapeutic effects.
The comprehensive analysis of the chemical constituents in Chinese herbal prescriptions and herbal medicines is hard work, on account of the fact they typically include extraordinarily complex components. LTQ-Orbitrap-MS as a high resolution mass spectrometry technique has proven to be an advanced, accurate and reliable tool for the comprehensive identification of compounds in complex systems such as TCMs [11,12,13,14,15]. It has high sensitivity, resolution and mass accuracy, which provides multi-stage MS analysis (MSn) information, acquisition of precursor ion data of high-resolution, determination of accurate mass charge ratios, structural information of fragment ions from MSn, and gives the elementary composition of compounds and allows the structure confirmation of trace components in complicated samples [16]. A C18 column with a 1.7 µm particle pore size is employed for UPLC, resulting in higher peak capacity, greater resolution, and faster and more sensitive separations in comparison with HPLC [17,18]. Therefore, the present study is aimed at establishing a rapid and sensitive method for making the constituents of WZYZW more clear and comprehensible, using the UPLC-LTQ-Orbitrap-MS technique in both negative and positive modes. As a result, a total of 106 constituents were screened and identified, and the developed method was proved efficient, giving accurate and information-rich results. To our knowledge, this is the first time a comprehensive analysis of the multiple class compounds of WZYZW has been reported.
2. Results and Discussion
2.1. Optimization of Analytical Conditions
To obtain better chromatographic separation and mass spectrometric detection, four mobile phase systems: methanol-aqueous solution, acetonitrile-aqueous solution, acetonitrile-formic acid aqueous solution and acetonitrile containing formic acid-formic acid aqueous solution were examined, the acetonitrile containing formic acid-formic acid aqueous solution showed a better degree of separation. Formic acid (0.1%) was added to the mobile phase to enhance the intensity of mass signals and improve the peak shape. In addition, the flow rate (0.25, 0.3, 0.35 mL/min), column temperature (25, 30, 40 °C) and injection volume (2, 3, 5 uL) were respectively optimized in this work. The chromatographic separation was better when the chromatographic conditions were as follows: acetonitrile containing formic acid-aqueous formic acid solution, flow rate of 0.3 mL/min, column temperature of 30 °C and injection volume of 3 μL.
2.2. Chemical Constituents Identification in WZYZW
Chemical profiles of WZYZW in both negative and positive modes were well separated and detected by using the established UPLC-ESI-LTQ-Orbitrap-MS method. The TIC chromatograms in both ESI modes are shown in Figure 1.
Figure 1.
Total ion chromatogram (TIC) of WZYZW in negative ion mode (a) and positive ion mode (b) using UPLC-ESI-LTQ-Orbitrap-MS.
In this study, for the compounds with available standards, the compound was identified by comparing the retention time and high-resolution accurate mass with the reference compound. Thus peaks 1, 5, 6, 10, 14, 17, 19, 20, 21, 27, 36, 51, 59, 60, 61, 64, 65, 68 and 75 were unambiguously identified as abromine, nicotinic acid, thiamine, riboflavin, taurine, quinic acid, atropine, ferulic acid, chlorogenic acid, scopoletin, rutin, esculin, apigenin, hesperidin, quercetin, kaempferol, luteolin, isorhamnetin and schisandrin, respectively. Meanwhile, the fragmentation patterns and pathways of the standards helped further confirm the structures of the derivatives of the reference compounds. When no standard compounds were available, the structures were elucidated by adopting the following measures to improve the quality of the identifications: firstly, based on the high-accuracy precursor ions obtained from the LTQ-Orbitrap-MS, using Xcalibur 2.1, the element compositions were calculated following some rules (C = 40, H = 100, O = 30, N = 5, Na = 2 and RDB equivalent value = 15). The maximum tolerance of mass error for all the precursor and product ions was set at 5 ppm, which can satisfy the requirements for positive identification. Besides, the most rational molecular formula was sought in different chemical databases such as the Spectral Database for Organic Compounds SDBS (http://sdbs.db.aist.go.jp) and Massbank (http://www.massbank.jp). When several matching compounds with the same formula were found, the structures were sought in the chemical information of the compounds from the five crude drugs of WZYZW. The component herb from which each compound was derived was confirmed by individually analyzing the herbs with the same UPLC-LTQ-Orbitrap-MS method. Finally, the chemical structures were further confirmed by combining the chromatographic data, the high resolution and accurate mass product ion information provided by data-dependent scan from MSn experiments and referring to literature references. Moreover, the fragmentation patterns and pathways of the standards contributed to further confirming the derivatives of the reference compounds, then, the structure could be tentatively identified. A total of 106 compounds of WZYYW were thus identified or tentatively identified, including 35 flavonoids, 34 phenylpropanoids, 17 organic acids, 8 alkaloids, 11 terpenoids and one miscellaneous ingredient; 14 ingredients from Fructus Lych were identified, 10 ingredients from Semen Cuscutae (fried), 33 ingredients from Fructus Rubi, 37 ingredients from Fructus Schisandrae chinensis (steamed), and 20 ingredients from Semen Plantaginis (fried with salt). Table 1 summarizes their information.
Table 1.
Identification of the chemical constituents of WZYZW by UPLC-ESI-LTQ-Obitrap-MS.
| No. | tR (min) | Molecular Formula | Caculated Mass (m/z) | Experimental Mass (m/z) | MS/MS Fragments | Identification Compound | Source | |||
|---|---|---|---|---|---|---|---|---|---|---|
| N-ion | ppm | P-ion | ppm | |||||||
| 1 | 0.81 | C5H11NO2 | 117.08626 | ― | ― | 118.08569 | −0.6 | 235[2M + H]+; | Abromine | a |
| 118[M + H]+; | ||||||||||
| 59[C3H9N]+; | ||||||||||
| 58[C3H8N]+ | ||||||||||
| 2 | 0.87 | C6H8O6 | 176.02371 | 175.02289 | −4.7 | ― | ― | 175[M − H]−; | Ascorbic acid | a |
| 115[C4H3O4]−; | ||||||||||
| 113[C5H5O3]− | ||||||||||
| 3 | 1.12 | C6H8O7 | 192.01863 | 191.0189 | 1.4 | ― | ― | 191[M − H]−; | Citric acid | d |
| 173[M − H − H2O]−; | ||||||||||
| 129[M − H − H2O − CO2]−; | ||||||||||
| 111[M − H − H2O − COOH − OH]− | ||||||||||
| 4 | 1.12 | C4H6O5 | 134.01315 | 133.01372 | 4.3 | ― | ― | 133[M + H]+; | Malic acid | d |
| 115[M + H − H2O]+ | ||||||||||
| 5 | 1.28 | C6H5NO2 | 123.03931 | 122.02417 | 4.2 | 124.03876 | −4.3 | 122[M − H]−; | Nicotinic acid | a |
| 124[M + H]+; | ||||||||||
