Abstract
Preimplantation genetic testing (PGT) is an early form of prenatal genetic diagnosis where abnormal embryos are identified, thereby allowing transfer of genetically normal embryos. This technology has become an integral part of Assisted Reproductive Technology (ART) procedures. Initial experiments with animals as early as 1890 and those in the mid and later part of the last century paved the forward path of ART and PGT. This review article covers the evolution of PGT and is a pointer toward current and fast-evolving technology, allowing scientists and doctors to better comprehend human reproduction, and ensure healthy pregnancy outcomes.
KEYWORDS: Assisted reproductive technology, preimplantation genetic diagnosis, preimplantation genetic screening, preimplantation genetic testing, fluorescence in situ hybridization, array comparative genomic hybridization, next-generation sequencing
INTRODUCTION
Preimplantation genetic testing (PGT) is an early form of prenatal genetic diagnosis where abnormal embryos are identified, and only genetically normal embryos are used for implantation. This has become an integral part of Assisted Reproductive Technology (ART) procedures.
Historically, the development of PGT technology dates back to 1890 with Walter Heape's experiments of successfully transferring embryos in the Belgian Hare doe rabbits.[1] Animal experiments continued through the first half of the 20th Century. In 1935, Gregory Pincus, inspired by Heape's results, was able to culture rabbit oocytes to the metaphase stage of meiosis II. Professor Robert Edwards discovered that human oocytes required 37 hours for polar body extrusion and having timed each stage of human oocyte maturation, he led the way for human in vitro fertilization (IVF). Bob Edwards was the one who ideated PGT in the mid 60s. Edwards and Gardner in 1967, using euchrysine 2GNX vital staining technique, stained rabbit blastocyst sex chromatin.[2] The preparation was observed under the fluorescence microscope. As this technique was potentially mutagenic, it was not compatible with embryo transfer. Hence, in 1968, they biopsied 200–300 rabbit trophoblast cells. These cells were further stained for sex chromatin. The biopsied blastocyst was implanted into a pseudopregnant female rabbit. At full term, the sex of the fetus was confirmed anatomically and histologically.[3] This experiment became the basis for PGT and its application to test for genetically inherited diseases.
Steptoe and Edwards, in their Manchester Laboratory, made many attempts since 1970 onwards to establish IVF in humans. Dr. Carl Wood of the Monash IVF team in Melbourne reported the first IVF pregnancy in 1973, although it resulted in an early miscarriage. In 1976, Steptoe and Edwards published a case of ectopic pregnancy following transfer of an early blastocyst. After several failed attempts, medical history was made on July 25, 1978, with the birth of the world's first “test tube baby” Louis Brown.[4] The reintroduction of ovarian stimulation by Trounson et al. in 1981 was a major breakthrough that increased the chances of pregnancy in IVF.[5]
Although pregnancy rates improved with time, results of IVF for male factor infertility remained very low with failed fertilization occurring commonly as the sperm of these men did not have the ability to perform all the steps needed for fertilization.
To characterize the fertilization potential of human sperm, the hamster egg-human sperm penetration assay was developed.[6] In their paper, Li et al. demonstrated Uehara and Yanagimachi's classic work in 1976 of injection of human sperm into hamster oocytes showing sperm nuclear decondensation [Figure 1].[7] The technique of sperm microinjection was pioneered by Hiramoto in the Sea Urchin in 1962[8] and by Lin in 1966 in mouse oocytes.[9]
Figure 1.
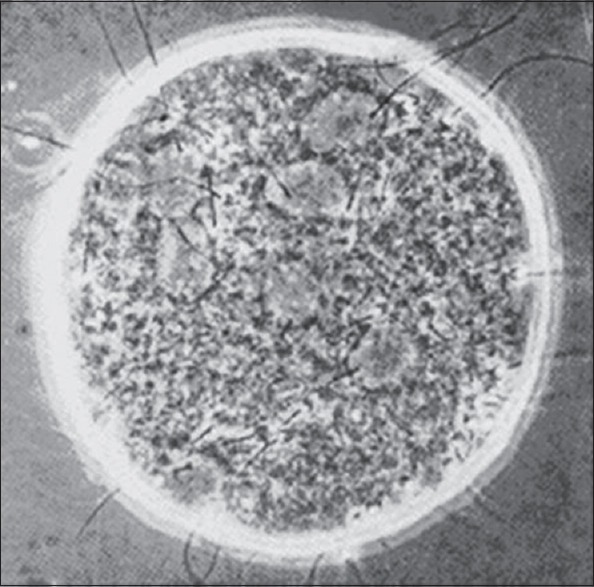
Sperm nuclear decondensation in zona-free hamster oocyte
The practical use of micromanipulation started in the mid 80's with zona drilling (ZD) and partial zona dissection (PZD) when the sperm count, motility, or morphology were low. Pioneering attempts of ZD on a mouse model were carried out by Gordon et al., using a micromanipulator to produce holes in the zona pellucida (ZP) of unfertilized mouse oocytes with acid Tyrode's solution.[10] The first attempts at ZD of human oocytes for the alleviation of male infertility resulted in fertilization, however, pregnancy did not ensue in the ten couples included in this report.[11]
The first live birth in the world with embryo micromanipulation techniques was reported by Ng and Bongso from Singapore, where insemination was done under the ZP.[12] This micro-insemination sperm transfer technique later became popularly known as subzonal injection of sperm (SUZI).[13] The earlier PZD technique did not give good results and was discontinued as it led to polyspermy, while SUZI gave better results and eventually led to the development of intracytoplasmic sperm injection (ICSI). Thus, by the end of the 1980's, several procedures of assisted fertilization had been developed and used where conventional IVF could not succeed. Microsurgical fertilization techniques helped to remove the barrier presented to the sperm by the ZP. Assisted hatching was pioneered by Cohen around the same time.[14]
To achieve fertilization in nature, the sperm has to penetrate the cumulus cells. This is followed by zona binding and penetration, egg-sperm membrane interaction, and oocyte activation. Lanzendorf initiated sperm microinjection. However, the fertilized oocyte only went up to the pronuclear stage. He, therefore, abandoned the technique.[15]
In 1992, Gianpiero Palermo, in Dr. André van Steirteghem's Laboratory in Brussels, created the first baby by sperm microinjection into the oocyte cytoplasm. The team called it ICSI [Figure 2].[16] This discovery got them international acclaim.
