Abstract
Background: Long noncoding RNAs (lncRNAs) are non-protein coding transcripts longer than 200 nucleotides in length. They drive many important cancer phenotypes through their interactions with other cellular macromolecules including DNA, RNA and protein. Recent studies have identified numerous lncRNAs active in colorectal cancer (CRC). The lncRNA small nucleolar RNA host gene 6 (SNHG6) has been reported to have an oncogenic role in multiple cancers. However, the biological role and mechanism of SNHG6 in the tumorigenesis of CRC has not been reported in-deep.
Methods: The Cancer Genome Atlas (TCGA) database and GEO database were used to identify SNHG6 expression in different human cancers and explore the relationship between SNHG6 expression and patient prognosis using Kaplan-Meier method analysis. SNHG6 expression in 77 pairs of clinical CRC tissues and different CRC cell lines were analyzed by quantitative real-time PCR (qRT-PCR). A CCK-8 assay was used to assess cell proliferation, transwell assay to detect the cell metastasis, and tumor growth was investigated with a nude mice model in vivo. Whether UPF1 and ZEB1 are downstream targets of SNHG6 was verified by bioinformatics target gene prediction, qRT-PCR and western blot.
Results: TCGA data showed that SNHG6 was significantly upregulated in colorectal cancer samples in comparison with healthy data samples (P < 0.01). CRC patients with high levels of SNHG6 had a significantly shorter overall survival than those with low levels of SNHG6 (P = 0.0162). qRT-PCR confirmed that the expression of SNHG6 was significantly upregulated in CRC tissues and cell lines. Upregulation of SNHG6 expression induced RKO and HCT116 cell proliferation as well as RKO cell metastasis, while downregulation of SNHG6 expression supressed the proliferation and metastasis of RKO cells and tumor growth in vivo. UPF1 was upregulated and ZEB1 was decreased when SNHG6 knockdown, regulating the TGF-β/Smad pathway and inducing EMT respectively.
Conclusions: SNHG6 may play an oncogenic role in CRC cells by activating TGF-β/Smad signaling pathway via targeting of UPF1 and inducing EMT via regulating ZEB1. This could be a prognostic biomarker and therapeutic target for CRC.
Keywords: Colorectal cancer, SNHG6, UPF1, EMT, ZEB1
Introduction
Colorectal cancer (CRC) is the third most common cancer and the fourth most common cause of cancer-related death worldwide.1, 2 CRC is caused by mutations that target oncogenes, tumor suppressor genes and genes related to DNA repair mechanisms. Interestingly, noncoding RNAs account for 90% of total transcribed RNAs in the human genome.3 Long noncoding RNAs (lncRNAs) are functionally defined as transcripts >200 nucleotides in length with no protein coding potential. They also number in the tens of thousands, many of which are uniquely expressed in differentiated tissues or specific cancer types.4 LncRNAs regulate cellular processes depending on their cellular localization: nuclear lncRNAs are enriched for functionality involving chromatin interactions, transcriptional regulation, and RNA processing, while cytoplasmic lncRNAs can modulate mRNA stability or translation and influence cellular signaling cascades.5 Since the lncRNA CCAT1 was identified in CRC, numerous lncRNAs have been characterized along with their oncogenic or tumor suppressor functions in CRC.6-9.
SNHG6 is a housekeeping gene from the 5'TOP family that encodes two non-coding RNAs (ncRNAs): U87 C/D box snoRNA (SNORD87),10 and lncRNA SNHG6,11 which has been demonstrated to be as a potential oncogene in various human cancers.12-14 In this study, we investigated SNGH6 expression in different human cancers using a TCGA dataset, and found that SNHG6 was highly expressed in CRC with a poor prognosis. Our study demonstrated that SNHG6 may act as an oncogene in CRC by activating the TGF-β/Smad signaling pathway via binding UPF1 and inducing epithelial-mesenchymal transition (EMT) through regulating ZEB1.
Materials and methods
The Cancer Genome Atlas (TCGA) database, GEO database, StarBase and bioinformatics analysis
TCGA and GEO data of different cancers was selected by GEPIA and UALCAN, so examine whether any significant differences in SNHG6 expression existed between paired normal and tumor tissues. Fold change > 1.5 and P-value < 0.01 between the tumor and normal tissues were considered as significant. The starBase v2.015 was used to selected downstream interacting protein.
Clinical specimens
Clinical CRC specimens and paired normal tissues were collected from 77 patients who underwent surgical treatment for CRC at Nanfang Hospital of Southern Medical University after obtaining informed consent. A diagnosis of CRC was histopathologically confirmed for each patient sample. Cancer tissues and matched normal tissues were stored at -80℃ until use. The protocols used in this study were approved by our hospital's Protection of Human Subjects Committee.
