Abstract
Purpose
To investigate the effects of sperm treatment medium—TCM199 or EGTA in Tris‐HCl buffer (TBS + EGTA)―for sonication of frozen‐thawed hamster spermatozoa in terms of sperm chromosome integrity and development of hamster oocytes injected with the sperm heads (ICSI).
Methods
Frozen‐thawed hamster spermatozoa were separated into heads and tails by sonication in TCM199 or TBS + EGTA. Sperm heads were injected into mouse oocytes to assess hamster sperm chromosomes. We further compared the development of hamster ICSI embryos produced by injecting sonicated sperm heads in TCM199 vs TBS + EGTA.
Results
Sperm chromosome integrity was greater following sonication of frozen‐thawed hamster spermatozoa in TBS + EGTA than in TCM199 (89.7% vs 69.0%). Embryonic development was improved following hamster oocyte injection with sperm heads sonicated in TBS + EGTA compared to in TCM199 (8‐cell: 84.1% vs 65.4%; morula: 78.4% vs 43.2%; blastocyst: 42.0% vs 17.3%). Gene expression of zygotic genome activation in 2‐cell embryos was significantly higher with TBS + EGTA than with TCM199. We transferred 43 morulae/blastocysts from the TBS + EGTA group to foster mothers, and 4 (9.3%) developed into live offspring.
Conclusion
These results showed that the rapid injection of hamster sperm heads separated by sonication in TBS + EGTA effectively produced more ICSI embryos during a short time.
Keywords: Hamster, intracytoplasmic sperm injection, live offspring, sonication, sperm chromosome
1. INTRODUCTION
Intracytoplasmic sperm injection (ICSI) is a method of producing fertilized oocytes. The first application of ICSI in mammals was performed with golden hamster in 1976.1 Since then, ICSI has successfully been performed to produce live offspring in many species, including humans.2 In human infertility clinics, ICSI is a powerful tool to overcome male infertility and produce fertilized oocyte.3
We previously reported the first birth of live offspring following ICSI in golden hamsters.4 Shortly thereafter, Haigo et al demonstrated that injection of hamster round spermatids into mature oocytes can produce live offspring.5 Later, Muneto and Horiuchi6 reported that injection of oocytes with freeze‐dried spermatozoa produced hamster embryos that could develop into live offspring after embryo transfer. However, only a low percentage of ICSI embryos exhibit in vitro and in vivo development.
Hamster oocytes are sensitive to short‐wave visible light emitted from ordinary fluorescence light sources, such that oocytes injected with spermatozoa under fluorescence light never develop beyond the 2‐cell stage.4, 7, 8, 9 In hamster zygotes, fluorescent light reportedly increases reactive oxygen species (ROS).9
Moreover, intracytoplasmic injection of fresh sperm heads with intact acrosomes dissolves the cytoplasm of hamster oocytes, leading to oocyte death.4, 10 Successful hamster ICSI requires the removal of acrosomes from spermatozoa. After injection of hamster or rabbit acrosome‐intact sperm heads into mouse oocytes, extrusion of the second polar body is followed by degeneration of the mouse oocyte cytoplasm, and cleavage never occurs.10 On the other hand, when a single acrosome‐intact spermatozoon is injected during human and mouse ICSI, the released acrosome enzymes do not cause any serious problem.11
Another complicating factor is that golden hamster oocytes show a high rate of spontaneous activation.12, 13, 14 Therefore, in hamsters, ICSI should be performed as soon as possible after ovulated oocyte collection. Yamauchi et al4 produced fertilized eggs by injecting acrosome‐free sperm heads that were separated from frozen‐thawed spermatozoa using a piezo pulse. However, compared to mouse spermatozoa, hamster spermatozoa show a relatively strong “neck” connection between the head and midpiece, such that it takes several minutes to detach sperm heads using a piezo pulse.
Sonication is an easy method for separating sperm heads from tails prior to ICSI. In early studies of hamster ICSI, Yanagida et al and Katayose et al used sperm heads detached by sonication.15, 16 They observed pronuclear formation following sperm head injection after heating or freeze‐drying, but did not demonstrate the developmental competence of zygotes following sonicated sperm head injection. In contrast, Kuretake et al17 performed mouse ICSI and showed higher percentages of blastocysts and normal offspring development following intracytoplasmic injection of sperm heads that were sonicated in nuclear isolation medium (NIM; a K‐rich and EDTA‐containing medium). Tateno et al demonstrated that sonication per se is not deleterious to sperm chromosomes, but that sperm suspension medium is easily damaged by sonication and chromosomal abnormalities can be caused by activation of Ca2+‐dependent DNase adjacent to the sperm chromosome.18
The solution used to freeze‐dry spermatozoa without chromosomal aberrations is also an attractive choice of solution for sonication. Suspension of mouse spermatozoa in a simple Tris‐HCl–buffered solution containing 50 mM EGTA and 50 mM NaCl enables maintenance of chromosome integrity during freeze‐drying or freezing without cryoprotection.19 Furthermore, Kusakabe et al20 showed that prior incubation of mouse spermatozoa in Tris‐HCl–buffered solution containing only 50 mM EGTA (without NaCl) increases the chromosomal normality after freeze‐drying and that these spermatozoa can support embryo development. However, it is presently unclear whether this solution can reduce chromosomal aberrations in sperm heads separated by sonication.
