Abstract
The clinical effectiveness of carbapenem antibiotics such as meropenem is becoming increasingly compromised by the spread of both metallo-β-lactamase (MBL) and serine-β-lactamase (SBL) enzymes on mobile genetic elements, stimulating research to find new β-lactamase inhibitors to be used in conjunction with carbapenems and other β-lactam antibiotics. Herein, we describe our initial exploration of a novel chemical series of metallo-β-lactamase inhibitors, from concept to efficacy, in a survival model using an advanced tool compound (ANT431) in conjunction with meropenem.
Keywords: antibiotic resistance, metallo-β-lactamase inhibitor, NDM-1, VIM-2, meropenem, efficacy
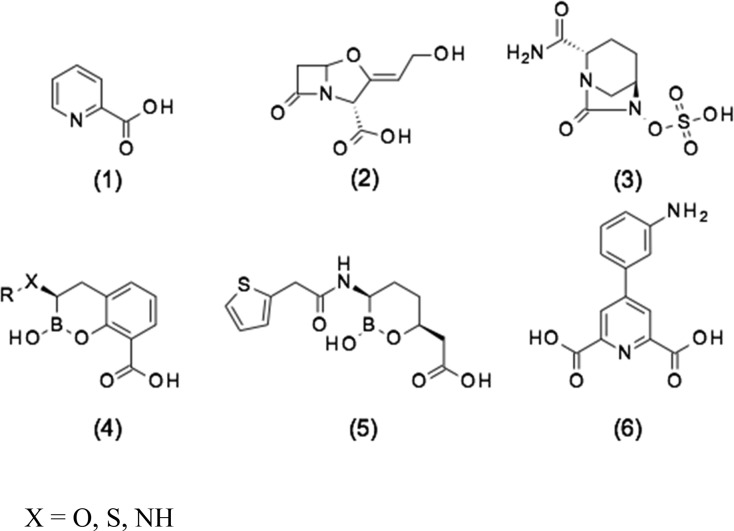
A major public health concern for our age is the increasing resistance to antibiotics. The U.K. government recently commissioned a review under Jim O’Neill1 that examined this issue in depth and proposed structural changes to industry practice, drug pricing, etc. in an attempt to redirect resources to the problem. Similarly, in the United States, the CARB-X2 initiative, a jointly funded public-private partnership between the Wellcome Trust and BARDA,3 is revitalizing research in this area through various funding initiatives. In particular, Gram-negative bacteria are becoming a major concern because not only are they inherently more difficult to treat with antibiotics due to entry and efflux issues but also the crucial carbapenem class of β-lactam antibiotics is becoming less clinically effective. As a consequence, major Gram-negative pathogens are currently evolving toward pan-drug-resistant phenotypes, whose potential global spread may lead to a dramatic return to the pre-antibiotic era. A major resistance mechanism to β-lactams antibiotics is represented by the production of one or more β-lactamase enzymes, which efficiently hydrolyze the amide bond of the β-lactam ring. β-Lactamases belong to two structurally and mechanistically unrelated families of enzymes,4 the serine-β-lactamases (SBLs; classes A, C, and D), which use an active serine to cleave the β-lactam in a covalent mechanism, and the metallo-β-lactamases (MBLs; class B), which use metal ion catalysis to directly hydrolyze the β-lactam without the formation of a covalent intermediate. To counter the threat of emerging resistance, the Streptomyces clavuligerus natural product clavulanic acid (2), an SBL inhibitor (Scheme 1), was introduced in 1981 as part of a combination together with the β-lactam antibiotic amoxicillin (as Augmentin),5 followed by tazobactam in 1992, combined with piperacillin.6 More recently, there has been renewed interest in the field of β-lactamase inhibitor discovery to counter the threat from newer β-lactamases, which are not inhibited by clavulanic acid or tazobactam, such as the extended spectrum β-lactamases (ESBLs) and carbapenemases. This has led to the development of two new synthetic classes of inhibitors, namely the diazabicyclooctane (DBO) series, as exemplified by avibactam7 (3), which is used in combination with ceftazidime, and the boronates of generalized bicyclic structure (4) plus monocyclic boronate vaborbactam8 (5), which is used in combination with meropenem. However, both 3 and 5 are exclusively SBL inhibitors, whereas examples of generalized structure (4) are known to have dual SBL and MBL inhibitory activity,9 a novel finding that can be rationalized at the molecular level.10 However, as of yet, there are no MBL inhibitors in clinical use despite there being a clear unmet medical need.11
Scheme 1. Representative Examples of β-Lactamase Inhibitors: (1) Pyridine-2-carboxylic Acid (Picolinic Acid), (2) Clavulanic Acid, (3) Avibactam, (4) Generalized Cyclic Boronate, (5) Vaborbactam, and (6) 2,6-Pyridine Dicarboxylic Acid Derivative.
The MBL family includes the clinically relevant B1 subfamily containing the acquired IMP, VIM, and NDM subgroups of allelic variants. Many classes of MBL inhibitors have been reported in the research stage including bisthiazolidines, thiols, tetrazoles, succinic acids, and hydroxamates.12
Recently Chen et al.13 reported novel MBL inhibitors (as exemplified by 6) based on a pyridine-2-carboxylic acid (1) scaffold, which was previously reported to be a modest inhibitor of the Aeromonas hydrophila CphA MBL (Ki [inhibition constant] = 6 μM).14 The CphA enzyme belongs to the divergent B2 subclass of MBLs, which have a single Zn ion in their active site and do not have the clinical significance of subclass B1 MBLs (which instead coordinate two Zn ions in the di-nuclear active site). These compounds have promising activity in terms of both NDM-1 inhibition and potentiation against clinical strains expressing NDM-1; however, the most-active compound from this work (compound 6 in Scheme 1) contains a lipophilic aniline, a moiety well-known to be associated with clastogenic and carcinogenic effects.15
In this study, we also report on the design and optimization of new MBL inhibitors starting from pyridine-2-carboxylic acid; however, we have taken a very different medicinal chemistry approach, leading to the identification of a chemically distinct series with promising activity against MBL-producing clinical isolates and drug-like properties, which makes it suitable for further pharmaceutical development.
Results and Discussion
Derivatives of Pyridine-2-carboxylic Acid, Pan-MBL Inhibitors
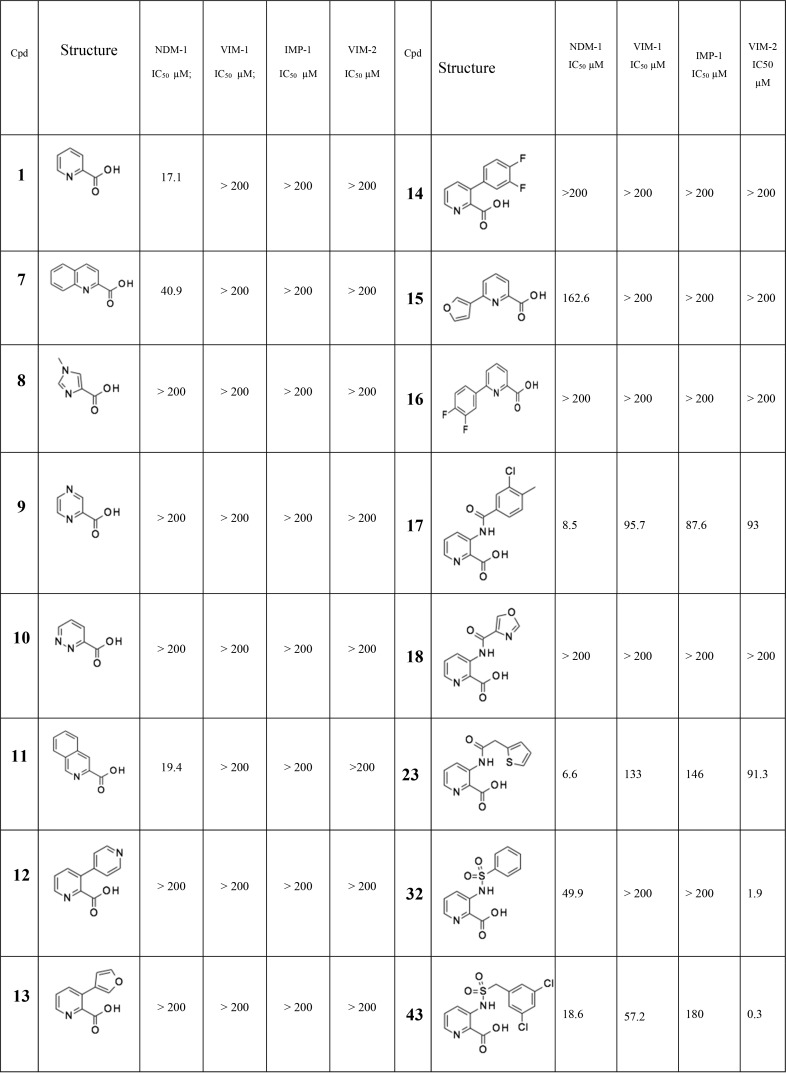
Although the biological activity of pyridine-2-carboxylic acid (1) is relatively modest, its low molecular weight and simple structure makes it an attractive starting point for medicinal chemistry. From the work of Horsfall et al.,14 biochemical and structural analyses clearly demonstrated that pyridine-2-carboxylic acid (1) was a competitive inhibitor of CphA. This mechanism of inhibition, relying on the formation of a stable enzyme–inhibitor complex, was different from that exhibited by its close bidentate analogue 2,6-dipicolinic acid, which instead acts as a strong zinc chelator and non-specifically inactivates metallo-enzymes. The latter mechanism of action, also shown by EDTA16 and some natural products,17 may produce significant off-target implications in clinical use18 and be less effective against newer NDM variants, which have evolved high-affinity Zn-binding and are therefore less prone to inhibition by metal-chelating agents.19 Using enzyme assays, we evaluated the potential of 1 to inhibit MBLs other than CphA and were encouraged to find that it exhibited an IC50 value of 17 μM against NDM-1, although there was no significant activity against VIM-1 or IMP-1 enzymes. However, 1 showed no reduction of imipenem MIC when tested at concentrations up to 1 mM on a laboratory strain of NDM-1-producing Escherichia coli (DH5α/pLBII-NDM-1), presumably due to lack of penetration into, or efflux from, the periplasm (data not shown).