| 106 [M + H − H2O]+ | ||||||||||
| 6 | 1.58 | C12H16N4OS | 264.11176 | ― | ― | 265.11071 | −3.9 | 265[M + H]+; | Thiamine | a |
| 156[C7H10NOS]+; | ||||||||||
| 144[C6H10NOS]+; | ||||||||||
| 122[C6H8N3]+ | ||||||||||
| 7 | 1.97 | C4H6O4 | 118.01824 | 117.01791 | −2.8 | ― | ― | 117[M − H]−; | Succinic acid | d |
| 99[M − H − H2O]−; | ||||||||||
| 73[M − H − CO2]− | ||||||||||
| 8 | 2.68 | C7H6O5 | 170.01315 | 169.01301 | −0.8 | ― | ― | 169[M − H]−; | Gallic acid | c |
| 125[M – H − CO2]− | ||||||||||
| 9 | 3.08 | C4H6O6 | 150.00806 | 149.00808 | 0.1 | ― | ― | 149[M − H]−; | Tartaric acid | d |
| 131[M − H − H2O]−; | ||||||||||
| 103[M − H − HCOOH]− | ||||||||||
| 10 | 3.28 | C17H20N4O6 | 376.12991 | 375.12988 | −0.1 | ― | ― | 375[M − H]−; | Riboflavin | a |
| 255[C13H11N4O2]−; | ||||||||||
| 212[C12H10N3O]− | ||||||||||
| 11 | 4.48 | C7H6O3 | 138.02332 | 137.02365 | 2.4 | ― | ― | 137[M − H]−; | Salicylic acid/ | c |
| 93[C6H5O]− | 4-Hydroxyben-zoic acid | |||||||||
| 12 | 4.66 | C7H6O4 | 154.01824 | 153.01862 | 2.5 | ― | ― | 153[M − H]−; | Protocatechuic acid | d |
| 109[M − H − CO2]− | ||||||||||
| 13 | 4.74 | C7H6O3 | 138.02332 | 137.02373 | 3 | ― | ― | 137[M − H]−; | Salicylic acid/ | c |
| 93[C6H5O]− | 4-Hydroxyben-zoic acid | |||||||||
| 14 | 7.2 | C2H7NO3S | 125.00629 | 124.00638 | 0.7 | ― | ― | 124[M − H]−; | Taurine | a |
| 80[SO3]− | ||||||||||
| 15 | 7.38 | C8H8O4 | 168.03389 | 167.0342 | 1.9 | ― | ― | 167[M − H]−; | Vanillic acid | a,c |
| 152[M − H − CH3]−; | ||||||||||
| 123[M − H − CO2]−; | ||||||||||
| 108[M − H − CH3 − CO2]− | ||||||||||
| 16 | 8.35 | C16H22O10 | 374.11293 | 373.11377 | 2.3 | 397.10892 | −4.0 | 373[M − H]−; | Geniposidic acid | e |
| 329[M − H − CO2]−; | ||||||||||
| 211[M − H − Glc]−; | ||||||||||
| 193 [M − H − Glc − H2O]−; | ||||||||||
| 167[M − H − Glc − CO2]−; | ||||||||||
| 149[M − H − Glc − CO2 − H2O]−; | ||||||||||
| 397[M + Na]+; | ||||||||||
| 353[M + Na − CO2]+; | ||||||||||
| 235[M + Na − Glc]+; | ||||||||||
| 217[M + Na − Glc − H2O]+; | ||||||||||
| 149[C7H10O2Na]+ | ||||||||||
| 17 | 8.79 | C7H12O6 | 192.05502 | 191.05503 | 0.1 | 193.06981 | −4.4 | 191[M − H]−; | Quinic acid | d |
| 173[M − H − H2O]−; | ||||||||||
| 155[M − H − 2H2O]−; | ||||||||||
| 127[M − H − 2H2O − CO]−; | ||||||||||
| 193 [M + H]+; | ||||||||||
| 175[M + H − H2O]+; | ||||||||||
| 157[M + H − 2H2O]+ | ||||||||||
| 18 | 9.68 | C9H8O4 | 180.03389 | 179.03429 | 2.2 | 181.04889 | −3.5 | 179[M − H]−; | Caffeic acid | a |
| 135[M − H − CO2]−; | ||||||||||
| 181[M + H]+; | ||||||||||
| 163[M + H − H2O]+; | ||||||||||
| 145[M + H − 2H2O]+ | ||||||||||
| 19 | 10.2 | C17H23NO3 | 289.17507 | ― | ― | 290.17578 | 2.4 | 290[M + H]+; | Atropine | a |
| 272[M + H − H2O]+; | ||||||||||
| 260[M + H − HCHO]+; | ||||||||||
| 242[M + H − HCHO − H2O]+; | ||||||||||
| 124[C8H14N]+; | ||||||||||
| 95[C7H11]+ | ||||||||||
| 20 | 11.91 | C10H10O4 | 194.04954 | 193.05001 | 2.4 | 195.06447 | −3.7 | 193[M − H]−; | Ferulic Acid | a,c |
| 175[M − H − H2O]−; | ||||||||||
| 149[M − H − CO2]−; | ||||||||||
| 134[M − H − CH3 − CO2]−; | ||||||||||
| 195[M + H]+; | ||||||||||
| 177[M + H − H2O]+; | ||||||||||
| 145[M + H − H2O − CH3 − OH]+ | ||||||||||
| 21 | 12.37 | C16H18O9 | 354.08671 | 353.08731 | 1.7 | 355.10097 | −3.9 | 353[M − H]−; | Chlorogenic acid | a |
| 191[C7H11O6]−; | ||||||||||
| 173[C7H9O5]−; | ||||||||||
| 127[C6H7O3]−; | ||||||||||
| 355[M + H]+; | ||||||||||
| 163[C9H7O3]+; | ||||||||||
| 117[C8H5O]+ | ||||||||||
| 22 | 13.1 | C9H8O3 | 164.03897 | 163.03905 | 0.5 | 165.05393 | −4.2 | 163[M − H]−; | p-Coumaric acid | a |
| 119[M − H − CO2]−; | ||||||||||
| 165 [M + H]+; | ||||||||||
| 147[M + H − H2O]+ | ||||||||||
| 23 | 13.64 | C7H10O5 | 174.04445 | 173.04381 | −3.7 | ― | ― | 173[M − H]−; | Shikimic acid | c |
| 155[M − H − H2O]−; | ||||||||||
| 137[M − H − 2H2O]−; | ||||||||||
| 128[M − H − COOH]− | ||||||||||
| 24 | 13.97 | C27H30O17 | 626.24908 | 625.25148 | 3.8 | ― | ― | 625[M − H]−; | Quercetin-3-O-β-d-galactopyra-noside-7-O-β-d-glucopyranoside | b |
| 463[M − H − Gal]−; | ||||||||||
| 301[M − H − Gal − Glc]−; | ||||||||||
| 300[M − H − Gal − Glc − H]−; | ||||||||||
| 273[M − H − Gal − Glc − CO]−; | ||||||||||
| 255[M − H − Gal − Glc − CO − H2O]− | ||||||||||
| 25 | 15.6 | C9H6O4 | 178.01824 | 177.01847 | 1.3 | 179.03317 | −4.0 | 177[M − H]−; | Esculetin | c |
| 149[M − H − CO]−; | ||||||||||
| 133[M − H − CO2]−; | ||||||||||
| 121[M − H − 2CO]−; | ||||||||||
| 105[M − H − CO2 − CO]−; | ||||||||||
| 93[M − H − 3CO]−; | ||||||||||
| 179[M + H]+; | ||||||||||
| 151[M + H − CO]+; | ||||||||||
| 133[M + H − CO − H2O]+ | ||||||||||
| 26 | 17.17 | C22H22O11 | 462.10784 | 461.10962 | 3.9 | ― | ― | 461[M − H]−; | Homoplantagini-n | e |
| 299[M − H − Glu]−; | ||||||||||
| 271[M − H − Glu − CO]−; | ||||||||||
| 181[C8H5O5]− | ||||||||||
| 27 | 19.43 | C10H8O4 | 192.03387 | 191.03419 | 1.6 | 193.04933 | −1.1 | 191[M − H]−; | Scopoletin | a |
| 193[M + H]+; | ||||||||||
| 178[M + H − CH3]+; | ||||||||||
| 165[M + H − CO]+; | ||||||||||
| 161[M + H − CH3OH]+; | ||||||||||
| 133[M + H − CH3OH − CO]+ | ||||||||||
| 28 | 19.6 | C21H20O12 | 464.08711 | 463.08829 | 2.6 | ― | ― | 463[M − H]−; | 6-Hydroxy-luteolin-7-O-glucoside | e |
| 301[M − H − Glc]−; | ||||||||||
| 273[M − H − Glc − CO]−; | ||||||||||
| 257[M − H − Glc − CO2]−; | ||||||||||
| 167[C7H3O5]− | ||||||||||
| 29 | 20.93 | C22H20O12 | 476.0871 | 475.08633 | −1.6 | ― | ― | 475[M − H]−; | Kaempferol-3-O-β-d-glucuronic acid methyl ester | c |
| 457[M − H − H2O]−; | ||||||||||
| 433[M − H − CO=CH2]−; | ||||||||||
| 415[M − H − CH3COOH]−; | ||||||||||
| 285[M − H − C7H10O6]− | ||||||||||
| 30 | 21.62 | C15H10O7 | 302.03428 | 301.03396 | −1.1 | ― | ― | 301[M − H]−; | 6-Hydroxy-luteolin | e |
| 273[M − H − CO]−; | ||||||||||
| 257[M − H − CO2]−; | ||||||||||
| 167[C7H3O5]− | ||||||||||
| 31 | 21,78 | C26H28O16 | 596.14463 | 595.14543 | 1.4 | ― | ― | 595[M − H]−; | Quercetin-3-O-β-d-galactosyl-(1→2)-β-d-apioside | b |
| 505[M − H – Api + C2H2O]−; | ||||||||||
| 463[M − H − Api]−; | ||||||||||
| 445[M − H − Api − H2O]−; | ||||||||||
| 301[M − H − Api − Gal]−; | ||||||||||
| 300[M − H − Api − Gal − H]−; | ||||||||||
| 273[M − H − Api − Gal − CO]−; | ||||||||||
| 257[M − H − Api − Gal − 2CO]−; | ||||||||||
| 179[C8H3O5]− | ||||||||||
| 32 | 21.88 | C14H6O8 | 302.01355 | ― | ― | 303.01321 | −1.1 | 303[M+H]+; | Ellagic acid | c |
| 285[M + H − H2O]+; | ||||||||||
| 275[M + H − CO]+; | ||||||||||
| 257[M + H − CO − H2O]+; | ||||||||||
| 247[M + H − 2CO]+; | ||||||||||
| 201[C11H5O4]+ | ||||||||||
| 33 | 22.26 | C29H28O9 | 520.18061 | ― | ― | 521.18042 | −0.4 | 521[M + H]+; | Schisantherin D | d |
| 399[M + H − C6H5COOH]+; | ||||||||||
| 355[M + H − C6H5COOH − C2H4O]+ | ||||||||||
