Transcription factor p53 induces cell cycle arrest and apoptosis in response to DNA damage1 and cellular stress,2 thereby playing a critical role in protecting cells from malignant transformation. The E3 ubiquitin ligase hDM2 controls p53 levels through a direct binding interaction that neutralizes p53 transactivation activity, exports nuclear p53, and targets it for degradation via the ubiquitylation-proteasomal pathway.3,4 Loss of p53 activity, either by deletion, mutation, or hDM2 overexpression, is the most common defect in human cancer.5 Tumors with preserved expression of wild type p53 are rendered vulnerable by pharmacologic approaches that stabilize native p53. In this context, hDM2 targeting has emerged as a validated approach to restore p53 activity and resensitize cancer cells to apoptosis in vitro and in vivo.6
The p53-hDM2 protein interaction is mediated by the 15-residue α-helical transactivation domain of p53, which inserts into a hydrophobic cleft on the surface of hDM2.7 Three residues within this domain (F19, W23, and L26) are essential for hDM2-binding.8,9 We sought to optimize the p53 peptide as a biological tool and prototype therapeutic by enforcing its α-helical structure while preserving the key interacting residues that enable specific hDM2 engagement. We previously developed a “peptide stapling” strategy in which an all-hydrocarbon cross-link is generated within natural peptides by ruthenium-catalyzed olefin metathesis of inserted α,α-disubstituted nonproteogenic amino acids bearing olefinic side chains (Figure 1A).10 We report the application of this strategy to the generation of stabilized alpha-helix of p53 (SAH-p53) peptides that exhibit high affinity for hDM2, and, in contrast to the corresponding unmodified p53 peptide, readily enter cells through an active uptake mechanism. SAH-p53 treatment reactivated the p53 tumor suppressor cascade by inducing the transcription of p53-responsive genes, providing the first example of a stapled peptide that kills cancer cells by targeting a transcriptional pathway.
Figure 1.
Synthesis, sequence, and biochemical analysis of SAH-p53 peptides. (A) Non-natural amino acids were synthesized as described and cross-linked by ruthenium-catalyzed ring-closing olefin metathesis. (B) SAH-p53 compounds were generated by stapling the p5314-29 sequence at the indicated positions. Charge, α-helicity, hDM2 binding affinity, cell permeability, and impact on cell viability are indicated for each compound. (C, E) Circular dichroism spectra revealed enhancement of α-helicity for SAH-p53 compounds. (D, F) Fluorescence polarization assays using FITC-peptides and hDM217-125 demonstrated subnanomolar hDM2-binding affinities for select SAH-p53 peptides. Note: UAH-p53-8 is the “unstapled” form of SAH-p53-8.
To design SAH-p53 compounds, we placed synthetic olefinic derivatives at positions that avoid critical hDM2-binding residues. Hydrocarbon staples spanning the i, i+7 positions were generated by olefin metathesis (Figure 1A). An initial panel of four SAH-p53 peptides was synthesized, each containing a distinctively localized cross-link (Figure 1B). To evaluate the structural impact of installing an all-hydrocarbon i, i+7 staple, we conducted circular dichroism (CD) experiments to determine α-helicity. While the wild type p53 peptide displayed 11% α-helical content in water at pH 7.0, SAH-p53s 1–4 demonstrated 10–59% α-helicity (Figure 1B,C). Fluorescence polarization binding assays using hDM217-125 and FITC-labeled derivatives of SAH-p53s 1–4 identified 4 as a subnanomolar interactor, having an affinity for hDM2 almost 3 orders of magnitude greater than that of the wild type peptide (Figure 1B,D). SAH-p53-4 also demonstrated improved proteolytic stability (Supporting Information, Figure 1).
We found that the initial SAH-p53 compounds generated were incapable of penetrating intact Jurkat T-cells (Figure 1B, Supporting Information, Figure 2). We noted that SAH-p53s 1–4 were negatively charged (−2) at physiological pH. Positive charge is a characteristic feature of certain classes of cell penetrating peptides.11 In developing a second generation of compounds, we replaced aspartic and glutamic acids with asparagines and glutamines to adjust peptide charge and mutated select amino acids previously reported to participate in p53 nuclear export (L14Q) and ubiquitylation (K24R)4,12 (Figure 1B). SAH-p53s 5–8 exhibited a 2-fold to 8.5-fold enhancement in α-helical content, retained high binding affinity for hDM2, and demonstrated time- and temperature-dependent cellular uptake by FACS and confocal microscopy (Figure 1B,E,F, Supporting Information, Figure 3). Cell viability assays using RKO or SJSA-1 cancer cells exposed to SAH-p53 peptides indicated that SAH-p53-8, which contained point mutations in both nuclear export and ubiquitylation sites, was the only structurally stabilized, cell-permeable, and high affinity hDM2 binder that adversely affected cell viability (Figures 1B and 4A).
