Abstract
Fibroblasts exhibit heterogeneous cell geometries in tissues and integrate both mechanical and biochemical signals in their local microenvironment to regulate genomic programs via chromatin remodelling. While in connective tissues fibroblasts experience tensile and compressive forces (CFs), the role of compressive forces in regulating cell behavior and, in particular, the impact of cell geometry in modulating transcriptional response to such extrinsic mechanical forces is unclear. Here we show that CF on geometrically well-defined mouse fibroblast cells reduces actomyosin contractility and shuttles histone deacetylase 3 (HDAC3) into the nucleus. HDAC3 then triggers an increase in the heterochromatin content by initiating removal of acetylation marks on the histone tails. This suggests that, in response to CF, fibroblasts condense their chromatin and enter into a transcriptionally less active and quiescent states as also revealed by transcriptome analysis. On removal of CF, the alteration in chromatin condensation was reversed. We also present a quantitative model linking CF-dependent changes in actomyosin contractility leading to chromatin condensation. Further, transcriptome analysis also revealed that the transcriptional response of cells to CF was geometry dependent. Collectively, our results suggest that CFs induce chromatin condensation and geometry-dependent differential transcriptional response in fibroblasts that allows maintenance of tissue homeostasis.
INTRODUCTION
Under physiological conditions, cells in our body are subjected to external tensile, shear, and compressive forces (Janmey and Miller, 2011; Wells, 2013). Fibroblasts, the most abundant type of cells in the connective tissue, among other signals, experience large compressive forces from the surrounding extracellular matrix as well as fluid in the order of kilopascals in magnitude and minutes-to-hours timescale during body movements such as walking, sitting, and sleeping (Langevin et al., 2013; Luboz et al., 2014). Compressive force could potentially regulate the behavior of cells by inducing cytoskeletal remodelling (Brangwynne et al., 2006; Ofek et al., 2009; Kanazawa et al., 2014), altering biochemical signalling processes (Kanjanamekanant et al., 2013; Kanazawa et al., 2014; Shimomura et al., 2014; Madden et al., 2015), modulating gene expression (Lee et al., 2007; Shieh and Athanasiou, 2007; Le Berre et al., 2012; Hara et al., 2014; Kanazawa et al., 2014; Chen et al., 2016; Chooi and Chan, 2016), and maintaining genomic integrity (Kumar et al., 2014). Such compressive force induced cellular behavior has implications in pathological conditions such as cancer, fibrosis, and pressure ulcer (Butcher et al., 2009; Janmey and Miller, 2011; Yu et al., 2011; Tse et al., 2012; Wells, 2013; Kanazawa et al., 2014). This is brought about by force-induced structural changes in chromatin assembly that are critical to the differential regulation of genes (Tessarz and Kouzarides, 2014; Heo et al., 2015, 2016) and has major implications in tissue homeostasis (Edsberg et al., 2001).
Importantly, fibroblasts in tissue exhibit distinct cell geometries that are determined by the force balance between the internal cytoskeletal structures and the extracellular matrix. The functional significance of this heterogeneity in cell geometric states has been described in recent studies by using cells cultured on defined geometric micropatterns (Thery, 2010). Such studies have highlighted the role of cell geometry in modulating cytoskeletal organization, nuclear morphology, three-dimensional chromosome organization and, importantly, transcription regulation (Versaevel et al., 2012; Jain et al., 2013; Wang et al., 2017). However, the mechanisms involved in controlling chromatin condensation states, in particular by compressive forces on different cell geometries and the regulation of their transcriptional response, has not been studied. In addition, mechanochemical models outlining the coupling between actomyosin contractility and chromatin condensation states have not been well developed.
In this study, using experiments and theoretical modelling, we show that static compressive force on mouse fibroblast cells results in the reduction of actomyosin contractility and increased nuclear localization of histone deacetylase 3 (HDAC3). This results in the removal of acetylation marks on histone tails and triggering increased heterochromatin assembly. Interestingly, on removal of the compressive force, cells reverted back to their original chromatin condensation. Such global chromatin condensation led to reduced transcriptional activity and inducing quiescence in these fibroblasts. The changes in transcription in response to compressive force were also found to be geometry dependent.
RESULTS
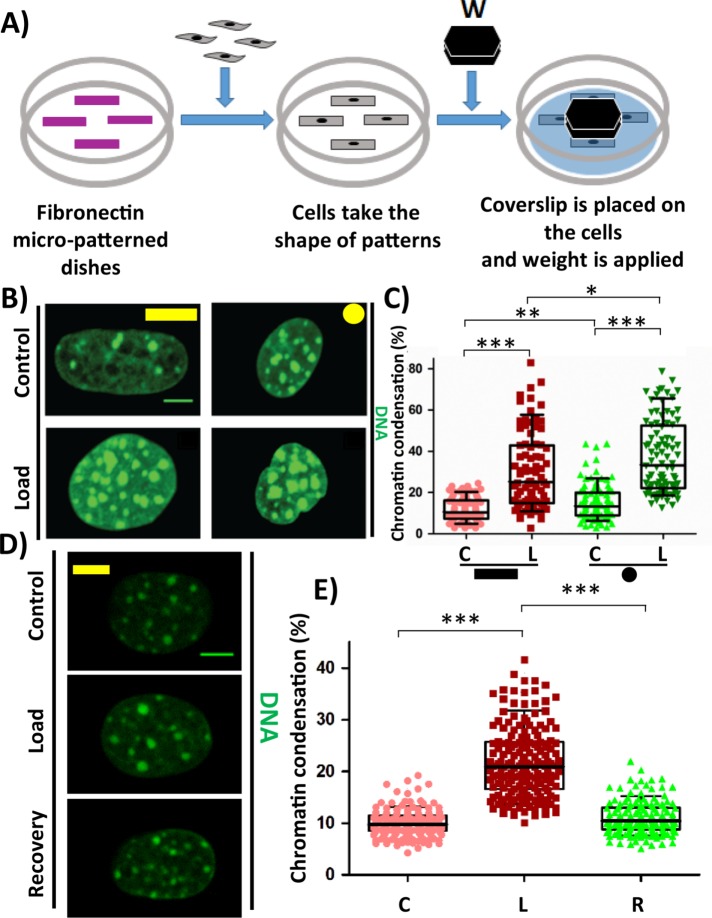
Compressive force induces reversible chromatin condensation
To study the role of compressive force on cells at single-cell resolution, NIH3T3 mouse fibroblasts were seeded onto fibronectin micropatterns of two extreme geometries, that is, a large rectangle (area: 1800 µm2) and a small circle (500 µm2). Once the cells take the given shape, compressive force in the order of micronewtons (for stress in the order of kPa) is applied to these cells for 1 h, which is then fixed and processed for immunofluorescence staining (Figure 1A). It was observed that on applying compressive force (CF), there was an increase in the heterochromatin content (HC) in both geometries, as seen by the increase in the chromatin condensation (Figure 1, B and C). The increase in chromatin condensation was due neither to the serum deprivation of cells nor to the sensing of the apical substrate (Supplemental Figure SE1). This increase in HC content originated from the pericentromeric region as these HC nodes were decorated with Centromeric protein A (CENP-A) which is known to replace Histone 3 only in the centromeric regions (Supplemental Figure SE2A). Further, Heterochromatin protein lα (HPlα), which stabilizes HC and also facilitates HC spreading by interacting with neighboring HPlα, was decorated in these HC nodes (also referred to as chromocenters) (Supplemental Figure SE2B). The increase in HC content was found to be reversible in nature (Figure lD). The chromatin condensation increased from 10% to 21% in response to CF and reduced to original level on recovery (Figure 1E). Recently, two reports suggested that there exists a phase separation in HP1α-mediated heterochromatin formation. HP1α is shown to have properties to form liquid droplets that favors phase separation and compaction of chromatin within the droplet (Larson et al., 2017; Strom et al., 2017). Since in our study we saw that there are visibly larger heterochromatin nodes labeled with HP1a, we hypothesized that in response to compressive force, the liquid-drop-like structures of HC nodes enriched with HP1α fuse with adjacent HC nodes to form larger HC nodes. To test this hypothesis, we measured the change in volume of each HC node as well as their numbers. We observed that in response to compressive force, there is an increase in the volume of HC nodes and a decrease in the number of HC nodes per nucleus, suggesting that adjacent HC nodes fuse with each other to form larger HC nodes (Supplemental Figures SE3A and SE3B). This is further supported by the observation that the average number of CENP-A in each heterochromatin node increases in response to compressive force (Supplemental Figure SE3C). There was also a reduction in nuclear height and nuclear volume in response to compressive force, but no change was observed in the nuclear projected area (Supplemental Figure SE4). These results demonstrate that on experiencing compressive force the cells condense their chromatin in a geometry-independent manner and that this process is reversible.