| 34 | 22.9 | C21H20O12 | 464.08711 | 463.08887 | 3.8 | ― | ― | 463[M − H]−; | Hyperoside | b,c |
| 301[M − H − Gal]−; | ||||||||||
| 255[M − H − Gal − CO − H2O]−; | ||||||||||
| 151[C7H3O4]−; | ||||||||||
| 107[C6H3O2]− | ||||||||||
| 35 | 23.02 | C16H12O6 | 300.05502 | 299.05453 | −1.6 | ― | ― | 299[M − H]−; | Hispidulin | e |
| 271[M − H − CO]−; | ||||||||||
| 255[M − H − CO2]−; | ||||||||||
| 227[M − H − CO − CO2]−; | ||||||||||
| 181[C8H5O5]− | ||||||||||
| 36 | 23.1 | C27H30O16 | 610.14501 | 609.1479 | 4.7 | ― | ― | 609[M − H]−; | Rutin | c |
| 343[C17H11O8]−; | ||||||||||
| 301[M − H − Rha − Glc]−; | ||||||||||
| 300[M − H − Rha − Glc − H]−; | ||||||||||
| 271[M − H − Rha − Glc − H − CO − H]−; | ||||||||||
| 255[M − H − Rha − Glc − H − CO − OH]−; | ||||||||||
| 179[C8H3O5]−; | ||||||||||
| 151[C7H3O4]− | ||||||||||
| 37 | 23.74 | C21H20O12 | 464.08711 | 463.0874 | 0.6 | ― | ― | 463[M − H]−; | Isoquercitrin | c |
| 301[M − H − Glc]−; | ||||||||||
| 255[M − H − Glc − CO − H2O]−; | ||||||||||
| 151[C7H3O4]−; | ||||||||||
| 107[C6H3O2]− | ||||||||||
| 38 | 24.2 | C29H36O16 | 640.19197 | 639.19263 | 1 | ― | ― | 639[M − H]−; | Plantamajoside | e |
| 477[M − H − C9H6O3]−; | ||||||||||
| 315[M − H − C9H6O3 − Glc]−; | ||||||||||
| 153[M − H − C9H6O3 − 2Glc]−; | ||||||||||
| 135[M − H − C9H6O3 − 2Glc − H2O]− | ||||||||||
| 39 | 24.95 | C27H30O15 | 594.15009 | 593.1507 | 1 | ― | ― | 593[M − H]−; | Kaempferol-3-O-rutinoside | c |
| 327[M − H − C10H18O8]−; | ||||||||||
| 285[M − H − Rha − Glc]−; | ||||||||||
| 284[M − H − Rha − Glc − H]−; | ||||||||||
| 257[M − H − Rha − Glc − CO]−; | ||||||||||
| 255[M − H − Rha − Glc − H − CO − H]−; | ||||||||||
| 229[M − H − Rha − Glc − 2CO]−; | ||||||||||
| 227[M − H − Rha − Glc − H − 2CO − H]− | ||||||||||
| 40 | 25.34 | C28H36O8 | 500.24829 | ― | ― | 501.25002 | 3.4 | 501[M + H]+; | Tigloylgomisin H/Angeloygomi-sin H | d |
| 484[M + H − OH]+; | ||||||||||
| 483[M + H − H2O]+; | ||||||||||
| 384[M + H − OH − C4H7COOH]+; | ||||||||||
| 357[M + H − C4H7COOH − C2H4O]+ | ||||||||||
| 41 | 25.95 | C21H20O11 | 448.09219 | 447.09378 | 3.6 | ― | ― | 447[M − H]−; | Luteoloside | e |
| 285[M − H − Glc]−; | ||||||||||
| 284[M − H − Glc − H]−; | ||||||||||
| 257[M − H − Glc − CO]−; | ||||||||||
| 243[M − H − Glc − C2H2O]−; | ||||||||||
| 241[M − H − Glc − CO2]−; | ||||||||||
| 199[M − H − Glc − C2H2O − CO2]−; | ||||||||||
| 197[M − H − Glc − 2CO2]−; | ||||||||||
| 151[C7H3O4]− | ||||||||||
| 42 | 26.1 | C28H36O8 | 500.24829 | ― | ― | 501.25071 | 4.8 | 501[M + H]+; | Tigloylgomisin H/Angeloygomi-sin H | d |
| 484[M + H − OH]+; | ||||||||||
| 483[M + H − H2O]+; | ||||||||||
| 384[M + H − OH − C4H7COOH]+; | ||||||||||
| 357[M + H − C4H7COOH − C2H4O]+ | ||||||||||
| 43 | 26.57 | C29H36O15 | 624.19707 | 623.19604 | −1.6 | ― | ― | 623[M − H]−; | Acteoside | e |
| 461[M − H − C9H6O3]−; | ||||||||||
| 315[M − H − C9H6O3 − Rha]−; | ||||||||||
| 179[C9H7O4]−; | ||||||||||
| 153[M − H − C9H6O3 − Rha − Glc]−; | ||||||||||
| 135[M − H − C9H6O3 − Rha − Glc − H2O]− | ||||||||||
| 44 | 27.19 | C21H20O11 | 448.09219 | 447.09225 | 0.1 | 449.10532 | −2.5 | 447[M − H]−; | Astragalin | b,c |
| 285[M − H − Glc]−; | ||||||||||
| 151[C7H3O4]−; | ||||||||||
| 449[M + H]+; | ||||||||||
| 287[M − H − Glc]+; | ||||||||||
| 241[M − H − Glc − CH2O2]+; | ||||||||||
| 213[M + H − Glc − C2H2O3]+ | ||||||||||
| 45 | 27.19 | C21H20O11 | 448.09219 | 447.09225 | 0.1 | 449.10699 | −1.9 | 447[M − H]−; | Plantaginin | e |
| 285[M − H − Glc]−; | ||||||||||
| 267[M − H − Glc − H2O]−; | ||||||||||
| 255[M − H − Glc − CH2O]−; | ||||||||||
| 239[M − H − Glc − H2O − CO]−; | ||||||||||
| 167[C7H3O5]−; | ||||||||||
| 449[M + H]+; | ||||||||||
| 287[M+H − Glc]+; | ||||||||||
| 269[M + H − Glc − H2O]+; | ||||||||||
| 169[C7H5O5]+ | ||||||||||
| 46 | 27.48 | C29H36O15 | 624.19705 | 623.19794 | 1.4 | 625.21005 | −4.2 | 623[M − H]−; | Methylhesperidin | c |
| 477[M − H − Rha]−; | ||||||||||
| 315[M − H − Rha − Glc]−; | ||||||||||
| 300[M − H − Rha − Glc − CH3]−; | ||||||||||
| 297[M − H − Rha − Gldc − H2O]−; | ||||||||||
| 285[M − H − Rha − Glc − 2CH3]−; | ||||||||||
| 282[M − H − Rha − Glc − H2O − CH3]−; | ||||||||||
| 272[M − H − Rha − Glc − CH3 − CO]−; | ||||||||||
| 257[M − H − Rha − Glc − 2CH3 − CO]−; | ||||||||||
| 229[M − H − Rha − Glc − 2CH3 − 2CO]−; | ||||||||||
| 625[M + H]+; | ||||||||||
| 479[M + H − Rha]+; | ||||||||||
| 317[M + H − Rha − Glc]+; | ||||||||||
| 299[M + H − Rha − Glc − H2O]+; | ||||||||||
| 281[M + H − Rha − Glc − 2H2O]+; | ||||||||||
| 193[C10H9O4]+; | ||||||||||
| 165[C9H9O3]+ | ||||||||||
| 47 | 27.72 | C21H20O11 | 448.09219 | 447.09402 | 4.1 | ― | ― | 447[M − H]−; | Quercitrin | c |
| 301[M − H − Rha]−; | ||||||||||
| 300[M − H − Rha − H]−; | ||||||||||
| 273[M − H − Rha − CO]−; | ||||||||||
| 255[M − H − Rha − CO − H2O]− | ||||||||||
| 48 | 27.86 | C22H22O12 | 478.10275 | 477.10461 | 3.9 | ― | ― | 477[M − H]−; | Nepetin-7-O-glucoside | e |
| 315[M − H − Glc]−; | ||||||||||
| 287[M − H − Glc − CO]−; | ||||||||||
| 271[M − H − Glc − CO2]−; | ||||||||||
| 181[C8H5O5]− | ||||||||||
| 49 | 27.93 | C21H20O10 | 432.09727 | 431.09805 | 1.8 | 433.11093 | −4.6 | 431[M − H]−; | Kaempferol-7-O-α-l-rhamnoside | c |
| 413[M − H − H2O]−; | ||||||||||
| 285[M − H − Rha]−; | ||||||||||
| 267[M − H − Rha − H2O]−; | ||||||||||
| 169[C12H9O]−; | ||||||||||
| 155[C11H7O]−; | ||||||||||
| 433[M + H]+ | ||||||||||
| 50 | 27.97 | C21H20O10 | 432.09727 | 431.09866 | 3.2 | 431[M − H]−; | Apigenin-7-O-glucoside | e | ||
| 269[M − H − Glc]−; | ||||||||||
| 201[M − H − Glc − C3O2]−; | ||||||||||
| 151[C7H3O4]− | ||||||||||
| 51 | 27.99 | C15H16O9 | 340.07106 | 339.07058 | −1.4 | ― | ― | 339[M − H]−; | Esculin | c |
| 177[M − H − Glc]−; | ||||||||||
| 133[M − H − Glc − CO2]−; | ||||||||||
| 105[M − H − Glc − CO2 − CO]−; | ||||||||||
| 89[C7H5]− | ||||||||||
| 52 | 28.28 | C27H30O14 | 578.15518 | 577.15662 | 2.5 | 579.17073 | −0.2 | 577[M − H]−; | Rhoifolin | e |
| 457[M − H − C4H8O4]−; | ||||||||||
| 431[M − H − Rha]−; | ||||||||||
| 413[M − H − Rha − H2O]−; | ||||||||||
| 269[M − H − Rha − Glc]−; | ||||||||||
| 225[M − H − Rha − Glc − CO2]−; | ||||||||||
| 183[C12H7O2]−; | ||||||||||
| 579[M + H]+; | ||||||||||
| 433[M + H − Rha]+; | ||||||||||
| 415[M + H − Rha − H2O]+; | ||||||||||
| 397[M + H − Rha − 2H2O]+; | ||||||||||
| 271[M + H − Rha − Glc]+; | ||||||||||
| 253[M + H − Rha − Glc − H2O]+; | ||||||||||
| 243[M + H − Rha − Glc − CO]+; | ||||||||||
| 225[M + H − Rha − Glc − H2O − CO]+; | ||||||||||
| 211[M + H − Rha − Glc − H2O − C2H2O]+; | ||||||||||
| 153[C7H5O4]+; | ||||||||||
| 119[C8H7O]+; | ||||||||||
| 91[C7H7]+ | ||||||||||
| 53 | 28.28 | C28H32O16 | 624.16066 | 623.16248 | 2.9 | 625.17896 | 4.2 | 623[M − H]−; | Isorhamnetin-3-O-β-d-rutinoside | a |
| 477[M − H − Rha]−; | ||||||||||
| 315[M − H − Rha − Glc]−; | ||||||||||
