Abstract
Cancer is a heterogeneous disease with unique genomic and phenotypic features that differ between individual patients and even among individual tumor regions. In recent years, large-scale genomic studies and new next-generation sequencing technologies have uncovered more scientific details about tumor heterogeneity, with significant implications for the choice of specific molecular biomarkers and clinical decision making. Genomic heterogeneity significantly contributes to the generation of a diverse cell population during tumor development and progression, representing a determining factor for variation in tumor treatment response. It has been considered a prominent contributor to therapeutic failure, and increases the likelihood of resistance to future therapies in most common cancers. The understanding of molecular heterogeneity in cancer is a fundamental component of precision oncology, enabling the identification of genomic alteration of key genes and pathways that can be targeted therapeutically. Here, we review the emerging knowledge of tumor genomics and heterogeneity, as well as potential implications for precision medicine in cancer treatment and new therapeutic discoveries. An analysis and interpretation of the TCGA database was included.
Keywords: Genomics, heterogeneity, next-generation sequencing, cancer treatment, precision medicine
Tumors are marked by high levels of heterogeneity and out-of-control cell growth. They encompass an competing ecosystem, comprising a remarkable number of cancerous and unaffected cell sub-populations, including tumor-cell-related epithelial cells, infiltrated immune cells, fibroblasts, mesenchymal stroma/stem cells (MSCs), along with the surrounding endothelium of blood vessels (Fig. 1).1 At the cellular level, gradual tumor development and progression appears to follow a classical evolution-like process following a step-wise accumulation of selected genomic or epigenetic alterations, which in turn lead to positive selection and expansion of certain cell lineages while other cell populations are depleted (Fig. 2 and Fig. 3).2,3 This process, termed clonal evolution of tumors, or the stochastic model, is a dynamic process leading to continuous tumor remodeling with distinct dimensions of heterogeneity (Fig. 2b).2,4,5 Conversely, the cancer stem cell model (CSC model or hierarchical model) is the other main mechanism that drives cancer progression, with either single or multiple progenitors (Fig. 2b). Interestingly, the heterogenetic patterns established with either the stochastic model or the CSC model are similar and hard to distinguish without identification, and in reality, they often co-occur. Indeed, CSCs usually are the cells that at a different lineage stage acquired mutations or epigenetic alternations that caused the cells to grow unchecked and form the tumor population at the clonal level.6
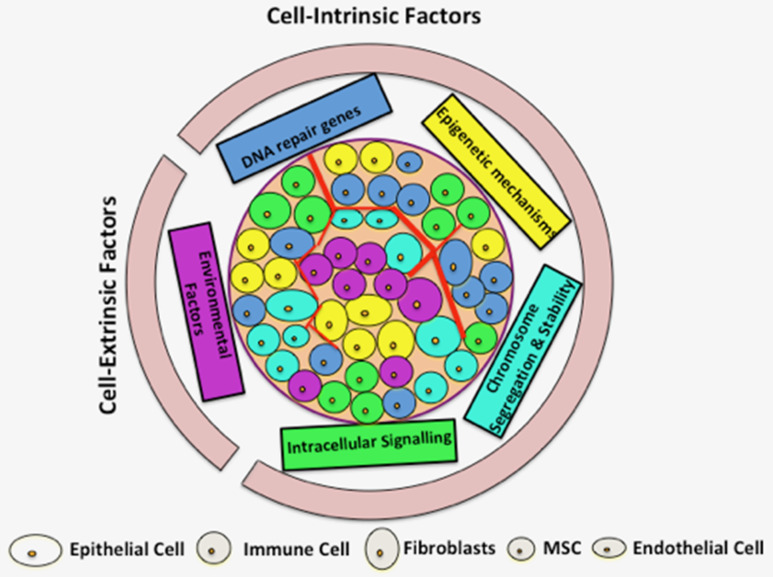
Figure 1.
Interplay of key contributing factors to tumor heterogeneity. Both cell-intrinsic and cell-extrinsic factors contribute to tumor heterogeneity. Key cell-intrinsic factors include mutation, DNA-repair genes, epigenetic mechanisms, chromosome segregation and stability, as well as intracellular signaling. Non-genetic or phenotypic variations as a result of contributing cell-intrinsic factors are depicted by different cytoplasmic colors. Cell-extrinsic mechanisms affect and contribute to the unequal microenvironment, indirectly contributing to tumor heterogeneity. Multiple cell types and different inter- and intra-cell interactions within a tumor may exist (only representatives are shown here), hence selectively contributing to tumor heterogeneity.
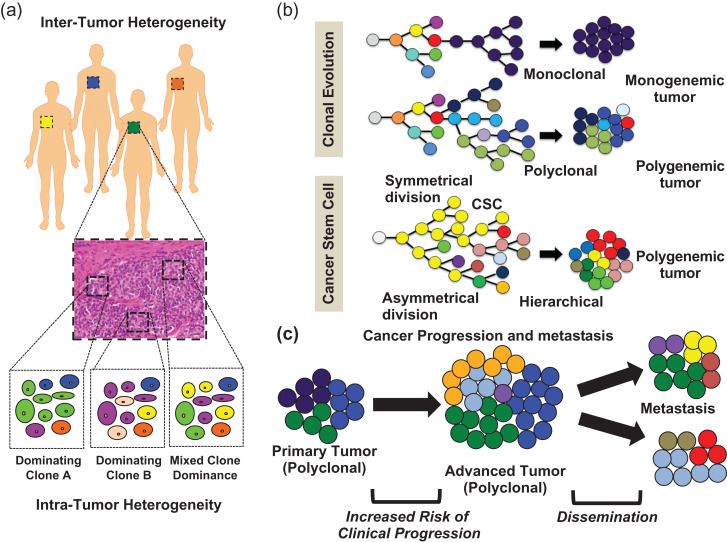
Figure 2.
Contribution of tumor heterogeneity in cancer progression and metastasis. (a) Graphical representation of inter- and intra-tumor heterogeneity origins at macroscopic and microscopic levels. (b) Graphical summary of the two recognized heterogeneity models: clonal (stochastic) evolution and cancer stem cell (CSC), involving either monoclonal evolution or single progenitor, and polyclonal evolution or multiple progenitors, linking tumor cellular paths to different tumor heterogeneity. (c) Contributing role of tumor heterogeneity with respect to cancer progression and metastasis.
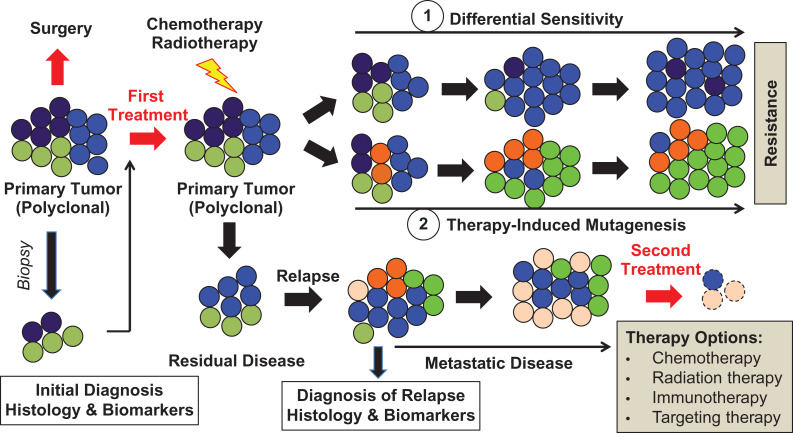
Figure 3.
Role of tumor heterogeneity in biomarker prediction and tumor resistance to clinical therapy. Initial cancer diagnosis and first treatment depends on initial cell and molecular characterization, derived from a small tumor fraction (biopsy, here figure shows a complete representation, but in some cases, it may be biased). In most cases, the current first-line treatments can successfully eliminate dominating cancer clones, with the cost of selecting resistant tumor clones through either differential sensitivity (1) or therapy-induced mutagenesis (2). These resistant clones are capable of driving disease progression and eventually metastasis. Hence, the clonal composition of metastatic lesions may significantly differ from clones in the primary tumor. As a result, initial treatment choice may not be effective in progressive metastatic disease. This necessitates a new diagnosis and additional comparative steps after relapse, prior to second and usually combined treatment options (i.e., immunotherapy, selective pathway component targeting and/or gene therapy) (Adapted from Tellez-Gabriel et al., 2016; doi:10.3390/ijms17122142).
There are numerous recognized heterogeneity contributing and/or selection factors. For example, artificial intervention by chemotherapy or radiotherapy can positively act on the cancer evolution process by reshaping tumor cell populations at genomic, epigenomic, transcriptomic, and proteomic levels, ultimately leading to different phenotypic properties (Fig. 3).7 Furthermore, cancer cell interaction with the surrounding microenvironment, given the complexity within and outside the cancer cell, is known to contribute to tumor heterogeneity.1
Based on numerous previous studies in multiple malignancies, tumor heterogeneity can be classified into inter-tumor (between tumors from different patients) or intra-tumor (within a single tumor, or tumor of a given patient) heterogeneity (Fig. 2a), based on specific molecular biomarker patterns (Table 1).4,8 Inter- or intra-tumor heterogeneity marks a key challenge in oncology, with significant implications for selecting specific biomarkers and/or primary gene mutations to guide clinical decisions for precision cancer therapies.2 The identification of alternatively expressed genes and multiple inter-/intra-cellular signaling pathways that drive phenotypic variation in multiple tumor types will also aid in the development of precise therapeutic approaches.
Table 1.
Identification of significant genomically altered genes from published TCGA data in 12 common cancer types.