FIGURE 1:
Compressive force causes chromatin condensation. (A) Experimental setup for Compressive force experiment. (B) Maximum intensity projection of confocal images of nucleus. DNA stained with DAPI (green). (C) Chromatin condensation in percentage based on DAPI stain of the nucleus. N: Rectangle (C# = 83, L# = 83); Circle (C# = 93 and L# = 90). (D) Maximum intensity projection of confocal images of nucleus. DNA stained with DAPI (green). (E) Chromatin condensation in percentage based on DAPI stain of the nucleus. C# = 138, L# = 144, R# = 124. Whisker box plotted from 10 to 90 percentile. Student’s t test, ***p < 0.0001, **p < 0.01, *p < 0.05, ns = not significant. Scale bar: 5 µm. #C = control; L = load, and R = load + recovery.
Compressive force modulates epigenetic marks
The large-scale changes in chromatin condensation in response to compressive force prompted us to characterize the changes in the epigenetic landscape. We observed an increase in both heterochromatin marks H3K27me3 (Figure 2, A and B, and Supplemental Figure SE5B) and H3K9me3 (Figure 2, C and D, and Supplemental Figure SE5A) in response to compressive force. However, their localization was different, that is, H3K9me3 was enriched in the bright HC nodes while H3K27me3 was enriched at the nuclear periphery. This was also observed in the circular geometry (Supplemental Figure SE6). Since the bulk change in the HC was found to be enriched with H3K9me3 and marked with HP1α, we, therefore, focused our investigation on how this type of HC increases on applying CF. An increase in H3K9me3 marked HC suggests that there should be a decrease in the H3K9ac that marks the EC since these modifications are occurring at the same residue. It was indeed observed that there was a decrease in H3K9ac levels in response to CF (Figure 2, E and F, and Supplemental Figure SE5C). Taken together, our observations indicate that compressive force–induced chromatin condensation is majorly facilitated due to epigenetic changes occurring at H3K9 residue. For H3K9 residue to be trimethylated by methyltransferase such as suppressor of variegation 3-9 homologue 1 (SUV39h1), it is important for that residue to be unmodified. Other modifications such as mono- or dimethylation were found to significantly impair the ability of SUV39h1 to trimethylate this residue (Rea et al., 2000). So it is clear that for HC to increase in size, the first step that is crucial is the removal of acetylation marks, which are characteristic of euchromatin, on the neighboring H3K9 residues. This is facilitated by a group of proteins called histone deactylases (HDACs). We therefore investigated whether HDACs are involved in this process in the next section.
FIGURE 2:
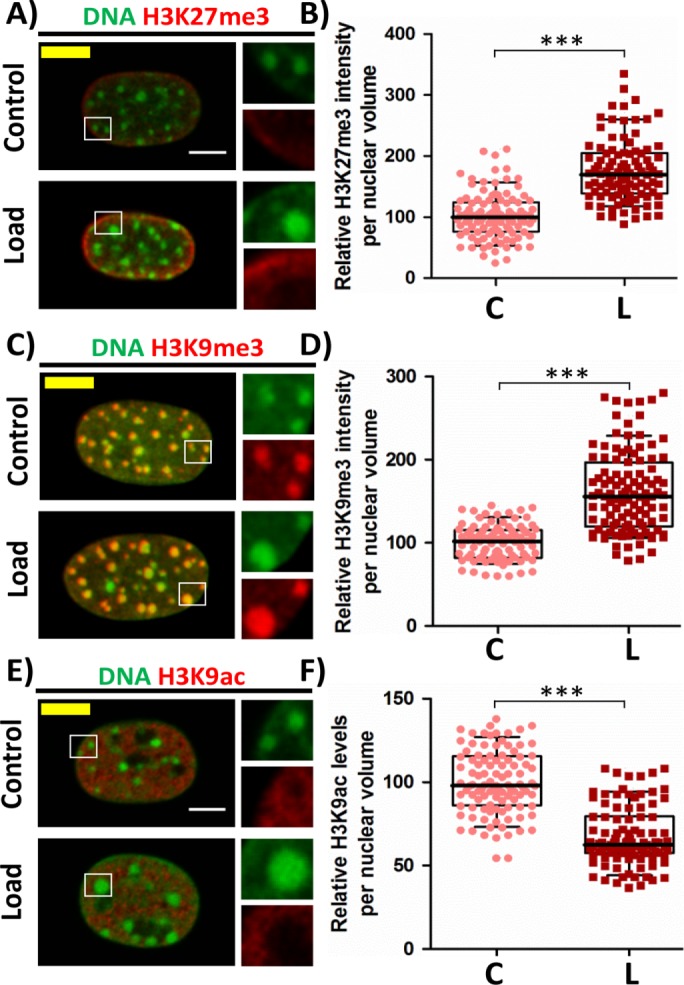
Increased heterochromatin marks and decreased euchromatin mark in response to CF. (A) Single cross-sectional slice of the nucleus. DNA stained with DAPI (green) and IF staining of H3K27me3 (red). (B) Relative H3K27me3 intensity levels per nuclear volume based on IF staining. N: C# = 107, L# = 99. (C) Maximum intensity projection of confocal images of nucleus. DNA stained with DAPI (green) and IF staining of H3K9me3 (red). (D) Relative H3K9me3 intensity levels per nuclear volume based on IF staining. N: C# = 86, L# = 111. (E) Single cross-sectional slice of the nucleus. DNA stained with DAPI (green) and IF staining of H3K9ac (red). (F) Relative H3K9ac intensity levels per nuclear volume based on IF staining. N: C# = 103, L# = 102. Whisker box plotted from 10 to 90 percentile. Student’s t test, ***p < 0.0001. Scale bar: 5 µm. #C = control and L = load.
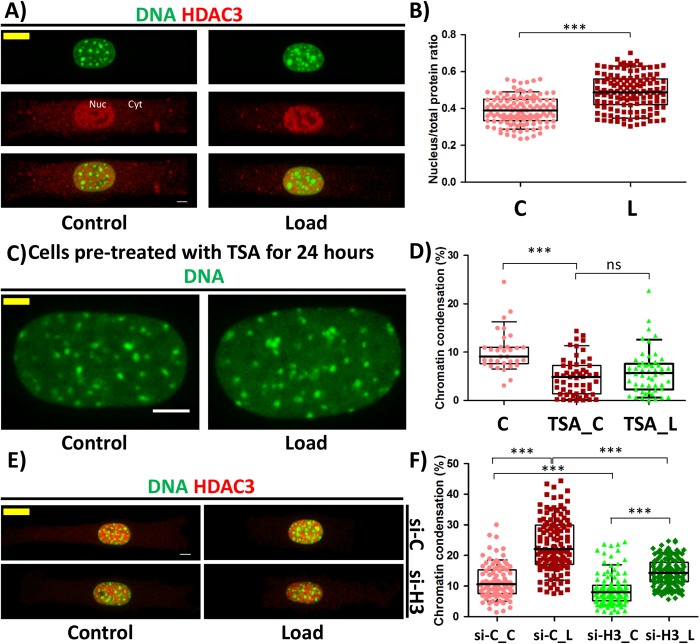
Increase in heterochromatin is driven by HDAC3
Previous studies in our lab had shown that when HDAC3 shuttles to the nucleus, there is a decrease in H3K9ac levels (Jain et al., 2013). We therefore hypothesized that HDAC3 might be involved in this process. It was observed that, on applying CF, HDAC3 shuttled into the nucleus, which is shown by an increase in the nucleus–to–total protein ratio of HDAC3 and an increase in the HDAC3 concentration in the nucleus (Figure 3, A and B, and Supplemental Figure SE7). To further investigate whether HDAC3 is important, we inhibited the HDAC3 activity using class I HDAC inhibitor Trichostatin A (TSA). Cells were pretreated with TSA for 24 h prior to the experiment, and it was observed that in the presence of TSA, there was no increase in the chromatin condensation on applying CF (Figure 3, C and D). Importantly, knockdown of HDAC3 by 60% inhibited the increase in chromatin condensation to only 5.6% as compared with 11.9% increase in control small interfering RNA (siRNA)-treated cells in response to CF. Further, we also observed 2.7% reduction in chromatin condensation in HDAC3 knockdown cells when compared with control siRNA-treated cells even without subjecting them to CF (Figure 3, E and F; Supplemental Figure SE8 and Supplemental Table TE1). These results suggest that HDAC3 is an important effector for chromatin condensation in response to CF.
FIGURE 3:
HDAC3 is a critical regulator of compressive force–induced heterochromatin. (A) Maximum intensity projection of confocal images of cell. DNA stained with DAPI (green) and IF staining of HDAC3 (red). Nuc = nucleus fraction and Cyt = cytoplasmic fraction (B) Nuclear to total protein ratio of HDAC3 based on IF staining. N: C# = 124, L# = 132. (C) Maximum intensity projection of confocal images of a nucleus from cells treated with Trichostatin A. DNA stained with DAPI (green). Scale bar: 5 µm. (D) Chromatin condensation in percentage based on DAPI stain of the nucleus. N: C# = 33, TSA_ C# = 55, TSA_ L# = 53. (E) Maximum intensity projection of confocal images of cell. DNA stained with DAPI (green) and IF staining of HDAC3 (red). Si-C = Cells transfected with control siRNA and si-H3 = cells transfected with siHDAC3. (F) Chromatin condensation in percentage based on DAPI stain of the nucleus. N: si-C_C = 96, si-C_L = 125, si-H3_C = 114, si-H3_L = 131. Whisker box plotted from 10 to 90 percentile. Student’s t test, ***p < 0.0001, ns = not significant. Scale bar: 5 µm. #C = control; L = load; TSA_C = TSA-treated control; TSA_L = TSA-treated load; si-C_C = control siRNA-treated rectangle control; si-C_L = control siRNA-treated rectangle load; si-H3_C = HDAC3 siRNA-treated rectangle control; si-H3_L = HDAC3 siRNA-treated rectangle load.