In the present study, we aimed to examine how the solution used during frozen‐thawing and sonication influenced hamster spermatozoa integrity, and the development of hamster zygotes produced by injecting sonicated sperm heads into oocytes. We also analyzed the expression of maternal effect genes (MEGs) and zygotic gene activation (ZGA) genes in the ICSI embryos.
2. MATERIALS AND METHODS
2.1. Chemicals and animals
Inorganic salts were purchased from Sigma‐Aldrich (St. Louis, MO, USA) or Nacalai Tesque Inc (Kyoto, Japan). Organic reagents were purchased from Sigma‐Aldrich unless otherwise stated.
Golden hamsters (Mesocricetus auratus) with brown coats and black eyes were purchased from Japan SLC Inc (Shizuoka, Japan). Mature females of 2‐4 months of age were used for the collection of unfertilized oocytes. Mature males of 2‐5 months of age were used for the collection of motile spermatozoa. All hamsters were fed a standard diet ad libitum and maintained in a temperature‐ and light‐controlled room at 25°C, with a 14‐hour light/10‐hour dark cycle (light starting at 06:00 hours). The experiments were approved by the Committee for Ethics on Animal Experiments of the Prefectural University of Hiroshima, Japan (16SA002).
2.2. Oocyte collection
Golden hamster females were induced to superovulate via an im injection of 20 IU eCG (Asuka Pharmaceuticals, Tokyo, Japan) during the morning of the day of postestrus discharge.21 At 56 hours after eCG injection, the hamsters received an im injection of 20 IU hCG (Asuka Pharmaceuticals, Tokyo, Japan).22 At approximately 15 hours hCG injection, mature unfertilized oocytes were collected from oviducts and were freed from cumulus cells by a 1‐minute treatment with 250 IU/mL hyaluronidase in M2 medium.23 Next, the oocytes were rinsed with TCM199TE and stored in fresh medium. All experiments were performed in a dark room with a small table light, and red filters were used on the microscope light source, as previously reported.4, 5, 6
2.3. Sperm preparation
From mature male hamster (2‐5 months of age), a dense sperm mass was collected from the cauda epididymis. A small drop of this sperm mass was placed in a 3.5‐mL dish containing 5 mL of TCM199 supplemented with 0.1% bovine serum albumin (Sigma‐Aldrich; A6003). Spermatozoa were allowed to swim through this medium for 5 minutes at 37.5°C, and then 700 µL aliquots of the medium were dispensed into 1.5‐mL centrifuge tubes. The collected sperm were washed by centrifugal separation and preserved with 50 mM EGTA in 100 mM Tris‐HCl–buffered solution (TBS + EGTA).20 Control sperm samples were stored in TCM199 supplemented with bovine serum albumin (1 mg/mL). As a negative control, we used TBS containing 2 mM CaCl2 (TBS + Ca), which is the same concentration as in TCM199. Sperm samples were frozen at −196°C with liquid nitrogen. Then, the frozen sperm were thawed in a water bath at 37°C and sonicated in the experimental or control medium using an ultra homogenizer (VP‐52; TITEC, Japan) for 1 minute just prior to ICSI. After the spermatozoa were frozen‐thawed and sonicated, they were immobilized, they lost their acrosomes, and the sperm heads and tails were separated.
2.4. Chromosomal analysis of hamster spermatozoa by injection into mouse oocytes
To analyze the sperm chromosomes, we performed intracytoplasmic injection of hamster spermatozoa into mouse oocytes, following the method described by Kishikawa et al24 B6D2F1 mouse females (CLEA Japan, Tokyo) at 8‐12 weeks of age were each injected with 5 IU eCG followed by 5 IU hCG 48 later. Oocytes were collected from oviducts between 15 and 16 hours after hCG injection. They were freed from cumulus cells by a 1‐minute treatment with 250 IU/mL hyaluronidase in M2 medium. These oocytes were kept in Global Total medium (Astec, Fukuoka, Japan) at 37°C for 30 minute before sperm injection.
About 5 hours after hamster sperm injection, mouse oocytes exhibiting two well‐developed pronuclei and a distinct second polar body were transferred to Global Total medium containing 0.006 µg/mL vinblastine. After 16‐19 hours of culture in the vinblastine solution, oocytes arrested at the first cleavage metaphase were treated with a hypotonic solution (1:1 mixture of 30% fetal bovine serum and 1% sodium citrate) for 7 minutes at 37°C. Chromosomes were spread on slides using the gradual‐fixation/air‐drying method and then stained with 2% Giemsa (Merck, Darmstadt, Germany) in buffered saline solution (pH 6.8) for 8 minutes for conventional chromosome analysis.25 An egg exhibiting 20 chromosomes (mouse) as well as another 22 chromosomes (hamster) with no structural and numerical abnormalities was recorded as karyotypically normal. We also recorded the number of aberrations per oocyte in the hamster sperm chromosomes.