| Cancer type | Sample size | Significantly altered genes | Reference |
|---|---|---|---|
| Glioblastoma | 206 | TP53,ERBB2,NF1,PARK2,AKT3,FGFR2,IRS2,PTPRD,MLH1,MSH2,MSH6,PMS2,PIK3R1 | doi:10.1038/nature07385 |
| Lung squamous cell carcinoma | 178 | TP53,CDKN2A,PTEN,PIK3CA,MLL2,NOTCH1,RB1,HLA-A,NFE2L2,KEAP1 | doi:10.1038/nature11404 |
| Lung adenocarcinoma | 230 | TP53,NF1,RIT1,RBM10,ERBB2,MAP2K1,NRAS,HRAS,NKX2-1,TERT,MDM2,KRAS,EGFR,BRAF,PIK3CA,STK11,KEAP1,MET,CCNE1 CCND1, TERC,MECOM | doi:10.1038/nature13385 |
| Colon rectal cancer | 276 | APC,TP53,KRAS,PIK3CA,FBXW7,SMAD4,TCF7L2,NRAS,CTNNB1 SMAD2,FAM123B,IGF2,NAV2,MYC,TGFBR2,BARF,MSH3,CASP8,CDC27,MAP7,PTEN,SOX9,ARID1A,FAM123B | doi:10.1038/nature11252 |
| Breast cancer | 510 | PIK3CA,PTEN,AKT1,TP53,GATA3,CDH1,RB1,MLL3,MAP3K1,CDKN1B,TBX3,RUNX1,CBFB,AFF2,PIK3R1,PTPN22,PTPRD,NF1,SF3B1,CCND3 | doi:10.1038/nature11412 |
| Ovarian carcinoma | 489 | TP53,BRCA1,BRCA2,RB1,NF1,FAT3,CSMD3,GABRA6,CDK12,NOTCH,FOXM1,BRAF,PIK3CA,KRAS,NRAS,CCNE,MYC,ZMYND8,IRF2BP2,PAX8,TERT,ID4 | doi:10.1038/nature10166 |
| Endometrial carcinoma | 373 | PTEN,CTNNB1,PIK3CA,ARID1A,PPP2R1A,KRAS,MYC,ERBB2,CTNNB1,CCNE1,FGFR3,SOX17,TP53,PTEN,ARID5B,PIK3R1,FBXW7,POLE | doi:10.1038/nature12113 |
| Urothelial bladder carcinoma | 131 | TP53,CDKN2A,FGFR3,PIK3CA,TSC1,RB1,HRAS,MLL2,CDKN1A,ERCC2,STAG2,RXRA,NFE2L2,ARID1A,KDM6A,EP300,FGFR3,PPARG E2F3, EGFR, CCND1,MDM2 | doi:10.1038/nature12965 |
| Clear cell renal cell carcinoma | 446 | VHL,PBRM1,BAP1,SETD2,HIF1A,PRKCI,MDS1,EVI1,MDM4,MYC,JAK2,CDKN2A,PTEN,NEGR1,QKI,CADM2,ARID1A,SMARCA4,PBAF | doi:10.1038/nature12222 |
| Gastric adenocarcinoma | 295 | TP53,KRAS,ARID1A,PIK3CA,ERBB3,HLA-B,JAK2,PD-L1,PDCD1LG2,PTEN,SMAD4,CDKN2A,CDH1,RHOA | doi:10.1038/nature13480 |
| Head and neck cancer | 279 | PIK3CA,TRAF3,E2F1,CDKN2A,HRAS,CASP8,NOTCH1,AJUBA,FAT1,NFE2L2,TP63,SOX2,EGFR,ERBB2,FGFR1 | doi:10.1038/nature14129 |
| Cervical cancer | 228 | APOBEC,SHKBP1,ERBB3,CASP8,HLA-A,TGFBR2,PD-L1,PDCD1LG2,BCAR4,KRAS,ARID1A,PTEN,PIK3CA,EP300,FBXW7,HLA-B,NFE2L2,MAPK1 | doi:10.1038/nature21386 |
Genomic profiling technology, that is genome-wide next-generation sequencing (NGS), is increasingly used to uncover different aspects of genomic heterogeneity in many types of human diseases, including cancer.9,10 The Cancer Genome Atlas (TCGA) project was a comprehensive and coordinated effort to accelerate our understanding of the molecular basis of multiple cancer types through application of genome-wide analysis technologies. The resulting data yielded insights into the close ties between tumor genetics and the evolutionary history of cellular processes across different cancer types.11 In addition, the above-mentioned discoveries have significantly expanded our understanding of cancer at the molecular level. Evidence of this can be seen in the extensive application of NGS in cancer diagnosis and prognosis prediction in clinical settings, as well as a dramatic increase in the number of new drug discoveries that target specific biological pathways and/or genes that are studied in ongoing clinical trials. Furthermore, there has also been a significant acceleration in the use of NGS to create genomic signatures for use in precision medicine.12,13
Highly promising and constantly refined single-cell sequencing (SCS) technologies offer an ultimate solution for tackling the previously encountered limitations of intra-tumor heterogeneity analysis.5 Simply put, SCS involves two major steps: (i) single- or multiple omics profiling of a large number of single cells, and (ii) classifying each tumor into different sub-populations from multiple spatial regions within a tumor biopsy with the use of sophisticated bioinformatics tools. These steps allow prediction of potential molecular relationships among these sub-populations within a single tumor biopsy. Combined with the serial spatial sampling set from a given tumor, SCS allows tracing of existing tumor cell lineages, and elucidation of potential therapeutic failure and resistance mechanisms, to further reveal the intricacies of tumor evolution.14,15 The aim of this review is to discuss the contribution of heterogeneity to cancer development and treatment, and to examine the potential implications and limitations of NGS in deciphering tumor biology, along with its clinical translation in precision medicine.
Articles associated with large-scale genomic studies and TCGA, reviews, and related new clinical trials for most common types of cancer published between January 1, 2012 and December 31, 2017 were collected using PubMed and accessible public databases. The cBioPortal web resource tool (http://cbioportal.org) was used for cancer genomic data evaluation, including somatic mutations, DNA copy-number alterations, mRNA and microRNA expression, DNA methylation, and protein and phosphoprotein abundance. This tool allows users to query genetic alterations for each gene and sample, as well as hypothesis testing concerning recurrence and genomic gene alteration events in various common cancers. ClinicalTrials.gov is the largest clinical trial database, currently holding registrations from more than 195 countries around the world, allowing insights into current ongoing clinical trials. Key word searches included genomics, heterogeneity, clinical features, drug resistance, clinical trials, and phase I-III. Further inclusion criteria for published genomic studies in this manuscript included: (i) a sample size of more than 120 patients for genomic studies, and (ii) at least 15 patient participants in clinical trials.
Cancer genomic heterogeneity associated with clinical features
Since the discovery by evolutionary biologist Julian Huxley in 1958, there have been remarkable advances in the knowledge of genomic diversity and single tumor heterogeneity.2 Today, there are many recognized factors contributing to genetic instability, including mutation of DNA repair genes directly or indirectly responsible for chromosomal stability, exposure to environmental mutagens, epigenetic mechanisms, as well as defects in chromosomal segregation, ultimately driving carcinogenesis (Fig. 1).16,17 Another important issue to mention is that there are different types of genetic instability (i.e., deletion, amplification, point-mutations, etc.), which contribute to the high variability of cancer genomes, such as promoting genetic heterogeneity and ultimately differences in treatment response.2,18,19
Identification of genomic heterogeneity in pan-cancer studies
Extensive cancer genome studies have established a comprehensive landscape of genomic and epigenetic heterogeneity, with a strong link to initiation and progression in major cancers.16 Recent data obtained from inter- and intra-tumor comparisons (Fig. 1a and b) link tumor heterogeneity to many types of malignant disease, that is lung,20 breast,21 prostate,22 myeloma,23 glioblastoma,24 and colorectal cancers (CRCs),25 as well as leukemia.26 Molecular and phenotypic aberrant variations are not only common between tumors of different tissue and cell types, but also within a tumor derived from the same tissue or cell type within an individual patient.12,16 For example, a study across 27 cancer types, including 3083 tumors and normal tissue pairs identified a total of 373 909 non-silent coding mutations by whole exome or whole genome sequencing.27 A subsequent comparison revealed a 1000-fold difference between individual patient mutation rates within or across selected cancer types.27 This evidence suggests that only a minority of these genes is essential for tumor development, with the majority having no significant biological impact (Table 1).27 Intriguingly, several recent studies highlighted remarkably divergent patterns of genetic alterations in primary tumors when compared with metastases obtained from the same patient, where the metastatic tumors had additional mutations that were not present in the primary tumor (Table 1).27–29 Essentially, the scientists hypothesized that all cells within a tumor have an equal potential to maintain and advance the tumor to metastasis, pending the acquisition of the necessary capability (Fig. 2c).30 A TCGA study comprising 178 lung squamous cell carcinoma (LSCC) and normal pairs, identified a total of 360 exonic mutations, 165 genomic rearrangements, and 323 segments of copy alteration within a given tumor.20 Statistical analysis uncovered 18 commonly mutated genes in 178 LSCCs, with TP53 being among them (Table 1).20 A very recent pan-cancer analysis, comprising over 3300 tumors, revealed a diverse genomic heterogeneity landscape across nine cancer types with a notable tendency for highly heterogeneous tumors to have lower levels of immune cell infiltration or T cell infiltration.31 Cancers arise when a sufficient number of mutations have occurred in any given tumor cell pool.32 These inevitably lead to accumulation of additional mutations within single cells that confer growth and survival advantages. Eventually, these cells will progressively give rise to new more aggressive progeny (Fig. 3).33,34 Furthermore, multiple studies also revealed that a single mutation in one gene (i.e., KRASG12D, BRAFV600E) could induce a quick and sufficient malignant transformation in corresponding tissues of several tumor types (Table 1).35,36 A significant association of high-level heterogeneity and poor survival was evident for lower grade glioma, prostate-, clear cell kidney carcinoma, head and neck-, as well as breast cancers, with borderline significance for melanoma.18,31,37
Identification of genomic heterogeneity in hematological malignancy
The molecular pathogenesis of acute myeloid leukemia (AML) has been studied by applying cytogenetic analysis tools for more than three decades.38 Characterization of AML genomes by next-generation sequencing has revealed that these tumors exhibit a relatively low recurrent somatic mutation rate, compared with most other cancers, with an average of only 13 identified mutations in AML associated genes (i.e., DNMT3A, FLT3, NPM1, IDH1/2, RUNX1, and CEBPA), along with recently discovered AML pathogenesis implicated genes (i.e., U2AF1, EZH2, SMC1A, and SMC3).38–40 These mutations mainly enhance proliferation and survival of hematopoietic progenitors through activation of signaling pathways (Fig. 4).39 A second class of mutated genes in AML includes transcription factors, such as CEBPA and RUNX1, which were found in ~20% of de novo normal cytogenetic AML, with short overall survival and relapse-free survival.40 Mutations in FLT3, NPM1, and CEBPA have been shown to have a significant prognostic impact, which ultimately resulted in their inclusion within the risk stratification system of European Leukemia patients and their use in standard-of-care testing.38,41TP53 mutation is frequently associated with therapy-related myeloid neoplasm and adverse prognostic impact.40 Somatic mutations in the epigenetic modifiers, DNMT3A, IDH1/2, and TET2, are considered initiating AML mutations.42NPM1 mutations confer a favorable prognosis only in the presence of a co-occurring IDH1 or IDH2 mutation. IDH1 and IDH2 inhibitors are currently being tested in clinical trials.18
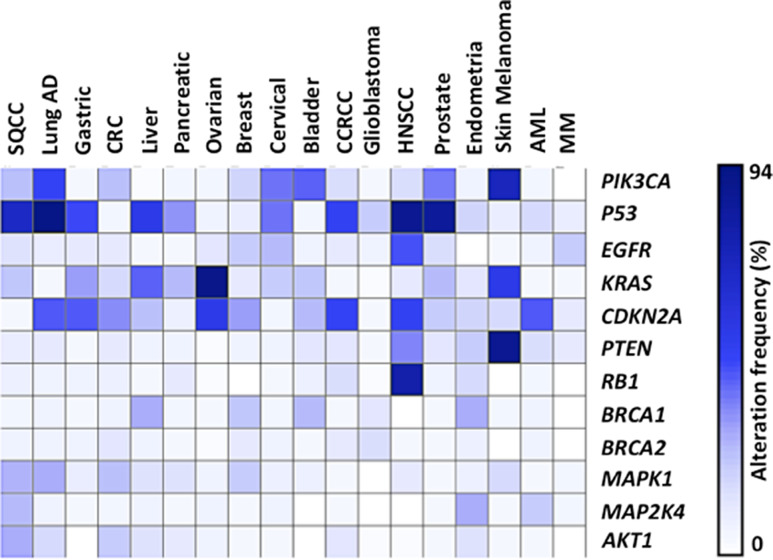
Figure 4.
Recurrent somatic alterations across common tumor types. Heatmap of significant genes that were genetically altered across the 18 most common cancers, as evaluated by the TCGA project. Percentage of alteration frequency (white = low to blue = high) for the genes is shown.