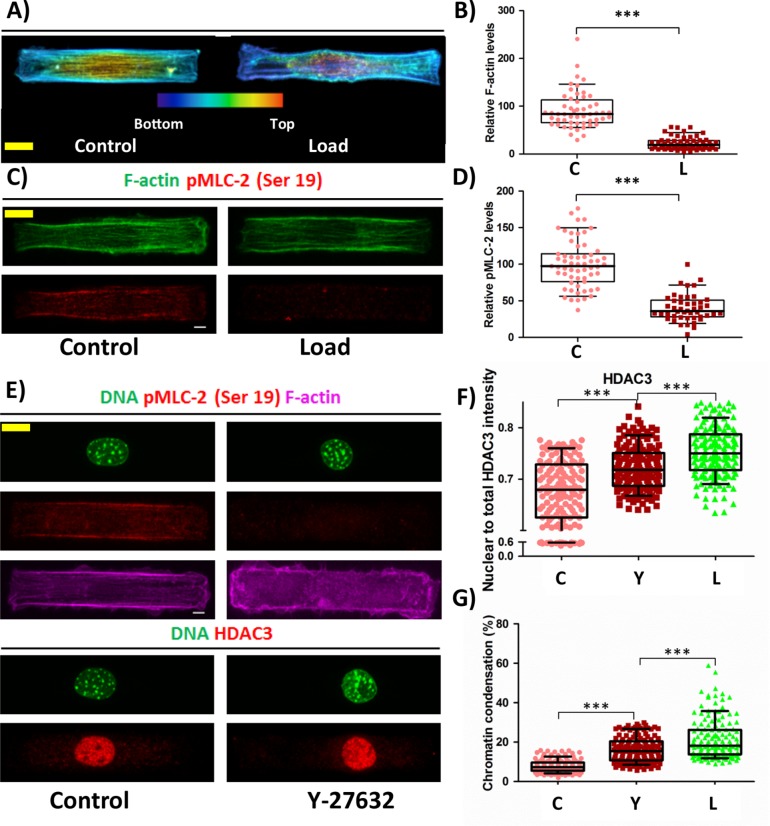
HDAC3 shuttling to the nucleus is driven by reduction in actomyosin contractility
We next asked how HDAC3 shuttles to the nucleus in response to CF. Previous results have shown that reduction in actomyosin contractility resulted in translocation of HDAC3 to the nucleus (Jain et al., 2013). Hence, we investigated the changes to the actin cytoskeleton and actomyosin contractility in response to CF. It was indeed observed that in response to CF, there was a reduction in the levels of polymerized actin. This reduction was majorly due to depolymerization of apical actin (Figure 4, A and B, and Supplemental Movies 1 and 2). In addition, there was a decrease in phosphorylated myosin light chain 2 at serine 19 (pMLC) on applying CF indicating a reduction in actomyosin contractility (Figure 4, C and D). Thus a reduction in actomyosin contractility could then facilitate the shuttling of HDAC3 into the nucleus and thereby increase HC. To validate this, we reduced actomyosin contractility using Y-27632, which is a ROCK inhibitor and is known to reduce actomyosin contractility. Treating the cells with Y-27632 for 1 h mimicked events similar to that in response to CF (Figure 4E). There was a reduction in pMLC levels as expected (Supplemental Figure SE9), resulting in increased nucleus to total protein ratio of HDAC3 (Figure 4F), increased HDAC3 concentration (Supplemental Figure SE9), and increased chromatin condensation (Figure 4G). Additionally, there was a decrease in nuclear volume, reduction in the nuclear projected area, and increase in nuclear height on treatment with Y-27632 (Supplemental Figure SE10). Consistent with this, we observed that reduced actomyosin contractility in circular geometry showed higher HDAC3 concentration in the nucleus as well as an increase in chromatin condensation when compared with rectangular geometry (Supplemental Figures SE11 and SE12 and Figure 1, B and C). These findings provide evidence to support that the loss of actomyosin contractility in response to CF results in HDAC3 shuttling to the nucleus and thereby trigger chromatin condensation.
FIGURE 4:
HDAC3 is shuttled to the nucleus on reduction in actomyosin contractility resulting in increased chromatin condensation. (A) Maximum intensity projection of confocal images of a cell. Actin stained by phalloidin. Color coded based on height. Bottom (blue) to top (red). (B) Relative F-actin intensity levels. N: C# = 53, L# = 60. (C) Maximum intensity projection of confocal images of a cell. F-actin stained with phalloidin (green) and IF staining of pMLC-2 (ser 19) (red). (D) Relative phosphorylated MLC-2 at Ser-19 levels based on IF staining. N: C# = 59, L# = 46. (E) Maximum intensity projection of confocal images of a cell. Top: DNA stained with DAPI (green), IF staining of pMLC (red), and F-actin stained with phalloidin (majenta). Bottom: DNA stained with DAPI (green) and IF staining of HDAC3 (red). (F) Nuclear to total protein ratio of HDAC3 based on IF staining. N: C# = 145, Y# = 157, L# = 165. (G) Chromatin condensation in percentage based on DAPI stain of the nucleus. N: C# = 150, Y# = 150, L# = 150. Whisker box plotted from 10 to 90 percentile. Student’s t test, ***p < 0.0001, **p < 0.01. Scale bar: 5 µm. #C = control; L = load, and Y = Y-27632.
Movie S1.
Shows a merged actin (green) and nucleus (red) stained control cell on the micropattern.
Movie S2.
Shows a merged actin (green) and nucleus (red) stained cell on the micropattern after compressive loading.
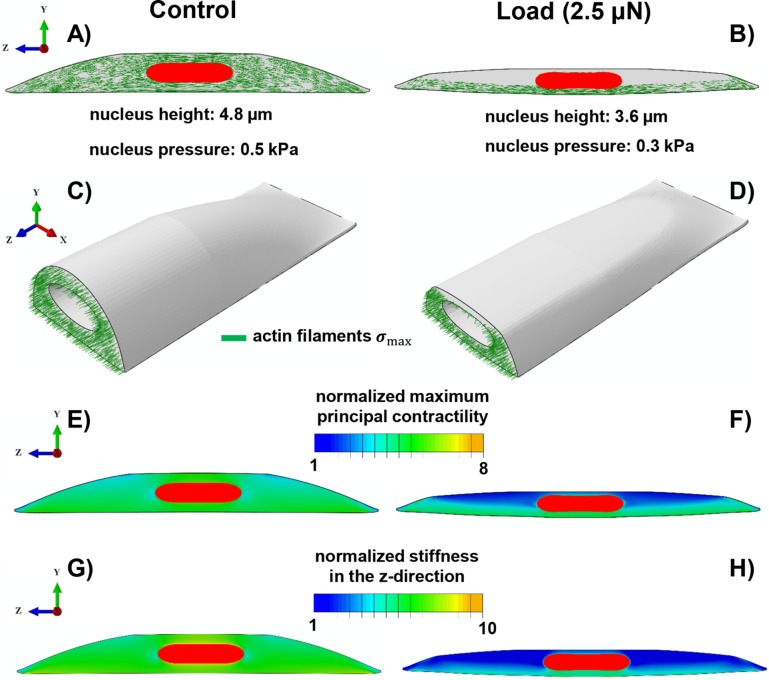
Mechanochemical model for CF-induced chromatin condensation
Here, we propose a three-dimensional mechanochemical model for the effect of compressive loads on chromatin condensation. Supplemental Figure SC1 shows different components of the proposed model, including 1) the cytoskeleton, 2) the nucleus, and 3) the focal adhesions. Figure 5 depicts our results for a polarized cell on an adhesive micropatterned substrate with an area of 95 × 19 µm2 before (control) and after (load) applying vertical compressive forces.
FIGURE 5:
Simulation results of the mechanochemical model depicting the role of compressive forces on apical actin and nuclear morphology. Starting with isotropic contractility (the same concentration of the phosphorylated myosin motors in all directions), our model predicts an increase in the density of the phosphorylated myosin motors (density of force dipoles) along the long axis of the cell (i.e., polarized contraction). The polarized contraction results in higher tensile stresses along the cell’s polarization direction and subsequently formation of actin filaments along the long axis of the cell (A, C) as observed in our experiments (see Figure 4A). A compressive force of 2.5 µN causes a significant reduction in the cell contractility (a decrease in the density of phosphorylated myosin motors) in the apical regions as shown in F. This reduction in the contractility leads to a decrease in the tensile stresses in the apical regions and subsequently depolymerization of apical actin filaments as shown in B and D. Our model also predicts a decrease in cell stiffness associated with the depolymerization of apical actin filaments in response to the compressive force as depicted in H when compared to G.
