Abstract
Background
Stool consistency is an important diagnostic criterion in both research and clinical medicine and is often used to define diarrheal disease.
Methods
We examine the pediatric enteric virome across stool consistencies to evaluate differences in richness and community composition using fecal samples collected from children aged 0 to 5 years participating in a clinical trial in the Amhara region of Ethiopia. The consistency of each sample was graded according to the modified Bristol Stool Form Scale for children (mBSFS-C) before a portion of stool was preserved for viral metagenomic analysis. Stool samples were grouped into 29 pools according to stool consistency type. Differential abundance was determined using negative-binomial modeling.
Results
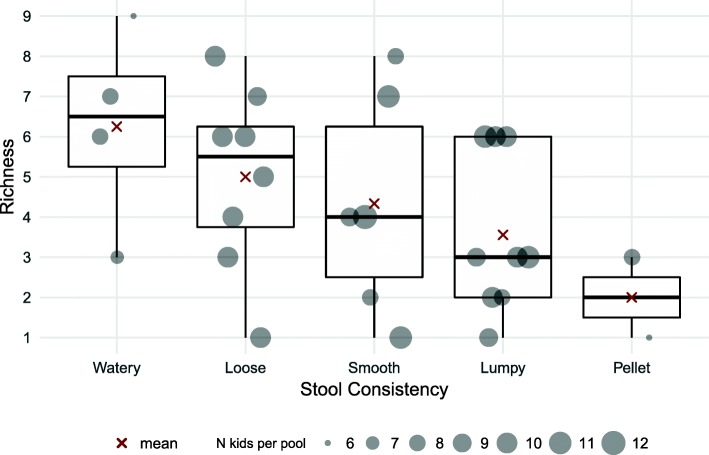
Of 446 censused children who were eligible to participate, 317 presented for the study visit examination and 269 provided stool samples. The median age of children with stool samples was 36 months. Species richness was highest in watery-consistency stool and decreased as stool consistency became firmer (Spearman’s r = − 0.45, p = 0.013). The greatest differential abundance comparing loose or watery to formed stool was for norovirus GII (7.64, 95% CI 5.8, 9.5) followed by aichivirus A (5.93, 95% CI 4.0, 7.89) and adeno-associated virus 2 (5.81, 95%CI 3.9, 7.7).
Conclusions
In conclusion, we documented a difference in pediatric enteric viromes according to mBSFS-C stool consistency category, both in species richness and composition.
Electronic supplementary material
The online version of this article (10.1186/s12879-019-3674-3) contains supplementary material, which is available to authorized users.
Keywords: Diarrheal disease, Virome, Viral infection, Norwalk virus
Background
Stool consistency is an important diagnostic criterion in both research and clinical medicine [1]. Changes in stool consistency are used to measure many gastrointestinal disorders such as ulcerative colitis, irritable bowel syndrome and diarrhea [2–6]. Most epidemiologic studies of diarrheal disease internationally use stool consistency, specifically ‘loose or watery stool’ to classify diarrhea cases [2, 7].
Visual and descriptive stool consistency scales may standardize and improve the accuracy of reported stool consistency. The most widely used stool form scale, The Bristol Stool Form Scale (BSFS), was developed in the late 1980s to measure gut transit time [8, 9]. The BSFS classifies stool form into seven categories according to stool cohesion, surface cracking and consistency.
The BSFS was later simplified to a five-level scale and renamed the modified Bristol Stool Form Scale for children (mBSFS-C) [10], ranging from type 1 (hard pellets) to type 5 (watery stool).
While recent studies have described the bacterial microbiome of the colon and feces, there have been few parallel investigations of the enteric and stool virome [11–14]. Healthy gut and fecal bacterial microbiomes are characterized as having higher species richness [15]. This relationship may not hold for the fecal virome, where higher species richness may signal more viral infections and disease.
Here we use metagenomics to examine the pediatric fecal virome across standardized stool consistency categories using stool samples from 269 children aged 0 to 5 years in rural Ethiopia. We evaluate potential associations between enteric virome composition, species richness and stool consistency.
Methods
Study design
This study was conducted during the final annual visit of a clinical trial evaluating a water improvement intervention in the Amhara region of Ethiopia (clinicaltrials.gov NCT02373657) [16–18]. Methods for the parent trial are described in detail elsewhere [19, 20]. A door-to-door population census was taken in all communities before the study visit. All children aged 0–5 years enumerated on the census were eligible to participate in the study and provide stool samples. The final study visit occurred in April 2016; April is the dry season in this region. The study visit is held in a central location in the community.
Stool sample collection and grading
Caregivers were instructed to have their child defecate in a plastic child’s potty chair lined with a black plastic bag. For children unable to produce a stool in the field, supplies were provided to the caregiver, with instructions to collect stool at home the following morning and bring it to a collection site the following day at a designated time. Stool samples that were collected at home were stored at room temperature.
When the stool was returned to the field station, it was independently inspected in the original collection container and graded according to the Modified Bristol Stool Form Scale for Children (mBSFS-C) by two trained laboratory technicians. Laboratory technicians underwent a two-day training on sample collection and consistency grading prior to data collection. The first laboratory technician’s grade was used to classify stool consistency for this analysis.
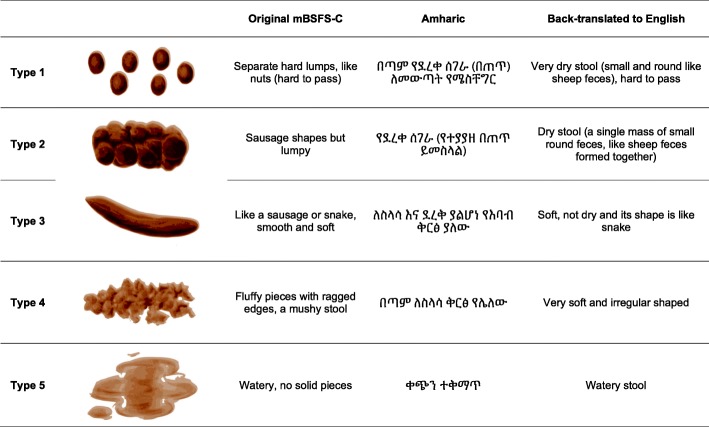
The mSFS-C was available as a laminated sheet with both the cartoon images and Amharic translations (Fig. 1). Methods describing the grading process and kappa evaluating agreement are described in detail elsewhere [21].
