Abstract
Self-assembly is a process of key importance in natural systems and in nanotechnology. Peptides are attractive building blocks due to their relative facile synthesis, biocompatibility, and other unique properties. Diphenylalanine (FF) and its derivatives are known to form nanostructures of various architectures and interesting and varied characteristics. The larger triphenylalanine peptide (FFF) was found to self-assemble as efficiently as FF, forming related but distinct architectures of plate-like and spherical nanostructures. Here, to understand the effect of triaromatic systems on the self-assembly process, we examined carboxybenzyl-protected diphenylalanine (z-FF) as a minimal model for such an arrangement. We explored different self-assembly conditions by changing solvent compositions and peptide concentrations, generating a phase diagram for the assemblies. We discovered that z-FF can form a variety of structures, including nanowires, fibers, nanospheres, and nanotoroids, the latter were previously observed only in considerably larger or co-assembly systems. Secondary structure analysis revealed that all assemblies possessed a β-sheet conformation. Additionally, in solvent combinations with high water ratios, z-FF formed rigid and self-healing hydrogels. X-ray crystallography revealed a “wishbone” structure, in which z-FF dimers are linked by hydrogen bonds mediated by methanol molecules, with a 2-fold screw symmetry along the c-axis. All-atom molecular dynamics (MD) simulations revealed conformations similar to the crystal structure. Coarse-grained MD simulated the assembly of the peptide into either fibers or spheres in different solvent systems, consistent with the experimental results. This work thus expands the building block library for the fabrication of nanostructures by peptide self-assembly.
Keywords: diphenylalanine, self-assembly, structural polymorphism, self-assembly mechanism, toroids, peptide nanotubes
Assembly of building blocks into supramolecular structures is a key process in nature. Many natural building blocks, such as nucleic acids, peptides, and phospholipids, undergo self-assembly to form functional structures through noncovalent interactions, including van der Waals (vdW) interactions, π−π stacking interactions, hydrogen-bonding, and electrostatic interactions.1,2 Although each of these forces is rather weak individually, the sum of molecular interactions is sufficient to form a stable self-organized system. For example, C−C and O−H covalent bonds have energies of 347−356 and ~110 kJ/mol, respectively. In comparison, OH⋯N, OH⋯O, and NH⋯O hydrogen bonds have energies of 29, 21, and 8 kJ/mol, respectively, and π−π stacking interactions have energies in the scale of several kJ/mol.1 Unlike covalent polymers, self-assembled materials are highly dynamic due to the noncovalent forces, resulting in a myriad of properties, such as dynamic structural transition, self-healing, and macro-structure phase separation.3–10 Self-assembly thus presents a relatively simple and easy method for the formation of nanomaterials of different architectures and “bottom-up” fabrication of supramolecular polymers with diverse physical, chemical, and biological properties.
Specifically, proteins and peptides are the main structural and functional compounds in nature. They self-assemble and serve as biological building blocks and molecular scaffolds in numerous processes, such as the organization of microtubules for cellular transportation,11 production of spider silk with extreme mechanical properties,12,13 fabrication of structural proteins in hair and nails, and the biomineralization of teeth and bone that is mediated by protein templates.14,15
Aromatic groups, together with hydrogen-bonding moieties (peptide bonds, N- and C-termini), present an attractive combination of molecular conditions for self-assembly. Diphenylalanine (FF), the core recognition motif in Alzheimer’s disease β-amyloid (Aβ) polypeptide, self-assembles into nanotubes when dissolved in 1,1,1,3,3,3-hexafluoroisopro-panol (HFIP) and diluted with water.16,17 The nanotubes show extraordinary rigidity, thermal and chemical stability, and interesting electronic and optical properties, such as semi-conductivity, blue luminescence, and piezoelectric and pyroelectric characteristics.18–25 The properties of FF and its analogs have been extensively studied toward utilization of self-assembly of short aromatic peptides into structures at various scales and for different applications, such as drug delivery, nanofabrication, energy storage and production, optical wave-guides, and piezoelectric sensors.4,6,24,26–33
The process of self-assembly is governed by both kinetics and thermodynamics, as the different states of the system, or phases, represent local minima in the potential energy. An assembly of many molecules will have a different potential energy profile than that of a monomer, and the different phases of the assembly might be kinetically trapped, even though they are energetically metastable.34 Therefore, the many parameters involved in the self-assembly process render the understanding of the effects of initial conditions and molecular interactions on the final structures highly challenging. For example, derivatives of FF were found to form different nanoscale morphologies under varying initial self-assembly conditions.3,23,35–41 Therefore, in order to design nanostructures by self-assembly, it is imperative to understand the underlying forces and dynamics that drive the process.