Figure 2.
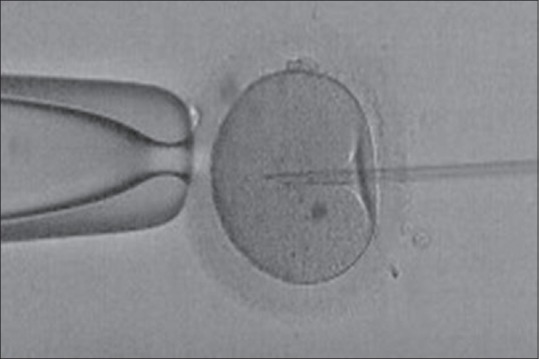
Intracytoplasmic sperm injection – Sperm is microinjected into the cytoplasm of the oocyte
In India, the first ICSI baby of South Asia “Luv Singh,” was created by our team at Jaslok Hospital, Mumbai, in 1994.[17]
BIOPSY TECHNIQUES
Parallel to the development of IVF technology, many experiments were being performed on animal models for obtaining a single cell from the growing embryos in vitro for future genetic analysis. Wilton and Trounson from Australia demonstrated the technique of removal of one blastomere from cleavage-stage embryos in the mouse.[18] In 1988, Marilyn Monk with Audrey Muggleton-Harris from UK developed the trophectoderm biopsy technique followed by Preimplantation Genetic Diagnosis (PGD) using biochemical microassay in a mouse model for Lesch–Nyhan disease.[19] Yury Verlinsky's group demonstrated the use of the first polar body biopsy to check for a maternal unaffected gene.[20] The Figures 3a–c show human gametes and embryos undergoing cleavage stage, tropehctoderm and polar body biopsy.
Figure 3.
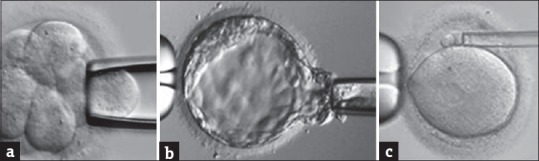
Biopsy techniques: (a) Blastomere biopsy (b) trophectoderm biopsy (c) polar body biopsy
DEVELOPMENT OF MOLECULAR TECHNOLOGY
After the presentation of the Watson and Crick model of DNA in 1953, attempts to develop synthetic oligonucleotides and to sequence the genomic DNA were made. The technique of polymerase chain reaction (PCR) was introduced by Saiki et al. in 1985.[21] This was a major breakthrough for the analysis of monogenic disorders in the field of molecular biology.
FIRST SUCCESSFUL PREIMPLANTATION GENETIC DIAGNOSIS ATTEMPTS
Successful attempts in the mouse model bore fruition in 1990, when Handyside et al. reported pregnancies after carrying out PGD for sex-linked disease and X-linked mental retardation on biopsied human preimplantation embryos. The PCR technique was carried out to detect male embryos free of the X-linked disease.[22]
Handyside's group included Wilton and Delhanty with her Ph.D. student Griffin. They introduced the world's first PGD cases where male, female, and Turner syndrome embryos could be easily identified using the fluorescence in situ hybridization (FISH) technique. The biopsy was carried out by Handyside; the cells were fixed on the slide by Wilton and FISH analysis was carried out by Griffin et al.[23] Simultaneously, Munné et al., from USA, applied PGD using the FISH technique for the first time using directly labeled probes.[24]
With this many groups started using PGD technology for testing for aneuploidy and translocations by FISH and monogenic disorders by PCR. By 2001, Verlinsky et al. from Chicago reported the first successful PGD with human leukocyte antigen matching for a sib with Fanconi anemia by haplotype analysis.[25] This led to the concept of “Savior Sib.” Using disease-free HLA-matched embryos for implantation, the previously affected child could be cured using the transplantation of cord stem cells, and bone marrow of the unaffected baby created free of disease by PGD.
The FISH technology was further improved using different probe mixtures for 5–12 chromosome pairs in multiple rounds. It was offered to women with advanced maternal age, with a history of recurrent abortions, implantation failures as well as inherited Robertsonian or reciprocal translocations and inversions. The main limitation of the FISH technology was that only around 5–12 pairs of chromosomes could be tested for aneuploidy from a total of 23 pairs of human chromosomes. Hence, further research was initiated for developing newer techniques which could test all chromosomes for aneuploidies using a single blastomere within 24-72 hours of the biopsy.
In 1999, two different groups, Wells et al. and Voullaire et al. demonstrated the use of Comparative Genomic Hybridization (CGH) technology on human blastomeres to check for aneuploidies of all chromosomes.[26,27] In 2000, Voullaire et al. did an extensive study of 12 human embryos using CGH technique on more than 60 blastomeres. The study demonstrated the presence of partial aneuploidy as well as gain and loss of fragments of chromosomes which were not previously identified using the FISH analysis.[28]
Wilton's group, in 2001, successfully applied PGD by CGH in a 38-year-old female with a history of primary infertility followed by an unsuccessful attempt at IVF by using FISH on IVF-PGD embryos. After testing by CGH, only one of five embryos turned out to be normal for every chromosome. This effort resulted in the birth of a healthy female child.[29] Thereafter, CGH technology was offered to many couples successfully for the detection of all chromosomal aneuploidies and unbalanced translocations. The major drawback of the technology was the need for cryopreservation of embryos as several days were required for testing. The other drawback was the inability of this technology to detect triploidy or tetraploidy. Based on all these attempts and results in the first decade of this century, FISH still remained the most popular technique for the detection of aneuploidies within 24-72 hours.