Fig. 1.
The modified Bristol Stool Form Scale for Children (mBSFS-C), translated into Amharic
After the stool sample consistency was graded, 0.5 mL of stool was placed in a 1 mL plastic tube, put on ice and transferred to a − 20 C freezer at the end of the day. At the completion of the sample collection, in early May 2016, all samples were transferred to Bahir Dar Regional Laboratory (Bahir Dar, Ethiopia) and kept at − 20 C until they were shipped to University of California, San Francisco in February 2017.
Laboratory methods
Stool specimens were combined into pools of 6 to 12, with sampling stratified by stool consistency grade. Pools were clarified using a tabletop microfuge at 15,000 rpm for ten minutes. Supernatants were then filtered using a 0.45-μm filter (Millipore). The filtrates were digested with a cocktail of nuclease enzymes. Viral nucleic acids protected from digestion within their protein capsids were then extracted using the automated extractor Maxwell 16 (Promega) [22]. Random RT-PCR was followed by the use of the Nextera™ XT Sample Preparation Kit (Illumina) to generate libraries for Illumina MiSeq (2 × 250 bases) using dual barcoding as described [23, 24].
Bioinformatic analyses
Human, bacterial, duplicate, and low-quality sequence reads were first removed. The remaining reads were then de novo assembled. BLASTx translated contigs and singlet reads were aligned against a customized viral proteome database. Candidate viral hit sequences were then aligned against a non-redundant non-virus protein database. Those candidates yielding E score higher (weaker) than those against viral proteins were removed to decrease false positive viral hits [23, 24]. A detailed description of the bioinformatics pipeline is available in the supplemental material (Additional file 1).
Statistical methods
All statistical analyses were performed in R version 3.4.2 (R Foundation for Statistical Computing, Vienna, Austria) using R Studio version 1.1.383. In the interest of reproducibility, an R markdown document containing complete commands for the analysis is available here: https://github.com/kaiemjoy/fecalvirome. The number of viral reads along with the taxonomic assignments and sample characteristics were assembled using the phyloseq package. We define absolute abundance as the raw number of reads for each species/genotype and relative abundance as the proportion of raw reads of each species/genotype in each pool. To calculate relative abundance, we divided the number or reads for each species/genotype in each pool by the total number of reads in that pool and multiplied by 100. We evaluated species richness (Observed, Chao1) and alpha diversity measures (Simpson, Shannon, and Fisher) using the estimate_richness function of phyloseq. We used Spearman’s rank order test to assess correlation between richness and mBSFS-C stool consistency category and determine statistical significance.
We determined differential abundance comparing loose and watery (mBSFS-C types 4&5) to formed stool (mBSFS-C types 1–3) at the species/genotype level using negative-binomial modeling in the DESeq2 package [25]. We used the Bonferroni method to adjust p-values for multiple comparisons. We explored both an unadjusted model and a model adjusted for age. To adjust for age, we calculated the median age for each pool and included it as a covariate in the model. Results are expressed as log2 fold change in loose or watery stool compared to formed stool.
Ethics statement
Ethical committees at the University of California (San Francisco, CA, USA); Emory University (Atlanta, GA, USA); The Food, Medicine and Health Care Administration and Control Authority of Ethiopia; and the Ethiopian Ministry of Science and Technology granted approval for this study. We obtained verbal informed consent in Amharic from all caregivers.
Results
Characteristics of study population
Of 446 censured children who were eligible to participate, 317 children presented for the study visit examination and 269 provided stool samples. The median age of children with stool samples was 36 months (IQR 12–48), 53.2% (143/269) of children were female. A detailed description of the study population characteristics by stool consistency category is presented in Table 1. The 269 samples were analyzed in 29 pools: 4 pools (29 samples) were watery/type 5, 8 pools (79 samples) were loose/type 4, 6 pools (59 samples) were smooth/type 3, 9 pools (88 samples) were lumpy/type 2 and 2 pools (12 samples) were hard pellets/type1. The median age for children with watery/type 5 stool was 7.2 months, younger than with loose/type 4 stool (36 months), smooth/type 3 (36 months), lumpy/type 2 (36 months) or pellet/type 1 (48 months). Caregivers reported observing blood in the stool during the past seven days in 4/29 (13.8%) of children with watery stool samples, 4/79 (5.1%) of children with loose stool samples, 2/59 (3.4%) of children with smooth stool samples, 5/88 (5.7%) of children with lumpy stool samples and 0/14 (0%) of children with pellet stool samples.
Table 1.
