Abstract
Background
Toxocara canis is quite closely related to Ascaris suum but its biology is more complex, involving a phase of arrested development (diapause or hypobiosis) in tissues as well as transplacental and transmammary transmission routes. In the present study, we explored and compared dauer-like signalling pathways of T. canis and A. suum to infer which components in these pathways might associate with, or regulate, this added complexity in T. canis.
Methods
Guided by information for Caenorhabditis elegans, we bioinformatically inferred and compared components of dauer-like signalling pathways in T. canis and A. suum using genomic and transcriptomic data sets. In these two ascaridoids, we also explored endogenous dafachronic acids (DAs), which are known to be critical in regulating larval developmental processes in C. elegans and other nematodes, by liquid chromatography-mass spectrometry (LC-MS).
Results
Orthologues of C. elegans dauer signalling genes were identified in T. canis (n = 55) and A. suum (n = 51), inferring the presence of a dauer-like signalling pathway in both species. Comparisons showed clear differences between C. elegans and these ascaridoids as well as between T. canis and A. suum, particularly in the transforming growth factor-β (TGF-β) and insulin-like signalling pathways. Specifically, in both A. suum and T. canis, there was a paucity of genes encoding SMAD transcription factor-related protein (daf-3, daf-5, daf-8 and daf-14) and insulin/insulin-like peptide (daf-28, ins-4, ins-6 and ins-7) homologues, suggesting an evolution and adaptation of the signalling pathway in these parasites. In T. canis, there were more orthologues coding for homologues of antagonist insulin-like peptides (Tc-ins-1 and Tc-ins-18), an insulin receptor substrate (Tc-ist-1) and a serine/threonine kinase (Tc-akt-1) than in A. suum, suggesting potentiated functional roles for these molecules in regulating larval diapause and reactivation. A relatively conserved machinery was proposed for DA synthesis in the two ascaridoids, and endogenous Δ4- and Δ7-DAs were detected in them by LC-MS analysis. Differential transcription analysis between T. canis and A. suum suggests that ins-17 and ins-18 homologues are specifically involved in regulating development and migration in T. canis larvae in host tissues.
Conclusion
The findings of this study provide a basis for functional explorations of insulin-like peptides, signalling hormones (i.e. DAs) and related nuclear receptors, proposed to link to development and/or parasite-host interactions in T. canis. Elucidating the functional roles of these molecules might contribute to the discovery of novel anthelmintic targets in ascaridoids.
Electronic supplementary material
The online version of this article (10.1186/s13071-018-3265-y) contains supplementary material, which is available to authorized users.
Keywords: Toxocara canis, Ascaris suum, Dauer signalling pathway, Dafachronic acid, Arrested development
Background
Toxocara canis is an important pathogen of both animal and human health importance worldwide [1]. This parasite, which is related to other ascaridoid nematodes such as Ascaris spp., can be directly transmitted to the human host via a faecal-oral route, and can cause toxocariasis and complications such as neurological and allergic diseases [1].
The biology of T. canis is complex and involves canids (e.g. dogs, wolves and foxes) as definitive hosts, paratenic hosts such as rodents, and accidental hosts including humans [2, 3]. The eggs of this parasite are expelled in the faeces from canids, embryonate and become infective in the environment. Following the ingestion of infective eggs by the canid host, infective, third-stage larvae (L3s) emerge from the eggs, penetrate the intestinal wall and migrate to the liver and lungs (hepato-pulmonary migration). In young dogs (< 12 weeks), larvae migrate to the airways and then get swallowed and make their way to the small intestine, where fourth-stage larvae (L4s) develop to adult worms (female and male), mate and reproduce. In dogs of ≥ 12 weeks, oral infection can occur, but larvae tend to encyst in various tissues (including muscles, brain and nerves), where they undergo hypobiosis (i.e. arrested development or diapause). In female dogs, encysted larvae become activated (in the last trimester of pregnancy), most of which undergo transplacental transmission to the foetuses in utero (~99%), and a minority of which (~1%) undergo transmammary (transcolostoral) transmission to newborn pups [4]. Adult worms eventually establish in the small intestine of pups, and reproductively-active female worms are a major source of egg contamination. Toxocara canis can also be transmitted to accidental (paratenic) hosts (e.g. rats, mice and rabbits) via accidental ingestion of infective eggs; in such hosts, larvae hatch from the eggs, penetrate the intestinal wall and then migrate to various organs and tissues, where they cause disease and/or encyst. If a canid eats infected tissues (containing larvae) from such a paratenic host, adult worms can develop in the small intestine [2, 3].
Humans are accidental hosts. They can become infected by ingesting infective eggs from contaminated soil, food or water, or larvae in tissues from infected paratenic hosts [1]. Following ingestion, infective larvae are released and invade the intestinal wall, and are then carried via the blood circulation to various tissues (including liver, lungs, muscles, central nervous system). Although these larvae undergo arrested development in these tissues, during prior migration they frequently cause pathogenic effects due to associated mechanical damage, inflammatory responses and granuloma formation, leading to the disease toxocariasis, of which there are four main clinical forms (visceral larva migrans, ocular larva migrans, neurotoxocariasis and covert toxocariasis) [1, 5]. Such larvae in tissues have also been implicated in neurodegenerative disorders (e.g. epilepsy, idiopathic Parkinson’s disease and dementia) and in allergic diseases (e.g. asthma and pruritus) [6–10].
Interestingly, T. canis has a marked tropism for the central nervous system (cerebrum) [11]. Larvae that migrate through the brain downregulate lipid/cholesterol biosynthesis [12]. There is evidence that prolactin (pituitary hormone) plays a role in activating arrested larvae of T. canis [13], and some studies have implicated this hormone in regulating larval growth and motility in T. canis, infection intensity and host immune responses [14, 15]. The information from these studies indicates that lipid or hormone signalling plays critical roles in the migration, diapause and host interplay of T. canis larvae. Roles of similar signalling pathways in regulating developmental processes have been described for the free-living nematode Caenorhabditis elegans [16–18]. Specifically, steroid hormone signalling, particularly the dafachronic acid-DAF-12 module, has been recognised as a ‘check-point’ for diapause (dauer) in this worm [19–22], and has been proposed to relate to an endocrine mechanism which appears to be conserved between free-living and parasitic nematodes [23–26]. However, little is known about the signalling pathways in parasitic nematodes and the differences in such pathways between Toxocara and other ascaridoids such as A. suum [27], which, unlike T. canis, does not undergo transplacental or transmammary transmission [28, 29]. Profound knowledge of dauer and associated signalling pathways in C. elegans enables comparative studies in parasitic nematodes utilising transcriptomic and genomic data sets and tools [30, 31], in the absence of functional genomic data. Here, guided by information and data for C. elegans, we inferred dauer-like signalling pathways of T. canis and A. suum, and identified unique components that we hypothesise regulate the development and/or other molecular processes in T. canis in the mammalian host.