Because the X-ray structure of 2,4-pyridine dicarboxylic acid in the MBL CphA clearly shows coordination of both the carboxylate group at position 2 and the pyridine nitrogen to the single Zn ion, coordinated by conserved residues Asp120, Cys221, and His263, which form the so-called “Zn2” site (Figure 1), we screened a number of related heterocyclic acids with this structural feature. The modest inhibition of NDM-1 was seen with quinoline acid (7) and isoquinoline acid (11), but no inhibition of the other two MBLs was detected at concentration as high as 200 μM. Surprisingly, other heterocyclic acids such as imidazole (8), pyrazine (9), and pyridazine (10) showed no detectable inhibition. Given the simplicity of the chemistry, compared with both the quinoline and isoquinoline series, we considered pyridine-2-carboxylic acid (1) to be a suitable starting point for a medicinal chemistry program. Initially, this involved some array chemistry, adding substituents to ethylpyridine-2-carboxylate followed ester hydrolysis to give the acid (Scheme 2). The short-term objective was to improve the inhibition spectrum of the molecule and obtain whole cell synergistic activity, with the longer-term objective being the discovery of a drug molecule with good PK and efficacy.
Figure 1.
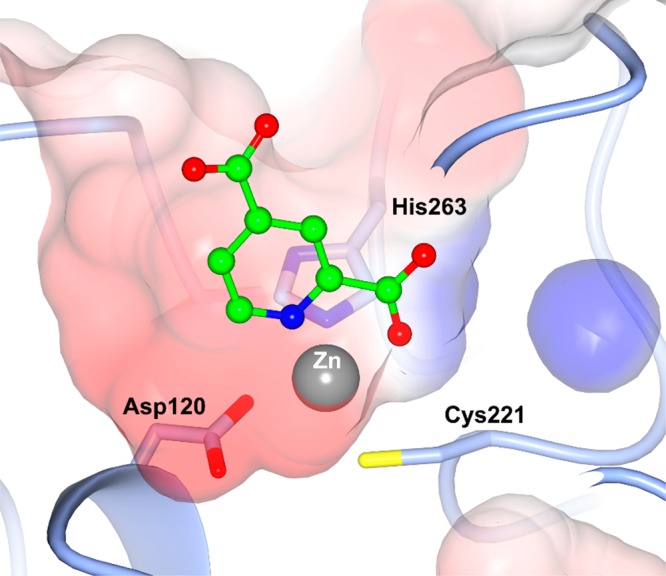
Mode of binding of 2,4-pyridine dicarboxylic acid (green) in the active site of CphA (PDB code 2GKL) showing the coordination to the single Zn ion.
Scheme 2. General Synthetic Routes Used to Prepare Compounds Based on the Pyridine-2-carboxylic Acid Scaffold.
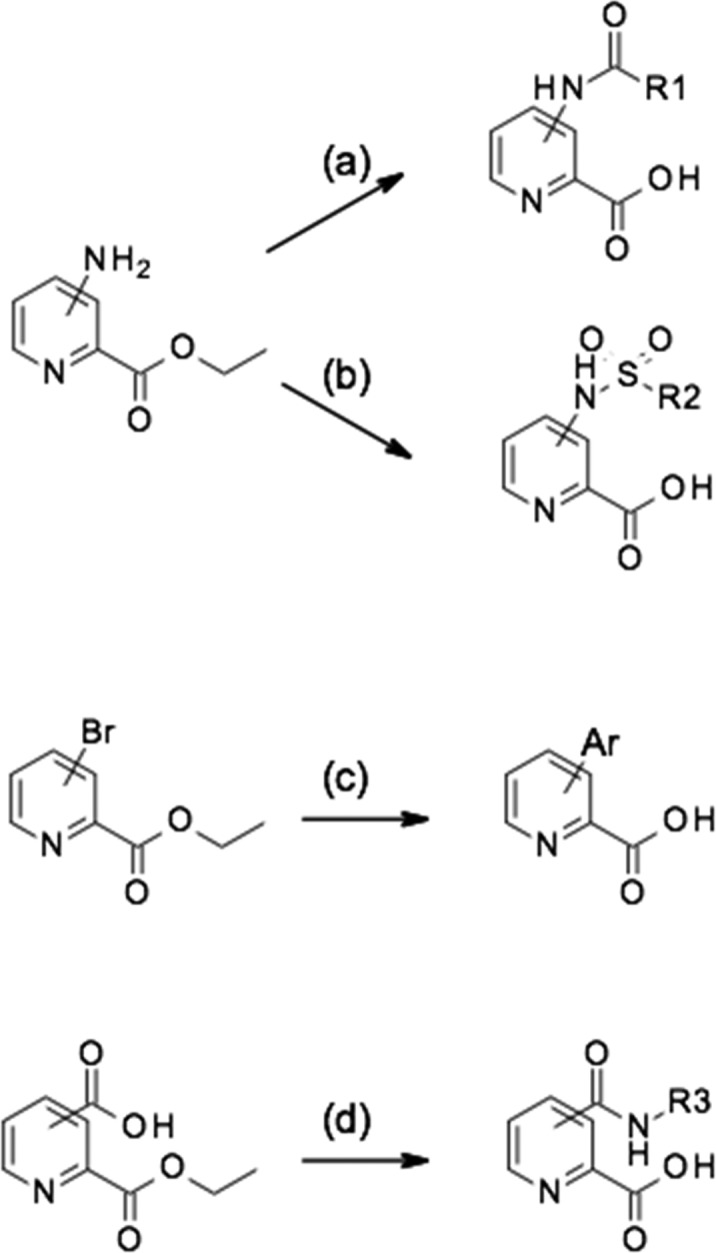
(a) R1CO2H, HATU, Et3N, then LiOH; (b) R2SO2Cl, Et3N, then LiOH; (c) ArB(OH)2 and (Ph3P)4P, then LiOH; (d) R3NH2, HATU, Et3N, then LiOH.
