Abstract
The earliest step in DNA replication is origin licensing, which is the DNA loading of minichromosome maintenance (MCM) helicase complexes. The Cdc10-dependent transcript 1 (Cdt1) protein is essential for MCM loading during the G1 phase of the cell cycle, but the mechanism of Cdt1 function is still incompletely understood. We examined a collection of rare Cdt1 variants that cause a form of primordial dwarfism (the Meier–Gorlin syndrome) plus one hypomorphic Drosophila allele to shed light on Cdt1 function. Three hypomorphic variants load MCM less efficiently than wild-type (WT) Cdt1, and their lower activity correlates with impaired MCM binding. A structural homology model of the human Cdt1–MCM complex positions the altered Cdt1 residues at two distinct interfaces rather than the previously described single MCM interaction domain. Surprisingly, one dwarfism allele (Cdt1-A66T) is more active than WT Cdt1. This hypermorphic variant binds both cyclin A and SCFSkp2 poorly relative to WT Cdt1. Detailed quantitative live-cell imaging analysis demonstrated no change in the stability of this variant, however. Instead, we propose that cyclin A/CDK inhibits the Cdt1 licensing function independent of the creation of the SCFSkp2 phosphodegron. Together, these findings identify key Cdt1 interactions required for both efficient origin licensing and tight Cdt1 regulation to ensure normal cell proliferation and genome stability.
INTRODUCTION
DNA replication must be tightly regulated to ensure normal cell proliferation throughout development. DNA damage arising from errors in DNA replication can lead to oncogenic transformation, developmental disorders, and aging (Arentson et al., 2002; Blow and Gillespie, 2008; Yekezare et al., 2013). The first essential DNA replication step is DNA helicase loading, which occurs in the G1 phase of the cell cycle through the nucleation of several protein components at presumptive replication origins. This process is known as “origin licensing.” DNA helicase loading renders origins competent for DNA replication in the subsequent S phase. Unscheduled origin licensing after G1 can lead to DNA rereplication, DNA damage, cell death, and genome instability (Vaziri et al., 2003; Melixetian et al., 2004; Li and Jin, 2010). For this reason, origin licensing is tightly restricted to G1 phase to ensure “once, and only once” genome duplication each cell cycle (Cook, 2009; Truong and Wu, 2011). On the other hand, insufficient licensing increases the probability of incomplete replication, another source of genome instability and proliferation failure (Shreeram et al., 2002; Machida et al., 2005; Nevis et al., 2009). To avoid incomplete replication, the length of the G1 phase is influenced by the status of origin licensing in normal mammalian cells (Shreeram et al., 2002; Vaziri et al., 2003; Liu et al., 2009; Nevis et al., 2009; Matson et al., 2017).
The Cdt1 (Cdc10-dependent transcript 1) protein is essential for origin licensing in eukaryotic cells. In coordination with ORC (origin recognition complex) and Cdc6 (cell division cycle 6), Cdt1 recruits and participates in loading the core of the replicative helicase MCM2–7 (minichromosome maintenance) at presumptive origins. Human Cdt1 licensing activity is restricted to G1 through combinations of transcriptional control, phosphorylation, ubiquitin-mediated degradation, and binding to a specialized inhibitor protein, geminin (Pozo and Cook, 2016). Unlike the ORC, Cdc6, and MCM ATPases, Cdt1 is not an enzyme. Moreover, the Cdt1 primary sequence is not as highly conserved among eukaryotic species as the other licensing factors, and the regulation of human Cdt1 is complex (Fujita, 2006). This complexity presumably arose because loss of proper human Cdt1 control is particularly genotoxic (Arentson et al., 2002; Liontos et al., 2007). Extensive biochemical assays of reconstituted yeast origin licensing reactions have demonstrated that Cdt1 is absolutely required for MCM loading (Evrin et al., 2009; Remus et al., 2009), but the precise role of Cdt1 remains relatively mysterious.
We sought additional insight into Cdt1 function by analyzing the consequences of naturally occurring Cdt1 missense alleles. The first partial-loss-of-function variant in a metazoan Cdt1 was described in Drosophila melanogaster (Whittaker et al., 2000). The orthologous vertebrate variant had low activity in vitro (De Marco et al., 2009; You et al., 2016), but the specific reason for weak origin-licensing activity was not determined. Importantly, several studies have found that inherited mutations in human origin-licensing factors, including Cdt1, can result in developmental disorders (Bicknell et al., 2011a,b; Burrage et al., 2015). Cdt1 mutations are one cause of a form of primordial dwarfism called Meier–Gorlin syndrome (MG). Patients are extraordinarily short with microcephaly, focal hypoplasias, and some characteristic facial features and tissue-specific phenotypes (Bicknell et al., 2011b; de Munnik et al., 2012); these phenotypes are consistent with cell proliferation defects. Indeed, primary fibroblasts from MG patients proliferate slowly in culture (Bicknell et al., 2011a). We hypothesized that each of these mutations perturbs at least one aspect of Cdt1 regulation or function. Our analyses of these alleles identifies a previously unappreciated MCM binding site and separately uncovers new features of cyclin A-dependent Cdt1 control to prevent genotoxic rereplication.
RESULTS
Comparative functional analysis of Cdt1 variants by rereplication induction
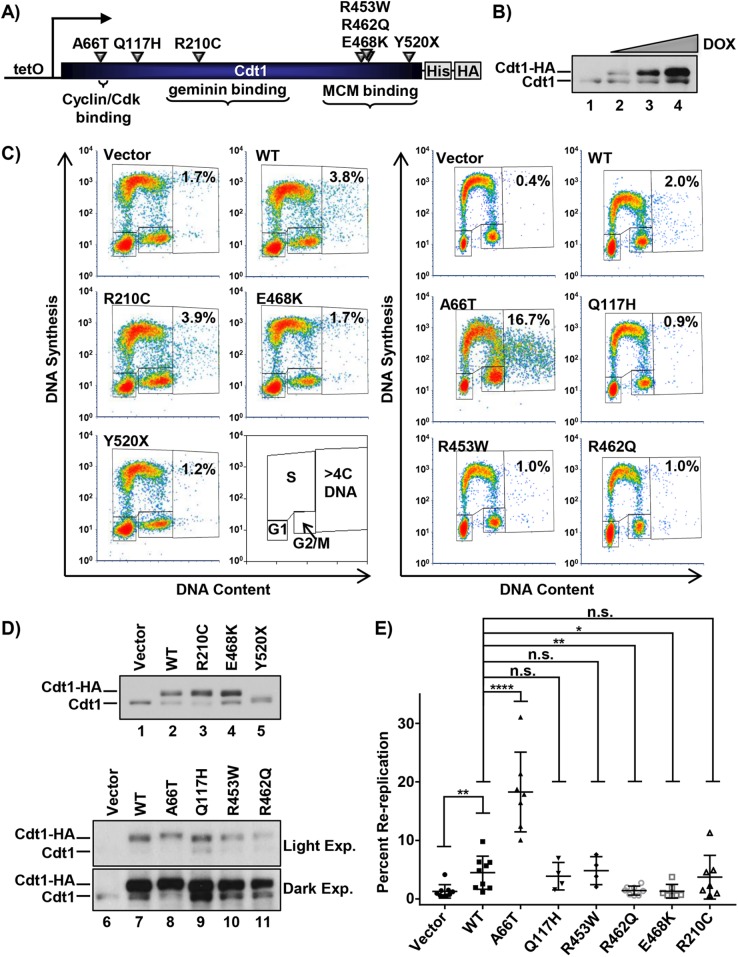
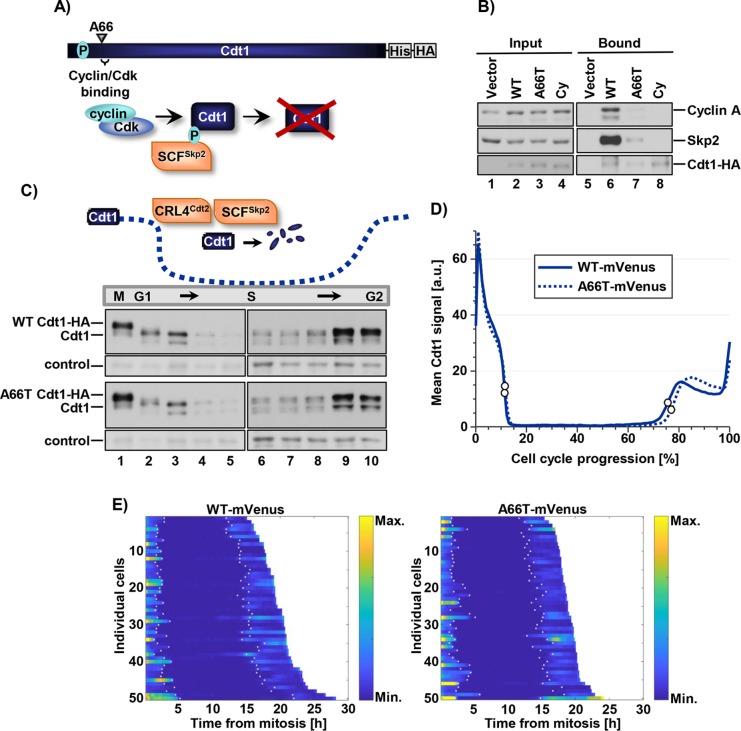
Bicknell et al. (2011b) reported eight Cdt1 alleles in MG patients; we marked the positions of the amino acids affected by all missense alleles and one of the three nonsense alleles in Figure 1A. All of the dwarfism patient genotypes were compound heterozygotes, and the most common combinations were a missense allele plus a nonsense allele predicted to encode a truncated Cdt1 protein. We included all missense mutations in our study. In addition, we included Cdt1-Y520X, because it encodes the longest of the predicted truncations, and we reasoned that if Cdt1-520X is null for function, then the lesser truncations are also null. We added Cdt1-R210C, a variant first discovered as a D. melanogaster partial–loss of function mutant (Whittaker et al., 2000). This analogous vertebrate variant has reduced origin licensing activity in vitro (De Marco et al., 2009; You et al., 2016), but the molecular mechanism of reduced activity is not known. We introduced each of these mutations into a vector encoding full-length Cdt1 under the control of a doxycycline-inducible promoter with a C-terminal extension that includes polyhistidine and HA epitope tags. We then generated derivatives of the U2OS osteosarcoma cell line by recombination of each expression construct into a chromosomal FRT site using Flp recombinase; the parent cell line constitutively expresses the Tet repressor (Malecki et al., 2006). Using this experimental approach, we achieved dose-responsive inducible ectopic Cdt1 expression (Figure 1B).