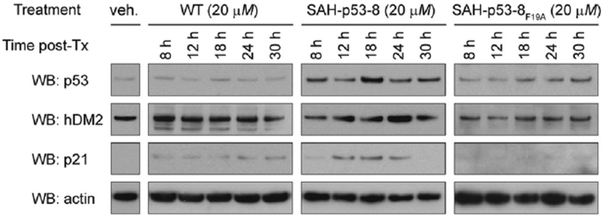
To determine if hDM2-targeting by SAH-p53-8 could specifically restore native p53 levels, we treated SJSA-1 cells with wild-type, 8, and 8F19A peptides for 8–30 h and monitored p53 protein levels by Western analysis (Figure 2). Cells exposed to SAH-p53-8 demonstrated increased p53 proteins levels that peaked at 18 h post-treatment. The p53 resuppression by 24 h correlated with the time-dependent upregulation of hDM2 by p53, consistent with an intact p53-hDM2 counter-regulatory mechanism.13 SAH-p53-8 likewise induced upregulation of the cyclin-dependent kinase inhibitor p21.14 The p21 upregulation in cells treated with 8 was detected at 12 h, reaching peak levels at 18 h. Baseline levels were restored by 30 h, consistent with resuppression of native p53. The hDM2 and p21 levels were unchanged in SJSA-1 cells treated with wild-type or 8F19A, highlighting the specificity of SAH-p53-8 modulation of the p53 signaling pathway.
Figure 2.
SAH-p53-8 reactivates the p53 transcriptional pathway. The hDM2 overexpressing SJSA-1 cells were exposed to the indicated peptides, and Western analyses for p53, hDM2, and p21 were performed at 8–30 h of treatment.
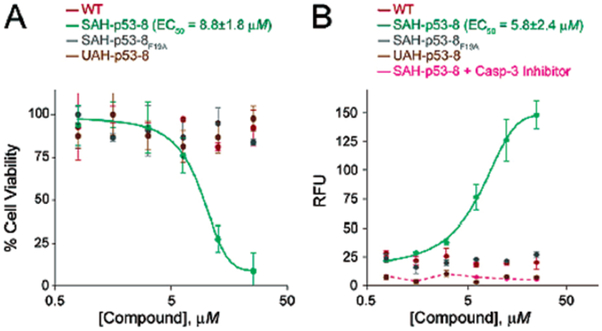
To examine whether SAH-p53-8-mediated stabilization of native p53 could inhibit cancer cells by reactivating the apoptotic pathway, we conducted viability and caspase-3 assays using SJSA-1 cells exposed to wild-type, 8, and 8F19A for 24 h (Figure 3). Whereas the wild-type and 8F19A peptides had no effect on cell viability, SAH-p53-8 exhibited dose-dependent inhibition of SJSA-1 cell viability (EC50 = 8.8 μM). Analysis of caspase-3 activation by fluorescence monitoring of the cleaved caspase-3 substrate Ac-DEVD-AMC15 showed that neither the wild-type nor the 8F19A peptides had any apoptotic effect; however, 8 triggered dose-dependent caspase-3 activation (EC50 = 5.8 μM) that was blocked by DEVD-CHO, a specific caspase-3 inhibitor, demonstrating that SAH-p53-8 specifically inhibited cell viability by activating apoptosis in hDM2-over-expressing SJSA-1 cells.
Figure 3.
Reactivation of apoptosis in SAH-p53-8-treated SJSA-1 cells. SAH-p53-8 demonstrated specific, dose-dependent cytotoxicity and apoptosis induction as measured by (A) CellTiter-Glo (Promega) and (B) Caspase-3 activation assays (Oncogene).
The identification of multiple organic compounds and p53 peptidomimetics with anti-hDM2 activity8,16 holds promise for achieving clinical benefit from manipulating the p53 pathway. By generating a stapled peptide-based hDM2 inhibitor, we have documented an in situ interaction between SAH-p53-8 and hDM2 (Supporting Information, Figure 4), confirming that its pro-apoptotic activity derives from restoration of the p53 pathway. The successful pharmacologic optimization of the transactivating α-helix of p53 suggests that this methodology may enable the development of a wide array of peptidic compounds to probe protein interactions and target deregulated transcriptional pathways for therapeutic benefit in cancer.
Supplementary Material
Acknowledgment.
This manuscript is dedicated to K. C. Nicolaou on the occasion of his 60th birthday and to Stanley J. Korsmeyer. We thank D. Brown, M. Salanga, and T. Diefenbach for confocal support and Eric Smith for editorial assistance. F.B. is the recipient of a Ruth L. Kirchstein NRSA F32CA103510. L.D.W. is supported by NHLBI Grant K08HL074049, a Burroughs-Wellcome Career Award, and a Culpeper Scholarship. This work was also supported by the Harvard/Dana-Farber Program in Cancer Chemical Biology.