We used standard synthetic routes to explore the activity of biaryl analogues, amides, and sulfonamides at all available positions of the parent structure 1 (Scheme 2 and Supplementary Data). These array compounds were screened against a panel of NDM-1, VIM-1, VIM-2, and IMP-1 enzymes. In terms of relative importance, we were most interested in NDM-1 and VIM-1 because of their widespread prevalence in Enterobacteriaceae (IMP-1 is not so widespread geographically, and VIM-2 is almost exclusively found in Pseudomonas).20 The results of this initial array work are reported in Table 1. Compounds of interest were progressed to a whole-cell meropenem potentiation assay to investigate if the inhibitor could restore the antibacterial activity of meropenem against resistant strains, where the resistance was attributable to the production of an MBL enzyme. Initially, we used laboratory strains transformed with plasmids expressing only the MBL resistance genes; however, early on, we switched to using clinical carbapenem-resistant Enterobacteriaceae (CRE) isolates to evaluate the synergistic activity of the compounds because these presented a greater challenge that was more representative of the clinical situation.
Table 1. Selected Pyridine-2-carboxylate Analogues (Full Table Can Be Found in the Supplementary Information)a.
All IC50 values reported herein are the average of at least two independent assays. All reported IC50 values have R2 values of >0.95 of the fitting curve.
Biaryl analogues such as compounds 12 to 16 were essentially inactive and were not pursued further. Encouragingly, amide groups were tolerated with respect to NDM-1 inhibition at all four positions on the pyridine ring. One of the earliest compounds prepared, amide 17, showed the promising low micromolar inhibition of NDM-1 as well as detectable inhibition of all three enzymes at higher concentrations, representing an interesting, although modest, broad-spectrum inhibitor. Related amides such as 18 proved, by contrast, inactive at a concentration of up to 200 μM on all tested enzymes, whereas 17 and 23 represented a significant advance from 1, which only had modest activity against the NDM-1 enzyme.
We also explored the sulfonamide substituent because, due to the different hybridization state of the sp3 sulfur compared to sp2 carbon, sulfonamides access alternative regions of space and H-bonding opportunities as compared with the amides. Surprisingly strong inhibition of VIM-2 was observed by certain sulfonamides such as 32 and 43. The modest but genuine MBL inhibition achieved against both NDM-1 and VIM-1 represented a significant advance over the starting position of 1, stimulating further efforts to improve the potency and spectrum of the series.
X-ray Crystallography Revealing of a Novel Binding Mode to the VIM-2 MBL
The mode of binding of certain inhibitors was ascertained by X-ray crystallography experiments. A crystal structure of VIM-2 in complex with the simple phenyl sulfonamide 32 revealed interesting features of its binding modality (Figure 2). Notably the pyridine nitrogen and the carboxylate group of the inhibitor were bound to a single Zn atom (Zn2, coordinated by conserved residues Asp120, Cys221, and His263). Inhibitor binding also resulted in the displacement of two crystallographic water molecules found in the native structure but, interestingly, not the so-called “bridging” water molecule coordinated by the two active site Zn ions, which provides the nucleophile attacking the carbonyl carbon of the β-lactam substrate. Taken with the enzyme inhibition data, this structural information is fully in accord with a reversible competitive mode of action and not a general chelation effect (whereby the ligand removes metal ions from solution eventually stripping metal ions from the enzyme through equilibrium processes). Although inhibitor binding did not alter the tertiary structure of the enzyme, several side chains movements were observed, such as the Arg228 side chain being displaced by 2.7 Å to create a strong salt-bridge interaction with inhibitor carboxylate, while the side chain of Tyr67 was folded over deeper into the active site cavity (∼60°) and rotated (∼75°) to align with the inhibitor’s phenyl moiety and create a strong aromatic interaction. Additional interactions were found to stabilize inhibitor binding, including a H-bond involving one sulfonamide oxygen with the backbone nitrogen of Asn233, a residue conserved in all subclass B1 MBLs and whose role is critical to properly orient the β-lactam substrate.
Figure 2.
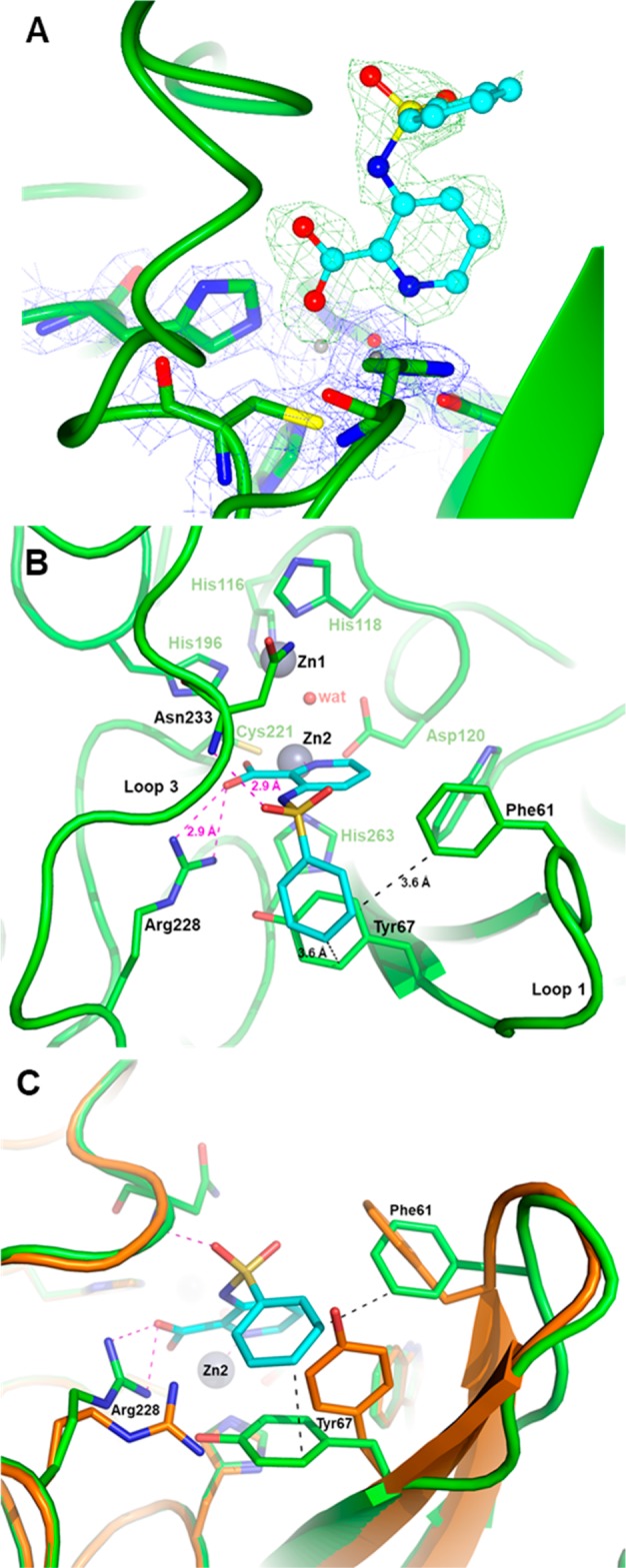
X-ray structure of VIM-2 inhibited by 32. (A) Close view of the VIM-2 active site (protein secondary structure elements and the active-site residues are shown in green, surrounded by the 2Fo–Fc Fourier map, blue meshes, contoured at 2 σ) in complex with compound 32 (shown in cyan and surrounded by the omit Fo–Fc map, green meshes, contoured at 3 σ). (B) Close-up view of the VIM-2 active site showing the network of interactions between the inhibitor (cyan), the Zn2 ion and residues Arg228 and Asn233. (C) Orthogonal view of panel B and superimposition with the VIM-2 native structure (PDB code 1KO3, orange) showing the significant movements of the side chains of residues Phe61, Tyr67, and Arg228 (green sticks) upon the binding of 32 (cyan).