FIGURE 1:
Functional analysis of Cdt1 variants by rereplication induction. (A) Illustration of the relative location and amino acid substitution of the alleles chosen in this study; polyhistidine and HA epitope tags and relevant binding domains are also marked. (B) Immunoblot of inducible expression of stably integrated HA-tagged WT Cdt1 in U2OS cells. Cells were grown in 0, 0.05, 0.1, and 1 µg/ml doxycycline (dox), respectively. (C) Analytical flow cytometry profiles of U2OS cells expressing vector, ectopic HA-tagged WT Cdt1, or the indicated HA-tagged Cdt1 variants. Cells were treated with 1 µg/ml dox for 72 h and pulse-labeled with EdU for 30 min before being harvested. An illustration of the gating scheme is also shown; “>4C DNA” are cells that have undergone DNA rereplication. (D) Immunoblots of Cdt1 expression from C. Light Exp., light exposure; Dark Exp., dark exposure. (E) The percentage of cells with >4C DNA content in at least four biological replicates. Bars represent mean and SD. ****p value < 0.0001; **p value < 0.005; *p value < 0.05; n.s. = not significantly different.
We first examined the effects of overexpressing each Cdt1 variant by high-dose doxycycline (dox) treatment. Cdt1 overexpression can induce DNA rereplication detectable as a population of cells with more than the normal G2 phase DNA content (i.e., >4C; Vaziri et al., 2003). We measured rereplication by analytical flow cytometric analysis after overproducing Cdt1-WT (wild-type) or Cdt1 variants in asynchronously proliferating cultures for 72 h (Figure 1, C and D). We scored the percentage of cells with >4C DNA content in multiple independent experiments (Figure 1E). Of note, Cdt1 overexpression to this degree had modest effects on the cell-cycle distributions among G1, S, and G2/M phases (Supplemental Figure S1). The more extensive the rereplication, the greater the down-regulation of endogenous Cdt1, which we had previously linked to Cul4-dependent Cdt1 degradation (for example, Figure 1D, endogenous Cdt1 in lanes 7 and 8; Hall et al., 2008).
After multiple independent tests, we noted that Q117H, R210C, and R453W had rereplication-inducing activity similar to WT, whereas R462Q and E468K were less active by this metric (Figure 1E). Y520X failed to accumulate to high levels at any doxycycline concentration (Figure 1D, lane 5, and unpublished data), which may indicate impaired protein folding and by extension, that all truncation alleles are likely null for Cdt1 biological activity. Given the proliferation defect associated with MG, we anticipated that most alleles encode partial–loss of function variants such as R462Q and E468K. Surprisingly, however, the A66T dwarfism variant consistently induced nearly four times more rereplication than Cdt1-WT did (Figure 1, C and E), even when produced at similar levels (Figure 1D), indicating that it is a gain-of-function allele. We focused our subsequent analyses on the subset of mutations with detectable effects on Cdt1 activity in vivo, that is, A66T, R462Q, E468K (Figure 1, C and E), and R210C (Whittaker et al., 2000; De Marco et al., 2009; You et al., 2016).
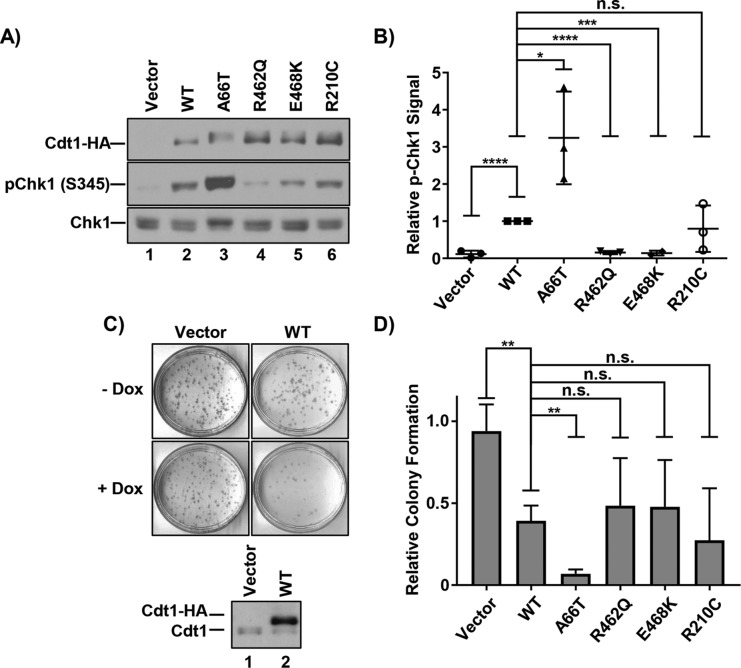
Given that DNA rereplication is associated with DNA damage and genomic stress, we assessed Cdt1-overproducing cells for activation of the DNA-damage response. We analyzed the activating Chk1 phosphorylation at S345 as a marker of replication stress and DNA damage 48 h after initiating Cdt1 overproduction (Figure 2, A and B). As expected (Vaziri et al., 2003; Davidson et al., 2006; Hall et al., 2008), Cdt1-WT overexpression induced Chk1 phosphorylation that correlated with the degree of rereplication induced by the different Cdt1 variants (Figure 2B). Of particular note, Cdt1-A66T induced significantly more rereplication and Chk1 activation than Cdt1-WT or Cdt1-R210C did, whereas Cdt1-R462Q and Cdt1-E468K induced significantly less rereplication and Chk1 activation than Cdt1-WT did (Figure 2, A and B).
FIGURE 2:
DNA damage and cell proliferation defects from Cdt1 variant overproduction. (A) Immunoblots of HA-tagged Cdt1 (anti-HA antibody), pChk1 (S345), and total Chk1 in U2OS cells grown in 1 µg/ml dox for 48 h. (B) Graph of pChk1 (S345) induction normalized to WT Cdt1. Bars represent mean and SD of three biological replicates. ****p value < 0.0001; ***p value = 0.0001; *p value < 0.05; n.s. = not significantly different. (C) Top, Representative vector and WT Cdt1 control colony-forming assays. Cells were plated at low density in the presence or absence of 1 µg/ml doxycycline (dox) and grown for ∼10 d. Bottom, A technical replicate plate was harvested after 72 h to assay for ectopic Cdt1 expression by immunoblotting with anti-Cdt1 antibody. (D) Relative colony formation normalized within each experiment to the vector control; values represent at least three biological replicates. Bars represent mean and SD. **p value < 0.005; n.s. = not significantly different.
Extensive rereplication, replication stress, and DNA damage can impair cell proliferation (Li and Jin, 2010; Truong and Wu, 2011). As a measure of the ability of each of the Cdt1 variants to impact proliferation, we plated each cell line in either high doxycycline or no doxycycline as a control and assessed colony formation over 10 d. Cdt1-WT overexpression strongly blocked colony formation (Figure 2, C and D). There was general correlation of the degree of rereplication and DNA damage response with the degree of toxicity induced by Cdt1 overproduction in the colony-forming assay (Figure 2D). In particular, A66T, which was hyperactive for rereplication, was even more toxic than WT Cdt1 in this assay (Figure 2D).
Comparative functional analysis of MCM loading
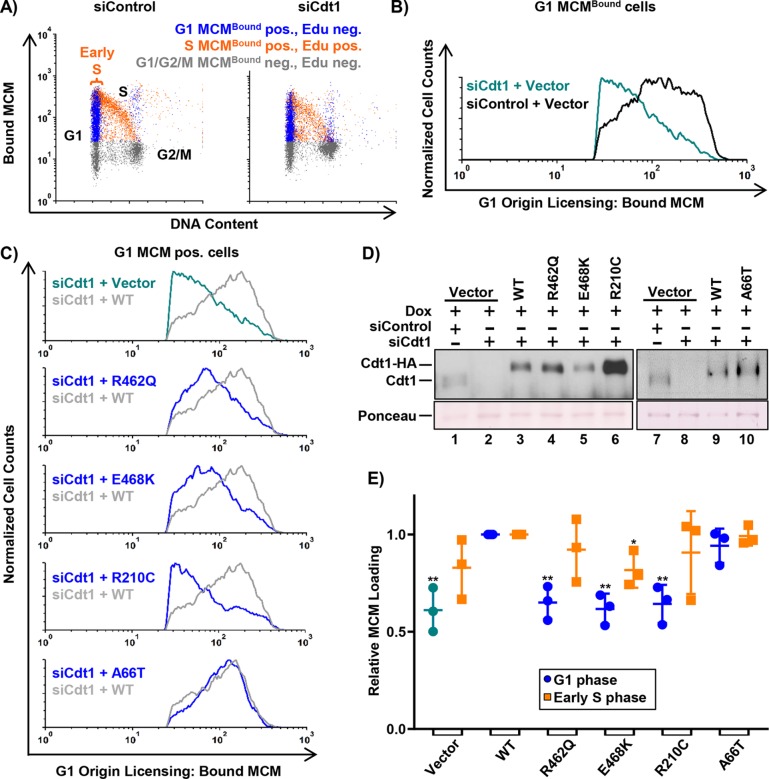
Given that most MG mutations affect genes encoding essential origin-licensing proteins (Cdt1, Cdc6, ORC, etc.), we hypothesized that the defects associated with Cdt1 hypomorphic variants are primarily related to MCM loading. To test this idea directly, we induced expression of the Cdt1 variants in asynchronously growing cells with low doxycycline to approximately match endogenous Cdt1 levels. We simultaneously depleted endogenous Cdt1 using a small interfering RNA (siRNA); the ectopic Cdt1 expression constructs bear synonymous mutations at the siRNA binding site and are thus resistant to depletion (Figure 3D). We then pulse-labeled the cells with EdU for 30 min before harvesting and extracted cells to release soluble MCM complexes, followed by fixation to retain loaded MCM complexes. We probed the extracted cells for Mcm2 as a marker of the MCM2–7 complex, stained for total DNA content, detected EdU incorporation, and analyzed the samples by flow cytometry (see Materials and Methods). We previously validated this assay for quantifying MCM loading rates in asynchronously proliferating individual cells (Matson et al., 2017). Figure 3A shows flow cytometry profiles of extracted cells with chromatin-bound MCM on the y-axis and DNA content on the x-axis. MCMBound-positive/EdU-negative G1 cells are shown in blue, MCMBound-positive/EdU-positive cells are shown in orange, and MCMBound-negative/EdU-negative cells are shown in gray. Using these analytical flow cytometry profiles, we isolated the G1 phase MCM positive cells in silico and plotted these data in histogram form as a measure of licensing activity (Figure 3B). In previous work, we demonstrated that these histograms reveal both the total amount of MCM loaded per cell and the rate of MCM loading within the G1 phase (Matson et al., 2017).