In spite of these pioneering attempts, why did PGD not become popular?
Mastenbroek's group, in 2007, published a paper of a multicentric, randomized, controlled trial (RCT) where they compared three cycles of IVF with and without Preimplantation Genetic Screening (PGS) in women in the age group of 35–41 years. They showed that the on-going pregnancy rates and live birth rates were 10% lower in women undergoing PGS by FISH, compared to the non-PGS group in cases of advanced maternal age.[30] This publication led to less use of PGS for the next few years.
The concept of better pregnancy outcome using this technology finally picked up when Munne and other scientists demonstrated its benefits.
New terminology was developed to differentiate between aneuploidy screening and detection of monogenic disorders. Aneuploidy screening was termed as “PGS” whereas testing for monogenic disorders was termed as “PGD.” The aneuploidy detection using FISH was termed as version 1 (PGS v1) whereas aneuploidy detection for all 24 chromosomes has now become version 2 (PGS v2). Recently, a new term Preimplantation Genetic Testing (PGT) has been introduced. The recently modified terminology is PGT-A for aneuploidy screening, PGT-SR for structural rearrangements (translocation or inversion), and PGT-M for monogenic disorders.
Considering the difficulties in the use of FISH technology, several groups perfected the long learning curve for PGT which included perfecting embryo biopsy techniques without harming the embryo and genetic diagnosis using different molecular techniques. Different groups studied the effect of day 3 cleavage-stage biopsy and day 5 blastocyst biopsy on embryo implantation and live birth outcomes. New methods were introduced for the detection of aneuploidy of all chromosomes within 24–48 hours.
Based on Wilton's use of CGH technology for preimplantation diagnosis for aneuploidy, Wells et al. with his team published a paper in 2008 including the use of microarray and CGH platforms for the detection of aneuploidy of all 23 chromosome pairs.[31] This technique was validated in 2011.[32] In the same year, Wells' group reported the first births after Preimplantation Genetic Diagnosis of structural chromosome abnormalities using array CGH (aCGH).[33]
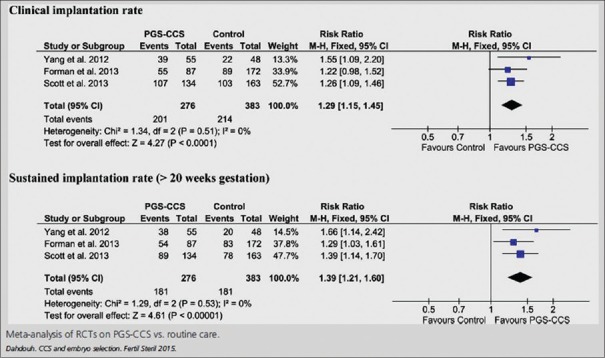
In 2013, Scott's group published their clinical trial showing that the biopsy of cleavage-stage embryos significantly impaired implantation potential; however, trophectoderm biopsy of blastocyst did not have any negative effect on implantation [Figure 4].[34] Capalbo et al. study showed that trophectoderm biopsy should be performed on all day 5, day 6, or day 7 blastocyst stage embryos to improve implantation outcome [Figure 5].[35] Several authors have shown the benefits of PGT to improve implantation rates (IRs). Dahdouh et al. carried out a meta-analysis of RCTs and observational studies to see whether PGS with comprehensive chromosome screening (CCS) improved clinical IR and sustained IR (beyond 20 weeks) compared with routine care for embryo selection in IVF cycles. They concluded that PGS with the use of CCS technology increases clinical and sustained IRs, thus improving embryo selection particularly in patients with normal ovarian reserve [Figure 6].[36] Other studies have shown an improvement in ICSI outcome with PGT.[37]
Figure 4.
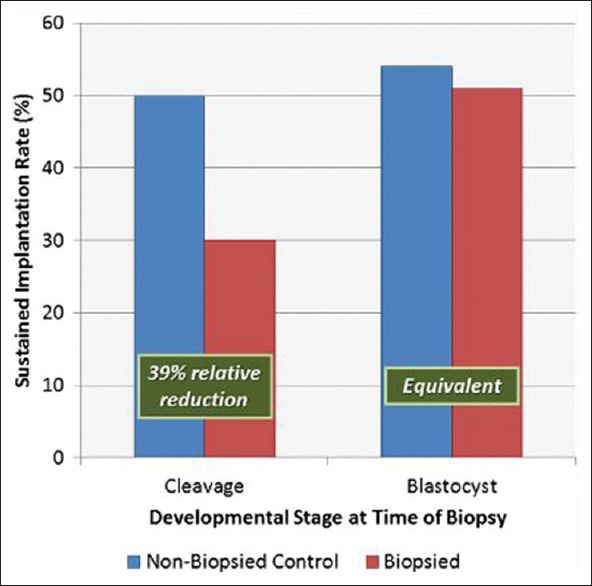
Day 3 versus day 5 biopsy. Implantation rates following a randomized paired analysis of the effects of cleavage-stage and blastocyst-stage biopsies on embryo reproductive potential. Sustained implantation and delivery of the biopsied embryo were significantly reduced compared with its control sibling, when the biopsy was performed on day 3 at the cleavage-stage (McNemar Chi-square: P < 0.03). A similar paired analysis demonstrated that the developmental potential of embryos undergoing trophectoderm biopsy at the blastocyst stage was equivalent to the nonbiopsied control sibling
Figure 5.