Scaffold-Hopping from Pyridine to Thiazole and Considerable Activity Improvement
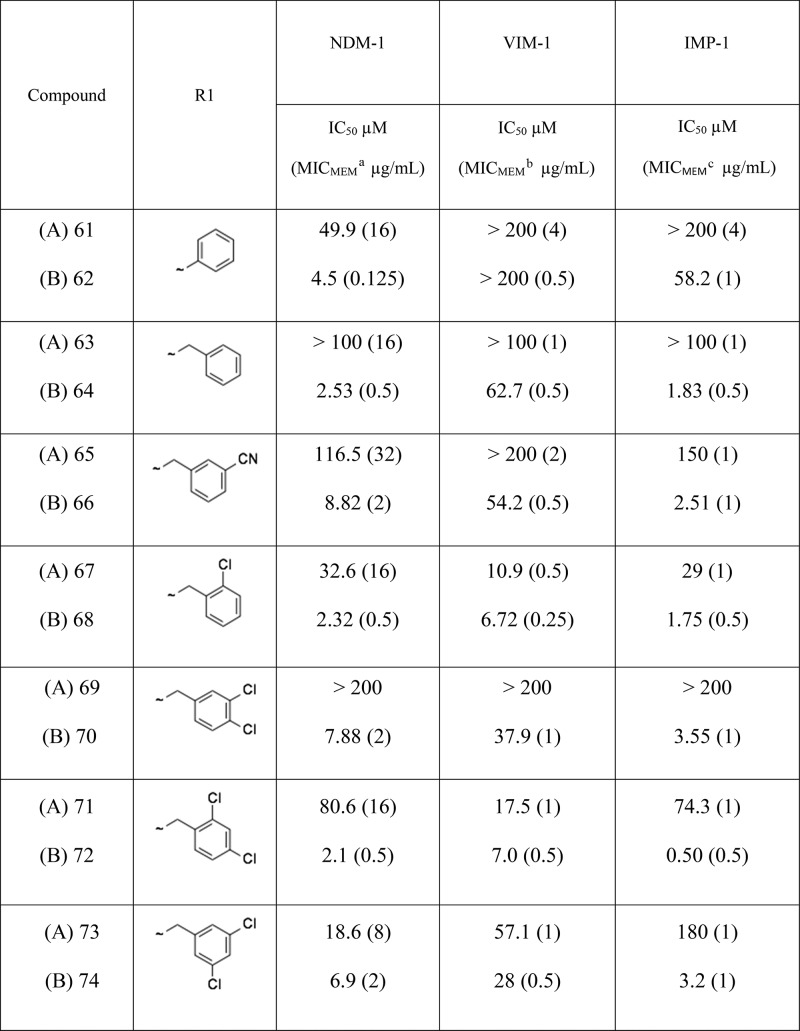
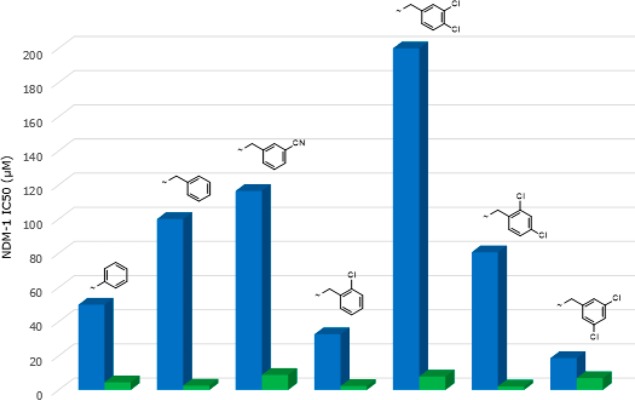
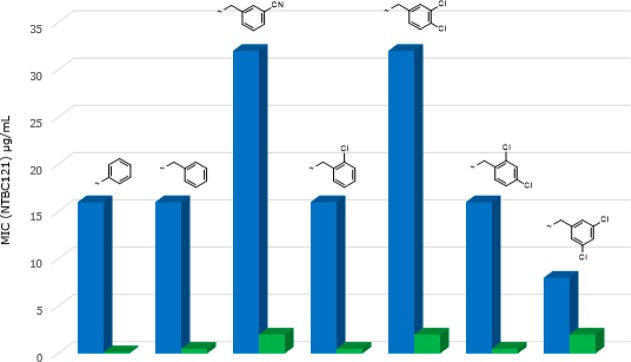
As mentioned above, as part of the initial chemistry exploration, we investigated alternatives to the pyridine ring while keeping a “pyridine-like” nitrogen adjacent to the carbon bearing the carboxy group. Several different heterocyclic replacements in combination with the best side chains were explored. All but one of the potential heterocyclic replacements showed worse activity than pyridine (data not shown); however, encouragingly, the thiazole analogues showed a significant and consistent improvement over the corresponding pyridine analogues, in terms of both in vitro enzyme inhibition and whole-cell meropenem potentiation (Table 2 and Figures 3 and 4)
Table 2. Comparison of IC50 Values and Meropenem Potentiation Data for Matched Pairs of Pyridyl (A) and Thiazolyl (B) Analogues.

NDM-1-producing strain Escherichia coli NTBC121 (MICMEM, 32 μg/mL).
VIM-1-producing strain Klebsiella pneumoniae NTBC055 (MICMEM, 16 μg/mL).
IMP-1-producing strain K. pneumoniae NTBC062 (MICMEM, 4 μg/mL).
The inhibitor was present at 100 μM.
Figure 3.
Comparison of NDM-1 IC50 values for matched pairs of pyridyl (blue) and thiazolyl (green) analogues.
Figure 4.
Comparison of meropenem potentiation data for matched pairs of pyridine (blue) and thiazole (green) analogues.
Several points emerge from this SAR analysis. First, trends are easier to identify in the activity observed against NDM-1 clinical strains because these are generally much more resistant than VIM-1- or IMP-1-producing clinical strains,19 and hence, there is a bigger window in which to observe changes in activity. Second, it is difficult to determine whether improvements are due solely to changes in enzyme inhibition or also due to better penetration into (or lack of efflux from) the periplasm. To help identify compounds that have an improved ability to penetrate into bacterial cells we have defined a new term we call the “potentiation efficiency” (PE) for each compound. This empirical term is calculated by dividing the potentiated MIC for an MBL-producing strain by the IC50 for the respective purified enzyme (PE = MIC/IC50), thus “normalizing” the whole cell potentiation with respect to enzyme activity such that the smaller the PE the better a compound is accessing the periplasm, independent of the actual enzyme inhibition. For example, if we consider the matched pair of 61 (pyridyl) and 62 (thiazolyl) analogues, then it is clear that the thiazolyl derivative has improved NDM-1 enzyme inhibitory activity (IC50 4.5 μM versus 49.9 μM) and also improved meropenem potentiation against a corresponding NDM-1 strain (MIC 0.125 μg/mL versus 16 μg/mL). The corresponding PE values for 61 and 62 are 0.32 and 0.027, respectively, showing that in this instance there is a 10-fold improvement in potentiation over and above what might be expected from the increase in enzyme inhibition alone, suggesting that introduction of the thiazole has a profound effect on both enzyme inhibition and bacterial penetration. Although we are using this metric in the situation in which the inhibitors have no intrinsic antibacterial activity, in principle, the PE function should have wide applicability in antibacterial research in which penetration into the periplasm or the cytoplasm is a prerequisite for antibacterial action. The limitation, of course, is that it does not give information as to whether penetration or efflux is an issue but it can be a useful empirical tool to facilitate SAR analysis and the direction of the medicinal chemistry. The reason for the differences between pyridyl and thiazolyl analogues is unclear at this juncture.
A crystal structure of VIM-2 in complex with thiazole (62) was also obtained, which confirmed a similar mode of inhibitor binding as 32, with the thiazole nitrogen atom interacting exclusively with Zn2 (Figure 5) and the carboxylate interacting with both Arg228 and Zn2. However, the substituted sulfonamide has adopted a slightly different conformation compared to that observed in the complex with (32) (Figure 5). In this alternative binding mode, one oxygen atom of the sulfonamide linker still interacts with the backbone nitrogen of Asn233 but the phenyl group is interacting with Phe61 instead of Tyr67. The side chain of the latter occupies a position similar to that found in the native structure, while Phe61 is slightly rotated and displaced upon binding of 62.
Figure 5.
