Abstract
The high incidence of Mycobacterium infection, notably multidrug-resistant M. tuberculosis infection, has become a significant public health concern worldwide. In this study, we isolate and analyze a mycobacteriophage, BTCU-1, and a foundational study was performed to evaluate the antimycobacterial activity of BTCU-1 and its cloned lytic endolysins. Using Mycobacterium smegmatis as host, a mycobacteriophage, BTCU-1, was isolated from soil in eastern Taiwan. The electron microscopy images revealed that BTCU-1 displayed morphology resembling the Siphoviridae family. In the genome of BTCU-1, two putative lytic genes, BTCU-1_ORF7 and BTCU-1_ORF8 (termed lysA and lysB, respectively), were identified, and further subcloned and expressed in Escherichia coli. When applied exogenously, both LysA and LysB were active against M. smegmatis tested. Scanning electron microscopy revealed that LysA and LysB caused a remarkable modification of the cell shape of M. smegmatis. Intracellular bactericidal activity assay showed that treatment of M. smegmatis—infected RAW 264.7 macrophages with LysA or LysB resulted in a significant reduction in the number of viable intracellular bacilli. These results indicate that the endolysins derived from BTCU-1 have antimycobacterial activity, and suggest that they are good candidates for therapeutic/disinfectant agents to control mycobacterial infections.
Keywords: mycobacteriophage, endolysin, antimycobacterial
1. Introduction
Mycobacterium tuberculosis (MTB) is the leading cause of tuberculosis which is a serious public health problem that results in millions of deaths around the world each year [1]. Approximately one-third of the world’s population is latently infected with MTB and at risk of reactivation [2]. The slow-growing MTB can persist in the latent state of asymptomatic infection for a long time, and the treatment of active diseases involves long multidrug regimens. Unfortunately, the world is at present struggling with multi-drug resistant (MDR) as well as extensively drug resistant (XDR) forms of MTB which threaten to make both the first and second line drugs ineffective [3,4]. The prevention of the spread of MTB is a continuing challenge, and finding novel agents to treat MDR and XDR MTB infections has become a priority.
Bacteriophages (phages) are viruses that infect and replicate within their bacterial hosts. The lytic phages have developed various ways to interfere with and kill their host cells. Therefore, phage therapy is being considered as a possible therapeutic alternative for the treatment of infections caused by MDR strains [5]. Particular attention has been paid to recombinant phage endolysins because of their potential to digest bacterial cell walls when applied exogenously, enabling their use as alternative antibacterials [6,7]. Presently, scientists have successfully tested the use of endolysins to control antibiotic-resistant bacterial pathogens in animal models [8].
Like other bacteriophages, mycobacteriophages were considered as an alternative therapy for Mycobacterium infection control [9]. Additionally, mycobacteriophage-encoded lytic endolysins have considerable potential to be effective antimicrobial agents-or enzybiotics-against a number of MDR and XDR MTB strains [10,11]. To date, several phages infecting Mycobacterium spp. have been reported [12,13,14]. Nonetheless, the isolation and genome description of mycobacteriophages in Taiwan still remain to be achieved.
In this study, we used Mycobacterium smegmatis as a rapidly growing host, and a mycobacteriophage was isolated from the soil near Buddhist Tzu Chi University and named BTCU-1. Electron microscopy images revealed morphology resembling the Siphoviridae family. The BTCU-1 genome is a linear double-stranded DNA of about 46 kb in length. To utilize mycobacteriophage lytic endolysins as therapeutic alternatives to antibiotics, we surveyed the genomic sequence of BTCU-1 and successfully identified two lytic-associated genes. Following cloning and expression/purification, various antimycobacterial activities of these two lytic proteins were determined in vitro.
2. Results and Discussion
2.1. Morphology of BTCU-1
To classify mycobacteriophage BTCU-1 into a morphotype-specific group, the phage particles were examined by TEM. The electron microscopy images revealed that BTCU-1 possessed an icosahedral head (65 nm) and a non-contractile long tail (diameter, 12 nm; length, 180 nm), displaying morphology resembling the Siphoviridae family (Figure 1).
Figure 1.
Electron micrograph of phage BTCU-1. Purified phage particles were negatively stained with 2% uranyl acetate. Bar 100 nm.
2.2. BTCU-1 Genome and Predicted Endolysins
The BTCU-1 genome is a linear double-stranded DNA of about 46 kb in length. The genomic sequence of BTCU-1 was deposited in GenBank (accession number KC172839). The complete genomic sequence of BTCU-1 showed more than 90% nucleotide identity to mycobacteriophages Rockstar and HelDan (Supplementary Figure S1). According to the functions predicted so far, in general, the structural and lysis proteins are encoded by one strand, and the proteins required for the maintenance of lysogeny are encoded by the opposite strand. One of the major differences of BTCU-1 from the other two genomes (Rockstar and HelDan) located at ORF4 and ORF5, with the limited knowledge that the product of ORF4 is a structural protein. In addition, about 2 Kb and 3.3 Kb extra genomic fragments were found after the repressor genes in the genomes of Rockstar and HelDan, respectively. Though the genome of BTCU-1 is suggested to contain genes related to lysogeny, the deletion of the genomic fragment in BTCU-1 may inactivate the repressor gene, causing the defect in establishing a lysogenic state. This presumption is supported by the clear plaques produced by BTCU-1 on M. smegmatis.
