Abstract
Stabilized alpha-helical (SAH) peptides are valuable laboratory tools to explore important protein–protein interactions. Whereas most peptides lose their secondary structure when isolated from the host protein, stapled peptides incorporate an all-hydrocarbon “staple” that reinforces their natural alpha-helical structure. Thus, stapled peptides retain their functional ability to bind their native protein targets and serve multiple experimental uses. First, they are useful for structural studies such as NMR or crystal structures that map and better define binding sites. Second, they can be used to identify small molecules that specifically target that interaction site. Third, stapled peptides can be used to test the importance of specific amino acid residues or posttranslational modifications to the binding. Fourth, they can serve as structurally competent bait to identify novel binding partners to specific alpha-helical motifs. In addition to markedly improved alpha-helicity, stapled peptides also display resistance to protease cleavage and enhanced cell permeability. Most importantly, they are useful for intracellular experiments that explore the functional consequences of blocking particular protein interactions. Because of their remarkable stability, stapled peptides can be applied to whole-animal, in vivo studies. Here we describe a protocol for the synthesis of a peptide that incorporates an all-hydrocarbon “staple” employing a ring-closing olefin metathesis reaction. With proper optimization, stapled peptides can be a fundamental, accurate laboratory tool in the modern chemical biologist’s armory.
Keywords: Stapled peptides, NMR, Protein–protein interactions, Olefin metathesis, Alpha helix
1. Introduction
Protein interactions are fundamental to the control of nearly every cellular function. The ability to manipulate these interactions is a highly desirable capability for both physiological experimentation and therapeutic benefit. Small molecules are particularly adept at targeting small hydrophobic pockets with high affinity and specificity [1]. However, the vast majority of protein interactions are mediated by relatively large and shallow interaction surfaces that are difficult to target with small molecules. Biologics, such as antibodies, are effective at recognizing large surface areas with high affinity, but their use is limited to targets that are extracellular [2]. Thus, many intracellular signaling pathways remain largely inaccessible to meaningful real-time manipulation.
Over billions of years of evolution Mother Nature has designed precise “lock-and-key” mechanisms to ensure specificity in the tremendous variety of intermolecular biologic recognition events. The most prevalent secondary structure element within proteins is the α-helix, which is employed for numerous protein–protein and protein–DNA interactions [3–5]. First modeled by Pauling and Corey, the alpha-helix contains 3.6 residues per turn with a rise of 0.15 nm per residue allowing for maximal reinforcement of hydrogen bonding between the amide protons and carbonyl oxygens [6]. The central core minimizes free space, while the amino acid side chains are displayed outwards to form unique topographies for interaction. Small peptides that correspond to a portion of a protein involved in such an interaction can potentially serve as effective binding partners because they accurately display a true mimic of its three-dimensional interaction surface. Nevertheless, taking a single helix out of the context of the entire protein has severe consequences on its structure and biological utility. In an aqueous environment, short peptides will often lose their alpha-helical secondary structure and unfold into random coil devoid of structure [7]. The loss of structure allows peptides to achieve an extended conformation making them prime targets for protease degradation [8]. Finally, with few exceptions, peptides cannot penetrate the cell membrane [9].
Many methods have been established for the enforcement of the alpha helical conformation, but many of these approaches make use of groups that are polar or pharmacologically labile [10, 11]. Each of these obstacles for native polypeptides can be overcome by the insertion of an all-hydrocarbon cross-link [12–15]. In this strategy, peptides are synthesized containing two α-methyl, α-alkenyl glycine substitutions positioned either one or two helix turns apart followed by a ruthenium-catalyzed, ring-closing olefin metathesis reaction which is compatible with modern solid-phase peptide synthesis protocols [16, 17]. Both α,α-disubstitution and macrocyclic bridge formation promote helix formation [18]. Likewise, the resulting amphipathic helix can promote cell permeability and precludes unfolding of the peptide enhancing its proteolytic stability [19].