Cell culture, plasmid construction, lentiviral construction and cell transfections
Human normal colon epithelial cell line (FHC) and human colorectal cancer cell lines (HT29, CaCO2, SW480, SW620, RKO, HCT116 and LoVo) were purchased from the Cell Bank of Type Culture Collection (CBTCC, Chinese Academy of Sciences, Shanghai, China) and were cultured in DMEM (Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum (Gibco, Carlsbad, CA). Cells were maintained at 37℃ in a water-saturated atmosphere with 5% CO2. In order to overexpress SNHG6, full-length SNHG6 was cloned into the expression vector pCMV (Vigene, Shandong, China) and transfected into RKO cells by using LipofectaminTM 3000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Knockdown of SNHG6 was accomplished using three different designed shRNAs (Cyagen, Guangzhou, China) that were transfected into RKO cells according to the manufacturer's instructions.
RNA isolation, cDNA synthesis, and quantitative real-time PCR
Total RNAs were extracted from cells or tissues with Trizol solution (TaKaRa, Dalian, China). Quantitative real-time polymerase chain reaction (qRT-PCT) was performed using the PrimeScript RT Reagent Kit and SYBR Premix Ex Taq (TaKaRa, Dalian, China) following the manufacturer's instructions. Our results were normalized to the expression of glyeraldehyde-3-phosphate dehydrogenase (GAPDH) or U6. The specific primers used are listed in Table 1. qRT-PCR results were analyzed to obtain Ct values of amplified products, and data was analyzed by the 2-ΔΔCt method.
Table 1.
List of qRT-PCR primers
| Gene | Forward primer (5'-3') | Reverse primer(5'-3') |
|---|---|---|
| SNHG6 | TTGAGGTGAAGGTGTATG | GGTAACGAAGCAGAAGTA |
| UPF1 | TCACGGCACAGCAGATCAACAAG | CGGCTTCTCCAGGTCCTCCAG |
| miR-101-3p | CGCGCGTACAGTACTGTGATAA-CTGAA | |
| ZEB1 | ACTGTTTGTAGCGACTGGATT | TAAAGTGGCGGTAGATGGTA |
| GAPDH | CGAGCCACATCGCTCAGACA | GTGGTGAAGACGCCAGTGGA |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
Cell proliferation assay
Cell proliferation was estimated using a Cell Counting Kit-8 (CCK-8) (Dojindo, Japan). Overexpression transfected RKO cells and HCT116 cells as well as RKO knockdown cells were seeded on the 96-well plates and each were cultured for 0h, 24h, 48h, 72h, 96h respectively. At the different time point, 10μL CCK-8 was added to the well and incubated for 2 hours. An absorbance value (OD) of 450nm was determined on the microplate reader.
Transwell assay
Cell migration and invasion assays were measured by trawnswell chamber (8μm pore size, Corning), and for cell invasion, the transwell chambers were also matrigel-coated. The lower chamber was filled with 500μL of 20% FBS medium. Transfected RKO cells (6×104) in 200μL of serum-free medium were gently loaded onto each filter insert (upper chamber) and then incubated at 37℃ for 48h. The filter inserts were removed from the chambers, fixed with methanol for 10min and stained with hematoxylin for 20 min. The samples were subsequently washed, dried and mounted onto slides. The migratory cells were stained blue, visualized under and inverted microscope and then counted in five random fields for statistical analysis.
Wound healing assay
Transfected overexpression and knockdown RKO cells were cultured in DMEM with 2% fetal bovine serum. Wounds were made in the cell monolayer using a 10-μl plastic pipette tip. The size of the wound was imaged and measured after 48h of wound formation. The cell migration area was measured with dashed areas and normalized to control cells.
In vivo experiments
4-week-old male nude mice were purchased from the Central Laboratory of Animal Science, Wuhan University (Wuhan, China) and were maintained in a specific pathogen-free facility. RKO cells stably transfected with SNHG6-shRNA or scramble-shRNA were harvested from 60mm plates and suspended at 5×106 cells/ml. The suspended cells (200μl) were subcutaneously injected into the left hip of 4 mice (4 weeks old) each group, and the mice were sacrificed 4 weeks after injection. The tumor volume (V) was obtained by measuring the length (L) and width (W) of the tumor with vernier calipers, and which was calculated using the formula V = (L×W2) × 0.5.
Western blot analysis
Total protein was extracted from cells using RIPA lysis buffer. Extracted proteins were mixed with loading buffer, separated by SDS-PAGE and transferred to PVDF membranes, which were subsequently blocked with a 5% solution of non-fat milk for 1h. Membranes were then incubated with primary antibody [GAPDH, UPF1, 1:5000, Proteintech; smad2, p-smad2, smad3, p-smad3, E-cadherin, N-cadherin, Vimentin, ZEB1, Slug, Snail, MMP9, MMP2, 1:1000, Cell Signaling Technology] according to the manufacturer's instructions. Then the membranes were washed three times with TBST and incubated with appropriate secondary antibodies for 1h at room temperature. The ECL chemiluminescence system was used to detect the signal.
