Abstract
Background
Around 1% of human proteins are predicted to contain a disordered and low complexity prion-like domain (PrLD). Mutations in PrLDs have been shown promote a transition towards an aggregation-prone state in several diseases.
Results
Recently, we have shown that an algorithm that considers the effects of mutations on PrLDs composition, as well as on localized amyloid propensity can predict the impact of these amino acid changes on protein intracellular aggregation. In this application note, we implement this concept into the AMYCO web server, a refined algorithm that forecasts the influence of amino acid changes in prion-like proteins aggregation propensity better than state-of-the-art predictors.
Conclusions
The AMYCO web server allows for a fast and automated evaluation of the effect of mutations on the aggregation properties of prion-like proteins. This might uncover novel disease-linked amino acid changes in the sequences of human prion-like proteins. Additionally, it can find application in the in silico design of synthetic prion-like proteins with tuned aggregation propensities for different purposes. AMYCO does not require previous registration and is freely available to all users at: http://bioinf.uab.cat/amyco/.
Electronic supplementary material
The online version of this article (10.1186/s12859-019-2601-3) contains supplementary material, which is available to authorized users.
Keywords: Prion-like domain, Protein aggregation, Amyloid, Protein mutation
Background
Prions are proteins able to adopt multiple structural conformations from which at least one has self-propagating properties [1]. Yeast prions are the best understood subset of functional prions. A common feature of most yeast prions is the presence of an intrinsically disordered and low complexity prion domain (PrD), which is necessary and sufficient for prion conversion and propagation. Proteins bearing prion-like domains (PrLDs) sharing these properties seem to exist in all kingdoms of life [2–6]. In particular, around 1% of the human proteome has been predicted to correspond to prion-like proteins [7]. This human protein subset is enriched in nucleic acid-binding proteins and involved in the formation of membraneless compartments through highly dynamic liquid-liquid demixing [7, 8]. A number of mutations in human PrLDs have been shown to convert these liquid compartments into solid aggregates, abolishing their dynamic nature and leading to the onset of neurodegenerative disorders [8, 9]. The development of tools able to anticipate the impact of such pathogenic amino acid changes is attracting increasing interest.
The self-assembling properties of prion-like proteins have been traditionally thought to rely on the biased amino acid composition of their PrLDs [10]. Disease-linked mutations would enhance the self-association of these domains, facilitating the transition to amyloid-like states [11, 12]. We have recently shown that the impact of point and multiple mutations or deletions on the aggregation of the model ALS-associated prion-like hnRNPA2 protein is best predicted by a function that takes into account both compositional features and amyloidogenic propensities [13]. Here we introduce the AMYCO (combined AMYloid and Composition based prediction of prion-like aggregation propensity) web server, which implements this approach to perform automated and fast predictions on top of prion-like protein sequences.
Implementation
AMYCO is written in Python and uses python2.7 as the interpreter (Anaconda distribution). The web interface has been build using html/css and inputs and outputs are processed by a cgi written in perl. It all runs in a CentOS 5 server with Apache 2.2.3 using Intel Xeon ‘Clovertown’ processors.
AMYCO pipeline
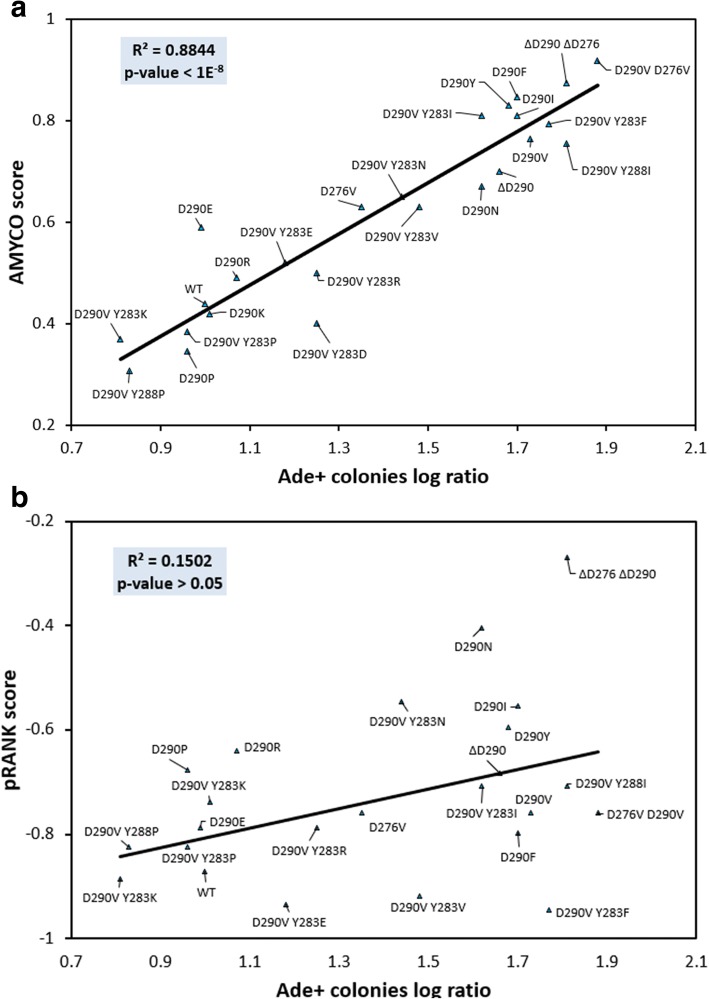