2.5. Microinjection of sperm heads into oocytes
One drop (15 µL) of sonicated sperm solution was thoroughly mixed with 30 µL of Ca‐ and Mg‐free M2 medium containing 12% (w/v) polyvinylpyrrolidone (PVP; molecular weight, ~360 000; MP Biochemicals, OH, USA). This sperm mixture was then transferred to the micromanipulation chamber on the microscope stage. A sperm head was drawn into a pipette and injected into an oocyte under TCM199TE.4, 5, 6
2.6. Embryo culture
Sperm‐injected oocytes were incubated for about 6 hours in 40 µL droplets of modified Hamster Embryo Culture Medium‐9 (mHECM‐9) supplemented with 5% fetal bovine serum (HyClone, UT, USA), under mineral oil and kept at 37.5°C in an atmosphere of 10% CO2, 10% O2%, and 80% N2. The oocytes were then examined using an inverted microscope (Diaphot TMD, Diaphot 300; Nikon, Tokyo, Japan) equipped with Hoffman modulation contrast optics.26 An oocyte exhibiting two distinct pronuclei and a clearly visible second polar body was considered normally fertilized. At 24 hours after sperm injection, all fertilized oocytes were transferred into 40 µL droplets of mHECM‐9 supplemented with 0.5 mg/mL human serum albumin (Sigma‐Aldrich; A‐1653), and cultured for 72 hours under the same gas mixture.
2.7. Quantitative RT‐PCR analysis in hamster embryos
At 6 hours post‐ICSI, 24 of pronuclear embryos and, at 44 hours post‐ICSI, 23 of 2‐cell embryos from the TCM199 or TBS + EGTA, respectively, were washed in PBS and collected in a small volume (~1 µL) of PBS as each sample. Total RNA was extracted from these embryos using the PureLink RNA Mini Kit (Invitrogen by Thermo Fisher Scientific, CA, USA) following the instruction manual. Extracted RNA was dissolved in 30 µL RNase‐free water.
Total RNA samples were reverse‐transcribed into cDNA using the Verso cDNA Synthesis Kit (Thermo Fisher Scientific, ABgene, UK) following the instruction manual. Gene transcripts were quantified in three replicates by quantitative RT‐PCR with specific primers (Table 1) using an Agilent Technologies Stratagene Mx3000P with Brilliant III (Agilent Technologies, CA, USA). The total reaction volume was 25 µL and contained Brilliant III, forward and reverse primers, and cDNA template. Reaction conditions were as follows: 2 minutes at 95°C, followed by 40 cycles of 5 seconds at 95°C and 20 seconds at 60°C. Data were normalized to the expression level of the 18s, ribosomal RNA gene.
Table 1.
Quantitative PCR primer sequences
| Gene | Primers (5′‐3′) | Product size (bp) | Annealing temperature (°C) | |
|---|---|---|---|---|
| Mater | Forward | CCGGCTTACAAAAGCTCAAG | 128 | 60 |
| Reverse | GACTTGGTTTTTGCGATGGT | |||
| Npm2 | Forward | CAGCCCTCAGGACAAGAGTC | 108 | 60 |
| Reverse | CCCTCGGACACAGAGATACC | |||
| Hsf1 | Forward | TCTCACTGGTGCAGTCGAAC | 154 | 60 |
| Reverse | GGAGACGGAGCTGAGTATGG | |||
| Hspa1a | Forward | CGACCTGAACAAGAGCATCA | 213 | 60 |
| Reverse | AAGATCTGCGTCTGCTTGGT | |||
| Myc | Forward | CGACTCCACAGCCTTCTCTC | 179 | 60 |
| Reverse | GCTGCCTCTTTTCCACAGAC | |||
| Hdac1 | Forward | CGCATGACTCACAATTTGCT | 134 | 60 |
| Reverse | CGAATGGAACGCAAGAATTT | |||
| 18 s | Forward | ATGGCCGTTCTTAGTTGGTG | 217 | 60 |
| Reverse | CGCTGAGCCAGTCAGTGTAG |
2.8. Embryo transfer
About 78 hours after ICSI, we randomly selected 43 morulae and blastocysts for transferred into recipient albino females that had been naturally mated with albino males 3 days previously. We transferred 4‐7 morulae/blastocysts into the uterus of each recipient hamster. The mothers were allowed to deliver and raise their own pups (albino) as well as the foster pups (brown coat and black eyes).4, 5, 6, 27
2.9. Statistical analysis
The chi‐square test was used to compare fertilization and embryo development rates, and chromosome analysis results. Quantitative PCR data were compared using Student's t test. Differences between means were evaluated using GraphPad PRISM 5 (GraphPad Software, La Jolla, CA, USA). Differences were considered to be significant at P < 0.05.