Statistical analysis
The SPSS 17.0 statistical analysis software was used for statistical analysis of experimental data. The significance of differences between groups was estimated by Student's t-test. Additionally, multiple group comparisons were analyzed with one-way ANOVA. Statistically significant correlation between SNHG6 and UPF1 expression levels in CRC tissues and cell lines was analyzed by Pearson's correlation analysis. The overall survival probability was analyzed using Kaplan-Meier method and calculated using the log-rank test. * P<0.05, **P<0.01, and ***P<0.001 were considered significant.
Results
SNHG6 is differentially expressed in CRC tumor and normal tissues and associated with CRC progression
According to TCGA, SNHG6 is significantly upregulated in colorectal cancer tissues in comparison with the normal counterparts (Fig. 1a-c, P < 0.01). Additionally, we used the Kaplan-Meier method analysis (log-rank test) to explore the relationship between SNHG6 expression and patient prognosis from GEO dataset (GSE17538). We found that patients with high levels of SNHG6 had a significantly shorter overall survival than those with low levels of SNHG6 (Fig. 1d, P = 0.0162).
Figure 1.
SNHG6 was upregulated in CRC tissues with a poor prognosis according to TCGA and GEO data. (a-c) GEPIA (http://gepia.cancer-pku.cn) and UALCAN (http://ualcan.path.uab.edu) showed that SNHG6 was highly expressed in CRC tissues compared to adjacent normal tissues (P < 0.01). (d) Kaplan-Meier method was used to analyze the GEO GSE17538 dataset. Patients with CRC are divided into a high-expression group (whose expression was higher than the median) and low-expression group (whose expression was lower than the median) (P = 0.0162).
SNHG6 is upregulated in colorectal cancer tissues and cell lines
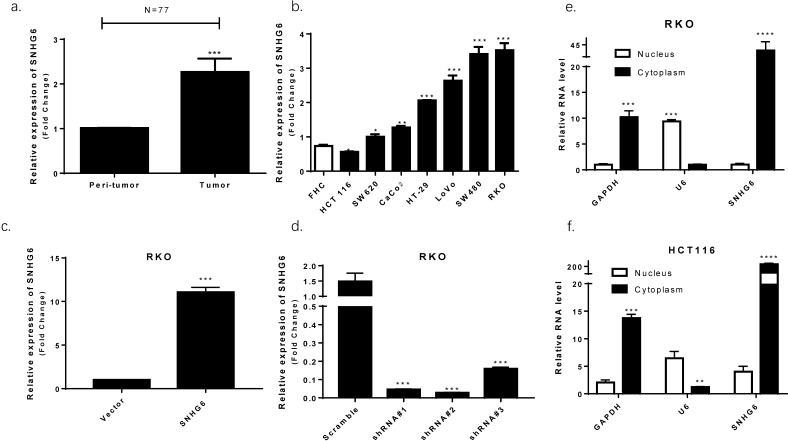
We used qRT-PCR to observe that SNHG6 was significantly upregulated in CRC tissues based on samples from 77 colorectal cancer patients (Fig. 2a, P < 0.001). High levels of SNHG6 was also confirmed in CRC cell lines (Fig. 2b). Furthermore, we detected SNHG6 localization because the activities of lncRNAs depended on their subcellular distribution. By analyzing cytoplasmic and nuclear RNA fractions from CRC cells, we found that SNHG6 was localized preferentially in the cytoplasm (Fig. 2e-f).
Figure 2.
SNHG6 overexpression in CRC tissues and cell lines localized to the cytoplasm. (a) qRT-PCR analysis of SNHG6 expression in 77 CRC patient samples, *** P<0.001, data was shown as the mean ± SD. (b) qRT-PCR analysis of SNHG6 expression in CRC cells and normal colon cells. * P<0.05, ** P<0.01, *** P<0.001, data was shown as the mean ± SD. (c-d) qRT-PCR analysis of SNHG6 expression level in RKO cells 48h after SNHG6-vector and SNHG6-specific shRNAs transfection. *** P<0.001, data was shown as the mean ± SD. (e-f) Nuclear and cytoplasmic RNA fractions were isolated from RKO cells and RKO cells, SNHG6 was located in the cytoplasm.* P<0.05, ** P<0.01, *** P<0.001, data was shown as the mean ± SD.
SNHG6 promotes CRC cell invasion and migration in vitro
The biological function of SNHG6 in CRC cells was determined by constructing plasmid vectors harboring SNHG6 or an empty vector. SNHG6 was examined in RKO cells with overexpression of SNHG6, and was then transfected with SNHG6-specific shRNAs to knockdown SNHG6 (Fig. 2c-d, P < 0.01). According to the knockdown efficiency of SNHG6, we chose shSNHG6#2 as functional shRNA.
Both transwell assays and wound healing assays showed that SNHG6 upregulation significantly promoted the invasion and migration of RKO cells compared with the control, and SNHG6 knockdown also reduced the metastasis ability in RKO cells (Fig. 3a-b). Finally, we also found that when SNHG6 knockdown in RKO cells, the levels of MMPs which are directly involved in the invasiveness of cells were downregulated (Fig. 5e).