| 300[M − H − Rha − Glc − CH3]−; | ||||||||||
| 272[M − H − Rha − Glc − CH3 − CO]−; | ||||||||||
| 271[M − H − Rha − Glc − CH3 − CO − H]−; | ||||||||||
| 243[M − H − Rha − Glc − CH3 − CO − H − CO]−; | ||||||||||
| 227[M − H − Rha − Glc − CH3 − CO − H − CO2]−; | ||||||||||
| 151[C7H3O4]−; | ||||||||||
| 625[M + H]+; | ||||||||||
| 479[M + H − Rha]+; | ||||||||||
| 317[M + H − Rha − Glc]+; | ||||||||||
| 302[M + H − Rha − Glc − CH3]+; | ||||||||||
| 285[M + H − Rha − Glc − CH3OH]+; | ||||||||||
| 274[M + H − Rha − Glc − CH3 − CO]+; | ||||||||||
| 257[M + H − Rha − Glc − CH3OH − CO]+; | ||||||||||
| 246[M + H − Rha − Glc − CH3 − 2CO]+; | ||||||||||
| 229[M + H − Rha − Glc − CH3OH − 2CO]+; | ||||||||||
| 153[C7H5O4]+ | ||||||||||
| 54 | 28.31 | C21H18O11 | 446.07654 | 445.07493 | −3.6 | ― | ― | 445[M − H]−; | Baicalin | e |
| 269[M − H − GluA]−; | ||||||||||
| 225[M − H − GluA − CO2]−; | ||||||||||
| 167[C7H3O5]− | ||||||||||
| 55 | 29.09 | C20H26O4 | 330.19039 | ― | ― | 331.18914 | −3.8 | 331[M + H]+; | Meso-dihydrog-uaiaretic acid | d |
| 313[M + H − H2O]+; | ||||||||||
| 301[M + H − 2CH3]+ | ||||||||||
| 56 | 29.58 | C15H10O5 | 270.0601 | 269.09566 | −1.6 | 271.05966 | −1.7 | 269[M − H]−; | Baicalein | e |
| 241[M − H − CO]−; | ||||||||||
| 225[M − H − CO2]−; | ||||||||||
| 197[M − H − CO − CO2]−; | ||||||||||
| 167[C7H3O5]−; | ||||||||||
| 271[M + H]+; | ||||||||||
| 253[M + H − H2O]+; | ||||||||||
| 169[C7H5O5]+ | ||||||||||
| 57 | 30.7 | C28H34O15 | 610.1814 | 609.18229 | 1.5 | 611.1994 | 3.9 | 609[M − H]−; | Neohesperidin | c |
| 505[M − H − C4H8O3]−; | ||||||||||
| 463[M − H − Rha]−; | ||||||||||
| 445[M − H − Rha − H2O]−; | ||||||||||
| 427[M − H − Rha − 2H2O]−; | ||||||||||
| 301[M − H − Rha − Glc]−; | ||||||||||
| 286[M − H − Rha − Glc − CH3]−; | ||||||||||
| 283[M − H − Rha − Glc − H2O]−; | ||||||||||
| 268[M − H − Rha − Glc − H2O − CH3]−; | ||||||||||
| 258[M − H − Rha − Glc − CH3 − CO]−; | ||||||||||
| 257[M − H − Rha − Glc − CO2]−; | ||||||||||
| 242[M − H − Rha − Glc − CH3 − CO2]−; | ||||||||||
| 239[M − H − Rha − Glc − H2O − CO2]−; | ||||||||||
| 199[C12H7O3]−; | ||||||||||
| 125[C6H5O3]−; | ||||||||||
| 611[M + H]+; | ||||||||||
| 303[M + H − Rha − Glc]+; | ||||||||||
| 285[M + H − Rha − Glc − H2O]+; | ||||||||||
| 179[C9H7O4]+; | ||||||||||
| 177[C10H9O3]+; | ||||||||||
| 151[C8H7O3]+ | ||||||||||
| 58 | 31.48 | C23H28O8 | 432.1857 | ― | ― | 433.18538 | −0.7 | 433[M + H]+; | Schizandradiol | d |
| 415[M + H − H2O]+; | ||||||||||
| 361[M + H − C4H8O]+; | ||||||||||
| 372[M + H − H2O − CH3CO]+; | ||||||||||
| 343[M + H − H2O − C4H7OH]+ | ||||||||||
| 59 | 31.54 39.44 | C15H10O5 | 270.04445 | 269.04353 | −3.4 | 271.06145 | 4.9 | 269[M − H]−; | Apigenin | e |
| 241[M − H − CO]−; | ||||||||||
| 227[M − H − C2H2O]−; | ||||||||||
| 225[M − H − CO2]−; | ||||||||||
| 201[M − H − C3O2]−; | ||||||||||
| 151[C7H3O4]−; | ||||||||||
| 271[M + H]+; | ||||||||||
| 253[M + H − H2O]+; | ||||||||||
| 225[M + H − H2O − CO]+ | ||||||||||
| 60 | 31.71 | C28H34O15 | 610.1814 | 609.18351 | 3.5 | 611.20001 | 4.8 | 609[M − H]−; | Hesperidin | c |
| 463[M − H − Rha]−; | ||||||||||
| 301[M − H − Rha − Glc]−; | ||||||||||
| 286[M − H − Rha − Glc − CH3]−; | ||||||||||
| 283[M − H − Rha − Glc − H2O]−; | ||||||||||
| 268[M − H − Rha − Glc − H2O − CH3]−; | ||||||||||
| 258[M − H − Rha − Glc − CH3 − CO]−; | ||||||||||
| 257[M − H − Rha − Glc − CO2]−; | ||||||||||
| 242[M − H − Rha − Glc − CH3 − CO2]−; | ||||||||||
| 239[M − H − Rha − Glc − H2O − CO2]−; | ||||||||||
| 199[C12H7O3]−; | ||||||||||
| 125[C6H5O3]−; | ||||||||||
| 611[M + H]+; | ||||||||||
| 303[M + H − Rha − Glc]+; | ||||||||||
| 285[M + H − Rha − Glc − H2O]+; | ||||||||||
| 179[C9H7O4]+; | ||||||||||
| 177[C10H9O3]+; | ||||||||||
| 151[C8H7O3]+ | ||||||||||
| 61 | 32.13 | C15H10O7 | 302.03428 | 301.03549 | 4 | 303.0488 | −1.1 | 301[M − H]−; | Quercetin | b,c |
| 273[M − H − CO]−; | ||||||||||
| 255[M − H − CO − H2O]−; | ||||||||||
| 151[C7H3O4]−; | ||||||||||
| 303[M + H]+; | ||||||||||
| 285[M + H − H2O]+; | ||||||||||
| 257[M + H − H2O − CO]+; | ||||||||||
| 247[M + H − 2CO]+; | ||||||||||
| 229[M + H − H2O − 2CO]+ | ||||||||||
| 62 | 33.02 | C30H26O13 | 594.12897 | 593.13049 | 2.6 | 595.14142 | −3.2 | 593[M − H]−; | Tiliroside | c |
| 447[M − H − C9H6O2]−; | ||||||||||
| 285[M − H − C9H6O2 − Glc]−; | ||||||||||
| 257[M − H − C9H6O2 − Glc − CO]−; | ||||||||||
| 229[M − H − C9H6O2 − Glc − 2CO]−; | ||||||||||
| 151[C7H3O4]−; | ||||||||||
| 617[M + Na]+; | ||||||||||
| 595[M + H]+; | ||||||||||
| 449[M + H − C9H6O2]+; | ||||||||||
| 287[M + H − C9H6O2 − Glc]+ | ||||||||||
| 63 | 34.12 | C15H12O6 | 288.05502 | 287.05549 | 1.7 | 289.07002 | −2.2 | 287[M − H]−; | Aromadendrin | c |
| 269[M − H − H2O]−; | ||||||||||
| 259[M − H − CO]−; | ||||||||||
| 243[M − H − CO2]−; | ||||||||||
| 225[M − H − H2O − CO2]−; | ||||||||||
| 215[M − H − CO − CO2]−; | ||||||||||
| 151[C7H3O4]−; | ||||||||||
| 107[C6H3O2]−; | ||||||||||
| 289[M + H]+; | ||||||||||
| 271[M + H − H2O]+; | ||||||||||
| 195[C9H7O5]+; | ||||||||||
| 145[C9H5O2]+ | ||||||||||
| 64 | 35.18 | C15H10O6 | 286.03936 | 285.03995 | 2.1 | 287.05359 | 4.9 | 285[M − H]−; | Kaempferol | b,c |
| 257[M − H − CO]−; | ||||||||||
| 229[M − H − 2CO]−; | ||||||||||
| 151[C7H3O4]−; | ||||||||||
| 309[M + Na]+; | ||||||||||
| 287[M + H]+; | ||||||||||
| 241[M + H − CH2O2]+; | ||||||||||
| 213[M + H − C2H2O3]+ | ||||||||||
| 65 | 35.31 | C15H10O6 | 286.03936 | 285.04019 | 2.9 | ― | ― | 571[2M − H]−; | Iuteolin | e |
| 285[M − H]−; | ||||||||||
| 243[M − H − C2H2O]−; | ||||||||||
| 241[M − H − CO2]−; | ||||||||||
| 199[M − H − C2H2O − CO2]−; | ||||||||||
| 197[M − H − 2CO2]−; | ||||||||||
| 171[M − H − C2H2O − CO2 − CO]−; | ||||||||||
| 151[C7H3O4]− | ||||||||||
| 66 | 35.58 | C16H12O7 | 316.04993 | 315.05117 | 3.9 | ― | ― | 315[M − H]−; | Nepetin | e |
| 287[M − H − CO]−; | ||||||||||
| 271[M − H − CO2]−; | ||||||||||
| 243[M − H − CO − CO2]−; | ||||||||||
| 181[C8H5O5]− | ||||||||||
| 67 | 35.61 | C15H10O6 | 286.03937 | 285.03989 | 1.8 | 287.05627 | 4.4 | 285[M − H]−; | Scutellarein | e |
| 257[M − H − CO]−; | ||||||||||
| 241[M − H − CO2]−; | ||||||||||
| 167[C7H3O5]−; | ||||||||||
| 287[M + H]+; | ||||||||||
| 269[M + H − H2O]+; | ||||||||||
| 241[M + H − H2O − CO]+; | ||||||||||
| 169[C7H5O5]+ | ||||||||||
| 68 | 35.61 | C16H12O7 | 316.04993 | 315.05027 | 1.1 | 317.06486 | −2.3 | 315[M − H]−; | Isorhamnetin | b |
| 300[M − H − CH3]−; | ||||||||||
| 272[M − H − CH3 − CO]−; | ||||||||||
| 271[M − H − CH3 − CO − H]−; | ||||||||||
| 243[M − H − CH3 − CO − H − CO]−; | ||||||||||
| 227[M − H − CH3 − CO − H − CO2]−; | ||||||||||
| 151[C7H3O4]−; | ||||||||||
| 107[C6H3O2]−; | ||||||||||
| 339[M + Na]+; | ||||||||||
| 317[M + H]+; | ||||||||||
| 302[M + H − CH3]+; | ||||||||||