Methods
Draft genomes and transcriptomes
Genome assemblies and annotations for C. elegans (BioProject PRJNA13758), T. canis (BioProjects PRJEB533 and PRJNA248777) and A. suum (BioProjects PRJNA80881 and PRJNA62057) were obtained from WormBase [32] and ParaSite at WormBase [33]. Transcriptomic data sets of T. canis and A. suum (BioProjects PRJNA248777 and PRJNA80881) were taken from NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) [34–37]. New versions of the transcriptomes of T. canis and A. suum were assembled de novo using the program Trinity v.2.4.0 [38, 39].
Identification of dauer signalling gene homologues
Genes (n = 107) representing the canonical dauer signalling pathway [i.e. cyclic guanosine monophosphate (cGMP), transforming growth factor-β (TGF-β), insulin/insulin-like growth factor (IGF), and steroid hormone signalling pathways] in C. elegans were available from published information [19, 27, 40]. Protein sequences (n = 182) and functional information were obtained from WormBase (WS261) [41]. Homologues were predicted by exhaustive homology searching of C. elegans protein sequences (using BLAT v.35 and tblastn v.2.5.1) against the genome and transcriptome assemblies of T. canis and A. suum. In addition, Pfam, PANTHER and SUPERFAMILY conserved domain architectures (using InterProScan v.5.15.54) [42, 43] of the dauer signalling gene products were used to predict homologues from the original gene predictions and de novo-assembled transcripts of these two ascaridoid species, using an established approach [44]. Predicted transcripts of potential homologues were matched (using blastx v.2.5.1, e-value: ≤ 10-5) to proteins of C. elegans (BioProject PRJNA13758.WS261) to verify their identity.
Curation of genes and classification of orthologues
Homologous sequences were manually curated using a recently described workflow [44]. In brief, identified gene and transcript sequences were mapped to the genome assemblies of each T. canis and A. suum using the program BLAT v.35 [45]. Transcripts that mapped to the same coding region were re-assembled using the program CAP3 for possible extensions [46]. Full-length transcripts were used to refine the corresponding gene models using the program Exonerate v.2.2.0 [47]. Gene products were predicted from the curated coding DNA sequences (CDSs) using ORFfinder [48]. Gene orthologues were classified according to groups inferred by OrthoMCL (e-value: ≤ 10-5; sequence similarity: ≥ 30%) [49].
Comparative analyses
Sequence similarities of the classified orthologues were compared by pairwise alignment using EMBOSS Needle (the Needleman-Wunsch algorithm) [50]. Domain architectures of inferred proteins were assigned using InterProScan v.5.15.54. Specifically, homologues of genes encoding insulin-like peptides were identified based on their sequence domain signatures. The inferred protein sequences were compared with the insulin/insulin-like peptides of C. elegans using blastp (e-value ≤ 10-5) to infer their identity. For inferred insulin-like peptides, conserved patterns and motifs were searched using the programs Pratt v.2.1 [51] and MEME v.5.0.2 [52]. The relationships of insulin-like peptides were verified manually using OrthoMCL (e-value: ≤ 10-5; sequence similarity: ≥ 50%).
Transcriptional analysis
Available RNA-seq reads from egg, and first- (L1), second- (L2), third- (L3, recovered from eggs, liver and lungs) and fourth-stage larvae and/or adults of T. canis and of A. suum [34–36] were used for transcriptional analyses and comparisons. In brief, paired-end reads were mapped to individual curated CDSs using Bowtie2 v.2.1.0 within the software package RSEM v.1.2.11 [53, 54]. Levels of messenger RNA (mRNA) transcription were recorded in transcripts per million (TPM). For individual developmental stages, transcription profiles for individual orthologues were displayed in a heat-map using the program heatmap.2 (in R v.3.5.1).
Liquid chromatography-mass spectrometry (LC-MS)
For each T. canis and A. suum, lipids were extracted from four individual male and four individual female adults using an established lipid extraction method [55, 56]. In brief, individual samples (1 mg dry weight; 4 replicates) were suspended in ice cold 40% methanol and homogenised using zirconium oxide beads (ZROB05, Next Advance, USA). A chloroform:methanol (2:1) mix was used to separate the aqueous and organic phases by centrifugation at 10,000× g for 10 min at room temperature (24 °C). The organic phase was retained, dried and resuspended in methanol for subsequent mass spectrometric analysis using an Orbitrap Fusion Lumos mass spectrometer coupled to an Ultimate 3000 UHPLC (Thermo Fisher Scientific, San Jose, CA, USA). Commercially available dafachronic acids (25S)-Δ7-DA and (25S)-Δ4-DA (exact mass: 413.3061) (cat no. 23017-97-2; Cayman Chemical Company) were used as reference standards for the identification of endogenous DAs.