Diffuse large B cell lymphoma (DLBCL) is the most common form of lymphoma in adults, accounting for 30-40% of newly diagnosed non-Hodgkin’s lymphomas (NHL).43 Historically, DLBCL has been divided into three molecular subtypes, including germinal center B cell-like (GBC), activated B cell-like (ABC), and the primary mediastinal B cell lymphoma (PMBL), with all exhibiting a striking heterogeneity in gene expression profiles as well as clinical outcomes.44 Deep sequencing identified 322 genes that were recurrently mutated in DLBCLs, including ARID1A, MEF2B, PIK3CD, and PIK3R1, with additional genes involved in the NF-kB pathway (i.e., TNFAIP3) and Wnt pathway (i.e., WIF1).45 The pathogenic driver status of CARD11 alteration was reported by the discovery of gain of function germline mutations that drive constitutive NF-kB activation.46 The GCB subtype was characterized by a more favorable outcome and a spectrum of genetic alterations, which include PTEN deletion and EZH2 and TP53 mutations.43 The ABC subtype has a less favorable outcome, being associated with a distinct genetic background, and marked by translocations, BCL2 amplification and MYD88 mutation, which occur in approximately 30% of patients.47 DLCBL patients with MYD88 mutations are significantly older than patients without these mutations. PMBL displays an amplification of JAK2 in 50% of cases and recurrent deletion of SOCS1, which is a suppressor of JAK signaling.44 The relationship between therapy and genetic alteration is likely to contribute to convergent evolution, where mutation-conferring resistance will become highly prevalent in subsequently relapsed disease (Fig. 2c). As aforementioned, the intensive application of high-throughput genomic analysis has enabled rapid progress in our understanding of genetic heterogeneity in hematologic malignancies. Altogether, these examples suggest that the promise of precision medicine is finally coming to fruition in the desired treatment of blood malignancies.
Identification of genomic heterogeneity in solid tumors
Lung cancer is the leading cause of solid cancer-related mortality worldwide.20 The discovery of recurrent mutations in EGFR kinase and ALK genes has led to a remarkable change in lung cancer treatment.48 Targeting mutations in BRAF, AKT1, ERBB2, and PIK3CA has achieved great success in cancer therapy.48 Recently, the comprehensive TCGA study of lung cancer from three large cohorts of patients comprising NSCLC, adenocarcinoma (AD), and squamous cell carcinoma (SQCC) characterized the presence of complex genomic alterations in these cancers.49 Differential activity of PI3K/AKT/mTOR and MAPK pathways was present across NSCLS genomic subtypes.49 The activation of p38/MAPK and mTOR pathways within a subset of lung AD, compared with other subtypes of lung AD and SQCC, was conducted. Significant somatic copy number alterations for the following genes, MDM2, KRAS, EGFR, MET, CCNE, were found in lung AD, with gene amplifications strongly dominating (Table 1 and Fig. 4). Interestingly, three AD associated subtypes expressed several immune checkpoint genes, commonly associated with tumor cells or gene products known to interact with T cells (i.e., PD1, PDL1, PDL2, CD3, and CTL4), and have been nominated as potential therapeutic targets. Gender in lung AD is significantly correlated with gene mutation patterns (RBM10) (Table 1). The lung AD subtype appears to share similar gene patterns with many other cancer types, including CRC, stomach, pancreatic, breast, and liver cancers (Table 1). As expected, lung SQCC cancers also share many alterations (i.e., PIK3CA, PTEN, TP53, CDKN2A, and RAS) with head and neck, bladder, as well as cervical cancers (Fig. 4). A partial sharing with a multi-tissue squamous molecular subtype (Table 1 and Fig. 4) is also evident, marked by high expression of both SOX1 and TP63 genes, providing further evidence of common dysfunction in cell cycle control. TCGA further revealed that PIK3CA is amplified or mutated in ~34% of HPV negative and 56% HPV positive head and neck squamous cell carcinoma (HNSCC) tumors (Table 1 and Fig. 4), implicating the PI3K pathway in promoting growth factor dependent or independent growth, as well as the commonly observed EGFR therapy resistance.50 It also promotes preferential expression of an oncogenic ΔNP63 gene isoform of TP63 encoded on chromosome 3 and involvement in squamous differentiation. Furthermore, a subset of ~22% of HPV positive HNSCC tumors had a notable 14q32.32 deletion or inactivating mutations in the TNFR associated factor (TRAF3) gene, with strong implications in suppressing survival of myeloid cancers and HPV positive HNSCC cell lines (Table 1).50–52
Approximately 15% of CRC display a high level of microsatellite instability (MSI), caused by germline mutations in one or more DNA mismatch repair (MMR) genes, as well as somatic inactivation of the same pathway.53 Patients with early stage MSI CRC tumors have a better prognosis, compared with those harboring microsatellite stable (MSS) tumors. It is widely recognized that multiple genetic pathways (i.e., Wnt, RAS-MAPK, PI3K, TGF, TP53, and DNA mismatch-repair) are altered between benign and malignant lesions in CRC.54 As expected, the TCGA project identified 24 significantly mutated genes, including APC, TP53, SMAD4, SOX9, and FAM123B (Table 1 and Fig. 4). Amplification of ERBB2 and the newly discovered IGF2 amplification was also observed, with promising drug-targetable potential. Mutated APC, TP53, KRAS, and SMAD4 genes revealed a strong association with metastasis.25 In early CRC stages, SMAD4, TP53, and APC appear to only display a very weak association with the disease outcome.25APC is a tumor suppressor gene and its mutation is known to regulate growth advantage in epithelial cells, ultimately leading to small adenoma formation.54 Subsequently, KRAS and BRAF mutations provide a second round of favorable cell expansion, resulting in large adenoma transformation.25,55 Eventually, mutations in PIK3CA, SMAD4, and TP53 genes generate a malignant tumor, with a high potential for invasion and metastasis.25,54
Genomic analysis of the main breast cancer subtypes revealed that its cause was also associated with different subsets of molecular heterogeneity (Table 2).56 Clinically, this heterogeneity of breast cancer can be broadly categorized into three basic therapeutic groups: (i) the estrogen receptor (ER) positive group is the most numerous and diverse, with several genomic tests (Ki-67) to assist in predicting the outcome for ER positive patients receiving endocrine therapy57; (ii) triple-negative breast cancer (ER-, progesterone receptor (PR)- and human epidermal growth factor receptor-2 (HER2)) is an optimal patient group for chemotherapy options only, marked by increased incidence of germline BRCA1 mutations58; and (iii) basal-like breast cancer typically lacks expression of the molecular targets that confer responsiveness to highly effective targeted therapies, such as Tamoxifen and Aromatase inhibitors or Trastuzumab. The TCGA project revealed that somatic mutations in only three genes (i.e., TP53, PIK3CA, and GATA3) occurred at >10% incidence across all breast cancer types (Table 2).56 Deletion or translocation events in tumor suppressor genes, such as AKT3 and MAG13, lead to functional abnormalities and initiate breast tumorigenesis.56 High levels of APOBEC3B gene expression have been shown to be associated with disease-free survival and overall survival outcomes in patients with ER+ breast cancer.59 Recent studies on breast cancer uncovered a list of driver genes, such as CCND1, RB1, ERBB1, FGFR1, MYC, and PTEN (Table 1 and Fig. 4).56 Variable frequencies of HER2 gene amplification between primary tumors and their metastatic tumor or circulating tumor cells in advanced breast cancer have also been reported.60,61 These studies suggest that a subset of patients with initial HER2 negative primary tumors may develop HER2 positive circulating tumor cells during disease progression, although the exact mechanism is still to be elucidated. Several studies also revealed a marked association between prior history of breast carcinoma and secondary acquired mutations in either primary or recurrent ovarian carcinomas, with breast carcinoma often preceding the ovarian carcinoma by many years.62,63 Therefore, increased focus on driver mutations in tumorigenesis would provide critical insights for personalized therapeutics in cancer treatment. The identification of significant genomic heterogeneity from published TCGA project data derived from 12 cancer types is summarized in Table 1.
Table 2.
Genomic heterogeneity in sub-types of breast-like cancer from the TCGA project.
| Mutated genes | Luminal A (%) | Luminal B (%) | HER2(+) (%) | Basal-like cancers (%) |
|---|---|---|---|---|
| PIK3CA | 45 | 29 | 39 | 9 |
| TP53 | 12 | 29 | 72 | 80 |
| MAPK1 | 13 | 5 | 4 | 0 |
| MAP2K4 | 7 | 2 | 2 | 0 |
| AKT1 | 4 | 2 | 2 | 0 |
| PTEN | 4 | 4 | 2 | 1 |
| RB1 | 0.40 | 3 | 0 | 4 |
Association of cancer genetic heterogeneity and therapeutic failures
Genetic heterogeneity is a prominent contributor to therapeutic failure, with increased likelihood of resistance to future therapies. This generates a diverse cell population during tumor development and progression, representing a key determining factor for variation in tumor therapeutic response (Fig. 3).8,16 Resistance to single drug targeting therapies is frequent in cancer, and near universal in major metastatic carcinomas. KRAS mutations are known to confer resistance to EGFR targeting in cancer treatment.64 The well-documented mechanisms of drug resistance to certain therapies are associated with alterations in signal transduction cascades, predominantly through activation of alternative or complementary pathways, often through molecular feedback loops.65 Insights into the genomic landscape of some cancers, such as NSCLCs and breast cancers, have fueled a shift in the treatment paradigm towards the use of precise treatments.49,66,67 Lung cancer patients with heterogeneous EGFR mutations appear to benefit less from the EGFR inhibitor Gefitinib than patients with homogeneous EGFR mutations.68 Mutated EGFR was shown to be mediated by selected cells that harbor the EGFR gatekeeper mutation (T790M) and/or MET gene amplification.69,70 The oncogenic BRAF amplification or MEK1 mutation associated with resistance to BRAF-specific inhibitors in melanomas is also well documented.71,72 However, BRAFV600E mutations are among the most commonly reported molecular alterations in melanomas, and BRAF is currently a promising therapeutic target.71 Successful clinical trials of selective BRAF inhibitors (i.e., ‘Vemurafenib’) in BRAFV600E mutated versus non-mutated patient melanoma tumors support their substantial potential and clinical significance, together with patient-derived tumor genotyping, prior to appropriate treatment selection.73 Chronic myeloid leukemia (CML) is a stem cell-like disease, marked by the presence of rare cell clones in about half of patients with unique BCR-ABL resistant mutations, possibly acquired after Imatinib treatment.74 Furthermore, impairment of apoptotic cell death plays a major role in therapy resistance and relapse in acute lymphoblastic leukemia (ALL).75 Recent studies have shown that apoptosis protein inhibitor interacting protein kinase 1 (RIP1) inhibitor ‘Birinapant’ potently induced cell death in patient-derived ALL cells both in vitro and in vivo.76,77 Patients in leukemic relapse are notoriously difficult to treat because drug resistance of leukemic clones is an insurmountable obstacle to effective chemotherapy in AML.78 Loss of tumor suppressor BRCA1/2 gene heterozygosity highly sensitizes patients to DNA cross-linking agents (platinum drugs) in ovarian or breast cancer.79 Predictive capability of platinum resistance through the presence of secondary BRCA1/2 mutations in ovarian cancer has been documented in in vitro and in vivo studies.63 CRC resistance to targeted therapy noted during disease progression is often occurring within 3-12 months after EGRF antagonist administration, demanding a change in the treatment choice.80 Proposed mechanisms associated with the failure of improved outcomes in CRC patients were linked to microscopic residual disease and the absence of tumor neoangiogenesis after ‘Bevacizumab’ (anti-EGFR antibody) application, as well as the epithelial to mesenchyme transition phenotype after ‘Cetuximab’ (anti-EGFR antibody) treatment.81 Furthermore, the expression of the excision repair cross-complementation group 1 (ERCC1) gene is under increased investigation as a potential resistance predictive marker to platinum compounds in CRC.82 The benefit of chemotherapy is further increased with combined targeting therapies (i.e., ‘Bevacizumab,’ ‘Cetuximab,’ or ‘Panitumumab’) in CRC patients with RAS-wild type harboring tumors.83 In addition, preliminary clinical data have revealed that HER2 is amplified in around 5% of patients with KRAS-wild type metastatic CRC, suggesting that this patient subset may benefit from dual HER2 inhibitors (i.e., ‘Trastuzuman’ and ‘Lapatinib’).84
Drug resistance mechanisms of selected tumor clones and the extremely intra-heterogeneous nature of the tumor are also widely accepted (Fig. 3).85 This leads to significant practical difficulties in identifying the most aggressive or drug resistant clones to deliver targeted therapy, through well-established conventional bulk sequencing approaches.11 At present, targeted therapeutic reagents are dependent upon biomarkers that are derived from primary tumor biopsies that are subjected to genomic sequencing. However, dramatic responses to initial therapy and relapse typically take place within 1-2 years following treatment initiation, which commonly arise from selective pressures created by the dynamic nature of the targeting agent.86 Furthermore, drug response failure of sub-clones within certain cancer tissues substantially limits the ability to predict treatment response.87 Our current understanding of heterogeneity extent in cancers is largely derived from bulk tumor specimen analysis. It should be noted that most bulk tumor samples are a mixture of non-malignant cells and diverse cancer cell sub-populations (Fig. 2 and Fig. 3). The implementation of single-cell analysis (SCA) technologies to study cancer heterogeneity has shown a strong potential to reveal genome-wide molecular profiles, regulation, and mechanisms, with unprecedented resolution.5,88 This state-of-the-art methodology allows, with reliable precision, the isolation and characterization of individual cells among a heterogeneous cell mixture. It further grants an opportunity for future breakthroughs in understanding the dynamic genetic heterogeneity in tumors, along with cancer origin, progression, and clinical management.