Characteristics of the study population by mBSFS-C stool consistency category
| N children (pools) | Watery 29 (4) |
Loose 79 (8) |
Smooth 59 (6) |
Lumpy 88 (9) |
Pellets 14 (2) |
|---|---|---|---|---|---|
| Median age in months (IQR) | 7.2 (4.8-12) | 36 (12-48) | 36 (24-48) | 36 (24-49.2) | 48 (36-57) |
| Female | 15 (51.7% ) | 39 (49.4% ) | 33 (55.9% ) | 48 (54.5% ) | 8 (57.1% ) |
| Blood in stoola | 4 (13.8% ) | 4 (5.1% ) | 2 (3.4% ) | 5 (5.7% ) | 0 (0% ) |
| Feverb | 9 (32.1% ) | 28 (36.8% ) | 20 (35.1% ) | 33 (38.8% ) | 6 (50% ) |
Numbers are N (%) unless otherwise indicated
aCaregiver reported observing blood in the stool any day in the past seven days
bCaregiver reported child had a fever any day in the past seven days
Prevalence
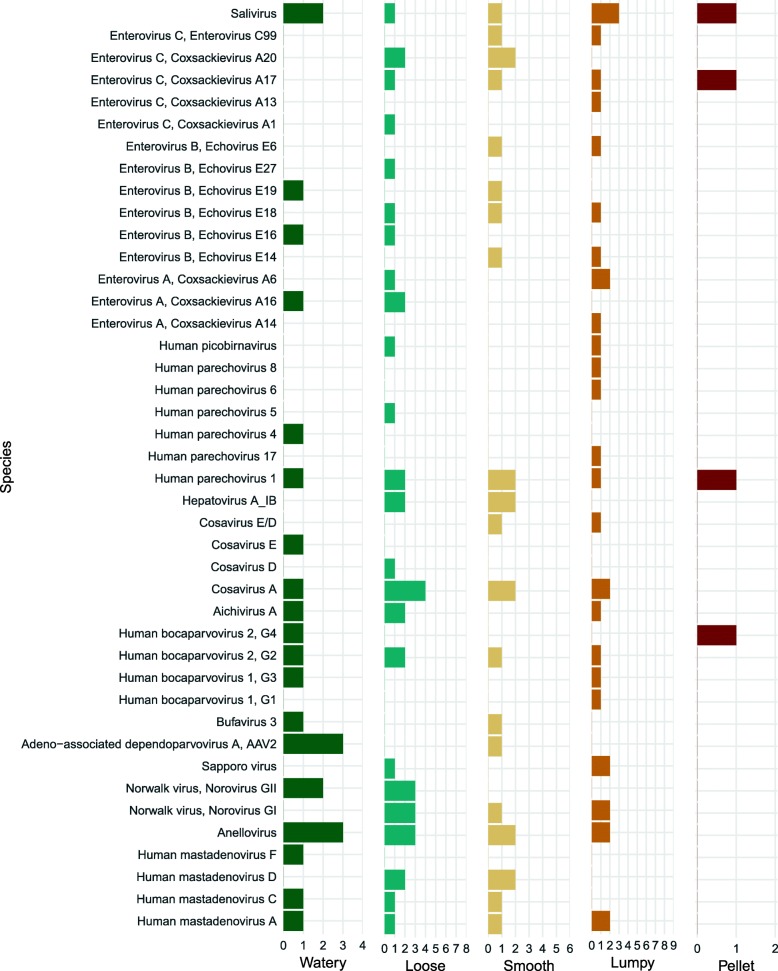
The most prevalent viral reads belonged to anelloviruses (10 of 29 pools), picornaviruses in the species cosavirus A (9 of 29 pools), and salivirus A (8 of 29 pools). Several viruses/genotypes were more prevalent in watery or loose stool compared to formed stool (Fig. 2). For example, Norovirus GII was detected in 5 of 12 (42%) loose or watery pools and 0 of 17 formed pools. Aichivirus A was detected in 3 of 12 (25%) loose or watery pools and 1 of 17 (6%) formed pools. Human mastadenovirus C was detected in 2 of 12 (17%) loose or watery pools and 1 of 17 (6%) formed pools. For some viruses, such as salivirus, Human parechovirus 1, and Saporro virus, there did not appear to be a difference in prevalence by stool consistency category. Human mastadenovirus D was only detected in 1 of 4 (25%) watery pools and no other pools. Rotavirus was not detected in any samples (Additional file 2: Table S1).
Fig. 2.
Prevalence of species/genotypes by mBSFS-C stool consistency category
Abundance
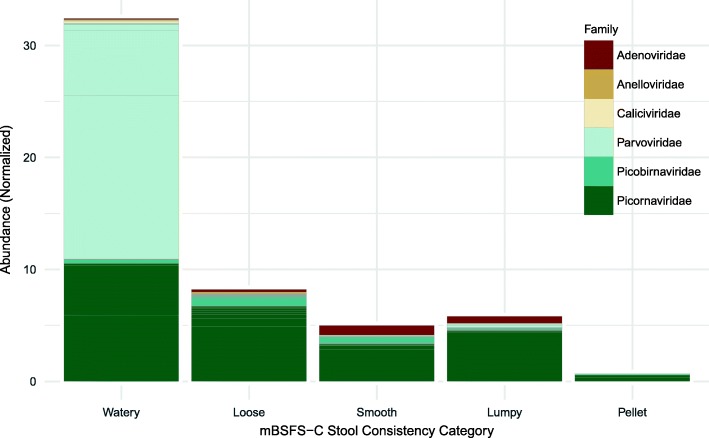
The most abundant reads belonged to the family Picornaviridae followed by Parvoviridae. Both Picornaviridae and Parvoviridae reads had the highest abundance in watery-consistency pools, followed by loose, smooth and lumpy consistency with the lowest abundance in pellet-consistency pools (Fig. 3).
Fig. 3.