This increasing interest in self-assembly processes has also been noticed in the computational community, with numerous studies on the self-assembly of phenylalanine (F),42,43 FF,21,44–46 FFF,44,47 and the co-assembly of FF and FFF.37 These studies provided insights into the mechanism of short aromatic peptide (and amino acid) self-assembly. Other studies explored the di- and tripeptide space, examining aggregation and gelation properties,39,48 pointing to the important role of aromatic moieties in peptide aggregation, mainly phenylalanine and FF peptides, and also tryptophan-based peptides and FFF analogs.
From all the studies noted above, it becomes clear that aromatic groups, together with hydrogen-bonding moieties (peptide bonds, N- and C-termini), present an attractive combination of molecular interactions for self-assembly. An interesting open question is related to the control of the self-assembly process toward different nanostructure architectures by a simple tuning of the assembly conditions, other than changing the molecular structure of the building blocks.
In this work, we focused on a tri-aromatic peptide system, comprised of a carboxybenzyl (Cbz)-protected diphenylalanine (z-FF, Figure 1a), as a minimal member of the tri-aromatic peptide family and as a building block for bottom-up nanofabrication. We examined the effects of initial self-assembly conditions on the resulting structures and their properties by changing the stock solvent (ethanol, HFIP, and acetic acid), ethanol−water ratios in the diluting solvent, as well as peptide concentration. We also explored the self-assembly mechanism of z-FF assemblies.
Figure 1.
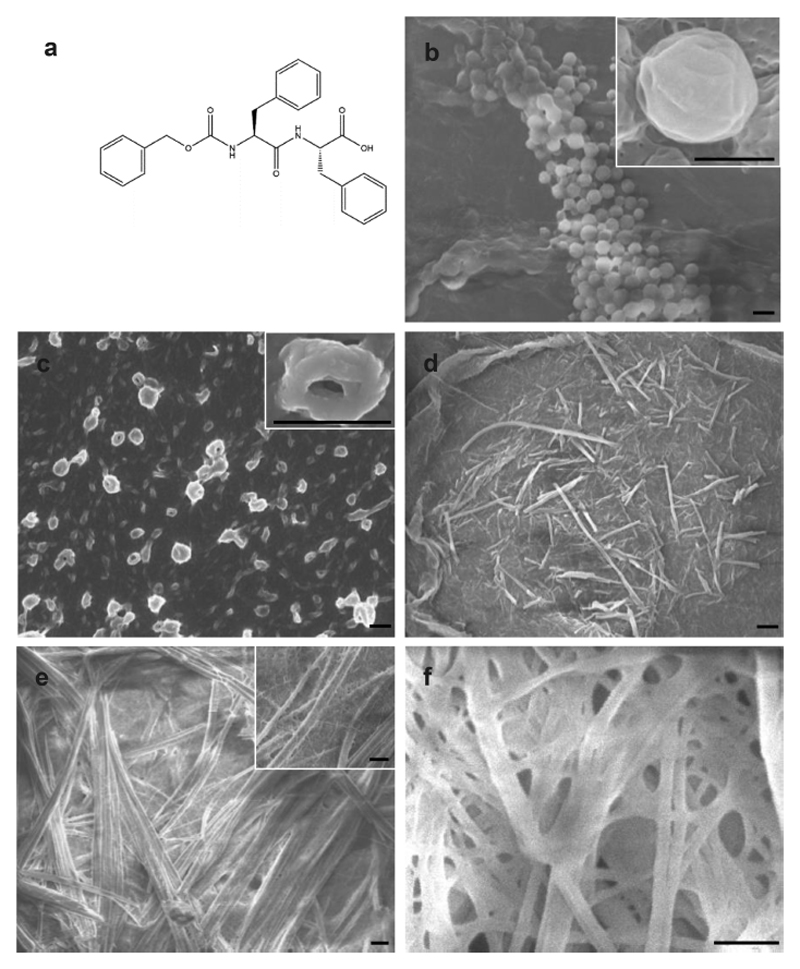
Structural analysis of z-FF assemblies. (a) Chemical structure of the z-FF molecule. (b-f) HR-SEM imaging of z-FF assemblies formed in (b) ethanol stock, 5 mg/mL, 50% ethanol; (c) ethanol stock, 0.5 mg/mL, 100% ethanol; (d) HFIP stock, 10 mg/mL, 50% ethanol; (e) methanol stock, 5 mg/mL, 50% methanol; and (f) methanol stock, 5 mg/mL, 25% methanol. Scale bar: 1 μm.