Advances of SCA technology to uncover dynamic genetic heterogeneity
Over the past few years, technological advances at the single-cell level have made high-throughput sequencing of tumor genomes possible. An increasing number of reported single-cell studies demonstrate considerable cell-to-cell variability in apparently homogeneous populations. Application of SCA has greatly enhanced the power of systematic cancer heterogeneity characterization, resulting in significant mechanistic insights into tumor progression.
Single-cell gene expression analysis dates back to the early 1990s89; however, it is in the last decade that a significant advance has been achieved in SCA technology development (refer to reviews14,90). To date, it is becoming possible to assay a substantial number (>100) of secreted proteins, cell surface markers, signaling pathway components, and even metabolites at the single-cell level.90 The most significant progress of SCA tool development is evident at the genomic, transcriptomic, epigenetic, and proteomic levels.5,90 SCA tools significantly contributed to the identification and characterization of cancer stem cells. The success of scRNA-seq in this area was marked by the discovery of ‘stemness’-like cells by analyzing transcriptome- and gene expression signatures of in vivo differentiated glioblastoma cells,91 as well as metastatic breast tumors.92 These observations support the theory that initial tumor stem cells may initiate and propagate metastatic cancer behavior.
As an application with a great clinical potential, SCA was applied to circulating tumor cells (CTCs). The previously impossible detection and characterization of CTCs, originating from tumors and at 1:109 ratio in the bloodstream, has been made possible through SCA techniques.92 The previously unreliable method of using magnetic beads coupled to a cell surface ECAM (Epithelial Cell Adhesion Molecule) recognizing antibody, has been optimized for single-cell CTC isolation from whole blood, by considering factors such as microscopic imaging, cell size, and passive capture.93,94 The elucidation of cancer progression through the comparison of genomic and transcriptomic derived CTC profiles has also been reported.95,96
By using single-cell adapted whole-genome sequencing, the discovery of the metastatic pathway (potentially because of differential CNV patterns) from lung cancer-derived CTCs was achieved.97,96 The promising applications of non-invasive SCAs to study cancer development, as well as cancer therapy resistance following chemotherapy are also evident.98 Currently, two prevalent therapy resistance theories exist: (i) adaptive resistance, where low frequency mutations in the original population are selected for and eventually rise in frequency during chemotherapy; or (ii) acquired resistance, where resistance-conferring mutations are directly linked to chemotherapy.90 As a consequence, the main goal and use of SCAs in clinical settings has been detection and evaluation of mutational differences over time in CTCs (i.e., before and after treatment). Eventually, this will lead to insights into the mechanisms of therapeutic resistance development in various cancer types, with subsequent validation of the aforementioned resistance theories. The use of SCA technology to study the response of mutated BRAFV600E melanoma to RAF- or RAF/MEK combined inhibitors in vitro or in vivo led to the discovery of an overexpressed, and well-known AXL resistance marker, which was also linked to the adaptive resistance mechanism.90,99 In another study, CTC tracking and subsequent whole-genome sequencing of prostate-derived cells, before or after androgen (AR)-targeted therapy, led to the discovery of two distinct resistant and AR amplified cell populations.100 One of the populations was shown to be closely related to the cells prior to initiated therapy, supporting the adaptive resistance theory.100 Furthermore, identification of heterogeneous resistance-conferring changes in the AR-independent Wnt signaling pathway could be derived using the scRNA-seq technique.101 In addition, following ‘Trastuzumab’ treatment of HER2 mutated breast tumor samples and subsequent STAR-FISH SCA, a close link between increased PIK3CA mutations and increased dispersion, as well as decreased frequency of HER2 amplification and chemotherapeutic resistance, was detected.102 The authors concluded that ‘Trastuzumab’ treatment has no benefit to patients who had already received chemotherapy and that the STAR-FISH approach could be used to predict poor prognosis.102
Clinical studies elucidating potential therapeutic significance of genetic heterogeneity for precision medicine
The subsequently mentioned studies further clarify the concept of tumor heterogeneity and its related pathogenesis, presenting a major area for new therapeutic approaches. Major subsets of molecular alterations in key pathways and driver mutations have interesting potential for targeting PI3K, mTOR, ERK/MAPK pathways, as well as checkpoint immunotherapy.
PI3K-AKT-mTOR inhibitors
The PI3K-AKT-mTOR pathway plays a critical role for many cellular functions, such as growth control, survival, and metabolism, and is known to be highly activated in human cancers.103 Previous studies have shown that PI3K-AKT-mTOR pathway over-activation is associated with mutations and amplification of genes encoding receptor tyrosine kinases (i.e., HER2 or EGFR), PIK3CA mutations, PTEN loss and/or mutation, and KRAS mutations during carcinogenesis (Table 1 and Fig. 4). A significant amount of effort has been put into development of drugs targeting several kinases throughout the phosphatidylinositol-3-kinase (PI3K) pathway for cancer therapy. Novel PI3K target inhibitors are currently being investigated in phase II and phase III clinical trials for various cancers (Table 3). There are different isoforms of PI3Ks, such as PI3K-α, β, γ, and δ, with studied inhibitors known to inhibit one or more isoforms.104 ‘Idelalisib’ (GS-1101 or CAL-101), a selective PI3K-δ inhibitor was approved in 2014 by the US FDA for treatment of various hematological malignancies, including chronic lymphocytic leukemia (CLL), relapsed B-cell non-Hodgkin’s lymphoma, and relapsed small lymphocytic lymphoma.105 ‘Copanlisib’ (BAY 80-6946), a PI3K inhibitor predominantly targeting PI3K-α and PI3K-γ isoforms was the second FDA-approved drug in 2017 to treat adult patients with relapsed lymphoma.106 The BMK120 and BYL719 compounds are pan-PI3K inhibitors that have demonstrated preliminary selective activity in preclinical models of solid tumors.104 Both compounds have shown favorable tolerability profiles with consistent on-target inhibition of PI3K. They have been studied as therapeutic targets either alone or in combination in phase II trials against solid tumors and hematologic malignancies (Table 3). PI3K-AKT-mTOR is the most frequently activated signaling pathway in breast cancer.56 ‘Everolimus’ (RAD-001), a selective inhibitor against mammalian target of rapamycin (mTOR), has been investigated in a phase III clinical trial and in combination with ‘Exemestane’ trials (funded by Novartis, NCT00863655) for ER positive advanced breast cancer.66 The results have shown that the median progression-free survival was 6.9 months with ‘everolimus’ plus ‘Exemestane’ and 2.8 months with placebo plus ‘Exemestane’.66 ‘Buparlisib’ (BKM120) is an oral pyrimidine-derived reversible pan-PI3K inhibitor with specific and potent activity against mutant PI3K-α, as well as wild-type PI3K-α, β, γ, and δ isoforms, but no inhibitory activity against the class III PI3K or mammalian target of Rapamycin (mTOR). A phase IB/II study has investigated combined ‘Buparlisib’ and ‘Trastuzumab’ (NCT01132664) treatment in relapsed HER2 (+) breast cancer that previously failed with ‘Trastuzumab’ alone (Table 3).107,108 The data revealed that ‘Buparlisib’ and ‘Trastuzumab’ were well tolerated, with preliminary signs of clinical activity being observed in two partial responders and seven patients with stable disease. This promising outcome has led to further ongoing investigations of PI3K inhibitors in patients with HER2 (+), HER2 (-), and/or AR (+) triple negative metastatic breast cancer (NCT01816594, NCT02379247, NCT02457910) (refer to Table 3). PI3K-AKT-mTOR pathway alterations associated with PIK3CA mutation are evident in almost a third of HNSCC (Table 1 and Fig. 4). A number of first generation PI3K and mTOR inhibitors (i.e., ‘Rapamycin’, ‘Temsorlimus’ (CCI-779), ‘Everolimus’ (RAD-001)) have shown activity in in vivo preclinical models.109 ‘Alpelisib’ (BYL719), specifically inhibits PIK3 in the PI3K/AKT kinase signaling pathway. ‘Alpelisib’ in combination with ‘Cetuximab’ have demonstrated synergistic activity in HNSCC cell lines, resulting in induced tumor regression in PIK3CA mutant HNSCC xenograft model.109 Currently, two ongoing phase II clinical trials are assessing ‘Buparlisib’ (BKM120) in recurrent or metastatic HNSCC on ‘Cisplatin’- and ‘Cetuximab’-based chemotherapy in PIK3CA-mutated and wild-type patient cohorts (NCT01737450, NCT01816984, refer to Table 3). Loss of PTEN is associated with increased PI3K-AKT pathway activation and is commonly observed in up to 30% of melanomas, and frequently also observed in tumors with a concurrent activating BRAF mutation (Table 1 and Fig. 4). A phase I/II clinical trial of GSK2636771 in combination with ‘pembrolizumab’ is currently ongoing in patients with refractory (non-responsive to treatment) metastatic melanoma (NCT03131908, refer to Table 3). A summary of previous and ongoing clinical trials of individual, as well as combined PI3K, mTOR and AKT inhibitors (as registered in ClinicalTrials.gov) is provided in Table 3.
Table 3.
