 A super-contrast NIR-II fluorophore IR-BEMC6P with enhanced quantum yield is developed and the excretion mechanism is identified.
A super-contrast NIR-II fluorophore IR-BEMC6P with enhanced quantum yield is developed and the excretion mechanism is identified.
Abstract
In vivo molecular imaging in the “transparent” near-infrared II (NIR-II) window has demonstrated impressive benefits in reaching millimeter penetration depths with high specificity and imaging quality. Previous NIR-II molecular imaging generally relied on high hepatic uptake fluorophores with an unclear mechanism and antibody-derived conjugates, suffering from inevitable nonspecific retention in the main organs/skin with a relatively low signal-to-background ratio. It is still challenging to synthesize a NIR-II fluorophore with both high quantum yield and minimal liver-retention feature. Herein, we identified the structural design and excretion mechanism of novel NIR-II fluorophores for NIR-II molecular imaging with an extremely clean background. With the optimized renally excreted fluorophore–peptide conjugates, superior NIR-II targeting imaging was accompanied by the improved signal-to-background ratio during tumor detection with reducing off-target tissue exposure. An unprecedented NIR-II imaging-guided microsurgery was achieved using such an imaging platform, which provides us with a great preclinical example to accelerate the potential clinical translation of NIR-II imaging.
Introduction
Biological imaging with fluorescence in the NIR-II region ranging from 1000 to 1700 nm (ref. 1–4) benefits from reduced body autofluorescence and light scattering, allowing for enhanced sub-centimeter penetration into biological systems.5–11 To achieve NIR-II molecular imaging with high contrast for preclinical/clinical use, both the fluorophore and conjugated targeting probe need to be renally excreted with low retention in normal tissues.12–14 Compared to antibodies and hormones, peptides provide high binding affinity, low immunogenicity and faster body clearance.15,16 So far, many kinds of peptides have been recognized and developed for specific tumor biomarkers,17 and over 150 peptides in active development have entered human clinical studies.18 For example, arginylglycylaspartic acid (RGD) motifs have been widely utilized as imaging-guided diagnostic probes,19–21 and radioactive isotope modified targeting peptides can reach high tumor-to-normal tissue (T/NT) ratios in positron emission tomography (PET) imaging.22,23
Fluorescent probes derived from specific peptides have been used clinically in NIR-I imaging-guided tumor surgery.24 Fluorescence imaging possesses an advantage in both spatial and temporal resolutions,25,26 and the bottleneck caused by penetration issues could be improved by imaging at longer wavelengths.27–30 Renally excreted conjugates in the NIR-II window profit from both the low uptake of normal tissue/immune systems and reduced tissue autofluorescence/scattering. As a result, NIR-II peptide conjugated dyes with high quantum yields and renal excretion ability afford great penetration depths and high signal/background ratios for preclinical and clinical settings. So far, NIR-II fluorophores have expanded to inorganic materials, polymer encapsulated organic dyes and water-soluble molecular fluorophores,29,31–47 which mostly suffer from high uptake and long-term retention in the immune system, causing considerable safety concerns in terms of clinical applications. It is of great importance to exploit bright NIR-II fluorophores with both high quantum yields and fast excretion ability.8,48–51
Although several NIR-II fluorophores with renal clearance ability have been exploited,16,35,46,52 they exhibit relatively low NIR-II quantum yields. Further, much less is known about the excretion mechanism in NIR-II fluorophores. It is generally considered that a small size less than the renal cutoff is one of the key factors to afford renal excretion ability.52–56 However, the experimental result showed that size is not the single determining factor that leads to such renal excretion.57 Other comparatively unexplored parameters limit the development of NIR-II fluorophores with renal excretion ability, and we lack a complete understanding of the mechanisms that contribute to renal and hepatobiliary excretion pathways.
Here, we screened and optimized a high-performance NIR-II fluorophore, IR-BEMC6P, with rapid renal excretion, minimal hepatic uptake and a relatively high quantum yield (QY) of 1.8%. By systematically comparing IR-BEMC6P and a series of NIR-II fluorophores, we found that a NIR-II fluorophore with renal excretion ability needs to possess a small size, fast dissociation with proteins, near-neutral functional groups, and low macrophage uptake. We next developed renally excreted targeting peptide-conjugated probes for in vivo NIR-II tumor imaging, exhibiting high tumor specificity and low off-target organ uptake. We further demonstrated that the IR-BEMC6P@peptide conjugate offered a competitive imaging quality to PET imaging with the same targeting peptide, allowing the possibility of NIR-II guided microsurgery. We expect that the renal excretion of NIR-II conjugates will accelerate NIR-II molecular imaging into clinical applications as new therapeutic agents.