Control.
The contractility of the cell is assumed to be initially isotropic as it is seeded on the micropattern. The stresses in the adhesion layer in response to the contractile forces exerted by the cell, in this case, are concentrated at the ends (Supplemental Figure SC1E). Subsequently, in response to these tensile forces at the ends, the stiffness of the connections to the substrate at the two ends increases representing the mature focal adhesion sites in agreement with our experiments (see Supplemental Figure SC1E). In turn, the mature focal adhesion sites (represented by the stiffened adhesion layer at the two ends) promote greater cell contractility and cell stiffening along the long axis of the cell. This positive feedback loop (mechanical cross-talk) between the cell contractility and the stiffening of the adhesion layer leads to the formation and alignment of stress fibers primarily along the long axis of the cell as shown in Figure 5A. The formation of these stress fibers provides physical links between the nucleus and focal adhesion sites. As depicted in Supplemental Figure SC3, our model also predicts high values of tensile stress along the polarization direction (z-direction) immediately above the nucleus in the cytoskeleton (apical regions). These tensile stresses lead to cytoskeletal stiffening along the long axis of the cell in our model representing the formation of apical stress fibers along the polarization direction as experimentally observed in Figure 4. Furthermore, our simulations show compressive stresses in the vertical direction (y-direction in Figure 5) in the apical regions. Consistent with our experiments and earlier study (Li et al., 2014), such vertical compressive stresses in our model show that the apical stress fibers form an arch above the nucleus and exert an active compressive load on it.
Load.
To study the effect of compressive forces on cell morphology, a vertical compressive force of 2.5 µN is applied to the top of the contractile cell by using a rigid flat plate. To include the effect of chromatin condensation in response to the compressive force, we also decrease the nuclear pressure (applied uniformly on the internal nuclear membrane) from 0.5 to 0.3 kPa (chromatin decondensation generates greater pressure on the nuclear membrane) (Mazumder et al., 2008). As reported in our previous study (Mazumder et al., 2008), the pressure applied on the nuclear membrane due to chromatin decondensation can be experimentally estimated using an atomic force microscope cantilever, where the pressure at which the nuclear scaffold ruptured (due to chromatin decondensation by enzymatic cleavage of histone tails) was reported to be 3 kPa. The measured pressure in these experiments ranges from ∼0.1 to 3 kPa during the nuclear expansion (nuclear swelling due to trypsin digestion). Note that the reported value of 3 kPa is the maximum pressure generated on the nuclear membrane (due to chromatin decompaction) after which the nucleus disintegrates. Therefore, the values of pressure used in our simulations for before (control) and after (load) chromatin condensation fall in the range of our previous measurements. Consistent with our experimental results in Supplemental Figure SE4, our model predicts a reduction in nuclear height from 4.8 to 3.6 µm in response to the compressive force 2.5 µN. Furthermore, this compressive force reduces the mean level of contractility, 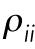 , in our model as experimentally shown in Figure 4, C and D (pMLC level decreases in the apical regions). This CF-induced reduction in cell contractility is visualized in Figure 5, E and F, where the maximum principal contractility
, in our model as experimentally shown in Figure 4, C and D (pMLC level decreases in the apical regions). This CF-induced reduction in cell contractility is visualized in Figure 5, E and F, where the maximum principal contractility 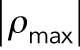 significantly decreases in the apical regions on applying the compressive force. Furthermore, we find that the compressive stresses applied on the top surface of the cells lead to the dissociation of the apical stress fibers as the magnitude of the maximum tensile principal stress decreases in the apical regions (Figure 5, B and D). Note that the vertical compressive force 2.5 µN causes a significant drop in tension along the cell’s polarization direction in the apical regions (Supplemental Figure SC3) where the stress fibers are depolymerized below a critical value of tension. Our model also predicts a reduction in cell stiffness associated with the depolymerization of apical actin in response to the compressive force (see Figure 5H). This reduction in cell stiffness agrees with the experimental studies in Rotsch and Radmacher (2000) and Haase and Pelling (2015), where drastic decreases in cell stiffness are observed on drug-induced actin depolymerization. Supplemental Movie 3 shows an animation of the actin depolymerization on applying the compressive force in our simulations.
significantly decreases in the apical regions on applying the compressive force. Furthermore, we find that the compressive stresses applied on the top surface of the cells lead to the dissociation of the apical stress fibers as the magnitude of the maximum tensile principal stress decreases in the apical regions (Figure 5, B and D). Note that the vertical compressive force 2.5 µN causes a significant drop in tension along the cell’s polarization direction in the apical regions (Supplemental Figure SC3) where the stress fibers are depolymerized below a critical value of tension. Our model also predicts a reduction in cell stiffness associated with the depolymerization of apical actin in response to the compressive force (see Figure 5H). This reduction in cell stiffness agrees with the experimental studies in Rotsch and Radmacher (2000) and Haase and Pelling (2015), where drastic decreases in cell stiffness are observed on drug-induced actin depolymerization. Supplemental Movie 3 shows an animation of the actin depolymerization on applying the compressive force in our simulations.
Movie S3.
Spatio-temporal visualization of actin stress fibres and the nucleus during the cell attachment and compressive loading using simulation results.
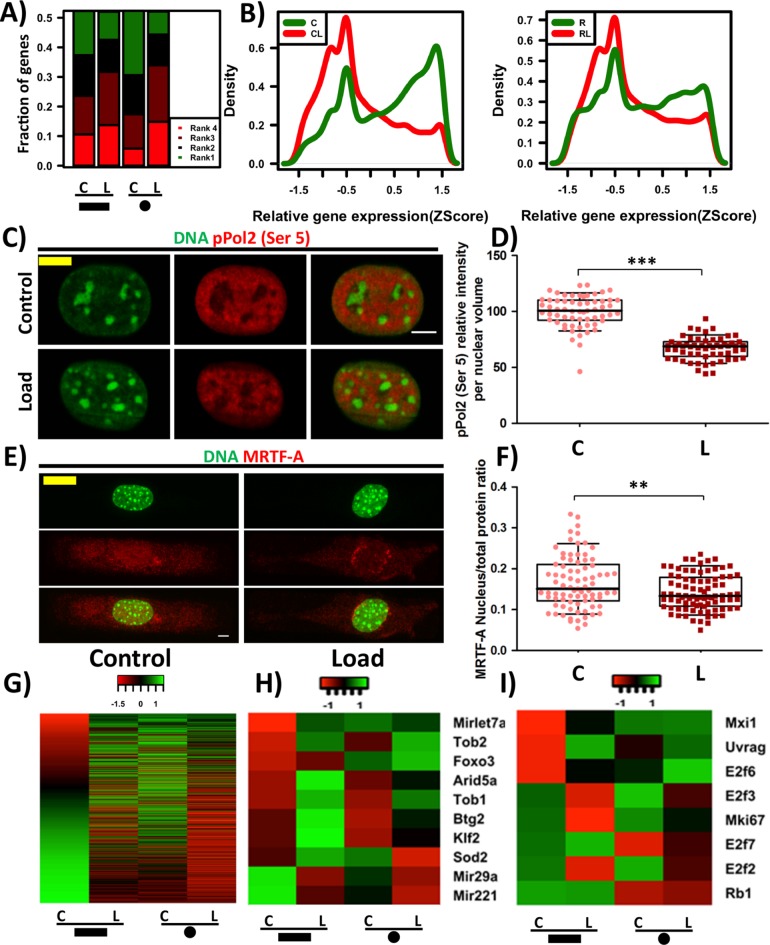
Transcriptome analysis reveals compressive force reduces transcriptional activity and induces cellular quiescence
To characterize the transcriptional changes as a result of compressive force–induced large-scale changes to chromatin organization, we quantified whole genome transcription profile of rectangular and circular cells exposed to and in the absence of compressive force using RNA Seq. Interestingly, we find that there is a large-scale reduction in the relative gene expression levels in response to compressive force in cells of both geometries (Figure 6, A and B). Consistent with this, there is a decrease in the levels of phosphorylated RNAPol2 (Ser5), a marker of transcription initiation, in the nucleus after cells experience compressive force (Figure 6, C and D). This suggests that cells in response to compressive force enter a transcriptionally less active state. Consistent with the previous observations (Figure 4), there is a reduction in the expression of genes regulating actin filament assembly and actin contractility (Supplemental Figure SE13).
FIGURE 6:
Compressive force–induced chromatin condensation results in reduced transcriptional activity and cellular quiescence. (A) A stacked bar plot that quantifies the frequency of the gene expression ranks in the various samples. Rank1 represents highest expression and Rank 4, the least for a particular gene. Note: This is a mere ranking of z-score values and the ranks do not indicate significant differences in the gene expression. (B) Probability density plots of relative expression of all genes in the genome, in rectangular and circular cells (with and without load). (C) Single cross-sectional slice of the nucleus. DNA stained with DAPI (green) and phosphorylated Pol2 (pS5) IF staining (red). (D) Relative phosphorylated Pol2 (pS5) intensity levels per nuclear volume based on IF staining. N: C# = 61, L# = 59. (E) Maximum intensity projection of confocal images of cell. DNA stained with DAPI (green) and IF staining of MRTF-A (red). (F) Nuclear to total protein ratio of MRTF-A based on IF staining. N: C# = 81, L# = 85. (G) Heat map for MRTF-A target genes. Heat map for quiescence- (H) and proliferative- (I) related genes. Whisker box plotted from 10 to 90 percentile. Student’s t test, ***p < 0.0001, **p < 0.01. Scale bar: 5 μm. #C = control and L = load.