AMYCO evaluates the impact of mutations on the aggregation propensity of PrLDs in prion-like proteins. They can be single or multiple residues substitutions, as well as deletions and insertions. It exploits the highly significant correlation between the scores obtained from a parameterized linear function, that balances the contribution of both PrLDs composition and amyloid propensity [13], and the intracellular aggregation of hnRNPA2 variants; the unique prion-like protein for which a large set of mutations, both natural and artificial have been experimentally validated (Fig. 1a). The contribution of PrLDs amino acid composition to prion-like proteins aggregation is calculated using the PAPA algorithm [10], a program that exploits a scale of prion propensity scores for natural amino acids derived from mutagenesis experiments in the Sup35 yeast prion domain [14]. The impact of amyloidogenic sequences within the PrLDs is calculated with pWALTZ [12], a program specially intended to identify short sequences of moderate amyloid propensity able to nucleate the aggregation reaction, as those found in yeast prion domains [15]. The outputs of the two different programs are combined in a linear manner, as described in detail in the Additional file 1.
Fig. 1.
Correlation between AMYCO and pRANK predictions and the aggregation propensity of human hnRNPA2 prion-like protein variants. a Graphic representation of the correlation between the mutants' AMYCO (a) and pRANK (b) scores and their ability to form prionic colonies (Ade+) when expressed in yeast, a direct reporter of their aggregation propensity [13]
The AMYCO web server is free and open to all users, and no previous login or registration is required.
The home page of AMYCO displays three clickable links in its upper margin: (i) a help page containing a brief description of the method, the output explanation and information on examples, (ii) references for the methodology and the web application and (iii) a contact e-mail. Immediately below two links allow to switch between the Compare Sequences mode in which multiple sequences can be compared to a reference one, and the Single Mutation mode in which all possible mutations for a given protein residue are evaluated. The adjacent “Example” button fills the input text areas with the full-length sequences of wild type (wt) human hnRNPA2 protein and its aggregation-prone D290V mutant [16] for Compare Sequences mode, or all its possible mutants for position 290 for Single Mutation mode.
The input interface allows two working modes. In Compare Sequences mode (default mode); the user should introduce a reference sequence and the mutated variants (one or several) in the left and right text boxes, respectively; all in FASTA format. In the Single Mutation mode, the user should introduce a single sequence as well as the position to be scanned. Protein sequences should be at least 60-residues long and only the 20 standard proteinogenic amino acids are allowed.
After submission, the output page will display a job identification number along with the names of the input sequences and the mutation position if applicable. The algorithm will return the AMYCO score for each sequence, together with a description of the mutations impact of the overall aggregation propensity. In addition, a graphical representation of the mutation/s effect will be displayed (Fig. 2). We set two arbitrary thresholds of low (< 0.45) and high (> 0.78) AMYCO scores to visualize better the overall aggregation propensities of the variants. hnRNPA2 mutants scoring < 0.45 were shown to decrease or increase < 5 times the propensity of the non-aggregating wild type protein, whereas, mutants scoring > 0.78 increased its aggregation by > 50 times [17]. Therefore, mutations rendering an AMYCO score < 0.45 are considered of low aggregation propensity and labeled in blue. Mutations that increase the aggregation propensity of the protein, but whose AMYCO score is below 0.78 are labeled in red, whereas mutations above this threshold are considered to be of high aggregation propensity are labeled in red and bold. Sequences might display AMYCO scores > 1.0, indicating that they are predicted to be more aggregation-prone than the highest scoring hnRNPA2 variant used in the parametrization of the prediction function. The output files can be downloaded for further analyses, as a ZIP file containing the resulting text explanation, a machine readable JSON file, the visualizations as a PNG file and FASTA files with the introduced sequences and the virtually generated mutants in Compare Sequences and Single Mutation modes, respectively.