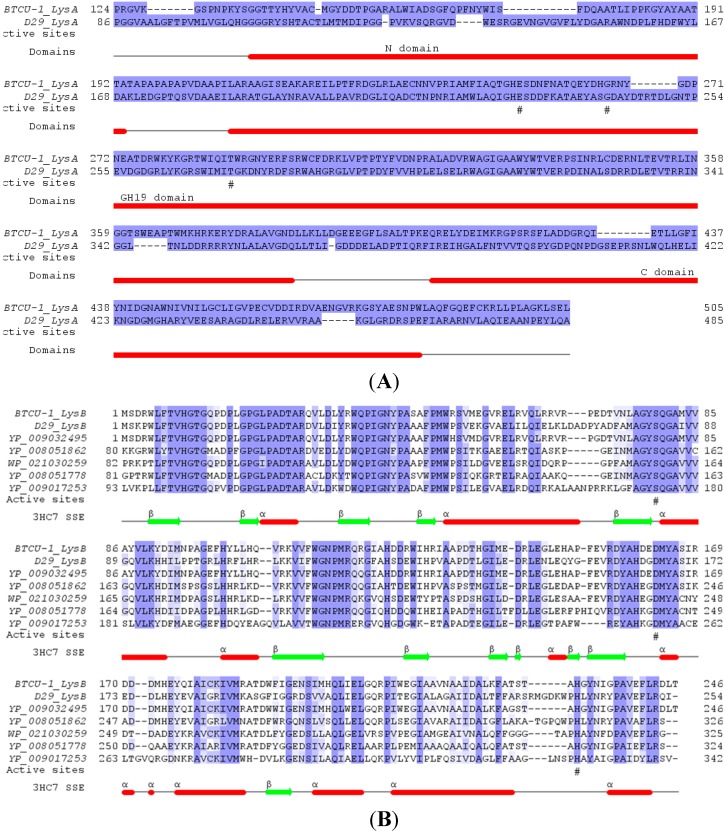
To utilize lytic endolysins as therapeutic alternatives to antibiotics, we surveyed the genomic sequence of BTCU-1 and successfully identified two lytic associated genes, BTCU-1_ORF7 and BTCU-1_ORF8 (termed lysA and lysB, respectively). LysA is a putative endolysin responsible for the cleavage of the peptidoglycan in the cell wall of mycobacteria. Sequence analysis suggests that LysA contains three domains: the N-terminal peptidase domain, the central catalytic GH19 (glycoside hydrolase family 19) domain, and the C-terminal cell wall binding domain (Figure 2A). LysB shares about 63% identity with the previously characterized LysB (gp12) in mycobacteriophage D29, and similarly lacks a possible peptidoglycan-binding domain at the N-terminal, while it presents in many D29 LysB homologues (Figure 2B) [15]. D29 LysB has been proved to be a mycolylarabinogalactan esterase required for the completion of lysis of the host mycobacterial cells [15]. It cleaves the ester linkage joining the mycolic acid-rich outer membrane to arabinogalactan, releasing free mycolic acids.
Figure 2.
Multiple sequence alignment of BTCU-1 lytic associated proteins, LysA and LysB. (A) Sequence alignment between BTCU-1 LysA and D29 LysA (gp10, NP_046825). The modular organization of LysA is composed of an N-terminal domain with putative peptidase activity, a central catalytic GH19 (glycoside hydrolase family 19) domain, and a C-terminal domain possibly responsible for cell wall binding. The predicted catalytic residues in GH19 domains are marked by hash signs under the alignment; (B) Multiple sequence alignment of BTCU-1 LysB and its homologues, including D29 LysB (gp12, NP_046827) whose structure has been resolved (PDB code 3HC7). The predicted residues involving in catalysis (Ser82-Asp166-His240 in 3HC7) are marked by hash signs under the alignment. 3HC7 SSE, resolved secondary structural elements for D29 LysB; ribbons indicate helices and arrows indicate strands.
2.3. Cloning, Expression and Purification of LysA and LysB
To confirm that LysA and LysB are mycobacteriophage lytic associated enzymes, their genes (BTCU-1_ORF7 and BTCU-1_ORF8) from the BTCU-1 genome were cloned for further characterization. After the PCR reaction, two respective amplified sequences from the BTCU-1 genome were inserted into the E. coli expression vector pET-30b, yielding pET30b-LysA and pET30b-LysB, respectively. In E. coli BL21 (DE3), pET30b-LysA directed the synthesis of a single individual protein with an apparent molecular mass of about 63 kDa on a sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (Figure 3A). Similarly, pET30b-LysB directed the synthesis of a single individual protein with an apparent molecular mass of about 34 kDa on an SDS gel (Figure 3A).