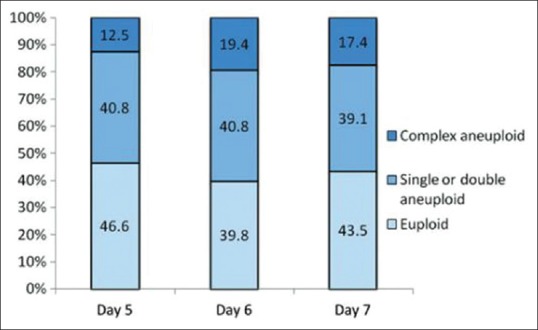
Day 5, day 6, and day 7 biopsies should be included for preimplantation genetic testing analysis. There is no significant difference between euploidy/aneuploidy rates in day 5, day 6, and day 7 blastocysts
Figure 6.
Meta-analysis of randomized controlled trials on preimplantation genetic screening with comprehensive chromosome screening versus routine care
After 2010, several other methods were developed for CCS such as single-nucleotide polymorphism (SNP) Testing, Quantitative Real-Time PCR (QT-PCR), and next-generation sequencing (NGS). Furthermore, the concept of euploid Single Embryo Transfer (eSET) was introduced. Forman et al. compared CCS-eSET group with non-CCS with SET group and showed higher on-going pregnancy rate (55% vs. 42%) and lower miscarriage rate (11% vs. 25%) in CCS-eSET group. They also showed that, in CCS-eSET group, overall IR was also higher compared to non-CCS with SET group irrespective of maternal age [Figure 7].[38] In 2013, several groups showed the successful use of NGS technology for PGT-A[39] and monogenic disorders.[40] NGS has become the most popular method due to the shorter testing time and cost-effectiveness. Recent studies by different groups for PGT-A outcomes comparing aCGH versus NGS showed marginally improved results with NGS with eSET.[41] Although there are several advantages of these new techniques for aneuploidy detection, due to the limitation of their sensitivity, FISH is still used for telomeric translocations and inversions.[42]
Figure 7.
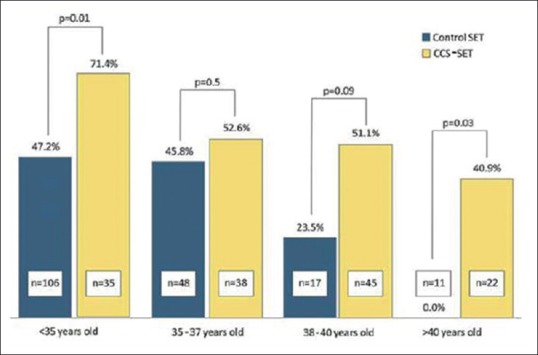
Overall implantation rate increases in comprehensive chromosome screening with eSET cases independent of age
The current indications for PGT include repeated implantation failures, repeated pregnancy loss, advanced maternal and paternal age, male factor infertility, and genetic disorders in the parents including mosaicism of sex chromosomes, structural rearrangements, and monogenic genetic diseases. Scott et al. published a paper in 2013 showing the analysis of an RCT. The trial showed that with CCS and fresh blastocyst transfer, sustained IR was significantly higher in the CCS group (66%) compared to control non-CCS group (48%). It also showed a higher delivery rate per cycle in CCS group (85%) compared to control non-CCS group (68%).[43]
BENEFITS OF PREIMPLANTATION GENETIC TESTING FOR ANEUPLOIDY (PGT-A)
Chromosomal aneuploidies are one of the major causes of infertility and maternal age-related reduced fertility potential. More than 70% of spontaneous miscarriages are due to chromosomal aneuploidies. PGT-A helps to shorten the time to a viable pregnancy by reducing the need of multiple IVF cycles. Euploid embryo transfer results in highest pregnancy rates and live birth rates reducing miscarriage risk independent of maternal age.
HOW MANY PREIMPLANTATION GENETIC TESTED EMBRYOS SHOULD BE TRANSFERRED?
Based on the recommendations given by the Practice Committee of the American Society for Reproductive Medicine (ASRM) and the Practice Committee of the Society for ART (SART) published in April 2017, single euploid cleavage-stage or blastocyst embryo should be transferred irrespective of the age group.[44]
However, PGT has its limitations under certain circumstances.[42] Subtelomeric deletions, mosaicism, small structural rearrangements, microdeletions, and microduplications may pose challenges. Furthermore, experienced laboratory personnel in IVF and genetics are important for a patient's success.
MITOCHONDRIAL DNA CONTENT
One of the new modalities for enhancing success is the evaluation of the mitochondrial DNA (mtDNA) content of the embryo.[45] The concept is that a high mtDNA copy number in euploid embryos is indicative of lower embryo viability and implantation.[46] This is still not the mainstay in the diagnosis of healthy energetic embryos. Victor et al. did not show any significant difference in mitochondrial levels in blastocysts irrespective of age, ploidy, or implantation potential.[47] As opposed to this, Fragouli et al. showed that no pregnancies resulted from blastocysts with elevated mtDNA levels.[48]
MOSAIC EMBRYO: TO TRANSFER OR NOT TO TRANSFER!