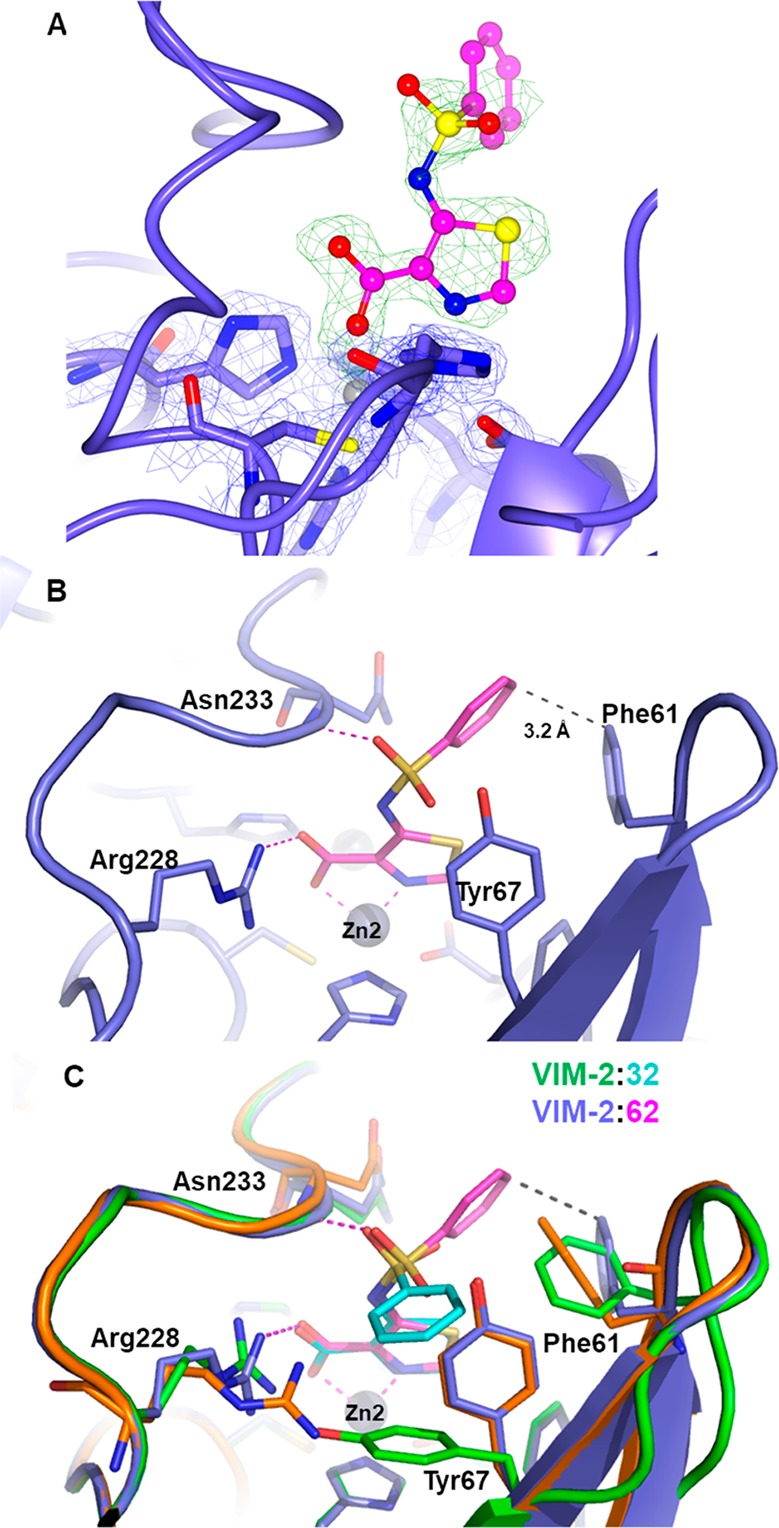
X-ray structure of VIM-2 inhibited by 62. (A) Close view of the VIM-2 active site (protein secondary structure elements and the active site residues are shown in blue, surrounded by the 2Fo–Fc Fourier map, blue meshes, contoured at 2 σ) in complex with 62 (shown in magenta and surrounded by the omit Fo–Fc map, green meshes, contoured at 3 σ). (B) Binding of 62 in the enzyme active site relies on a similar set of interactions between the thiazole moiety, Zn2, and the Arg228 side chain. (C) Comparison of the substituted sulfonamide conformation of 62, thiazole (magenta) and 32, pyridine (cyan); the position of Phe61 and Tyr67 in the native VIM-2 is shown in orange.
Moving from Phenyl Sulfonamide to Pyridyl Sulfonamide and Improvement of Solubility of the Acid Form
Alongside improving compound activity, we also focused on improving the physicochemical profile and in particular aqueous solubility. The aqueous solubility of the key compound 62 was determined as 1.1 mg/mL, whereas a solubility of at least 10 mg/mL would be required to deliver a therapeutically effective intravenous dose in an acceptable volume of the aqueous vehicle. A standard approach to increasing polarity and aqueous solubility is to add nitrogen atoms into a phenyl ring. Consequently, we prepared a small set of analogues set of side-chain pyridyl analogues of 62. A total of two of the three aza analogues had improved enzyme inhibition against NDM-1 (Table 3), and furthermore, the most active compound (76) facilitated a meropenem MIC of 0.06 μg/mL against one of our NDM-1 producing clinical strains (E. coli NTBC121) when tested in the presence of 100 μM inhibitor compared to an MIC of 0.125 μg/mL for 62. The solubility of the side-chain pyridyl analogue (2.4 mg/mL) was double that of the phenyl analogue 62, and conversion to the sodium salt improved the solubility to almost 20 mg/mL (Table 3), achieving the target solubility for testing in animal efficacy studies.
Table 3. Enzyme Inhibition, Potentiation, and Solubility of Side-Chain Phenyl and Pyridyl Analogues.
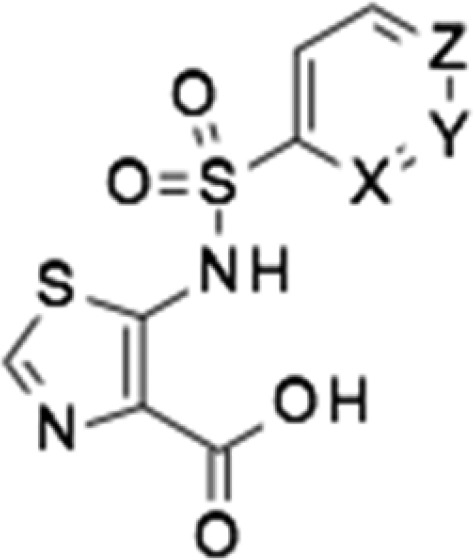
| compound | X | Y | Z | NDM-1 IC50 μM (MEM MIC, μg/mL)a | VIM-1 IC50 μM | IMP-1 IC50 μM | VIM-2 IC50 μM | solubility, mg/mLb (Na salt) |
|---|---|---|---|---|---|---|---|---|
| 75 | N | CH | CH | 12.9 | >200 | 113.7 | 16.1 | N/D |
| 76 | CH | N | CH | 2.67 (0.06) | >200 | 54.0 | 6.7 | 2.4 (16.9) |
| 77 | CH | CH | N | 3.4 | 180 | 23.4 | 1.8 | ND |
| 62 | CH | CH | CH | 4.5 (0.125) | >200 | 58.2 | 6.2 | 1.1 (18.9) |
NDM-1-producing strain E. coli NTBC121 (MEM MIC, 32 μg/mL). The inhibitor was present at 100 μM.
Solubility was determined in phosphate-buffered saline (PBS) at pH 7.4.
The X-ray structure of VIM-2 bound to (76) showed a broadly similar set of interactions to that of compounds 32 and 62 (Figure 6), with the sulfonamide substituent located between residues Phe61 and Tyr67, inducing a significant rotation of the side chain of the latter. Overall, these structural data, which highlight a salt bridge interaction between the negatively charged carboxylate group of the inhibitor and the positively charged side chain of Arg228 could provide, to some extent, a rationale for the overall lower activity of these compounds (Table 3) on VIM-1. Indeed, Arg228 (whose role can be assumed by Lys224 present in NDM-1 and IMP-1) is replaced by a serine residue in VIM-1, in which this interaction will be very likely abolished.
Figure 6.