Results and discussion
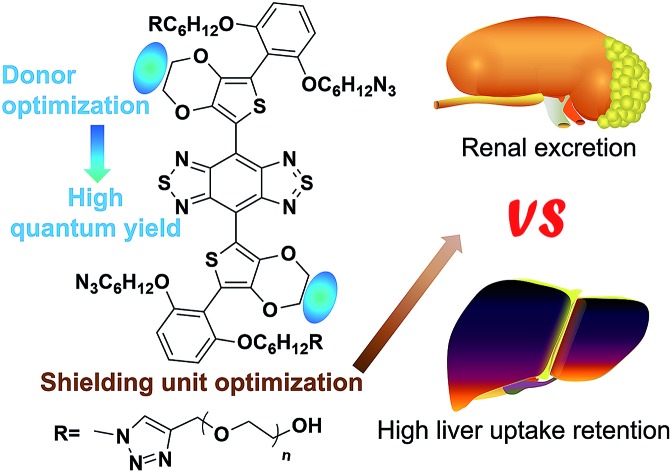
Rational design of minimal liver-uptake and super high-contrast NIR-II fluorophores with donor/shielding unit engineering of S-D-A-D-S molecules
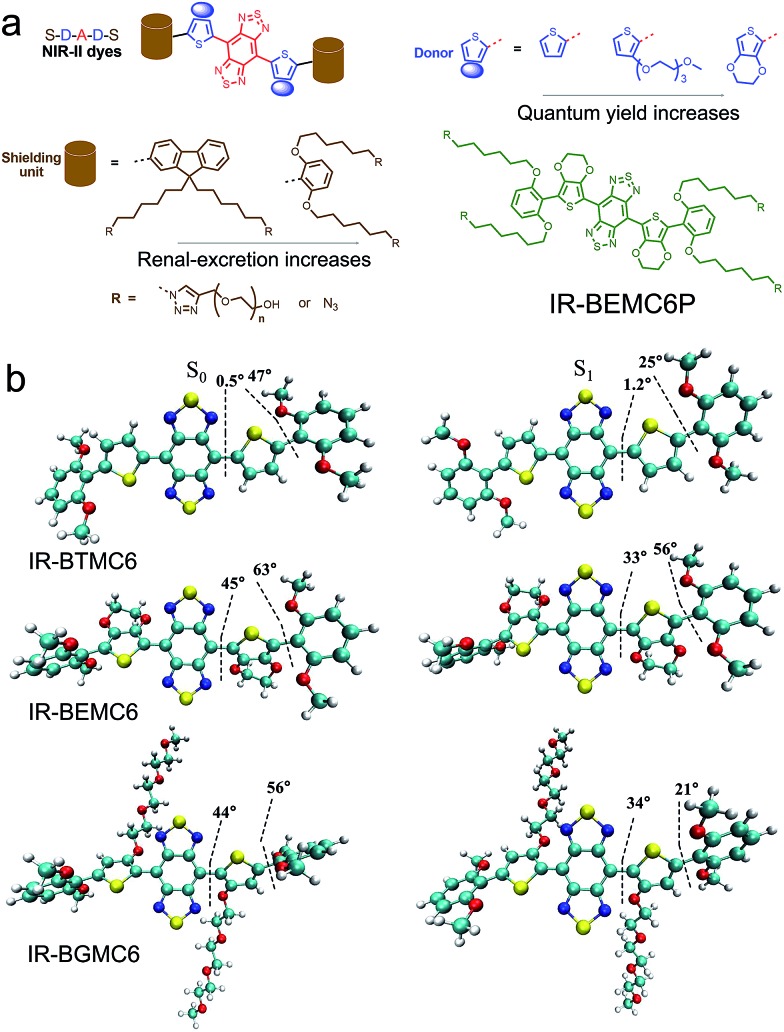
We first synthesized a new NIR-II molecular fluorophore, IR-BEMC6P, based on the shielding–donor–acceptor–donor–shielding (S–D–A–D–S) structure (Fig. 1a, see the synthesis optimization process and structural characterization in Fig. S1–S10†). Benzo[1,2-c:4,5-c′]bis([1,2,5] thiadiazole) (BBTD) was employed as the acceptor unit58 with 3,4-ethylenedioxythiophene (EDOT) substituted thiophene as the bridging donor unit (Fig. 1a). Compared to IR-BTMC6P with solely thiophene as the π unit, EDOT can distort the conjugated backbone and lower molecular interactions to reduce non-radiative transitions (Fig. 1b, Tables S1 and S2†).57 Dialkoxy substituted benzene modules were introduced as the shielding units, as the stretching dialkoxy chain on the conjugated backbone could prevent intermolecular interactions to enhance QY. Further, the dialkoxy substituted benzene shielding unit can enable renal excretion ability.52 The termini of the two side chains were modified by polyethylene glycol (PEG) for good biocompatibility, while the two side chains were functionalized with azide groups for bioconjugation. IR-BEMC6P showed an absorption peak at 725 nm, and its fluorescence peaked at 1025 nm and ranged from 900 to 1400 nm (Fig. S10†). The QY of IR-BEMC6P was determined to be 1.8% with HiPCO SWCNT as the reference fluorophore (Table S1†).59
Fig. 1. Design of a bright renal-excretion dye with shielding/donor group optimization. (a) BBTD serves as the acceptor unit with modified thiophene functioning as the bridging donor unit. For the shielding unit, dialkoxyl-benzene rather than dialkoxyl-fluorene endows NIR-II dyes with renal excretion ability. For donor groups, EDOT substituted thiophene affords improved quantum yield compared with solely thiophene. (b) Dihedral angles and molecular shapes from the simulated results of the NIR-II molecular fluorophores.