Methodical examination of stereochemistry and chain length revealed that amino acids containing 5 carbon atoms in their alkenyl side chain with S stereochemistry (S5) placed one α-helical turn apart or, in the i and i+4 positions, is ideal [13]. Alternatively, to span two α-helical turns, the R stereoisomer with an 8 carbonatom alkenyl chain (R8) is placed at the i position and S5 in the i+ 7 position [20].
One of the benefits of synthesizing a peptide is the flexibility to include select modifications. For example, incorporating amino acids that are phosphorylated, methylated, or glycosylated easily explores posttranslational modifications. Moreover, peptides can be easily tracked in microscopy experiments by synthesizing them with specific labels such as FITC or various Alexa Fluor dyes [12]. Addition of a biotin moiety bestows a useful handle to pull down the peptide and any interacting partners with streptavidin [21]. A photoreactive group for UV light-induced covalent cross-linking can be used to capture interacting proteins and map interaction surfaces [21, 22]. Therefore, in addition to simply blocking a specific interaction, stapled peptides are valuable tool compounds in exploration of protein–protein interaction mechanisms in general. The methods subsequently described are broadly applicable and have been used to successfully manipulate numerous and diverse protein targets.
2. Materials
Rink amide MBHA resin.
1-Methyl-2-pyrrolidinone (NMP).
Piperidine.
2-(6-Chloro-1H-benzotriazole-1-yl)-1,1,3,3-tetramethylaminium hexaflurophosphate (HCTU).
N,N-diisopropyl ethylamine (DIEA).
1,2-Dichloroethane (DCE).
Grubbs catalyst (Grubbs I (bis(tricyclohexylphosphine)-benzylidene ruthenium (IV) dichloride)).
Dichloromethane (DCM).
Methanol.
Methyl-tert-butyl ether.
Acetic anhydride.
Trifluoroacetic acid (TFA).
Triisopropylsilane (TIS).
1,2-Ethanedithiol (EDT).
Diethyl ether.
Acetonitrile.
Fmoc amino acids.
Fritted reaction vessel (e.g., disposable chromatography columns or peptide synthesis glassware).
Machines: Peptide synthesizer.
Machines: HPLC/LC/MS.
3. Methods
3.1. Design Considerations
In the design of stapled alpha-helical peptides, it is crucial to mimic as closely as possible the original sequence of the protein in question. Residues known or presumed to be involved in the binding interaction being targeted should remain intact. In turn, the staple should be located away from the binding interface to avoid interference with the interaction surface. Because the location and size of the staple are crucial to the successful stabilization of the helix, panels of compounds with staples at different allowed locations with different chain lengths should be designed and synthesized. Once the ideal position and size of the staple have been found (based on in vitro binding studies with its target), substitutions to the native sequence can be undertaken in order to optimize the peptide for improved binding and potency.
3.2. Peptide Synthesis
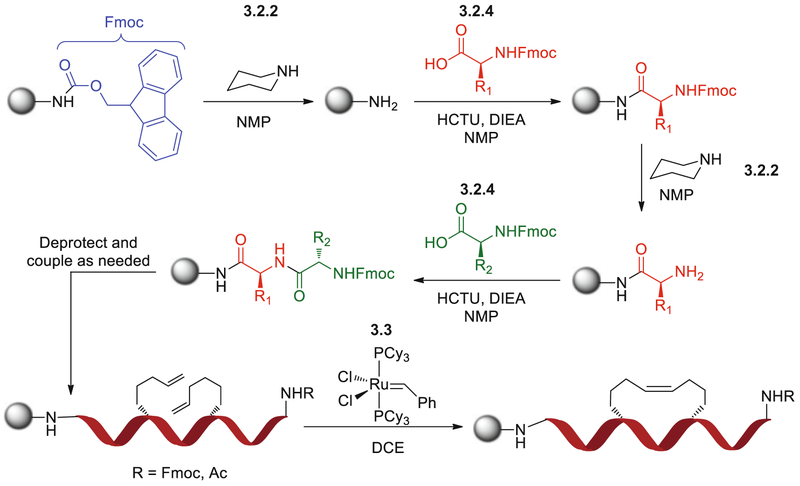
The synthesis of stapled peptides is carried out using standard Fmoc (fluorenylmethoxycarbonyl) solid-phase peptide synthesis protocols (Fig. 1). The procedure outlined below is suitable for the synthesis of a single stapled peptide in a 25 μmol scale.