Embryonic chromosomal mosaicism is a condition in which more than one cell line is present, where one has a normal chromosomal constituent and others have abnormalities in chromosome number. It is assumed that mosaicism has adverse effects to the implantation and development of the embryo. Munné et al. reported that 41% of mosaic embryos resulted in on-going implantation. Complex mosaic blastocysts and embryos with >40% abnormal cells had a lower on-going IR than other mosaics.[49] Spinella et al. in their study showed that the extent of mosaicism influences the success rate of IVF.[50] Here, they used mosaic embryos with low aneuploidy percentage for implantation with higher chances of healthy live births compared to embryos with a higher percentage of mosaicism. Kushnir et al., from their study, concluded that there was a higher on-going pregnancy rate and a lower miscarriage rate when euploid embryos were used for implantation compared to the use of mosaic embryos. However, there was no significant difference in the on-going pregnancy rates or miscarriage rates among mosaic embryo transfers at any threshold of aneuploidy, and the degree of trophectoderm mosaicism was a poor predictor of on-going pregnancy and miscarriage.[51]
NEWER TECHNOLOGY IN PREIMPLANTATION GENETIC TESTING-M: KARYOMAPPING: BEYOND STANDARD NEXT-GENERATION SEQUENCING
In 2010, Alan Handyside with his group described the concept of karyomapping. It is genome-wide parental haplotyping using high-density SNP genotyping. Here, a linkage-based diagnosis is carried out for any single-gene defect. By knowing the genotyping of the parents and a close relative of known disease status, generally a previously affected child, this technology eliminates the need for customized test development. Karyomapping identifies informative loci for each of the four parental haplotypes across each chromosome and maps the inheritance of these haplotypes and the position of any crossovers in the proband as well as in the preimplantation embryos. Thus, it identifies the embryo-carrying normal chromosome copies.[52]
NONINVASIVE PREIMPLANTATION GENETIC TESTING TECHNIQUES
As embryo biopsy is an invasive procedure, efforts are being made to find different embryonic samples which do not require embryo biopsy. One of the novel approaches is the use of noninvasive PGS. Palini et al., in 2013, attempted isolation of cell-free DNA from blastocoel fluid (BF) for aneuploidy testing using the microarray technique.[53] Gianaroli et al. compared ploidy status of BF with trophectoderm cells, whole embryo, polar body, and/or blastomere and concluded that BF could be used as an alternative source for aneuploidy testing.[54] Lane et al. tried aneuploidy detection using DNA isolated from the spent culture medium.[55] Kuznyetsov et al. tried a combination of blastocyst culture-conditioned medium (BCCM) and BF to obtain sufficient embryonic DNA for whole genome amplification and accurate aneuploidy screening.[56] All these approaches are still under research.
OUR EXPERIENCE WITH PREIMPLANTATION GENETIC TESTING
We initiated PGT at Jaslok Hospital, Mumbai, in 1999, using the FISH technique.[57] Ours was the first center in India to offer PGT by FISH for various genetic disorders in the early 2000s.[58,59,60,61] With PGT-A by FISH, our clinical pregnancy rate was 36% per patient and 28% per cycle. Our team reported the first live births in India for a Robertsonian[62] and reciprocal translocation,[63] inversion with a cryptic translocation picked up on pre-PGT-A workup[64] and pregnancy after PGT for a complex translocation.[65] Currently, we use the NGS platform to offer PGT-A. In our latest series of 197 cycles, our pregnancy rate was 40%. We have also successfully carried out PGT-M for the first time, in India, for genetic disorders such as Duchenne muscular dystrophy, neurofibromatosis, sickle cell anemia, Leigh syndrome, retinoblastoma, hereditary inclusion body myopathy, cardiac disorders, and carriers of BRCA1.[66] We recently reported for the first time, in India, twin babies born free of the autosomal dominant BRCA1 mutation to a woman who was a BRCA1 mutation carrier[67] and had familial hereditary cancer syndrome in herself and her close family. We also have several pregnancies in couples carrying mutations for beta thalassemia.
CONCLUSION
From the above review, we can conclude that PGT is a major diagnostic tool to prevent transmission of any known genetic disorder. It also helps in populations which are at high risk of having babies with certain genetic aberrations. PGT reduces the trauma of multiple failed IVF cycles, early miscarriages, and helps in cases of advanced maternal age to prevent the birth of a syndromic child. PGT-M protects the child from inherited monogenic disorders. With the concept of savior sibling, PGT-M is useful in some of the hematological disorders to cure an affected child.
PGT technology should be integrated into ART to offer the best outcomes to patients. However, this technology should be used judiciously, and its pitfalls should be understood.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
The authors would like to thank the IVF and Genetics teams of FertilTree-Jaslok International Fertility Centre.
REFERENCES
- 1.Heape W. Preliminary note on the transplantation and growth of mammalian ova within a uterine foster-mother. Proc R Soc Lond B Biol Char. 1890;48:457–8. [Google Scholar]