3. RESULTS
3.1. Chromosome analysis of hamster spermatozoa after sonication in TCM199 or TBS + EGTA
Table 2 summarizes the results of chromosomal analysis of hamster spermatozoa following injection into mouse oocytes. The percentage of normal karyotypes was significantly higher in spermatozoa that were frozen‐stored and sonicated in TBS + EGTA (89.7%) than in TCM199 (69.0%) or TBS + Ca (68.0%; P < 0.05). Chromosome‐type breaks were the chromosome aberration most commonly observed after sonication (Figure 1A,B).
Table 2.
Chromosome analysis of hamster spermatozoa that were frozen‐stored; separated by sonication in TCM199, TBS + EGTA, or TBS + Ca; and then injected into mouse oocytes
| Medium used for storage and sonication | No. of zygotes analyzed | No. (%) of oocytes with normal sperm chromosomes | No. of spermatozoa with chromosome aberrations | |
|---|---|---|---|---|
| Structural* | Aneuploidy | |||
| TCM199 | 58 | 40 (69.0)b | 18 | 0 |
| TBS + EGTA | 58 | 52 (89.7)a | 6 | 0 |
| TBS + Ca | 50 | 34 (68.0)b | 16 | 0 |
TBS + EGTA, 100 mM Tris‐HCl–buffered solution +50 mM EGTA; TBS + Ca, 100 mM Tris‐HCl–buffered solution +2 mM CaCl2 .
Significantly different values (P < 0.05) between different superscripts. Percentages of oocytes with normal sperm chromosomes were analyzed by Fisher's exact probability test and chi‐square test.
Structural chromosome aberrations included chromosome/chromatid‐type breaks and exchanges.
Figure 1.
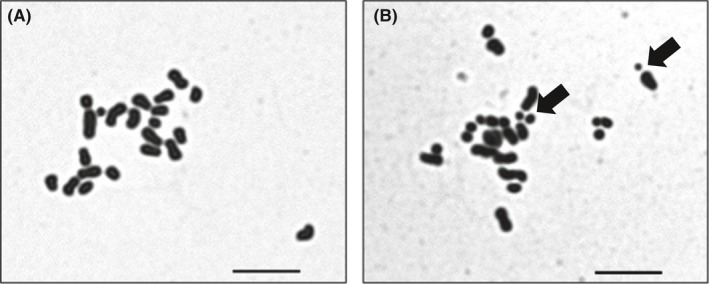
Chromosome spreads of hamster spermatozoa, prepared by sonicating the spermatozoa in TCM199 or TBS + EGTA, and then injecting sperm heads into mouse oocytes. Representative normal (A) and abnormal (B) hamster sperm chromosomes are shown. Arrows indicate chromosome fragments. n = 22 normal, n > 22 abnormal. Bars = 10 μm
3.2. In vitro development of hamster embryos after injecting sonicated sperm heads into oocytes
Table 3 summarizes the results regarding the in vitro development of hamster embryos produced by microinjection of sperm heads that were sonicated in TCM199 or TBS + EGTA. The percentages of normally fertilized oocytes and 2‐cell embryos did not significantly differ between the TCM199 and TBS + EGTA conditions. However, compared to TCM199, TBS + EGTA yielded significantly higher percentages of 8‐cell embryos (84.1% vs 65.4%), morulae (78.4% vs 43.2%), and blastocysts (42.0% vs 17.3%).
Table 3.
In vitro development of hamster oocytes injected with sperm heads separated by sonication in TCM199 or TBS + EGTA
| Medium used for storage and sonication | No. of oocytes injected (no. of replicates) | No. of oocytes with 2PN (%) | No. (%) of embryos developed to | |||
|---|---|---|---|---|---|---|
| ≥2‐cell | ≥8‐cell | ≥Morulae | Blastocysts | |||
| TCM199 | 81 (4) | 75 (92.6)a | 72 (88.9)a | 53 (65.4)a | 35 (43.2)a | 14 (17.3)a |
| TBS + EGTA | 88 (4) | 80 (90.9)a | 80 (90.9)a | 74 (84.1)b | 69 (78.4)b | 37 (42.0)b |
TBS + EGTA, 100 mM Tris‐HCl–buffered solution +50 mM EGTA; 2PN, two well‐developed pronuclei.
Significantly different values (P < 0.05) between different superscripts within the same column. Percentage of embryo development was analyzed by chi‐square test.
3.3. Gene expression of MEGs (Mater, Npm2, and Hsf1) in pronuclear embryos and ZGA (Hspa1a, Myc, and Hdac1) in 2‐cell embryos
Figure 2 shows the differences in expression of MEGs and ZGA genes between ICSI embryos in TCM199 and TBS + EGTA. Expression of maternal effect genes (Mater, Npm2, and Hsf1) in pronuclear embryos did not significantly differ between TCM199 and TBS + EGTA. In contrast, the marker genes for zygotic genome activation showed significantly upregulated the transcription levels in 2‐cell embryos with TBS + EGTA compared to TCM199.
Figure 2.