Fig. 1.
Scheme for Fmoc-based solid-phase peptide synthesis with ruthenium-catalyzed olefin metathesis. Reactions are numbered by step within the protocol, where Subheading 3.2, step 2 represents the deprotection reaction, Subheading 3.2, step 4 the coupling reaction, and Subheading 3.3 the metathesis reaction. The chemical structure in Subheading 3.2, step 2 is that of piperidine, Subheading 3.2, step 4 the Fmoc-protected amino acid, and Subheading 3.3 the Grubbs catalyst. Gray ball represents rink amide MBHA resin
Swell Rink Amide MBHA resin (100–200 mesh, preferably with a substitution lower than 0.4 mmol/g) with 1 mL NMP for 15 min.
To remove the Fmoc-protecting group, wash the resin with 1 mL of a 20 % (v/v) solution of piperidine in NMP for 15 min and drain. Repeat once.
Wash resin with 1 mL NMP for 1 min draining to waste. Repeat five times.
Couple the Fmoc amino acid onto the drained resin by addition of 10 equivalents N-α-Fmoc-protected amino acid in NMP, 9.9 equivalents of HCTU in NMP, and 20 equivalents of DIEA. The reaction is shaken at room temperature for 45 min. If coupling a sterically hindered amino acid (His, Ile, Pro, Thr, Trp, Val) shake for 60 min. To couple an olefinic amino acid, add the cross-linker, HCTU, and DIEA in a molar ratio of 4:3.8:8 to the resin and shake for 60 min.
Wash resin with 1 mL NMP for 1 min, five times draining to waste after each wash.
Go back to step 2, and repeat for each amino acid to add.
Once the synthesis has been completed, the resin is washed twice with DCM (1 mL × 3 min) and shrunk with methanol (1 mL × 5 min).
3.3. Olefin Metathesis
In order to carry out the olefin metathesis reaction, the N-terminus of the peptide must not be exposed as the free amine. The reaction may be carried out with the peptide protected as the Fmoc carbamate.
Swell resin by adding 1 mL dry DCE for 30 min.
Add 1 mL of freshly dissolved 10 mM Grubbs catalyst in DCE for 2 h with constant bubbling under nitrogen. Add additional DCE to avoid loss of volume due to evaporation.
Wash once with DCE (1 mL × 1 min). Repeat step 2.
Wash three times with DCE (1 mL × 1 min).
Shrink resin with methanol (1 mL × 5 min).
3.4. N-Terminus Modifications
The N-terminus of the peptide can be modified using amine-reactive agents. Acetylation of the N-terminus is carried out by treating the pre-swollen and deprotected (i.e., free amine at the N-terminus) resin with 1 mL of a 2:1 (v/v) mixture of acetic anhydride and DIEA.
To attach other modifications such as fluoresceination or biotinylation, the swollen and deprotected resin is exposed to fluorescein isothiocyanate (FITC) or biotinyl-O-succinimide (Biotin-OSu) in the presence of DIEA.
3.5. Cleavage and Deprotection
Freshly make a cleavage cocktail that is 95 % TFA, 2.5 % TIS, and 2.5 % water on ice (CAUTION: mixture is exothermic). If the peptide has sulfur-containing amino acids or modifications (e.g., cysteine, methionine, biotin) then use 94 % TFA, 1 % TIS, 2.5 % water, and 2.5 % EDT.