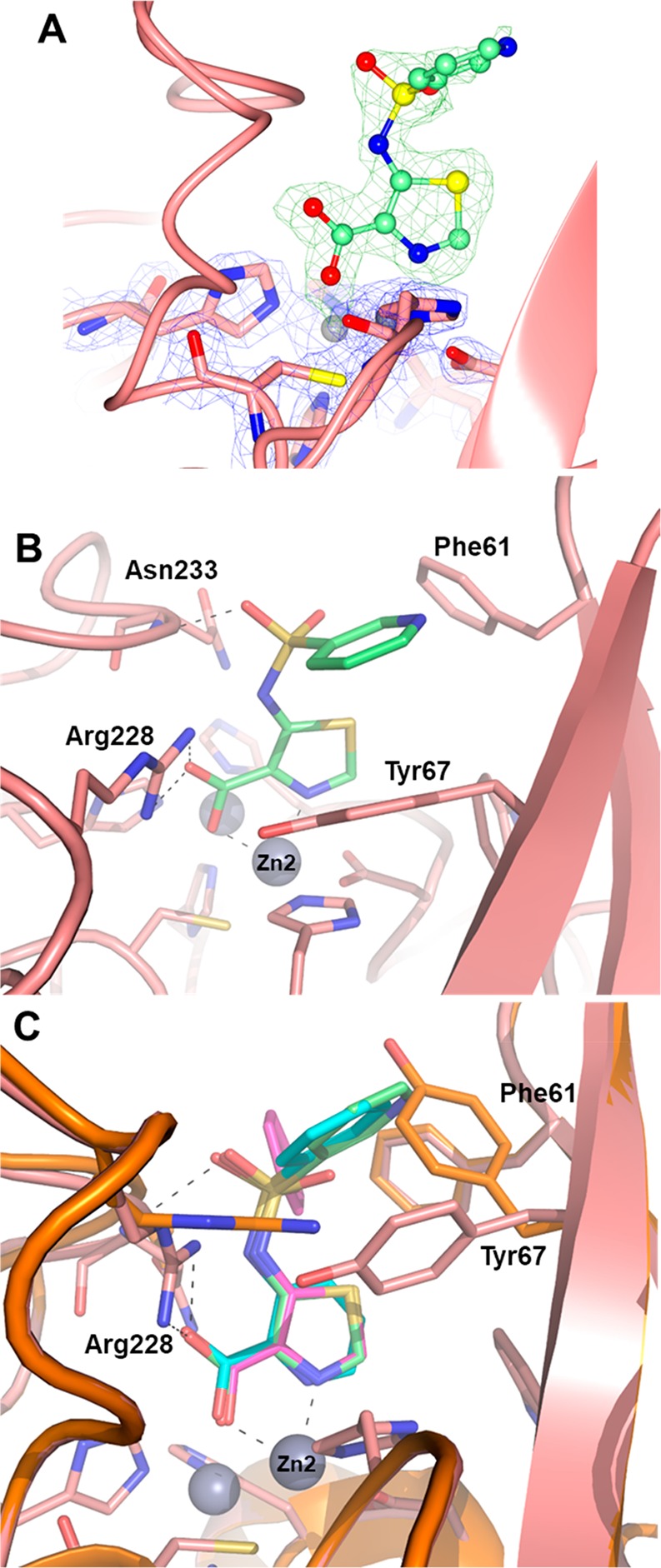
X-ray structure of VIM-2 inhibited by 76. (A) Close view of the VIM-2 active site (protein secondary structure elements and the active site residues are shown in pink, surrounded by the 2Fo–Fc Fourier map, blue meshes, contoured at 2 σ) in complex with 62 (shown in green and surrounded by the omit Fo–Fc map, green meshes, contoured at 3 σ). (B) Close view of the VIM-2 active site (pink) inhibited by 76 (light green). (C) Overlay of inhibitors 32 (cyan), 62 (magenta), and 76 (light green) in the VIM-2 active site, showing the conserved interactions between the inhibitor and Zn2, Arg228, and Asn233; the binding of 76 also induces the rotation of the Tyr67 side chain to accommodate the pyridyl sulfonamide substituent (native VIM-2 shown in orange).
Conclusions
Our approach to the problem of antibiotic resistance due to the emergence of MBL-producing Gram-negative pathogens has been to undertake a medicinal chemistry campaign to develop MBL inhibitors starting from pyridine-2-carboxylic acid (1). Early array chemistry expanded the MBL coverage from NDM-1 to encompass VIM-1, VIM-2, and IMP-1 while also introducing whole cell activity such as to restore the antibiotic activity of meropenem against clinical strains of MBL-producing Enterobacteriaceae. Key benefits of this approach over a nonspecific metal chelation approach (such as using EDTA) are that off-target human toxicities are minimized and the fact that newer NDM variants, due to increased Zn affinity, are less susceptible to compounds that depend on stripping Zn from the enzyme active site.20−22
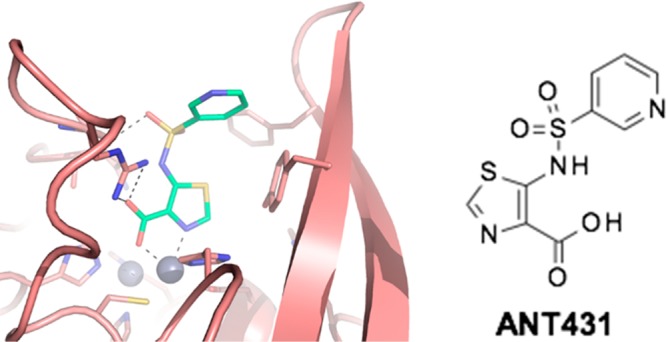
A leap in both enzyme inhibition and potentiation was achieved by switching from pyridine to the thiazole scaffold. As shown from its increased PE value, the thiazolyl series exhibits improved penetration into the bacterial periplasm as well as improved enzyme inhibition. Additionally, although a modest improvement to kinetic solubility was obtained by moving from a side-chain phenyl sulfonamide to a side-chain pyridyl sulfonamide, greater improvements were achieved on making the sodium salt, enabling formulation at sufficiently high concentrations in aqueous buffers to perform efficacy testing in animal models. Full biological characterization of compound (76) (designated as ANT431) has recently been reported elsewhere, demonstrating it to potentiate meropenem activity against a panel of 94 randomly selected NDM- and VIM-producing clinical strains and restore the efficacy of meropenem in an E. coli (IR3) murine thigh infection model.23 It should be pointed out that the potentiation of meropenem by ANT431 is reported here at 100 μM inhibitor concentration because that was the concentration we routinely operated at during the early stages of this project. In fact, ANT431 is effective at 28 μM (8 μg/mL).23 Further research is underway to optimize this promising chemical series and deliver a preclinical development candidate.
Methods and Materials
Antimicrobial Susceptibility Testing
Minimal inhibitory concentrations (MIC) were determined using the Clinical Laboratory Standards Institute (CLSI) broth microdilution procedure. MBL-positive Enterobacteriaceae clinical isolates tested in the susceptibility study included NDM-1-producing strain E. coli NTBC121, VIM-1-producing strain K. pneumoniae NTBC055, and IMP-1-producing strain K. pneumoniae NTBC062. (Individual MICs for all strains tested are presented in Table 2 and Table 1 in the Supporting Information).
In Vitro Enzyme Inhibition Assays
Inhibition against purified MBLs24 were determined by following hydrolysis of 300 μM imipenem in 10 mM HEPES (pH 7.5) buffer (25 °C) in the presence of 0.025 to 500 μM inhibitor using a PerkinElmer Envision (UV absorbance: 290 nm). Percentage of enzymatic inhibition is calculated for each concentration tested and reported on a nonlinear regression versus the logarithmic concentration of inhibitors. The IC50 value is determined from the regression curve only when the R2 of the fitting curve is >0.95. The average of a minimum of at least 2 replicates is therefore reported.
Kinetic Solubility
This was determined by vigorous stirring (1200 rpm) of a 20 mg/mL mixture of the test compound in phosphate-buffered saline (PBS) at pH 7.4 for 4 h. The mixture was then centrifuged for 20 min at 10 000 rpm and the supernatant further processed by filtration. The concentration of the test compound in the filtrate was then determined by high-performance liquid chromatography (UV analysis) by comparison to a standard calibration curve of the test compound.
X-ray Crystallography
The VIM-2 MBL was produced, purified, and crystallized essentially as previously described.25,26 The experimental details of protein crystallization, crystal soaking, data collection, reduction, and refinement (performed using the procedures described elsewhere)26 are reported in the Supporting Information. Omit maps of the three inhibitors bound to VIM-2 are shown in Figures 2, 5 and 6. Coordinates and structure factors of VIM-2 in complex with inhibitors ANT-090 (32), ANT-330 (62), and ANT-431 (76) were deposited in the RCSB Protein Data Bank under the accession codes 5MXQ, 5MXR, and 6HF5, respectively.