| 285[M + H − CH3OH]+; | ||||||||||
| 274[M + H − CH3 − CO]+; | ||||||||||
| 257[M + H − CH3OH − CO]+; | ||||||||||
| 246[M + H − CH3 − 2CO]+; | ||||||||||
| 229[M + H − CH3OH − 2CO]+; | ||||||||||
| 153[C7H5O4]+ | ||||||||||
| 69 | 35.76 | C30H46O4 | 470.34689 | 469.33251 | 2.7 | 471.34509 | −3.8 | 469[M − H]−; | Nigranoic acid | d |
| 423[M − H − HCOOH]−; | ||||||||||
| 378[M − H − HCOOH − HCOO]−; | ||||||||||
| 471[M + H]+; | ||||||||||
| 456[M + H − CH3]+; | ||||||||||
| 453[M + H − H2O]+; | ||||||||||
| 390[M + H − C6H9]+ | ||||||||||
| 70 | 35.89 | C28H24O16 | 616.09807 | 615.10097 | 4.7 | ― | ― | 615[M − H]−; | 2″-O-Galloylhyperoside | c |
| 463[M − H − C7H4O4]−; | ||||||||||
| 445[M − H − C7H6O5]−; | ||||||||||
| 301[M − H − C7H4O4 − Glc]− | ||||||||||
| 71 | 36.79 | C23H30O7 | 418.20643 | ― | ― | 419.20477 | −4.0 | 419[M + H]+; | Gomisin H | d |
| 401[M + H − H2O]+; | ||||||||||
| 384[M + H − OH − H2O]+; | ||||||||||
| 383[M + H − 2H2O]+; | ||||||||||
| 369[M + H − OH − CH3 − H2O]+; | ||||||||||
| 353[M + H − OH − OCH3 − H2O]+ | ||||||||||
| 72 | 37.08 | C28H34O8 | 498.23264 | ― | ― | 499.23119 | −2.9 | 499[M + H]+; | Angeloygomisin O/Angeloylisogomisin O | d |
| 399[M + H − C4H7COOH]+; | ||||||||||
| 369[M + H − C4H7COOH − CH2O]+; | ||||||||||
| 357[M + H − C4H7COOH − C3H6]+; | ||||||||||
| 343[M + H − C4H7COOH − C4H8]+ | ||||||||||
| 73 | 38.12 | C28H34O8 | 498.23264 | ― | ― | 499.23026 | −4.8 | 499[M + H]+; | Angeloygomisin O/Angeloylisogomisin O | d |
| 399[M + H − C4H7COOH]+; | ||||||||||
| 369[M + H − C4H7COOH − CH2O]+; | ||||||||||
| 357[M + H − C4H7COOH − C3H6]+; | ||||||||||
| 343[M + H − C4H7COOH − C4H8]+ | ||||||||||
| 74 | 39.28 | C23H28O7 | 416.19078 | ― | ― | 417.18903 | −4.2 | 417[M + H]+; | Gomisin O/Epigomisin O | d |
| 402[M + H − CH3]+; | ||||||||||
| 399[M + H − H2O]+; | ||||||||||
| 385[M + H − CH3OH]+; | ||||||||||
| 373[M + H − C2H4O]+ | ||||||||||
| 75 | 39.38 | C24H32O7 | 432.22209 | ― | ― | 433.22103 | −2.4 | 433[M + H]+; | Schisandrin | d |
| 415[M + H − H2O]+; | ||||||||||
| 400[M + H − H2O − CH3]+; | ||||||||||
| 384[M + H − H2O − OCH3]+; | ||||||||||
| 373[M + H − H2O − C3H6]+; | ||||||||||
| 359[M + H − H2O − C4H8]+ | ||||||||||
| 76 | 39.44 | C30H46O4 | 470.34689 | ― | ― | 471.34551 | −2.9 | 471[M + H]+; | Kadsuric acid | d |
| 453[M + H − H2O]+; | ||||||||||
| 398[M + H − CH2CH2COOH]+; | ||||||||||
| 370[M + H − CH2CH2COOH − C2H4]+ | ||||||||||
| 77 | 39.6 | C23H28O7 | 416.19078 | ― | ― | 417.19103 | 0.6 | 417[M + H]+; | Gomisin O/Epigomisin O | d |
| 402[M + H − CH3]+; | ||||||||||
| 399[M + H − H2O]+; | ||||||||||
| 385[M + H − CH3OH]+; | ||||||||||
| 373[M + H − C2H4O]+ | ||||||||||
| 78 | 40.63 | C28H34O10 | 530.22247 | ― | ― | 531.22218 | −0.6 | 531[M + H]+; | Gomisin D | d |
| 485[M + H − CH2O2]+; | ||||||||||
| 401[M + H − C6H10O3]+; | ||||||||||
| 383[M + H − C6H10O3 − H2O]+ | ||||||||||
| 79 | 40.98 | C22H28O6 | 388.19586 | ― | ― | 389.19503 | −2.1 | 389[M + H]+; | Gomisin J | d |
| 374[M + H − CH3]+; | ||||||||||
| 357[M + H − CH3OH]+; | ||||||||||
| 342[M + H − CH3OH − CH3]+ | ||||||||||
| 80 | 41.02 | C30H48O5 | 488.3418 | 487.3439 | 4.3 | 489.35526 | −4.5 | 487[M − H]−; | Rosolic acid/2α,3α,19α-Trihydroxyolean-12-ene-28-oic acid/Arjunolic acid | c |
| 469[M − H − H2O]−; | ||||||||||
| 443[M − H − CO2]−; | ||||||||||
| 425[M − H − CO2 − H2O]−; | ||||||||||
| 407[M − H − CO2 − 2H2O]−; | ||||||||||
| 391[M − H − CO2 − 2H2O − CH4]−; | ||||||||||
| 489[M + H]+; | ||||||||||
| 453[M + H − 2H2O]+ | ||||||||||
| 81 | 41.22 | C30H48O5 | 488.3418 | 487.34378 | 4 | 489.35544 | −4.1 | 487[M − H]−; | Rosolic acid/2α,3α,19α-Trihydroxyolean-12-ene-28-oic acid/Arjunolic acid | c |
| 469[M − H − H2O]−; | ||||||||||
| 443[M − H − CO2]−; | ||||||||||
| 425[M − H − CO2 − H2O]−; | ||||||||||
| 407[M − H − CO2 − 2H2O]−; | ||||||||||
| 391[M − H − CO2 − 2H2O − CH4]−; | ||||||||||
| 489[M + H]+; | ||||||||||
| 453[M + H − 2H2O]+ | ||||||||||
| 82 | 41.75 | C15H24N2O2 | 264.10889 | ― | ― | 265.18997 | −4.0 | 265[M + H]+; | Sophoranol | b |
| 248[M + H − OH]+; | ||||||||||
| 247[M + H − H2O]+ | ||||||||||
| 83 | 42.18 | C30H46O3 | 454.35198 | ― | ― | 455.35025 | −3.8 | 455[M + H]+; | Ganwuweizic acid | d |
| 409[M + H − HCOOH]+; | ||||||||||
| 313[M + H − C8H14O2]+ | ||||||||||
| 84 | 42.32 | C30H48O5 | 488.3418 | 487.34302 | 2.5 | 489.35529 | −4.4 | 487[M − H]−; | Rosolic acid/2α,3α,19α-Trihydroxyolean-12-ene-28-oic acid/Arjunolic acid | c |
| 469[M − H − H2O]−; | ||||||||||
| 443[M − H − CO2]−; | ||||||||||
| 425[M − H − CO2 − H2O]−; | ||||||||||
| 407[M − H − CO2 − 2H2O]−; | ||||||||||
| 391[M − H − CO2 − 2H2O − CH4]−; | ||||||||||
| 489[M + H]+; | ||||||||||
| 453[M + H − 2H2O]+ | ||||||||||
| 85 | 42.89 | C12H16N2O | 204.13354 | ― | ― | 205.13409 | 2.6 | 205[M + H]+; | N-methylcytisine | b |
| 146[M + H − C3H9N]+; | ||||||||||
| 108[C6H6NO]+ | ||||||||||
| 86 | 43.27 | C23H28O6 | 400.19587 | ― | ― | 401.19434 | −3.8 | 401[M + H]+; | Schizandrin B | d |
| 386[M + H − CH3]+; | ||||||||||
| 370[M + H − OCH3]+; | ||||||||||
| 355[M + H − CH3 − OCH3]+; | ||||||||||
| 339[M + H − 2CH3 − CH3OH]+ | ||||||||||
| 87 | 43.3 | C30H34O8 | 522.23265 | ― | ― | 523.23095 | −3.2 | 523[M + H]+; | Benzoylgomisin H | d |
| 505[M + H − H2O]+; | ||||||||||
| 401[M + H − C6H5COOH]+; | ||||||||||
| 383[M + H − C6H5COOH − H2O]+ | ||||||||||
| 88 | 45.42 | C23H30O6 | 402.21151 | ― | ― | 403.21011 | −3.5 | 403[M + H]+; | Schisanhenol | d |
| 372[M + H − OCH3]+; | ||||||||||
| 371[M + H − CH3OH]+; | ||||||||||
| 356[M + H − CH3OH − CH3]+; | ||||||||||
| 340[M + H − OCH3 − CH3OH]+ | ||||||||||
| 89 | 45.44 | C23H28O7 | 416.19078 | ― | ― | 417.18957 | −2.9 | 417[M + H]+; | Schisandrol B | d |
| 399[M + H − H2O]+; | ||||||||||
| 343[M + H − C4H8 − H2O]+; | ||||||||||
| 307[M + H − 2CH3 − 2OCH3 − H2O]+ | ||||||||||
| 90 | 45.67 | C28H34O9 | 514.22755 | ― | ― | 515.22559 | −3.8 | 515[M + H]+; | Schisantherin B/Schisantherin C | d |
| 415[M + H − C4H7COOH]+; | ||||||||||
| 385[M + H − C4H7COOH − CH2O]+; | ||||||||||
| 367[M + H − C4H7COOH − CH2O − H2O]+; | ||||||||||
| 353[M + H − C4H7COOH − CH2O − CH3OH]+ | ||||||||||
| 91 | 46.06 | C30H32O9 | 536.21191 | ― | ― | 537.20981 | −3.9 | 537[M + H]+; | Schisantherin A | d |
| 415[M + H − C6H5COOH]+; | ||||||||||
| 397[M + H − C6H5COOH − H2O]+; | ||||||||||
| 373[M + H − C6H5COOH − C3H6]+ | ||||||||||
| 92 | 46.58 | C30H48O4 | 472.34689 | 471.34872 | 3.9 | 473.30911 | −3.7 | 471[M − H]−; | Corosolic acid/Maslinic acid | c |
| 453[M − H − H2O]−; | ||||||||||
| 427[M − H − CO2]−; | ||||||||||
| 391[M − H − CO2 − 2H2O]−; | ||||||||||
| 473[M + H]+; | ||||||||||
| 455[M + H − H2O]+; | ||||||||||
| 203[C15H23]+; | ||||||||||
| 105[C8H9]+ | ||||||||||
| 93 | 46.73 | C15H24N2O | 248.19614 | ― | ― | 249.19699 | 2.2 | 271[M + Na]+; | Matrine | b |