- 2.Edwards RG, Gardner RL. Sexing of live rabbit blastocysts. Nature. 1967;214:576–7. doi: 10.1038/214576a0. [DOI] [PubMed] [Google Scholar]
- 3.Gardner RL, Edwards RG. Control of the sex ratio at full term in the rabbit by transferring sexed blastocysts. Nature. 1968;218:346–9. doi: 10.1038/218346a0. [DOI] [PubMed] [Google Scholar]
- 4.Steptoe PC, Edwards RG. Birth after the reimplantation of a human embryo. Lancet. 1978;2:366. doi: 10.1016/s0140-6736(78)92957-4. [DOI] [PubMed] [Google Scholar]
- 5.Trounson AO, Leeton JF, Wood C, Webb J, Wood J. Pregnancies in humans by fertilization in vitro and embryo transfer in the controlled ovulatory cycle. Science. 1981;212:681–2. doi: 10.1126/science.7221557. [DOI] [PubMed] [Google Scholar]
- 6.Binor Z, Sokoloski JE, Wolf DP. Penetration of the zona-free hamster egg by human sperm. Fertil Steril. 1980;33:321–7. doi: 10.1016/s0015-0282(16)44602-9. [DOI] [PubMed] [Google Scholar]
- 7.Li CY, Jiang LY, Chen WY, Li K, Sheng HQ, Ni Y, et al. CFTR is essential for sperm fertilizing capacity and is correlated with sperm quality in humans. Hum Reprod. 2010;25:317–27. doi: 10.1093/humrep/dep406. [DOI] [PubMed] [Google Scholar]
- 8.Hiramoto Y. Microinjection of the live spermatozoa into sea urchin eggs. Exp Cell Res. 1962;27:416–26. doi: 10.1016/0014-4827(62)90006-x. [DOI] [PubMed] [Google Scholar]
- 9.Lin TP. Microinjection of mouse eggs. Science. 1966;151:333–7. doi: 10.1126/science.151.3708.333. [DOI] [PubMed] [Google Scholar]
- 10.Gordon JW, Talansky BE. Assisted fertilization by zona drilling: A mouse model for correction of oligospermia. J Exp Zool. 1986;239:347–54. doi: 10.1002/jez.1402390306. [DOI] [PubMed] [Google Scholar]
- 11.Gordon JW, Grunfeld L, Garrisi GJ, Talansky BE, Richards C, Laufer N, et al. Fertilization of human oocytes by sperm from infertile males after zona pellucida drilling. Fertil Steril. 1988;50:68–73. doi: 10.1016/s0015-0282(16)60010-9. [DOI] [PubMed] [Google Scholar]
- 12.Ng SC, Bongso A, Chang SI, Sathananthan H, Ratnam S. Transfer of human sperm into the perivitelline space of human oocytes after zona-drilling or zona-puncture. Fertil Steril. 1989;52:73–8. doi: 10.1016/s0015-0282(16)60792-6. [DOI] [PubMed] [Google Scholar]
- 13.Laws-King A, Trounson A, Sathananthan H, Kola I. Fertilization of human oocytes by microinjection of a single spermatozoon under the zona pellucida. Fertil Steril. 1987;48:637–42. doi: 10.1016/s0015-0282(16)59478-3. [DOI] [PubMed] [Google Scholar]
- 14.Cohen J. Assisted hatching of human embryos. J In Vitro Fert Embryo Transf. 1991;8:179–90. doi: 10.1007/BF01130802. [DOI] [PubMed] [Google Scholar]
- 15.Lanzendorf SE, Maloney MK, Veeck LL, Slusser J, Hodgen GD, Rosenwaks Z, et al. Apreclinical evaluation of pronuclear formation by microinjection of human spermatozoa into human oocytes. Fertil Steril. 1988;49:835–42. doi: 10.1016/s0015-0282(16)59893-8. [DOI] [PubMed] [Google Scholar]
- 16.Palermo G, Joris H, Devroey P, Van Steirteghem AC. Pregnancies after intracytoplasmic injection of single spermatozoon into an oocyte. Lancet. 1992;340:17–8. doi: 10.1016/0140-6736(92)92425-f. [DOI] [PubMed] [Google Scholar]
- 17.Parikh FR, Kodwaney G, Kamat S, Parikh RM. Successful human micromanipulation with subzonal sperm Insemination and intracytoplasmic sperm injection. J Obstet Gynaecol India. 1994;44:458–63. [Google Scholar]
- 18.Wilton L, Trounson AO. Viability of mouse embryos and blastomeres following biopsy of a single cell. Proceedings of the 18th Annual Conference, Australian Society for Reproductive Biology; Brisbane, Australia. 1986. [Google Scholar]
- 19.Monk M, Muggleton-Harris AL, Rawlings E, Whittingham DG. Pre-implantation diagnosis of HPRT-deficient male and carrier female mouse embryos by trophectoderm biopsy. Hum Reprod. 1988;3:377–81. doi: 10.1093/oxfordjournals.humrep.a136711. [DOI] [PubMed] [Google Scholar]
- 20.Verlinsky Y, Ginsberg N, Lifchez A, Valle J, Moise J, Strom CM, et al. Analysis of the first polar body: Preconception genetic diagnosis. Hum Reprod. 1990;5:826–9. doi: 10.1093/oxfordjournals.humrep.a137192. [DOI] [PubMed] [Google Scholar]
- 21.Saiki R, Scharf S, Faloona F, Mullis K, Horn G, Erlich H, et al. A novel method for the prenatal diagnosis of sickle cell anemia. Am J Hum Genet. 1985;37:a172. [Google Scholar]
- 22.Handyside AH, Kontogianni EH, Hardy K, Winston RM. Pregnancies from biopsied human preimplantation embryos sexed by Y-specific DNA amplification. Nature. 1990;344:768–70. doi: 10.1038/344768a0. [DOI] [PubMed] [Google Scholar]
- 23.Griffin D, Wilton L, Handyside A, Winston R, Delhanty J. Pregnancies following the diagnosis of sex in preimplantation embryos by fluorescent in situ hybridisation. Br Med J. 1993;306:1382–3. doi: 10.1136/bmj.306.6889.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munné S, Weier HU, Stein J, Grifo J, Cohen J. A fast and efficient method for simultaneous X and Y in situ hybridization of human blastomeres. J Assist Reprod Genet. 1993;10:82–90. doi: 10.1007/BF01204446. [DOI] [PubMed] [Google Scholar]
- 25.Verlinsky Y, Rechitsky S, Schoolcraft W, Strom C, Kuliev A. Preimplantation diagnosis for fanconi anemia combined with HLA matching. JAMA. 2001;285:3130–3. doi: 10.1001/jama.285.24.3130. [DOI] [PubMed] [Google Scholar]