Results and Discussion
Nanostructure Formation
Several self-assembly conditions known to affect the resulting structures, namely stock solution, final peptide concentration, and ethanol−water solution ratio, were examined (Figure S1). High-resolution scanning electron microscopy (HR-SEM) imaging showed that in ethanol, spheres ~1 μm in diameter were formed at various z-FF concentrations and ethanol−water ratios, without any recognizable consistency (Figure 1b). Noncanonical toroids 100−1000 nm across were formed in ethanol stock at a peptide concentration of 0.5 mg/mL diluted with 100% ethanol (Figure 1c). These findings were further examined under high-resolution transmission electron microscopy (HR-TEM, Figure S2). Nanofibers 50−1000 nm across were also formed at peptide concentrations of 1 and 5 mg/mL, in 75 and 50% ethanol (Figure 1d,e). Higher water ratios resulted in the formation of hydrogels, regardless of peptide concentration (Figure 1f).
To the best of our knowledge, toroids formed from short aromatic peptides were only documented in the co-assembly of FF and FFF.37 The z-FF toroids are made up of a ring of nanofibers (Figure 1c), implying that the fibers are first formed and are subsequently assembled into the macrostructures. This mechanism has been previously demonstrated for various FF derivatives.29,36
HFIP stock shows similar spherical and nanofibrillar structures, at slightly different conditions. Similar to the ethanol stock, hydrogels were also formed at all peptide concentrations at high water ratios. The use of acetic acid as a stock solvent resulted in a similar pattern to the ethanol stock, except that the toroids were formed at higher peptide concentrations and in a 75% ethanol−water ratio.
Taken together, similar spherical (~1 μm in diameter) and nanofibrillar (50−100 nm in width) structures are observed in all stock solutions under different conditions. Hydrogels are formed in high water ratios, regardless of the stock solution type and peptide concentration. Toroids are formed under different conditions in either ethanol or acetic acid stock, but not in HFIP stock. We therefore hypothesize that ethanol and acetic acid stock solutions promote the formation of a mixed population of z-FF monomers, dimers, and higher oligomers, which then assemble into the various nanostructures. However, HFIP stock solution may promote the formation of a different population of oligomers, or mainly monomers. These differences in the initial building blocks formed in the stock solvents may result in the different nanostructure architectures.
Secondary Structure Analysis
The secondary structures of the peptide assemblies were analyzed by circular dichroism (CD) and Fourier-transformed infrared (FTIR) spectroscopy.