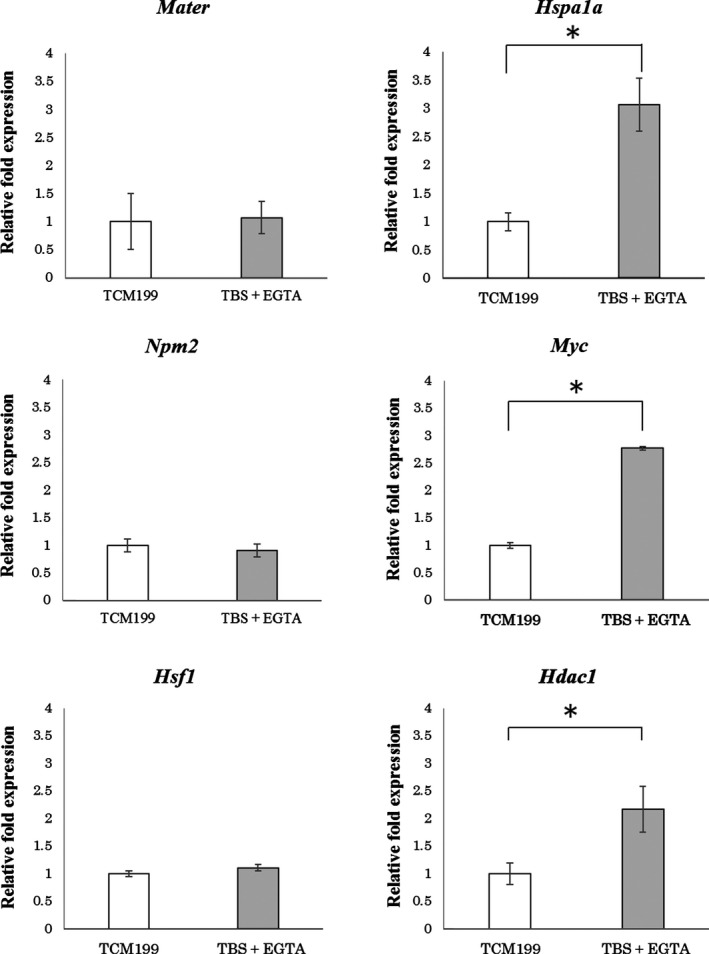
Expression of maternal effect genes (MEGs; Mater, Npm2, and Hsf1) in pronuclear embryos, and zygotic gene activation genes (ZGA; Hspa1a, Myc, and Hdac1) in 2‐cell embryos. ICSI embryos were produced by sonication of hamster spermatozoa in TCM199 or TBS + EGTA. Analyses were performed using primer sets Mater, Npm2, and Hsf1 at 6 h post‐ICSI, and Hspa1a, Myc, and Hdac1 at 44 h post‐ICSI. *P < 0.05, Student's t test
3.4. Full‐term development of embryos fertilized by injection of sperm heads sonicated in TBS + EGTA
Table 4 summarizes the result of embryo transfer. A total of 43 embryos derived from the injection of sperm heads sonicated in TBS + EGTA were transferred to recipients, of which 4 (9.3%) developed into live young. Figure 3 shows a recipient mother that gave birth to one pup (brown fur) that developed from the oocytes fertilized by injection of sonicated sperm heads in TBS + EGTA.
Table 4.
In vivo development of golden hamster zygotes that were injected with sperm heads separated by sonication in TBS‐EGTA and then transferred into pregnant albino hamsters
| Recipient ID | No. of embryos transferred | No. (%) of live offspringa | ||
|---|---|---|---|---|
| Recipient's own (albino) |
Foster (%) (golden) |
Sex of foster offspringb | ||
| a | 9 | 6 | 1 (11.1) | M |
| b | 13 | 8 | 1 (7.7) | M |
| c | 11 | 9 | 1 (9.1) | M |
| d | 10 | 13 | 1 (10.0) | F |
| Total | 43 | 36 | 4 (9.3) | 3M, 1F |
Offspring origin was determined by eye and fur color at 3 weeks after birth.
Babies were sexed at 3 weeks after birth.
Figure 3.

Pups with brown coat (indicated by arrow) were conceived from the injection of hamster oocytes injected with sperm heads separated by sonication of frozen‐thawed spermatozoa in TBS + EGTA. The recipient's own pups were albino
4. DISCUSSION
Our present results showed that TBS + EGTA was effective for cryopreservation and sonication of hamster spermatozoa, enabling the injection of sperm heads with a higher proportion of normal chromosomes (Table 2). The percentages of 8‐cell embryos, morulae, and blastocysts were significantly higher in TBS + EGTA than in TCM199 (Table 3). Furthermore, after transfer of 43 ICSI embryos, 4 developed into live offspring (Table 4). These results indicated that the use of TBS + EGTA for cryopreservation and sonication was effective for maintaining sperm chromosomal normality, and supported the development of zygotes fertilized by sperm head injection into oocytes.