Results
Dauer signalling orthologues
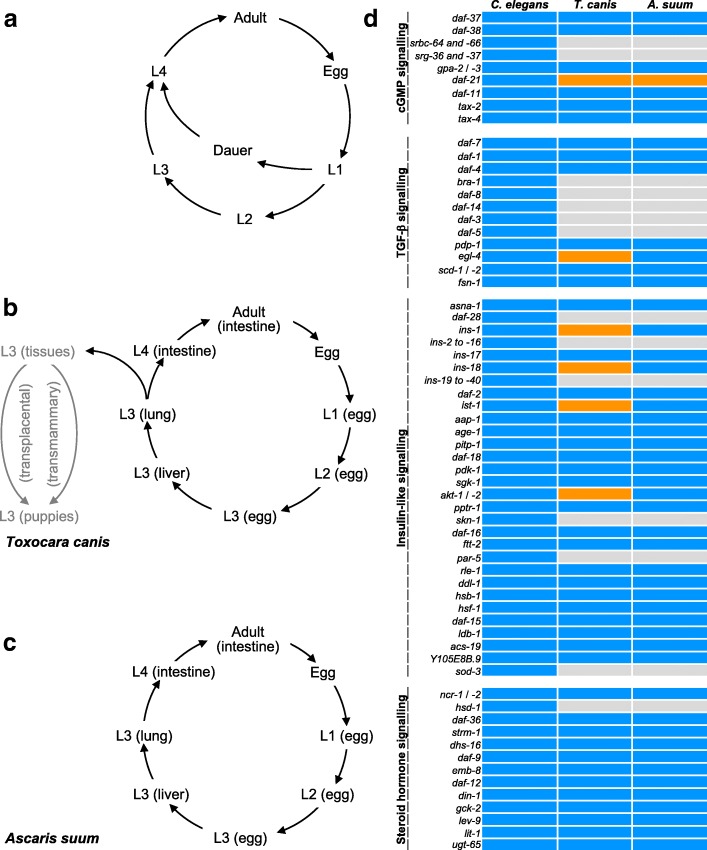
Based on the information available for C. elegans, we identified 55 and 51 orthologues encoding signalling molecules in T. canis and A. suum, respectively (Additional file 1: Tables S1-S4). These numbers are markedly lower than for C. elegans (n = 107) and relate mainly to less TGF-β and insulin/insulin-like signalling components in the ascaridoids. Specifically, orthologues inferred to represent SMAD transcription factors (daf-3, daf-8 and daf-14), SKI family transcriptional co-repressor (daf-5), insulin (daf-28), insulin-like peptides (e.g. ins-4, ins-6 and ins-7), serine/threonine-protein kinase (akt-2), bZip transcription factor (skn-1), 14-3-3 protein (par-5) and iron/manganese superoxide dismutase (sod-3) were not detected in either T. canis or A. suum, whereas two orthologues encoding heat-shock protein 90 (daf-21) were identified in both species (Fig. 1d). A comparison indicated more orthologues coding for a cGMP-dependent protein kinase, insulin-like peptides, an insulin receptor substrate and a serine/threonine-protein kinase (egl-4, ins-1, ins-18, ist-1 and akt-1) in T. canis compared with A. suum (see Fig. 1d). More transcript isoforms were predicted for T. canis orthologues, such as Tc-akt-1 (n = 5) and Tc-daf-12 (n = 18), than for A. suum (see Additional file 1: Tables S3 and S4).
Fig. 1.
Life-cycle and dauer signalling orthologues of Caenorhabditis elegans, Toxocara canis and Ascaris suum. Schematic representation of the life-cycles of C. elegans (a), T. canis (b) and A. suum (c). Specifically, the third-stage larvae (L3s) of T. canis can arrest in the tissues for years, and can be reactivated in the female dog (bitch) in the last trimester of pregnancy and then migrate to uterus or mammary glands (post partum), leading to transplacental or transmammary transmission to offspring (grey arrows). d Dauer signalling gene orthologues in T. canis and A. suum are indicated (in blue) and compared to C. elegans genes. Increased numbers of orthologues are identified in T. canis and/or A. suum (orange). Orthologues not inferred for T. canis and/or A. suum are indicated in grey
Insulin-like peptide-coding genes and their relationships
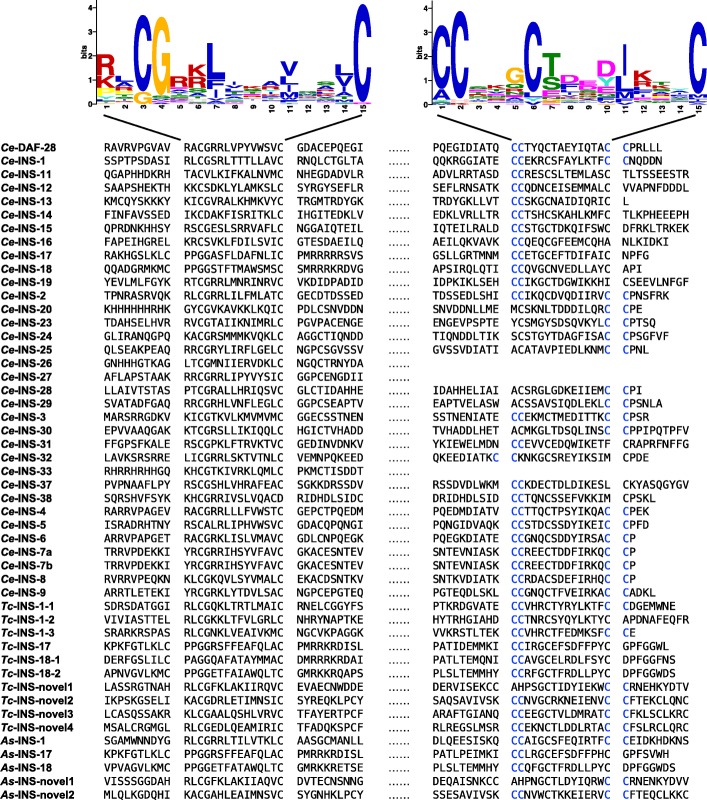
We identified 10 and 5 sequences encoding signatures characteristic of the insulin-like superfamily in T. canis and A. suum, respectively (Additional file 1: Table S5). Although marked sequence diversity (23-77%) was seen among the inferred insulin-like peptides of C. elegans, T. canis and A. suum (Additional file 1: Table S5), two characteristic motifs (RLCGRKLIKAVQSLC and CCSKGCTDEDJKKYC; P-value: ≤ 10-5) and one conserved sequence pattern (C-C; fitness = 8.34) were discovered (Fig. 2). Specifically, the proteins inferred for T. canis and A. suum had significant sequence similarity (blastp e-value cut-off: ≤ 10-5) to Ce-INS-1, Ce-INS-12, Ce-INS-17, Ce-INS-18 or Ce-INS-32 (Additional file 1: Table S5). Relationships among the insulin-like peptides for these species were supported by their orthologous groups (n = 12; e-value: ≤ 10-5; sequence similarity: ≥ 50%) (Additional file 1: Table S5). Apart from the orthologues inferred (i.e. INS-1, INS-17 and INS-18), one more homologue of Ce-ins-1 was inferred for T. canis, and novel insulin-like peptides were predicted for T. canis (n = 4) and A. suum (n = 2) (Fig. 2; Additional file 1: Table S5).
Fig. 2.