Figure 3.
SNHG6 promotes CRC cell metastasis in vitro. (a) Both transwell assays regarding invasion and migration revealed that SNHG6 overexpression promoted RKO cell metastasis and reversed with SNHG6 knockdown. * P<0.05, ** P<0.01, *** P<0.001, data is shown as the mean ± SD. (b) Wound healing assays showed that SNHG6 overexpression can promote RKO cell migration and repress when SNHG6 knockdown. ** P<0.01, *** P<0.001, data was shown as the mean ± SD.
Figure 5.
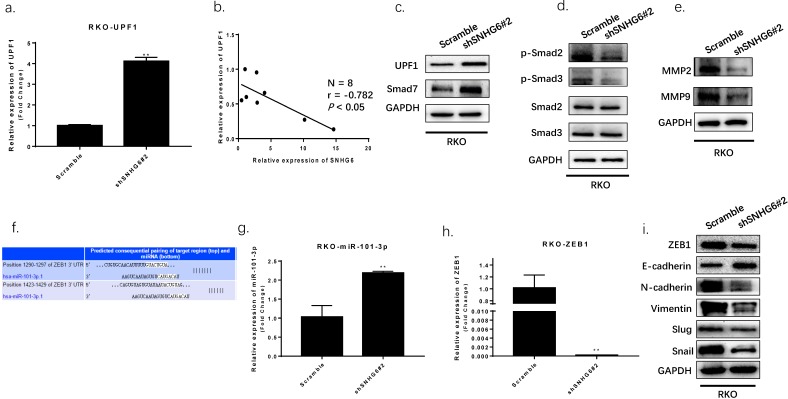
SNHG6 activated TGF-β/Smad signaling pathway via targeting of UPF1 and induced EMT via regulating of ZEB1. (a-d) The expression of UPF1 protein and downstream effectors (p-Smad2 and p-Smad3) were detected by qRT-PCR and western blot analysis. Our findings indicated UPF1was upregulated with SNHG6-knockdown in RKO cells, and UPF1 has a inverse correlation with SNHG6 in CRC tissues, ** P<0.01. (f) miR-101-3p and ZEB1 predicted consequential paring of target regions from TargetScan database (http://www.targetscan.org/vert_71/). (g-h) qRT-PCR analysis of miR-101-3p and ZEB1 when SNHG6 knockdown in RKO cells, ** P<0.01. (i) western blot analysis of ZEB1 and EMT proteins following the transfection of RKO cells, ** P<0.01.
SNHG6 promotes CRC cell proliferation in vitro and knockdown of SNHG6 represses tumor growth in vivo
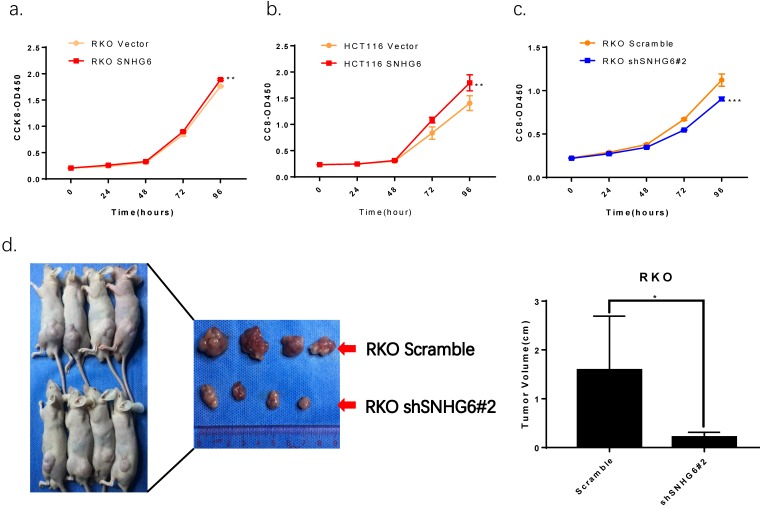
CCK-8 assays demonstrated that overexpression of SNHG6 resulted in a higher proliferative capacity in RKO cells and HCT116 cells compared with that of parallel stable cell lines containing the empty vector; SNHG6 knockdown significantly decreased RKO cells growth (Fig.4a-c). In order to investigate the roles of SNHG6 in tumorigenesis in vivo, RKO cells were stably transfected with SNHG6-shRNA # 2 and control cells were injected into the left hips of male nude mice. We found that after 25 days, SNHG6-shRNA # 2 inhibited tumor growth compared to the control group (Fig.4c, P < 0.05).
Figure 4.