Viral abundance by mBSFS-C stool consistency category
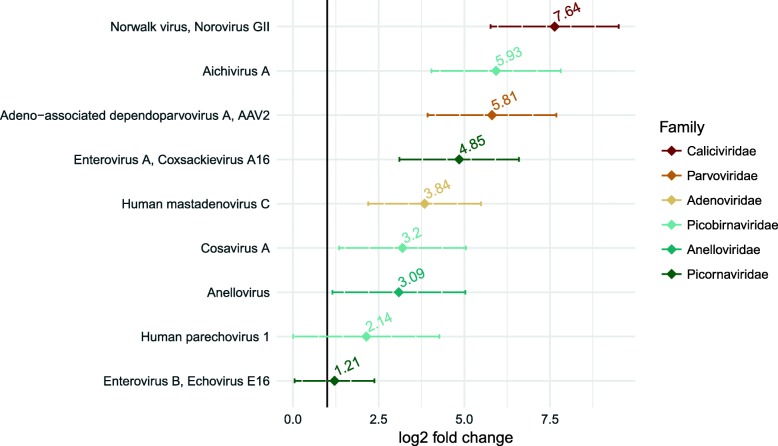
The most abundant species/genotype, both in terms of absolute and relative abundance, was adeno-associated dependoparvovirus A, adeno-associated virus 2 (AAV2) followed by saliviruses, human bocavirus 2, and echovirus E19. The greatest differential abundance comparing loose or watery stool to formed stool was for norovirus GII, followed by aichivirus A, AAV2, coxsackievirus A16, human mastadenovirus, cosavirus A and anellovirus (Fig. 4). For norovirus GII, the abundance was 2656 reads in loose in watery consistency pools and 0 reads in formed consistency with a differential abundance of 7.64 (95% CI 5.8–9.5, p = < 0.001) comparing loose or watery stool to formed stool. For aichivirus A, the abundance was 5659 reads in loose or watery stool compared to 86 reads in formed stool, with a differential abundance of 5.93 (95% CI 4.0–7.8, p = < 0.001). The abundance of AAV2 was 115,946 in loose or watery-consistency pools and 222 in formed-consistency pools with a differential abundance of 5.81 (95%CI 16.0–27.6, p = < 0.001) (Table 2).
Fig. 4.
Differential abundance: Loose/watery stool compared to formed stool
Table 2.
Absolute abundance and log2 fold change
| Species | Abundance in loose stool | Abundance in formed stool | log2 fold change (95% CIs) |
p-value | adjusted p-value* |
|---|---|---|---|---|---|
| Norwalk virus, Norovirus GII | 2656 (0.26) | 0 (0) | 7.64 (5.8 , 9.5) | <0.001 | <0.001 |
| Aichivirus A | 5659 (0.38) | 86 (0.01) | 5.93 (4.0 , 7.8) | <0.001 | <0.001 |
| Adeno-associated dependoparvovirus A, AAV2 | 115946 (14.73) | 222 (0.03) | 5.81 (3.9 , 7.7) | <0.001 | <0.001 |
| Enterovirus A, Coxsackievirus A16 | 2561 (0.22) | 0 (0) | 4.85 (3.1 , 6.6) | <0.001 | <0.001 |
| Human mastadenovirus C | 3023 (0.27) | 449 (0.03) | 3.84 (2.2 , 5.5) | <0.001 | <0.001 |
| Cosavirus A | 932 (0.09) | 122 (0.02) | 3.2 (1.3 , 5) | 0.001 | 0.003 |
| Anellovirus | 2515 (0.28) | 427 (0.06) | 3.09 (1.2 , 5) | 0.002 | 0.006 |
| Parechovirus A, Human parechovirus 1 | 3545 (0.53) | 1986 (0.24) | 2.14 (0 , 4.3) | 0.049 | 0.071 |
| Enterovirus B, Echovirus E16 | 71447 (4.44) | 0 (0) | 1.21 (0 , 2.4) | 0.040 | 0.064 |
*p-value adjusted for multiple comparisons using the Bonferroni method
Richness and alpha diversity
Species richness was highest in watery-consistency stool and decreased consistently as stool consistency became firmer (Spearman’s r = − 0.45, p = 0.013). The median number of distinct viruses was 6.5 (IQR 2.25) for watery stool, 5.5 (IQR 2.5) for loose stool, 4.0 (IQR 3.75) for smooth-consistency stool, 3.0 (IQR 4) for lumpy-consistency stool and 2.0 (IQR 1) for pellet-consistency stool (Fig. 5).
Fig. 5.
Species richness according to mBSFS-C stool consistency category
There was no association between stool consistency and Simpson alpha diversity (Spearman’s r = − 0.05, p = 0.80) or Shannon alpha diversity (Spearman’s r = − 0.08, p = 0.67). With the Fisher alpha diversity metric, loose and watery stool appeared to be more diverse than formed stool, but the difference was not statistically significant (Spearman’s r = − 22, p = 0.238) (Additional file 3: Figure S1).
Discussion
We document differences in the richness and composition of the pediatric enteric virome by stool consistency. There was a marked and statistically significant decrease in the number of distinct virus species as stool consistency became firmer, with the highest number of distinct species in watery (mBSFS-C type 5) stool and the lowest number of distinct species in firm pellet stool (mBSFS-C type 1). Loose and watery stool types were more likely to have norovirus GII, aichivirus A, adeno-associated dependoparvovirus A - AAV2, coxsackievirus A16, human mastadenovirus C, cosavirus A and annelovirus compared to formed stool.
Our findings regarding species richness contrast with many studies in the bacterial microbiome literature which report elevated species richness in healthy individuals and stool. For example, a study of the fecal bacterial microbiome according to self-reported BSFS stool consistency and found that women with looser stool had fewer distinct bacterial species compared to women with firmer stool [13]. Virus richness, unlike bacterial richness, may be a marker for disease rather than health. Stool consistency is often used in symptomatic definitions of diarrhea, in particular, the World Health Organization definition (“three or more loose or watery stools in a 24 h period [2]”). Our findings of elevated species richness in watery and loose stool support the content validity of consistency-based definitions as they relate to infectious episodes of diarrhea.
Noroviruses are the leading cause of epidemic viral gastroenteritis globally [26]. We detected norovirus genotype II and genotype I in a fifth of pools, with genotype II showing the highest differential abundance in loose or watery stools compared to formed stools. Norovirus genotype II has been shown to be elevated in children with diarrhea and or gastroenteritis in many regions, including Africa [27, 28].