Motifs and conserved pattern of insulin-like peptides inferred for Caenorhabditis elegans, Toxocara canis and Ascaris suum. Motifs RLCGRKLIKAVQSLC and CCSKGCTDEDJKKYC in the inferred protein sequences are aligned and indicated in sequence logos. The conserved pattern C-C in these sequences is indicated in blue
Dafachronic acid biosynthesis machinery
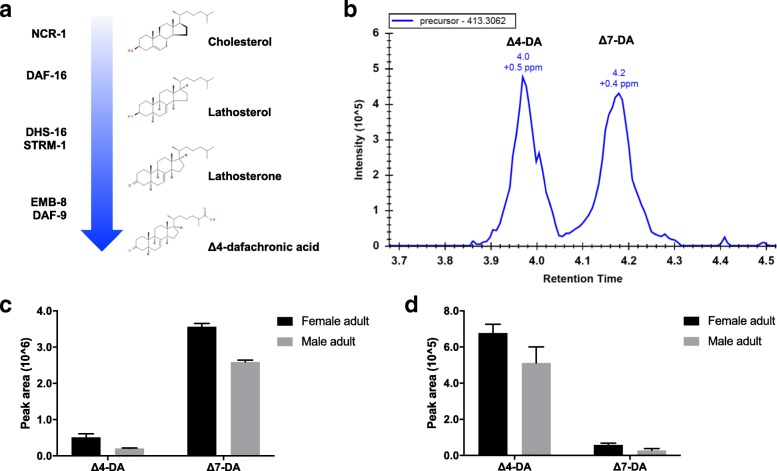
We inferred 6 orthologues to be involved in the biosynthesis of DAs. Specifically, the identification of orthologues ncr-1, daf-36, dhs-16, strm-1, emb-8 and daf-9 indicated a relatively conserved biosynthetic pathway for DA (i.e. Δ7) among C. elegans, T. canis and A. suum (Fig. 3a), although the orthologue of Ce-hsd-1 was not identified in either T. canis or A. suum.
Fig. 3.
Dafachronic acids (DAs) in Toxocara canis and Ascaris suum. a Proposed biosynthetic pathway of Δ7-DA in T. canis and A. suum, which involves NCR-1, DAF-36, DHS-16, STRM-1, EMB-8 and DAF-9. b Using Δ4- and Δ7-DAs (exact mass = 413.3061) as standards, endogenous DAs, with mass errors estimated at 0.5 and 0.4 parts per million (ppm), are present in the adults (female and male) of T. canis and A. suum, respectively. c and d Relative abundances of Δ4- and Δ7-DAs in the adult (female and male) stages of T. canis and A. suum, respectively
Endogenous DAs (Δ4 and Δ7) were detected by LC-MS in the female and male adults of T. canis and A. suum, with mass errors estimated at 0.5 and 0.4 parts per million (ppm), respectively (Fig. 3b). Specifically, Δ7-DA was dominant in T. canis, whereas Δ4-DA was in A. suum (Fig. 3). For both species, the relative abundance of DAs in female adults was higher than in male adults (P > 0.05) (Fig. 3c).
Developmental transcription of dauer signalling components
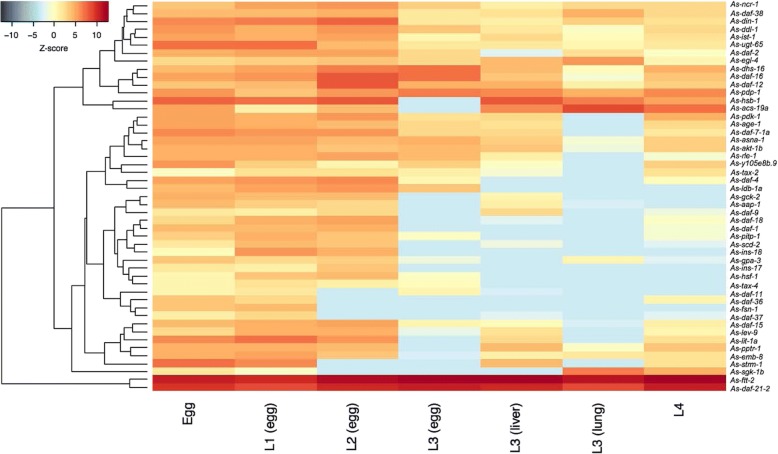
We compared levels of mRNA transcription of individual dauer signalling orthologues in L3 and adult (female and male) stages of T. canis (Additional file 1: Table S6). For some orthologues, including Tc-gpa-3, Tc-daf-11 and Tc-tax-4 (cGMP signalling), Tc-asna-1 and Tc-ins-18 (insulin-like signalling), and Tc-daf-9 (steroid hormone signalling), transcription was higher in the L3 than the adult stage (Additional file 1: Table S6). We also compared mRNA transcription levels of individual dauer signalling orthologues in 7 distinct developmental stages of A. suum (Additional file 1: Table S6). Although similar transcriptional profiles were observed in egg and L1 stages, marked variation was seen among L2, L3 (recovered from eggs, liver and lungs) and L4 stages of A. suum, particularly for the genes As-gpa-3, As-daf-11 and As-tax-4 (cGMP signalling), As-asna-1 and As-daf-2 (insulin-like signalling), and As-daf-9 (steroid hormone signalling) (Fig. 4). A comparative analysis showed high levels of transcription for the orthologues daf-21 and ftt-2 in the stages of T. canis and A. suum studied, and low levels for ins-17 and ins-18 in the latter species (Additional file 1: Table S6).
Fig. 4.
Transcriptional profiles of dauer signalling genes in different developmental stages of Ascaris suum. Transcriptional levels (transcripts per million, TPM) of dauer signalling orthologues in egg, the first- and second-stage larvae recovered from eggs; third-stage larvae from eggs, host liver or lung, and the fourth-stage larvae of A. suum are indicated in the heat map. Colour scales and Z-scores indicate scaled TPMs
Discussion
Based on current genomic and transcriptomic datasets, we identified orthologues of the dauer signalling genes in T. canis and compared them with those of A. suum using a bioinformatic strategy for gene curation and classification. We compared these components, proposed the biosynthetic pathways of DA and investigated developmental transcription of identified orthologues.
The identification of key dauer-associated signalling gene orthologues implies dauer-like signalling pathways in T. canis and A. suum. Although dauer signalling gene homologues had been identified previously in a range of parasitic nematodes, including A. suum and T. canis [27], orthologues were defined here using an effective approach for curation and classification [44]. Specifically, the sequence re-assembly strategy used increased the likelihood of identification. For instance, we identified orthologues of Ce-ins-1 and Ce-dhs-16 in both T. canis and A. suum, which were not reported previously [27]. In addition, the present sequence-based classification strategy improved the accuracy of annotation. Although Ce-bra-1 homologues had been identified [27], the orthologue of this gene was not identified herein in T. canis or A. suum. Specifically, orthologues of Ce-ftt-2 and Ce-par-5 (encoding 14-3-3 proteins) were classified based on reciprocal BLAST searches and OrthoMCL grouping. Thus, the accurate identification and classification of orthologues provided a basis for reliable comparative studies.