Fig. 2.
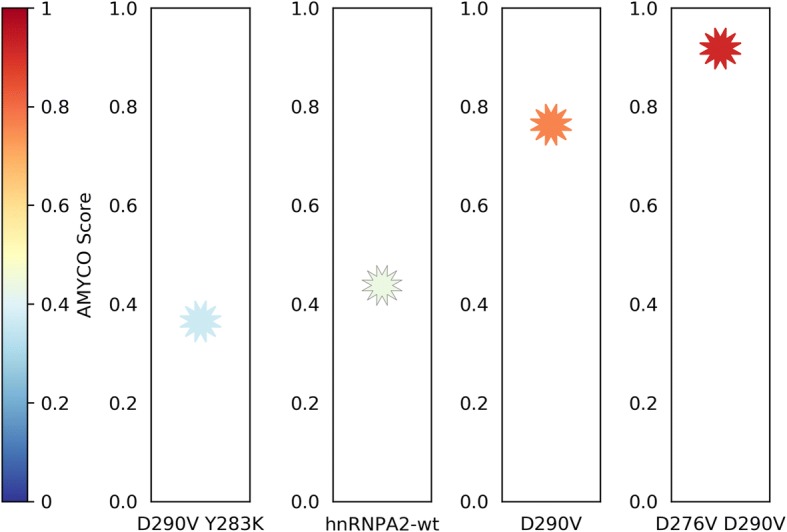
Graphical representation of the AMYCO score. AMYCO output representation of a low aggregation-prone (D290V Y283K), the wild type, the natural pathogenic mutant D290V and a high aggregation-prone (D290V D276V) hnRNPA2 prion-like protein variants
Results
Performance
pRANK is a novel multiple-instance machine learning method aimed to predict prion propensity based on amino acid composition alone [18]. We compared the performance of AMYCO and pRANK web servers in predicting the impact of mutations on human hnRNPA2 aggregation propensity (Fig. 1). AMYCO clearly outperforms pRANK (Table 1), an observation which is consistent with the important influence that sequential features exert on protein aggregation [19].
Table 1.
Performance of pRANK and AMYCO approaches in the prediction of mutation impact upon the aggregation of the human prion-like protein hnRNPA2
| pRANK | AMYCO | |
|---|---|---|
| Sensitivity | 0 | 1 |
| Specificity | 1 | 1 |
| Precision | – | 1 |
| Accuracy | 0.45 | 1 |
| MCC | – | 1 |
| Mean % error | −7.08 | −1.25 |
| Standard Deviation (%) | 37.71 | 12.19 |
| SEM (%) | 8.04 | 2.44 |
| Coefficient of Determination | 0.152 | 0.882 |
| P-value (two tailed test) | 0.468 | < 1.00E-08 |
| Rho (ρ) | 0.334 | 0.929 |
The best performance according to each particular parameter is shown in bold. Details on the calculation of the different statistic parameters are provided in the Additional file 1
AMYCO was further assayed on known mutations promoting the apparition of a de novo prion-like behavior (Table 2). It was able to predict a large increase in aggregation propensity for mutations that convert the non-prionic PrLDs of PUF4, YLR177W, KC11 and PDC2 yeast proteins into prionic when expressed in yeast [20] (Table 2). Importantly, according to AMYCO, five out of the eight variants were predicted to have acquired a very high aggregation propensity. These variants are exactly the ones experimentally shown to induce a prionic phenotype with basal protein levels, without a need for overexpression [20] (Table 2).
Table 2.
AMYCO correctly predicts prion converting mutations on yeast proteins
| Protein variant | AMYCO score |
|---|---|
| PUF4 wt | 0 |
| PUF4 mut | 0.69 |
| *PUF4 6PP,1N | 0.93 |
| PUF4 4PP | 0.60 |
| YLR177W wt | 0 |
| *YLR177W mut | 0.85 |
| *YLR177W 4PP,1N | 1.23 |
| *YLR177W 4PP | 1.03 |
| KC11 wt | 0 |
| *KC11 mut | 0.97 |
| PDC2 wt | 0 |
| PDC2 mut | 0.78 |
AMYCO correctly predicts mutations that induce prionic phenotypes. Mutations predicted to increase and highly increase aggregation propensity are shown in italics and bold, respectively. Variants that do not need overexpression to generate a prionic phenotype in yeast are indicated with an asterisk [20]
Finally, AMYCO is able to predict an increase in aggregation propensity for a series of disease-linked mutations occurring in different human prion-like proteins. In particular, mutations in hnRNPA1 associated to ALS [16], mutations in hnRNP D0/AUF1 identified in familiar cases of Crohn Disease [21] and mutations in hnRNP DL causing limb-girdle muscular dystrophy 1G [22] (Table 3). In contrast, natural variants of these proteins bearing mutations in the PrLDs not associated to clinical phenotypes [23] do not have any significant impact in the polypeptides predicted aggregation propensity.