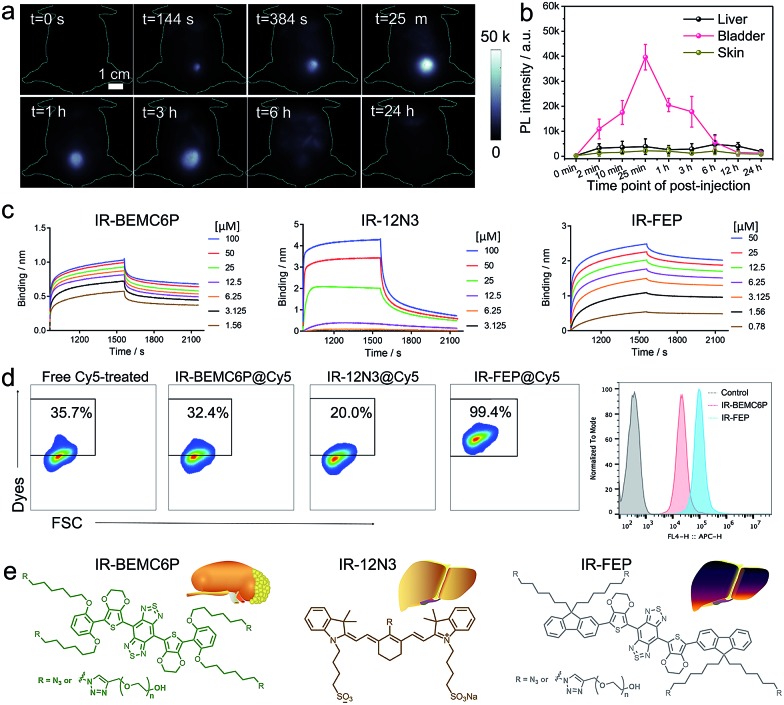
The screened dialkoxy substituted benzene shielding groups served as a key factor to endow the IR-BEMC6P with renal excretion ability. Although previously studied NIR-II fluorophores IR-FEP and IR-FGP with fluorene as the shielding group exhibited similar quantum yields,57,59 they lacked the renal excretion ability and suffered from high liver uptake and long retention (Fig. S11†). To confirm the excretion route of IR-BEMC6P, shaved CD-1 mice were intravenously injected with IR-BEMC6P at a dose of 1 mg kg–1 (Fig. 2a). The blood clearance of IR-BEMC6P was observed with 70% ID per g at 3 minutes post injection (p.i.) and decreased to <1% ID per g at 12 hours p.i. (Fig. S12a†). The accumulated signal of IR-BEMC6P in the bladder indicated a very fast renal clearance pathway (Fig. S12b†), affording the super clean skin/normal tissue background (Fig. 2b). Biodistribution at 24 h p.i. showed that most organs presented extremely low fluorescence signals (Fig. S13a†). As a result, IR-BEMC6P was proven to be a bright NIR-II imaging contrast agent with fast clearance behavior post-imaging. In vivo toxicity of IR-BEMC6P in the administrated mouse cohorts was evaluated by appearance (Fig. S13b†), hematology and serum biochemical analysis (Table S3†), as well as the pathology analysis of the main organs (Fig. S14†).15 Overall, the IR-BEMC6P fluorophore did not cause toxic reactions, demonstrating favorable biocompatibility in both research and clinical workflows.56,60,61
Fig. 2. The interaction between the NIR-II fluorophores and plasma protein, innate immune cells. (a) NIR-II imaging of the IR-BEMC6P injected mouse showed high bladder fluorescence signals at different p.i. time points. Injected dose: 1 mg kg–1. Imaging details: 1100 nm long pass filter, 808 nm laser. (b) Representative fluorescence signal intensity of the liver, bladder, and skin regions for IR-BEMC6P. (c) Kinetic binding assay of IR-BEMC6P, IR-12N3, and IR-FEP to albumin was measured by bio-layer interferometry. Long liver uptake dye (IR-FEP) has slower dissociating speed than short liver uptake dye (IR-12N3) and renal excretion dye (IR-BEMC6P). (d) Flow cytometry result of Cy5 labeled IR-BEMC6P, IR-12N3, and IR-FEP uptake by macrophage cells. IR-12N3 and IR-BEMC6P have much lower macrophage uptake than IR-FEP. (e) The chemical structure of renalexcretion and liver-uptake NIR-II fluorophores.
Excretion mechanism of NIR-II fluorophores
After observing that IR-BEMC6P is endowed with renal excretion ability, we further aimed to study the relationship between the chemical structure and excretion behavior of this type of S–D–A–D–S dye. Generally, small and hydrophilic molecules are mainly excreted via the kidneys, whereas large and amphipathic molecules are preferentially excreted via the liver.62 By additionally investigating the whole-body imaging of non-renal excreted IR-FGP and IR-FEP,57,59 we confirmed that the S–D–A–D–S dyes with fluorene groups suffered from long-term liver retention (Fig. S11†). With this preliminary excretion data for S–D–A–D–S dyes, we chose three types of NIR-II dyes, IR-BEMC6P with fast renal excretion, IR-12N3 with fast hepatobiliary clearance,63 and IR-FEP/IR-FGP with long liver retention, for further investigation.
To understand the excretion mechanism of these types of NIR-II dyes, we tested the size, protein binding affinity, and macrophage uptake of the screened NIR-II dyes. Previous experimental evidence suggested that a small size less than the renal cutoff (5 nm) was one of the most favorable factors to endow organic fluorophores with renal excretion ability.54 In the present study, the size of the renal excretion dye IR-BEMC6P was assuredly found to be less than 5 nm (Table S4†), compared to the high liver-uptake dye IR-FGP with a size of over 50 nm. However, the size of another high liver-uptake dye IR-FEP was also found to be less than 5 nm. The deviation of size-controlled excretion pathways indicates that other unexplored parameters rather than size, such as interaction with serum plasma and the immune system, have to be involved. Studying these parameters is likely essential to identify the excretion mechanism of the NIR-II fluorophores.
We then studied the interaction with the serum plasma and macrophage of these fluorophores. The dyes' binding affinities with albumin (the most abundant protein in plasma) were first tested. The Kd values of IR-BEMC6P and IR-FEP are 1.2 μM and 2.5 nM, respectively, indicating the stronger albumin binding for long-liver-retention dyes. Although IR-12N3 and IR-FGP also showed high binding affinities to albumin in the range of 1–30 μM, both IR-BEMC6P and IR-12N3 showed rapid de-binding behavior (sharp dissociation curves), while the long liver retention dyes IR-FGP/IR-FEP showed very slow de-binding ability (flat dissociation curves in Fig. 2c and S15†). This suggested that fast excretion dyes were primarily transported by serum proteins and could be quickly released when they reached the excretory system.15 What's more, when we incubated the investigated fluorophores with macrophages, IR-FEP and IR-FGP showed a much higher macrophage uptake (99.4% for IR-FEP and 73.6% for IR-FGP) compared to fast excretion dyes (32.4% for IR-BEMC6P and 20.0% for IR-12N3), indicating that long liver-uptake dyes were preferentially endocytosed by macrophages (Fig. 2d and S16†).64 Finally, we verified that the surface chemistry was also able to affect the renal excretion behaviors. By changing the azide group of IR-BEMC6P to amine and carboxyl groups, the fast renal excretion dyes became high liver-uptake dyes (Fig. S17†). Based on the above evidence, we concluded that NIR-fluorophores with renal excretion ability possessed small size, near-neutral functional groups, fast dissociation with proteins, and low uptake by macrophages (Fig. 2e).