Summary of small molecule inhibitor clinical trials in human cancers. Data taken from http://clinicaltrials.gov/.
| Drug | Combination | Sponsor | Tumor type | Sample size | Status | Recruitment Status | Clinical trial ID |
|---|---|---|---|---|---|---|---|
| PI3K inhibitors | |||||||
| BAY80-6946 (Copanlisib) | - | Bayer | Lymphoma, Non-Hodgkin’s | 227 | Phase II | Active | NCT01660451 |
| BKM120 | - | Hospices Civils de Lyon | Thyroid Cancers | 47 | Phase II | Active | NCT01830504 |
| BKM120 | - | SOLTI Breast Cancer Research Group | Triple Negative Metastatic Breast Cancer | 50 | Phase II | Completed | NCT01629615 |
| BKM120 | Centre Leon Berard | Metastatic Head and Neck Cancer Recurrent or Progressive | 70 | Phase II | Recruiting | NCT01737450 | |
| BKM120 | Cetuximab | University of Chicago | Recurrent or Metastatic Head and Neck Cancer | 30 | Phase II | Active | NCT01816984 |
| PQR309 | - | PIQUR Therapeutics AG | Lymphoma, Malignant | 72 | Phase II | Recruiting | NCT02249429 |
| - | Endometrial Clear Cell Adenocarcinoma | ||||||
| - | Endometrial Adenosquamous Carcinoma | ||||||
| BKM120 | Trastuzumab | Novartis Pharmaceuticals | HER2-positive Primary Breast Cancer | 50 | Phase I/II | Completed | NCT01816594 |
| BYL719 | Paclitaxel | Priyanka Sharma | HER-2 Negative Breast Cancer | 44 | Phase I/II | Active | NCT02379247 |
| Taselisib | Enzalutamide | Vanderbilt-Ingram Cancer Center | AR Positive Triple-Negative Metastatic Breast Cancer | 73 | Phase I/II | Recruiting | NCT02457910 |
| Idelalisib | Entospletinib | Hematologic Malignancies | 66 | Phase I/II | Completed | NCT01796470 | |
| GSK2636771 | Pembrolizumab | M.D. Anderson Cancer Center | Metastatic Melanoma and PTEN Loss | 41 | Phase I/II | Recruiting | NCT03131908 |
| Everolimus | Exemestane | Novartis Pharmaceuticals | Metastatic Breast Cancer with ER+ | Phase III | Completed | NCT00863655 | |
| Akt inhibitors | |||||||
| Akt Inhibitor MK2206 | - | National Cancer Institute (NCI) | Endometrial Adenocarcinoma | 37 | Phase II | Completed | NCT01307631 |
| Akt Inhibitor MK2206 | - | National Cancer Institute (NCI) | CRC | 18 | Phase II | Completed | NCT01802320 |
| MK-2206 + AZD6244 | - | National Cancer Institute (NCI) | Colorectal Neoplasms | 21 | Phase II | Completed | NCT01333475 |
| mTOR inhibitors | |||||||
| Everolimus | Vinorebine | AIO-Studien-gGmbH | Advanced Breast Cancer | 139 | Phase II | Completed | NCT01520103 |
| BEZ235 | - | Novartis Pharmaceuticals | Pancreatic Neuroendocrine Tumors (pNET) | 31 | Phase II | Completed | NCT01658436 |
| Rapamycin | - | The University of Texas Health Science Center at San Antonio | Cancer of Breast | 60 | Phase II | Recruiting | NCT02642094 |
| Everolimus | - | M.D. Anderson Cancer Center | Endometrial Cancer | 270 | Phase II | Recruiting | NCT02397083 |
| Everolimus | - | University of Texas Southwestern Medical Center | Children With Recurrent or Progressive Ependymoma | 18 | Phase II | Recruiting | NCT02155920 |
| Everolimus | - | National Cancer Institute (NCI) | Kidney Cancer or Renal Cancer | 18 | Phase II | Recruiting | NCT02504892 |
| TAK-228 | - | Fox Chase Cancer Center | Soft Tissue Sarcomas | 33 | Phase II | Recruiting | NCT02987959 |
| AZD2014 | - | Canadian Cancer Trials Group | Glioblastoma Multiforme | 52 | Phase II | Recruiting | NCT02619864 |
| Everolimus | Cisplatin | Jenny C. Chang, MD | Triple Negative Breast Cancer | 32 | Phase I/II | Recruiting | NCT01931163 |
| Everolimus | Sorafenib Tosylate | Alliance for Clinical Trials in Oncology | Thyroid Cancer | 34 | Phase I/II | Recruiting | NCT02143726 |
| Everolimus | LEE011 | Memorial Sloan Kettering Cancer Center | Neuroendocrine Tumors | 41 | Phase I/II | Recruiting | NCT03070301 |
| Sirolimus+ | Cisplatin | University of Washington | Bladder Cancer | 21 | Phase I/II | Completed | NCT01938573 |
| Enzalutamide | LY3023414 | Eli Lilly and Company | Prostate Cancer | 144 | Phase I/II | Recruiting | NCT02407054 |
| ERK1/2 and MAPK inhibitors | |||||||
| Regorafenib | - | Gerald Batist | Metastatic Colorectal Cancer | 52 | Phase II | Recruiting | NCT01949194 |
| Vandetanib | - | Ronald Weigel | Invasive Breast Cancer | 100 | Phase II | Recruiting | NCT01934335 |
| BVD-523 | - | BioMed Valley Discoveries, Inc | Myelodysplastic Syndrome | 53 | Phase II | Completed | NCT02296242 |
| TDM1 | Abraxane, Lapatinib | Jenny C. Chang, MD | Metastatic HER2 Positive Breast Cancer | 45 | Phase I/II | Recruiting | NCT02073916 |
| LY2228820 | Radiotherapy + TMZ | Centre Jean Perrin | Newly Diagnosed Glioblastoma | 50 | Phase I/II | Recruiting | NCT02364206 |
| Dabrafenib | Pazopanib hydrochloride | Manisha Shah | Unspecified Adult Solid Tumor | 56 | Phase 1 | Active, not recruiting | NCT01713972 |
| GSK2118436 | GSK1120212 | Novartis Pharmaceuticals | Cancer | 430 | Phase 2 | Active, not recruiting | NCT01072175 |
| NFκB inhibitors | |||||||
| Pentoxifylline | - | Ramón Óscar González-Ramella, Ph.D | Pediatric Acute Lymphoblastic Leukemia | 44 | Phase II | Recruiting | NCT02451774 |
| Dexamethasone | - | Emory University | Plasma Cell Myeloma | 90 | Phase II | Recruiting | NCT02765854 |
| Ibrutinib | - | Icahn School of Medicine at Mount Sinai | Multiple Myeloma Patients | 36 | Phase II | Recruiting | NCT02943473 |
| Lansoprazole | - | National Health Research Institutes, Taiwan | Early-stage HP(+) Gastric Pure DLBCL | 30 | Phase II | Recruiting | NCT02388581 |
| Ibrutinib | Rituximab | Samsung Medical Center | EB+ Diffuse Large B-cell Lymphoma | 24 | Phase I/II | Recruiting | NCT02670616 |
MARK/MEK inhibitors
The mitogen-activated protein kinase/extracellular signal regulated kinase (MAPK/ERK) signaling cascade is tightly regulated by phosphatases and bi-directional communication with other pathways, such as the AKT/mTOR pathway.110 This pathway is vital for human cancer cell survival, dissemination, and drug resistance development. It is known to be frequently activated by a wide variety of receptors, including upstream genomic events and/or activation of multiple signaling events in solid and hematological malignancies.103 Genomic tumor profiling has identified amplifications of several growth factor receptor genes, including EGFR, ERBB2, IGF1R, FGFR1, and mutations in RAS, BRAF and MAPK/ERK pathway genes that are ready for targeting in cancer treatment (Table 1, Table 3, and Fig. 4). Currently approved B-RAF kinase inhibitors (BRAFi) for melanoma treatment are being investigated either alone or in combination with other agents in many other tumor types (refer to Table 3). The clinical trial using CDK inhibitor ‘LEE011’ in combination with phase II BRAF inhibitor ‘Encorafenib’ (LGX818), with the aim to target key enzymes in the MAPK signaling pathway of BRAF mutant melanoma patients, was abandoned because of safety concerns (NCT01777776, refer to Table 3).111 However, therapies targeting MAPK/ERK components appear to have variable responses, when used in different solid tumors, including breast cancer, CRC, and glioblastoma (Table 3). BRAF and MEK inhibition results in increased melanoma antigen expression, as observed in melanoma cell lines.110 This phenomenon may increase tumor recognition by T-cells, with a strong potential to develop into a successful immunotherapeutic approach, warranting further exploration into combined approaches of immunotherapies and MAPK/ERK inhibitors.112 Currently ongoing Phase II clinical trials using agents targeting BRAF and MEK kinases are summarized in Table 3.
Nuclear factor kappa-B (NF-κB) inhibitors
The implication of the NF-κB signaling pathway has been well established in recent decades in both physiologic and pathologic conditions, including cancer.113 The role of NF-κB in human cancer initiation, metastasis, and resistance to treatment has been exclusively investigated and has drawn particular attention. A significant number of human cancer genomic studies have revealed that NK-κB activation is highly associated with an inflammatory microenvironment and various oncogenic mutations.114 It appears to be a key mediator in the crosstalk between inflammation and carcinogenesis. The NK-κB family consists of five master transcription factors, including NF-κB1, NF-κB2, RelA, RelB, and c-Rel, which bind to DNA and regulate gene transcription. The role of NF-κB in cancer development started to be closely investigated when several NF-kB family genes were found to harbor rare mutations in certain types of cancers, especially in hematopoietic malignancies.114 As such, a large cohort study in DLBCL was characterized by preferential activation of the NF-κB pathway and subsequent nuclear expression of p50/p65 and p50/c-Rel dimers, compared with germinal B cell lymphocytes.115 Amplifications and rearrangements in c-Rel genes are often detected in various non-Hodgkin’s B cell lymphomas.116 NF-κB also plays a significant role in metastasis of several solid tumors, including breast cancer, HNSCC, and lung cancer.113 It has been shown that inhibition of NF-κB abolishes VEGF production and subsequent angiogenesis in a variety of conditions.117 In addition, NF-κB induces the expression of anti-apoptotic genes, such as caspase-8 inhibitor FLIP, inhibitor of apoptosis genes c-IAP1/2 and XIAP, as well as apoptosis regulating genes belonging to the Bcl-2 family. Furthermore, many oncogenic mutations in EGFR, Ras, PI3K, and TP53 genes are known to contribute to NF-κB activation in cells derived from pancreatic, colorectal, and lung tumors, further warranting NF-kB targeting as a cancer therapy.117,118 Recently, therapeutic agents specifically targeting the NF-kB pathway have been considered to be front-line therapy. ‘Pentoxifylline’ specifically targets c-Rel nuclear translocation and also inhibits NFAT, which is currently under investigation in a phase II clinical trial involving pediatric ALL patients (NCT02451774, refer to Table 2). ‘Ibrutinib’ selectively inhibits BCR and NF-kB singling, hence reducing cell proliferation in CLL patients that is characterized by prominent activation of NF-kB and BCR.43,74 The phase I/II clinical trial applying ‘Ibrutinib’ either alone or in combination with EGFR inhibitor ‘Rituximab’ is being currently tested in DLBCL patients (NCT02670616, NCT02388581, refer to Table 3). Hundreds of natural and synthetic compounds have been reported to selectively inhibit NF-kB; however, their clinical application has shown little efficacy, except for certain types of lymphoma and leukemia.119 There is evidence that various NF-kB inhibitors prolonged survival in NSCLC mouse tumor models, induced by KRAS and TP53 compound mutations; however, resistant tumors appeared within several weeks.120 Mechanisms that led to this resistance remain unclear. Nevertheless, NF-κB inhibitors still appear attractive, although combinations with other chemotherapies are currently considered a better choice.121 Furthermore, NF-kB activation has been linked to sensitization to chemo- or radiation therapies, with a strong potential to serve as a biomarker.121 Thus, clinical trials are currently investigating NF-kB activation as a biomarker in response to radio- and chemotherapies in patients with rectal- (NCT00280761) and gastric carcinomas (NCT01905969).