FIGURE 3:
Functional analysis of MCM loading. (A) Analytical flow cytometry profiles of chromatin-bound MCM in U2OS cells treated with 100 nM control siRNA (left) or Cdt1 siRNA (right). Cells were pulse-labeled with 10 µM EdU for 30 min before harvesting and extraction of soluble MCM. Bound (unextracted) MCM was detected with anti-MCM2 antibody, and cells were stained with DAPI for total DNA content. Blue: MCMBound positive, EdU negative, G1 DNA content; orange: EdU positive, MCMBound positive; gray: EdU negative, MCMBound negative. (B) Histograms of the G1 MCMBound positive, EdU negative (i.e., blue in A) cells from both samples in A. Bound MCM on the x-axis and normalized cell counts on the y-axis (counts normalized to siControl). (C) Histograms of G1 MCMBound positive cells depleted of endogenous Cdt1 and expressing each Cdt1 variant compared with WT Cdt1 as in B. siRNA transfected cells were cultured in 0.002–0.006 µg/ml doxycycline for 72 h before EdU labeling and processing as in A. (D) Immunoblot of endogenous and ectopic Cdt1 from C detected with anti-Cdt1 antibody. (E) Complementation of G1(green/blue) and early S (orange) MCM loading normalized to WT Cdt1. Mean MCMBound loading intensity of each variant was divided by the mean MCM loading intensity of WT Cdt1 within each experiment. Early S phase is defined as G1 DNA content and EdU-positive indicated by the bracket in A; see also Supplemental Figure S3B. Bars represent mean and SD of three biological replicates. *p value < 0.05; **p value < 0.005 where indicated; otherwise the difference between WT Cdt1 and variant was not significant.
As expected, Cdt1 depletion without ectopic Cdt1 expression resulted in defective MCM chromatin loading in G1 (Figure 3B, green trace), but expression of the epitope-tagged Cdt1-WT complemented this MCM loading defect (Figure 3C; compare gray and green traces). We quantified the G1 origin-licensing dynamics for multiple replicates and plotted the average amount of MCM loaded in G1 relative to Cdt1-WT controls in Figure 3E (green dots for vectors and blue dots for cells expressing ectopic Cdt1). By this measure, the R462Q and E468K variants were significantly impaired for MCM loading in G1 (Figure 3, C and E, blue traces and blue dots). Strikingly, the R210C variant was also significantly impaired for MCM loading even when it accumulated to higher levels than in Cdt1-WT (Figure 3, C; D, lane 6; and E). On the basis of these complementation assays, we interpret the relative activity of the hypomorphs as R462Q=E468K>R210C (i.e., Cdt1-R210C is the weakest for G1 MCM loading when expressed at normal levels), whereas R210C is more active for inducing rereplication than R462Q and E468K when overproduced (Figure 1).
Unlike the hypomorphic alleles, Cdt1-A66T showed no MCM loading defect in G1, which is consistent with the idea that this variant is not a loss-of-function allele (Figure 3, C and E). Interestingly, cells expressing Cdt1-A66T rereplicated more than Cdt1 WT–expressing cells, even at expression levels that were as low as endogenous Cdt1 (Figure 3D; Supplemental Figure S2B). These results suggest that Cdt1-A66T hyperactivity does not require artificial overproduction.
The defects in origin licensing exhibited by the hypomorphic variants prompted us to ask whether these cells enter S phase with underlicensed chromosomes. To directly determine whether these cells routinely enter the S phase with lower levels of MCM loading, we isolated the early S-phase cells (Figure 3A) in silico from the analytical flow cytometry profiles (Supplemental Figure S3) and quantified the amount of MCM loaded in early S for each variant (Figure 3E, orange squares). Interestingly, cells expressing the hypomorphic variants entered the S phase with amounts of loaded MCM similar to those in cells expressing WT Cdt1 (Figure 3E, orange squares). Moreover, a consequence of underlicensed S phase is hypersensitivity to replication stress (Woodward et al., 2006; Blow et al., 2011; McIntosh and Blow, 2012), but cells depleted of endogenous Cdt1 and expressing the hypomorphic variants showed no hypersensitivity to low doses of hydroxyurea (a source of exogenous replication stress) compared with controls (unpublished data). We noted, however, that these cells did proliferate a little more slowly over a 3-d time course (Supplemental Figure S2A). Instead of entering the S phase with too little MCM loaded, cells expressing hypomorphic Cdt1 variants spent slightly more time in the G1 phase compared with cells expressing WT Cdt1 (Supplemental Figure S2). Together, these results suggest that the slower MCM loading in G1 can delay S-phase entry due to the activity of a previously documented origin-licensing checkpoint (Shreeram et al., 2002; Liu et al., 2009; Nevis et al., 2009). The outcome is that most cells enter S with sufficient licensed chromosomes to complete a normal S phase, but the cell population proliferates more slowly.
Comparative analysis of MCM binding
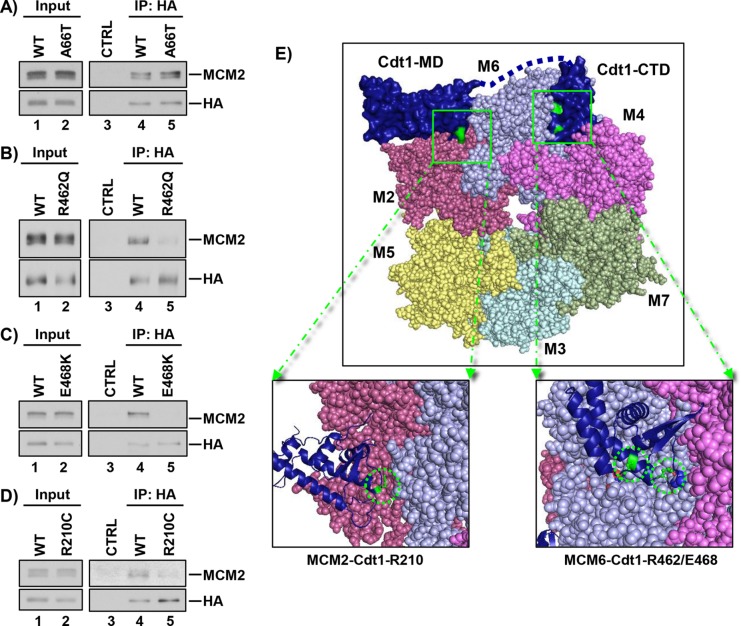
We considered that the functional effects of Cdt1 mutations may be linked to key protein–protein interactions, specifically Cdt1-MCM binding. To test this notion, we immunoprecipitated WT or variant Cdt1 using the HA epitope tag and probed for MCM2–7 interaction using Mcm2 as a marker of the complex. Previous studies have mapped the Cdt1-MCM binding domain to a C-terminal region, and some mutations in this domain of metazoan Cdt1 impair binding to a partial MCM complex (Ferenbach et al., 2005; Jee et al., 2010). It was not surprising, then, that Cdt1-A66T, which is located near the N-terminus of Cdt1 and not the C-terminal domain, bound MCM as well as WT Cdt1 did (Figure 4A). Also consistent with prior studies of the Cdt1-MCM interaction, the two hypomorphic variants located in the C-terminal domain of Cdt1, Cdt1-R462Q and Cdt1-E468K, were consistently impaired for MCM binding (Figure 4, B and C). On the other hand, Cdt1-R210 is located in the middle domain of Cdt1 and not in the previously described Cdt1-MCM–binding domain. It was thus unexpected that, like Cdt1-R462Q and Cdt1-E468K, Cdt1-R210C was also impaired for MCM interaction (Figure 4D). This result suggests the existence of an MCM–binding domain in Cdt1 distinct from the C-terminal domain.
FIGURE 4:
Relative MCM binding. (A–D) WT and the indicated Cdt1 variants were transiently expressed in HEK 293T cells and immunoprecipitated using the HA epitope tag. Portions (2%) of whole-cell lysates and bound proteins were probed for HA-Cdt1 (anti-HA antibody) and for MCM2 as a marker of the MCM complex; immunoglobulin G or beads were used as controls (CTRL). (E) Top, Homology model of the human MCM2-7-Cdt1 complex. The yeast OCCM structure (PDB ID: 5UDB) was used as a template to model the human MCM2-7 complex; numbers indicate individual MCM subunits, and colors are similar to those in Yuan et al. (2017). The structures of the human C-terminal Cdt1 winged helix, “Cdt1-CTD” (PDB ID: 2WVR), and the mouse Cdt1 central/middle domain, “Cdt1-MD” (PDB ID: 3A4C), were used to model hCdt1–hMCM interactions. Bottom, left, Magnified view of the proposed interacting surfaces with R210 highlighted in green. Bottom, right, Magnified view of the proposed interacting surfaces with R462 and E468 highlighted in green.