Add 1 mL of cleavage cocktail per 20 mg peptide-bound resin, and shake at room temperature for 2–3 h (NOTE: the longer reaction times are necessary for peptides containing multiple arginines).
Filter the reaction mixture through a fritted disposable chromatography column, and collect the filtrate in a chilled, 15 mL conical tube containing 5–6 mL of cold diethyl ether. The peptide should precipitate immediately upon contact with the solvent.
Pellet the peptide by centrifugation at 3,000 ×g for 15 min at 4 °C.
Decant the ether supernatant, and air-dry the pelleted stapled peptide.
3.6. Purification
Dissolve the peptide in approximately 1 mL of 50 % acetonitrile in water.
Inject the peptide on a reverse-phase high-performance liquid chromatography with a C18 column and a mobile-phase gradient of water and acetonitrile, each with 0.1 % TFA.
Monitor the HPLC fractions by LC/MS, pooling fractions.
Pooled fractions are lyophilized overnight.
The lyophilized peptide is dissolved in DMSO and quantified by amino acid analysis.
Prepare stock solutions in DMSO at 1–10 mM and store at 4 °C or −20 °C.
4. Notes
Be sure to take proper precautions before using any chemicals. This includes appropriate safety goggles, lab coats, and closed toe shoes. All reactions should be carried out in a chemical fume hood.
Piperidine should be aliquoted using a disposable syringe and needle as the acidic fumes will corrode pipettes.
Make sure that the frit is firmly in place and flush against the bottom before adding any reagents.
Optimization of the peptide through iterative syntheses that explore alternative staple sties and start and stop positions is essential in the process. These factors can be modified in order to avoid helix breakers (e.g., proline and glycine). Each compound should be tested for helicity, mechanism of action, and cell permeability.
In general, sulfUr-containing amino acids should be avoided because they may inactivate the ruthenium-containing Grubbs catalyst, leading to potentially low yields of product. If the use of a cysteine is necessary, then the trityl-protected variant Fmoc-Cys(Tr)-OH should be employed during coupling. Methionines can be substituted with the isosteric amino acid norleucine.
It is important to design a negative control stapled peptide to confirm target-specific effects. Ensure that the negative control mutation does not appreciably affect helicity or cell penetration.
References
- 1.Hopkins AL, Groom CR (2002) The druggable genome. Nat Rev Drug Discov 1(9):727–730, PubMed PMID: 12209152 [DOI] [PubMed] [Google Scholar]
- 2.Buss NA, Henderson SJ, McFarlane M, Shenton JM, de Haan L (2012) Monoclonal antibody therapeutics: history and future. Curr Opin Pharmacol 12(5):615–622, PubMed PMID: 22920732 [DOI] [PubMed] [Google Scholar]
- 3.Guharoy M, Chakrabarti P (2007) Secondary structure based analysis and classification of biological interfaces: identification of binding motifs in protein-protein interactions. Bioinformatics 23(15):1909–1918, PubMed PMID: 17510165 [DOI] [PubMed] [Google Scholar]
- 4.Jochim AL, Arora PS (2009) Assessment of helical interfaces in protein-protein interactions. Mol BioSyst 5(9):924–926, PubMed PMID: 19668855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones S, Thornton JM (1996) Principles of protein-protein interactions. Proc Natl Acad Sci U S A 93(1):13–20, PubMed PMID: 8552589. Pubmed Central PMCID: 40170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pauling L, Corey RB, Branson HR (1951) The structure of proteins; two hydrogen-bonded helical configurations of the polypeptide chain. Proc Natl Acad Sci U S A 37(4):205–211, PubMed PMID: 14816373. Pubmed Central PMCID: 1063337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kéri GR, Tóth IN. (2003) Molecular pathomechanisms and new trends in drug research. London; New York: Taylor & Francis xiv, 635 [Google Scholar]