MRTF-A, a transcription cofactor for serum response factor (SRF), is known to be retained in the cytoplasm when it is bound to G-actin. Hence, we hypothesized that, in response to compressive force–induced actin depolymerization, MRTF-A will shuttle to the cytoplasm and result in reduced transcription of its target genes. We indeed observed that there was a decrease in the nuclear to total protein ratio of MRTF-A in response to the compressive force resulting in decreased MRTF-A concentration in the nucleus (Figure 6, E and F, and Supplemental Figure SE14). As a result, we find a reduction in the expression of several MRTF-A-dependent SRF target genes (Figure 6G). In addition, we confirm the down-regulation of the SRF target gene α-SMA (Acta-2) by reverse transcription PCR (Supplemental Figure SE15).
This is also observed when a cell enters into quiescent state under serum-deprived conditions. In line with this, we find that the expression of quiescent characteristic genes Tob1, Tob2, Arid5a, and Btg2 (Tzachanis et al., 2001; Coller et al., 2006; Anwar et al., 2018) and transcription factors Klf2 (Buckley et al., 2001) and Foxo3 (Gopinath et al., 2014) increased in both geometries in response to compressive force. In addition, the expression of miR29a and miR221, microRNA genes reported to be down-regulated during quiescence (Medina et al., 2008; Suh et al., 2012) also reduced in response to compressive force (Figure 6H). Additionally, we find a reduction in the expression of cell proliferative marker Mki67 (Scholzen and Gerdes, 2000) along with E2f2 and E2f3, markers for DNA synthesis. Consistently, we find an increase in the DNA synthesis inhibitors E2f6 and E2f7 along with an increase in the expression of cell-cycle inhibitors such as Mxi1 (Taj et al., 2001) and Uvrag (Figure 6I). Several reports have indicated a reduction in Lamin B1 levels when a cell undergoes senescence (Freund et al., 2012; Ivanov et al., 2013). We observe a decrease in Lamin B1 levels in both the geometries (Supplemental Figure SE17). In line with this, we also see a decrease in Lamin B1 using immunofluorescence staining (Supplemental Figure SE17). Collectively, these results suggest that compressive force leads to cells in transcriptionally less active, quiescent, and senescent states.
Cell geometry modulates transcriptional response to compressive force
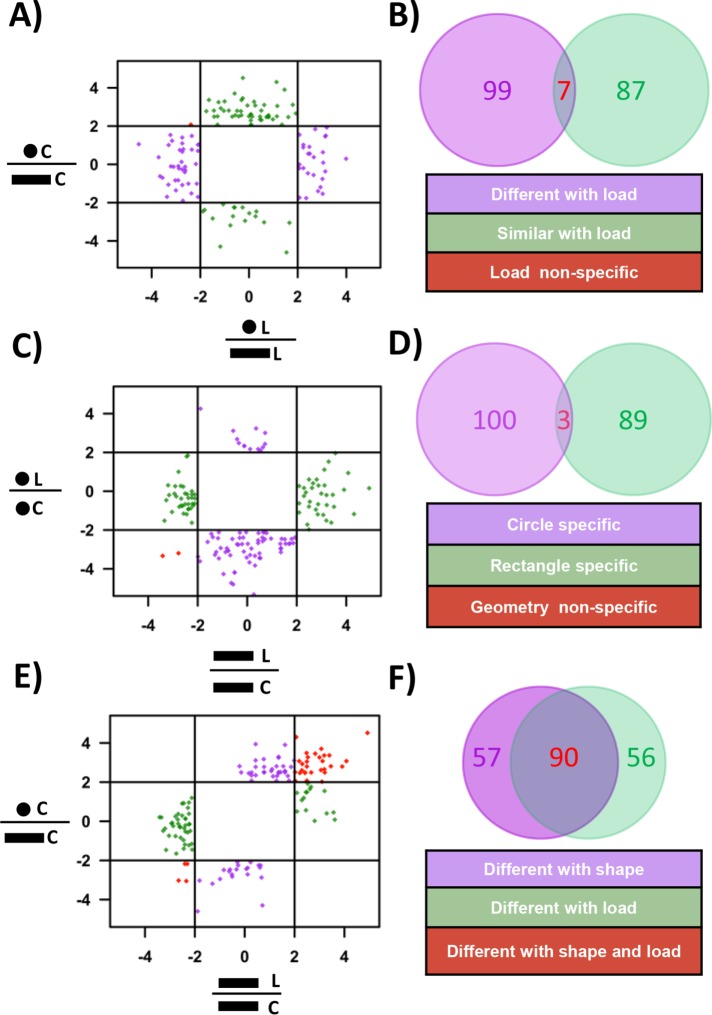
We next examined whether cell geometry plays a role in regulating transcriptional response to compressive forces. As reported earlier (Mitra et al., 2017), the transcription profile of circular and rectangular cells were different (Supplemental Figure SE16A). To identify similarities between the expression patterns of genes in the two geometries with and without compressive force, genes whose expression is more than fourfold different between rectangular and circular cells unique to control and CF conditions were calculated. We find ∼100 genes whose expression is different in control cells but are similar after compressive force and vice versa (Figure 7, A and B, and Supplemental Section SD).
FIGURE 7:
Geometry specific global gene expression changes in response to compressive force. (A) Plot between the expression ratio log2 (fold change) of genes between circular and rectangular cells with and without exposure to compressive force. (B) Venn diagram representing the number of genes that are either similar or different with load. (C) Plot between the expression ratio log2 (fold change) of genes with and without exposure to compressive force in circular and rectangular cells. (D) Venn diagram representing the number of genes whose expression is unique to either of the two geometries. (E) Plot between the gene expression ratio log2 (fold change) of genes before and after applying compressive force on rectangular cells and expression ratio between circular and rectangular cells. (F) Venn diagram representing the number of genes whose expression is unique to compressive force or changing geometries. C = control and L = load.
We next looked at the genes that were differentially altered in both the geometries in response to compressive force. Exposure to compressive force led to a differential expression of 182 genes in rectangles and 193 genes in circles (Supplemental Figure SE16A). Consistent with the previous observations, most of the genes that were altered in response to compressive force were found to be down-regulated (Supplemental Figure SE16, B and C). Genes that are differentially expressed in response to compressive force in a geometry specific manner were identified as those genes whose expression ratio with and without compressive force is more than fourfold in one geometry and not the other. We find that genes which have the highest change in expression are highly specific to the cell geometry with a few geometry-independent genes (Figure 7, C and D, and Supplemental Section SD). These results show that there are subset of genes whose expression changes in a geometry specific manner.
Since changing both the shape of the cell from a rectangular cell to a circular cell and applying compressive force results in cells with lower actomyosin contractility, we tested the effects of these two conditions in transcriptional control. Geometry and compressive force specific genes are identified as those that show more than fourfold difference in their expression ratio in one condition and not the other. We find ∼50 genes that are distinct to either changing shape or compressive force and around 90 genes that are common to both conditions (Figure 7, E and F, and Supplemental Section SD). These results show that reduction in actomyosin contractility by two different methods results in differential transcriptional response. Collectively, these observations demonstrate the importance of cell geometry in modulating the transcriptional response to compressive forces.
DISCUSSION
Intrinsic cellular states of fibroblasts such as cell geometry, nuclear morphology, and transcription are set up by the local mechano-chemical signals in the connective tissue. Owing to localized variations in the balance of these signals, fibroblasts in different regions of the tissue exhibit different cell geometry. In addition, they also experience extrinsic forces such as compressive force that further regulate their behavior. These mechanical forces have been shown to govern cellular contractility, chromatin condensation states, and thereby differential gene expression. However, these studies have been done on a population of cells with heterogeneous cell geometries with the assumption that population means are descriptive of cellular phenomena. Recent studies have challenged this by demonstrating that cells of different geometries differentially modulate nuclear morphology, chromosome organization, and, importantly, transcription regulation (Versaevel et al., 2012; Jain et al., 2013; Wang et al., 2017). However, the importance of cell geometry in determining cellular response to extrinsic signals is relatively unknown. In this study, we micro patterned fibroblasts into two extreme geometries (polarized and isotropic) and looked at their response to compressive force.