Structures of budding yeast Cdt1 in complex with other licensing proteins have recently been reported, and although two domains of mammalian Cdt1 orthologs have been structurally characterized (Frigola et al., 2017; Yuan et al., 2017; Zhai et al., 2017), there is no complete structure of metazoan Cdt1 available. Therefore, to visualize the locations of these mutations relative to the human MCM2–7 complex, we generated a homology model using publicly available crystal, cryo-EM, and NMR structures of Cdt1-MCM2–7 complexes. We derived our model using the recent cryo-EM yeast ORC1–6-Cdc6-Cdt1-MCM2–7 (OCCM) complex (Yuan et al., 2017) as a template for modeling the human MCM complex (see Materials and Methods). We superimposed structures for the human Cdt1 C-terminal domain (De Marco et al., 2009) and mouse C-terminal (Khayrutdinov et al., 2009) on the OCCM structure to model the mammalian Cdt1-MCM2–7 interaction (Figure 4E, top). The Cdt1 C-terminal domain adopts a winged helix fold of the type predicted to mediated protein–protein rather than protein–DNA interactions (Khayrutdinov et al., 2009). In this model, residues R462 and E468 are at the interface between the Cdt1 C-terminal domain and the Mcm6 subunit of the MCM2–7 heterohexamer (Figure 4E, bottom right). Mutating these residues likely disrupts the binding surface between Cdt1 and Mcm6, resulting in defective Cdt1–MCM interactions.
We also consistently observed weak binding between Cdt1-R210C and MCM (Figure 4D), but Cdt1 R210 is not in the C-terminal MCM-binding domain. Our homology model positions this residue near the Mcm2 subunit of the MCM2–7 heterohexamer (Figure 4E, bottom left). This proposed interface is distinct from the suggested interface between the Cdt1 C-terminal domain and Mcm6. In addition, cryo-EM structures of the yeast Cdt1–MCM2–7 complex predict multiple contact points between Cdt1 and the MCM2–7 complex (Sun et al., 2013; Yuan et al., 2017). The functional defects of Cdt1-R210C, Cdt1-R462Q, and Cdt1-E468K in cells support the notion that Cdt1 requires multiple binding interfaces with the MCM2–7 complex. Our functional and binding analysis suggests that disrupting either of these interfaces is sufficient to impair both MCM2–7 binding and MCM2–7 loading. We note that this domain, rather than the C-terminal domain, is the primary site of Cdt1 interaction with its inhibitor, geminin (Lee et al., 2004; Ferenbach et al., 2005).
Cdt1-A66T impairs cyclin A and Skp2 binding but does not stabilize Cdt1 in the S phase
The unexpected gain-of-function phenotype of the A66T dwarfism-associated variant prompted us to explore this variant in more detail. Cdt1 A66 is just N-terminal to a previously identified negative regulatory domain in Cdt1 (Coulombe et al., 2013) and very close to the well-defined cyclin/CDK binding motif (“Cy motif”) at positions 68–70. On the basis of the close proximity, we hypothesized that A66T perturbs the Cdt1–cyclin/CDK interaction (Figure 5A). To test this idea, we isolated Cdt1-A66T, Cdt1-WT, and Cdt1-Cy, a bona fide mutational disruption in the Cy motif (alanines at positions 68, 69, and 70), from cell lysates using the C-terminal tag and probed the complexes for cyclin A; previous studies identified cyclin A as the primary cyclin that interacts with the Cdt1 Cy motif (Sugimoto et al., 2004). Cdt1-WT bound cyclin A by this assay, but the Cy motif mutant did not (Figure 5B). Interestingly, Cdt1-A66T bound cyclin A very poorly in comparison with Cdt1-WT and only slightly better than the Cy motif mutant (Figure 5B; compare lane 7 with lanes 6 and 8).
FIGURE 5:
Cdt1-A66T impairs cyclin A and Skp2 binding but does not stabilize Cdt1 in S phase. (A) Illustration of SCFSkp2-dependent degradation of WT Cdt1 via CDK-mediated phosphorylation at threonine 29. (B) Cells were cultured in 1 µg/ml dox (high) for 18 h, lysed, and incubated with nickel–agarose to retrieve His-tagged Cdt1. Portions of whole cell lysates (2%) and bound complexes were probed for the indicated proteins (bottom panel anti-HA antibody). (C) Top, Illustration of Cdt1 degradation and accumulation during the cell cycle. Bottom, U2OS cells expressing Cdt1-WT and Cdt1-A66T were synchronized by double-thymidine/nocodazole (lanes 1–5) or double-thymidine (lanes 6–10) block and released into fresh medium. Time points were taken after release and analyzed by immunoblotting with anti-Cdt1 antibody. A nonspecific band serves as a loading control. (D) The intensity of WT or A66T Cdt1-Venus expressed in U2OS cells imaged during asynchronous proliferation every 10 min. Traces are the average Venus intensity in arbitrary units from mitosis (0%) to mitosis (100% cell-cycle progression); n = 50 cells. White circles denote the beginning and end of the S phase as determined by the localization of stably coexpressed fluorescently tagged PCNA. (E) Heat map of fluorescence intensity of Cdt1 WT-Venus (left) and Cdt1-A66T- Venus (right) in 50 randomly selected U2OS cells. Maps from individual cells are arranged according to the duration of the cell cycles; colors indicate differences in fluorescence levels. White dots in each track denote the beginning and end of the S phase as determined by the localization of stably coexpressed fluorescently tagged PCNA.
The consequences of cyclin A binding to Cdt1 have been linked to CDK-mediated Cdt1 phosphorylation at T29. Cdt1 phosphorylation at T29 creates a binding site for the Skp2 substrate adapter of the SCF E3 ubiquitin ligase, SCFSkp2 (Figure 5A; Takeda et al., 2005). We therefore tested whether the weak cyclin A binding by Cdt1-A66T also resulted in weak Skp2 binding; indeed, Cdt1-A66T bound very poorly to Skp2 in comparison with Cdt1-WT and slightly better than the Cdt1-Cy variant (Figure 5B, lanes 6–8). For this reason, we tested the stability of Cdt1-A66T relative to Cdt1-WT during S phase. We synchronized cells in mitosis and released them to progress from G1 into S phase, taking time points until mid-S. We also blocked cells in the early S phase and released them to progress into G2, and then monitored endogenous and ectopic Cdt1 by immunoblotting. We found no detectable differences between Cdt1-A66T and Cdt1-WT or endogenous Cdt1 in degradation in early S or in Cdt1 reaccumulation as the S phase ends (Figure 5C). SCFSkp2-mediated Cdt1 ubiquitylation cooperates with a second E3 ubiquitin ligase, CRL4Cdt2, to destroy Cdt1 during the S phase (Abbas and Dutta, 2011; Havens and Walter, 2011; Figure 5C illustration), and this targeting does not require CDK-mediated Cdt1 phosphorylation (Arias and Walter, 2005). CRL4Cdt2 ubiquitylates Cdt1 both in the S phase and after DNA damage (Arias and Walter, 2006; Jin et al., 2006; Havens and Walter, 2009). We observed no effect of the A66T mutation on Cdt1 degradation after UV irradiation (Supplemental Figure S4); thus the change has no effect on CRL4Cdt2 targeting.
Nonetheless, we assumed that Cdt1-A66T could be slightly more stable at specific cell-cycle times or in other settings in a manner that increases the likelihood of origin relicensing and subsequent rereplication. We therefore added a C-terminal fluorescent tag to both Cdt1-WT and Cdt1-A66T (Supplemental Figure S5) and carried out live cell imaging of asynchronously proliferating U2OS cells after doxycycline-induced expression. We tracked individual cells with similar maximum fluorescence intensities for both Cdt1-WT and Cdt1-A66T and plotted both the mean (Figure 5D) and the intensity values of 50 individual proliferating cells (Figure 5E). Importantly, we observed no statistically significant differences in the dynamics of Cdt1 degradation and reaccumulation during the cell cycle for those cells that successfully divided. Moreover, those A66T-expressing cells that arrested with large nuclei (presumably from rereplication; Melixetian et al., 2004) had normal degradation and accumulation in the S phase before the arrest (unpublished data).
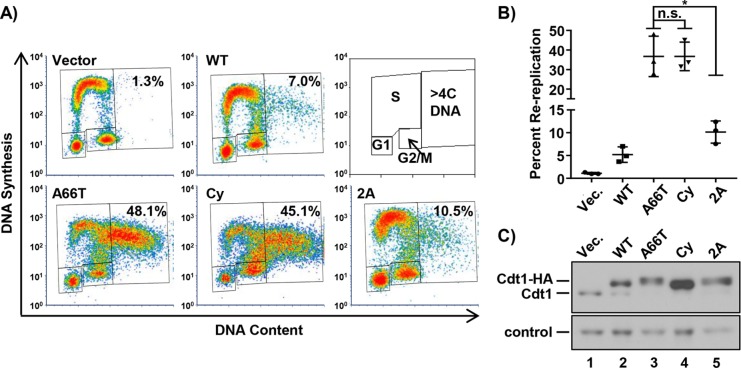
Cdt1-A66T is largely impaired for SCFSkp2 binding, but there were no detectable consequences for Cdt1 stability, since Cdt1-A66T levels are still subject to CRL4Cdt2 control. Nonetheless, Cdt1-A66T is a potent rereplication inducer. We thus assumed that the mutation has consequences for Cdt1 activity beyond phosphorylation at T29. To test that idea directly, we expressed a Cdt1 phosphorylation–site mutant in which T29 is converted to unphosphorylatable alanine. Cdt1 is also phosphorylated at S31 (Hornbeck et al., 2015), and although this phosphorylation has minimal impact on Skp2 binding compared with T29 phosphorylation (Takeda et al., 2005), we also converted S31 to alanine to avoid possible compensatory effects at this position; this double alanine mutant is “Cdt1-2A.” If the primary effect of Cdt1-A66T is to prevent phosphorylation at T29 (and S31), then we predicted that the phenotypes of cells overexpressing Cdt1-A66T, the Cdt1-Cy motif mutant, and Cdt1-2A should be similar, since each alteration blocks CDK-mediated T29 phosphorylation. We compared the DNA rereplication activity induced by overproducing each of these Cdt1 variants. Strikingly, both Cdt1-A66T and Cdt1-Cy induced significantly more rereplication than Cdt1-2A did (Figure 6, A and B). In these longer-expression experiments, Cdt1-Cy accumulates to higher levels than WT (Figure 6C), though we note that in shorter experiments such as that in Figure 5B, Cdt1-Cy levels are similar to those in Cdt1-WT and Cdt1-A66T. This hyperaccumulation may be a consequence of cell-cycle phase distribution from long-term expression (Supplemental Figure S6) and/or of the apparent complete defect in SCFSkp2 binding. Nonetheless, Cdt1-A66T and Cdt1-2A routinely accumulate to similar levels (Figure 6C; compare lanes 3 and 5), yet Cdt1-A66T induces significantly more rereplication than Cdt1-2A does (Figure 6, A and B). We thus conclude that Cdt1-A66T disrupts cyclin A binding as a near-mimic of the engineered Cdt1-Cy motif mutant, and that cyclin A binding to Cdt1 negatively regulates Cdt1 function by at least one mechanism that is independent of simply creating a phosphodegron for the SCFSkp2 E3 ubiquitin ligase.