NIR-II renally excreted fluorophore–peptide conjugates and NIR-II molecular imaging guided microsurgery
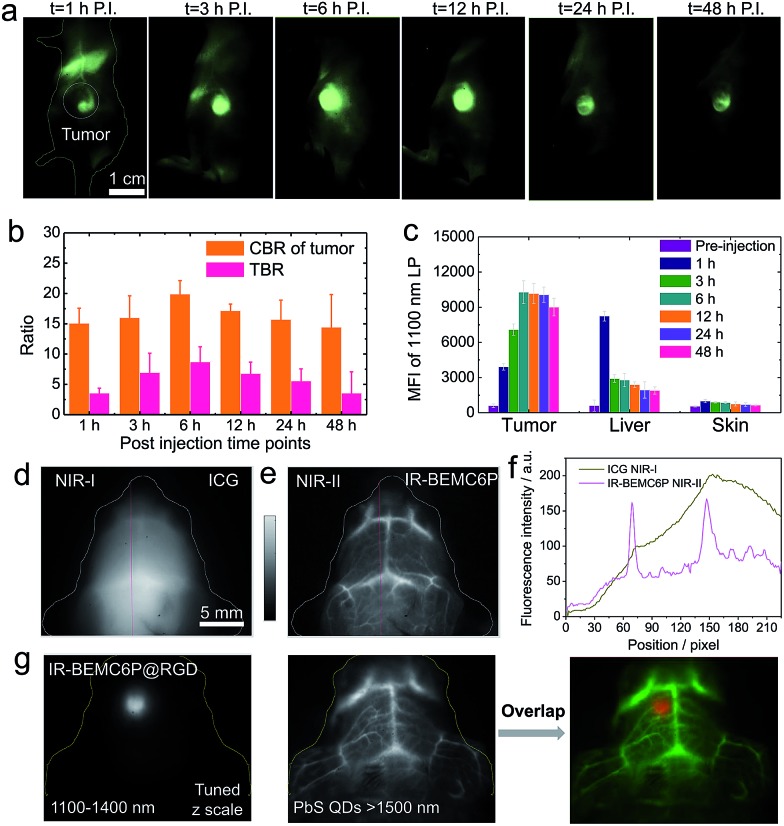
We then aimed to develop a high-performance renal-excretion peptide bioconjugate for tumor imaging based on the optimized renal excretion NIR-II fluorophore and targeting peptides. Although the NIR-II fluorophore–peptide conjugate has been reported with renally excreted behavior,16 the liver also had non-negligible uptake given the unoptimizable structural design. We first conjugated IR-BEMC6P to an RGD peptide and proved the superior excretion behavior of the IR-BEMC6P@RGD conjugate p.i. (Fig. S18†). To test the efficiency of conjugate targeting, we investigated molecular imaging in vivo with U87 tumor-bearing mice. The tumor mice were intravenously injected with IR-BEMC6P@RGD probes in PBS. NIR-II imaging over 1200 nm indicated that IR-BEMC6P@RGD accumulated in the U87 tumor in less than 1 h (Fig. 3a), with the T/NT ratio reaching a maximum of ∼9 at 6 h p.i. (Fig. 3b) and low accumulation in other organs (Fig. 3c).46 In contrast, the non-targeted IR-BEMC6P fluorophore showed a relatively weaker signal in the U87 tumor and the maximum T/NT ratio was less than ∼4 (Fig. S19†), due to the nonspecific accumulation of the free IR-BEMC6P.
Fig. 3. NIR-II targeting imaging of the U87 tumor model with IR-BEMC6P@RGD. (a) Imaging of the U87 tumor-bearing mouse intravenously treated with IR-BEMC6P@RGD in the NIR-II window. Injected dose: 200 μL of 25–75 μM conjugate solution. Imaging details: 1200 nm long pass filter, 808 nm laser. (b) CBR and TBR statistics of U87 tumor signal: contrast-to-background ratio (CBR) = (fluorescence – background)/background. Tumor-to-background ratio (TBR) = CBR of tumor/CBR of normal tissue. (c) Ex vivo scanning of the organs (tumor, liver, skin) after 24 h post injection of IR-BEMC6P@RGD. (d and e) Brain vessel imaging in C57 mice with shaved heads by injecting either ICG at 850–900 nm or IR-BEMC6P with 1300 nm long pass filter. (f) Section profile curve of vessels in both NIR-I and NIR-II windows. (g) High magnification NIR-II fluorescence imaging showing strong tumor fluorescence detectable through the intact scalp/skull at 12 h post-injection in an orthotopic brain tumor model. PbS quantum dots were intravenously injected as an additional channel over 1500 nm emission to visualize the vessels.65,66 .
We further investigated the advantage of NIR-II bioimaging by imaging the brain vessels in C57 mice with intact skin/scalp using either ICG or IR-BEMC6P, respectively (Fig. 3d–f).53 To demonstrate the benefit of the developed renal excretion conjugate in NIR-II molecular imaging, a glioblastoma brain tumor was surgically inoculated through the U87MG cells in the left hemisphere of the mouse brain. By intravenously injecting IR-BEMC6P@RGD, the NIR-II imaging of the brain tumor through a non-invasive route was performed through both the scalp and the skull. 6–12 h after administration of IR-BEMC6P@RGD, the tumor was distinguished with a T/NT ratio of ∼6 with NIR-II whole body imaging (Fig. 3g). By subsequently injecting PbS@PEG quantum dots with over 1500 nm emission,65 two-color imaging of both the brain tumor and vessel was obtained at the same time, affording multicolor live molecular imaging across the NIR-II window in a deep tumor model.