We demonstrate that independent of the cellular mechanical state, exposure to compressive force results in global chromatin condensation that is reversible on removal of CF. The bulk increase in chromatin condensation was found to occur at the pericenteromeric heterochromatin nodes as marked with CENP-A and enriched with H3K9me3. In line with recent findings, we find that the increase in the individual heterochromatin node volume was a result of both heterochromatin spreading as well as fusion of adjacent heterochromatin nodes (Larson et al., 2017; Strom et al., 2017). The increase in the heterochromatin spreading was facilitated by a decrease in H3K9ac (euchromatin marker), which was in turn mediated by nuclear enrichment of HDAC3 in response to CF. The importance of HDAC3 in this process was further elucidated by the inhibition and a decrease in the degree of CF-induced chromatin condensation in the presence of TSA (HDAC inhibitor) and on down-regulation of HDAC3 (siRNA), respectively. Consistent with CF-induced global chromatin condensation, we find that in response to CF, cells independent of their geometry enter into a transcriptionally less-active state as seen by the large-scale reduction in the gene expression levels and global reduction in active pol2 levels in response to compressive force. Earlier reports have also shown that formation of hyper chromatin condensed regions due to hyperosmotic pressure (Irianto et al., 2013) leads to relocalization of RNA pol2 to interchromatin regions and at the border of hyper chromatin condensed regions (Albiez et al., 2006). Additionally, exposure to CF results in depolymerization of actin filaments and global reduction in actomyosin contractility. Interestingly, in rectangular cells we find a preferential depolymerization of apical actin. In line with this, we find that MRTF-A which binds to g-actin was found to be enriched in the cytoplasm in response to CF. Consequently, there was a decrease in the expression levels of MRTF-A target genes and that of positive regulators of actin polymerization, which is a characteristic feature of cells under serum deprivation. Actin cytoskeletal structure and chromatin condensation state are very important determinants of nuclear morphology (Li et al., 2014; Mazumder et al., 2008; Ramdas et al., 2015). Hence, as a result of CF-induced actin depolymerization and chromatin condensation, there was a change in the nuclear morphology, notably a decrease in nuclear volume and height. Further, global reduction in actomyosin contractility in response to CF was found to be the driving force for the nuclear enrichment of HDAC3 and subsequent increase in chromatin condensation as shown by the Y-27632 experiment.
Further, we developed a mechanochemical model that captures all the essential features of the experiment, that is, the polarization of cells and the formation of actin stress fibers on micropatterned substrates, the depolymerization of actin on compressive force and finally the changes in nuclear prestress arising due to increased chromatin condensation. Our model also does well in predicting changes in actomyosin contractility for cells on circular geometry and on Y-27632 treatment of polarized cells. When the cells are polarized, the nucleus is highly flattened by both the actomyosin contractility and the apical stress fibers. The internal pressure of the nucleus is balanced by both the cytoskeletal and chromatin condensation forces (Mazumder et al., 2008; Mazumder and Shivashankar, 2010; Li et al., 2014). The application of compressive forces results in altering this force balance due to reduced contractile forces and increased chromatin condensation.
Since the geometric state of cells dictates the spatial organization of the chromatin, a critical determinant of gene expression, we conjectured that downstream transcriptional response may be different between cells of two different geometries. Indeed, we found that the transcriptional profiles of cells in two geometries states with different three-dimensional chromosome architecture (Wang et al., 2017), are distinct from each other as seen by ∼200 genes that are geometry specific and this is in line with our previous study (Jain et al., 2013). In tissues, since individual cells have to integrate mechanical and biochemical signals to regulate their cellular responses, we hypothesize that the transcriptional response to these signals will depend on the cell geometric state. Such cell geometry-dependent transcriptional response to the inflammatory cytokine has recently been described (Mitra et al., 2017). In this study, we explored whether mechanical forces such as compression can elicit differential transcriptional responses based on cell geometry. Incredibly, we found that the genes, whose expression undergoes large-scale change in response to CF, are geometry dependent with a handful of genes showing geometry-independent response. Additionally, such changes to the transcriptional profile of rectangular and circular cells could either drive the aforementioned geometry-specific genes to become similar after CF or it could lead to the geometry-independent genes to become similar after CF, and, intriguingly, we observed both. Collectively, these results indicate a complex relationship among cell geometry, gene expression, and transcriptional response to CF.
Such differential transcription profiles of genes in response to CF in the two geometries resulted in modular changes in the transcriptional activity of certain functional pathways. We find that in response to CF, the expression profile of cells in both geometry indicate that the cells are entering quiescence. This is supported by the increase in the expression of quiescence marker genes and cell-cycle inhibitors and a decrease in cell proliferation genes. This is further supported by the geometry-independent decrease in the expression of genes participating in the AKT-mTOR pathway, which indicates a reduction in cell metabolic activity. Further, our study also shows that fibroblasts may enter into senescence, which has implications in pressure ulcer. Patient-derived fibroblasts from pressure ulcer regions show replicative senescence much earlier when compared with fibroblasts from the adjacent normal region (Vande Berg et al., 1998, 2005). Studies have reported that autophagy is activated during quiescence and is an important characteristic of serum starvation-induced cell-cycle arrest, but it is not essential for survival of quiescent fibroblast (Valentin and Yang, 2008). Here we see an up-regulation of autophagy genes (Munson and Ganley, 2015) in response to compressive force, but surprisingly, only in rectangular cells but not circular cells.
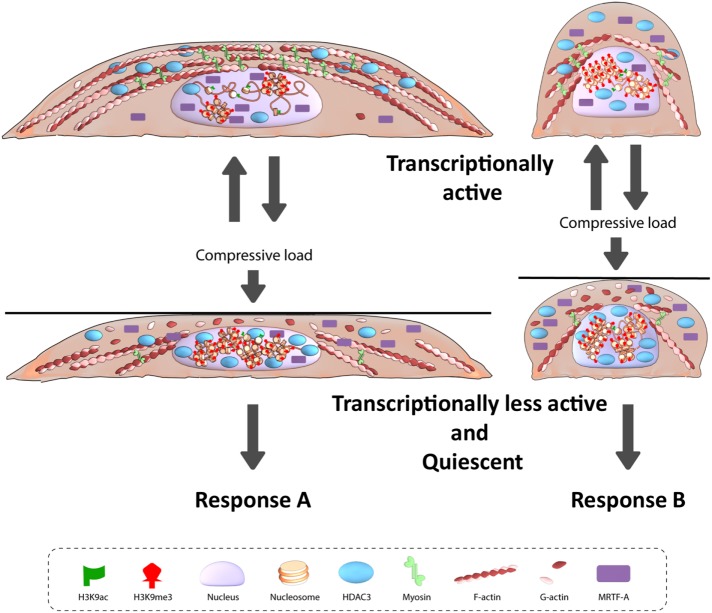
Taken together, our results demonstrate that static CF induces actin depolymerization-driven nuclear entry of HDAC3, which results in the global increase in chromatin condensation levels. Extraordinarily, we find that cells with different geometries and three-dimensional chromosome organization have different transcriptional response to the same CF (model summarized in Figure 8). We hypothesize that in cells with different cell geometries different regions of the genome are compacted in response to CF and this in combination with the differential nuclear-cytoplasmic localization of transcription factors give rise to geometry-dependent differential transcriptional output. Collectively, our observations highlight the coupling between the intrinsic cellular properties such as cellular mechanical state, chromatin condensation states, and transcriptional output with the extrinsic mechanical signal, compression that results in context-dependent gene expression.
FIGURE 8:
Model for geometry-dependent response to compressive force.
MATERIALS AND METHODS
Cell culture and pharmacological treatments
NIH3T3 (CRL-1658) cells were obtained from the American Type Culture Collection. They were cultured in DMEM (ThermoFisher Scientific 11885092) supplemented with 10% FBS (ThermoFisher Scientific 16000044) and 1% pen-strep (Sigma P4333) antibiotic. For Y-27632 experiments, cells were treated with 100 µM of Y-27632 (Sigma Y0503-1MG) for 1 h. For TSA experiments, cells were treated with 3 µM TSA (Sigma T8552-1MG) for 24 h, and the compressive force assay was performed in the presence of TSA. For experiments involving measurements of nuclear morphometry, cells were serum starved using DMEM + 1% serum overnight prior to the experiments. Cell culture and experiments were all performed in 37°C, 5% CO2, and humid conditions.
Micro contact printing
Fibronectin micropatterning was performed as described previously (Makhija et al., 2016). Briefly, rectangular fibronectin (Sigma F1141-2MG) micropatterns (area = 1800 µm2 and aspect ratio = 1:5) were made on uncoated Ibidi dishes (81151). These micropatterned dishes were then passivated with 0.2% pluronic acid (Sigma P2443) for 10 min and washed with phosphate-buffered saline (PBS).
Compressive force assay
NIH3T3 cells were trypsinized (ThermoFisher Scientific 15400054) and 7000 cells were seeded onto each of these fibronectin micropatterned dishes and allowed to spread for 3 h. A pluronic acid–treated coverslip was then placed on the cells. CF was applied on cells for 1 h by placing parafilm-wrapped metallic nuts (mass = 2 g) on the coverslip. This corresponds to each NIH3T3 fibroblast experiencing a force in the order of micronewtons (calculation shown in the Supplemental Methods). For recovery, the weight was removed and the cells were allowed to recover for 1 h before being fixed for immunofluorescence staining.