FIGURE 6:
CDK-Cdt1 binding suppresses rereplication independent of the Cdt1 phosphodegron. (A) Analytical flow cytometry profiles of cells treated with 1 µg/ml doxycycline for 48 h and analyzed as in Figure 1. Cy: cyclin/CDK binding motif mutant; 2A: Cdt1 T29A, S31A. (B) The percentage of cells with >4C DNA content in at least three biological replicates. Bars represent mean and SD. *p value < 0.05; n.s. = not significantly different. (C) HA-tagged Cdt1 was detected by immunoblotting whole-cell lysates from A with anti-Cdt1 antibody; a nonspecific band serves as a loading control; Vec. = Vector.
DISCUSSION
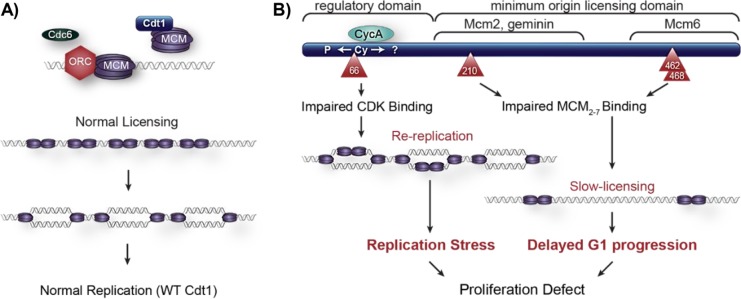
In this study, we analyzed naturally arising mutations in Cdt1 and demonstrated that hypo- and hypermorphic variants cause defects in cell proliferation through distinct molecular mechanisms. Specifically, Cdt1 mutations found in humans afflicted with MG result in either Cdt1/MCM binding or cyclin/CDK binding defects. Both scenarios can lead to proliferation defects from either changes in cell-cycle length or problems in DNA replication control attributed to perturbed Cdt1 activity (Figure 7).
FIGURE 7:
Model of cell proliferation defects from both hypomorphic or hypermporphic Cdt1 variants. (A) WT Cdt1 supports normal MCM loading/origin licensing and normal DNA replication in the S phase. (B) The A66T variant is impaired for CDK-mediated repression, resulting in relicensing and rereplication. The R210C, R462Q, and E468K variants are impaired for MCM2-7 binding in the G1 phase, leading to slow origin licensing and thus to delayed G1 progression. Both scenarios ultimately lead to proliferation defects.
Hypomorphic alleles
MG syndrome is a form of primordial dwarfism characterized by growth retardation beginning in utero and continuing through adolescence. On the basis of the patient phenotypes, we hypothesized that all MG Cdt1 alleles are hypomorphic. Indeed, two of the alleles analyzed here, including Cdt1-R462Q, which was present in most of the MG patients with Cdt1 mutations reported thus far, are hypomorphic for Cdt1 function (Bicknell et al., 2011a,b; Guernsey et al., 2011; de Munnik et al., 2012). Q117 is poorly conserved among Cdt1 sequences, suggesting that it is not critical for Cdt1 function. On the basis of its apparently normal ability to induce rereplication and the poor conservation of Q117, we infer that Cdt1-Q117H is hypomorphic for Cdt1 expression in the MG patient rather than for function—possibly from inefficient mRNA splicing (Bicknell et al., 2011b). The mutation may reduce overall Cdt1 expression in vivo rather than impacting Cdt1 activity per se. R453 is buried in the winged-helix domain core of the human Cdt1 C-terminal domain (Khayrutdinov et al., 2009; Jee et al., 2010). Introducing a bulky aromatic tryptophan may globally disrupt folding rather than altering Cdt1 interactions or function.
The two functionally hypomorphic MG alleles in this study, Cdt1-R462Q and Cdt1-E468K, encode substitutions of conserved solvent-exposed amino acids in the C-terminal Cdt1 winged helix domain (Khayrutdinov et al., 2009; Jee et al., 2010). By analytical flow cytometry, we found that these variants support slower MCM loading than WT Cdt1. The hypomorphic nature of these alleles induces slow proliferation through a modest increase in cell-cycle length (Supplemental Figure S2A). Furthermore, our homology model places R462 and E468 at the interface between the Cdt1 C-terminal domain and the Mcm6 subunit of the MCM2–7 heterohexamer, close to the Mcm6–Mcm4 interface. Thus, mutations in this region understandably impair MCM binding.
Cdt1-R210C is orthologous to a mutation in the Drosophila melanogaster Cdt1 gene, Double-Parked (Dup). Whittaker et al. (2000) characterized this variant as hypomorphic, resulting in DNA replication defects and female sterility. Previous studies reported that this Cdt1 variant supported less DNA synthesis in vitro (De Marco et al., 2009), and had a modest effect on migration of a Cdt1–MCM2–7 complex by native gel electrophoresis (You et al., 2016). We found that this variant has impaired Cdt1–MCM2–7 binding by coimmunoprecipitation from human cell lysates. It supports slow origin licensing that is nearly as slow as with the two dwarfism hypomorphic alleles in the C-terminal domain. The similarity in both cellular and molecular phenotypes of Cdt1-R462Q, Cdt1-E468K, and Cdt1-R210C suggests that both the central domain and C-terminal domain are equally important for Cdt1–-MCM binding.
Jee et al. (2010) suggested the existence of cooperation between the central domain of Cdt1 and the C-terminal domain in origin licensing. Yanagi et al. (2002) found that a fragment of murine Cdt1 including the central domain but lacking the Cdt1 C-terminal domain can associate with a subcomplex of three subunits, MCM4/6/7. The notion of multiple contacts between Cdt1 and MCM2–7 is consistent with recent structural and functional analysis of yeast Cdt1–MCM2–7 (Frigola et al., 2017). In this model, Cdt1 must engage the MCM2–7 complex at two distinct points to serve as a brace to keep the Mcm2/Mcm5 “gate” open for DNA entry during MCM loading.
The existence of a second MCM2–7 binding site in the central region of Cdt1 sheds light on the mechanism of Cdt1 inhibition by the origin-licensing inhibitor protein, Geminin. Previous studies have shown that Geminin inhibits Cdt1 by blocking its interaction with the MCM complex (Yanagi et al., 2002; Cook et al., 2004). The molecular mechanism of that interference cannot be easily explained if the only place MCM binds Cdt1 is the C-terminal domain. The cocrystal structure of Cdt1 in complex with geminin includes only the central region of Cdt1 (including R210) and not the C-terminal domain (Lee et al., 2004). If the central domain is also essential for MCM2–7 binding, then we postulate that it is only this interaction that geminin targets. Moreover, poor binding at either interface is sufficient to impair overall MCM2–7 binding and therefore MCM2–7 loading.
Dwarfism hypermorphic allele
Cdt1 is tightly regulated throughout the cell cycle to ensure once, and only once DNA replication. One of the mechanisms for restricting Cdt1 activity outside of G1 phase and avoiding rereplication is ubiquitin-mediated proteolysis. This process is carried out by two E3 ubiquitin ligases, CRL4Cdt2 and SCFSkp2 (Nishitani et al., 2006; Figure 5C, top). CRL4Cdt2 relies on chromatin-bound proliferating cell nuclear antigen (PCNA) to ubiquitylate its PIP-degron-containing substrates (Arias and Walter, 2006; Jin et al., 2006; Havens and Walter, 2011). On the other hand, ubiquitylation of Cdt1 by SCFSkp2 is dependent on cyclin/CDK phosphorylation generating a phosphodegron that is recognized by the Skp2 adapter subunit (Li et al., 2003; Liu et al., 2004). Given the proximity of the A66T mutation to the cyclin/CDK binding motif, coupled with the defect in cyclin A and Skp2 binding, we first reasoned the hyperactivity of this variant was due to increased protein stability. Cdt1-A66T is not more stable than WT Cdt1, however, so impaired degradation does not explain this variant’s phenotype.
Our comparison of Cdt1-A66T, an engineered null for cyclin/CDK binding (Cdt1-Cy), and a variant that can bind cyclin but cannot generate a phosphodegron (Cdt1-2A) directly demonstrated that cyclin A–dependent regulation of Cdt1 involves more than just degradation, because mutating the phosphodegron had less impact than mutating the cyclin/CDK binding site (Figure 6). We thus postulate that cyclin/CDK also inhibits Cdt1 by nondegradation mechanisms. Coulombe et al. (2013) described a negative-regulatory PEST domain (a.a. 74–108) in mammalian Cdt1 that contains multiple candidate CDK phosphorylation sites. This domain functions independent of either geminin or the E3 ubiquitin ligase system. Deleting the PEST domain induced DNA rereplication similarly to Cdt1-A66T. Cyclin/CDK may phosphorylate any of the other CDK target residues—either in the PEST domain or elsewhere—which could inhibit Cdt1 activity. A total of 20 candidate CDK phosphorylation sites have been detected in human Cdt1 by mass spectrometry, and only seven of these have been functionally tested so far (Pozo and Cook, 2016). Given the apparent efficient interaction of Cdt1 with cyclin A/CDK, it is also possible that cyclin binding itself inhibits Cdt1 activity independently of phosphoregulation. We are actively pursuing a molecular explanation for nondegradative Cdt1 inhibition by cyclin A/CDK.