Figure 3.
SDS-PAGE of purified LysA and LysB and their antimycobacterial activity. (A) Purified LysA and LysB were electrophoresed on a 12% SDS gel, followed by staining with Coomassie brilliant blue R-250. Lane M, molecular weight markers; lane 2, purified LysA; lane 4, purified LysB; Lanes 1 and 3 indicate crude extract of individual encoding strains about four hours after induction; (B) The minimal bactericidal concentration (MBC) of LysA was over 80 μg·mL−1, and the MBC of LysB was between 20 to 40 μg·mL−1.
2.4. Antimycobacterial Activity of BTCU-1 and Purified LysA and LysB
We first tested the lytic activity of BTCU-1 against two M. smegmatis strains and seven M. tuberculosis isolates listed in Table 1. BTCU-1 showed broad lytic host range, affecting almost all the tested Mycobacterium strains except TCGH59490 (Supplementary Figure S2). To study the antibacterial activity of LysA and LysB, M. smegmatis ATCC 14468 was examined for its susceptibility to these proteins. As shown in Figure 3B, the MBC of LysA was over 80 μg·mL−1, and the MBC of LysB was between 20 and 40 μg·mL−1. To further analyze the antibacterial spectra of LysA and LysB, another three Gram-negative (E. coli ATCC 25922; Acinetobacter baumannii ATCC 17978 and Salmonella enteric BCRC 10746) and two Gram-positive bacteria (Bacillus subtilis BCRC 10447 and Staphylococcus aureus ATCC 25923) were used for the MBC assay. The MBC of LysA and LysB towards these five bacteria were higher than 200 μg·mL−1. We noticed that LysA and LysB had narrow spectra of antimicrobial activity against only Mycobacteria spp., with relatively lower bactericidal activity against other non-Mycobacteria species.
Table 1.
Bacterial strains, plasmids, and oligonucleotide primers used in this study.
| Strain, Plasmid, or Primer | Relevant Characteristics, Description, or Sequence | Source |
|---|---|---|
| Bacterial Strains | ||
| M. smegmatis ATCC 14468 | reference strain | American Type Culture Collection (ATCC) |
| M. smegmatis mc2155 (ATCC 70084) | reference strain | ATCC |
| M. tuberculosis TCGH 57612 | Clinical strain | Buddhist Tzu Chi General Hospital (BTCGH) |
| M. tuberculosis TCGH 58339 | Clinical strain | BTCGH |
| M. tuberculosis TCGH 58821 | Clinical strain | BTCGH |
| M. tuberculosis TCGH 59490 | Clinical strain | BTCGH |
| M. tuberculosis TCGH 62046 | Clinical strain | BTCGH |
| M. tuberculosis TCGH 45545 | MDR Clinical strain | BTCGH |
| M. tuberculosis TCGH 17037 | XDR Clinical strain | BTCGH |
| E. coli Top10 | Laboratory strain for TA cloning use | Invitrogen, San Diego, CA, USA |
| E. coli BL21 (DE3) | Laboratory strain for protein expression | Invitrogen |
| E. coli ATCC 25922 | Gram-negative reference strain | ATCC |
| A. baumannii ATCC 17978 | Gram-negative reference strain | ATCC |
| S. enteric BCRC 10746 | Gram-negative reference strain | Bioresource Collection and Research Center (BCRC) |
| B. subtilis BCRC 10447 | Gram-positive reference strain | BCRC |
| S. aureus ATCC 25923 | Gram-positive reference strain | ATCC |
| Plasmids | ||
| pGEM-T-easy | 3015-bp E. coli vector, Ampr, Plac, lacZ | Promega, San Diego, CA, USA |
| pET30b | 5421-bp E. coli vector, Kmr, PT7, His-Tag | Novagen, Madison, WI, USA |
| pGEM-LysA | pGEM-T-easy::1566 bp of lysA | This study |
| pGEM-LysB | pGEM-T-easy::741 bp of lysB | This study |
| pET30b-LysA | pET-30b::1566 bp of lysA; His-Tag-LysA under PT7 | This study |
| pET30b-LysB | pET-30b:: 741 bp of lysB; His-Tag-LysB under PT7 | This study |
| Primers | ||
| BTCU-1_lysA-FP | 5ʹ-ATGACGGAACGGGTACTCCC-3ʹ | This study |
| BTCU-1_lysA-RP | 5ʹ-TCACTGCTCGATGACCCTGTT-3ʹ | This study |
| BTCU-1_lysB-FP | 5'-GTGAGCGACCGCTGGCT-3ʹ | This study |
| BTCU-1_lysB-RP | 5ʹ-TCATGTCAAGTCGCGTAGAAACT-3ʹ | This study |
2.5. Electron Microscopy Experiments
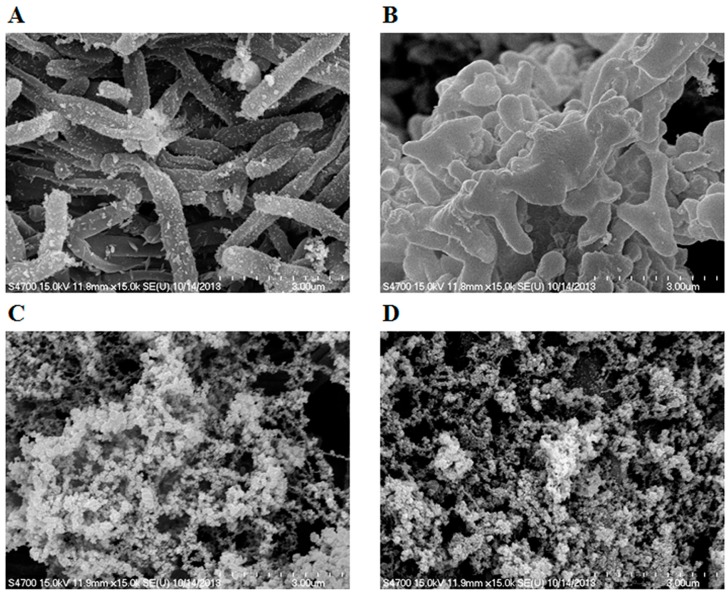