SNHG6 promotes CRC cell proliferation in vitro and represses tumor growth in vivo. (a-c) CCK-8 assays showed that SNHG6 overexpression stimulated RKO cells and HCT116 cells proliferation, while silencing of SNHG6 inhibited RKO cell proliferation. ** P<0.01, *** P<0.001. (d) Images of tumor formation in nude mice (n=4) injected subcutaneously with RKO cells silencing SNHG6 (lower side) and scramble (upper side) after 4 weeks. Tumor volume in SNHG6 knockdown cells was lower than those of control cells. * P<0.05.
SNHG6 regulates TGF-β/Smad by targeting UPF1 and inducing EMT by ZEB1
In order to understand the mechanism by which SNHG6 contributed to CRC, we performed bioinformatic analysis using StarBase v2.0 (Table 2) and found that Up-frameshift Protein 1 (UPF1) may be a target gene of SNHG6; a function which has been demonstrated in HCC. 11
Table 2.
Partial Human RBP-LncRNA interactions of SNHG6 from StarBase v2.0 (target sites ≥ 5)
| Name | LncRNA Name | Target Sites | Clip-seq Read Number |
|---|---|---|---|
| UPF1 | SNHG6 | 48 | 218 |
| U2AF65 | SNHG6 | 20 | 1128 |
| eIF4AIII | SNHG6 | 11 | 413 |
| IGF2BP1 | SNHG6 | 5 | 41 |
| FMRP | SNHG6 | 5 | 5000 |
| LIN28 | SNHG6 | 5 | 148 |
Then we used qRT-PCR and western blot to determine SNHG6 and UPF1 expression. We found that UPF1 was upregulated in RKO cells when SNHG6 was knocked down, and it has an inverse correlation with SNHG6 in CRC tissues (Fig.5a-b). UPF1 was already reported as a tumor suppressor gene for HCC by targeting Smad7 and affecting the TGF-β pathway.16 Thus, we hypothesized that SNHG6 promoted CRC cells tumorigenesis by regulating the ability of UPF1 to mediate the TGF-β/Smad pathway. We further used western blot to investigate the relationship between SNHG6 and UPF1. Our results demonstrated that the expression of UPF1 protein, and the Smad7 downstream TGF-β pathway proteins, such as p-Smad2 and p-Smad3, were decreased with SNHG6 knockdown whereas total Smad2 and Smad3 expression level was not significantly altered (Fig. 5c). We were able to conclude that SNHG6 regulated the expression of UPF1 and affected the TGF-β pathway.
Furthermore, a previous study demonstrated that SNHG6 could affect ZEB1 through sponging miR-101-3p.11 As previously reported, ZEB1 is a crucial transcription factor of EMT which can regulate the expression of many EMT markers.17 We speculated that SNHG6 could also induce EMT by upregulating ZEB1 expression in CRC via miR-101-3p. We searched TargetScan database finding that miR-101-3p has two predicted binding sites with ZEB1 (Fig.5f). We detected miR-101-3p and ZEB1 expression by qRT-PCR, finding that miR-101-3p was upregulated and ZEB1 was downregulated while SNHG6 knockdown in RKO cells (Fig.5g-h). To further explore the effect of targeting SNHG6 on EMT, we found that knockdown of SNHG6 in RKO cells resulted in increased expression of E-cadherin but decreased expression of ZEB1, N-cadherin, Vimentin, Slug and Snail compared to control cells by western blot(Fig.5i). The above data indicated that SNHG6 may induce EMT by regulating ZEB1 via sponging miR-101-3p.
Discussion
LncRNAs are involved in numerous biological and cellular pathways by interacting with various macromolecules such as DNA, chromatin, proteins, and various RNA species; including mRNAs, microRNAs, and other lncRNAs.18 Recent studies have implied that lncRNAs are widely involved in proliferation, invasion, and metastasis and thus represent potential prognostic biomarkers in colorectal cancer, such as CCAT,19 DANCR,20 CRNDE.21
Small nucleolar RNAs (snoRNAs) are another class of small non-coding RNA molecules, which are concentrated in the nucleoli and have a stable metabolism.22 Their main function is to participate in the post-transcriptional modification of rRNA and other RNAs in the cytoplasm.23 Most snoRNAs are encoded by host genes and are processed from the introns of pre-mRNAs. However, recent studies have also indicated that snoRNA host genes could affect cell proliferation, transformation and tumorigenesis in a variety of human cancers, such as SNGH1 in HCC24, 25 and CRC,26, 27 SNGH5 in GC28, 29 and CRC.30.
SNHG6 has been reported to have an oncogenic role in tumors such as HCC,11, 12, 31 glioma,32, esophageal squamous cell carcinoma33 and osteosarcoma.14 The current study showed that SNHG6 was significantly overexpressed in human CRC tissues as well, with poor prognosis. Moreover, we applied in vitro and in vivo methods to reveal the involvement of SNHG6 in CRC tumorigenesis, such as CRC cellular growth and metastasis.