It is surprising that mutational alterations that lead to similar phenotypes in MG dwarfism patients behave differently at the molecular level with respect to Cdt1. In the case of Cdt1-R462Q and Cdt1-E468K, impaired Cdt1-MCM interactions can lead to G1 phase lengthening and thus slower proliferation, because G1 length and origin licensing status are coordinated by an origin-licensing checkpoint. Of note, the transformed U2OS cells used in this study have a less active licensing checkpoint than untransformed fibroblasts (Shreeram et al., 2002; Nevis et al., 2009). Thus, otherwise normal MG patient cells may have experienced even longer G1 phases in vivo from a more robust checkpoint. Indeed, MG patient-derived cells proliferate slowly in culture (Bicknell et al., 2011a). Over the full course of development, the accumulated effects of slightly longer G1 phases could explain the overall short stature and hypoplasias associated with these hypomorphic alleles of cdt1 and likely other genes encoding licensing proteins. Perhaps the tissue-specific phenotypes reflect differences in the severity of the licensing defect in those cell types or alternatively, differences in the execution of the cellular response to impaired licensing. On the other hand, Cdt1-A66T dysregulation by cyclin/CDK results in rereplication-induced stress, which can also lead to proliferation failure, but in this case by checkpoint activation in S or G2 phase rather than G1. Even cells expressing endogenous levels of Cdt1-A66T in place of endogenous Cdt1 rereplicated and spent more time in S and G2 phases due to the replication stress (Supplemental Figure S2). The ultimate outcome from either hypo- or hypermorphic mutations, however, is impaired overall proliferation (Figure 7). Continual improvements in our understanding of the molecular mechanisms governing origin licensing are essential to link processes of cell proliferation, genome stability, and development.
MATERIALS AND METHODS
Cell culture and manipulations
U2OS Flp-in Trex (Malecki et al., 2006) cells bearing a single FRT site (gift of J. Aster, Brigham and Women’s Hospital, Harvard Medical School) and HEK 293T cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) and 1× penicillin/streptomycin (complete medium); cell line identity was verified by STR profiling, and the cells tested negative for mycoplasma. To generate stable isogenic cell lines, U2OS cells were cotransfected with flippase recombinase (Flp) and a Cdt1 expression vector derived from pcDNA5/FRT/TO-Venus-Flag-Gateway (1124), a gift from Jonathon Pines (The Institute of Cancer Research, London) (Addgene plasmid # 40999), using X-tremeGENE HP DNA transfection reagent (Roche). The Cdt1 cDNAs encode normal Cdt1 or harbor a single point mutation and a drug resistance cassette. At 48 h posttransfection, cells were selected for resistance to either 150 µg/ml hygromycin B (Roche) or 1 µg/ml puromycin (Sigma), depending on the Cdt1 vector used. For inducible expression of Cdt1 variants, U2OS cells were treated with varying concentrations of doxycycline ranging from 0.003 to 1 µg/ml (CalBiochem) by either media exchange or adding directly into cell culture plates. For colony-forming assays, U2OS cells harboring Cdt1 mutant alleles were plated at a density of ∼500 cells/6-cm dish in the presence or absence of doxycycline. Cells were grown for 10 d, changing media every 3 d, and stained using 0.4% crystal violet (Fisher Scientific). Colony numbers and size were quantified using ImageJ (National Institutes of Health [NIH]). A technical replicate plate was harvested after 72 h to assay for immunoblot analysis.
For G1- to S-phase synchronization, U2OS cells were treated with 2.5 mM thymidine for 24 h, followed by release into complete medium containing 100 ng/ml nocodazole plus 0.05 µg/ml doxycycline for 16 h. Cells were then harvested by mitotic shake-off and replated in complete medium plus 0.05 µg/ml doxycycline for each time point. For S- to G2/M-phase synchronization, U2OS cells were treated with 2.5 mM thymidine for 18 h followed by release into complete medium for 8 h. Cells were then treated with 2.5 mM thymidine plus doxycycline for 18 h followed by release into complete medium plus doxycycline for each time point. To transiently express Cdt1 variants, HEK 293T cells were transfected with Cdt1 expression vectors using PEI Max (Sigma) according to the manufacturer’s instructions. HEK 293T cells were harvested 16 h posttransfection and processed for subsequent coimmunoprecipitation assays.
All cell lines were validated by STR profiling and tested mycoplasma-negative.
Plasmids
Cdt1 mutations (Cdt1-A66T, -Q117H, -R210C, -R453W, -R462Q, -E468K) were generated by PCR-based mutagenesis from a WT Cdt1 coding sequence template. The resulting PCR products were cloned into pENTR vectors harboring the full-length Cdt1 sequence with C-terminal polyhistidine (His) and hemagglutinin (HA) epitope tags. The Cdt1-Y520X truncation was generated using Gibson assembly (NEB) from a pENTR plasmid harboring a WT version of Cdt1 with C-terminal polyhistidine and HA epitope tags, following the manufacturer’s protocols. The pENTR-EGFP (vector control) plasmid was generated by subcloning EGFP from an EGFP-bearing plasmid into pENTR via Gateway cloning (Invitrogen). EGFP, Cdt1-WT-His-HA, Cdt1-Y520X-His-HA, Cdt1-Mutant-His-HA, Cdt1-Cy-His-HA, and Cdt1-2A-His-HA versions were transferred from pENTR plasmids into derivatives of pcDNA5/FRT/TO-Venus-Flag-Gateway (1124), harboring either hygromycin B (Roche) or puromycin (Sigma) selection cassettes, via Gateway cloning. The mVenus-tagged constructs were constructed by subcloning mVenus into the Cdt1-WT-His-HA or the Cdt1-A66T-His-HA pENTR plasmids before Gateway cloning into pcDNA5/FRT/TO-Venus-Flag-Gateway (1124).
Flow cytometry analysis for DNA rereplication
U2OS cell lines harboring stably integrated individual Cdt1 alleles were cultured in complete medium plus doxycycline for either 48 or 72 h. Cell were pulse-labeled with 10 µM EdU (Sigma) for 30 min before being harvested by trypsinization. Approximately 20% of this suspension was reserved for subsequent immunoblotting analysis. The remaining 80% was fixed in 1× PBS plus 4% paraformaldehyde (Sigma) at room temperature for 15 min. Cells were permeabilized in 1% BSA plus 0.5% Triton X-100 for 15 min and then processed for EdU detection by conjugation to Alexa Fluor 647 azide (Life Technologies) in 1 mM CuSO4 and 100 mM ascorbic acid; total DNA was detected by staining with 1 µg/ml 4’,6-diamidino-2-phenylindole (DAPI; Life Technologies) in 100 µg/ml RNase A (Sigma). Samples were analyzed on a Beckman Coulter CyAn ADP cytometer and data were analyzed using FCS Express 6 (De Novo Software) software.
MCM-loading analysis by flow cytometry
U2OS cell lines harboring stably integrated individual Cdt1 alleles were plated into dishes containing a mixture of siRNA (100 nM final concentration), Dharmafect 1 (Dharmacon), and antibiotic-free media plus doxycycline for 72 h. Cells were pulse-labeled with 10 µM EdU (Sigma) for 30 min before harvesting by trypsinization. Approximately 20% of this suspension was reserved for subsequent immunoblotting analysis, while the remaining 80% was analyzed for bound MCM as described and validated in Haland et al. (2015), Moreno et al. (2016), and Matson et al. (2017). Briefly, cells were extracted in cold CSK buffer (10 mM Pipes, pH 7.0, 300 mM sucrose, 100 mM NaCl, 3 mM MgCl2) supplemented with 0.5% Triton X-100, protease inhibitors (0.1 mM AEBSF, 1 µg/ml pepstatin A, 1 µg/ml leupeptin, 1 µg/ml aprotinin), and phosphatase inhibitors (10 µg/ml phosvitin, 1 mM β-glycerol phosphate, 1 mM Na-orthovanadate). Cells were washed with PBS plus 1% BSA and then fixed in 4% paraformaldehyde (Sigma) followed by processing for EdU conjugation to Alexa Fluor 647 azide (Life Technologies). Bound MCM was detected by incubation with anti-MCM2 primary antibody at 1:200 dilution and anti-mouse-488 at 1:1,000 dilution at 37°C for 1 h. Total DNA was detected by incubation in 1 µg/ml DAPI (Life Technologies) and 100 µg/ml RNase A (Sigma). Samples were processed on a Beckman Coulter CyAn ADP cytometer and data were analyzed using FCS Express 6 (De Novo Software) software. Control samples omitting primary antibody or EdU detection were prepared to define thresholds of detection as in Matson et al. (2017).
Antibodies
The following antibodies were purchased from Cell Signaling Technologies: anti-pChk1 S345 (Cat# 2341), anti-Chk1 (Cat# 2345), anti-Cdt1 (Cat# 8064), and anti-Skp2 (Cat# 4313). Anti-HA used for immunoblotting was purchased from Roche (Cat# 11867423001). Anti-HA used for coimmunoprecipitation was purchased from Santa Cruz Biotechnology (Cat# SC-805). Anti-cyclin A was purchased from Santa Cruz Biotechnology (Cat# SC-596). Anti-MCM2 was purchased from BD Biosciences (San Jose, CA, Cat#610700). Anti-mouse Alexa 488 (Jackson ImmunoResearch) and Alexa 647-azide (Life Technologies) were used in flow cytometry analyses. Secondary antibodies for immunoblotting were purchased from Jackson ImmunoResearch.