The incidence of structural chromosome aberrations in hamster spermatozoa was significantly reduced when the spermatozoa were frozen‐stored and sonicated in TBS + EGTA compared to in TCM199. Moreover, sonication in TBS + Ca (TBS containing 2 mM calcium chloride which is the same concentration as in TCM199) led to a significantly increased incidence of sperm chromosome aberrations. The percentages of chromosome aberrations were similar between TCM199 and TBS + Ca. These findings suggest that the presence of calcium ions during spermatozoa sonication activates DNase in sperm heads, thus increasing structural chromosome aberration. Since divalent cations are essential for DNase activity, chelating agents (eg, EGTA) would suppress their activity.28
To analyze hamster sperm chromosomes, we performed xenogeneic injection of hamster sperm heads separated by sonication into mouse oocytes. This method of interspecies sperm chromosome analysis using mouse oocytes is systematic, and the procedure is completely controlled.18, 24, 25 The xenogeneic system makes it easy to identify the hamster sperm chromosomes. However, future studies should include homogeneous sperm chromosome analysis using hamster oocytes to determine the relationship between sperm chromosome normality and ICSI embryo development.
The sonication solution contained 50 mM EGTA to prevent chromosome breakage by DNase in the sperm heads. Prior to sperm head injection, the EGTA solution with sperm heads was mixed and diluted by one‐third with 12% PVP solution (final volume ~17 mM). With an estimated injection volume of ~1 pL, the injected amount of EGTA is estimated to be ~17 fmol. Changes in the intercellular calcium concentration underlie the initiation, progression, and completion of fertilization.29 In our present study, hamster ICSI with EGTA did not disturb pronuclear formation, nor did the amount of EGTA injected into the ooplasm with the sperm head seem to affect the fertilization process. However, the present data are insufficient to confirm the minimal EGTA concentration for sonication and sperm injection, and further investigations are needed to optimize ICSI.
Mouse and hamster embryonic genome activation occur at the 2‐cell embryo stage.30, 31 Bovine embryonic genome is activated at the 8‐cell embryo stage, and embryo arrest reportedly occurs mainly during embryonic gene activation.32, 33 Hspa1a and Myc are ZGA marker genes, and the gene expression in 2‐cell embryos derived from ICSI is significantly lower than in vivo.34, 35, 36, 37 The histone deacetylase gene Hdac1 is one factor that controls transcription, and Hdac1 knockout leads to arrested embryonic development in mice.38 The mRNA expression of Hdac1 in hamster 2‐cell embryos derived from ICSI is lower than in vivo.37 In our present study, MEGs expression in pronuclear embryos did not significantly differ between the TBS + EGTA and TCM199 groups, suggesting oocyte quality did not differ between the two groups. On the other hand, expression of ZGA genes in 2‐cell embryos derived from ICSI was significantly higher in TBS + EGTA than in TCM199. This indicates that zygotes fertilized by injection of sonicated sperm heads with normal chromosomes exhibited embryonic gene expression (HSPa1a, Myc, and Hdac1), with appropriate timing.
The percentage of blastocysts developed at 96 hours after sperm head injection was higher in TBS + EGTA than in TCM199 (42.0% vs 17.3%; Table 3). To evaluate in vivo development of ICSI embryos generated using TBS + EGTA, at about 78 hours after injection, we transferred morulae and blastocysts (mostly morulae) to each uterine horn of day 3 pregnant albino females. Sato and Yanagimachi and Bavister et al report that on day 3 of pregnancy, hamster embryos develop to the 8‐cell stage at 12:00, to morulae at 18:00, and to early blastocysts at 24:00.39, 40 In hamster embryo transfer, synchronous timing between donor embryos and recipient females is important for in vivo embryo development.27, 39, 40 In our present study, at about 78 hours after injection, we transferred mainly morulae into albino recipient hamsters at day 3 of pregnancy. The development speed of ICSI embryos was delayed by about a half day, and the percentage of blastocysts developed from morulae was much lower among the ICSI embryos than the in vivo‐fertilized embryos (unpublished). Therefore, the ICSI embryos developed into live offspring at a lower rate, as has been described in previous reports.4 To produce more offspring, we must further improve the developmental speed and quality of ICSI embryos.
It is important to be able to produce hamster ICSI embryos using sonicated sperm heads and to enhance the efficiency of sperm injection per unit time. The rapid ICSI system is necessary to reduce the time from oocyte collection to injection, since spontaneous oocyte activation is common in hamsters. To produce higher quality hamster ICSI embryos, we must improve the manipulation of sonicated sperm heads to enable smooth fertilization, optimize the culture medium for ICSI embryos, and regulate spontaneous oocyte activation.
In conclusion, here we found that the use of TBS + EGTA for sonication of hamster frozen‐thawed spermatozoa reduced sperm chromosome aberrations. Upon injecting hamster oocytes with sonicated sperm heads having less chromosome aberrations, we found that the ZGA gene expression at the two‐cell stage was relatively higher than with TCM199. Moreover, the ICSI zygotes produced using TBS + EGTA developed to morulae and blastocysts at a higher rate compared to with TCM199. Finally, ICSI embryos produced with TBS + EGTA successfully developed to live offspring after embryo transfer. Overall, these results suggest that rapid ICSI using sperm heads separated by sonication in TBS + EGTA leads to increased chromosome normality and effective production of live offspring.