The effects of LysA and LysB on the morphology of M. smegmatis were visualized using scanning electron microscopy (SEM). The exposure of bacteria to 100 μg·mL−1 LysA or LysB for 24 hour caused a remarkable modification of their cell shape, as shown by SEM. Untreated bacteria displayed a rough bright surface with no obvious cellular debris (Figure 4A). The crinkled and irregular spheroidal cell morphology was observed in the cells of M. smegmatis after exposed to LysA as shown in Figure 4B. The treatment with LysB resulted in notable bacterial rupture and breakdown in M. smegmatis (Figure 4C), and treatment with LysA combined LysB exhibited a wide range of significant abnormalities, including the collapse of the cell structure and notable cracked cells in M. smegmatis (Figure 4D).
Figure 4.
Scanning electron microscopy of M. smegmatis treated with LysA or LysB. (A) Untreated M. smegmatis cells; (B) M. smegmatis treated with 100 μg·mL−1 LysA for 24 h; (C) M. smegmatis treated with 100 μg·mL−1 LysB for 24 h; (D) M. smegmatis treated with 100 μg·mL−1 LysA combined LysB for 24 h.
2.6. LysA and LysB Kill Intracellular M. smegmatis
To determine the effect of BTCU-1 derived endolysins on intracellular M. smegmatis, RAW 264.7 macrophage cell lines were infected with M. smegmatis ATCC 14468, and 2 h after infection, treated with LysA or LysB for 12 h. As shown in Figure 5, the viabilities of intracellular M. smegmatis dropped to <40% following incubation with LysA compared to the negative control (incubation with DMEM). However, the viabilities of M. smegmatis dropped to <20% following incubation with LysB for the same period of time. These results suggest that BTCU-1 derived endolysins can effectively decrease the number of intracellular M. smegmatis.
Figure 5.
The intracellular survival of M. smegmatis after 12 h treated with LysA or LysB. Survival was expressed as the percentage in the recovered number of viable intracellular M. smegmatis compared with the negative control (incubation with DMEM). The differences in percentage in the recovered number between treated with LysA or LysB and negative control were analyzed by the Student’s t-test. Each bar represents the average value of three independent experiments, and the error bars represent the standard deviations. * p < 0.05.
2.7. Discussion
The prevention of the spread of MTB is a continuing challenge, and finding novel agents to treat MDR and XDR MTB infections has become a priority. Mycobacteriophages own the abilities to attack and kill mycobacteria and can potentially serve as an alternative option to combat mycobacteria infection.
In this work, a mycobacteriophage was isolated from the soil near Buddhist Tzu Chi University, Taiwan and was named BTCU-1. To our knowledge, this is the first detailed study of a mycobacteriophage isolated from Taiwan. Similar to other mycobacteriophages reported, BTCU-1 showed a broad host range, being able to lyse almost all the tested Mycobacterium strains. This suggests its use as an alternative sanitation or disinfectant agent will be highly feasible for application in controlling MTB infections [16]. In order to have a better understanding about mycobacteriophage distribution in Taiwan, more work on the isolation and characterization of mycobacteriophages is still needed.
Mycobacterium spp. possess unusual cell wall core, which consists of a peptidoglycan layer covalently attached to a mycolic acid layer via the polysaccharide arabinogalactan [17]. Upon infection, mycobacteriophages produce lysins to digest the peptidoglycan and mycolic acid layers of the host cell wall, causing the rupture of the bacterial cells and the release of the phage particles. The ability to lyse mycobacterial cells makes lysins extremely important. There have been hundreds of mycobacteriophage genomes sequenced [18], and the sequence features of the major types of lysins have been analyzed [11,15]. Nevertheless, the evaluation of their antimycobacterial effect, particularly applied exogenously, is still very limited. In the example of mycobacteriophage Ms6, its LysA was characterized as a peptidoglycan amidase; however, in culture, it showed no antibacterial effect on either M. tuberculosis or M. smegmatis [19].