Protein–protein interaction assays
For HEK 293T coimmunoprecipitation assays, cells were transiently transfected using expression vectors harboring individual Cdt1 alleles. Cells were harvested by trypsinization, pelleted, and resuspended in co-IP buffer (50 mM HEPES, pH 7.2, 33 mM KAc, 1 mM MgCl2, 0.5% Triton X-100, 10% glycerol) containing protease inhibitors (0.1 mM AEBSF, 10 µg/ml pepstatin A, 10 µg/ml leupeptin, 10 µg/ml aprotinin), phosphatase inhibitors (5 µg/ml phosvitin, 1 mM β-glycerol phosphate, 1 mM Na-orthovanadate), and 1 mM ATP, and supplemented with 5 mM CaCl2 and 15 U of S7 micrococcal nuclease (Roche). Lysates were sonicated for 10 s at low power followed by incubation on ice for 20 min and clarification by centrifugation at 13,000 × g at 4°C. Supernatants were precleared with Protein A-Agarose (Roche) then incubated with 1 µg antibody at 4°C overnight with rotation. Antibody–antigen complexes were collected on Protein A beads at 4°C for 1 h with rotation. Complexes were washed three times rapidly with 1 ml ice-cold co-IP buffer and then eluted by boiling in SDS sample buffer supplemented with 10% β-ME and 100 mM dithiothreitol (DTT) for subsequent immunoblot analysis.
For polyhistidine pull-down assays, U2OS cells harboring each individual allele were plated in complete medium plus 1 µg/ml doxycycline for 16 h, and then lysed in 50 mM HEPES, pH 8.0, 33 mM KAc, 117 mM NaCl, 20 mM imidazole, 0.5% Triton X-100, 10% glycerol containing protease inhibitors (0.1 mM AEBSF, 10 µg/ml pepstatin A, 10 µg/ml leupeptin, 10 µg/ml aprotinin), phosphatase inhibitors (5 µg/ml phosvitin, 1 mM β-glycerol phosphate, 1 mM Na-orthovanadate), 1 mM ATP, and 1 mM MgCl2, and supplemented with 5 mM CaCl2 and 15 U of S7 micrococcal nuclease (Roche). Clarified lysates were incubated with nickel NTA agarose (Qiagen) for 2 h at 4°C with rotation. Beads were washed four times rapidly with 1 ml ice-cold lysis buffer and then boiled in sample buffer before immunoblot analysis.
Live-cell imaging and analysis
U2OS cells stably expressing a PCNA-mTurquoise2 fusion (introduced by retroviral transduction) were plated on glass-bottomed plates (Cellvis) #1.5 in FluoroBrite DMEM (Invitrogen) supplemented with FBS, l-glutamine, and penicillin/streptomycin and kept in a humidified chamber (Okolabs) at 37°C with 5% CO2. A Nikon Ti Eclipse inverted microscope with Plan Apochromat dry objective lenses 20× (NA 0.75), Nikon Perfect Focus System, and Andor Zyla 4.2 sCMOS detector with 12-bit resolution was used for imaging. For fluorescence imaging, Chroma filters were optimized for YFP spectral range—excitation: 500/20 nm, beam splitter: 515 nm, and emission: 535/30 nm. Images were collected every 10 min using NIS-Elements AR software. No photobleaching or phototoxicity was observed in imaged cells. Expression of Cdt1 (WT/A66T)–mVenus was induced with 50 ng/ml Dox (A66T) or 100 ng/ml Dox (WT).
Image and data analysis were performed using Fiji, ImageJ NIH software (version 1.51n; Schindelin et al., 2012), and Matlab (R2017b MathWorks). Briefly, asynchronous colonies of cells were followed in time-lapse experiments and individual cells were tracked, segmented, and synchronized in silico. Before the analysis, images were background-corrected using rolling-ball subtraction. Individual cells were tracked in a user-assisted way and nuclear regions were segmented based on PCNA images. These regions of interest were used to measure Cdt1 (WT/A66T)–mVenus intensity. Cells in the S phase were detected based on the S-phase punctate pattern of PCNA by calculating the variance of fluorescence intensity of PCNA on a spatial scale corresponding to focus size. Cells lacking sufficient PCNA contrast for confident detection of S-phase boundaries were manually removed from the analysis set. To visualize the dynamics of the mean Cdt1 (WT/A66T) signal throughout the cell cycle in a population of cells, the signals collected from individual cells were normalized to cell-cycle type in this manner: cell-cycle phases were defined for individual cells based on PCNA localization, and traces of Cdt1 intensity were linearly interpolated over the expected number of time points in each cell-cycle phase (based on measurements of median cell-cycle phase lengths in the population). This in silico alignment emphasized the sharp changes in protein abundance at the boundaries of cell-cycle phases rather than the smoothing from averaging cells with different lengths of individual phases.
Structural model of hMCM complex with hCdt1 middle domain and C-terminal winged-helix domain
The atomic resolution structure of the yeast MCM2–7, Cdc6, ORC1-6, and Cdt1 complex was determined by electron microscopy at a resolution of 3.9 Å (PDB ID 5udb; Yuan et al., 2017). This structure was the template used for modeling the atomic structures of the human MCM (hMCM) complex as well as the interaction of human Cdt1 (hCdt1) with hMCM. MCM subunits are highly conserved during evolution, with yeast and human subunits sharing 46–50% sequence identity. Human and yeast MCM2, MCM4, and MCM6 subunits, in particular, share 50%, 47%, and 47% sequence identity, respectively. Modeller v9.16 (Marti-Renom et al., 2000) was used to generate the structural models of human MCM subunits using the yeast MCM subunits (PDB ID 5udb) as a template. No modeling of hCdt1 was needed, as x-ray crystallography had been used to determine the structure of the N-terminal winged helix domain of hCdt1 at a resolution of 3.3 Å (PDB ID 2wvr; De Marco et al., 2009), while the C-terminal winged helix domain of mouse Cdt1 was determined at 1.89 Å resolution (PDB ID 3a4c; Khayrutdinov et al., 2009). Due to the low sequence conservation between yeast and human Cdt1, the two mammalian winged helix domains were superimposed on the corresponding yeast Cdt1 winged helix domains in 5udb using the sequence-independent and structure-based dynamic programming alignment method accessed through the align command in the PyMOL molecular vision system (PyMOL Molecular Graphics System, Version 2.0, Schrödinger).
Quantification and statistical analyses
Data were analyzed using GraphPad Prism 7.0 and MATLAB and Statistics Toolbox Release 2017b, MathWorks, Natick, MA.
Supplementary Material
Acknowledgments
We thank J. Aster for the generous gift of U2OS FRT Flp-in Trex cells and all members of the Cook laboratory for their insightful discussions in preparing this article. This project was supported by National Institutes of Health F31GM121073 to P.N.P., National Science Foundation Graduate Student Fellowship DGE-1144081 to J.P.M., and National Institutes of Health R01GM102413 and R25GM055336 and a grant from the W. M. Keck Foundation to J.G.C. National Cancer Institute, National Institutes of Health T32CA009156 supported G.D.G. The UNC Flow Cytometry Core Facility is supported in part by P30 CA016086 Cancer Core Support Grant to the UNC Lineberger Comprehensive Cancer Center.
Abbreviations used:
- CDK
cyclin-dependent kinase
- Cdt1
Cdc10-dependent transcript 1
- MCM
minichromosome maintenance
- MG
Meier–Gorlin syndrome
- SCF
Skp1-Cullin1-F-box protein
- Skp2
S-phase kinase-associated protein 2.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-04-0242) on October 3, 2018.