Synthesis of Sodium 5-(Pyridine-3-sulfonamido)-1,3-thiazole-4-carboxylate (ANT431, Na Salt)
Ethyl 5-amino-1,3-thiazole-4-carboxylate,
pyridine-3-sulfonyl chloride and all other reagents and solvents were
purchased and used as such. 1H nuclear magnetic resonance
(NMR) spectra were obtained at 400 MHz in deuterated CDCl3 or DMSO-d6 solutions
(reported in ppm). 13C NMR spectra were obtained at 100
MHz in DMSO-d6, When
peak multiplicities are reported, the following abbreviations are
used: s (singlet), d (doublet), t (triplet), m (multiplet), dd (doublet
of doublets), dt (doublet of triplets), and q (quartet). Coupling
constants are reported in hertz (Hz).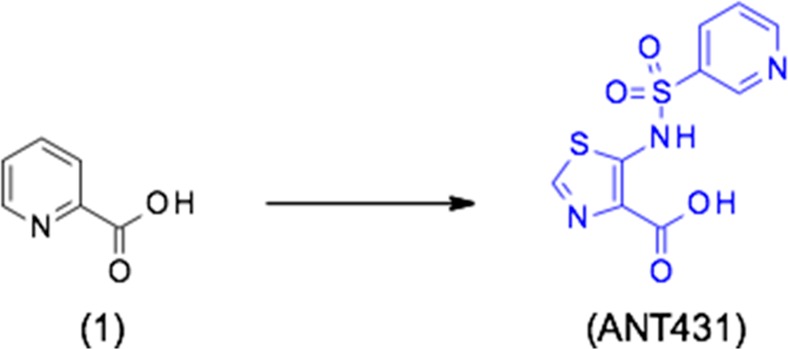
Ethyl 5-(Pyridine-3-sulfonamido)-1,3-thiazole-4-carboxylate
To a stirred suspension of sodium hydride (60% dispersion in oil) (929 mg, 23.22 mmol) in tetrahydrofuran (50 mL) was added a solution of ethyl 5-amino-1,3-thiazole-4-carboxylate (2 g, 11.61 mmol) in tetrahydrofuran (50 mL) at 0 °C under N2 atmosphere. The resulting reaction mixture was stirred for 30 min at 0 °C and then treated with pyridine-3-sulfonyl chloride (2.47 g, 13.93 mmol) in tetrahydrofuran (3 mL) in a drop-wise manner at 0 °C. The resulting reaction mixture was stirred at room temperature for 3 h, then quenched with ice, diluted with water, and washed with 50% ethyl acetate in hexane (100 mL). The aqueous layer was separated, neutralized with 1 N HCl solution, and extracted with ethyl acetate (2 × 100 mL). The combined organic layers were dried over anhydrous Na2SO4, filtered, and concentrated to obtain the crude product. This was purified by column chromatography on silica gel using 1–10% methanol in dichloromethane (Rf 0.1 with 10% methanol in dichloromethane) as eluent to give the title compound as a yellow solid (1.0 g, 27%). 1H NMR (CDCl3, 400 MHz, δ): 10.26 (s, 1H), 9.12 (d, J = 2.0 Hz, 1H), 8.83 (dd, J = 4.8, 1.2 Hz, 1H), 8.31 (s, 1H), 8.19–8.16 (m, 1H), 7.46 (dd, J = 8.0, 4.8 Hz, 1H), 4.39 (q, J = 7.2 Hz, 2H), 1.39 (t, J = 7.2 Hz, 3H). m/z: 314.0 [M + H]+.
5-(Pyridine-3-sulfonamido)-1,3-thiazole-4-carboxylic acid (ANT431)
To a stirred solution of ethyl 5-(pyridine-3-sulfonamido)-1,3-thiazole-4-carboxylate (1.0 g, 3.19 mmol) in a mixture of tetrahydrofuran and water (10:5 in milliliters) was added LiOH.H2O (1.33 g, 31.9 mmol) at room temperature. The resulting reaction mixture was stirred at room temperature for 48 h and then concentrated under vacuum. The crude residue was diluted with water (30 mL) and washed with ethyl acetate (30 mL). The aqueous layer was acidified with 1 N HCl solution to pH4. The resulting precipitation was filtered and dried under high vacuum to obtain the title compound as a yellow solid (0.85g, 93%). 1H NMR (DMSO-d6, 400 MHz, δ): 8.94 (d, J = 2.0 Hz, 1H), 8.79 (dd, J = 4.8, 1.2 Hz, 1H), 8.50 (s, 1H), 8.18 (td, J = 8.0, 2.0 Hz, 1H), 7.61 (dd, J = 8.0, 4.8 Hz, 1H). m/z: 286.0 [M + H]+.
Sodium 5-(Pyridine-3-sulfonamido)-1,3-thiazole-4-carboxylate (ANT431, Na Salt)
To a solution of 5-(pyridine-3-sulfonamido)-1,3-thiazole-4-carboxylic acid (850 mg, 2.98 mmol) in tetrahydrofuran (20 mL) was added 0.1 M sodium hydroxide (29.8 mL, 2.98 mmol) solution and sonicated for 10 min to get a clear solution. Water (60 mL) was added, and the mixture was lyophilized to obtain the title compound as a light brown solid (890 mg, 97%) with a melting point of 297–300 °C. 1H NMR (DMSO-d6, 400 MHz, δ): 13.33 (s, 1H), 8.88 (s, 1H), 8.65 (d, J = 4 Hz, 1H), 8.08 (d, J = 8 Hz, 2H), 7.49 (dd, J = 8.0, 4.8 Hz, 1H); 13C NMR (DMSO-d6, 100 MHz, δ): 163.94, 155.34, 151.77, 146.56, 140.16, 139.54, 133.88, 128.77, 123.95. m/z: 284.0 [M – Na]−. High-resolution electrospray ionization mass spectrometry (m/z): [M – Na] calcd for C9H6O4N3S2, 283.9794; found, 283.9735.
Acknowledgments
The authors acknowledge the support of the Wellcome Trust (grant no. 099212/Z/12) through the provision of a Seeding Drug Discovery Initiative award to Antabio SAS. We thank Dr. Rod Porter (rod.porter@rodporterconsultancy.com) for constructive discussions relating to potentiation efficiency. We acknowledge the Diamond Light Source (Didcot, U.K.) for providing access to beamlines.
Glossary
ABBREVIATIONS
- CphA
carbapenem-hydrolyzing metallo-β-lactamase from Aeromonas hydrophila
- MBL
metallo-β-lactamase
- NDM
New Delhi metallo-β-lactamase
- PE
potentiation efficiency
- SBL
serine-β-lactamase
- VIM
Verona integron-encoded metallo-β-lactamase
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsinfecdis.8b00246.
Details on the synthesis of representative examples of the array and X-ray data collection, structure solution, and refinement (PDF)
Author Present Address
∇ Laurus Laboratories Limited, Plot No. DS1, IKP Knowledge Park, Shameerpet, Hyderabad 500078, Telangana, India
Author Contributions
S. Leiris, D. T. Davies, T. D. Pallin, M. C. Cramp, N. Jennings, G. Raphy, M. W. Jones, S. Pothukanuri, R. Pattipati, B. Shankar, R. S. Sivasubrahmanyam, A. K. Soodhagani, R. R. Juventhala, and N. Pottabathini designed and synthesized the molecules and performed the medicinal chemistry. A. Coelho, J. Castander, M. Bayet, C. Lozano, J. Bougnon, J. Bousquet, M. Zalacain, M. Everett, M. Lemonnier, and N. Sprynski carried out the in vitro enzyme assays and MIC testing. M. Benvenuti, C. Pozzi, S. Mangani, F. de Luca, G. Cerboni, and J.-D. Docquier prepared the MBL-producing laboratory strains and the MBL proteins and carried out the X-ray studies. All authors analyzed the data, and D. T. Davies, J.-D. Docquier, and M. Everett wrote the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- O’Neill J. (2016) Review on Antimicrobial Resistance, Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. For this seminal review of antibacterial resistance, see https://amr-review.org/Publications.html (accessed June 28, 2018).
- CARB-X. https://www.phe.gov/about/barda/CARB-X/Pages/default.aspx (accessed June 28, 2018).