In this study, we surveyed the genomic sequence of BTCU-1 and identified two lytic associated genes, BTCU-1_ORF7 and BTCU-1_ORF8 (termed lysA and lysB, respectively). Sequence analysis suggests that LysA is a putative endolysin responsible for the cleavage of the peptidoglycan in the cell wall of mycobacteria, whereas LysB probably acts at the mycolylarabinogalactan bond to liberate free mycolic acid. To our surprise, either LysA or LysB alone, while applied exogenously, could cause a remarkable modification of the cell shape of M. smegmatis and also cause cell death. Scanning electron microscopy (SEM) result showed that LysA-treated M. smegmatis displayed crinkled and irregular spheroidal morphology. A simple explanation for this is that the peptidoglycan in the cell wall of mycobacteria was partially removed by the action of LysA, and the membrane tension caused the cell to form a characteristic spherical shape. However, how the exogenous LysA permeates through the mycolic acid-rich outer membrane to reach the substrates remains to be discovered. Furthermore, in vitro and in vivo antimicrobial assays showed that LysB possessed better bactericidal capacity than LysA while applied exogenously. Based on the results of SEM and sequence analysis, we can only presume that LysB proteins reach their host mycolylarabinogalactan targets from the outside, and decrease the integrity of the mycolic acid linkage to the arabinogalactan-peptidoglycan layer. This may directly affect the cell wall permeability barrier in mycobacteria. However, the mechanism of LysB that ultimately results in the bacteriolysis seen in these experiments is a question worthy of investigation.
3. Experimental Section
3.1. Bacteria, Vector and Growth Conditions
The bacterial strains and plasmids used in this study are listed in Table 1. M. smegmatis strains were grown at 37 °C in Middlebrook 7H9 medium (Difco, Detroit, MI, USA) with shaking, or on Middlebrook 7H11 agar (Difco) supplemented with 0.5% glycerol. E. coli strains were grown at 37 °C in Luria-Bertani (LB) broth or agar supplemented with 100 μg·mL−1 ampicillin or 50 μg·mL−1 kanamycin, when appropriate.
3.2. Isolation and Characterization of the Mycobacteriophage
In this study, M. smegmatis ATCC 14468 is a non-virulent mycobacterial strain and was used as a host for phage isolation. The soil sample was collected from sites near Buddhist Tzu Chi University, Hualien, Taiwan. The land is owned by Tzu-Chi Foundation, the founder of Tzu Chi University, and no specific permission was required to access the land or collect the samples. No endangered or protected species were found or are registered in this area. Each time, about 300–500 g of soil was collected from a surface of 100 cm2, at a depth of 3–5 cm. We believe the environmental impact is minimal. The mycobacteriophage isolation and purification process was performed according to the procedures described by Pope et al. [12]. Phage morphology was examined by TEM of negatively stained preparations [20]. A drop of approximately 1010 PFU/mL was applied to the surface of a formvar-coated grid (200 mesh copper grids), negatively stained with 2% uranyl-acetate and then examined in a Hitachi H-7500 transmission electron microscope (Hitachi Company, Tokyo, Japan) operated at 80 kV.
3.3. Phage DNA Preparation and Genome Sequencing
Phage DNA isolation was performed as previously described in detail [21]. The genome of mycobacteriophage was sequenced using the Ion Torrent PGM 314 chip (approximately 500 × coverage rate, Thermo Fisher Scientific, Waltham, MA, USA). The order of assembled contigs was predicted from the genomic sequences of Mycobacteriophage Rockstar (GenBank accession number JF704111), and Mycobacteriophage HelDan (accession no. JF957058) and confirmed by PCR. Primer walking was used to fill the gaps. Gene prediction was performed using GenMark.hmm [22], GenMarkS and Glimmer [23], followed by a manual correction where needed. All genes were annotated by BLAST searches of the GenBank databases. tRNA genes were predicted using the tRNAscan-SE tools [24].