- 26.Wells D, Sherlock JK, Handyside AH, Delhanty JD. Detailed chromosomal and molecular genetic analysis of single cells by whole genome amplification and comparative genomic hybridisation. Nucleic Acids Res. 1999;27:1214–8. doi: 10.1093/nar/27.4.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voullaire L, Wilton L, Slater H, Williamson R. Detection of aneuploidy in single cells using comparative genomic hybridization. Prenat Diagn. 1999;19:846–51. [PubMed] [Google Scholar]
- 28.Voullaire L, Slater H, Williamson R, Wilton L. Chromosome analysis of blastomeres from human embryos by using comparative genomic hybridization. Hum Genet. 2000;106:210–7. doi: 10.1007/s004390051030. [DOI] [PubMed] [Google Scholar]
- 29.Wilton L, Williamson R, McBain J, Edgar D, Voullaire L. Birth of a healthy infant after preimplantation confirmation of euploidy by comparative genomic hybridization. N Engl J Med. 2001;345:1537–41. doi: 10.1056/NEJMoa011052. [DOI] [PubMed] [Google Scholar]
- 30.Mastenbroek S, Twisk M, van Echten-Arends J, Sikkema-Raddatz B, Korevaar JC, Verhoeve HR, et al. In vitro fertilization with preimplantation genetic screening. N Engl J Med. 2007;357:9–17. doi: 10.1056/NEJMoa067744. [DOI] [PubMed] [Google Scholar]
- 31.Wells D, Alfarawati S, Fragouli E. Use of comprehensive chromosomal screening for embryo assessment: Microarrays and CGH. Mol Hum Reprod. 2008;14:703–10. doi: 10.1093/molehr/gan062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gutiérrez-Mateo C, Colls P, Sánchez-García J, Escudero T, Prates R, Ketterson K, et al. Validation of microarray comparative genomic hybridization for comprehensive chromosome analysis of embryos. Fertil Steril. 2011;95:953–8. doi: 10.1016/j.fertnstert.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 33.Alfarawati S, Fragouli E, Colls P, Wells D. First births after preimplantation genetic diagnosis of structural chromosome abnormalities using comparative genomic hybridization and microarray analysis. Hum Reprod. 2011;26:1560–74. doi: 10.1093/humrep/der068. [DOI] [PubMed] [Google Scholar]
- 34.Scott RT, Jr, Upham KM, Forman EJ, Zhao T, Treff NR. Cleavage-stage biopsy significantly impairs human embryonic implantation potential while blastocyst biopsy does not: A randomized and paired clinical trial. Fertil Steril. 2013;100:624–30. doi: 10.1016/j.fertnstert.2013.04.039. [DOI] [PubMed] [Google Scholar]
- 35.Capalbo A, Rienzi L, Cimadomo D, Maggiulli R, Elliott T, Wright G, et al. Correlation between standard blastocyst morphology, euploidy and implantation: An observational study in two centers involving 956 screened blastocysts. Hum Reprod. 2014;29:1173–81. doi: 10.1093/humrep/deu033. [DOI] [PubMed] [Google Scholar]
- 36.Dahdouh EM, Balayla J, García-Velasco JA. Comprehensive chromosome screening improves embryo selection: A meta-analysis. Fertil Steril. 2015;104:1503–12. doi: 10.1016/j.fertnstert.2015.08.038. [DOI] [PubMed] [Google Scholar]
- 37.Chen M, Wei S, Hu J, Quan S. Can comprehensive chromosome screening technology improve IVF/ICSI outcomes? A Meta-analysis. PLoS One. 2015;10:e0140779. doi: 10.1371/journal.pone.0140779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Forman EJ, Tao X, Ferry KM, Taylor D, Treff NR, Scott RT, Jr, et al. Single embryo transfer with comprehensive chromosome screening results in improved ongoing pregnancy rates and decreased miscarriage rates. Hum Reprod. 2012;27:1217–22. doi: 10.1093/humrep/des020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin X, Tan K, Vajta G, Jiang H, Tan Y, Zhang C, et al. Massively parallel sequencing for chromosomal abnormality testing in trophectoderm cells of human blastocysts. Biol Reprod. 2013;88:69. doi: 10.1095/biolreprod.112.106211. [DOI] [PubMed] [Google Scholar]
- 40.Treff NR, Fedick A, Tao X, Devkota B, Taylor D, Scott RT, Jr, et al. Evaluation of targeted next-generation sequencing-based preimplantation genetic diagnosis of monogenic disease. Fertil Steril. 2013;99:1377–84. doi: 10.1016/j.fertnstert.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 41.Friedenthal J, Maxwell SM, Munné S, Kramer Y, McCulloh DH, McCaffrey C, et al. Next generation sequencing for preimplantation genetic screening improves pregnancy outcomes compared with array comparative genomic hybridization in single thawed euploid embryo transfer cycles. Fertil Steril. 2018;109:627–32. doi: 10.1016/j.fertnstert.2017.12.017. [DOI] [PubMed] [Google Scholar]
- 42.Morin SJ, Eccles J, Iturriaga A, Zimmerman RS. Translocations, inversions and other chromosome rearrangements. Fertil Steril. 2017;107:19–26. doi: 10.1016/j.fertnstert.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 43.Scott RT, Jr, Upham KM, Forman EJ, Hong KH, Scott KL, Taylor D, et al. Blastocyst biopsy with comprehensive chromosome screening and fresh embryo transfer significantly increases in vitro fertilization implantation and delivery rates: A randomized controlled trial. Fertil Steril. 2013;100:697–703. doi: 10.1016/j.fertnstert.2013.04.035. [DOI] [PubMed] [Google Scholar]
- 44.Practice Committee of the American Society for Reproductive Medicine. Electronic address: ASRM@asrm.org, Practice Committee of the Society for Assisted Reproductive Technology. Guidance on the limits to the number of embryos to transfer: A committee opinion. Fertil Steril. 2017;107:901–3. doi: 10.1016/j.fertnstert.2017.02.107. [DOI] [PubMed] [Google Scholar]
- 45.Diez-Juan A, Rubio C, Marin C, Martinez S, Al-Asmar N, Riboldi M, et al. Mitochondrial DNA content as a viability score in human euploid embryos: Less is better. Fertil Steril. 2015;104:534–410. doi: 10.1016/j.fertnstert.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 46.de Los Santos MJ, Diez Juan A, Mifsud A, Mercader A, Meseguer M, Rubio C, et al. Variables associated with mitochondrial copy number in human blastocysts: What can we learn from trophectoderm biopsies? Fertil Steril. 2018;109:110–7. doi: 10.1016/j.fertnstert.2017.09.022. [DOI] [PubMed] [Google Scholar]