The subcellular localization of lncRNAs is also a critical factor to determine their functions by providing them different opportunities to interact with different molecules.34. For instance, lncRNAs localized in nucleus tend to be involved in transcriptional and epigenetic regulations by interacting with genomic DNA, chromatin, transcription factors, chromatin regulators, spliceosomes and other nuclear proteins.18 Meanwhile, cytosolic lncRNAs are frequently implicated in post-transcriptional, translational, and posttranslational regulatory processes through interactions with various key factors in epigenetic and signaling pathways.35 Based on the findings of previous study suggested that SNHG6 was mostly located in cytoplasm, where it could bind to proteins and microRNAs.
UPF1, a part of the human nonsense-mediated mRNA decay (NMD) substrate, could mediate RNA decay processes and destabilize the encoding TGF-β inhibitor, Smad7, stimulating TGF-β signaling.36 This gene was reported to be a tumor suppressor gene regulating cell proliferation and differentiation in HCC.16 Moreover, UPF1 gene was found to be commonly mutated in pancreatic adenosquamous carcinoma (ASC), which represents the first known example of genetic alterations in a NMD gene in human tumors.37 In this study, we used StarBase v2.0 finding that UPF1 may be a target gene of SNGH6. Our results confirmed that knockdown SNHG6 by shRNA increased UPF1 and overexpression of SNHG6 decreasing UPF1, while regulating TGF-β/Smad signaling pathway in CRC cells. Meanwhile, SNHG6 has been reported to regulate ZEB1 by sponging miR-101-3p in gastric cancer.13 We used qRT-PCR and western blot confirmed that SNHG6 could regulate ZEB1 and induce EMT in CRC cells.
Conclusion
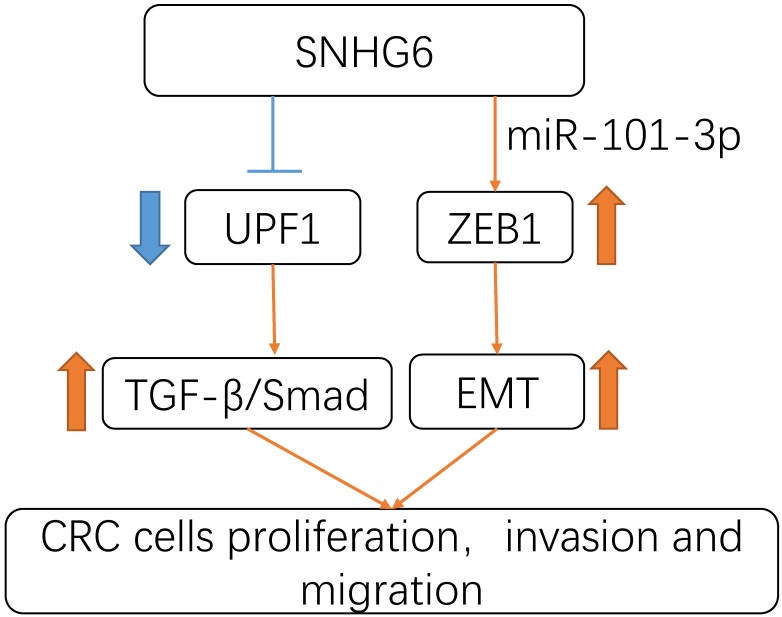
In summary, our study revealed that SNHG6 could play an oncogenic role in CRC. SNHG6 promoted tumor cell proliferation and metastasis by activating the TGF-β/Smad pathway via binding UPF1. Meanwhile SNHG6 could regulate ZEB1 by inducing EMT via miR-101-3p (Fig.6). These findings suggest that SNHG6 is an important prognostic factor and therapeutic target for CRC.
Figure 6.
Schematic model of SNHG6 in CRC cells. SNHG6 promotes tumor cell roliferation, invasiveness and metastasis by activating the TGF-β/Smad pathway via binding UPF1, meanwhile SNHG6 could regulate ZEB1 inducing EMT via miR-101-3p.
Acknowledgments
This study was supported by Guangzhou Pilot Project of Clinical and Translational Research Center (early gastrointestinal cancer, No. 7415696196402), Guangdong gastrointestinal disease research center (No.2017B02029003).