Table 3.
AMYCO predicts disease-causing mutations on human prion-like proteins
| Protein variant | AMYCO score |
|---|---|
| hnRNPA1 wt | 0.34 |
| hnRNPA1 Q277K | 0.34 |
| hnRNPA1 G283R | 0.34 |
| hnRNPA1 P340S | 0.36 |
| hnRNPA1 D314V | 0.59 |
| hnRNPA1 D314N | 0.53 |
| hnRNP DL wt | 1.18 |
| hnRNP DL D378H | 1.26 |
| hnRNP DL D378N | 1.30 |
| hnRNP D0 wt | 1.13 |
| hnRNP D0 F225 L | 1.13 |
| hnRNP D0 D319V | 1.33 |
| hnRNP D0 isoform-2 D300V | 1.33 |
AMYCO identifies multisystem proteinopathy and ALS causing mutations on hnRNPA1 [16], Crohn Disease causing mutations on both isoforms of hnRNP D0/AUF1 [21] and limb-girdle muscular dystrophy 1G (LGMD1G) on hnRNP DL [22] are shown in bold. Natural variants not associated to a clinical phenotype are shown in italics
Conclusion
AMYCO has been developed as a web application to assess the impact of mutations on the aggregation propensity of prion-like proteins, allowing a fast and accurate evaluation of the effect of disease-associated mutations in these polypeptides; as well as engineering novel variants with designed aggregation propensities for different applications.
Availability and requirements
Project name: AMYCO.
Project home page: http://bioinf.uab.cat/amyco/.
Operating system(s): Platform independent.
Programming language: A computing core coded in Python and a front end written in a combination of html and perl cgi.
Other requirements: A web browser with a working internet connection.
License: None.
Any restrictions to use by non-academics: None.
Additional file
Dataset obtention and performance analysis. (PDF 219 kb)
Acknowledgments
Funding
This work was funded by the Spanish Ministry of Economy and Competitiveness (BIO2016–783-78310-R to SV). SV has been granted an ICREA ACADEMIA award.
Availability of data and materials
All data analyzed during this study are included in articles [16, 17, 20–23].
Abbreviations
- MCC
Matthews correlation coefficient
- PrD
Prion domain
- PrLD
Prion like domain
- SEM
Standard error of the mean
Authors’ contributions
VI and OC analyzed the data and implemented the web server. CB analyzed the data. VI drafted the manuscript. SV designed the research, analyzed the data and wrote the final version of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Valentin Iglesias, Email: valentin.iglesias.mas@gmail.com.
Oscar Conchillo-Sole, Email: ocs@bioinf.uab.es.
Cristina Batlle, Email: Cristina.Batlle@uab.cat.
Salvador Ventura, Email: salvador.ventura@uab.cat.