Footnotes
Supporting Information Available: Complete ref 16h, peptide characterization, experimentals, and supplementary figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Kastan MB; Onyekwere O; Sidransky D; Vogelstein B; Craig RW Cancer Res. 1991, 51, 6304–6311. [PubMed] [Google Scholar]
- (2).Wu X; Bayle JH; Olson D; Levine AJ Genes Dev. 1993, 7, 1126–1132. [DOI] [PubMed] [Google Scholar]; Yonish-Rouach E; Resnftzky D; Lotem J; Sachs L; Kimchi A; Oren M Nature 1991, 352, 345–347. [DOI] [PubMed] [Google Scholar]; Momand J; Zambetti GP; Olson DC; George D; Levine AJ Cell 1992, 69, 1237. [DOI] [PubMed] [Google Scholar]
- (3).Levine AJ; Hu W; Feng Z Cell Death Differ. 2006, 13, 1027. [DOI] [PubMed] [Google Scholar]; Honda R; Tanaka H; Yasuda H FEBS Lett. 1997, 420, 25. [DOI] [PubMed] [Google Scholar]; Tao W; Levine AJ Proc. Nat. Acad. Sci. U.S.A 1999, 96, 3077–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Li M; Brooks CL; Wu-Baer F; Chen D; Baer R; Gu W Science 2003, 302, 1972–1975. [DOI] [PubMed] [Google Scholar]
- (5).Hollstein M; Sidransky D; Vogelstein B; Harris CC Science 1991, 253, 49–53. [DOI] [PubMed] [Google Scholar]
- (6).Chène P Nat. Rev. Cancer 2003, 3, 102–109. [DOI] [PubMed] [Google Scholar]
- (7).Kussie PH; Gorina S; Marechal V; Elenbaas B; Moreau J; Levine AJ; Pavletich NP Science 1996, 274, 948–953. [DOI] [PubMed] [Google Scholar]
- (8).Sakurai K; Chung HS; Kahne DJ Am. Chem. Soc 2004, 126, 16288–16289. [DOI] [PubMed] [Google Scholar]; Vassilev LT; Vu BT; Graves B; Carvajal D; Podlaski F; Filipovic Z; Kong N; Kammlott U; Lukacs C; Klein C; Fotouhi N; Liu EA Science 2004, 303, 844–848. [DOI] [PubMed] [Google Scholar]
- (9).Lin J; Chen J; Elenbaas B; Levine AJ Genes Dev. 1994, 8, 1235–1246. [DOI] [PubMed] [Google Scholar]
- (10).Schafmeister CE; Po J; Verdine GL J. Am. Chem. Soc 2000, 122, 5891–5892. [Google Scholar]; Walensky LD; Kung AL; Escher I; Malia TJ; Barbuto S; Wright RD; Wagner G; Verdine GL; Korsmeyer SJ Science 2004, 305, 1466–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Vivès E; Lebleu B The Tat-Derived Cell-Penetrating Peptide In Cell-Penetrating Peptides: Processes and Applications; Langel Ü, Ed.; CRC Press: Boca Raton, 2002; pp 3–21. [Google Scholar]
- (12).Zhang Y; Xiong Y Science 2001, 292, 1910–1915. [DOI] [PubMed] [Google Scholar]
- (13).Barak Y; Juven T; Haffner R; Oren M EMBO J. 1993, 12, 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]; Juven T; Barak Y; Zauberman A; George DL; Oren M Oncogene 1993, 8, 3411–3416. [PubMed] [Google Scholar]
- (14).Dulic V; Kaufmann WK; Wilson SJ; Tisty TD; Lees E; Harper JW; Elledge SJ; Reed SI Cell 1994, 76, 1013. [DOI] [PubMed] [Google Scholar]; El-Deiry WS; Tokino T; Velculescu VE; Levy DB; Parsons R; Trent JM; Lin D; Mercer WE; Kinzler KW; Vogelstein B Cell 1993, 75, 817. [DOI] [PubMed] [Google Scholar]
- (15).Pochampally R; Fodera B; Chen L; Lu W; Chen JJ Biol. Chem 1999, 274, 15271–15277. [DOI] [PubMed] [Google Scholar]
- (16).(a) Duncan SJ; Gruschow S; Williams DH; McNicholas C; Purewal R; Hajek M; Gerlitz M; Martin S; Wrigley SK; Moore MJ Am. Chem. Soc 2001, 123, 554–560. [DOI] [PubMed] [Google Scholar]; (b) Chène P; Fuchs J; Bohn J; Garcia-Echeverria C; Furet P; Fabbro DJ Mol. Biol 2000, 299, 245. [DOI] [PubMed] [Google Scholar]; (c) Sakurai K; Schubert C; Kahne DJ Am. Chem. Soc 2006, 128, 11000–11001. [DOI] [PubMed] [Google Scholar]; (d) Kritzer JA; Hodsdon ME; Schepartz AJ Am. Chem. Soc 2005, 127, 4118–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Kritzer JA; Lear JD; Hodsdon ME; Schepartz AJ Am. Chem. Soc 2004, 126, 9468–9469. [DOI] [PubMed] [Google Scholar]; (f) Wasylyk C; Salvi R; Argentini M; Dureuil C; Delumeau I; Abecassis J; Debussche L; Wasylyk B Oncogene 1999, 18, 1921–1934. [DOI] [PubMed] [Google Scholar]; (g) Garcia-Echeverria C; Chene P; Blommers MJJ; Furet PJ Med. Chem 2000, 43, 3205–3208. [DOI] [PubMed] [Google Scholar]; (h) Grasberger BL; et al. J. Med. Chem 2005, 48, 909–912. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.