- 8.Tyndall JD, Nall T, Fairlie DP (2005) Proteases universally recognize beta strands in their active sites. Chem Rev 105(3):973–999, PubMed PMID: 15755082 [DOI] [PubMed] [Google Scholar]
- 9.Madani F, Lindberg S, Langel U, Futaki S, Graslund A (2011) Mechanisms of cellular uptake of cell-penetrating peptides. J Biophys 2011:414729, PubMed PMID: 21687343. Pubmed Central PMCID: 3103903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garner J, Harding MM (2007) Design and synthesis of alpha-helical peptides and mimetics. Organic Biomol Chem 5(22):3577–3585, PubMed PMID: 17971985 [DOI] [PubMed] [Google Scholar]
- 11.Henchey LK, Jochim AL, Arora PS (2008) Contemporary strategies for the stabilization of peptides in the alpha-helical conformation. Curr Opin Chem Biol 12(6):692–697, PubMed PMID: 18793750. Pubmed Central PMCID: 2650020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walensky LD, Pitter K, Morash J, Oh KJ, Barbuto S, Fisher J et al. (2006) A stapled BID BH3 helix directly binds and activates BAX. Mol Cell 24(2): 199–210, PubMed PMID: 17052454 [DOI] [PubMed] [Google Scholar]
- 13.Schafmeister CE, Po J, Verdine GL (2000) An all-hydrocarbon cross-linking system for enhancing the helicity and metabolic stability of peptides. J Am Chem Soc 122(24):5891–5892, PubMed PMID: WOS:000087845700030. English [Google Scholar]
- 14.Brown CJ, Cheok CF, Verma CS, Lane DP (2011) Reactivation of p53: from peptides to small molecules. Trends Pharmacol Sci 32(1): 53–62, PubMed PMID: 21145600 [DOI] [PubMed] [Google Scholar]
- 15.Meyers RA (2004) Encyclopedia of molecular cell biology and molecular medicine, 2nd edn. Weinheim, Wiley-VCH Verlag [Google Scholar]
- 16.Blackwell HE, Grubbs RH (1998) Highly efficient synthesis of covalently cross-linked peptide helices by ring-closing metathesis. Angew Chem Int Edit 37(23):3281–3284, PubMed PMID: WOS:000077806300017. English [DOI] [PubMed] [Google Scholar]
- 17.Blackwell HE, Sadowsky JD, Howard RJ, Sampson JN, Chao JA, Steinmetz WE et al. (2001) Ring-closing metathesis of olefinic peptides: design, synthesis, and structural characterization of macrocyclic helical peptides. J Organic Chem 66(16):5291–5302, PubMed PMID: 11485448 [DOI] [PubMed] [Google Scholar]
- 18.Venkatraman J, Shankaramma SC, Balaram P (2001) Design of folded peptides. Chem Rev 101(10):3131–3152, PubMed PMID: 11710065 [DOI] [PubMed] [Google Scholar]
- 19.Shepherd NE, Hoang HN, Abbenante G, Fairlie DP (2005) Single turn peptide alpha helices with exceptional stability in water. J Am Chem Soc 127(9):2974–2983, PubMed PMID: 15740134 [DOI] [PubMed] [Google Scholar]
- 20.Bernal F, Tyler AF, Korsmeyer SJ, Walensky LD, Verdine GL (2007) Reactivation of the p53 tumor suppressor pathway by a stapled p53 peptide. J Am Chem Soc 129(9):2456–2457, PubMed PMID: 17284038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Braun CR, Mintseris J, Gavathiotis E, Bird GH, Gygi SP, Walensky LD (2010) Photoreactive stapled BH3 peptides to dissect the BCL-2 family interactome. Chem Biol 17(12): 1325–1333, PubMed PMID: 21168768. Pubmed Central PMCID: 3048092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leshchiner ES, Braun CR, Bird GH, Walensky LD (2013) Direct activation of full-length proapoptotic BAK. Proc Natl Acad Sci U S A 110(11):E986–E995, PubMed PMID: 23404709. Pubmed Central PMCID: 3600461 [DOI] [PMC free article] [PubMed] [Google Scholar]