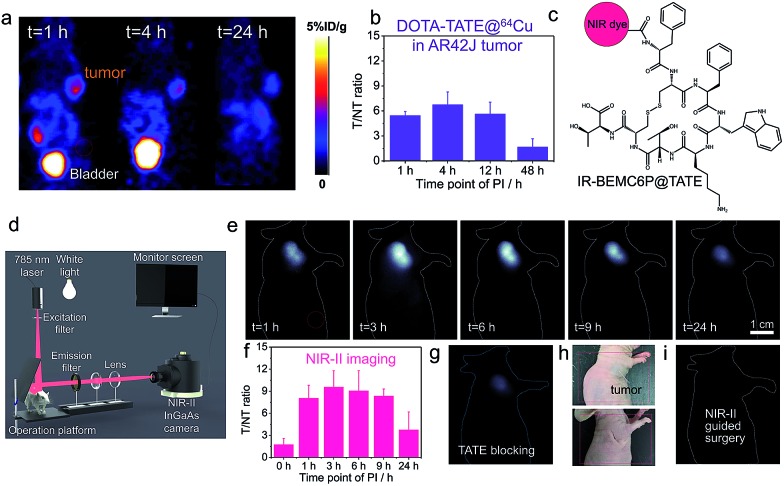
We subsequently used the developed strategy to improve the imaging contrast of the octreotate (TATE) peptide. TATE has somatostatin receptors (SSTRs) in gastroenteropancreatic neuroendocrine tumors, in which PET imaging is usually applied with a T/NT around 6, given the high normal tissue signal (Fig. 4a and b).15 The NIR-II conjugate IR-BEMC6P@TATE was synthesized with a TATE peptide containing a Dde protecting group (Fig. 4c). The renal excretion ability of IR-BEMC6P@TATE was confirmed by intravenous injection of IR-BEMC6P@TATE in CD-1 mice with high signal in the bladder but very low signal in the liver (Fig. S20†). The IR-BEMC6P@TATE was then injected through the tail vein in the immune-deficient mice bearing xenograft AR42J tumors. The tumor fluorescence distinctly showed up at 1 h p.i., followed by the T/NT ratio reaching ∼10 at 3 h p.i. (Fig. 4e–f). The improved T/NT ratio compared to PET imaging indicates that such a renal excretion conjugate truly benefited from the reduced normal tissue exposure of the conjugate. By injecting a blocking dose of the free, unconjugated TATE together with the IR-BEMC6P@TATE, a very low tumor signal proved the specificity of IR-BEMC6P@TATE on the AR42J tumor model (Fig. 4g). Further, we performed a tumor excision surgery under both NIR-II imaging and white light guidance (Fig. 4d). White light was selected for less interference with NIR-II monitoring, and the NIR-II imaging allowed for clear distinction of the tumor margin (Fig. 4h and i). Moreover, NIR-II guided micro-surgery was additionally performed utilizing an IR-BEMC6P@follicle-stimulating hormone (FSH) conjugate, allowing us to cut single follicles that are hundreds of μm in size (Fig. S21†). Only mild alterations in behavior post-surgery were observed, indicating the high safety of micro-surgery. Although fluorescence-guided surgery was achieved in the NIR-I systems,67 it is still critically important to further improve the detection depth and sensitivity, to yield a comprehensive fluorescence-guided platform with adjustable imaging speed (real-time) and contrast (S/B ratio), and multi-targeted imaging for visualizing complex diseases simultaneously.30,68
Fig. 4. In vivo NIR-II imaging of the AR42J tumor-bearing mice intravenously injected with IR-BEMC6P@TATE. (a) Typical PET imaging by DOTA-TATE@64Cu in the AR42J (SSTR2+) xenograft tumor model. Red dashed circle indicates the signal statistics position of the normal tissue. (b) Variation of T/NT signal ratio as a function of p.i. time points with DOTE@TATE@64Cu. (c) Conjugate scheme of IR-BEMC6P@TATE. (d) Scheme of NIR-II set-up for guided surgery. (e) NIR-II imaging of the AR42J tumor mice after tail vein injection of IR-BEMC6P@TATE. Injected dose: 200 μL of 25–75 μM conjugate solution. Imaging details: 1200 nm long pass filter, 808 nm laser. (f) Variation of T/NT signal ratio on the basis of p.i. time points with IR-BEMC6P@TATE. (g) NIR-II imaging of the IR-BEMC6P@TATE injection after blocking by TATE peptide only. (h) Digital photograph of the tumor xenograft mouse before and after NIR-II guided surgery. (i) NIR-II imaging-guided surgery to excise the tumor.
Conclusions
In summary, we developed a minimal hepatic-uptake NIR-II dye (IR-BEMC6P) with enhanced QY and derived renal excretion peptide conjugations. The excretion mechanism was exploited through comprehensive factors, and we found that NIR-II fluorophores with renal excretion ability possessed small size, near-neutral functional groups, fast de-binding with proteins, and low uptake by macrophages. This well-established rule for excretion behavior will guide future NIR-II dye synthesis and enrich the NIR-II fluorophore library with the clinical choice of NIR-II imaging, as one of the important features of potential NIR fluorophores for clinically translatable imaging is timely clearance from the body with minimum retention in major organs after administration.5 The renal excretion fluorophore derived peptide–dye conjugate provided efficient tumor targeting ability in mice with a high T/NT ratio in two different tumor models, affording great ability for NIR-II guided microsurgery. Importantly, the peptide IR-BEMC6P@(RGD, TATE) probes were rapidly renally excreted, in sharp contrast to long-term liver accumulation of typical antibody–dye conjugates. These rapidly excreted, highly safe integrin/somatostatin receptor targeting NIR-II probes will accelerate the clinical translation of NIR-II molecular imaging in cancer diagnosis and imaging navigation therapy.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported by the Shenzhen Key Lab Funding Grant ZDSYS201505291525382, the Shenzhen Peacock Program Grant KQTD20140630160825828, the Shenzhen Basic Research funding (JCYJ20170307151634428) and the Intramural Research Program of the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the National Institutes of Health (NIH). S. Z. thanks Dr Xue Zhang in Jilin Agricultural University for scheme designing. All animal studies were conducted in accordance with the principles and procedures outlined in the National Institutes of Health (NIH) Guide for the Care and Use of Animals, and under protocols approved by the NIH Clinical Center Animal Care and Use Committee (CC-ACUC, protocol number: NIBIB 16-03).