Disclosures
Conflict of interest: The authors declare no conflict of interest. Human rights statement and informed consent: This article does not contain any study with human participants that has been performed by any of the authors. Animal studies: All the experiments in this research were approved by the Committee for Ethics on Animal Experiments of the Prefectural University of Hiroshima, Japan (16SA002).
Morishita N, Ochi M, Horiuchi T. Development of golden hamster embryos effectively produced by injection of sperm heads sonicated in Tris‐HCl buffer with EGTA. Reprod Med Biol. 2019;18:83–90. 10.1002/rmb2.12253
REFERENCES
- 1. Uehara T, Yanagimachi R. Microsurgical injection of spermatozoa into hamster eggs with subsequent transformation of sperm nuclei into male pronuclei. Biol Reprod. 1976;15:467‐470. [DOI] [PubMed] [Google Scholar]
- 2. Yanagimachi R. Intracytoplasmic injection of spermatozoa and spermatogenic cells: its biology and applications in humans and animals. RBM Online. 2005;10:247‐286. [DOI] [PubMed] [Google Scholar]
- 3. Van Steirteghem A, Nagy Z, Joris H, et al. High fertilization and implantation rates after intracytoplasmic sperm injection. Hum Reprod. 1993;8:1061‐1066. [DOI] [PubMed] [Google Scholar]
- 4. Yamauchi Y, Yanagimachi R, Horiuchi T. Full‐term development of golden hamster oocytes following intracytoplasmic sperm head injection. Biol Reprod. 2002;67:534‐539. [DOI] [PubMed] [Google Scholar]
- 5. Haigo K, Yamauchi Y, Yazama F, Yanagimachi R, Horiuchi T. Full‐term development of hamster embryos produced by injection of round spermatids into oocytes. Biol Reprod. 2004;71:194‐198. [DOI] [PubMed] [Google Scholar]
- 6. Muneto T, Horiuchi T. Full‐term development of hamster embryos produced by injecting freeze‐dried spermatozoa into oocytes. J Mamm Ova Res. 2011;28:32‐39. [Google Scholar]
- 7. Hirao Y, Yanagimachi R. Detrimental effect of visible light on meiosis of mammalian eggs in vitro. J Exp Zool. 1978;206:365‐369. [DOI] [PubMed] [Google Scholar]
- 8. Umaoka Y, Noda Y, Nakayama T, Narimoto K, Mori T, Iritani A. Effect of visual light on in vitro embryonic development in the hamster. Theriogenology. 1992;38:1043‐1054. [DOI] [PubMed] [Google Scholar]
- 9. Takenaka M, Horiuchi T, Yanagimachi R. Effects of light on development of mammalian zygotes. PNAS. 2007;104:14289‐14293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kimura Y, Yanagimachi R, Kuretake S, Bortkiewicz H, Perry A, Yanagimachi H. Analysis of mouse oocyte activation suggests the involvement of sperm perinuclear material. Biol Reprod. 1998;58:1407‐1415. [DOI] [PubMed] [Google Scholar]
- 11. Morozumi K, Yanagimachi R. Incorporation of the acrosome into the oocyte during intracytoplasmic sperm injection could be potentially hazardous to embryo development. PNAS. 2005;102:14209‐14214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yoo E. Spontaneous activation and cleavage in unfertilized ova of the golden hamster (Mesocricetus auratus). Biol Reprod. 1990;42:93 (abstract). [Google Scholar]
- 13. He F, Liow S, Chen M, Ng S, Ge Q. Spontaneous activation of hamster oocytes in vitro. Chin Med J. 1999;112:1105‐1108. [PubMed] [Google Scholar]
- 14. Sun XS, Yue KZ, Zhou JB, Chen QX, Tan JH. In vitro spontaneous parthenogenetic activation of golden hamster oocytes. Theriogenology. 2002;57:845‐851. [DOI] [PubMed] [Google Scholar]
- 15. Yanagida K, Yanagimachi R, Perreault SD, Kleinfeld RG. Thermostability of sperm nuclei assessed by microinjection into hamster oocytes. Biol Reprod. 1991;44:440‐447. [DOI] [PubMed] [Google Scholar]
- 16. Katayose H, Matsuda J, Yanagimachi R. The ability of dehydrated hamster and human sperm nuclei to develop into pronuclei. Biol Reprod. 1992;47:277‐284. [DOI] [PubMed] [Google Scholar]
- 17. Kuretake S, Kimura Y, Hoshi K, Yanagimachi R. Fertilization and development of mouse oocytes injected with isolated sperm heads. Biol Reprod. 1996;55:789‐795. [DOI] [PubMed] [Google Scholar]
- 18. Tateno H, Kimura Y, Yanagimachi R. Sonication Per Se Is Not as Deleterious to Sperm Chromosomes as Previously Inferred. Biol Reprod. 2000;63:341‐346. [DOI] [PubMed] [Google Scholar]
- 19. Kusakabe H, Szczygiel MA, Whittingham DG, Yanagimachi R. Maintenance of genetic integrity in frozen and freeze‐dried mouse spermatozoa. PNAS. 2001;98:13501‐13506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kusakabe H, Yanagimachi R, Kamiguchi Y. Mouse and human spermatozoa can be freeze‐dried without damaging their chromosomes. Hum Reprod. 2008;23:233‐239. [DOI] [PubMed] [Google Scholar]