Immunofluorescence staining
After treating cells with cytokine or compressive force, cells were fixed with 4% paraformaldehyde (Sigma; 252549-500 ml) for 10 min. For compressive force samples, cells were fixed in the presence of the load. The load and coverslip were removed after the fixation step. Cells were then washed with PBS and permeabilized with 0.5% triton (Sigma; X100-100 ml) for 10 min and with PBS washes in between. The cells were then blocked with 5% bovine serum albumin for 30 min and incubated with primary antibody as per the manufacturer’s instructions. After secondary antibody incubation, the cells were labeled with 4′,6-diamidino-2-phenylindole (DAPI) solution (ThermoFisher Scientific R37606) in PBS as recommended by the supplier. Each experiment was performed at least three times, and the measurements were combined together and plotted unless stated otherwise.
Antibodies.
CENPA (Cell Signalling Technology 2048S), H3K27me3 (EMD Millipore 07-449), H3K9me3 (Abcam ab8898), H3K9ac (Abcam ab4441), HP1α (Abcam ab109028), HDAC3 (Cell Signalling Technology 3949S), pMLC-2 (Ser 19), pPol2 (Abcam ab5131), MRTF-A (Santa Cruz Biotechnology sc-21558), and Lamin B1 (Abcam ab16048).
HDAC3 knockdown experiment
siRNA-mediated knockdown of HDAC3 was performed as per manufacturer’s instructions. Cells (2 × 105) were seeded in each well of a six-well plate. After 18 h, cells were transfected with 80 pmol of control siRNA (Santa Cruz Biotechnology sc37007) and HDAC3 siRNA (Santa Cruz Biotechnology sc35539). Transfection was performed using siRNA transfection reagent (Santa Cruz Biotechnology sc29528) and siRNA transfection medium (Santa Cruz Biotechnology sc36868). After 24 h, the transfection medium was replaced with fresh culture medium, and the cells were further incubated for 48 h (72 h posttransfection). Cells were then used for compressive force assay.
Imaging
Images were obtained at single-cell resolution using Nikon A1R confocal microscope. Z-stack images were captured using a 100 × objective with a pixel size of 0.1-µm and 0.5-µm depth.
Real-time PCR
Changes in α-SMA gene expression in response to CF was measured using real-time PCR. Briefly, RNA was extracted using RNeasy Mini kit (Qiagen). cDNA was obtained using iScript cDNA Synthesis kit (Bio-Rad). Quantitative PCR was performed using SsoFast qPCR (Bio-Rad) for 40 cycles in a Bio-Rad CFX96. Cq value obtained were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and used to plot relative expression level with respect to control.
α-SMA primers
Forward: 5′-GCT GAC AGG ATG CAG AAG GAG A-3′
Reverse: 5′-CGT CGT ATT CCT GTT TGC TGA T-3′
GAPDH primers
Forward: 5′-CAG GCA TCT GAG GGC CCA CTG AA-3′
Reverse: 5′-CGT ATT CAT TGT CAT ACC AGG AA-3′
Analysis
Measuring nuclear morphometric features.
All the measurements were done using FijiImageJ (Schindelin et al., 2012). Measurement for chromatin condensation is shown in the Supplemental Methods. For other nuclear morphometric features, the nucleus is first thresholded using the Otsu method. The height is measured as the number of slices multiplied by 0.5-µm Z-depth. Nuclear projected area is measured using Z-projected images where the number of pixels is multiplied by 0.01 µm2 (pixel size = 0.1 µm). For volume measurements, the area of the nucleus in each slice is multiplied by 0.5-µm Z-depth. The total volume is then measured by adding all the slices. The relative change is then calculated and plotted for these parameters by setting the mean value of control nuclei equal to 100%.
Immunofluorescence intensity measurements.
The total intensity is measured for each protein in their respective channel. The nuclear fraction in nuclear to total protein ratio measurements is obtained by creating a mask for nucleus using the DAPI channel and then using this mask to measure the total intensity in the corresponding channel of the protein of interest. The relative change is then calculated and plotted for these parameters by setting the mean value of control nuclei equal to 100%.
RNA Seq.
RNA-seq libraries were sequenced in HiSeq 2000 platform at the Genome Institute Singapore. The Sequence Quality was accessed by FASTQC (Andrews, 2010). These reads were of high quality with a Phred score of above +27 and length of 70 bases. For the assembly of RNA-seq reads into the genome TopHat (Kim et al., 2013) was used and the number of aligned sequences to a transcript was quantified using HT-Seq (Anders et al., 2015) based on the Ensemblev79 for Mus musculus. These steps were carried out in Galaxy (Afgan et al., 2016). Other statistical analyses were carried out in R (R Development Core Team, 2008). The read counts were normalized by calculating the transcript per kilobasepair per million (TPKM).
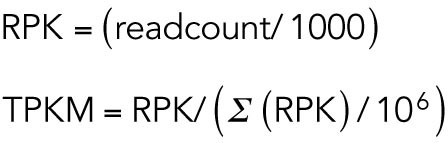 |
Other statistical analysis was carried out in R (R Development Core Team, 2008). Pathway and transcription factor target genes were obtained from gene set data for pathway analysis in mouse (Bares and Ge, 2015). For each pathway or transcription factor, the sum of all participating genes in each sample was used to plot heat maps.
Statistical test
Student’s unpaired two-tailed t test was performed.
Supplementary Material
Acknowledgments
We thank the Mechanobiology Institute, National University of Singapore, Singapore, and the Ministry of Education Tier-3 Grant Program for funding. The theoretical work was supported by National Cancer Institute awards U01CA202177 and U54CA193417 (to V.B.S.), National Institute of Biomedical Imaging and Bioengineering award R01EB017753 (to V.B.S.), and the NSF Center for Engineering Mechanobiology (CMMI-154857). We also thank Melanie Lee Wei Ting for the illustration.
Abbreviations used:
- α-SMA
alpha-smooth muscle actin
- CENP-A
centromeric protein A
- CF
compressive force
- DAPI
4′,6-diamidino-2-phenylindole
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- HC
heterochromatin content
- HDAC
histone deacetylase
- HDAC3
histone deacetylase 3
- HPlα
heterochromatin protein lα
- IF
immunofluorescence
- MRTF-A
myocardin-related transcription factor A
- PBS
phosphate-buffered saline
- pMLC
phosphorylated myosin light chain
- RNAPol2
RNA polymerase II
- ROCK
rho-associated protein kinase
- SRF
serum response factor
- SUV39H1
suppressor of variegation 3-9 homologue 1
- TSA
trichostatin A
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-04-0256) on September 26, 2018.
REFERENCES
- Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Eberhard C, et al (2016). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res , W3–W10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albiez H, Cremer M, Tiberi C, Vecchio L, Schermelleh L, Dittrich S, Kupper K, Joffe B, Thormeyer T, von Hase J, et al (2006). Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chromosome Res , 707–733. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. (2015). HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics , 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. (2010). FastQC: a quality control tool for high throughput sequence data, version 0.11.4. Available online at www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed April 2017). [Google Scholar]
- Anwar T, Sen B, Aggarwal S, Nath R, Pathak N, Katoch A, Aiyaz M, Trehanpati N, Khosla S, Ramakrishna G. (2018). Differentially regulated gene expression in quiescence versus senescence and identification of ARID5A as a quiescence associated marker. J Cell Physiol , 3695–3712. [DOI] [PubMed] [Google Scholar]
- Bares V, Ge X. (2015). gskb: Gene Set data for pathway analysis in mouse. R package version 1.8.0.