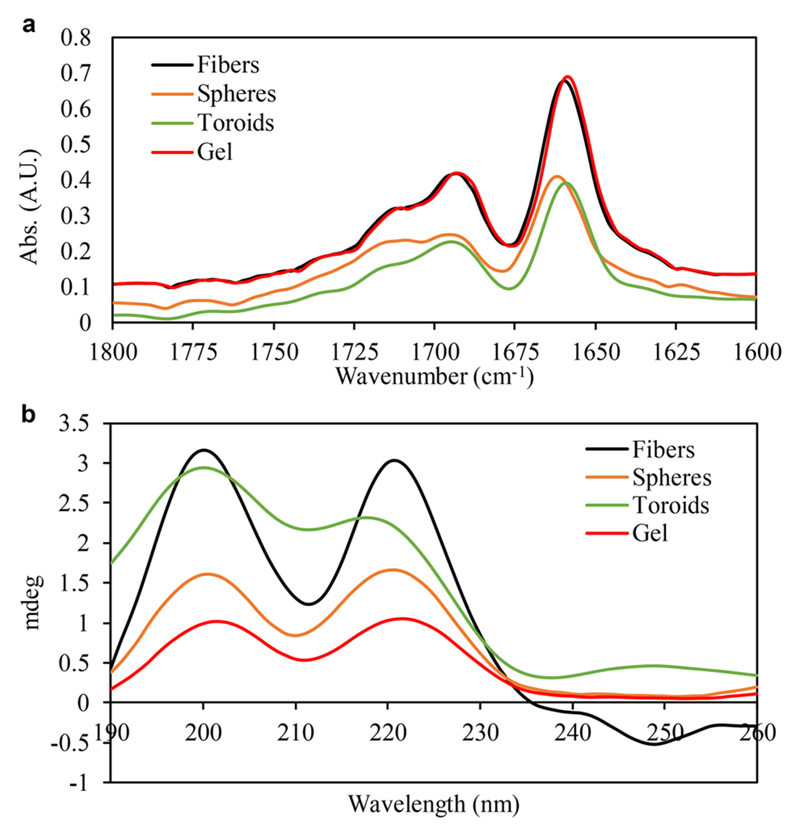
FTIR spectroscopy applied for all samples resulted in two major amide-I peaks at 1660 and 1694 cm−1 (Figure 2a). Second derivative analysis revealed additional peaks at 1624 and 1636 cm−1. These peaks are indicative of a β-sheet secondary structure.49,50 CD spectra showed peaks at 202 and 222 nm, corresponding to π−π* and n−π* transitions (Figure 2b).51,52 These peaks were previously reported for phenyl-alanine-based compounds, such as FF and its analogs, and Phe-Glu-Phe.22,53
Figure 2.
Secondary structure analysis of z-FF assemblies. (a) FTIR spectra of z-FF at representative conditions. (b) CD spectra of z-FF at representative conditions.
Hydrogel Rheology
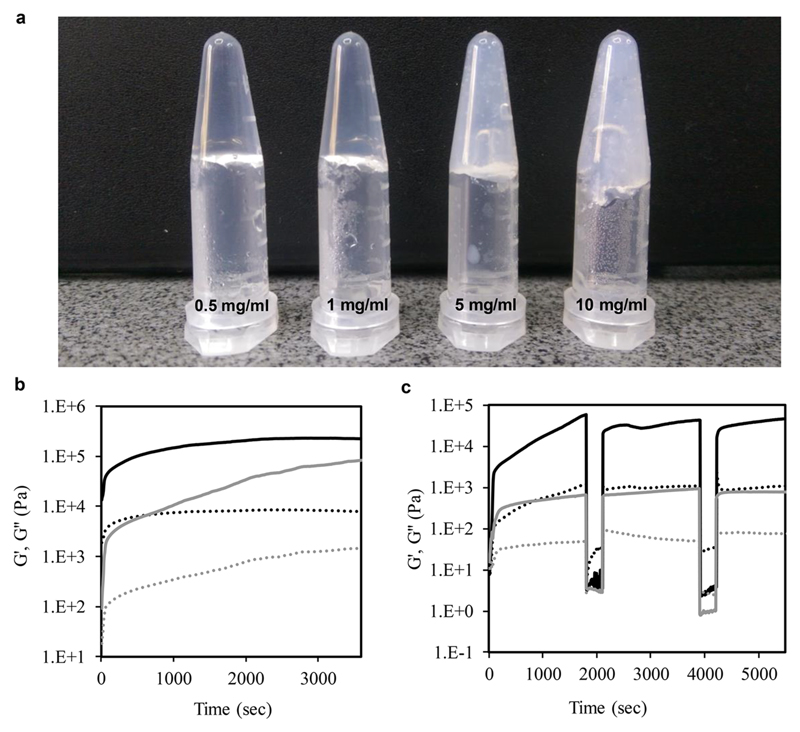
As noted above, z-FF was found to form hydrogels when all stock solutions were diluted to a solution of 75−100% water, in all peptide concentrations (Figure 1f). The gels became white and opaque as peptide concentration was increased (Figure 3a).
Figure 3.
Rheological properties of z-FF hydrogels. (a) z-FF gels. (b) Time sweep rheology showing G′ and G″ (solid and dashed lines, respectively) of 10 and 5 mg/mL gels (black and gray lines, respectively). (c) Time sweep for thixotropic properties.