The present findings indicated a divergent evolution and adaptation of the dauer-like signalling pathway among the three nematodes studied. Compared with C. elegans, the reduced number of genes coding for SMAD-related transcription factors and insulin/insulin-like peptides in T. canis and A. suum suggest divergences in both TGF-β and insulin-like signalling between the free-living and the two parasitic nematodes. Specifically, orthologues of Ce-daf-3, Ce-daf-5, Ce-daf-8 and Ce-daf-14 were not detected in T. canis or A. suum, suggesting a uniqueness in TGF-β signalling in these two species (clade III), as suggested previously by other workers for other parasitic nematodes such as Trichinella spiralis and Trichuris suis (clade I) [27, 57]. In addition, apart from ins-1, ins-17 and ins-18, no orthologue of any of the other insulin/insulin-like peptide-coding genes (n = 37) of C. elegans (clade V) was found, implying a contraction of the gene family representing these signalling molecules in the two ascaridoids (clade III) studied here. Reduced numbers of genes encoding insulin-like peptides have been reported also in other parasitic nematodes, including Strongyloides stercoralis (clade IV) [40].
Distinctiveness in signalling between T. canis and A. suum might relate to differences in biology and/or developmental regulation. Although the dauer-like signalling pathways of T. canis and A. suum were similar, the increased number of orthologues in the former species indicates a distinction in TGF-β and insulin-like signalling (Fig. 1d) which might associate with entry into and exit from arrested larval development in T. canis. First, the three Tc-ins-1 and two Tc-ins-18 paralogues encode antagonist insulin-like peptides, which are recognised to function in promoting dauer formation in C. elegans [58, 59]. The novel insulin-like peptides (n = 4; Tc-ins-1n to Tc-ins-4n; n = novel) predicted for T. canis might also be involved in regulating larval development, but this proposal needs to be assessed. Secondly, the two Tc-ist-1 paralogues likely encode insulin receptor substrates, which function together with phosphoinositide 3-kinase (PI3K) adaptor/regulatory subunit to potentiate dauer-associated daf-2/insulin-like signalling, as known for C. elegans [60]. Thirdly, the two Tc-akt-1 orthologues encode serine/threonine kinases Akt/protein kinase B, which play a role in antagonising the fork head transcription factor DAF-16 when activated by phospholipid products from PI3K [61]. Therefore, compared with A. suum, it appears that T. canis produces more antagonist molecules (INS-1 and INS-18) which may trigger a stronger inhibitive effect on the fork head transcription factor DAF-16 (via phosphorylation of a serine/threonine kinase) to enable arrested development, particularly when migrating through and/or encysting in host tissues. In addition, the relatively high numbers of transcript isoforms for Tc-akt-1 (n = 5) and Tc-daf-12 (n = 18) might indicate rapid adaptive expression and/or plastic functional roles, similar to those of daf-16 reported for C. elegans [62]. However, although two Tc-egl-4 paralogues were identified in T. canis, their functions are unclear, because orthologues coding for SMAD-related transcription factors [63] were not identified in this nematode. Clearly, detailed functional explorations of these molecules need to be undertaken to explain their roles in the development, migration and/or hypobiosis of T. canis larvae. Significant progress has been made through the development of a functional genomics platform for A. suum [64], which suggests that a similar system might be established for T. canis, in order to explore the functional roles of signalling molecules in this species.
To investigate dauer-like signalling pathway components in T. canis and A. suum, we also analysed transcription profiles across developmental stages, employing RNA-sequence data for pooled worms. The consistently high messenger RNA levels of daf-21 and ftt-2 suggest key roles for these genes and their products in larval development, supported, to some extent, by information for C. elegans [19]. By contrast, variable transcription levels for the genes gpa-3, daf-11 and tax-4 (cGMP signalling), asna-1 (insulin-like signalling) and daf-9 (steroid hormone signalling) among developmental stages, particularly L3 stages in eggs, liver and lung, might suggest plastic, but crucial roles in regulating developmental processes and/or host-parasite interactions. In addition, differential transcriptional levels of the genes ins-17 and ins-18 in the L3 stage between T. canis and A. suum suggest a distinctive signalling mechanism in the host animal, since Ce-ins-17 and Ce-ins-18 promote dauer formation in C. elegans [59, 65]. This difference might suggest unique roles for selected insulin-like peptides in regulating arrested development in host tissues or parasite-host interactions [57, 66]. This hypothesis warrants testing through large-scale, integrative ‘omic investigations.
Comparisons with C. elegans implied relative conservation in the biosynthetic machinery for DA between T. canis and A. suum. The orthologues of Ce-ncr-1, Ce-daf-36, Ce-dhs-16, Ce-emb-8 and Ce-daf-9 likely function in the trafficking, catalysation and modification of cholesterol destined for the biosynthesis of DAs, known to be critical for regulating larval development in C. elegans and S. stercoralis [16, 67–69]. The biosynthetic machinery proposed for ascaridoids is supported by the identification of endogenous Δ4- and Δ7-DAs in T. canis and A. suum. Interestingly, although Δ4- and Δ7-DAs are isomers, differential abundance in the adult stages of T. canis and A. suum might suggest functional distinctions in specific biological processes between these species. Based on the information for C. elegans and some parasitic nematodes [40, 70], the endogenous biosynthesis of Δ7-DA might be regulated by the strm-1 gene in A. suum. Although similar components of the DA synthesis were predicted for both T. canis and A. suum, the functionality of this machinery in these species needs to be shown. By contrast, an orthologue of Ce-hsd-1, which likely functions in the biosynthesis of other DAs, was not detected in either T. canis or A. suum [71], suggesting that both of these ascaridoid nematodes have a simplified machinery to synthesise these signalling hormones. Although it seems that there is a relatively conserved steroid hormone signalling module in these ascaridoids, future work should focus on verifying the biosynthesis signalling module and the functional roles of DAs in these worms. Specifically, although it has been reported that prolactin plays a role in re-activating arrested larvae of T. canis [13], the proposed role for DAs (“bile acid-like steroids”) in larval reactivation (relating particularly to ensuing transplacental and/or transmammary transmission) and their associated signalling pathway(s) remain to be explored [21, 68, 72]. A better understanding of these areas, particularly signalling hormones and their nuclear receptors, would provide insight into regulatory processes in larval development and might enable the discovery of new anthelmintic targets [22, 73, 74].
Conclusions
The present study reveals distinctiveness in the TGF-β and insulin-like signalling pathways among C. elegans, T. canis and A. suum, but indicates similarity in the steroid hormone signalling pathway between the two ascaridoid nematodes. Inferring the elements of the dauer-like signalling pathway in these ascaridoids provides a basis for future explorations of the functional roles of these elements in each species using a reliable functional genomics platform. Understanding these processes could enlighten developmental processes and host-parasite interactions for these ascaridoids and facilitate the discovery of novel intervention strategies against the diseases that these parasites cause.