3.4. Cloning/Purification of Endolysins
Genomic DNA of the mycobacteriophage BTCU-1 was extracted as previously described in detail [21]. Plasmid DNA was purified from E. coli using a QIAprep Spin MiniPrep Kit (Qiagen, Hilden, Germany). Polymerase chain reaction (PCR) amplification, with the DNA of BTCU-1 as a template, was carried out to produce two putative endolysin genes lysA and lysB. To construct the LysA expression vector, a 1566-base pair DNA fragment of lysA was amplified by PCR with BTCU-1_lysA-FP and BTCU-1_lysA-RP primers and cloned into a TA cloning site of pGEM-T-easy (Promega); the resulting recombinant DNA (BTCU-1_lysA) was digested with EcoRI and cloned into the EcoRI sites of pET-30b (Novagen). The resulting plasmid, pET30b-LysA, was then used to transform E. coli strain BL21 (DE3) for expression. The pET30b-LysB expression vector was constructed in a similar approach to pET30b-LysA, using BTCU-1_lysB-FP and BTCU-1_lysB-RP as primers. All of the constructed plasmids for expression were confirmed by DNA sequencing. The subsequent expression and purification procedures of the recombinant proteins were carried out according to the processes as described by Lai et al. [21]. The E. coli BL21 (DE3) strains, harboring the plasmids pET30b-LysA or pET30b-LysB were grown overnight in LB medium containing kanamycin (50 μg/mL). The overnight culture of the transformed E. coli was diluted 100 times with the same medium and incubated at 37 °C with shaking (150 rpm) until the optical density of the medium at OD600nm reached 0.5. The expression of the target gene was induced by the addition of isopropyl-l-d-thiogalacto pyranoside at a final concentration of 0.1 mM. After further incubation for 3 h, the cells were harvested by centrifugation. The cell pellet was suspended in 10 mL of lysisequilibration-wash (LEW) buffer containing 50 mM NaH2PO4/300 mM NaCl (pH 8.0), disrupted by sonication and centrifuged at 10,000 g for 15 min to remove debris. Crude supernatant was loaded onto Protino Ni-TED packed columns (MACHEREY-NAGEL, Düren, Germany) equilibrated with LEW buffer. The fractions were eluted with the elution buffer containing 50 mM NaH2PO4/300 mM NaCl/250 mM imidazole (pH 8.0). Active fractions were pooled and dialyzed against the elution buffer and concentrated by Amicon Ultra-0.5 centrifugal filter (MILLPORE, Bedford, MA, USA). The concentration of each purified protein was determined by the Bradford assay using bovine serum albumin as a standard.
3.5. Antimycobacterial Assays
The host-range of BTCU-1 was determined by the plaque-forming method, adapting a spot-test technique described by Rybniker et al. [25], with some modifications as detailed below. BTCU-1 was prepared in a phage buffer (10 mM Tris, pH 7.5, 1 mM MgSO4, and 70 mM NaCl) and diluted to 106 PFU/mL. The mycobacterial strains used in this test were grown overnight in Middlebrook 7H9 broth (Difco) (for M. smegmatis) or grown on 7H11 agar (for 7 M. tuberculosis clinical strains) at 37 °C. Until the bacteria grew to an appropriate concentration, the mycobacterial strains were diluted to an OD600 of 0.1. Two hundred microliters (µL) of mycobacteria were mixed with 200 µL of prepared BTCU-1 and added to 3 mL top agar containing 1 mM CaCl2 and poured onto 7H11 agar plates. Plates were incubated at 37 °C for four days for the fast-growing Mycobacteria, and for up to 4–6 weeks for the slow-growing strains.
In vitro antimicrobial assays were performed for the purified LysA and LysB. Antimicrobial activity was determined as described by Chen et al. [26], with some modifications as detailed below. Bacteria were grown overnight in Middlebrook 7H9 medium or Mueller–Hinton broth (Difco) at 37 °C, and during the mid-logarithmic phase, bacteria were diluted to 106 colony-forming units (CFU)/mL in phosphate buffer. LysA and LysB were serially diluted in the same buffer to the concentration range from 10 to 400 μg·mL−1. Fifty microliter (µL) of bacteria was mixed with fifty µL of LysA and LysB at varying concentrations followed by incubation at 37 °C for 24 h without shaking. At the end of incubation, bacteria were inoculated on Middlebrook 7H11 agar or Mueller-Hinton agar, and allowed growth at 37 °C for four days. The lowest concentration of LysA or LysB on the agar plate which displayed no bacterial growth is defined as the minimal bactericidal concentration (MBC). All experiments were performed in triplicate.
3.6. Scanning Electron Microscopy
The scanning electron microscopy images were prepared as previously described in detail [16]. M. smegmatis ATCC 14468 was grown in Middlebrook 7H9 medium to a log phase, harvested by centrifugation, washed twice with deionized water and resuspended in the same water. Approximately 108 cells were incubated at 37 °C for 24 h with 100 μg·mL−1 LysA or LysB. Negative controls were run in the presence of phosphate buffer. The volume was adjusted to 100 µL. The treated cells were fixed with 2.5% (w/v) glutaraldehyde in 0.1 M cacodylate buffer and 1% tannic acid, thoroughly washed with the phosphate buffer and dehydrated with a graded ethanol series. After critical-point drying and gold coating, the samples were observed with a HITACHI S-4700 instrument (Hitachi Company, Tokyo, Japan) operated at 15 kV.