To determine the effect of peptide concentration on the viscoelastic properties, rheology was employed. In a time-sweep measurement, the elastic response of the gel (G′) exceeded the viscosity response (G″) by at least 1 order of magnitude after full gelation, indicating the formation of a solid-like hydrogel supported matrix (Figure 3b). The storage modulus of the 10 mg/mL gel reached a plateau slightly higher than 105 Pa after about 2000 s. However, for the 5 mg/mL gel, a consistent increase of G′ was observed throughout the experiment, until reaching a value slightly lower than 105 Pa, indicating that the final peptide concentration affects the gel stiffness. When compared to G′ of Fmoc-FF, a well-studied short-peptide-based hydrogelator,23 the final 5 mg/mL z-FF gel is of similar stiffness, about 104 Pa, while the 10 mg/mL z-FF gel is stiffer by about 1 order of magnitude. Frequency and strain sweeps were conducted to examine the performance of the gel, showing that in frequencies of 1−100 Hz, the gel remained intact. At strain values higher than about 1%, the gels broke, regardless of the peptide concentration (Figure S4).
The thixotropic properties were measured in a time sweep experiment. Increased strain resulted in gel breakage, as indicated by the immediate and significant decrease and inversion of G′ and G″ values, showing the domination of the viscous response, implying the liquid state of the system. Decreased strain resulted in immediate restoration of the G′ and G″ values observed before the gel breakage, indicating thixotropic properties and self-healing (Figure 3c), which are important for tissue engineering as cell culture scaffolding.41,54
Crystal Structure
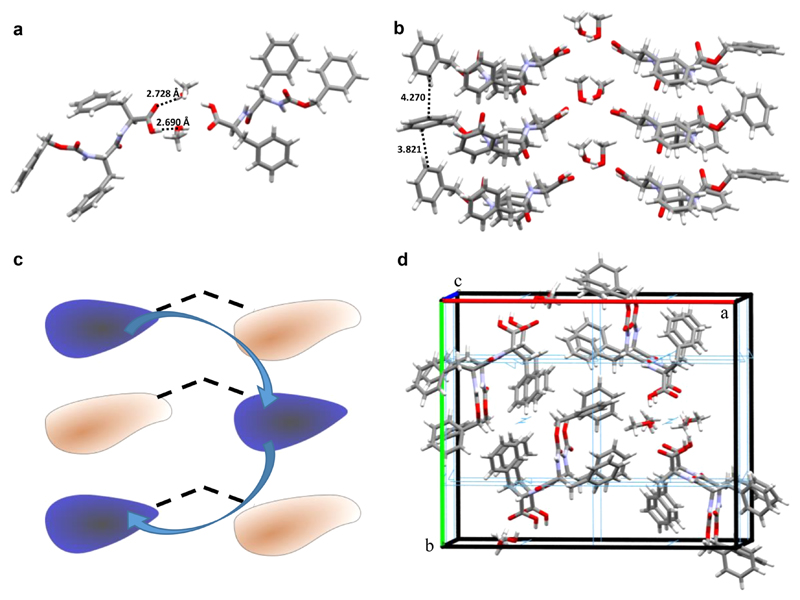
We next aimed to characterize the structure of z-FF assemblies by X-ray diffraction. After examining various crystallization conditions, single z-FF crystals were obtained by slow evaporation of a z-FF solution in methanol. X-ray diffraction analysis of the crystal structure revealed two independent z-FF molecules per asymmetric unit, showing different conformations with the carboxylic termini pointing toward each other. The two z-FF molecules are linked by methanol-bridged hydrogen bonds (Figure 4a). The molecules’ conformation alternates along the c-axis, and the opposing z-FF molecules inhabit the opposite conformation (Figure 4b,c), corresponding to a space group symmetry of P 212121, resembling a “wishbone-shaped” organization. An alternative explanation for the lattice organization would be that the basic dimeric unit-cell repeats along the c-axis, in 180° rotations along the axis, effectively generating a 2-fold screw symmetry. The β-sheet structure is stabilized by face-to-edge π−π interactions with minimal distances of 3.8−4.3 Å (Figure 4d), consistent with the FTIR and CD studies.