Additional file
Table S1. Dauer signalling gene homologues of Toxocara canis and Ascaris suum. Table S2. Orthologous groups of dauer signalling genes in Toxocara canis and Ascaris suum inferred from Caenorhabditis elegans. Table S3. Salient information on dauer signalling genes of Toxocara canis predicted from Caenorhabditis elegans orthologues. Table S4. Salient information on dauer signalling genes of Ascaris suum predicted from Caenorhabditis elegans orthologues. Table S5. Classification of insulin-like peptide homologues in Toxocara canis and Ascaris suum. Table S6. Transcription of dauer-like signalling gene orthologues in key developmental stages of Toxocara canis and Ascaris suum. (XLSX 418 kb)
Acknowledgements
We would like to thank Professor David Mangelsdorf and Dr Zhu Wang (Department of Pharmacology, University of Texas Southwestern Medical Center, USA) for providing dafachronic acid, and Miss Haoyun Fang (Department of Biochemistry and Molecular Biology, The University of Melbourne, Australia) for her kind support with lipid extraction.
Funding
Funding from the National Health and Medical Research Council (NHMRC) of Australia, the Australian Research Council, Melbourne Water Corporation, Yourgene Bioscience and The University of Melbourne is gratefully acknowledged (RBG). NDY holds an NHMRC Career Development Fellowship (CDF1), and PKK holds an NHMRC Early Career Researcher Fellowship (ECRF). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
Data supporting the conclusions of this article are included within the article. Nucleotide and inferred amino acid sequence data reported in this article can be found in Additional file 1.
Abbreviations
- DAs
Dafachronic acids
- LC-MS
Liquid chromatography-mass spectrometry
- TGF-β
Transforming growth factor-β
- cGMP
Cyclic guanosine monophosphate
- IGF
Insulin/insulin-like growth factor
- TPM
Transcripts per million
- PI3K
Phosphoinositide 3-kinase
Authors’ contributions
GM, TW and SN undertook the laboratory and analytical work. GM, TW and RBG wrote the manuscript, with inputs from PKK, GER, AJS, AVK, BCHC, AH and NDY. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Guangxu Ma, Email: guangxum@student.unimelb.edu.au.
Tao Wang, Email: tao.wang1@unimelb.edu.au.
Pasi K. Korhonen, Email: pasi.korhonen@unimelb.edu.au
Shuai Nie, Email: shuai.nie@unimelb.edu.au.
Gavin E. Reid, Email: Gavin.reid@unimelb.edu.au
Andreas J. Stroehlein, Email: andreas.stroehlein@unimelb.edu.au
Anson V. Koehler, Email: anson.koehler@unimelb.edu.au
Bill C. H. Chang, Email: bchang@yourgene.com.tw
Andreas Hofmann, Email: a.hofmann@structuralchemistry.org.
Neil D. Young, Email: nyoung@unimelb.edu.au
Robin B. Gasser, Email: robinbg@unimelb.edu.au
References
- 1.Ma G, Holland CV, Wang T, Hofmann A, Fan C-K, Maizels RM, et al. Human toxocariasis. Lancet Infect Dis. 2018;18:e14–e24. doi: 10.1016/S1473-3099(17)30331-6. [DOI] [PubMed] [Google Scholar]
- 2.Schnieder T, Laabs E-M, Welz C. Larval development of Toxocara canis in dogs. Vet Parasitol. 2011;175:193–206. doi: 10.1016/j.vetpar.2010.10.027. [DOI] [PubMed] [Google Scholar]
- 3.Strube C, Heuer L, Janecek E. Toxocara spp. infections in paratenic hosts. Vet Parasitol. 2013;193:375–389. doi: 10.1016/j.vetpar.2012.12.033. [DOI] [PubMed] [Google Scholar]
- 4.Burke TM, Roberson EL. Prenatal and lactational transmission of Toxocara canis and Ancylostoma caninum: experimental infection of the bitch before pregnancy. Int J Parasitol. 1985;15:71–75. doi: 10.1016/0020-7519(85)90104-3. [DOI] [PubMed] [Google Scholar]
- 5.Maizels RM. Toxocara canis: molecular basis of immune recognition and evasion. Vet Parasitol. 2013;193:365–374. doi: 10.1016/j.vetpar.2012.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gavignet B, Piarroux R, Aubin F, Millon L, Humbert P. Cutaneous manifestations of human toxocariasis. J Am Acad Dermatol. 2008;59:1031–1042. doi: 10.1016/j.jaad.2008.06.031. [DOI] [PubMed] [Google Scholar]
- 7.Çelik T, Kaplan Y, Ataş E, Öztuna D, Berilgen S. Toxocara seroprevalence in patients with idiopathic Parkinson’s disease: chance association or coincidence? Biomed Res Int. 2013;2013:685196–685194. doi: 10.1155/2013/685196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan C-K, Holland CV, Loxton K, Barghouth U. Cerebral toxocariasis: silent progression to neurodegenerative disorders? Clin Microbiol Rev. 2015;28:663–666. doi: 10.1128/CMR.00106-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aghaei S, Riahi SM, Rostami A, Mohammadzadeh I, Javanian M, Tohidi E, et al. Toxocara spp. infection and risk of childhood asthma: a systematic review and meta-analysis. Acta Trop. 2018;182:298–304. doi: 10.1016/j.actatropica.2018.03.022. [DOI] [PubMed] [Google Scholar]
- 10.Fialho PMM, Correa CRS, Lescano SZ. Asthma and seroconversion from Toxocara spp. infection: which comes first? Biomed Res Int. 2018;2018:4280792–4280796. doi: 10.1155/2018/4280792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janecek E, Beineke A, Schnieder T, Strube C. Neurotoxocarosis: marked preference of Toxocara canis for the cerebrum and T. cati for the cerebellum in the paratenic model host mouse. Parasit Vectors. 2014;7:194. doi: 10.1186/1756-3305-7-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janecek E, Wilk E, Schughart K, Geffers R, Strube C. Microarray gene expression analysis reveals major differences between Toxocara canis and Toxocara cati neurotoxocarosis and involvement of T. canis in lipid biosynthetic processes. Int J Parasitol. 2015;45:495–503. doi: 10.1016/j.ijpara.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 13.Jin Z, Akao N, Ohta N. Prolactin evokes lactational transmission of larvae in mice infected with Toxocara canis. Parasitol Int. 2008;57:495–498. doi: 10.1016/j.parint.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 14.Chávez-Güitrón LE, Morales-Montor J, Muñoz-Guzmán MA, Nava-Castro KE, Ramírez-Álvarez H, Moreno-Méndoza NA, et al. The in vitro effect of prolactin on the growth, motility and expression of prolactin receptors in larvae of Toxocara canis. Vet Parasitol. 2016;224:33–38. doi: 10.1016/j.vetpar.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Del Río-Araiza VH, Nava-Castro KE, Alba-Hurtado F, Quintanar-Stephano A, Muñoz-Guzmán MA, Cuenca-Micò O, et al. Endocrine immune interactions during chronic toxocariasis caused by Toxocara canis in a murine model: new insights into the pathophysiology of an old infection. Vet Parasitol. 2018;252:173–179. doi: 10.1016/j.vetpar.2018.01.026. [DOI] [PubMed] [Google Scholar]