The second highest differential abundance we detected was for aichivirus A, which was detected 25% of loose or watery pools and 6% of formed pools. Aichivirus A has been detected in stool samples from children with diarrhea in France [29], Brazil [30], Tunisia [31] and several countries in Asia [32, 33]. We likewise found an elevated abundance of adeno-associated dependoparvovirus A - AAV2 in loose or watery consistency pools compared to formed consistency pools. AAV2 needs a helper virus, commonly an adenovirus, to infect the gastrointestinal tract and it is not thought to be pathogenic or cause diarrheal disease [34]. However, adeno-associated viruses have been detected in children with diarrhea using metagenomic sequencing [35]. Indeed, we detected adenovirus in three of the four pools with detected AAV2 reads.
We also found an increased abundance of coxsackievirus A16 in loose or watery consistency pools compared to formed pools. Coxsackievirus A16 is a leading cause of hand foot and mouth disease. Hand-foot and mouth infections are common in young children and we suspect that age may confound the association between stool consistency and coxsackie A16 abundance. In the median age-adjusted model, the differential abundance of coxsackie A16 in loose or watery stools compared to formed stools decreased to 1.78, and the 95% confidence intervals included the null effect of 1.0.
Enteric human adenoviruses, primarily human mastadenovirus F, are considered the third leading cause of nonbacterial diarrhea globally [27, 36]. We detected human mastadenovirus F in just one watery pool with too few reads to detect a statistically significant difference. We did detect a signal for higher differential abundance of human mastadenovirus C in loose or watery stool compared to formed stool although the p-value was not statistically significant. Studies in Albania, Korea and Asia have all detected human mastadenovirus C in children with diarrhea and gastroenteritis, but these studies lacked healthy controls to compare prevalence [37–39]. A study of human adenovirus in diarrhea children in Tanzania did not find a difference in the prevalence of human mastadenovirus F between children with and without diarrhea but did report a human mastadenovirus C prevalence of 12.5% in diarrhea cases and 7.7% in controls (prevalence ratio of 1.6) [40]. Human mastadenovirus C may be an unrecognized cause of pediatric diarrhea in Africa and warrants additional research with a larger sample size.
Cosavirus A was first identified 2008 and has since been recognized as a cause of diarrhea in children [41–44]. In our sample, Cosavirus A was 3.2 times more abundant in loose or watery stool compared to formed stool. Additional un-pooled studies would help confirm if Cosavirus A may be a cause of loose or watery stools in this population. Annelovirus is a nearly ubiquitous virus in human blood and its presence in the pooled samples may indicate blood in the feces, particularly in loose and watery stool samples [45, 46]. We did not detect any rotavirus, a leading cause of pediatric diarrhea. Ethiopia initiated a country-wide vaccination campaign in November of 2013, reaching a coverage of 85% by 2015 [47]. Successful rotavirus vaccination may explain why no rotavirus was detected in loose, watery or formed stool.
There are several limitations of this study that are important to consider. Our samples were pooled, reducing the effective sample size from 269 individual stool specimens to 29 pools. With a larger sample size (smaller pools) or individually run samples, we would have had more statistical precision for evaluating differences in the fecal virome by measured stool consistency. However, pooling has been shown to be an efficient strategy to accurately estimate prevalence when resources are limited [48–50]. The kappa measuring agreement between the laboratory technicians stool consistency grades was .72, introducing some possible misclassification of stool consistency into our analyses. This misclassification, likely non-differential, would on average bias any associations towards the null. Laboratory staff were not masked to the consistency of the pools when running the analyses. However, the samples were processed in an arbitrary order, not according to stool consistency. This study was conducted at the final study visit of a trial evaluating the effect of a water-improvement intervention on ocular chlamydia. However, we did not find any measurable effect of the water intervention on the enteric virome, as reported in a companion paper [51]. We did not run any of the pools in duplicate to assess repeatability. Another potential limitation is selection bias as only 269 of the 317 eligible children were able to provide a stool sample. However, we found no statistically significant difference in the age or gender of children who provided stool samples compared to those who did not.
Finally, fecal specimens were stored without media, although this should not affect identification of viruses as it would for bacteria.
Despite these limitations, our study had several strengths. Unlike many comparable studies using targeted PCR to detect specific pre-specified viruses, we used a metagenomic approach allowing systematic and unbiased characterization of the stool virome. We applied modern statistical tools developed for microbiome analysis, enabling a comprehensive view of the entire fecal viral community. Our study was also population-based, enrolling a representative sample of all children in the community, both healthy and sick.
Conclusions
We found differences in the pediatric fecal virome by stool sample consistency as measured by the modified Bristol Stool Form Scale for Children (mBSFS-C), both in terms of species richness and composition. Watery and loose stools had greater species richness compared to formed stools and were more abundant in norovirus GII, aichivirus A, AAV2 and human mastadenovirus C.
Additional files
Bioinformatics Pipeline. (PDF 29 kb)
Table S1. Prevalence of species/genotype by mBSFS-C stool consistency category. (PDF 58 kb)
Figure S1. Alpha Diversity according to mBSFS-C stool consistency category. (PDF 19 kb)
Acknowledgements
We gratefully acknowledge the participation of communities, households and individuals in this study. We are thankful for collaboration with The Carter Center of Ethiopia and the Amhara Regional Health Bureau and are very appreciative of the time, effort and commitment of all field teams, drivers, and laboratory technicians.
Funding
This study was supported by the National Institute of Health [NEI U10 EY016214], [NICHD F31 HD088070-01A1 (to KA)], That Man May See and The Sara & Evan Williams Foundation; and Research to Prevent Blindness. National Heart, Lung, and Blood Institute R01 HL105770 grant and Blood Systems Research Institute for further support to EA, TG, PXD, & ED.
Availability of data and materials
The datasets generated and/or analysed during the current study will be available in an open science framework repository {link}. The code used to analyze the data is available in the dollowing github repository: https://github.com/kaiemjoy/fecalvirome.