- Biomedical Advanced Research and Development Authority. http://www.phe.gov/about/barda/Pages/default.aspx (accessed June 28, 2018).
- Bush K.; Jacoby G. A. (2010) Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother. 54, 969–976. 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Koning G. A.; Tio D.; Coster J. F.; Coutinho R. A.; Ansink-Schipper M. C. (1981) The combination of clavulanic acid and amoxycillin (Augmentin) in the treatment of patients infected with penicillinase producing gonococci. J. Antimicrob. Chemother. 8, 81–82. 10.1093/jac/8.1.81. [DOI] [PubMed] [Google Scholar]
- Bryson H. M.; Brogden R. N. (1994) Piperacillin/tazobactam. A review of its antibacterial activity, pharmacokinetic properties and therapeutic potential. Drugs 47 (3), 506–35. 10.2165/00003495-199447030-00008. [DOI] [PubMed] [Google Scholar]
- Mawal Y.; Critchley I. A.; Riccobene T. A.; Talley A. K. (2015) Ceftazidime-avibactam for the treatment of complicated urinary tract infections and complicated intra-abdominal infections. Expert Rev. Clin. Pharmacol. 8 (6), 691–707. 10.1586/17512433.2015.1090874. [DOI] [PubMed] [Google Scholar]
- Castanheira M.; Huband M. D.; Mendes R. E.; Flamm R. K. (2017) Meropenem-Vaborbactam Tested against Contemporary Gram-Negative Isolates Collected Worldwide during 2014, Including Carbapenem-Resistant, KPC-Producing, Multidrug-Resistant, and Extensively Drug-Resistant Enterobacteriaceae. Antimicrob. Agents Chemother. 61 (9), e00567–17. 10.1128/AAC.00567-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackel M., and Sahm D. (2018) In vitro activity of cefepime in combination with VNRX-5133 against meropenem and/or cefepime resistant clinical isolates of Pseuodomonas aeruginosa; Poster Presentation ECCMID 2018, April 21−24; Madrid, Spain, Abstract P1543 (European Society of Clinical Microbiology and Infectious Diseases; ).
- Brem J.; Cain R.; Cahill S.; McDonough M. A.; Clifton I. J.; Jimenez-Castellanos J.-C.; Avison M. B.; Spencer J.; Fishwick C. W. G.; Schofield C. J. (2016) Structural basis of metallo-β-lactamase, serine-β-lactamase and penicillin-binding protein inhibition by cyclic boronates. Nat. Commun. 7, 12406. 10.1038/ncomms12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dortet L.; Poirel L.; Nordmann P. (2014) Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. BioMed Res. Int. 2014, 249856. 10.1155/2014/249856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docquier J. D.; Mangani S. (2018) An update on β-lactamase inhibitor discovery and development. Drug Resist. Updates 36, 13–29. 10.1016/j.drup.2017.11.002. [DOI] [PubMed] [Google Scholar]
- Chen A. Y.; Thomas P. W.; Stewart A. C.; Bergstrom A.; Cheng Z.; Miller C.; Bethel S. H.; Credille C. V.; Riley C. L.; Page R. C.; Bonomo R. A.; Crowder M. W.; Tierney D. L.; Fast W.; Cohen S.; Marshall S. H. (2017) Dipicolinc acid derivatives as inhibitors of New Delhi Metallo-betalactamase-1. J. Med. Chem. 60 (7), 7267–7283. 10.1021/acs.jmedchem.7b00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsfall L. E.; Garau G.; Lienard B. M. R.; Dideberg O.; Schofield C. J.; Frere J. M.; Galleni M. (2007) Competitive inhibitors of the CphA metallo-β-lactamase from Aeromonas hydrophila. Antimicrob. Agents Chemother. 51 (6), 2136–2142. 10.1128/AAC.00866-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomhard E. M. (2003) High-dose clastogenic activity of aniline in the rat bone marrow and its relationship to the carcinogenicity in the spleen of rats. Arch. Toxicol. 77 (5), 291–297. 10.1007/s00204-003-0443-1. [DOI] [PubMed] [Google Scholar]
- Aoki N.; Ishii Y.; Tateda K.; Saga T.; Kimura S.; Kikuchi Y.; Kobayashi T.; Tanabe Y.; Tsukada H.; Gejyo F.; Yamaguchi K. (2010) Efficacy of Calcium-EDTA as an Inhibitor for Metallo-β-Lactamase in a Mouse Model of Pseudomonas aeruginosa Pneumonia. Antimicrob. Agents Chemother. 54 (11), 4582–4588. 10.1128/AAC.00511-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A. M.; Reid-Yu S. A.; Wang W.; King D. T.; De Pascale G.; Strynadka N. C.; Walsh T. T.; Coombes B. K.; Wright G. D. (2014) Aspergillomarasmine A overcomes metallo-β-lactamase antibiotic resistance. Nature 510, 503–506. 10.1038/nature13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashikawa N.; Arai K. (1993) Pharmacological profiles of aspergillomarasmines as endothelin converting enzyme inhibitors. Jpn. J. Pharmacol. 63, 187–193. [DOI] [PubMed] [Google Scholar]
- Kazmierczak K. M.; Rabine S.; Hackel M.; McLaughlin R. E.; Biedenbach D. J.; Bouchillon S. K.; Sahm D. F.; Bradford P. A. (2016) Multiyear, Multinational Survey of the Incidence and Global Distribution of Metallo-β-Lactamase-Producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 60, 1067–1078. 10.1128/AAC.02379-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahr G.; Vitor-Horen L.; Bethel C. R.; Bonomo R. A.; González L. J.; Vila A. J. (2017) Clinical Evolution of New Delhi Metallo-β-Lactamase (NDM) Optimizes Resistance under Zn(II) Deprivation. Antimicrob. Agents Chemother. 62 (1), e01849–17. 10.1128/AAC.01849-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z.; Thomas P. W.; Ju L.; Bergstrom A.; Mason K.; Clayton D.; Miller C.; Bethel C. R.; VanPelt J.; Tierney D. L.; Page R. C.; Bonomo R. A.; Fast W.; Crowder M. W. (2018) Evolution of New Delhi metallo-β-lactamase (NDM) in the clinic: effects of NDM mutations on stability, zinc affinity, mono-zinc activity. J. Biol. Chem. 293, 12606–12618. 10.1074/jbc.RA118.003835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju L. C.; Cheng Z.; Fast W.; Bonomo R. A.; Crowder M. W. (2018) The Continuing Challenge of Metallo-β-Lactamase Inhibition: Mechanism Matters. Trends Pharmacol. Sci. 39 (7), 635–647. 10.1016/j.tips.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett M.; Sprynski N.; Coelho A.; Castandet J.; Bayet M.; Lozano C.; Davies D. T.; Leiris S.; Zalacain M.; Morrissey I.; Magnet S.; Holden K.; Warn P.; De Luca F.; Docquier J.-D.; Lemonnier M.; Bougnon J. (2018) Discovery of a novel metallo-β-lactamase inhibitor, which can potentiate Meropenem activity against carbapenem-resistant Enterobacteriaceae. Antimicrob. Agents Chemother. 62 (5), e00074–18. 10.1128/AAC.00074-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docquier J.-D.; Lamotte-Brasseur J.; Galleni M.; Amicosante G.; Frère J. M.; Rossolini G. M. (2003) On functional and structural heterogeneity of VIM-type metallo-β-lactamases. J. Antimicrob. Chemother. 51, 257–266. 10.1093/jac/dkg067. [DOI] [PubMed] [Google Scholar]
- Garcia-Saez I.; Docquier J.-D.; Rossolini G. M.; Dideberg O. (2008) The three-dimensional structure of VIM-2, a Zn-β-lactamase from Pseudomonas aeruginosa in its reduced and oxidised form. J. Mol. Biol. 375, 604–611. 10.1016/j.jmb.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Docquier J.-D.; Benvenuti M.; Calderone V.; Stoczko M.; Menciassi N.; Rossolini G. M.; Mangani S. (2010) High-resolution crystal structure of the subclass B3 metallo-β-lactamase BJP-1: rational basis for substrate specificity and interaction with sulfonamides. Antimicrob. Agents Chemother. 54, 4343–4351. 10.1128/AAC.00409-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.