Abbreviations
- lncRNA
long noncoding RNA
- SNHG6
small nucleolar RNA host gene 6
- CRC
colorectal cancer
- TCGA
The Cancer Genome Atlas
- qRT-PCR
quantitative real-time PCR
- UPF1
Up-frameshift Protein 1
- HCC
Hepatocellular carcinoma
- EMT
epithelial-mesenchymal transition
- ZEB1
zinc finger E-box binding homeobox 1
- CCK-8
Cell Counting Kit-8.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017[J] CA: a cancer journal for clinicians. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Marmol I, Sanchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer[J] International journal of molecular sciences; 2017. p. 18. (1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patrushev LI, Kovalenko TF. Functions of noncoding sequences in mammalian genomes[J] Biochemistry Biokhimiia. 2014;79(13):1442–69. doi: 10.1134/S0006297914130021. [DOI] [PubMed] [Google Scholar]
- 4.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y. et al. The landscape of long noncoding RNAs in the human transcriptome[J] Nature genetics. 2015;47(3):199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways[J] Cancer cell. 2016;29(4):452–63. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ozawa T, Matsuyama T, Toiyama Y, Takahashi N, Ishikawa T, Uetake H. et al. CCAT1 and CCAT2 long noncoding RNAs, located within the 8q.24.21 'gene desert', serve as important prognostic biomarkers in colorectal cancer[J] Ann Oncol. 2017;28(8):1882–8. doi: 10.1093/annonc/mdx248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma Y, Yang Y, Wang F, Moyer M, Wei Q, Zhang P. et al. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/β-catenin signalling pathway via suppression of activator protein 2α[J] Gut. 2016;65(9):1494–504. doi: 10.1136/gutjnl-2014-308392. [DOI] [PubMed] [Google Scholar]
- 8.Tsai K, Lo Y, Liu H, Yeh C, Chen Y, Hsu C. et al. Linc00659, a long noncoding RNA, acts as novel oncogene in regulating cancer cell growth in colorectal cancer[J] Mol Cancer. 2018;17(1):72. doi: 10.1186/s12943-018-0821-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang C, Qiu S, Sun F, Li W, Wang Z, Yue B. et al. Long non-coding RNA HNF1A-AS1 mediated repression of miR-34a/SIRT1/p53 feedback loop promotes the metastatic progression of colon cancer by functioning as a competing endogenous RNA[J] Cancer Lett. 2017;410:50–62. doi: 10.1016/j.canlet.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 10.Gogolevskaya IK, Makarova JA, Gause LN, Kulichkova VA, Konstantinova IM, Kramerov DA. U87 RNA, a novel C/D box small nucleolar RNA from mammalian cells[J] Gene. 2002;292(1-2):199–204. doi: 10.1016/s0378-1119(02)00678-9. [DOI] [PubMed] [Google Scholar]
- 11.Chang L, Yuan Y, Li C, Guo T, Qi H, Xiao Y. et al. Upregulation of SNHG6 regulates ZEB1 expression by competitively binding miR-101-3p and interacting with UPF1 in hepatocellular carcinoma[J] Cancer Lett. 2016;383(2):183–94. doi: 10.1016/j.canlet.2016.09.034. [DOI] [PubMed] [Google Scholar]
- 12.Cao C, Zhang T, Zhang D, Xie L, Zou X, Lei L. et al. The long non-coding RNA, SNHG6-003, functions as a competing endogenous RNA to promote the progression of hepatocellular carcinoma[J] Oncogene. 2017;36(8):1112–22. doi: 10.1038/onc.2016.278. [DOI] [PubMed] [Google Scholar]
- 13.Yan K, Tian J, Shi W, Xia H, Zhu Y. LncRNA SNHG6 is Associated with Poor Prognosis of Gastric Cancer and Promotes Cell Proliferation and EMT through Epigenetically Silencing p27 and Sponging miR-101-3p[J] Cell Physiol Biochem. 2017;42(3):999–1012. doi: 10.1159/000478682. [DOI] [PubMed] [Google Scholar]
- 14.Ruan J, Zheng L, Hu N, Guan G, Chen J, Zhou X. et al. Long noncoding RNA SNHG6 promotes osteosarcoma cell proliferation through regulating p21 and KLF2[J] Arch Biochem Biophys. 2018;646:128–36. doi: 10.1016/j.abb.2018.03.036. [DOI] [PubMed] [Google Scholar]
- 15.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data[J] Nucleic acids research. 2014;42(Database issue):D92–7. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang L, Li C, Guo T, Wang H, Ma W, Yuan Y. et al. The human RNA surveillance factor UPF1 regulates tumorigenesis by targeting Smad7 in hepatocellular carcinoma[J] J Exp Clin Cancer Res. 2016;35:8. doi: 10.1186/s13046-016-0286-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan X, Banerjee P, Liu X, Yu J, Gibbons D, Wu P. et al. The epithelial-to-mesenchymal transition activator ZEB1 initiates a prometastatic competing endogenous RNA network[J] J Clin Invest. 