- 21. Orsini MW. The external vaginal phenomena characterizing the stage of the estrous cycle, pregnancy, pseudopregnancy, lactation, and the anestrous hamster, Mesocricetus auratus waterhouse. Proc Anim Care Panel. 1961;11:193‐206. [Google Scholar]
- 22. Yanagimachi R. Developmental ability of precociously superovulated golden hamster eggs. Gamete Res. 1984;9:231‐237. [Google Scholar]
- 23. Quinn P, Barros C, Whittingham DG. Preservation of hamster oocytes to assay the fertilizing capacity of human spermatozoa. J Reprod Fertil. 1982;66:161‐168. [DOI] [PubMed] [Google Scholar]
- 24. Kishikawa H, Tateno H, Yanagimachi R. Chromosome analysis of BALB/c mouse spermatozoa with normal and abnormal head morphology. Biol Reprod. 1999;61:809‐812. [DOI] [PubMed] [Google Scholar]
- 25. Kamiguchi Y, Mikamo K. An improved, efficient method for analyzing human sperm chromosomes using zona‐free hamster ova. Am J Hum Genet. 1986;38:724‐740. [PMC free article] [PubMed] [Google Scholar]
- 26. Mckiernan SH, Bavister BD. Culture of one‐cell hamster embryos with water soluble vitamins: pantothenate stimulates blastocyst production. Hum Reprod. 2000;15:157‐164. [DOI] [PubMed] [Google Scholar]
- 27. Barnett DK, Bavister BD. Hypotaurine requirement for in vitro development of golden hamster one‐cell embryos into morulae and blastocysts, and production of term offspring from in vitro‐fertilized ova. Biol Reprod. 1992;47:297‐304. [DOI] [PubMed] [Google Scholar]
- 28. Clark P, Eichhorn GL. A predictable modification of enzyme specificity. Selective alteration of DNA bases by metal ions to promote cleavage specificity by deoxyribonuclease. Biochemistry. 1974;13:5098‐5102. [DOI] [PubMed] [Google Scholar]
- 29. Wakai T, Vanderheyden V, Fissore R. Ca2+ signaling during mammalian fertilization: Requirements, players, and adaptations. Cold Spring Harbor Prespect Biol. 2011;3:a006767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Flach G, Johnson MH, Braude PR, Traylor RA, Bolton VN. The transition from maternal to embryonic control in the 2‐cell mouse embryo. EMBO J. 1982;1:681‐686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Seshagiri PB, McKenzie DI, Bavister BD, Williamson JL, Aiken JM. Golden hamster embryonic genome activation occurs at the two‐cell stage: correlation with major developmental changes. Mol Reprod Dev. 1992;32:229‐235. [DOI] [PubMed] [Google Scholar]
- 32. Frei RE, Schultz GA, Church RB. Qualitative and quantitative changes in protein synthesis occur at the 8–16‐cell stage of embryogenesis in the cow. J Reprod Fertil. 1989;86:637‐641. [DOI] [PubMed] [Google Scholar]
- 33. Meirelles FV, Caetano AR, Watanabe YF, et al. Genome activation and developmental block in bovine embryos. Anim Reprod Sci. 2004;82–83:13‐20. [DOI] [PubMed] [Google Scholar]
- 34. Sonehara H, Nagata M, Aoki F. Roles of the first and second round of DNA replication in the regulation of zygotic gene activation in mice. J Reprod Dev. 2008;54(5):381‐384. [DOI] [PubMed] [Google Scholar]
- 35. Shin SW, Tokoro M, Nishikawa S, et al. Inhibition of the ubiquitin‐proteasome system leads to delay of the onset of ZGA gene expression. J Reprod Dev. 2010;56(6):655‐663. [DOI] [PubMed] [Google Scholar]
- 36. Christians E, Campion E, Thompson EM, Renard JP. Expression of the HSP 70.1 gene, a landmark of early zygotic activity in the mouse embryo, is restricted to the first burst of transcription. Development. 1995;121:113‐122. [DOI] [PubMed] [Google Scholar]
- 37. Pan X, Delong Kong D, Liu L, et al. Development block of golden hamster ICSI embryos is associated with decreased expression of HDAC1, HSPA1A and MYC. Cell Bio Int. 2014;38:1280‐1290. [DOI] [PubMed] [Google Scholar]
- 38. Montgomery RL, Davis CA, Potthoff MJ, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790‐1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sato A, Yanagimachi R. Transplantation of preimplantation hamster embryos. J Reprod Fert. 1971;30:329‐332. [DOI] [PubMed] [Google Scholar]
- 40. Bavister BD, Leibfried ML, Liebferman G. Development of preimplantation embryos of the golden hamster in a defined culture medium. Biol Reprod. 1983;28:235‐247. [DOI] [PubMed] [Google Scholar]