Immune checkpoint inhibitors
Recent exciting advancements in cancer treatment have been achieved in the field of immunotherapy. Vital fundamental discoveries over the last few decades have shown that the immune cells play a critical role in maintaining an equilibrium between immune recognition and tumor development, with the dual capacity of promoting and suppressing tumor growth.122 It is well accepted that tumor cells derive from genetic instability, uncontrolled cell division, and reduced immunogenicity that allows tumors to evade the immune system.123 These processes enable tumor cells to impair the immune system’s capacity to eradicate them by immune suppressive effects or by loss of targetable antigen expression. Therefore, cancer immunotherapy involves use of naturally derived or synthetically generated components with the goal of activating the immune system to target the cancer.123 Immune checkpoint inhibitors have demonstrated a considerably important breakthrough in the recent approval to treat solid tumor and hematologic malignancies in cancer immunotherapy.124 The main concept of immune checkpoint targeting is to prevent receptors on the T cells and cancer cell ligands from binding to each other, hence disrupting signaling cascades that help cancer cells evade T cell-mediated cell death. Immune checkpoint inhibitors modulate interactions between tumor cells and cytotoxic T lymphocytes within the tumor environment, which are exhausted in their function.124 Currently, two immune checkpoint proteins, cytotoxic T lymphocyte-associated 4 (CTLA-4) and programmed cell death protein 1 (PD-1) or its ligand (PD-L1) have been evaluated. They have been found to positively influence cancer treatment outcomes, disease progression-free and/or overall survival, compared with chemotherapy-based treatment.124,125 CTLA-4 and PD-1 are known to mediate immunological homeostasis by acting as downregulators of T cell activity after pathogen elimination. The FDA has already approved anti-CTLA-4 antibodies (i.e., ‘Ipilimumab’), PD1 antibodies (i.e., ‘Nivolumab’ and ‘Pembrolizumab’) and PD-L1 antibodies (i.e., ‘Atezolizumab’). Since 2011, these have demonstrated remarkable results either alone or in combination with other drugs or surgery for cancer treatment. This has been observed for many malignancies, including melanoma, Hodgkin’s lymphoma, bladder, kidney, and/or lung cancer.125 Many clinical trials involving combinations of these promising targeting agents are currently under investigation in various cancer types (liver, renal, ovarian, HNSCC, and pancreatic cancers) (refer to Table 4). Further approaches to be applied in the field of hematological malignancies involve combining immune checkpoint inhibitors with chimeric antigen receptor T cells (CAR-T cells). In 2017, CD19-targeting CARs T cell therapy was approved by the FDA. The first one being ‘Kymriah™,’ which was used for ALL treatment in children. This therapy achieved complete remission in 83% of patients with B cell ALL, although 49% of them suffered from strong cytokine release adverse effects.126 Similarly, ‘YescartaTM’ is applicable for adult advanced lymphomas. Initial results show that 72% of patients positively responded to this therapy, with 51% even showing complete remission of cancer after a single infusion.127 Currently, most of the available checkpoint inhibitor trials are in phase I/II clinical trial stages (Table 4). There are a few ongoing phase III clinical trials, which are investigating checkpoint inhibitors in combination with a single agent (i.e ‘Docetaxel,’ ‘Ipilimumab,’ Cisplatin, 5-Fu, ‘Cetuximab,’ etc.) in SCLC and HNSCC (refer to Table 4). A summary of currently ongoing clinical trials of individual, as well as combined immune checkpoint inhibitors (as registered in ClinicalTrials.gov) is provided in Table 4.
Table 4.
Summary of checkpoint inhibitor clinical trials for human cancers. Data taken from http://clinicaltrials.gov/.
| Drug | Combination | Sponsor | Tumor types | Sample size | Phases | Recruitment Status | Clinical trial ID |
|---|---|---|---|---|---|---|---|
| Anti-PD1 antibody | |||||||
| Nivolumab | Tetrahydrouridine | Yogen Saunthararajah | Non Small Cell Lung Cancer | 60 | II | Recruiting | NCT02664181 |
| - | National Cancer Institute (NCI) | Ependymoma, Meningioma, Chordoma | 180 | II | Recruiting | NCT03173950 | |
| TIL infusion | Inge Marie Svane | Metastatic Ovarian Cancer | 12 | I/II | Recruiting | NCT03287674 | |
| - | Hospital Moinhos de Vento | Prostate Cancer | 29 | II | Recruiting | NCT03040791 | |
| Denosumab | Australia and New Zealand Melanoma Trials Group | Metastatic Melanoma | 72 | I/II | Recruiting | NCT03161756 | |
| TAE | Teclison Ltd. | Liver Cancer | 40 | II | Recruiting | NCT03259867 | |
| Viagenpumatucel-L | Heat Biologics | Non Small Cell Lung Cancer | 120 | I/II | Recruiting | NCT02439450 | |
| Radiation | Giuseppe Giaccone | Small Cell Lung Cancer | 56 | I/II | Recruiting | NCT03325816 | |
| Ipilimumab | Bristol-Myers Squibb | Recurrent or Metastatic HNSCC | III | Recruiting | NCT02741570 | ||
| Interleukin-2 | University of Michigan Cancer Center | Metastatic Clear Cell Renal Cell Cancer | 23 | I/II | Recruiting | NCT02989714 | |
| Omaveloxolone or Ipilimumab | Reata Pharmaceuticals, Inc. | Melanoma | 102 | I/II | Recruiting | NCT02259231 | |
| Pembrolizumab | Gemcitabine or Cisplatin | Cedars-Sinai Medical Center | Recurrent Platinum-resistant Ovarian Cancer | 25 | II | Recruiting | NCT02608684 |
| Idelalisib | Zhonglin Hao | Non Small Cell Lung Cancer | 40 | I | Recruiting | NCT03257722 | |
| Docetaxel | Medical University of Vienna | Recurrent or Metastatic Head and Neck Cancer | 22 | I/II | Recruiting | NCT02718820 | |
| INCB001158 | Incyte Corporation | Advanced/Metastatic Solid Tumors | 346 | I/II | Recruiting | NCT02903914 | |
| Vitamin D | Translational Genomics Research Institute | Pancreatic Cancer | 24 | II | Recruiting | NCT03331562 | |
| B-701 | BioClin Therapeutics, Inc. | Advanced or Metastatic Urothelial Cell Carcinoma | 74 | I/II | Recruiting | NCT03123055 | |
| Methotrexate/Docetaxel/Cetuximab | Merck Sharp & Dohme Corp. | Recurrent or Metastatic Head and Neck Cancer | 495 | III | Active, not recruiting | NCT02252042 | |
| Cisplati/Carboplatin/5-FU/Cetuximab | Merck Sharp & Dohme Corp. | Recurrent or Metastatic HNSCC | 825 | III | Active, not recruiting | NCT02358031 | |
| - | Kindai University | Hepatocellular Carcinoma | 50 | II | Not yet recruiting | NCT03337841 | |
| - | Biothera | Advanced MelanomaTriple-Negative Breast Cancer | 95 | II | Recruiting | NCT02981303 | |
| Olaptesed | NOXXON Pharma AG | Colorectal and Pancreatic Cancer | 20 | I/II | Recruiting | NCT03168139 | |
| Laser Interstitial Thermotherapy | Comprehensive Cancer Center | Recurrent Glioblastoma | 34 | I/II | Recruiting | NCT03277638 | |
| Pembrolizumab or Nivolumab | HyperAcute®-Melanoma | NewLink Genetics Corporation | Metastatic Melanoma | 100 | II | Unknown | NCT02054520 |
| IBI308 | Docetaxel | Innovent Biologics (Suzhou) Co., Ltd. | Squamous Cell Lung Carcinoma | 266 | III | Recruiting | NCT03150875 |
| JS001 | - | Shanghai Junshi Bioscience Co., Ltd. | Advanced or Metastatic Bladder Urothelial Carcinoma | 370 | II | Recruiting | NCT03113266 |
| JS001 | - | Shanghai Junshi Bioscience Co., Ltd. | Mucosal Melanoma | 220 | II | Recruiting | NCT03178123 |
| PD-1 Antibodies | - | University Hospital Heidelberg | Melanoma | 40 | II | Recruiting | NCT03171064 |
| Anti-PD-L1 antibody | |||||||
| Atezolizumab | Radiotherapy | Gustave Roussy, Cancer Campus, Grand Paris | Metastatic Tumors | 180 | II | Recruiting | NCT02992912 |
| Guadecitabine | University of Southern California | Acute Myeloid Leukemia | 72 | I/II | Recruiting | NCT02935361 | |
| Atezolizumab | Immune Design | Sarcoma | 88 | II | Active, not recruiting | NCT02609984 | |
| Avelumab | CMB305 | Clinique Neuro-Outaouais | Glioblastoma Multiforme of Brain | 30 | II | Recruiting | NCT03047473 |
| Blocking interaction of PD1 and PDL1 | |||||||
| Durvalumab | Tremelimumab | Samsung Medical Center | Inoperable Esophageal Cancer | 40 | II | Recruiting | NCT03377400 |
| PDR001 | - | Novartis Pharmaceuticals | Advanced Malignancies | 318 | I/II | Recruiting | NCT02404441 |
| Anti-CTLA-4 antibody | |||||||
| Ipilimumab | Nivolumab | Olivia Newton-John Cancer Research Institute | Gastrointestinal Cancer and Neuroendocrine Tumors | 60 | II | Recruiting | NCT02923934 |
| Olaparib | Cediranib | National Cancer Institute (NCI) | Advanced Solid Tumors | 421 | I/II | Recruiting | NCT02484404 |
| CDK4/6 inhibitor | |||||||
| Trilaciclib | Atezolizumab | G1 Therapeutics, Inc. | Small Cell Lung Cancer | 105 | II | Active, not recruiting | NCT03041311 |
| Others | |||||||
| Enfortumab vedotin | Astellas Pharma Global Development, Inc. | Advanced or Metastatic Urothelial Bladder Cancer | 120 | II | Recruiting | NCT03219333 | |
| PV-10 | Dacarbazine | Provectus Biopharmaceuticals, Inc. | Advanced Cutaneous Melanoma | 225 | III | Recruiting | NCT02288897 |
| Anti-OX40 Antibody PF-04 518 600 | Axitinib | University of Southern California | Metastatic Kidney Cancer | 104 | II | Recruiting | NCT03092856 |
Concluding remarks
The way to reach the ultimate goal of precision treatment of cancer, that is the delivery of effective drugs to each individual patient, based on their characterized molecular profiles requires a considerable amount of basic research to understand the fundamentals of cancer heterogeneity. Cancer is an evolutionary complex, dynamic, and genetically heterogeneous disease, with multiple contributing factors and cellular components involved in its initiation, progression, and metastasis. This immense complexity, together with increasing resistance of tumors against currently implemented therapeutic interventions in clinical settings, requires a thorough understanding of cancer evolution at the cellular, genomic, transcriptomic, epigenetic, and proteomic levels.
In addition to conventional tools used to study cancer heterogeneity in bulk tissues, the recent development and constant optimization of more sophisticated sequencing tools at the single-cell level will continue to advance our insight and knowledge into tumor evolutionary origins, unique microenvironments, as well as metastasis. Studying intra-tumor heterogeneity and the spatial orientation of sub-clones within the primary tumor, via novel spatial transcriptomic methods together with simultaneous multiple ‘omic’-sequencing, will promote specific drug targeting of individual tumor sub-clones in the near future. Examining the nature of stem-like tumor cells and the transcriptomic mechanisms required to give rise to new tumor populations, will give clarity to the origin of various metastatic disease states. Targeting these stem-like cells could hamper the spread of cancer throughout the body. Being able to longitudinally isolate and sample CTCs will permit non-invasive diagnosis and monitoring, hence enabling highly personalized treatment. Treatment approaches can be constantly modified upon tracking the response and evolution of CTCs throughout the treatment. Finally, treatment resistance can be prevented through more accurate modeling of tumor resistance development to current drugs or radiotherapy. Much work still remains to make these goals a reality, but as single-cell sequencing methods continue to become cheaper, capable of achieving higher coverage, enabling multi-omic analyses, have higher fidelity and the ability to process a greater number of cells at faster rates, there is no doubt that these goals are attainable. Thus, we are coming closer to a promising future with the enhanced ability to generate new personalized therapeutic strategies in our constant fight against cancer.