Abbreviations
- AAV2
Adeno-associated dependoparvovirus A
- BSFS
Bristol Stool Form Scale
- CI
Confidence interval
- IQR
Inter-Quartile Range
- mBSFS-C
Modified Bristol Stool Form Scale for Children
- PCR
Polymerase Chain Reaction
- RT-PCR
Real Time Polymerase Chain Reaction
Authors’ contributions
KA designed the study, analyzed the data and wrote the manuscript. EA, JK and ED contributed significantly to the design and analysis of the study and writing of the manuscript. EA, ED, TP, and XD conducted the laboratory and bioinformatic analysis of the stool specimens. SA and DF were responsible for data collection. SA, DF, MC, ZT & KC coordinated the study implementation. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Ethical committees at the University of California (San Francisco, CA, USA); Emory University (Atlanta, GA, USA); The Food, Medicine and Health Care Administration and Control Authority of Ethiopia; and the Ethiopian Ministry of Science and Technology granted approval for this study. We obtained verbal informed consent in Amharic from all caregivers. The procedures for verbal consent were approved by all ethical committees.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kristen Aiemjoy, Phone: 808.227.7633, Email: kristen.aiemjoy@ucsf.edu.
Eda Altan, Email: ealtan@bloodsystems.org.
Solomon Aragie, Email: Solomon.Aragie@cartercenter.org.
Dionna M. Fry, Email: dionna.fry@ucsf.edu
Tung G. Phan, Email: tuphan@bloodsystems.org
Xutao Deng, Email: xdeng@bloodsystems.org.
Melsew Chanyalew, Email: yeshiwork97@yahoo.com.
Zerihun Tadesse, Email: Zerihun.Tadesse@cartercenter.org.
E. Kelly Callahan, Email: kelly.callahan@cartercenter.org.
Eric Delwart, Email: eric.delwart@ucsf.edu.
Jeremy D. Keenan, Email: jeremy.keenan@ucsf.edu
References
- 1.Longstreth GF, Thompson WG, Chey WD, Houghton LA, Mearin F, Spiller RC. Functional bowel disorders. Gastroenterology. 2006;130(5):1480–1491. doi: 10.1053/j.gastro.2005.11.061. [DOI] [PubMed] [Google Scholar]
- 2.Diarrhea (definition and sequelae) [http://www.who.int/topics/diarrhoea/en/].
- 3.Austin GL, Dalton CB, Hu YM, Morris CB, Hankins J, Weinland SR, Westman EC, Yancy WS, Drossman DA. A very low-carbohydrate diet improves symptoms and quality of life in diarrhea-predominant irritable bowel syndrome. Clin Gastroenterol Hepatol. 2009;7(6):706–708. doi: 10.1016/j.cgh.2009.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tinmouth J, Tomlinson G, Kandel G, Walmsley S, Steinhart HA, Glazier R. Evaluation of stool frequency and stool form as measures of HIV-related diarrhea. HIV Clin Trials. 2015;8(6):421–8. [DOI] [PubMed]
- 5.Shulman RJ, Eakin MN, Czyzewski DI, Jarrett M, Ou CN. Increased gastrointestinal permeability and gut inflammation in children with functional abdominal pain and irritable bowel syndrome. J Pediatr. 2008;153(5):646–650. doi: 10.1016/j.jpeds.2008.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heaton KW, Odonnell LJD. An office guide to whole-gut transit - time patients recollection of their stool form. J Clin Gastroenterol. 1994;19(1):28–30. doi: 10.1097/00004836-199407000-00008. [DOI] [PubMed] [Google Scholar]
- 7.Johnston BC, Shamseer L, da Costa BR, Tsuyuki RT, Vohra S. Measurement issues in trials of pediatric acute diarrheal diseases: a systematic review. Pediatrics. 2010;126(1):e222–e231. doi: 10.1542/peds.2009-3667. [DOI] [PubMed] [Google Scholar]
- 8.Odonnell L, Virjee J, Heaton K. Pseudo-diarrhea in the irritable bowel syndrome-patients records of stool form reflect transit-time while stool frequency does not. In: Gut: 1988: BMJ. 1988;29:1455.
- 9.Lewis SJ, Heaton KW. Stool form scale as a useful guide to intestinal transit time. Scand J Gastroenterol. 1997;32(9):920–924. doi: 10.3109/00365529709011203. [DOI] [PubMed] [Google Scholar]
- 10.Chumpitazi BP, Lane MM, Czyzewski DI, Weidler EM, Swank PR, Shulman RJ. Creation and initial evaluation of a stool form scale for children. J Pediatr. 2010;157(4):594–597. doi: 10.1016/j.jpeds.2010.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gevers D, Knight R, Petrosino JF, Huang K, McGuire AL, Birren BW, Nelson KE, White O, Methé BA, Huttenhower C. The human microbiome project: a community resource for the healthy human microbiome. PLoS Biol. 2012;10(8):e1001377. doi: 10.1371/journal.pbio.1001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326(5960):1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vandeputte D, Falony G, Vieira-Silva S, Tito RY, Joossens M, Raes J. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut. 2015;65(1):57–62. doi: 10.1136/gutjnl-2015-309618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marchesi JR, Adams DH, Fava F, Hermes GD, Hirschfield GM, Hold G, Quraishi MN, Kinross J, Smidt H, Tuohy KM. The gut microbiota and host health: a new clinical frontier. Gut. 2016;65(2):330–339. doi: 10.1136/gutjnl-2015-309990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aiemjoy K, Stoller NE, Gebresillasie S, Shiferaw A, Tadesse Z, Sewnet T, Ayele B, Chanyalew M, Callahan K, Stewart A, et al. ‘If an eye is washed properly, it means it would see clearly’: a mixed methods study of face washing knowledge, attitudes, and behaviors in rural Ethiopia. PLoS Negl Trop Dis. 2016;10(10):e0005099. doi: 10.1371/journal.pntd.0005099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aiemjoy K, Stoller NE, Gebresillasie S, Shiferaw A, Tadesse Z, Sewent T, Ayele B, Chanyalew M, Aragie S, Callahan K. Is using a latrine “a strange thing to do”? A mixed-methods study of sanitation preference and behaviors in rural Ethiopia. Am J Trop Med Hyg. 2016:96(1):65–73. 10.4269/ajtmh.16-0541 [DOI] [PMC free article] [PubMed]