Footnotes
†Electronic supplementary information (ESI) available: Synthesis optimization process and structure characterization, excretion study, binding affinity, etc. See DOI: 10.1039/c8sc03751e
References
- Jin R., Zeng C., Zhou M., Chen Y. Chem. Rev. 2016;116:10346–10413. doi: 10.1021/acs.chemrev.5b00703. [DOI] [PubMed] [Google Scholar]
- Lyu Y., Fang Y., Miao Q., Zhen X., Ding D., Pu K. ACS Nano. 2016;10:4472–4481. doi: 10.1021/acsnano.6b00168. [DOI] [PubMed] [Google Scholar]
- Lyu Y., Xie C., Chechetka S. A., Miyako E., Pu K. J. Am. Chem. Soc. 2016;138:9049–9052. doi: 10.1021/jacs.6b05192. [DOI] [PubMed] [Google Scholar]
- Shi B., Jie K., Zhou Y., Zhou J., Xia D., Huang F. J. Am. Chem. Soc. 2016;138:80–83. doi: 10.1021/jacs.5b11676. [DOI] [PubMed] [Google Scholar]
- Hong G., Antaris A. L., Dai H. Nature Biomedical Engineering. 2017;1:0010. [Google Scholar]
- Wang R., Zhang F. J. Mater. Chem. B. 2014;2:2422. doi: 10.1039/c3tb21447h. [DOI] [PubMed] [Google Scholar]
- He S., Song J., Qu J., Cheng Z. Chem. Soc. Rev. 2018;47:4258–4278. doi: 10.1039/c8cs00234g. [DOI] [PubMed] [Google Scholar]
- Qi J., Sun C., Zebibula A., Zhang H., Kwok R. T. K., Zhao X., Xi W., Lam J. W. Y., Qian J., Tang B. Z. Adv. Mater. 2018;30:e1706856. doi: 10.1002/adma.201706856. [DOI] [PubMed] [Google Scholar]
- Woo S. J., Park S., Jeong J. E., Hong Y., Ku M., Kim B. Y., Jang I. H., Heo S. C., Wang T., Kim K. H., Yang J., Kim J. H., Woo H. Y. ACS Appl. Mater. Interfaces. 2016;8:15937–15947. doi: 10.1021/acsami.6b04276. [DOI] [PubMed] [Google Scholar]
- Shou K., Tang Y., Chen H., Chen S., Zhang L., Zhang A., Fan Q., Yu A., Cheng Z. Chem. Sci. 2018;9:3105–3110. doi: 10.1039/c8sc00206a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhong D., Zhou S. J. Mater. Chem. B. 2018;6:349–365. doi: 10.1039/c7tb02573d. [DOI] [PubMed] [Google Scholar]
- Winter G. E., Buckley D. L., Paulk J., Roberts J. M., Souza A., Dhe-Paganon S., Bradner J. E. Science. 2015;348:1376–1381. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfbeis O. S. Chem. Soc. Rev. 2015;44:4743–4768. doi: 10.1039/c4cs00392f. [DOI] [PubMed] [Google Scholar]
- Dai Y., Xu C., Sun X., Chen X. Chem. Soc. Rev. 2017;46:3830–3852. doi: 10.1039/c6cs00592f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian R., Jacobson O., Niu G., Kiesewetter D. O., Wang Z., Zhu G., Ma Y., Liu G., Chen X. Theranostics. 2018;8:735–745. doi: 10.7150/thno.23491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Ma Z., Zhu S., Wan H., Yue J., Ma H., Ma R., Yang Q., Wang Z., Li Q., Qian Y., Yue C., Wang Y., Fan L., Zhong Y., Zhou Y., Gao H., Ruan J., Hu Z., Liang Y., Dai H. Adv. Mater. 2018;30:e1800106. doi: 10.1002/adma.201800106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T., Nishikawa H., Wada H., Nagano Y., Sugiyama D., Atarashi K., Maeda Y., Hamaguchi M., Ohkura N., Sato E., Nagase H., Nishimura J., Yamamoto H., Takiguchi S., Tanoue T., Suda W., Morita H., Hattori M., Honda K., Mori M., Doki Y., Sakaguchi S. Nat. Med. 2016;22:679. doi: 10.1038/nm.4086. [DOI] [PubMed] [Google Scholar]
- Lau J. L., Dunn M. K. Bioorg. Med. Chem. 2018;26:2700–2707. doi: 10.1016/j.bmc.2017.06.052. [DOI] [PubMed] [Google Scholar]
- Chen H., Niu G., Wu H., Chen X. Theranostics. 2016;6:78–92. doi: 10.7150/thno.13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Wu J., Liu Y., Yu M., Zhao L., Zhu X., Bhasin S., Li Q., Ha E., Shi J., Farokhzad O. C. Angew. Chem., Int. Ed. 2016;55:7091–7094. doi: 10.1002/anie.201601273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curnis F., Fiocchi M., Sacchi A., Gori A., Gasparri A., Corti A. Nano Res. 2016;9:1393–1408. doi: 10.1007/s12274-016-1035-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aioub M., El-Sayed M. A. J. Am. Chem. Soc. 2016;138:1258–1264. doi: 10.1021/jacs.5b10997. [DOI] [PubMed] [Google Scholar]
- Zou Q., Abbas M., Zhao L., Li S., Shen G., Yan X. J. Am. Chem. Soc. 2017;139:1921–1927. doi: 10.1021/jacs.6b11382. [DOI] [PubMed] [Google Scholar]
- Li D., Zhang J., Chi C., Xiao X., Wang J., Lang L., Ali I., Niu G., Zhang L., Tian J., Ji N., Zhu Z., Chen X. Theranostics. 2018;8:2508–2520. doi: 10.7150/thno.25599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Wang J., Chen C. Adv. Mater. 2013;25:3869–3880. doi: 10.1002/adma.201301890. [DOI] [PubMed] [Google Scholar]