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China (Grant No. 81770173), and the National Institutes of Health (Grant No. R01 DK100858).
Conflict of interest statement
None declared.
Authors’ contributions
Conception development and article design: Xinghua Pan and Jialing Zhang. Acquisition of data: Jialing Zhang and Stephan Stanislaw Späth. Writing and/or revision the manuscript: Jialing Zhang, Stephan Stanislaw Späth, Sadie L. Marjani, Wengeng Zhang and Xinghua Pan.
References
- 1. Heppner GH, Shekhar M: Tumor Heterogeneity Is Fundamental to the Tumor Ecosystem. Oncology-Ny 2014, 28:780–1. [PubMed] [Google Scholar]
- 2. McGranahan N, Swanton C: Biological and Therapeutic Impact of Intratumor Heterogeneity in Cancer Evolution (vol 27, pg 15, 2015). Cancer Cell 2015, 28:141–1. [DOI] [PubMed] [Google Scholar]
- 3. Tellez-Gabriel M, Ory B, Lamoureux F, et al. Tumour Heterogeneity: The Key Advantages of Single-Cell Analysis. Int J Mol Sci 2016, 17. pii: E2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Meacham CE, Morrison SJ: Tumour heterogeneity and cancer cell plasticity. Nature 2013, 501:328–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Litzenburger UM, Buenrostro JD, Wu B, et al. Single-cell epigenomic variability reveals functional cancer heterogeneity. Genome Biol 2017, 18:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kreso A, Dick JE: Evolution of the cancer stem cell model. Cell Stem Cell 2014, 14:275–91. [DOI] [PubMed] [Google Scholar]
- 7. Liu L, Liu J, Shao D, et al. Comprehensive genomic profiling of lung cancer using a validated panel to explore therapeutic targets in East Asian patients. Cancer Sci 2017, 108:2487–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shackleton M, Quintana E, Fearon ER, et al. Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell 2009, 138:822–9. [DOI] [PubMed] [Google Scholar]
- 9. Shyr D, Liu Q: Next generation sequencing in cancer research and clinical application. Biol Proced Online 2013, 15:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. New Engl J Med 2012, 366:883–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Akbani R, Ng KS, Werner HM, et al. A pan-cancer proteomic analysis of The Cancer Genome Atlas (TCGA) project. [abstract]. In: Proceedings of the 105th Annual Meeting of the American Association for Cancer Research; 2014 Apr 5–9; San Diego, CA. Philadelphia (PA): AACR; Cancer Res 2014, 74 (19 Suppl): Abstract nr 4262. [Google Scholar]
- 12. Lee H, Palm J, Grimes SM, et al. The Cancer Genome Atlas Clinical Explorer: a web and mobile interface for identifying clinical-genomic driver associations. Genome Med 2015, 7:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nakagawa H, Fujita M: Whole genome sequencing analysis for cancer genomics and precision medicine. Cancer Sci 2018, 109:513–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang XY, Marjani SL, Hu ZY, et al. Single-Cell Sequencing for Precise Cancer Research: Progress and Prospects. Cancer Res 2016, 76:1305–12. [DOI] [PubMed] [Google Scholar]
- 15. Dalerba P, Kalisky T, Sahoo D, et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol 2011, 29:1120–U1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013, 339:1546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Weinstein JN, Collisson EA, Mills GB, et al. Network CGAR: The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013, 45:1113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Patel JP, Gonen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med 2012, 366:1079–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shah SP, Roth A, Goya R, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 2012, 486:395–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012, 489:519–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koboldt DC, Fulton RS, McLellan MD, et al. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wyatt AW, Mo F, Wang K, et al. Heterogeneity in the inter-tumor transcriptome of high risk prostate cancer. Genome Biol 2014, 15: 426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lohr JG, Stojanov P, Carter SL, et al. Widespread Genetic Heterogeneity in Multiple Myeloma: Implications for Targeted Therapy. Cancer Cell 2014, 25:91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Network TC: Corrigendum: Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2013, 494:506. [DOI] [PubMed] [Google Scholar]
- 25. Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487:330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li S, Garrett-Bakelman FE, Chung SS, et al. Distinct evolution and dynamics of epigenetic and genetic heterogeneity in acute myeloid leukemia. Nat Med 2016, 22:792–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013, 499:214–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kalikaki A, Koutsopoulos A, Trypaki M, et al. Comparison of EGFR and K-RAS gene status between primary tumours and corresponding metastases in NSCLC. Brit J Cancer 2008, 99:923–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Albanese I, Scibetta AG, Migliavacca M, et al. Heterogeneity within and between primary colorectal carcinomas and matched metastases as revealed by analysis of Ki-ras and p53 mutations. Biochem Bioph Res Commun 2004, 325:784–91. [DOI] [PubMed] [Google Scholar]
- 30. Sottoriva A, Kang H, Ma ZC, et al. A Big Bang model of human colorectal tumor growth. Nat Genet 2015, 47:209–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morris LGT, Riaz N, Desrichard A, et al. Pan-cancer analysis of intratumor heterogeneity as a prognostic determinant of survival. Oncotarget 2016, 7:10051–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Merlo LMF, Pepper JW, Reid BJ, et al. Cancer as an evolutionary and ecological process. Nat Rev Cancer 2006, 6:924–35. [DOI] [PubMed] [Google Scholar]
- 33. Korolev KS, Xavier JB, Gore J: Turning ecology and evolution against cancer. Nat Rev Cancer 2014, 14:371–80. [DOI] [PubMed] [Google Scholar]
- 34. Taylor TB, Johnson LJ, Jackson RW, et al. First steps in experimental cancer evolution. Evol Appl 2013, 6:535–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shain AH, Yeh I, Kovalyshyn I, et al. The Genetic Evolution of Melanoma from Precursor Lesions. New Engl J Med 2015, 373:1926–36. [DOI] [PubMed] [Google Scholar]
- 36. Nguyen LV, Pellacani D, Lefort S, et al. Barcoding reveals complex clonal dynamics of de novo transformed human mammary cells. Nature 2015, 528:267–71. [DOI] [PubMed] [Google Scholar]
- 37. Sievers CK, Leystra AA, Clipson L, et al. Understanding Intratumoral Heterogeneity: Lessons from the Analysis of At-Risk Tissue and Premalignant Lesions in the Colon. Cancer Prev Res 2016, 9:638–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dohner H, Estey EH, Amadori S, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood 2010, 115:453–74. [DOI] [PubMed] [Google Scholar]
- 39. Shin SY, Lee ST, Kim HJ, et al. Mutation profiling of 19 candidate genes in acute myeloid leukemia suggests significance of DNMT3A mutations. Oncotarget 2016, 7:54825–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ok CY, Patel KP, Garcia-Manero G, et al. TP53 mutation characteristics in therapy-related myelodysplastic syndromes and acute myeloid leukemia is similar to de novo diseases. J Hematol Oncol 2015, 8: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ley TJ: Genomic and Epigenomic Landscapes of Adult De Novo Acute Myeloid Leukemia (vol 368, pg 2059, 2013). New Engl J Med 2013, 369:98–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Duncavage EJ, Abel HJ, Szankasi P, et al. Targeted next generation sequencing of clinically significant gene mutations and translocations in leukemia. Modern Pathol 2012, 25:795–804. [DOI] [PubMed] [Google Scholar]
- 43. Bohers E, Mareschal S, Bouzelfen A, et al. Targetable Activating Mutations are Very Frequent in GCB and ABC Diffuse Large B-Cell Lymphoma. Gene Chromosome Canc 2014, 53:144–53. [DOI] [PubMed] [Google Scholar]
- 44. Jardin F: Next generation sequencing and the management of diffuse large B-cell lymphoma: from whole exome analysis to targeted therapy. Discov Med 2014, 18:51–65. [PubMed] [Google Scholar]
- 45. Zhang J, Grubor V, Love CL, et al. Genetic heterogeneity of diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A 2013, 110:1398–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Compagno M, Lim WK, Grunn A, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature 2009, 459:717–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gonzalez-Aguilar A, Idbaih A, Boisselier B, et al. Recurrent mutations of MYD88 and TBL1XR1 in primary central nervous system lymphomas. Clin Cancer Res 2012, 18:5203–5211. [DOI] [PubMed] [Google Scholar]
- 48. Araujo LH, Timmers C, Bell EH, et al. Genomic Characterization of Non-Small-Cell Lung Cancer in African Americans by Targeted Massively Parallel Sequencing. J Clin Oncol 2015, 33:1966–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen F, Zhang Y, Parra E, et al. Multiplatform-based molecular subtypes of non-small-cell lung cancer. Oncogene 2017, 36:1384–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lawrence MS, Sougnez C, Lichtenstein L, et al. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517:576–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hajek M, Sewell A, Kaech S, et al. TRAF3/CYLD mutations identify a distinct subset of human papillomavirus-associated head and neck squamous cell carcinoma. Cancer 2017, 123:1778–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang J, Cheng H, Yang X, et al. Alternative NF-κB pathway activation enhanced by deficient TRAF3 in human papillomavirus (HPV)-associated head and neck cancer. [abstract]. In: Proceedings of the 105th Annual Meeting of the American Association for Cancer Research, 2014 Apr 5–9; San Diego, CA. Philadelphia (PA): AACR; Cancer Res 2014, 74 (19 Suppl): Abstract 3170. [Google Scholar]
- 53. Gatalica Z, Vranic S, Xiu J, et al. High microsatellite instability (MSI-H) colorectal carcinoma: a brief review of predictive biomarkers in the era of personalized medicine. Fam Cancer 2016, 15:405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lochhead P, Chan AT, Giovannucci E, et al. Progress and Opportunities in Molecular Pathological Epidemiology of Colorectal Premalignant Lesions. Am J Gastroenterol 2014, 109:1205–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Guglielmini PF, Rossi M, Grosso F, et al. Double KRAS and BRAF mutations in colorectal cancer in a single oncologic department series. J Clin Oncol 2013, 31 (15 Suppl): Abstract e14657. [Google Scholar]
- 56. The Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature 2012, 490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dubsky P, Brase JC, Jakesz R, et al. The EndoPredict score provides prognostic information on late distant metastases in ER+/HER2- breast cancer patients. Brit J Cancer 2013, 109:2959–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zonno KD, Kaldate RR, Arnell C, et al. BRCA1/2 mutation prevalence among triple-negative breast cancer patients from a large commercial testing cohort. J Clin Oncol 2013, 31 (15 Suppl): Abstract 1544. [Google Scholar]
- 59. Wen WX, Soo JSS, Kwan PY, et al. Germline APOBEC3B deletion is associated with breast cancer risk in an Asian multi-ethnic cohort and with immune cell presentation. Breast Cancer Res 2016, 18: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ellsworth RE, Blackburn HL, Shriver CD, et al. Molecular heterogeneity in breast cancer: State of the science and implications for patient care. Semin Cell Dev Biol 2017, 64:65–72. [DOI] [PubMed] [Google Scholar]
- 61. Seol H, Lee HJ, Choi Y, et al. Intratumoral heterogeneity of HER2 gene amplification in breast cancer: its clinicopathological significance. Modern Pathol 2012, 25:938–48. [DOI] [PubMed] [Google Scholar]
- 62. George E, Kim H, Krepler C, et al. A patient-derived-xenograft platform to study BRCA-deficient ovarian cancers. JCI Insight 2017, 2:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang WX, Figg WD: Secondary BRCA1 and BRCA2 alterations and acquired chemoresistance. Cancer Biol Ther 2008, 7:1004–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lopez-Chavez A, Carter CA, Giaccone G: The role of KRAS mutations in resistance to EGFR inhibition in the treatment of cancer. Curr Opin Investig Drugs 2009, 10:1305–14. [PubMed] [Google Scholar]