| 249[M+H]+; | ||||||||||
| 231[M + H − H2O]+; | ||||||||||
| 150[C10H16N]+; | ||||||||||
| 148[C10H14N]+ | ||||||||||
| 94 | 47.28 | C22H26O6 | 386.18022 | ― | ― | 387.17992 | −0.8 | 387[M + H]+; | Gomisin L1/Gomisin L2/Gomisin M1/Gomisin M2 | d |
| 372[M + H − CH3]+; | ||||||||||
| 369[M + H − H2O]+; | ||||||||||
| 357[M + H − CH2O]+; | ||||||||||
| 329[M + H − C2H2O2]+ | ||||||||||
| 95 | 47.52 | C30H48O4 | 472.34689 | 471.34863 | 3.7 | 473.36044 | −4.4 | 471[M − H]−; | Corosolic acid/Maslinic acid | c |
| 453[M − H − H2O]−; | ||||||||||
| 427[M − H − CO2]−; | ||||||||||
| 391[M − H − CO2 − 2H2O]−; | ||||||||||
| 473[M + H]+; | ||||||||||
| 455[M + H − H2O]+; | ||||||||||
| 203[C15H23]+; | ||||||||||
| 105[C8H9]+ | ||||||||||
| 96 | 47.53 | C22H26O6 | 386.18022 | ― | ― | 387.17847 | −4.5 | 387[M + H]+; | Gomisin L1/Gomisin L2/Gomisin M1/Gomisin M2 | d |
| 372[M + H − CH3]+; | ||||||||||
| 369[M + H − H2O]+; | ||||||||||
| 357[M + H − CH2O]+; | ||||||||||
| 329[M + H − C2H2O2]+ | ||||||||||
| 97 | 47.76 | C28H34O9 | 514.22755 | ― | ― | 515.22716 | −0.7 | 515[M + H]+; | Schisantherin B/Schisantherin C | d |
| 415[M + H − C4H7COOH]+; | ||||||||||
| 385[M + H − C4H7COOH − CH2O]+; | ||||||||||
| 367[M + H − C4H7COOH − CH2O − H2O]+; | ||||||||||
| 353[M + H − C4H7COOH − CH2O − CH3OH]+ | ||||||||||
| 98 | 48.01 | C30H48O3 | 456.36763 | 455.35424 | 4.9 | 457.36636 | −2.7 | 455[M − H]−; | Oleanolic acid/Ursolic acid | c,e |
| 407[M − H − HCHO − H2O]−; | ||||||||||
| 391[M − H − HCHO − H2O − CH4]−; | ||||||||||
| 479[M + Na]+; | ||||||||||
| 457[M + H]+; | ||||||||||
| 439[M + H − H2O]+; | ||||||||||
| 393[M + H − HCOOH − H2O]+ | ||||||||||
| 99 | 48.24 | C22H26O6 | 386.18022 | ― | ― | 387.17883 | −3.6 | 387[M + H]+; | Gomisin L1/Gomisin L2/Gomisin M1/Gomisin M2 | d |
| 372[M + H − CH3]+; | ||||||||||
| 369[M + H − H2O]+; | ||||||||||
| 357[M + H − CH2O]+; | ||||||||||
| 329[M + H − C2H2O2]+ | ||||||||||
| 100 | 48.4 | C30H48O3 | 456.36763 | 455.35394 | 4.3 | 457.36612 | −3.3 | 455[M − H]−; | Oleanolic acid/Ursolic acid | c,e |
| 407[M − H − HCHO − H2O]−; | ||||||||||
| 391[M − H − HCHO − H2O − CH4]−; | ||||||||||
| 479[M + Na]+; | ||||||||||
| 457[M + H]+; | ||||||||||
| 457[M + H − H2O]+; | ||||||||||
| 393[M + H − HCOOH − H2O]+ | ||||||||||
| 101 | 48.65 | C22H26O6 | 386.18022 | ― | ― | 387.17877 | −3.7 | 387[M + H]+; | Gomisin L1/Gomisin L2/Gomisin M1/Gomisin M2 | d |
| 372[M + H − CH3]+; | ||||||||||
| 369[M + H − H2O]+; | ||||||||||
| 357[M + H − CH2O]+; | ||||||||||
| 329[M + H − C2H2O2]+ | ||||||||||
| 102 | 49.79 | C24H32O6 | 416.22714 | ― | ― | 417.22649 | −1.6 | 417[M + H]+; | Deoxyschizan-drin | d |
| 402[M + H − CH3]+; | ||||||||||
| 386[M + H − OCH3]+; | ||||||||||
| 370[M + H − CH3 − CH3OH]+; | ||||||||||
| 347[M + H − CH3 − C4H7]+; | ||||||||||
| 316[M + H − C6H13O]+; | ||||||||||
| 286[M + H − C6H13O − 2CH3]+; | ||||||||||
| 273[M + H − C6H13O − CH3 − CO]+; | ||||||||||
| 227[M + H − C6H13O − CO − OCH3 − 2CH3]+ | ||||||||||
| 103 | 52.02 | C16H14O4 | 270.09649 | ― | ― | 271.09595 | −2.0 | 271[M + H]+; 203[M + H − C5H8]+; | Imperatorin | c |
| 175[M + H − C5H8 − CO]+; | ||||||||||
| 159[M + H − C5H8 − CO2]+ | ||||||||||
| 104 | 53.29 | C30H32O8 | 520.21701 | ― | ― | 521.21559 | −2.7 | 521[M + H]+; | Benzoylgomisin O/Benzoyisolgomisin O | d |
| 399[M + H − C6H5COOH]+; | ||||||||||
| 369[M + H − C6H5COOH − CH2O]+; | ||||||||||
| 357[M + H − C6H5COOH − C3H6]+; | ||||||||||
| 343 M + H − C6H5COOH − C4H8]+ | ||||||||||
| 105 | 53.43 | C22H24O6 | 384.16457 | ― | ― | 385.16406 | −1.3 | 385[M + H]+; | Schizandrin C | d |
| 370[M + H − CH3]+; | ||||||||||
| 355[M + H − CH2O]+ | ||||||||||
| 106 | 54.08 | C30H32O8 | 520.21701 | ― | ― | 521.21611 | −1.7 | 521[M + H]+; | Benzoylgomisin O/Benzoyisolgomisin O | d |
| 399[M + H − C6H5COOH]+; | ||||||||||
| 369[M + H − C6H5COOH − CH2O]+; | ||||||||||
| 357[M + H − C6H5COOH − C3H6]+; | ||||||||||
| 343 M + H − C6H5COOH − C4H8]+ | ||||||||||
a: Fructus Lych, b: Semen Cuscutae (fried), c: Fructus Rubi, d: Fructus Schisandrae chinensis (steamed), e: Semen Plantaginis (fried with salt); Glc : β-d-glucose, GluA: Glucuronic acid, Xyl: β-d-xylose, Rha: l-rhamnose; Gal: d-galactose; Api: d-apiose, ppm: difference between calculated and found mass.
2.2.1. Flavonoids
Flavonoids have a diphenylpropane skeleton bearing two benzene rings (A and B) connected by a pyran ring attached to the A ring, and are further divided into several subclasses (flavones, flavonols, flavanones, flavanonols, anthocyanidins, aurone, halcones and isoflavonoids). In this work, four types of flavonoids were found in WZYZW by UPLC–ESI-LTQ-Orbitrap-MS. Taking compound 53 as an example, the precise molecular weight is 623.16248 (C28H31O16) and in the negative ion spectrum, the main fragment ions were observed at m/z 623 [M − H]−, 477 [M − H − Rha]−, 315 [M − H − Rha − Glc]−, 300 [M − H − Rha − Glc − CH3]−, 272 [M − H − Rha − Glc − CH3 − CO]−, 271 [M − H − Rha − Glc − CH3 − CO − H]−, 243 [M − H − Rha − Glc − CH3 − CO − H − CO]−, 227 [M − H − Rha − Glc − CH3 − CO − H − CO2]−, and 151 [C7H3O4]−, thus, compound 53 was identified as isorhamnetin-3-O-β-d-rutinoside. Its mass spectrum and proposed fragmentation pathways in negative mode are shown in Figure 2.
Figure 2.
ESI-CID-MS3 spectra and proposed fragmentations for isorhamnetin-3-O-β-d-rutinoside (53) from WZYZW.
Compounds 34 and 37 were considered to be isomers as they displayed the same [M − H]− ions at m/z 463.08887 (C21H19O12) and 463.08740 (C21H19O12). Moreover, they had the similar fragment ions of m/z 301, 255 and 151 in the MSn spectra, but different max UV absorption wavelengths (λmax), as the λmax of compound 34 were 254 and 356 nm, and compound 37 showed λmax peaks at 254 and 346 nm. Considering the different retention times, compounds 34 and 37 were tentatively identified as hyperoside and isoquercitrin, respectively (see Table 1).
Similarly, based on the chromatographic behavior and MSn spectrometry for further confirming the fragmentation patterns as shown above, a total of 35 flavonoids were tentatively identified, including 15 flavones and derivatives, 16 flavonols and derivatives, three flavanone glycosides, and one flavanonol, which are summarized in Figure 3.
Figure 3.
Flavonoids identified in WZYZW by UPLC-ESI-LTQ-Orbitrap-MS.
2.2.2. Phenylpropanoids
Phenylpropanoids have one or more C6-C3 structures and there are three main subclasses: simple phenylpropanoids, coumarins and lignans. In the present study, 34 phenylpropanoids were found in WZYZW, including two simple phenylpropanoids, four coumarins and 28 lignans. The classification of these compounds is shown in Figure 4.
Figure 4.
Phenylpropanoids identified in WZYZW by UPLC-ESI-LTQ-Orbitrap-MS.