3.7. Intracellular Bactericidal Assay
Infection of macrophages and quantitation of intracellular bacteria were performed as already described with some modifications [27]. Mouse peritoneal macrophage cell line, RAW 264.7, was obtained from the American Tissue Culture Collection. Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Difco Laboratories) supplemented with 10% fetal bovine serum and 2 mM l-glutamine. For the assays described in this article, RAW 264.7 macrophages (106) were treated with trypsin, washed, and seeded on a six-well tissue culture plate (Corning Costar, Cambridge, MA, USA) and allowed to grow overnight at 37 °C with an atmosphere of 5% CO2. M. smegmatis ATCC 14468 were used to infect RAW 264.7 macrophages. Bacteria were grown overnight in Middlebrook 7H9 medium at 37 °C, and during the mid-logarithmic phase, bacteria were diluted to 108 CFU/mL in DMEM. Monolayers (∼106 cells) were incubated with M. smegmatis at a ratio of 100 bacteria to 1 cell. Infection was allowed to occur for 2 h, and then the monolayers were washed with Hank’s Balanced Salt Solution (HBSS) twice and reincubated for two hours with medium containing kanamycin 50 µg·mL−1 to kill extracellular bacteria. After washed with HBSS three times, the M. smegmatis—infected monolayers were then incubated for 12 h with either 5 µg·mL−1 LysA and 5 µg·mL−1 LysB, and the control experiment was performed with DMEM or 5 µg·mL−1 Rifampicin. After the treatment of the phage-related endolysins, cell monolayers were washed three times and the medium was replaced with 1 mL of sterile distilled water to lyse the macrophages. After vigorous pipetting to ensure complete cell lysis, viable intracellular M. smegmatis were determined by quantitative plating of serial dilutions of the lysates on Middlebrook 7H11 agar. Each test was done three times in independent experiments, and the number of CFU recovered per well (mean number ± S.D.) was determined.
4. Conclusions
Our results indicate that BTCU-1 derived endolysins have antimycobacterial activity. These results also suggest that a variety of the genes encoding mycobacteriophage-related lytic endolysins can be readily isolated from the mycobacteriophage genomes. Use of these lytic endolysins as alternative sanitation or disinfectant agents will be highly feasible for application in controlling mycobacterium infections.
Acknowledgments
This work was supported partly by grants (contract number TCIRP99002-03 and TCRPP101011) from Tzu Chi University and partly by grants NSC101-2320-B-320-003 from National Science Council of Taiwan. We thank the Electron Microscopy Laboratory of the Department of Anatomy of Tzu Chi University for technical assistance.
Supplementary Materials
Supplementary materials can be accessed at: http://www.mdpi.com/1420-3049/20/10/19277/s1.
Author Contributions
Kai-Chih Chang and Meng-Jiun Lai conceived and designed the experiments; Shinn-Jong Jiang, Meng-Hsuan Tu, Jen-Jyh Lee and Ying-Huei Chen performed the experiments; Chih-Chin Liu and Meng-Jiun Lai analyzed the BTCU-1 genome and predicted endolysins; Po-Chi Soo contributed reagents/materials; Kai-Chih Chang and Meng-Jiun Lai wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the title compounds are available from the authors.
References
- 1.Haydel S.E. Extensively drug-resistant tuberculosis: A sign of the times and an impetus for antimicrobial discovery. Pharmaceuticals. 2010;3:2268–2290. doi: 10.3390/ph3072268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diaz E., Lopez R., Garcia J.L. Chimeric pneumococcal cell wall lytic enzymes reveal important physiological and evolutionary traits. J. Biol. Chem. 1991;266:5464–5471. [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention Emergence of Mycobacterium tuberculosis with extensive resistance to second-line drugs—Worldwide, 2000–2004. MMWR Morb. Mortal. Wkly. Rep. 2006;55:301–305. [PubMed] [Google Scholar]
- 4.Raviglione M.C., Smith I.M. XDR tuberculosis—Implications for global public health. N. Engl. J. Med. 2007;356:656–659. doi: 10.1056/NEJMp068273. [DOI] [PubMed] [Google Scholar]
- 5.Kutateladze M., Adamia R. Bacteriophages as potential new therapeutics to replace or supplement antibiotics. Trends Biotechnol. 2010;28:591–595. doi: 10.1016/j.tibtech.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Borysowski J., Weber-Dabrowska B., Gorski A. Bacteriophage endolysins as a novel class of antibacterial agents. Exp. Biol. Med. 2006;231:366–377. doi: 10.1177/153537020623100402. [DOI] [PubMed] [Google Scholar]