- 47.Victor AR, Brake AJ, Tyndall JC, Griffin DK, Zouves CG, Barnes FL, et al. Accurate quantitation of mitochondrial DNA reveals uniform levels in human blastocysts irrespective of ploidy, age, or implantation potential. Fertil Steril. 2017;107:34–42.e3. doi: 10.1016/j.fertnstert.2016.09.028. [DOI] [PubMed] [Google Scholar]
- 48.Fragouli E, McCaffrey C, Ravichandran K, Spath K, Grifo JA, Munné S, et al. Clinical implications of mitochondrial DNA quantification on pregnancy outcomes: A blinded prospective non-selection study. Hum Reprod. 2017;32:2340–7. doi: 10.1093/humrep/dex292. [DOI] [PubMed] [Google Scholar]
- 49.Munné S, Blazek J, Large M, Martinez-Ortiz PA, Nisson H, Liu E, et al. Detailed investigation into the cytogenetic constitution and pregnancy outcome of replacing mosaic blastocysts detected with the use of high-resolution next-generation sequencing. Fertil Steril. 2017;108:62–71.e8. doi: 10.1016/j.fertnstert.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 50.Spinella F, Fiorentino F, Biricik A, Bono S, Ruberti A, Cotroneo E, et al. Extent of chromosomal mosaicism influences the clinical outcome of in vitro fertilization treatments. Fertil Steril. 2018;109:77–83. doi: 10.1016/j.fertnstert.2017.09.025. [DOI] [PubMed] [Google Scholar]
- 51.Kushnir VA, Darmon SK, Barad DH, Gleicher N. Degree of mosaicism in trophectoderm does not predict pregnancy potential: A corrected analysis of pregnancy outcomes following transfer of mosaic embryos. Reprod Biol Endocrinol. 2018;16:6. doi: 10.1186/s12958-018-0322-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Handyside AH, Harton GL, Mariani B, Thornhill AR, Affara N, Shaw MA, et al. Karyomapping: A universal method for genome wide analysis of genetic disease based on mapping crossovers between parental haplotypes. J Med Genet. 2010;47:651–8. doi: 10.1136/jmg.2009.069971. [DOI] [PubMed] [Google Scholar]
- 53.Palini S, Galluzzi L, De Stefani S, Bianchi M, Wells D, Magnani M, et al. Genomic DNA in human blastocoele fluid. Reprod Biomed Online. 2013;26:603–10. doi: 10.1016/j.rbmo.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 54.Gianaroli L, Magli MC, Pomante A, Crivello AM, Cafueri G, Valerio M, et al. Blastocentesis: A source of DNA for preimplantation genetic testing. Results from a pilot study. Fertil Steril. 2014;102:1692–9. doi: 10.1016/j.fertnstert.2014.08.021. [DOI] [PubMed] [Google Scholar]
- 55.Lane M, Zander-Fox DL, Hamilton H, Jasper MJ, Hodgson BL, Fraser M, et al. Ability to detect aneuploidy from cell free DNA collected from media is dependent on the stage of development of the embryo. Fertil Steril. 2017;108:e61. [Google Scholar]
- 56.Kuznyetsov V, Madjunkova S, Antes R, Abramov R, Motamedi G, Ibarrientos Z, et al. Evaluation of a novel non-invasive preimplantation genetic screening approach. PLoS One. 2018;13:e0197262. doi: 10.1371/journal.pone.0197262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parikh FR, Naik NJ, Uttamchandani SA, Nadkarni SG, Athalye AS, Madon PF, et al. The Indo-European Seminar cum Workshop on Advances in Human Cytogenetics. Lucknow: SGPGI; 1999. Preimplantation genetic diagnosis using the diode laser in a couple with a family history of haemophilia. [Google Scholar]
- 58.Parikh FR, Madon PF, Athalye AS, Naik NJ, Gada SD, Ganla KN, et al. Preimplantation genetic diagnosis of chromosomal abnormalities by multicolour fluorescence in situ hybridisation. J Indian Med Assoc. 2001;99:441–4. [PubMed] [Google Scholar]
- 59.Parikh FR, Madon PF. Preimplantation genetic diagnosis: PGD. Mod Medicare. 2004;1:24–6. [Google Scholar]
- 60.Athalye AS, Parikh FR, Naik NJ, Madon PF. Preimplantation genetic diagnosis. Health Screen. 2006;2:19,34–6. [Google Scholar]
- 61.Madon PF, Athalye AS, Naik NJ, Parikh FR. Preimplantation genetic diagnosis. In: Telang M, editor. Atlas of Human Assisted Reproductive Technologies. India: Jaypee; 2007. pp. 167–74. [Google Scholar]
- 62.Madon PF, Athalye AS, Naik NJ, Naik DJ, Parikh FR. PGD for a Robertsonian translocation by FISH: first successful pregnancy from India. J Prenat Diag Ther. 2010;1:20–2. [Google Scholar]
- 63.Naik DJ, Madon PF, Naik NJ, Athalye AS, Parikh FR. PGD by FISH for a reciprocal translocation:First baby from India. J Fetal Med. 2014;1:41–3. [Google Scholar]
- 64.Sanap RR, Athalye AS, Madon PF, Naik NJ, Naik DJ, Mehta TV, et al. First successful pregnancy by pre-implantation genetic diagnosis by FISH for an inversion together with a cryptic translocation in India. J Fetal Med. 2016;3:25–30. [Google Scholar]
- 65.Athalye AS, Sanap RR, Madon PF, Naik DJ, Warang DJ, Padyal PM, et al. Preimplantation genetic testing for a complex chromosome rearrangement, case report of a cryptic translocation detected on pre-PGT workup. Obstet Gynecol Int J. 2018;9:138–41. [Google Scholar]
- 66.Madon PF, Athalye AS, Parikh FR. Principles and Practice of Fetal Medicine. New Delhi: Jaypee Brothers Publishers; 2016. Preimplantation genetic diagnosis; pp. 210–7. [Google Scholar]
- 67.Parikh F, Naik D, Naik N, Sanap R, Madon P, Sanap M, et al. First successful twin delivery in India after preimplantation genetic diagnosis for BRCA1 mutation. Indian J Appl Res. 2018;8:338–9. [Google Scholar]