- Xie C., Zhen X., Miao Q., Lyu Y., Pu K. Adv. Mater. 2018;30:e1801331. doi: 10.1002/adma.201801331. [DOI] [PubMed] [Google Scholar]
- Michalet X., Pinaud F. F., Bentolila L. A., Tsay J. M., Doose S., Li J. J., Sundaresan G., Wu A. M., Gambhir S. S., Weiss S. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F., Zhan Y., Lu X., Sun Y. Chem. Sci. 2018;9:4370–4380. doi: 10.1039/c8sc01153b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Zhu S., Antaris A. L., Chen H., Xiao Y., Lu X., Jiang L., Diao S., Yu K., Wang Y., Herraiz S., Yue J., Hong X., Hong G., Cheng Z., Dai H., Hsueh A. J. Chem. Sci. 2017;8:3703–3711. doi: 10.1039/c6sc04897h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B., Sheng Z., Kenry K., Hu D., Lin X., Xu S., Liu C., Zheng H., Liu B. Mater. Horiz. 2017;4:1151–1156. [Google Scholar]
- Antaris A. L., Chen H., Diao S., Ma Z., Zhang Z., Zhu S., Wang J., Lozano A. X., Fan Q., Chew L. Nat. Commun. 2017;8:15269. doi: 10.1038/ncomms15269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y., Ma Z., Zhu S., Yue J., Zhang M., Antaris A. L., Yuan J., Cui R., Wan H., Zhou Y. Nat. Commun. 2017;8:737. doi: 10.1038/s41467-017-00917-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Yung B. C., Chandra S., Niu G., Antaris A. L., Chen X. Theranostics. 2018;8:4141–4151. doi: 10.7150/thno.27995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong G., Diao S., Antaris A. L., Dai H. Chem. Rev. 2015;115:10816. doi: 10.1021/acs.chemrev.5b00008. [DOI] [PubMed] [Google Scholar]
- Antaris A. L., Chen H., Cheng K., Sun Y., Hong G. S., Qu C. R., Diao S., Deng Z. X., Hu X. M., Zhang B., Zhang X. D., Yaghi O. K., Alamparambil Z. R., Hong X. C., Cheng Z., Dai H. J. Nat. Mater. 2016;15:235. doi: 10.1038/nmat4476. [DOI] [PubMed] [Google Scholar]
- Wang R., Li X., Zhou L., Zhang F. Angew. Chem., Int. Ed. 2014;53:12086–12090. doi: 10.1002/anie.201407420. [DOI] [PubMed] [Google Scholar]
- Franke D., Harris D. K., Chen O., Bruns O. T., Carr J. A., Wilson M. W., Bawendi M. G. Nat. Commun. 2016;7:12749. doi: 10.1038/ncomms12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K., Chen H., Jenkins C. H., Zhang G., Zhao W., Zhang Z., Han F., Fung J., Yang M., Jiang Y., Xing L., Cheng Z. ACS Nano. 2017;11:12276–12291. doi: 10.1021/acsnano.7b05966. [DOI] [PubMed] [Google Scholar]
- Wan H., Yue J., Zhu S., Uno T., Zhang X., Yang Q., Yu K., Hong G., Wang J., Li L., Ma Z., Gao H., Zhong Y., Su J., Antaris A. L., Xia Y., Luo J., Liang Y., Dai H. Nat. Commun. 2018;9:1171. doi: 10.1038/s41467-018-03505-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H., Lin Y., Tian Z. Q., Zhu D. L., Zhang Z. L., Pang D. W. Small. 2018;14:e1703296. doi: 10.1002/smll.201703296. [DOI] [PubMed] [Google Scholar]
- Diao S., Blackburn J. L., Hong G., Antaris A. L., Chang J., Wu J. Z., Zhang B., Cheng K., Kuo C. J., Dai H. Angew. Chem., Int. Ed. 2015;54:14758–14762. doi: 10.1002/anie.201507473. [DOI] [PubMed] [Google Scholar]
- Graf A., Tropf L., Zakharko Y., Zaumseil J., Gather M. C. Nat. Commun. 2016;7:13078. doi: 10.1038/ncomms13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naczynski D. J., Tan M. C., Zevon M., Wall B., Kohl J., Kulesa A., Chen S., Roth C. M., Riman R. E., Moghe P. V. Nat. Commun. 2013;4:2199. doi: 10.1038/ncomms3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang X., Gu L., Qi J., Correa S., Zhang G., Belcher A. M., Hammond P. T. Proc. Natl. Acad. Sci. U. S. A. 2016;113:5179–5184. doi: 10.1073/pnas.1521175113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsher K., Liu Z., Sherlock S. P., Robinson J. T., Chen Z., Daranciang D., Dai H. Nat. Nanotechnol. 2009;4:773–780. doi: 10.1038/nnano.2009.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Ding M., Zeng X., Xiao Y., Wu H., Zhou H., Ding B., Qu C., Hou W., Er-Bu A., Zhang Y., Cheng Z., Hong X. Chem. Sci. 2017;8:3489–3493. doi: 10.1039/c7sc00251c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Qu C., Chen H., He M., Tang C., Shou K., Hong S., Yang M., Jiang Y., Ding B., Xiao Y., Xing L., Hong X., Cheng Z. Chem. Sci. 2016;7:6203–6207. doi: 10.1039/c6sc01561a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosco E. D., Caram J. R., Bruns O. T., Franke D., Day R. A., Farr E. P., Bawendi M. G., Sletten E. M. Angew. Chem., Int. Ed. 2017;56:13126–13129. doi: 10.1002/anie.201706974. [DOI] [PubMed] [Google Scholar]