- 65. Housman G, Byler S, Heerboth S, et al. Drug Resistance in Cancer: An Overview. Cancers 2014, 6:1769–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Baselga J, Campone M, Piccart M, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med 2012, 366:520–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Beck JT: Potential role for mammalian target of rapamycin inhibitors as first-line therapy in hormone receptor-positive advanced breast cancer. Oncotargets Ther 2015, 8:3629–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Uchibori K, Inase N, Araki M, et al. Brigatinib combined with anti-EGFR antibody overcomes osimertinib resistance in EGFR-mutated non-small-cell lung cancer. Nat Commun 2017, 8:14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Li H, Hu HC, Wang R, et al. Primary concomitant EGFR T790M mutation predicted worse prognosis in non-small cell lung cancer patients. Oncotargets Ther 2014, 7:513–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Turke AB, Zejnullahu K, Wu YL, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell 2010, 17:77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Flaherty KT, Infante JR, Daud A, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med 2012, 367:1694–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature 2002, 417:949–54. [DOI] [PubMed] [Google Scholar]
- 73. Ascierto PA, McArthur GA, Dreno B, et al. Cobimetinib combined with vemurafenib in advanced BRAF(V600)-mutant melanoma (coBRIM): updated efficacy results from a randomised, double-blind, phase 3 trial. Lancet Oncol 2016, 17:1248–60. [DOI] [PubMed] [Google Scholar]
- 74. Valent P: Imatinib-resistant chronic myeloid leukemia (CML): Current concepts on pathogenesis and new emerging pharmacologic approaches. Biologics 2007, 1:433–48. [PMC free article] [PubMed] [Google Scholar]
- 75. Holleman A, Cheok MH, den Boer ML, et al. Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. New Engl J Med 2004, 351:533–42. [DOI] [PubMed] [Google Scholar]
- 76. McComb S, Aguade-Gorgorio J, Harder L, et al. Activation of concurrent apoptosis and necroptosis by SMAC mimetics for the treatment of refractory and relapsed ALL. Sci Transl Med 2016, 8:339ra70. [DOI] [PubMed] [Google Scholar]
- 77. Nugues AL, El Bouazzati H, Hetuin D, et al. RIP3 is downregulated in human myeloid leukemia cells and modulates apoptosis and caspase-mediated p65/RelA cleavage. Cell Death Dis 2014, 5:e1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ding L, Ley TJ, Larson DE, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 2012, 481:506–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Dhillon KK, Swisher EM, Taniguchi T: Secondary mutations of BRCA1/2 and drug resistance. Cancer Sci 2011, 102:663–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Van Cutsem E, Tabernero J, Lakomy R, et al. Addition of aflibercept to fluorouracil, leucovorin, and irinotecan improves survival in a phase III randomized trial in patients with metastatic colorectal cancer previously treated with an oxaliplatin-based regimen. J Clin Oncol 2012, 30:3499–3506. [DOI] [PubMed] [Google Scholar]
- 81. Leto SM, Trusolino L: Primary and acquired resistance to EGFR-targeted therapies in colorectal cancer: impact on future treatment strategies. J Mol Med (Berl) 2014, 92:709–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Smith DH, Fiehn AM, Fogh L, et al. Measuring ERCC1 protein expression in cancer specimens: validation of a novel antibody. Sci Rep 2014, 4:4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Van Emburgh BO, Arena S, Siravegna G, et al. Acquired RAS or EGFR mutations and duration of response to EGFR blockade in colorectal cancer. Nat Commun 2016, 7:13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sartore-Bianchi A, Trusolino L, Martino C: Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial (vol 17, pg 738, 2016). Lancet Oncol 2016, 17:E420–0. [DOI] [PubMed] [Google Scholar]
- 85. Friedman R: Drug resistance in cancer: molecular evolution and compensatory proliferation. Oncotarget 2016, 7:11746–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ellis LM, Hicklin DJ: Resistance to Targeted Therapies: Refining Anticancer Therapy in the Era of Molecular Oncology. Clin Cancer Res 2009, 15:7471–8. [DOI] [PubMed] [Google Scholar]
- 87. Izar B, Rotow J, Gainor J, et al. Pharmacokinetics, clinical indications, and resistance mechanisms in molecular targeted therapies in cancer. Pharmacol Rev 2013, 65:1351–95. [DOI] [PubMed] [Google Scholar]
- 88. Navin N, Kendall J, Troge J, et al. Tumour evolution inferred by single-cell sequencing. Nature 2011, 472:90–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Eberwine J, Yeh H, Miyashiro K, et al. Analysis of gene expression in single live neurons. Proc Natl Acad Sci U S A 1992, 89:3010–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Gawad C, Koh W, Quake SR: Single-cell genome sequencing: current state of the science. Nat Rev Genet 2016, 17:175–88. [DOI] [PubMed] [Google Scholar]
- 91. Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014, 344:1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Jordan NV, Bardia A, Wittner BS, et al. HER2 expression identifies dynamic functional states within circulating breast cancer cells. Nature 2016, 537:102–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Gorner K, Bachmann J, Schiemann M, et al. Circulating Tumour Cells: Analysis of Single Cells Isolated from Ductal Pancreatic Adenocarcinoma Patients. Tumor Biol 2010, 31:S81–1. [Google Scholar]
- 94. Chen XX, Bai F: Single-cell analyses of circulating tumor cells. Cancer Biol Med 2015, 12:184–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Ni XH, Zhuo ML, Su Z, et al. Single Cell Genomic Analyses of Circulating Tumor Cells from Lung Cancer Patients. J Thorac Oncol 2013, 8:S493–3. [Google Scholar]
- 96. Wu CP, Wu P, Zhao HF, et al. Clinical Applications of and Challenges in Single-Cell Analysis of Circulating Tumor Cells. DNA Cell Biol 2018, 37:78–89. [DOI] [PubMed] [Google Scholar]
- 97. Taenzer A, Alix-Panabieres C, Wikman H, et al. Circulating tumor-derived biomarkers in lung cancer. J Thorac Dis 2012, 4:448–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Van Loo P, Voet T: Single cell analysis of cancer genomes. Curr Opin Genet Dev 2014, 24:82–91. [DOI] [PubMed] [Google Scholar]
- 99. Fallahi-Sichani M, Moerke NJ, Niepel M, et al. Systematic analysis of BRAF(V600E) melanomas reveals a role for JNK/c-Jun pathway in adaptive resistance to drug-induced apoptosis. Mol Syst Biol 2015, 11: 797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Galletti G, Portella L, Tagawa ST, et al. Circulating Tumor Cells in Prostate Cancer Diagnosis and Monitoring: An Appraisal of Clinical Potential. Mol Diagn Ther 2014, 18:389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Miyamoto DT, Zheng Y, Wittner BS, et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science 2015, 349:1351–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Janiszewska M, Liu L, Almendro V, et al. In situ single-cell analysis identifies heterogeneity for PIK3CA mutation and HER2 amplification in HER2-positive breast cancer. Nat Genet 2015, 47:1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Porta C, Paglino C, Mosca A: Targeting PI3K/Akt/mTOR Signaling in Cancer. Front Oncol 2014, 4:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Wang X, Ding J, Meng LH: PI3K isoform-selective inhibitors: next-generation targeted cancer therapies. Acta Pharmacol Sin 2015, 36:1170–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Guarinos C, Juarez M, Egoavil C, et al. Prevalence and characteristics of MUTYH-associated polyposis in patients with multiple adenomatous and serrated polyps. Clin Cancer Res 2014, 20:1158–68. [DOI] [PubMed] [Google Scholar]
- 106. Markham A: Copanlisib: First Global Approval. Drugs 2017, 77:2057–62. [DOI] [PubMed] [Google Scholar]
- 107. Massacesi C, Di Tomaso E, Urban P, et al. PI3K inhibitors as new cancer therapeutics: implications for clinical trial design. Oncotargets Ther 2016, 9:203–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Brufsky AM: Current Approaches and Emerging Directions in HER2-resistant Breast Cancer. Breast Cancer (Auckl) 2014, 8:109–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Van Waes C, Musbahi O: Genomics and advances towards precision medicine for head and neck squamous cell carcinoma. Laryngoscope Investig Otolaryngol 2017, 2:310–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Mendoza MC, Er EE, Blenis J: The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem Sci 2011, 36:320–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Tripathy D, Bardia A, Sellers WR: Ribociclib (LEE011): Mechanism of Action and Clinical Impact of This Selective Cyclin-Dependent Kinase 4/6 Inhibitor in Various Solid Tumors. Clinical Cancer Research 2017, 23:3251–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Eroglu Z, Ribas A: Combination therapy with BRAF and MEK inhibitors for melanoma: latest evidence and place in therapy. Ther Adv Med Oncol 2016, 8:48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Karin M: Nuclear factor-kappaB in cancer development and progression. Nature 2006, 441:431–6. [DOI] [PubMed] [Google Scholar]
- 114. Park MH, Hong JT: Roles of NF-kappaB in Cancer and Inflammatory Diseases and Their Therapeutic Approaches. Cells 2016, 5:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Cai Q, Tu M, Xu-Monette ZY, et al. NF-kappaB p50 activation associated with immune dysregulation confers poorer survival for diffuse large B-cell lymphoma patients with wild-type p53. Mod Pathol 2017, 30:854–76. [DOI] [PubMed] [Google Scholar]
- 116. Hunter JE, Butterworth JA, Zhao B, et al. The NF-kappaB subunit c-Rel regulates Bach2 tumour suppressor expression in B-cell lymphoma. Oncogene 2016, 35:3476–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Tabruyn SP, Griffioen AW: NF-kappa B: a new player in angiostatic therapy. Angiogenesis 2008, 11:101–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Prabhu L, Mundade R, Korc M, et al. Critical role of NF-kappaB in pancreatic cancer. Oncotarget 2014, 5:10969–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Gupta SC, Sundaram C, Reuter S, et al. Inhibiting NF-kappaB activation by small molecules as a therapeutic strategy. Biochim Biophys Acta 2010, 1799:775–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Xue W, Meylan E, Oliver TG, et al. Response and resistance to NF-kappaB inhibitors in mouse models of lung adenocarcinoma. Cancer Discov 2011, 1:236–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Godwin P, Baird AM, Heavey S, et al. Targeting nuclear factor-kappa B to overcome resistance to chemotherapy. Front Oncol 2013, 3:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Liu Y, Zeng G: Cancer and innate immune system interactions: translational potentials for cancer immunotherapy. J Immunother 2012, 35:299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Vinay DS, Ryan EP, Pawelec G, et al. Immune evasion in cancer: Mechanistic basis and therapeutic strategies. Semin Cancer Biol 2015, 35S185–98. [DOI] [PubMed] [Google Scholar]
- 124. Azoury SC, Straughan DM, Shukla V: Immune Checkpoint Inhibitors for Cancer Therapy: Clinical Efficacy and Safety. Curr Cancer Drug Targets 2015, 15:452–62. [DOI] [PubMed] [Google Scholar]
- 125. Dine J, Gordon R, Shames Y, et al. Immune Checkpoint Inhibitors: An Innovation in Immunotherapy for the Treatment and Management of Patients with Cancer. Asia Pac J Oncol Nurs 2017, 4:127–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Tasian SK, Gardner RA: CD19-redirected chimeric antigen receptor-modified T cells: a promising immunotherapy for children and adults with B-cell acute lymphoblastic leukemia (ALL). Ther Adv Hematol 2015, 6:228–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Makita S, Yoshimura K, Tobinai K: Clinical development of anti-CD19 chimeric antigen receptor T-cell therapy for B-cell non-Hodgkin lymphoma. Cancer Sci 2017, 108:1109–18. [DOI] [PMC free article] [PubMed] [Google Scholar]