Figure 4.
z-FF crystal structure. (a) A z-FF dimer with H-bonds bridged by methanol. (b) “Wishbone” structure stacking between unit cells. (c) Schematic representation of the “wishbone” structure. (d) View down the c-axis, rotated by 10°, of a high-order assembly. A 2-fold screw axis symmetry element is seen in light blue.
Ramachandran plot analysis indicated that the z-FF crystal lattice had a β-sheet structure with torsion angles of ϕ = −126.5° and ψ = 108.0° for one conformer (Figure S3) and ϕ = −151.2° and ψ = 108.0° for the other conformer. This is also in agreement with the FTIR and CD spectra.
All-Atom Molecular Dynamics Simulations
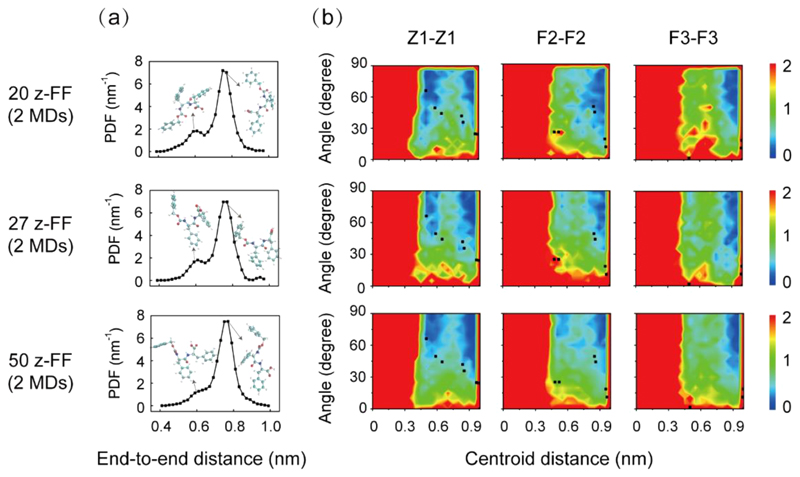
MD simulations were performed to understand the dynamics and thermodynamics of the building blocks assembly into ordered structures. Two independent all-atom MD simulations were carried out for z-FF oligomers (20-, 27-, and 50-mer) in methanol, and quantitatively similar results were obtained for each system. The probability distribution of the end-to-end distance of z-FF revealed two peaks: a small peak located at 0.60 nm and a dominant peak centered at 0.77 nm (Figure 5a). The latter peak is close to the end-to-end distance (0.82 nm) of the z-FF peptide in the wishbone crystal organization. To examine whether the stacking patterns of the intermolecular rings in the z-FF oligomers are similar to those in the wishbone structure, we calculated the free energy surface of z-FF oligomers as a function of the centroid distance and the angle between intermolecular aromatic ring pairs of the Z1−Z1 (Cbz protecting group), F2−F2 (N-terminus Phe), and F3−F3 (C-terminus Phe) groups, and projected the stacking patterns of the aromatic rings in the crystal structure onto the free energy surface (black dots in Figure 5b). The aromatic rings were arranged in conformations similar to those in the wishbone crystal structure, except for the F3−F3 ring pairs, indicating that the conformations of the z-FF peptide in the crystal were sampled in the MD simulations, even though the well-organized wishbone structure was not observed. This can be explained by the relatively short simulation time scale and the small number of molecules, compared to those of the experimental system.
Figure 5.
All-atom MD simulations of the self-assembly of 20, 27, and 50 z-FF molecules in methanol (left panel, middle panel, and right panel, respectively). All simulations started from disordered states. (a) The probability density function (PDF) of end-to-end distance of z-FF molecules. The structures of two representative z-FF peptides with their end-to-end distance located at the two peaks are also shown. (b) The free energy surface of z-FF oligomers as a function of the centroid distance and the angle between two aromatic rings of intermolecular Z1−Z1, F2−F2, and F3−F3 pairs.