- 18.Aiemjoy KG, Gebresillasie S, Stoller N, Shiferaw A, Tadesse Z, Aragie S, Callahan K, Chanyalew M, Keenan J. Epidemiology of Soil-Transmitted Helminth and Intestinal Protozoa Infections in Pre-School Aged Children in the Amhara region of Ethiopia. Am J Trop Med Hyg. 2017;96(4):866–872. 10.4269/ajtmh.16-0800. [DOI] [PMC free article] [PubMed]
- 19.Gebre T, Ayele B, Zerihun M, Genet A, Stoller NE, Zhou Z, House JI, Yu SN, Ray KJ, Emerson PM, et al. Comparison of annual versus twice-yearly mass azithromycin treatment for hyperendemic trachoma in Ethiopia: a cluster-randomised trial. Lancet. 2012;379(9811):143–151. doi: 10.1016/S0140-6736(11)61515-8. [DOI] [PubMed] [Google Scholar]
- 20.Aiemjoy K, Aragie S, Gebresillasie S, Fry D, Chanyalew M, Dagnew A, Hailu D, Tadesse Z, Callahan K, Freeman M, et al. Defining Diarrhea: A population-based validation study of caregiver-reported stool consistency in the Amhara region of Ethiopia. Am J Trop Med Hyg. 2018; Accepted December 2017(In Press). [DOI] [PMC free article] [PubMed]
- 21.Aiemjoy K, Aragie S, Gebresillasie S, Fry DM, Dagnew A, Hailu D, Chanyalew M, Tadesse Z, Stewart A, Callahan K. Defining diarrhea: a population-based validation study of caregiver-reported stool consistency in the Amhara region of Ethiopia. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phan TG, da Costa AC, Del Valle MJ, Bucardo-Rivera F, Nordgren J, O'Ryan M, Deng X, Delwart E. The fecal virome of south and central American children with diarrhea includes small circular DNA viral genomes of unknown origin. Arch Virol. 2016;161(4):959–966. doi: 10.1007/s00705-016-2756-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L, Deng X, Mee ET, Collot-Teixeira S, Anderson R, Schepelmann S, Minor PD, Delwart E. Comparing viral metagenomics methods using a highly multiplexed human viral pathogens reagent. J Virol Methods. 2015;213:139–146. doi: 10.1016/j.jviromet.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phan TG, Mori D, Deng X, Rajindrajith S, Ranawaka U, Fan Ng TF, Bucardo-Rivera F, Orlandi P, Ahmed K, Delwart E. Small circular single stranded DNA viral genomes in unexplained cases of human encephalitis, diarrhea, and in untreated sewage. Virology. 2015;482:98–104. doi: 10.1016/j.virol.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siebenga JJ, Vennema H, Zheng D-P, Vinjé J, Lee BE, Pang X-L, Ho ECM, Lim W, Choudekar A, Broor S, et al. Norovirus illness is a global problem: emergence and spread of norovirus GII.4 variants, 2001–2007. J Infect Dis. 2009;200(5):802–812. doi: 10.1086/605127. [DOI] [PubMed] [Google Scholar]
- 27.Liu J, Platts-Mills JA, Juma J, Kabir F, Nkeze J, Okoi C, Operario DJ, Uddin J, Ahmed S, Alonso PL, et al. Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: a reanalysis of the GEMS case-control study. Lancet. 2016;388(10051):1291–1301. doi: 10.1016/S0140-6736(16)31529-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ouedraogo N, Kaplon J, Bonkoungou IJ, Traore AS, Pothier P, Barro N, Ambert-Balay K. Prevalence and genetic diversity of enteric viruses in children with diarrhea in Ouagadougou, Burkina Faso. PLoS One. 2016;11(4):e0153652. doi: 10.1371/journal.pone.0153652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ambert-Balay K, Lorrot M, Bon F, Giraudon H, Kaplon J, Wolfer M, Lebon P, Gendrel D, Pothier P. Prevalence and genetic diversity of Aichi virus strains in stool samples from community and hospitalized patients. J Clin Microbiol. 2008;46(4):1252–1258. doi: 10.1128/JCM.02140-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oh D-Y, Silva P, Hauroeder B, Diedrich S, Cardoso D, Schreier E. Molecular characterization of the first Aichi viruses isolated in Europe and in South America. Arch Virol. 2006;151(6):1199–1206. doi: 10.1007/s00705-005-0706-7. [DOI] [PubMed] [Google Scholar]
- 31.Sdiri-Loulizi K, Hassine M, Gharbi-Khelifi H, Sakly N, Chouchane S, Guediche MN, Pothier P, Aouni M, Ambert-Balay K. Detection and genomic characterization of Aichi viruses in stool samples from children in Monastir, Tunisia. J Clin Microbiol. 2009;47(7):2275–2278. doi: 10.1128/JCM.00913-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pham NTK, Khamrin P, Nguyen TA, Kanti DS, Phan TG, Okitsu S, Ushijima H. Isolation and molecular characterization of Aichi viruses from fecal specimens collected in Japan, Bangladesh, Thailand, and Vietnam. J Clin Microbiol. 2007;45(7):2287–2288. doi: 10.1128/JCM.00525-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamashita T, Sakae K, Kobayashi S, Ishihara Y, Miyake T, Mubina A, Isomura S. Isolation of cytopathic small round virus (Aichi virus) from Pakistani children and Japanese travelers from Southeast Asia. Microbiol Immunol. 1995;39(6):433–435. doi: 10.1111/j.1348-0421.1995.tb02225.x. [DOI] [PubMed] [Google Scholar]