- Brangwynne CP, MacKintosh FC, Kumar S, Geisse NA, Talbot J, Mahadevan L, Parker KK, Ingber DE, Weitz DA. (2006). Microtubules can bear enhanced compressive loads in living cells because of lateral reinforcement. J Cell Biol , 733–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley AF, Kuo CT, Leiden JM. (2001). Transcription factor LKLF is sufficient to program T cell quiescence via a c-Myc-dependent pathway. Nat Immunol , 698–704. [DOI] [PubMed] [Google Scholar]
- Butcher DT, Alliston T, Weaver VM. (2009). A tense situation: forcing tumour progression. Nat Rev Cancer , 108–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Wei X, Wang S, Jiao Q, Zhang Y, Du G, Wang X, Wei F, Zhang J, Wei L. (2016). Compression regulates gene expression of chondrocytes through HDAC4 nuclear relocation via PP2A-dependent HDAC4 dephosphorylation. Biochim Biophys Acta , 1633–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chooi WH, Chan BP. (2016). Compression loading-induced stress responses in intervertebral disc cells encapsulated in 3D collagen constructs. Sci Rep , 26449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller HA, Sang L, Roberts JM. (2006). A new description of cellular quiescence. PLoS Biol , e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edsberg LE, Natiella JR, Baier RE, Earle J. (2001). Microstructural characteristics of human skin subjected to static versus cyclic pressures. J Rehabil Res Dev , 477–486. [PubMed] [Google Scholar]
- Freund A, Laberge RM, Demaria M, Campisi J. (2012). Lamin B1 loss is a senescence-associated biomarker. Mol Biol Cell , 2066–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath SD, Webb AE, Brunet A, Rando TA. (2014). FOXO3 promotes quiescence in adult muscle stem cells during the process of self-renewal. Stem Cell Rep , 414–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase K, Pelling AE. (2015). Investigating cell mechanics with atomic force microscopy. J R Soc Interface , 20140970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara M, Fujii T, Hashizume R, Nomura Y. (2014). Effect of strain on human dermal fibroblasts in a three-dimensional collagen sponge. Cytotechnology , 723–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo SJ, Han WM, Szczesny SE, Cosgrove BD, Elliott DM, Lee DA, Duncan RL, Mauck RL. (2016). Mechanically induced chromatin condensation requires cellular contractility in mesenchymal stem cells. Biophys J , 864–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo SJ, Thorpe SD, Driscoll TP, Duncan RL, Lee DA, Mauck RL. (2015). Biophysical regulation of chromatin architecture instills a mechanical memory in mesenchymal stem cells. Sci Rep , 16895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irianto J, Swift J, Martins RP, McPhail GD, Knight MM, Discher DE, Lee DA. (2013). Osmotic challenge drives rapid and reversible chromatin condensation in chondrocytes. Biophys J , 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A, Pawlikowski J, Manoharan I, van Tuyn J, Nelson DM, Rai TS, Shah PP, Hewitt G, Korolchuk VI, Passos JF, et al (2013). Lysosome-mediated processing of chromatin in senescence. J Cell Biol , 129–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N, Iyer KV, Kumar A, Shivashankar G. (2013). Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. Proc Natl Acad Sci , 11349–11354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmey PA, Miller RT. (2011). Mechanisms of mechanical signaling in development and disease. J Cell Sci , 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa T, Nakagami G, Minematsu T, Yamane T, Huang L, Mugita Y, Noguchi H, Mori T, Sanada H. (2014). Biological responses of three-dimensional cultured fibroblasts by sustained compressive loading include apoptosis and survival activity. PLoS One , e104676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanjanamekanant K, Luckprom P, Pavasant P. (2013). Mechanical stress-induced interleukin-1beta expression through adenosine triphosphate/P2X7 receptor activation in human periodontal ligament cells. J Periodontal Res , 169–176. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol , R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Mazzanti M, Mistrik M, Kosar M, Beznoussenko GV, Mironov AA, Garre M, Parazzoli D, Shivashankar GV, Scita G, et al (2014). ATR mediates a checkpoint at the nuclear envelope in response to mechanical stress. Cell , 633–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langevin HM, Nedergaard M, Howe AK. (2013). Cellular control of connective tissue matrix tension. J Cell Biochem , 1714–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, Narlikar GJ. (2017). Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature , 236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Berre M, Aubertin J, Piel M. (2012). Fine control of nuclear confinement identifies a threshold deformation leading to lamina rupture and induction of specific genes. Integr Biol (Camb) , 1406–1414. [DOI] [PubMed] [Google Scholar]
- Lee YH, Nahm DS, Jung YK, Choi JY, Kim SG, Cho M, Kim MH, Chae CH, Kim SG. (2007). Differential gene expression of periodontal ligament cells after loading of static compressive force. J Periodontol , 446–452. [DOI] [PubMed] [Google Scholar]
- Li Q, Kumar A, Makhija E, Shivashankar GV. (2014). The regulation of dynamic mechanical coupling between actin cytoskeleton and nucleus by matrix geometry. Biomaterials , 961–969. [DOI] [PubMed] [Google Scholar]
- Luboz V, Petrizelli M, Bucki M, Diot B, Vuillerme N, Payan Y. (2014). Biomechanical modeling to prevent ischial pressure ulcers. J Biomech , 2231–2236. [DOI] [PubMed] [Google Scholar]
- Madden RM, Han SK, Herzog W. (2015). The effect of compressive loading magnitude on in situ chondrocyte calcium signaling. Biomech Model Mechanobiol , 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhija E, Jokhun DS, Shivashankar GV. (2016). Nuclear deformability and telomere dynamics are regulated by cell geometric constraints. Proc Natl Acad Sci USA , E32–E40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder A, Roopa T, Basu A, Mahadevan L, Shivashankar GV. (2008). Dynamics of chromatin decondensation reveals the structural integrity of a mechanically prestressed nucleus. Biophys J , 3028–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder A, Shivashankar GV. (2010). Emergence of a prestressed eukaryotic nucleus during cellular differentiation and development. J R Soc Interface (Suppl 3), S321–S330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina R, Zaidi SK, Liu CG, Stein JL, van Wijnen AJ, Croce CM, Stein GS. (2008). MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Res , 2773–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra A, Venkatachalapathy S, Ratna P, Wang Y, Jokhun DS, Shivashankar GV. (2017). Cell geometry dictates TNFalpha-induced genome response. Proc Natl Acad Sci USA , E3882–E3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson MJ, Ganley IG. (2015). MTOR, PIK3C3, and autophagy: Signaling the beginning from the end. Autophagy , 2375–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofek G, Wiltz DC, Athanasiou KA. (2009). Contribution of the cytoskeleton to the compressive properties and recovery behavior of single cells. Biophys J , 1873–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramdas NM, Shivashankar GV. (2015). Cytoskeletal control of nuclear morphology and chromatin organization. J Mol Biol , 695–706. [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2008). R: A language and environment for statistical computing, Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. (2000). Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature , 593–599. [DOI] [PubMed] [Google Scholar]
- Rotsch C, Radmacher M. (2000). Drug-induced changes of cytoskeletal structure and mechanics in fibroblasts: an atomic force microscopy study. Biophys J , 520–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods , 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholzen T, Gerdes J. (2000). The Ki-67 protein: from the known and the unknown. J Cell Physiol , 311–322. [DOI] [PubMed] [Google Scholar]
- Shieh AC, Athanasiou KA. (2007). Dynamic compression of single cells. Osteoarthritis Cartilage , 328–334. [DOI] [PubMed] [Google Scholar]
- Shimomura K, Kanamoto T, Kita K, Akamine Y, Nakamura N, Mae T, Yoshikawa H, Nakata K. (2014). Cyclic compressive loading on 3D tissue of human synovial fibroblasts upregulates prostaglandin E2 via COX-2 production without IL-1beta and TNF-alpha. Bone Joint Res , 280–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH. (2017). Phase separation drives heterochromatin domain formation. Nature , 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh EJ, Remillard MY, Legesse-Miller A, Johnson EL, Lemons JM, Chapman TR, Forman JJ, Kojima M, Silberman ES, Coller HA. (2012). A microRNA network regulates proliferative timing and extracellular matrix synthesis during cellular quiescence in fibroblasts. Genome Biol , R121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taj MM, Tawil RJ, Engstrom LD, Zeng Z, Hwang C, Sanda MG, Wechsler DS. (2001). Mxi1, a Myc antagonist, suppresses proliferation of DU145 human prostate cells. Prostate , 194–204. [DOI] [PubMed] [Google Scholar]
- Tessarz P, Kouzarides T. (2014). Histone core modifications regulating nucleosome structure and dynamics. Nat Rev Mol Cell Biol , 703–708. [DOI] [PubMed] [Google Scholar]
- Thery M. (2010). Micropatterning as a tool to decipher cell morphogenesis and functions. J Cell Sci , 4201–4213. [DOI] [PubMed] [Google Scholar]
- Tse JM, Cheng G, Tyrrell JA, Wilcox-Adelman SA, Boucher Y, Jain RK, Munn LL. (2012). Mechanical compression drives cancer cells toward invasive phenotype. Proc Natl Acad Sci USA , 911–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzachanis D, Freeman GJ, Hirano N, van Puijenbroek AA, Delfs MW, Berezovskaya A, Nadler LM, Boussiotis VA. (2001). Tob is a negative regulator of activation that is expressed in anergic and quiescent T cells. Nat Immunol , 1174–1182. [DOI] [PubMed] [Google Scholar]
- Valentin M, Yang E. (2008). Autophagy is activated, but is not required for the G0 function of BCL-2 or BCL-xL. Cell Cycle , 2762–2768. [DOI] [PubMed] [Google Scholar]
- Vande Berg JS, Rose MA, Haywood-Reid PL, Rudolph R, Payne WG, Robson MC. (2005). Cultured pressure ulcer fibroblasts show replicative senescence with elevated production of plasmin, plasminogen activator inhibitor-1, and transforming growth factor-beta1. Wound Repair Regen , 76–83. [DOI] [PubMed] [Google Scholar]
- Vande Berg JS, Rudolph R, Hollan C, Haywood-Reid PL. (1998). Fibroblast senescence in pressure ulcers. Wound Repair Regen , 38–49. [DOI] [PubMed] [Google Scholar]
- Versaevel M, Grevesse T, Gabriele S. (2012). Spatial coordination between cell and nuclear shape within micropatterned endothelial cells. Nat Commun , 671. [DOI] [PubMed] [Google Scholar]
- Wang Y, Nagarajan M, Uhler C, Shivashankar GV. (2017). Orientation and repositioning of chromosomes correlate with cell geometry-dependent gene expression. Mol Biol Cell , 1997–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells RG. (2013). Tissue mechanics and fibrosis. Biochim Biophys Acta , 884–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Mouw JK, Weaver VM. (2011). Forcing form and function: biomechanical regulation of tumor evolution. Trends Cell Biol , 47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.