- Li B., Lu L., Zhao M., Lei Z., Zhang F. Angew. Chem., Int. Ed. 2018;57:7483–7487. doi: 10.1002/anie.201801226. [DOI] [PubMed] [Google Scholar]
- Xu G., Yan Q., Lv X., Zhu Y., Xin K., Shi B., Wang R., Chen J., Gao W., Shi P., Fan C., Zhao C., Tian H. Angew. Chem., Int. Ed. 2018;57:3626–3630. doi: 10.1002/anie.201712528. [DOI] [PubMed] [Google Scholar]
- Carr J. A., Franke D., Caram J. R., Perkinson C. F., Saif M., Askoxylakis V., Datta M., Fukumura D., Jain R. K., Bawendi M. G., Bruns O. T. Proc. Natl. Acad. Sci. U. S. A. 2018;115:4465–4470. doi: 10.1073/pnas.1718917115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. D., Wang H., Antaris A. L., Li L., Diao S., Ma R., Nguyen A., Hong G., Ma Z., Wang J., Zhu S., Castellano J. M., Wyss-Coray T., Liang Y., Luo J., Dai H. Adv. Mater. 2016;28:6872–6879. doi: 10.1002/adma.201600706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antaris A. L., Chen H., Cheng K., Sun Y., Hong G., Qu C., Diao S., Deng Z., Hu X., Zhang B., Zhang X., Yaghi O. K., Alamparambil Z. R., Hong X., Cheng Z., Dai H. Nat. Mater. 2016;15:235–242. doi: 10.1038/nmat4476. [DOI] [PubMed] [Google Scholar]
- Choi H. S., Liu W., Misra P., Tanaka E., Zimmer J. P., Itty Ipe B., Bawendi M. G., Frangioni J. V. Nat. Biotechnol. 2007;25:1165–1170. doi: 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., Gravier J., Bao K., Wada H., Lee J. H., Baek Y., El Fakhri G., Gioux S., Rubin B. P., Coll J. L., Choi H. S. Adv. Mater. 2016;28:8162–8168. doi: 10.1002/adma.201601101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Liu J., Ning X., Zheng J. Angew. Chem., Int. Ed. 2015;54:15434–15438. doi: 10.1002/anie.201507868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Ma Z., Wang H., Zhou B., Zhu S., Zhong Y., Wang J., Wan H., Antaris A., Ma R., Zhang X., Yang J., Zhang X., Sun H., Liu W., Liang Y., Dai H. Adv. Mater. 2017;29:1605497. doi: 10.1002/adma.201605497. [DOI] [PubMed] [Google Scholar]
- Qian G., Gao J. P., Wang Z. Y. Chem. Commun. 2012;48:6426–6428. [Google Scholar]
- Zhu S., Yang Q., Antaris A. L., Yue J., Ma Z., Wang H., Huang W., Wan H., Wang J., Diao S., Zhang B., Li X., Zhong Y., Yu K., Hong G., Luo J., Liang Y., Dai H. Proc. Natl. Acad. Sci. U. S. A. 2017;114:962–967. doi: 10.1073/pnas.1617990114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H. S., Nasr K., Alyabyev S., Feith D., Lee J. H., Kim S. H., Ashitate Y., Hyun H., Patonay G., Strekowski L., Henary M., Frangioni J. V. Angew. Chem., Int. Ed. 2011;50:6258–6263. doi: 10.1002/anie.201102459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Hao G., Thomas P., Liu J., Yu M., Sun S., Oz O. K., Sun X., Zheng J. Angew. Chem., Int. Ed. 2012;51:10118–10122. doi: 10.1002/anie.201203031. [DOI] [PubMed] [Google Scholar]
- Hagenbuch B. Clin. Pharmacol. Ther. 2010;87:39–47. doi: 10.1038/clpt.2009.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Hu Z., Tian R., Yung B. C., Yang Q., Zhao S., Kiesewetter D. O., Niu G., Sun H., Antaris A. L., Chen X. Adv. Mater. 2018;30:e1802546. doi: 10.1002/adma.201802546. [DOI] [PubMed] [Google Scholar]
- Weissleder R., Nahrendorf M., Pittet M. J. Nat. Mater. 2014;13:125–138. doi: 10.1038/nmat3780. [DOI] [PubMed] [Google Scholar]
- Zhu S., Herraiz S., Yue J., Zhang M., Wan H., Yang Q., Ma Z., Wang Y., He J., Antaris A. L., Zhong Y., Diao S., Feng Y., Zhou Y., Yu K., Hong G., Liang Y., Hsueh A. J., Dai H. Adv. Mater. 2018;30:e1705799. doi: 10.1002/adma.201705799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Yue J., Cui R., Ma Z., Wan H., Wang F., Zhu S., Zhou Y., Kuang Y., Zhong Y., Pang D. W., Dai H. Proc. Natl. Acad. Sci. U. S. A. 2018;115:6590–6595. doi: 10.1073/pnas.1806153115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi C., Du Y., Ye J., Kou D., Qiu J., Wang J., Tian J., Chen X. Theranostics. 2014;4:1072–1084. doi: 10.7150/thno.9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Mao D., Hu F., Xu S., Chen C., Zhang C. J., Cheng X., Yuan Y., Ding D., Kong D., Liu B. Adv. Mater. 2017;29:1700548. doi: 10.1002/adma.201700548. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.