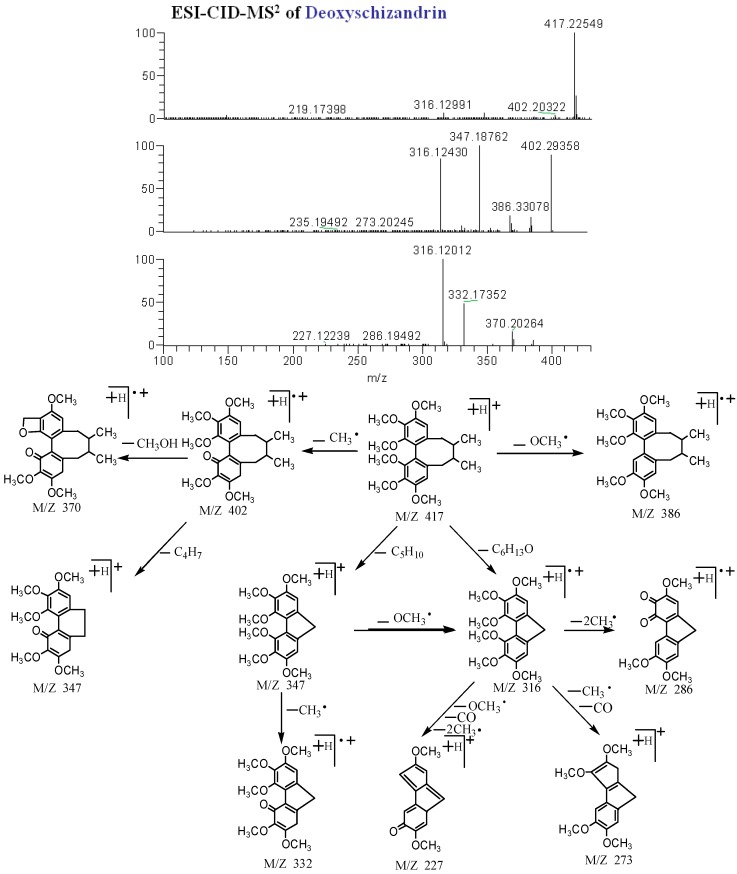
Compound 102 afforded a quasi-molecular ion at m/z 417.22649 (C24H33O6) in positive mode, and its MSn spectra showed representative ions at m/z 417 [M + H]+, 402 [M + H − CH3]+, 386 [M + H − OCH3]+., 370 [M + H − CH3 − CH3OH]+, 347 [M + H − CH3 − C4H7]+, 316 [M + H − C6H13O]+, 286 [M + H − C6H13O − 2CH3]+, 273 [M + H − C6H13O − CH3 − CO]+, and 227 [M + H − C6H13O – CO − OCH3 − 2CH3]+. Consequently, compound 102 was tentatively identified as deoxyschizandrin. The MSn mass spectra and the fragmentation pathways of deoxyschizandrin are shown in Figure 5. As with the fragmentations described above, the other phenylpropanoid compounds were also tentatively identified. However, six groups of isomers (compounds 40 and 42, 72 and 73, 74 and 77, 90 and 97, 94, 96, 99 and 101, 104 and 106) had the same molecular weights in the MS spectra, similar fragment ions in the MSn spectra and similar max UV absorption wavelengths. Thus we could not distinguish them, and these structures will require further confirmation (see Table 1).
Figure 5.
ESI-CID-MS3 spectra and proposed fragmentations for deoxyschizandrin (102) from WZYZW.
2.2.3. Organic Acids
The UPLC-ESI-LTQ-Orbitrap method was applied to comprehensively characterize the organic acids in this study, and the results showed that three types of organic acids could be detected in WZYZW: four aliphatic organic acids, three alicyclic organic acids and 10 phenolic acids. The specific details are displayed in Figure 6.
Figure 6.
Organic acids identified in WZYZW by UPLC-ESI-LTQ-Orbitrap-MS.
Compound 15 produced a [M − H]− ion at m/z 167.03420 (C8H7O4), and its fragment ions showed the losses of CH3, CO2 and CH3 + CO2, respectively. Thus, compound 15 was identified as vanillic acid, which was confirmed by MSn experiments. Similarly, the above fragmentation patterns were applied to further confirm the identities of the other organic acids compound, and finally, 17 compounds were identified as organic acids.
2.2.4. Terpenoids
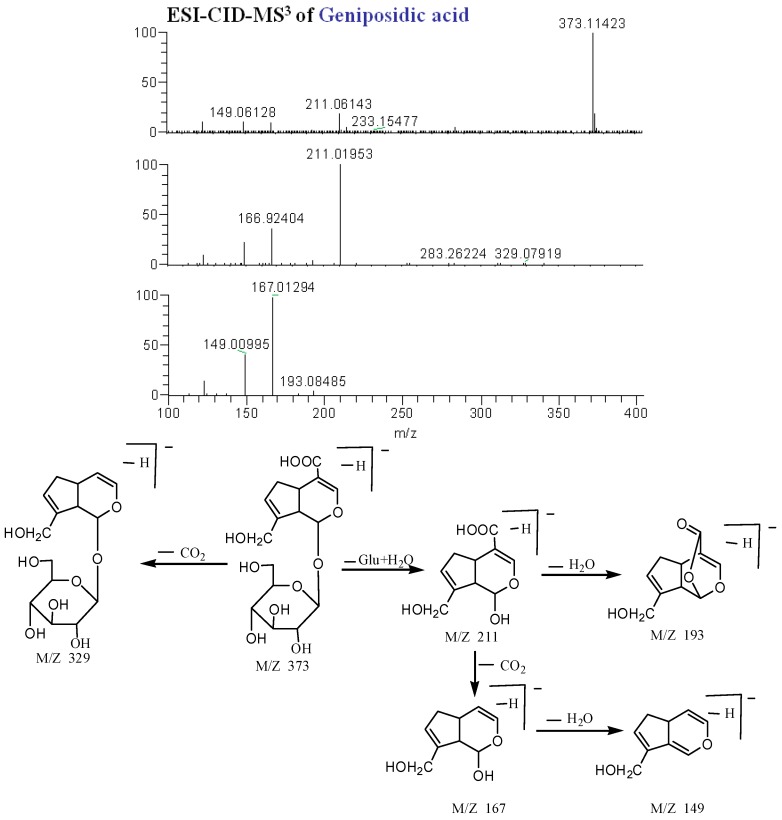
As displayed in Figure 7, the precise molecular weight of compound 16 is 373.11377 (C16H21O10) in the negative ion spectrum, and the main fragment ions were observed at m/z 373 [M − H]−, 329 [M – H − CO2]−, 211 [M − H − Glc]−, 193 [M − H − Glc − H2O]−, 167 [M − H − Glc − CO2]− and 149 [M − H − Glc − CO2 − H2O]−. Therefore, compound 16 was tentatively identified as geniposidic acid.
Figure 7.
ESI-CID-MS3 spectra and proposed fragmentations for geniposidic acid (16) from WZYZW.
Using high-resolution MSn mass spectrometry and the similar method of analysis of the aforementioned fragmentations, a total of 11 terpenoids were found: one iridoid (compound 16) was identified as geniposidic acid; 10 triterpenes—compounds 69, 76, 80, 81, 83, 84, 92, 95, 98 and 100 were identified as nigranoic acid, kadsuric acid, rosolic acid, 2α,3α,19α-trihydroxyolean-12-ene-28-oic acid, ganwuweizic acid, arjunolic acid, corosolic acid, maslinic acid, oleanolic acid and ursolic acid. Compounds 80, 81 and 84, 98 and 100, and 92 and 95, constituted three groups of isomers with similar fragmentation pathways in their MS spectra, so we could not precisely identify them by Mass (see Table 1).
2.2.5. Alkaloids
Compound 5 exhibited a [M + H]+ ion and a [M − H]− at m/z 124.03876 (C6H6NO2) and 122.02417 (C6H4NO2), respectively, and positive product ions at m/z 124 [M + H]+ and 106 [M + H − H2O]+ were detected. Thus, compound 5 was inferred as nicotinic acid (see Table 1). Similarly, eight alkaloids were identified in WZYZW and confirmed using MSn data: abromine (1), nicotinic acid (5), thiamine (6), taurine (14), atropine (19), sophoranol (82), n-methylcytisine (85), and matrine (93).
2.2.6. Miscellaneous
Riboflavin (compound 10) was also identified in WZYZW by its [M − H]− ion at m/z 375.12988 (C17H19N4O6). The corresponding MSn spectra showed a peak at m/z 255 [C13H11N4O2]−, and another fragment ion was also observed at m/z 212 [C12H10 N3O]−.
To summarize, in this study, a reliable and rapid UPLC-ESI-LTQ-Orbitrap-MS method has been established for the first comprehensive analysis of the phytochemical constituents of the Chinese herbal prescription WZYZW. This method revealed that UPLC-ESI-LTQ-Orbitrap-MS was useful for screening and identifying the complex constituents of WZYZW. Based on the method, a total of 106 compounds were tentatively characterized. In contrast to the published papers, it is noteworthy that this work detected more components. Our results provide essential data for further pharmacological studies and clinical evaluation of WZYZW and be useful for quality control of WZYZW, so as to guarantee its safe use in the clinic.
3. Experimental Section
3.1. Materials and Reagents
Fructus Lych, Semen Cuscutae (fried), Fructus Rubi, Fructus Schisandrae chinensis (steamed), and Semen Plantaginis (fried with salt) were supplied by the Tongrentang Drug Store in Beijing (China). The origins of the crude drugs were identified by Professor Rui Chao Lin of the Department of Pharmacognosy, Beijing University of Chinese Medicine. Voucher specimens were deposited at the authors’ laboratory. Reference standards of abromine, nicotinic acid, thiamine, riboflavin, taurine, quinic acid, atropine, ferulic acid, chlorogenic acid, scopoletin, rutin, esculin, apigenin, hesperidin, quercetin, kaempferol, luteolin, isorhamnetin, schisandrin were purchased from the National Institute for the Control of Pharmaceutical and Biological Products (Beijing, China). Acetonitrile and methanol (HPLC grade) were purchased from Fisher Scientific Co. (Waltham, MA, USA). Distilled water was purchased from Watson’s Food & Beverage Co. (Guangzhou, China). Formic acid, analytical grade, was obtained from Beijing Reagent Company (Beijing, China). Other reagents were HPLC grade or analytical reagent grade. High-purity nitrogen (99.9%) and helium (99.99%) were purchased from Gas Supplies Center of Peking University Health Science Center (Beijing, China).
3.2. Preparation of WZYZW Samples
A standard preparation of WZYZW was made in accordance with the 2015 version of the Chinese Pharmacopoeia. Fructus Lych, Semen Cuscutae (fried), Fructus Rubi, Fructus Schisandrae chinensis (steamed), and Semen Plantaginis (fried with salt) (8:8:4:2:1) were weighed, then crushed to powder (40 mesh size) and immersed in 10-fold volume of water for 1 h and heated. After boiling, heating was ontinued until the volume was reduced by 5-fold. The mixture was filtered through gauze while hot, concentrated to 1 g crude drug/mL, and freeze-dried to a powder. Finally refined honey (85 g) was mixed with freeze-dried powder (100 g) to make boluses of WZZYW for the experiments. Freeze-dried single herb extract powders were prepared as described for WZYZW. Freeze-dried samples of WZYZW (250 mg) were extracted with 50 mL of methanol with the aid of ultrasound for 60 min. The extracts were centrifuged at 10,000 rpm for 15 min at 4 °C, the supernatant was collected and filtered through a filter (0.22 μm), and the filtrate was collected for UPLC-LTQ-Orbitrap analysis. Respective standard stock solutions of 19 components were prepared at concentrations of 50 ng/mL by weighing the desired amount of each component into a volumetric flask and dissolving it in 100% ethanol; the 19 samples were filtered through a filter (0.22 μm), and the filtrates were analyzed by UPLC-LTQ-Orbitrap.