- 7.Hermoso J.A., Garcia J.L., Garcia P. Taking aim on bacterial pathogens: From phage therapy to enzybiotics. Curr. Opin. Microbiol. 2007;10:461–472. doi: 10.1016/j.mib.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 8.Daniel A., Euler C., Collin M., Chahales P., Gorelick K.J., Fischetti V.A. Synergism between a novel chimeric lysin and oxacillin protects against infection by methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2010;54:1603–1612. doi: 10.1128/AAC.01625-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trigo G., Martins T.G., Fraga A.G., Longatto-Filho A., Castro A.G., Azeredo J., Pedrosa J. Phage therapy is effective against infection by Mycobacterium ulcerans in a murine footpad model. PLoS Negl. Trop. Dis. 2013;7:e2183. doi: 10.1371/journal.pntd.0002183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pohane A.A., Joshi H., Jain V. Molecular dissection of phage endolysin: An interdomain interaction confers host specificity in lysin a of Mycobacterium phage D29. J. Biol. Chem. 2014;289:12085–12095. doi: 10.1074/jbc.M113.529594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Payne K.M., Hatfull G.F. Mycobacteriophage endolysins: Diverse and modular enzymes with multiple catalytic activities. PLoS ONE. 2012;7:e34052. doi: 10.1371/journal.pone.0034052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pope W.H., Jacobs-Sera D., Russell D.A., Peebles C.L., Al-Atrache Z., Alcoser T.A., Alexander L.M., Alfano M.B., Alford S.T., Amy N.E., et al. Expanding the diversity of mycobacteriophages: Insights into genome architecture and evolution. PLoS ONE. 2011;6:e16329. doi: 10.1371/journal.pone.0016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatfull G.F. The secret lives of mycobacteriophages. Adv. Virus Res. 2012;82:179–288. doi: 10.1016/B978-0-12-394621-8.00015-7. [DOI] [PubMed] [Google Scholar]
- 14.Jacobs-Sera D., Marinelli L.J., Bowman C., Broussard G.W., Guerrero Bustamante C., Boyle M.M., Petrova Z.O., Dedrick R.M., Pope W.H., Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES) program et al. On the nature of mycobacteriophage diversity and host preference. Virology. 2012;434:187–201. doi: 10.1016/j.virol.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Payne K., Sun Q., Sacchettini J., Hatfull G.F. Mycobacteriophage Lysin B is a novel mycolylarabinogalactan esterase. Mol. Microbiol. 2009;73:367–381. doi: 10.1111/j.1365-2958.2009.06775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai M.J., Soo P.C., Lin N.T., Hu A., Chen Y.J., Chen L.K., Chang K.C. Identification and characterisation of the putative phage-related endolysins through full genome sequence analysis in Acinetobacter baumannii ATCC 17978. Int. J. Antimicrob. Agents. 2013;42:141–148. doi: 10.1016/j.ijantimicag.2013.04.022. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann C., Leis A., Niederweis M., Plitzko J.M., Engelhardt H. Disclosure of the mycobacterial outer membrane: Cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc. Natl. Acad. Sci. USA. 2008;105:3963–3967. doi: 10.1073/pnas.0709530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischetti V.A. Bacteriophage lysins as effective antibacterials. Curr. Opin. Microbiol. 2008;11:393–400. doi: 10.1016/j.mib.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahapatra S., Piechota C., Gil F., Ma Y., Huang H., Scherman M.S., Jones V., Pavelka M.S., Jr., Moniz-Pereira J., Pimentel M., et al. Mycobacteriophage ms6 LysA: A peptidoglycan amidase and a useful analytical tool. Appl. Environ. Microbiol. 2013;79:768–773. doi: 10.1128/AEM.02263-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin N.T., Chiou P.Y., Chang K.C., Chen L.K., Lai M.J. Isolation and characterization of ϕAB2: A novel bacteriophage of Acinetobacter baumannii. Res. Microbiol. 2010;161:308–314. doi: 10.1016/j.resmic.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 21.Lai M.J., Lin N.T., Hu A., Soo P.C., Chen L.K., Chen L.H., Chang K.C. Antibacterial activity of Acinetobacter baumannii phage ϕAB2 endolysin (LysAB2) against both gram-positive and gram-negative bacteria. Appl. Microbiol. Biotechnol. 2011;90:529–539. doi: 10.1007/s00253-011-3104-y. [DOI] [PubMed] [Google Scholar]
- 22.Lukashin A.V., Borodovsky M. Genemark.Hmm: New solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delcher A.L., Bratke K.A., Powers E.C., Salzberg S.L. Identifying bacterial genes and endosymbiont DNA with glimmer. Bioinformatics. 2007;23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lowe T.M., Eddy S.R. Trnascan-se: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rybniker J., Kramme S., Small P.L. Host range of 14 mycobacteriophages in Mycobacterium ulcerans and seven other mycobacteria including Mycobacterium tuberculosis—Application for identification and susceptibility testing. J. Med. Microbiol. 2006;55:37–42. doi: 10.1099/jmm.0.46238-0. [DOI] [PubMed] [Google Scholar]
- 26.Chen H.L., Su P.Y., Chang Y.S., Wu S.Y., Liao Y.D., Yu H.M., Lauderdale T.L., Chang K., Shih C. Identification of a novel antimicrobial peptide from human hepatitis B virus core protein arginine-rich domain (ARD) PLoS Pathog. 2013;9:e1003425. doi: 10.1371/journal.ppat.1003425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Broxmeyer L., Sosnowska D., Miltner E., Chacon O., Wagner D., McGarvey J., Barletta R.G., Bermudez L.E. Killing of Mycobacterium avium and Mycobacterium tuberculosis by a mycobacteriophage delivered by a nonvirulent mycobacterium: A model for phage therapy of intracellular bacterial pathogens. J. Infect. Dis. 2002;186:1155–1160. doi: 10.1086/343812. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.