REFERENCES
- Abbas T, Dutta A. (2011). CRL4Cdt2: master coordinator of cell cycle progression and genome stability. Cell Cycle , 241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arentson E, Faloon P, Seo J, Moon E, Studts JM, Fremont DH, Choi K. (2002). Oncogenic potential of the DNA replication licensing protein CDT1. Oncogene , 1150–1158. [DOI] [PubMed] [Google Scholar]
- Arias EE, Walter JC. (2005). Replication-dependent destruction of Cdt1 limits DNA replication to a single round per cell cycle in Xenopus egg extracts. Genes Dev , 114–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias EE, Walter JC. (2006). PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat Cell Biol , 84–90. [DOI] [PubMed] [Google Scholar]
- Bicknell LS, Walker S, Klingseisen A, Stiff T, Leitch A, Kerzendorfer C, Martin CA, Yeyati P, Al Sanna N, Bober M, et al. (2011a). Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier–Gorlin syndrome. Nat Genet , 350–355. [DOI] [PubMed] [Google Scholar]
- Bicknell LS, Bongers EM, Leitch A, Brown S, Schoots J, Harley ME, Aftimos S, Al-Aama JY, Bober M, Brown PA, et al. (2011b). Mutations in the pre-replication complex cause Meier–Gorlin syndrome. Nat Genet , 356–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Ge XQ, Jackson DA. (2011). How dormant origins promote complete genome replication. Trends Biochem Sci , 405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Gillespie PJ. (2008). Replication licensing and cancer—a fatal entanglement? Nat Rev Cancer , 799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrage LC, Charng WL, Eldomery MK, Willer JR, Davis EE, Lugtenberg D, Zhu W, Leduc MS, Akdemir ZC, Azamian M, et al. (2015). De novo GMNN mutations cause autosomal-dominant primordial dwarfism associated with Meier–Gorlin syndrome. Am J Hum Genet , 904–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook JG. (2009). Replication licensing and the DNA damage checkpoint. Front Biosci (Landmark Ed) , 5013–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook JG, Chasse DA, Nevins JR. (2004). The regulated association of Cdt1 with minichromosome maintenance proteins and Cdc6 in mammalian cells. J Biol Chem , 9625–9633. [DOI] [PubMed] [Google Scholar]
- Coulombe P, Gregoire D, Tsanov N, Mechali M. (2013). A spontaneous Cdt1 mutation in 129 mouse strains reveals a regulatory domain restraining replication licensing. Nat Commun , 2065. [DOI] [PubMed] [Google Scholar]
- Davidson IF, Li A, Blow JJ. (2006). Deregulated replication licensing causes DNA fragmentation consistent with head-to-tail fork collision. Mol Cell , 433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marco V, Gillespie PJ, Li A, Karantzelis N, Christodoulou E, Klompmaker R, van Gerwen S, Fish A, Petoukhov MV, Iliou MS, et al. (2009). Quaternary structure of the human Cdt1–geminin complex regulates DNA replication licensing. Proc Natl Acad Sci USA , 19807–19812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Munnik SA, Bicknell LS, Aftimos S, Al-Aama JY, van Bever Y, Bober MB, Clayton-Smith J, Edrees AY, Feingold M, Fryer A, et al. (2012). Meier-Gorlin syndrome genotype-phenotype studies: 35 individuals with pre-replication complex gene mutations and 10 without molecular diagnosis. Eur J Hum Genet , 598–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C. (2009). A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci USA , 20240–20245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenbach A, Li A, Brito-Martins M, Blow JJ. (2005). Functional domains of the Xenopus replication licensing factor Cdt1. Nucleic Acids Res , 316–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigola J, He J, Kinkelin K, Pye VE, Renault L, Douglas ME, Remus D, Cherepanov P, Costa A, Diffley JFX. (2017). Cdt1 stabilizes an open MCM ring for helicase loading. Nat Commun , 15720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M. (2006). Cdt1 revisited: complex and tight regulation during the cell cycle and consequences of deregulation in mammalian cells. Cell Div , 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guernsey DL, Matsuoka M, Jiang H, Evans S, Macgillivray C, Nightingale M, Perry S, Ferguson M, LeBlanc M, Paquette J, et al. (2011). Mutations in origin recognition complex gene ORC4 cause Meier–Gorlin syndrome. Nat Genet , 360–364. [DOI] [PubMed] [Google Scholar]
- Haland TW, Boye E, Stokke T, Grallert B, Syljuasen RG. (2015). Simultaneous measurement of passage through the restriction point and MCM loading in single cells. Nucleic Acids Res , e150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JR, Lee HO, Bunker BD, Dorn ES, Rogers GC, Duronio RJ, Cook JG. (2008). Cdt1 and Cdc6 are destabilized by rereplication-induced DNA damage. J Biol Chem , 25356–25363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havens CG, Walter JC. (2009). Docking of a specialized PIP Box onto chromatin-bound PCNA creates a degron for the ubiquitin ligase CRL4Cdt2. Mol Cell , 93–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havens CG, Walter JC. (2011). Mechanism of CRL4(Cdt2), a PCNA-dependent E3 ubiquitin ligase. Genes Dev , 1568–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. (2015). PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res , D512–D520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jee J, Mizuno T, Kamada K, Tochio H, Chiba Y, Yanagi K, Yasuda G, Hiroaki H, Hanaoka F, Shirakawa M. (2010). Structure and mutagenesis studies of the C-terminal region of licensing factor Cdt1 enable the identification of key residues for binding to replicative helicase Mcm proteins. J Biol Chem , 15931–15940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Arias EE, Chen J, Harper JW, Walter JC. (2006). A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell , 709–721. [DOI] [PubMed] [Google Scholar]
- Khayrutdinov BI, Bae WJ, Yun YM, Lee JH, Tsuyama T, Kim JJ, Hwang E, Ryu KS, Cheong HK, Cheong C, et al. (2009). Structure of the Cdt1 C-terminal domain: conservation of the winged helix fold in replication licensing factors. Protein Sci , 2252–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Hong B, Choi JM, Kim Y, Watanabe S, Ishimi Y, Enomoto T, Tada S, Kim Y, Cho Y. (2004). Structural basis for inhibition of the replication licensing factor Cdt1 by geminin. Nature , 913–917. [DOI] [PubMed] [Google Scholar]
- Li C, Jin J. (2010). DNA replication licensing control and rereplication prevention. Protein Cell , 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhao Q, Liao R, Sun P, Wu X. (2003). The SCF(Skp2) ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation. J Biol Chem , 30854–30858. [DOI] [PubMed] [Google Scholar]
- Liontos M, Koutsami M, Sideridou M, Evangelou K, Kletsas D, Levy B, Kotsinas A, Nahum O, Zoumpourlis V, Kouloukoussa M, et al. (2007). Deregulated overexpression of hCdt1 and hCdc6 promotes malignant behavior. Cancer Res , 10899–10909. [DOI] [PubMed] [Google Scholar]
- Liu E, Li X, Yan F, Zhao Q, Wu X. (2004). Cyclin-dependent kinases phosphorylate human Cdt1 and induce its degradation. J Biol Chem , 17283–17288. [DOI] [PubMed] [Google Scholar]
- Liu P, Slater DM, Lenburg M, Nevis K, Cook JG, Vaziri C. (2009). Replication licensing promotes cyclin D1 expression and G1 progression in untransformed human cells. Cell Cycle , 125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida YJ, Teer JK, Dutta A. (2005). Acute reduction of an origin recognition complex (ORC) subunit in human cells reveals a requirement of ORC for Cdk2 activation. J Biol Chem , 27624–27630. [DOI] [PubMed] [Google Scholar]
- Malecki MJ, Sanchez-Irizarry C, Mitchell JL, Histen G, Xu ML, Aster JC, Blacklow SC. (2006). Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol Cell Biol , 4642–4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. (2000). Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct , 291–325. [DOI] [PubMed] [Google Scholar]
- Matson JP, Dumitru R, Coryell P, Baxley RM, Chen W, Twaroski K, Webber BR, Tolar J, Bielinsky AK, Purvis JE, Cook JG. (2017). Rapid DNA replication origin licensing protects stem cell pluripotency. Elife , e30473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh D, Blow JJ. (2012). Dormant origins, the licensing checkpoint, and the response to replicative stresses. Cold Spring Harb Perspect Biol , a012955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melixetian M, Ballabeni A, Masiero L, Gasparini P, Zamponi R, Bartek J, Lukas J, Helin K. (2004). Loss of geminin induces rereplication in the presence of functional p53. J Cell Biol , 473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno A, Carrington JT, Albergante L, Al Mamun M, Haagensen EJ, Komseli ES, Gorgoulis VG, Newman TJ, Blow JJ. (2016). Unreplicated DNA remaining from unperturbed S phases passes through mitosis for resolution in daughter cells. Proc Natl Acad Sci USA , E5757–E5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevis KR, Cordeiro-Stone M, Cook JG. (2009). Origin licensing and p53 status regulate Cdk2 activity during G(1). Cell Cycle , 1952–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H, Sugimoto N, Roukos V, Nakanishi Y, Saijo M, Obuse C, Tsurimoto T, Nakayama KI, Nakayama K, Fujita M, et al. (2006). Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J , 1126–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo PN, Cook JG. (2016). Regulation and function of Cdt1: a key factor in cell proliferation and genome stability. Genes (Basel) , E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JF. (2009). Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell , 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods , 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shreeram S, Sparks A, Lane DP, Blow JJ. (2002). Cell type-specific responses of human cells to inhibition of replication licensing. Oncogene , 6624–6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto N, Tatsumi Y, Tsurumi T, Matsukage A, Kiyono T, Nishitani H, Fujita M. (2004). Cdt1 phosphorylation by cyclin A-dependent kinases negatively regulates its function without affecting geminin binding. J Biol Chem , 19691–19697. [DOI] [PubMed] [Google Scholar]
- Sun J, Evrin C, Samel SA, Fernandez-Cid A, Riera A, Kawakami H, Stillman B, Speck C, Li H. (2013). Cryo-EM structure of a helicase loading intermediate containing ORC-Cdc6-Cdt1-MCM2-7 bound to DNA. Nat Struct Mol Biol , 944–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda DY, Parvin JD, Dutta A. (2005). Degradation of Cdt1 during S phase is Skp2-independent and is required for efficient progression of mammalian cells through S phase. J Biol Chem , 23416–23423. [DOI] [PubMed] [Google Scholar]
- Truong LN, Wu X. (2011). Prevention of DNA re-replication in eukaryotic cells. J Mol Cell Biol , 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri C, Saxena S, Jeon Y, Lee C, Murata K, Machida Y, Wagle N, Hwang DS, Dutta A. (2003). A p53-dependent checkpoint pathway prevents rereplication. Mol Cell , 997–1008. [DOI] [PubMed] [Google Scholar]
- Whittaker AJ, Royzman I, Orr-Weaver TL. (2000). Drosophila Double Parked: a conserved, essential replication protein that colocalizes with the origin recognition complex and links DNA replication with mitosis and the down-regulation of S phase transcripts. Genes Dev , 1765–1776. [PMC free article] [PubMed] [Google Scholar]
- Woodward AM, Gohler T, Luciani MG, Oehlmann M, Ge X, Gartner A, Jackson DA, Blow JJ. (2006). Excess Mcm2-7 license dormant origins of replication that can be used under conditions of replicative stress. J Cell Biol , 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagi K, Mizuno T, You Z, Hanaoka F. (2002). Mouse geminin inhibits not only Cdt1-MCM6 interactions but also a novel intrinsic Cdt1 DNA binding activity. J Biol Chem , 40871–40880. [DOI] [PubMed] [Google Scholar]
- Yekezare M, Gomez-Gonzalez B, Diffley JF. (2013). Controlling DNA replication origins in response to DNA damage—inhibit globally, activate locally. J Cell Sci , 1297–1306. [DOI] [PubMed] [Google Scholar]
- You Z, Ode KL, Shindo M, Takisawa H, Masai H. (2016). Characterization of conserved arginine residues on Cdt1 that affect licensing activity and interaction with geminin or Mcm complex. Cell Cycle , 1213–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Riera A, Bai L, Sun J, Nandi S, Spanos C, Chen ZA, Barbon M, Rappsilber J, Stillman B, et al. (2017). Structural basis of Mcm2-7 replicative helicase loading by ORC-Cdc6 and Cdt1. Nat Struct Mol Biol , 316–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y, Cheng E, Wu H, Li N, Yung PY, Gao N, Tye BK. (2017). Open-ringed structure of the Cdt1-Mcm2-7 complex as a precursor of the MCM double hexamer. Nat Struct Mol Biol , 300–308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.