3.3. Instrumentation and Conditions
3.3.1. Liquid Chromatography Conditions
The chromatographic separation was performed on an ACQUITY UPLCTM BEH C18 column (1.7 μm, 2.1 mm × 100 mm) using an ACQUITY UPLCTM system (Waters Corporation, Milford, MA, USA) equipped with quaternary pump, vacuum degasser, autosampler and photodiode array detector. A linear gradient elution of A (HCOOH:H2O = 0.1:100) and B (HCOOH:CH3CN = 0.1:100) was used. The optimized gradient program is shown in Table 2. The flow rate was 0.3 mL/min and column temperature was set at 30 °C, the injection volume was 3 μL. The effluent was roughly split at a ratio of 3:1 (v/v) before entering the ESI source.
Table 2.
Solvent gradient program of UPLC analysis.
| Time (min) | Flow (mL/min) | A (%) | B (%) |
|---|---|---|---|
| 0 | 0.300 | 99.0 | 1.0 |
| 1.0 | 0.300 | 99.0 | 1.0 |
| 12 | 0.300 | 91.0 | 9.0 |
| 17 | 0.300 | 88.0 | 12.0 |
| 17.5 | 0.300 | 86.5 | 13.5 |
| 23 | 0.300 | 86.5 | 13.5 |
| 33 | 0.300 | 70 | 30.0 |
| 35 | 0.300 | 60 | 40.0 |
| 50 | 0.300 | 35 | 65.0 |
| 52 | 0.300 | 35 | 65.0 |
| 55 | 0.300 | 1.0 | 99.0 |
3.3.2. ESI-MS/MS Detection
The LTQ/Orbitrap mass spectrometer (Thermo Scientific, Bremen, Germany) was equipped with an ESI source operating in positive and negative ESI mode. The negative ion mode operation parameters were as follows: capillary voltage, 35 V; electrospray voltage, 3.0 kV; capillary temperature, 350 °C; sheath gas, 30 (arbitrary units); auxiliary gas, 10 (arbitrary units); tube lens, 110 V. The positive ion mode operation parameters: capillary voltage, 25 V; electrospray voltage, 4.0 kV; capillary temperature, 350 °C; sheath gas, 30 (arbitrary units); auxiliary gas, 5(arbitrary units ); tube lens, 110 V. Samples were detected by full-scan mass analysis from m/z 100 to 1000 at a resolving power of 30,000 with data-dependent MS2 analysis triggered by the three most-abundant ions from the predicted precursor list followed by MS3 analysis of the most-abundant product ions. To avoid performing many repeated data acquisitions on the same sample, dynamic exclusion is used for the data collection with an exclusion duration of 60s and the repeat count was set at 5 with a dynamic repeat time at 30 s. Collision-induced dissociation (CID) was performed with an isolation width of 2 Da. The collision energy was set to 35%. An external calibration for mass accuracy was carried out before the analysis. The measured masses were within 5ppm of the theoretical masses. The data analysis was achieved using XCalibur softwarev2.0.7 (Thermo Fisher Scientific).
3.4. Data Processing
The data analysis was processed using Thermo Xcaliber 2.1 workstation (Thermo Fisher Scientific) for peak detection and peak alignment, the raw data were processed by the computer-based NMDF approach.
4. Conclusions
In this study, a rapid, sensitive and reliable UPLC-ESI-LTQ-Orbitrap-MS method was established for screening and identifying the chemical constituents of WZYZW in both the positive and negative ion modes. Based on the chromatographic and spectrometric data, and referring to the literature, we were able to tentatively characterize 109 compounds, including organic acids, flavonoids, phenylpropanoids, alkaloids and terpenoids. In all 14 ingredients from Fructus Lych were identified, 11 ingredients from Semen Cuscutae (fried), 35 ingredients from Fructus Rubi, 37 ingredients from Fructus Schisandrae chinensis (steamed), and 21 ingredients from Semen Plantaginis (fried with salt). Our results broadens the chemical knowledge of WZYZW, which should be helpful for the quality control of WZYZW, and for further research on the pharmacokinetic studies and the health and medical properties of WZYZW. Moreover, with the successful application of the UPLC-ESI-LTQ-Orbitrap-MS to characterizing the constituents of WZYZW, it is suggest that this method offer a rapid, sensitive and high throughput methodology for the identification of constituents of TCM prescriptions and herbal medicines.
Acknowledgments
This research was financially supported by the Program of the Key Laboratory of Beijing City (NO. BZ0386).
Author Contributions
Dixin Zou and Ruichao Lin designed the research, performed the experimental work, analyzed data and wrote the manuscript. Jinfeng Wang, Suhua Xie and Kexin Xu prepared the samples. Bo Zhang and Qing Wang drew the chemical structures and coordinated the experiments. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Not available.
References
- 1.Wu W.Y., Hou J.J., Long H.L., Yang W.Z., Liang J., Guo D.A. TCM-base new drug discovery and development in China. Chin. J. Nat. Med. 2014;12:241–250. doi: 10.1016/S1875-5364(14)60050-9. [DOI] [PubMed] [Google Scholar]
- 2.Li W.F., Jiang J.G., Chen J. Chinese medicine and its modernization demands. Arch. Med. Res. 2008;39:246–251. doi: 10.1016/j.arcmed.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 3.Chan K. Chinese medicinal materials and their interface with Western medical concepts. J. Ethnopharmacol. 2005;96:1–18. doi: 10.1016/j.jep.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 4.Duan H., Zhu C.Y., Yuan W.C., Ma J., Yu R., Guo J. Advances in research on pharmacological effects and clinical application of Wuzi Yanzong Pill. Liaoning J. Tradit. Chin. Med. 2015;42:1814–1816. [Google Scholar]
- 5.An Q., Zou L. A meta-analysis of the Chinese medicine Wuziyanwan for treatment of oligospermia and asthenospermia. Chin. J. Hum. Sex. 2015;24:84–89. [Google Scholar]
- 6.Sun Z.W., Li Y.Z., Zhang C.C. Research advances on pharmacological effects and clinical applications of Wu Zi Yan Zong Wan. Asia-Pac. Tradit. Med. 2010;6:179–181. [Google Scholar]
- 7.Zhou J.L., Liu W., Tan C.M., Zhu M., Ma S.C. Quality control study of Wuziyanzong Pills. Chin. Pharm. J. 2015;50:125–130. [Google Scholar]
- 8.Zhou J.L., Liu W., Chen B.L., Zhu M. Determination of scopoletin in Lycii Fructus and Wuzi Yanzong Pill by HPLC-Fluorescence detection. Chin. J. Mod. Appl. Pharm. 2015;32:482–486. [Google Scholar]
- 9.Miao L., Chen M.L., Cao J., Sun M.Q., Liu J.X. LC-MS determination of 9 constituents in active fraction of Wuziyanzong. Chin. J. Pharm. Anal. 2011;31:659–663. [Google Scholar]
- 10.Liu W., Zhou J.L., Chen B.L., Zhu M. Simultaneous determination of five ingredients in Wuzi Yanzong Pill by HPLC. Chin. J. Exp. Tradit. Med. Formulae. 2014;20:74–78. [Google Scholar]
- 11.Pan H.Q., Yang W.Z., Zhang Y.B., Yang M., Feng R.H., Wu W.Y., Guo D.A. An integrated strategy for the systematic characterization and discovery of new indole alkaloids from Uncaria rhynchophylla by UHPLC/DA-D/LTQ-Orbitrap-MS. Anal. Bioanal. Chem. 2015;407:6057–6070. doi: 10.1007/s00216-015-8777-0. [DOI] [PubMed] [Google Scholar]
- 12.Wang S.S., Xu H.Y., Ma Y.N., Wang X.G., Shi Y., Huang B., Tang S.H., Zhang Y., Li D.F., Liang R.X., et al. Characterization and rapid identification of chemical constituents of NaoXinTong capsules by UHPLC-linear ion trap/Orbitrap mass spectrometry. J. Pharm. Biomed. Anal. 2015;111:104–118. doi: 10.1016/j.jpba.2015.01.020. [DOI] [PubMed] [Google Scholar]
- 13.Kouloura E., Skaltsounis A.L., Michel S., Halabalaki M. Ion tree-based structure elucidation of acetophenone dimers (AtA) from Acronychia pedunculata and their identification in extracts by liquid chromatography electrospray ionization LTQ-Orbitrap mass spectrometry. J. Mass. Spectrom. 2015;50:495–512. doi: 10.1002/jms.3556. [DOI] [PubMed] [Google Scholar]
- 14.Vallverdú-Queralt A., Boix N., Piqué E., Gómez-Catalan J., Medina-Remon A., Sasot G., Mercader-Martí M., Llobet J.M., Lamuela-Raventos R.M. Identification of phenolic compounds in red wine extract samples and zebrafish embryos by HPLC-ESI-LTQ-Orbitrap-MS. Food. Chem. 2015;181:146–151. doi: 10.1016/j.foodchem.2015.02.098. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J.Y., Wang F., Zhang H., Lu J.Q., Qiao Y.J. Rapid identification of polymethoxylated flavonoids in traditional Chinese medicines with a practical strategy of stepwise mass defect filtering coupled to diagnostic product ions analysis based on a hybrid LTQ-Orbitrap mass spectrometer. Phytochem. Anal. 2014;25:405–414. doi: 10.1002/pca.2508. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y.W. The performance and featured applications of LTQ Orbitrap velos, a hybrid high resolution mass spectrometer using electrostatic orbital mass analyzer coupled with dual pressure ion trap. Mod. Instrum. Med. Treat. 2010;5:5–19. [Google Scholar]
- 17.Kumar A., Saini G., Nair A., Sharma R. UPLC: A preeminent technique in pharmaceutical analysis. Acta Pol. Pharm. 2012;69:371–380. [PubMed] [Google Scholar]
- 18.Jin G.W., Zhang F.F., Xue X.Y., Xiao Y.S., Xu Q., Liang X.M. Application of Ultra-performance Liquid Chromatography in the separation and analysis of complicated system-Traditional Chinese Medicine. Mod. Tradit. Chin. Med. Mater. Med. World Sci. Technol. 2006;8:106–111. [Google Scholar]