2018;128(4):1267–82. doi: 10.1172/JCI97225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmitt AM, Chang HY. Long Noncoding RNAs: At the Intersection of Cancer and Chromatin Biology[J] Cold Spring Harbor perspectives in medicine; 2017. p. 7. (7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xin Y, Li Z, Zheng H, Chan M, Ka Kei Wu W. CCAT2: A novel oncogenic long non-coding RNA in human cancers[J] Cell Prolif; 2017. p. 50. (3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Lu Z, Wang N, Feng J, Zhang J, Luan L. et al. Long noncoding RNA DANCR promotes colorectal cancer proliferation and metastasis via miR-577 sponging[J] Exp Mol Med. 2018;50(5):57. doi: 10.1038/s12276-018-0082-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding J, Li J, Wang H, Tian Y, Xie M, He X. et al. Long noncoding RNA CRNDE promotes colorectal cancer cell proliferation via epigenetically silencing DUSP5/CDKN1A expression[J] Cell Death Dis. 2017;8(8):e2997. doi: 10.1038/cddis.2017.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rederstorff M, Huttenhofer A. Small non-coding RNAs in disease development and host-pathogen interactions[J] Current opinion in molecular therapeutics. 2010;12(6):684–94. [PubMed] [Google Scholar]
- 23.Khalaj M, Park C. snoRNAs contribute to myeloid leukaemogenesis[J] Nat Cell Biol. 2017;19(7):758–60. doi: 10.1038/ncb3566. [DOI] [PubMed] [Google Scholar]
- 24.Zhang M, Wang W, Li T, Yu X, Zhu Y, Ding F. et al. Long noncoding RNA SNHG1 predicts a poor prognosis and promotes hepatocellular carcinoma tumorigenesis[J] Biomed Pharmacother. 2016;80:73–9. doi: 10.1016/j.biopha.2016.02.036. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H, Zhou D, Ying M, Chen M, Chen P, Chen Z. et al. Expression of Long Non-Coding RNA (lncRNA) Small Nucleolar RNA Host Gene 1 (SNHG1) Exacerbates Hepatocellular Carcinoma Through Suppressing miR-195[J] Med Sci Monit. 2016;22:4820–9. doi: 10.12659/MSM.898574. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Zhao Y, Qin Z, Feng Y, Tang X, Zhang T, Yang L. Long non-coding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) promote cell proliferation in colorectal cancer by affecting P53[J] Eur Rev Med Pharmacol Sci. 2018;22(4):976–84. doi: 10.26355/eurrev_201802_14379. [DOI] [PubMed] [Google Scholar]
- 27.Sun X, Wang Z, Yuan W. Down-regulated long non-coding RNA SNHG1 inhibits tumor genesis of colorectal carcinoma[J] Cancer Biomark. 2017;20(1):67–73. doi: 10.3233/CBM-170112. [DOI] [PubMed] [Google Scholar]
- 28.Zhao L, Guo H, Zhou B, Feng J, Li Y, Han T. et al. Long non-coding RNA SNHG5 suppresses gastric cancer progression by trapping MTA2 in the cytosol[J] Oncogene. 2016;35(44):5770–80. doi: 10.1038/onc.2016.110. [DOI] [PubMed] [Google Scholar]
- 29.Zhao L, Han T, Li Y, Sun J, Zhang S, Liu Y. et al. The lncRNA SNHG5/miR-32 axis regulates gastric cancer cell proliferation and migration by targeting KLF4[J] FASEB J. 2017;31(3):893–903. doi: 10.1096/fj.201600994R. [DOI] [PubMed] [Google Scholar]
- 30.Damas N, Marcatti M, Côme C, Christensen L, Nielsen M, Baumgartner R. et al. SNHG5 promotes colorectal cancer cell survival by counteracting STAU1-mediated mRNA destabilization[J] Nat Commun. 2016;7:13875. doi: 10.1038/ncomms13875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Birgani M, Hajjari M, Shahrisa A, Khoshnevisan A, Shoja Z, Motahari P, Long Non-Coding RNA SNHG6 as a Potential Biomarker for Hepatocellular Carcinoma[J] Pathol Oncol Res; 2017. [DOI] [PubMed] [Google Scholar]
- 32.Cai G, Zhu Q, Yuan L, Lan Q. LncRNA SNHG6 acts as a prognostic factor to regulate cell proliferation in glioma through targeting p21[J] Biomed Pharmacother. 2018;102:452–7. doi: 10.1016/j.biopha.2018.03.083. [DOI] [PubMed] [Google Scholar]
- 33.Fan R, Guo J, Yan W, Huang M, Zhu C, Yin Y. et al. Small nucleolar host gene 6 promotes esophageal squamous cell carcinoma cell proliferation and inhibits cell apoptosis[J] Oncol Lett. 2018;15(5):6497–502. doi: 10.3892/ol.2018.8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen LL. Linking Long Noncoding RNA Localization and Function[J] Trends in biochemical sciences. 2016;41(9):761–72. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 35.Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ. Targeting noncoding RNAs in disease[J] The Journal of clinical investigation. 2017;127(3):761–71. doi: 10.1172/JCI84424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lou C, Shao A, Shum E, Espinoza J, Huang L, Karam R. et al. Posttranscriptional control of the stem cell and neurogenic programs by the nonsense-mediated RNA decay pathway[J] Cell Rep. 2014;6(4):748–64. doi: 10.1016/j.celrep.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu C, Karam R, Zhou Y, Su F, Ji Y, Li G. et al. The UPF1 RNA surveillance gene is commonly mutated in pancreatic adenosquamous carcinoma[J] Nat Med. 2014;20(6):596–8. doi: 10.1038/nm.3548. [DOI] [PMC free article] [PubMed] [Google Scholar]