- 16.Motola DL, Cummins CL, Rottiers V, Sharma KK, Li T, Li Y, et al. Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell. 2006;124:1209–1223. doi: 10.1016/j.cell.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 17.Bethke A, Fielenbach N, Wang Z, Mangelsdorf DJ, Antebi A. Nuclear hormone receptor regulation of microRNAs controls developmental progression. Science. 2009;324:95–98. doi: 10.1126/science.1164899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hochbaum D, Zhang Y, Stuckenholz C, Labhart P, Alexiadis V, Martin R, et al. DAF-12 regulates a connected network of genes to ensure robust developmental decisions. PLoS Genet. 2011;7:e1002179. doi: 10.1371/journal.pgen.1002179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu PJ. Dauer. WormBook. 2007. pp. 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fielenbach N, Antebi A. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 2008;22:2149–2165. doi: 10.1101/gad.1701508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogawa A, Streit A, Antebi A, Sommer RJ. A conserved endocrine mechanism controls the formation of dauer and infective larvae in nematodes. Curr Biol. 2009;19:67–71. doi: 10.1016/j.cub.2008.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, Zhou XE, Motola DL, Gao X, Suino-Powell K, Conneely A, et al. Identification of the nuclear receptor DAF-12 as a therapeutic target in parasitic nematodes. Proc Natl Acad Sci USA. 2009;106:9138–43. [DOI] [PMC free article] [PubMed]
- 23.Hotez P, Hawdon J, Schad GA. Hookworm larval infectivity, arrest and amphiparatenesis: the Caenorhabditis elegans Daf-c paradigm. Parasitol Today. 1993;9:23–26. doi: 10.1016/0169-4758(93)90159-D. [DOI] [PubMed] [Google Scholar]
- 24.Bento G, Ogawa A, Sommer RJ. Co-option of the hormone-signalling module dafachronic acid-DAF-12 in nematode evolution. Nature. 2010;466:494–497. doi: 10.1038/nature09164. [DOI] [PubMed] [Google Scholar]
- 25.Lee SS, Schroeder FC. Steroids as central regulators of organismal development and lifespan. PLoS Biol. 2012;10:e1001307. doi: 10.1371/journal.pbio.1001307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crook M. The dauer hypothesis and the evolution of parasitism: 20 years on and still going strong. Int J Parasitol. 2014;44:1–8. doi: 10.1016/j.ijpara.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilabert A, Curran DM, Harvey SC, Wasmuth JD. Expanding the view on the evolution of the nematode dauer signalling pathways: refinement through gene gain and pathway co-option. BMC Genomics. 2016;17:476. doi: 10.1186/s12864-016-2770-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crompton DW. Ascaris and ascariasis. Adv Parasitol. 2001;48:285–375. doi: 10.1016/S0065-308X(01)48008-0. [DOI] [PubMed] [Google Scholar]
- 29.Holland CV. Ascaris: The Neglected Parasite. Amsterdam Boston: Academic Press; 2013.
- 30.Gasser RB. A perfect time to harness advanced molecular technologies to explore the fundamental biology of Toxocara species. Vet Parasitol. 2013;193:353–364. doi: 10.1016/j.vetpar.2012.12.031. [DOI] [PubMed] [Google Scholar]
- 31.Gasser RB, Korhonen PK, Zhu XQ, Young ND. Harnessing the Toxocara genome to underpin toxocariasis research and new interventions. Adv Parasitol. 2016;91:87–110. doi: 10.1016/bs.apar.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 32.Lee RYN, Howe KL, Harris TW, Arnaboldi V, Cain S, Chan J, et al. WormBase 2017: molting into a new stage. Nucleic Acids Res. 2018;46:D869–D874. doi: 10.1093/nar/gkx998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Howe KL, Bolt BJ, Cain S, Chan J, Chen WJ, Davis P, et al. WormBase 2016: expanding to enable helminth genomic research. Nucleic Acids Res. 2016;44:D774–D780. doi: 10.1093/nar/gkv1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jex AR, Liu S, Li B, Young ND, Hall RS, Li Y, et al. Ascaris suum draft genome. Nature. 2011;479:529–533. doi: 10.1038/nature10553. [DOI] [PubMed] [Google Scholar]
- 35.Zhu X-Q, Korhonen PK, Cai H, Young ND, Nejsum P, von Samson-Himmelstjerna G, et al. Genetic blueprint of the zoonotic pathogen Toxocara canis. Nat Commun. 2015;6:6145. doi: 10.1038/ncomms7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou RQ, Ma GX, Korhonen PK, Luo YL, Zhu HH, Luo YF, et al. Comparative transcriptomic analyses of male and female adult Toxocara canis. Gene. 2017;600:85–89. doi: 10.1016/j.gene.2016.11.024. [DOI] [PubMed] [Google Scholar]
- 37.Wang J, Gao S, Mostovoy Y, Kang Y, Zagoskin M, Sun Y, et al. Comparative genome analysis of programmed DNA elimination in nematodes. Genome Res. 2017;27:2001–2014. doi: 10.1101/gr.225730.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al. De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stoltzfus JD, Minot S, Berriman M, Nolan TJ, Lok J. RNAseq analysis of the parasitic nematode Strongyloides stercoralis reveals divergent regulation of canonical dauer pathways. PLoS Negl Trop Dis. 2012;6:e1854. doi: 10.1371/journal.pntd.0001854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.WormBase: https://www.wormbase.org. Accessed 22 Oct 2018.