- 34.Schmaljohn C, Fields S. Fields virology. 5. Philadelphia: Lippincott Williams & Wilkins; 2007. Virology; pp. 1741–1789. [Google Scholar]
- 35.Finkbeiner SR, Allred AF, Tarr PI, Klein EJ, Kirkwood CD, Wang D. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog. 2008;4(2):e1000011. doi: 10.1371/journal.ppat.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chhabra P, Payne DC, Szilagyi PG, Edwards KM, Staat MA, Shirley SH, Wikswo M, Nix WA, Lu X, Parashar UD, et al. Etiology of viral gastroenteritis in children <5 years of age in the United States, 2008-2009. J Infect Dis. 2013;208(5):790–800. doi: 10.1093/infdis/jit254. [DOI] [PubMed] [Google Scholar]
- 37.La Rosa G, Della Libera S, Petricca S, Iaconelli M, Donia D, Saccucci P, Cenko F, Xhelilaj G, Divizia M. Genetic diversity of human adenovirus in children with acute gastroenteritis, Albania, 2013–2015. Biomed Res Int. 2015;2015:142912. doi: 10.1155/2015/627201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee JI, Lee GC, Chung JY, Han TH, Lee YK, Kim MS, Lee CH. Detection and molecular characterization of adenoviruses in Korean children hospitalized with acute gastroenteritis. Microbiol Immunol. 2012;56(8):523–528. doi: 10.1111/j.1348-0421.2012.00469.x. [DOI] [PubMed] [Google Scholar]
- 39.Li L, Phan TG, Nguyen TA, Kim KS, Seo JK, Shimizu H, Suzuki E, Okitsu S, Ushijima H. Molecular epidemiology of adenovirus infection among pediatric population with diarrhea in Asia. Microbiol Immunol. 2005;49(2):121–128. doi: 10.1111/j.1348-0421.2005.tb03711.x. [DOI] [PubMed] [Google Scholar]
- 40.Moyo SJ, Hanevik K, Blomberg B, Kommedal O, Nordbø SA, Maselle S, Langeland N. Prevalence and molecular characterisation of human adenovirus in diarrhoeic children in Tanzania; a case control study. BMC Infect Dis. 2014;14:666. doi: 10.1186/s12879-014-0666-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holtz LR, Finkbeiner SR, Kirkwood CD, Wang D. Identification of a novel picornavirus related to cosaviruses in a child with acute diarrhea. Virol J. 2008;5(1):159. doi: 10.1186/1743-422X-5-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ayouni S, Estienney M, Hammami S, Guediche MN, Pothier P, Aouni M, Belliot G, De Rougemont A. Cosavirus, Salivirus and Bufavirus in diarrheal Tunisian infants. PLoS One. 2016;11(9):e0162255. doi: 10.1371/journal.pone.0162255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kapoor A, Victoria J, Simmonds P, Slikas E, Chieochansin T, Naeem A, Shaukat S, Sharif S, Alam MM, Angez M. A highly prevalent and genetically diversified Picornaviridae genus in south Asian children. Proc Natl Acad Sci. 2008;105(51):20482–20487. doi: 10.1073/pnas.0807979105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kapusinszky B, Phan TG, Kapoor A, Delwart E. Genetic diversity of the genus Cosavirus in the family Picornaviridae: a new species, recombination, and 26 new genotypes. PLoS One. 2012;7(5):e36685. doi: 10.1371/journal.pone.0036685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kapusinszky B, Minor P, Delwart E. Nearly constant shedding of diverse enteric viruses by two healthy infants. J Clin Microbiol. 2012;50(11):3427–3434. doi: 10.1128/JCM.01589-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun G, Zang Q, Gu Y, Niu G, Ding C, Zhang P. Viral metagenomics analysis of picobirnavirus-positive feces from children with sporadic diarrhea in China. Arch Virol. 2016;161(4):971–975. doi: 10.1007/s00705-015-2726-2. [DOI] [PubMed] [Google Scholar]
- 47.Weldegebriel G, Mwenda JM, Chakauya J, Daniel F, Masresha B, Parashar UD, Tate JE. Impact of rotavirus vaccine on rotavirus diarrhoea in countries of East and Southern Africa. Vaccine. 2017;36(47):7124–7130. 10.1016/j.vaccine.2017.10.050. [DOI] [PubMed]
- 48.Diamant J, Benis R, Schachter J, Moncada J, Pang F, Jha HC, Bhatta RC, Porco T, Lietman T. Pooling of chlamydia laboratory tests to determine the prevalence of ocular chlamydia trachomatis infection. Ophthalmic Epidemiol. 2001;8(2–3):109–117. doi: 10.1076/opep.8.2.109.4156. [DOI] [PubMed] [Google Scholar]
- 49.Sacks JM, Bolin SR, Crowder SV. Prevalence estimation from pooled samples. Am J Vet Res. 1989;50(2):205–206. [PubMed] [Google Scholar]
- 50.Sinha I, Jones L, Smyth RL, Williamson PR. A systematic review of studies that aim to determine which outcomes to measure in clinical trials in children. PLoS Med. 2008;5(4):569–578. doi: 10.1371/journal.pmed.0050096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altan E, Aiemjoy K, Phan TG, Deng X, Aragie S, Tadesse Z, Callahan KE, Keenan J, Delwart E. Enteric virome of Ethiopian children participating in a clean water intervention trial. PLoS One. 2018;13(8):e0202054. doi: 10.1371/journal.pone.0202054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bioinformatics Pipeline. (PDF 29 kb)
Table S1. Prevalence of species/genotype by mBSFS-C stool consistency category. (PDF 58 kb)
Figure S1. Alpha Diversity according to mBSFS-C stool consistency category. (PDF 19 kb)
Data Availability Statement
The datasets generated and/or analysed during the current study will be available in an open science framework repository {link}. The code used to analyze the data is available in the dollowing github repository: https://github.com/kaiemjoy/fecalvirome.