References
- 1.Aguzzi A, Calella AM. Prions: protein aggregation and infectious diseases. Physiol Rev. 2009;89:1105–1152. doi: 10.1152/physrev.00006.2009. [DOI] [PubMed] [Google Scholar]
- 2.Malinovska L, Palm S, Gibson K, Verbavatz JM, Alberti S. Dictyostelium discoideum has a highly Q/N-rich proteome and shows an unusual resilience to protein aggregation. Proc Natl Acad Sci U S A. 2015;112(20):E2620–E2629. doi: 10.1073/pnas.1504459112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iglesias V, de Groot NS, Ventura S. Computational analysis of candidate prion-like proteins in bacteria and their role. Front Microbiol. 2015;6:1–13. doi: 10.3389/fmicb.2015.01123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pallarès I, Iglesias V, Ventura S. The rho termination factor of Clostridium botulinum contains a prion-like domain with a highly Amyloidogenic Core. Front Microbiol. 2016;6:1–12. doi: 10.3389/fmicb.2015.01516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan AH, Hochschild A. A bacterial global regulator forms a prion. Science. 2017;355(6321):198–201. doi: 10.1126/science.aai7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakrabortee S, Kayatekin C, Newby GA, Mendillo ML, Lancaster A, Lindquist S. Luminidependens (LD) is an Arabidopsis protein with prion behavior. Proc Natl Acad Sci. 2016;113(21):6065–6070. doi: 10.1073/pnas.1604478113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King OD, Gitler AD, Shorter J. The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res. 2012;1462:61–80. doi: 10.1016/j.brainres.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162(5):1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 9.Polymenidou M, Cleveland DW. Prion-like spread of protein aggregates in neurodegeneration. J Exp Med. 2012;209(5):889–893. doi: 10.1084/jem.20120741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toombs JA, Petri M, Paul KR, Kan GY, Ben-Hur A, Ross ED. De novo design of synthetic prion domains. Proc Natl Acad Sci U S A. 2012;109(17):6519–6524. doi: 10.1073/pnas.1119366109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryan VH, Dignon GL, Zerze GH, Chabata CV, Silva R, Conicella AE, Amaya J, Burke KA, Mittal J, Fawzi NL. Mechanistic view of hnRNPA2 low-complexity domain structure, interactions, and phase separation altered by mutation and arginine methylation. Mol Cell. 2018;69(3):465–479. doi: 10.1016/j.molcel.2017.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabate R, Rousseau F, Schymkowitz J, Ventura S. What makes a protein sequence a prion? PLoS Comput Biol. 2015;11(1):e1004013. doi: 10.1371/journal.pcbi.1004013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Batlle C, Fernandez MR, Iglesias V, Ventura S. Perfecting prediction of mutational impact on the aggregation propensity of the ALS-associated hnRNPA2 prion-like protein. FEBS Lett. 2017;591(13):1966–1971. doi: 10.1002/1873-3468.12698. [DOI] [PubMed] [Google Scholar]
- 14.Toombs JA, McCarty BR, Ross ED. Compositional determinants of prion formation in yeast. Mol Cell Biol. 2010;30(1):319–332. doi: 10.1128/MCB.01140-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sant’Anna R, Fernandez MR, Batlle C, Navarro S, de Groot NS, Serpell L, Ventura S. Characterization of amyloid cores in prion domains. Sci Rep. 2016;6:34274. doi: 10.1038/srep34274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495(7442):467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paul KR, Molliex A, Cascarina S, Boncella AE, Taylor JP, Ross ED. Effects of mutations on the aggregation propensity of the human prion-like protein hnRNPA2B1. Mol Cell Biol. 2017;37(8):e00652. doi: 10.1128/MCB.00652-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Afsar Minhas FU, Ross ED, Ben-Hur A. Amino acid composition predicts prion activity. PLoS Comput Biol. 2017;13(4):e1005465. doi: 10.1371/journal.pcbi.1005465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabate R, Espargaro A, de Groot NS, Valle-Delgado JJ, Fernandez-Busquets X, Ventura S. The role of protein sequence and amino acid composition in amyloid formation: scrambling and backward reading of IAPP amyloid fibrils. J Mol Biol. 2010;404(2):337–352. doi: 10.1016/j.jmb.2010.09.052. [DOI] [PubMed] [Google Scholar]
- 20.Paul KR, Hendrich CG, Waechter A, Harman MR, Ross ED. Generating new prions by targeted mutation or segment duplication. Proc Natl Acad Sci U S A. 2015;112(28):8584–8589. doi: 10.1073/pnas.1501072112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prakash T, Veerappa A. N BR: complex interaction between HNRNPD mutations and risk polymorphisms is associated with discordant Crohn’s disease in monozygotic twins. Autoimmunity. 2017;50(5):275–276. doi: 10.1080/08916934.2017.1300883. [DOI] [PubMed] [Google Scholar]
- 22.Vieira NM, Naslavsky MS, Licinio L, Kok F, Schlesinger D, Vainzof M, Sanchez N, Kitajima JP, Gal L, Cavacana N, et al. A defect in the RNA-processing protein HNRPDL causes limb-girdle muscular dystrophy 1G (LGMD1G) Hum Mol Genet. 2014;23(15):4103–4110. doi: 10.1093/hmg/ddu127. [DOI] [PubMed] [Google Scholar]
- 23.UniProt C UniProt: a hub for protein information. Nucleic Acids Res. 2015;43(Database issue):D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dataset obtention and performance analysis. (PDF 219 kb)
Data Availability Statement
All data analyzed during this study are included in articles [16, 17, 20–23].