- 42.Zdobnov EM, Apweiler R. InterProScan - an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 43.Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stroehlein AJ, Young ND, Gasser RB. Improved strategy for the curation and classification of kinases, with broad applicability to other eukaryotic protein groups. Sci Rep. 2018;8:6808. doi: 10.1038/s41598-018-25020-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kent WJ. BLAT - the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Slater GS, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rombel IT, Sykes KF, Rayner S, Johnston SA. ORF-FINDER: a vector for high throughput gene identification. Gene. 2002;282:33–41. doi: 10.1016/S0378-1119(01)00819-8. [DOI] [PubMed] [Google Scholar]
- 49.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li W, Cowley A, Uludag M, Gur T, McWilliam H, Squizzato S, et al. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 2015;43:W580–W584. doi: 10.1093/nar/gkv279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jonassen I, Collins JF, Higgins DG. Finding flexible patterns in unaligned protein sequences. Protein Sci. 1995;4:1587–1595. doi: 10.1002/pro.5560040817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bailey TL, Johnson J, Grant CE, Noble WS. The MEME suite. Nucleic Acids Res. 2015;43(W1):W39–W49. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. [DOI] [PMC free article] [PubMed]
- 55.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 56.Wang T, Nie S, Ma G, Korhonen PK, Koehler AV, Ang C-S, et al. The developmental lipidome of Haemonchus contortus. Int J Parasitol. 2018;48:887–895. doi: 10.1016/j.ijpara.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 57.Beall MJ, Pearce EJ. Transforming growth factor-beta and insulin-like signalling pathways in parasitic helminths. Int J Parasitol. 2002;32:399–404. doi: 10.1016/S0020-7519(01)00348-4. [DOI] [PubMed] [Google Scholar]
- 58.Pierce SB, Costa M, Wisotzkey R, Devadhar S, Homburger SA, Buchman AR, et al. Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes Dev. 2001;15:672–686. doi: 10.1101/gad.867301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matsunaga Y, Gengyo-Ando K, Mitani S, Iwasaki T, Kawano T. Physiological function, expression pattern, and transcriptional regulation of a Caenorhabditis elegans insulin like peptide INS-18. Biochem Biophys Res Commun. 2012;423:478–483. doi: 10.1016/j.bbrc.2012.05.145. [DOI] [PubMed] [Google Scholar]
- 60.Wolkow CA, Muñoz MJ, Riddle DL, Ruvkun G. Insulin receptor substrate and p55 orthologous adaptor proteins function in the Caenorhabditis elegans daf-2/insulin-like signaling pathway. J Biol Chem. 2002;277:49591–49597. doi: 10.1074/jbc.M207866200. [DOI] [PubMed] [Google Scholar]
- 61.Paradis S, Ruvkun GB. Caenorhabditis elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes Dev. 1998;12:2488–2498. doi: 10.1101/gad.12.16.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kwon E, Narasimhan SD, Yen K, Tissenbaum HA. A new DAF-16 isoform regulates longevity. Nature. 2010;466:498–502. doi: 10.1038/nature09184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Daniels SA, Ailion M, Thomas JH, Sengupta P. egl-4 acts through a transforming growth factor-beta/SMAD pathway in Caenorhabditis elegans to regulate multiple neuronal circuits in response to sensory cues. Genetics. 2000;156:123–141. doi: 10.1093/genetics/156.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McCoy CJ, Warnock ND, Atkinson LE, Atcheson E, Martin RJ, Robertson AP, et al. RNA interference in adult Ascaris suum - an opportunity for the development of a functional genomics platform that supports organism-, tissue- and cell-based biology in a nematode parasite. Int J Parasitol. 2015;45:673–678. doi: 10.1016/j.ijpara.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matsunaga Y, Nakajima K, Gengyo-Ando K, Mitani S, Iwasaki T, Kawano TA. Caenorhabditis elegans insulin-like peptide, INS-17: its physiological function and expression pattern. Biosci Biotechnol Biochem. 2012;76:2168–2172. doi: 10.1271/bbb.120540. [DOI] [PubMed] [Google Scholar]
- 66.Lok JB. Signaling in parasitic nematodes: physicochemical communication between host and parasite and endogenous molecular transduction pathways governing worm development and survival. Curr Clin Microbiol Rep. 2016;3:186–197. doi: 10.1007/s40588-016-0046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gerisch B, Rottiers V, Li D, Motola DL, Cummins CL, Lehrach H, et al. A bile acid-like steroid modulates Caenorhabditis elegans lifespan through nuclear receptor signaling. Proc Natl Acad Sci USA. 2007;104:5014–9. [DOI] [PMC free article] [PubMed]
- 68.Albarqi MM, Stoltzfus JD, Pilgrim AA, Nolan TJ, Wang Z, Kliewer SA, et al. Regulation of life cycle checkpoints and developmental activation of infective larvae in Strongyloides stercoralis by dafachronic acid. PLoS Pathog. 2016;12:e1005358. doi: 10.1371/journal.ppat.1005358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Butcher RA. Small-molecule pheromones and hormones controlling nematode development. Nat Chem Biol. 2017;13:577–586. doi: 10.1038/nchembio.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hannich JT, Entchev EV, Mende F, Boytchev H, Martin R, Zagoriy V, et al. Methylation of the sterol nucleus by STRM-1 regulates dauer larva formation in Caenorhabditis elegans. Dev Cell. 2009;16:833–843. doi: 10.1016/j.devcel.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 71.Patel DS, Fang LL, Svy DK, Ruvkun GB, Li W. Genetic identification of HSD-1, a conserved steroidogenic enzyme that directs larval development in Caenorhabditis elegans. Development. 2008;135:2239–2249. doi: 10.1242/dev.016972. [DOI] [PubMed] [Google Scholar]
- 72.Tissenbaum HA, Hawdon J, Perregaux M, Hotez P, Guarente L, Ruvkun G. A common muscarinic pathway for diapause recovery in the distantly related nematode species Caenorhabditis elegans and Ancylostoma caninum. Proc Natl Acad Sci USA. 2000;97:460–5. [DOI] [PMC free article] [PubMed]
- 73.Wang Z, Schaffer NE, Kliewer SA, Mangelsdorf DJ. Nuclear receptors: emerging drug targets for parasitic diseases. J Clin Invest. 2017;127:1165–1171. doi: 10.1172/JCI88890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Patton JB, Bonne-Année S, Deckman J, Hess JA, Torigian A, Nolan TJ, et al. Methylprednisolone acetate induces, and Δ7-dafachronic acid suppresses Strongyloides stercoralis hyperinfection in NSG mice. Proc Natl Acad Sci USA. 2018;115:204–9. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Dauer signalling gene homologues of Toxocara canis and Ascaris suum. Table S2. Orthologous groups of dauer signalling genes in Toxocara canis and Ascaris suum inferred from Caenorhabditis elegans. Table S3. Salient information on dauer signalling genes of Toxocara canis predicted from Caenorhabditis elegans orthologues. Table S4. Salient information on dauer signalling genes of Ascaris suum predicted from Caenorhabditis elegans orthologues. Table S5. Classification of insulin-like peptide homologues in Toxocara canis and Ascaris suum. Table S6. Transcription of dauer-like signalling gene orthologues in key developmental stages of Toxocara canis and Ascaris suum. (XLSX 418 kb)
Data Availability Statement
Data supporting the conclusions of this article are included within the article. Nucleotide and inferred amino acid sequence data reported in this article can be found in Additional file 1.