Coarse-Grained Molecular Dynamics (CG-MD) Simulations
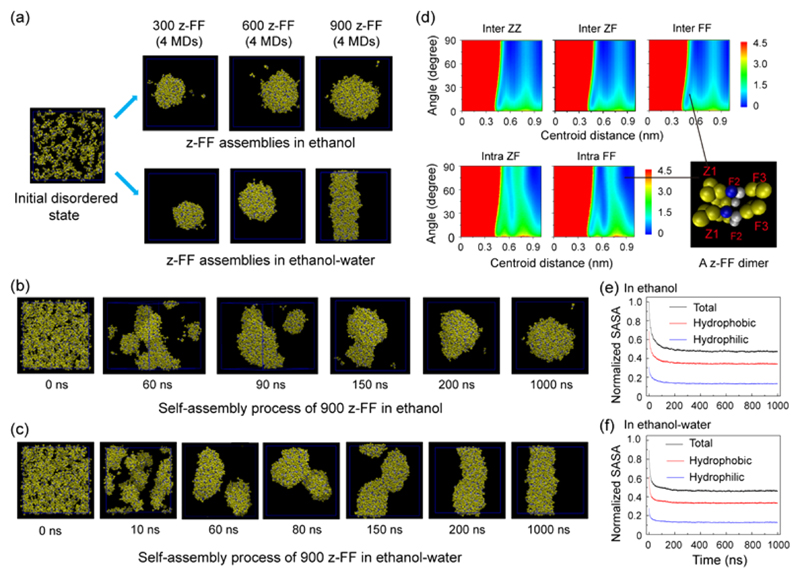
To understand the self-assembly of larger sizes of z-FF architectures, we performed multiple microsecond CG-MD simulations on systems consisting of 300, 600, or 900 z-FF molecules in ethanol and a 1:1 ethanol:water mixture. Starting from a disordered state, z-FF self-assembled into solid spheres in ethanol at all peptide concentrations, whereas in the ethanol−water mixture, z-FF assembled into solid spheres at low peptide concentrations and into fibrils at high concentrations (Figure 6). Nine hundred z-FF molecules assemble into spheres in ethanol (Figure 6b) and into fibrils in an ethanol−water mixture (Figure 6c). The mechanism of assembly begins by rapid aggregation into clusters of various sizes, within 60 ns. Following the aggregation, after 200 ns, the clusters start to fuse and merge into one large aggregate. Finally, after 1 μs, spherical or cylindrical structures are formed in ethanol or in the ethanol−water mixture, respectively. This self-assembly process is quite similar to the process previously reported for FFF peptides,37 indicating that z-FF may be used as a minimal member of the tri-aromatic peptide family. The simulated z-FF spheres and fibrils have a similar topology to the assemblies obtained experimentally. However, toroids were not observed in the CG-MD simulations. Again, this discrepancy might be due to the short time scale and smaller number of molecules in the simulations, compared to those of the experimental system, as the toroids are several micrometers in diameter and are composed of bundled fibers. Thus, a much larger number of z-FF molecules is needed for the formation of toroids, which is not feasible with the currently available computational power.
Figure 6.
Multiple microsecond CG-MD simulations of the self-assembly of 300, 600, and 900 z-FF molecules in ethanol and an ethanol−water mixture. All simulations were initiated from disordered states. (a) The z-FF assemblies generated at t = 1 μs of CG-MD simulations in ethanol (upper panel) and in an ethanol−water mixture (lower panel). (b,c) Representative CG-MD self-assembly process of 900 z-FF molecules leading to (b) the formation of spheres in ethanol and (c) the formation of fibrils in an ethanol−water mixture. (d) The free energy surface of z-FF assemblies as a function of the centroid distance and the angle between two aromatic rings of inter/intra-molecular ZZ, ZF, and FF pairs. A parallel z-FF dimer from the z-FF spheres/fibrils in the bottom right shows the parallel/T-shaped stacking pattern between two inter/intra-molecular FF aromatic rings. (e,f) Normalized solvent accessible surface area (SASA) of z-FF assemblies in (e) ethanol and (f) an ethanol−water mixture as a function of simulation time. The SASAs include total, hydrophobic, and hydrophilic SASA.
Previous experimental and computational studies reported that aromatic stacking interactions may play an important role in peptide self-assembly.45,55–57 To investigate the aromatic stacking interactions in the z-FF assemblies, we calculated the free energy surface of z-FF structures as a function of the centroid distance and the angle between two aromatic rings of intermolecular ZZ (Z1−Z1), ZF (Z1−F2 and Z1−F3), and FF (F2−F2, F2−F3, and F3−F3) pairs and intramolecular ZF (Z1−F2 and Z−Figure) and FF (F2−F3) pairs (Figure 6d). There are many possible stacking patterns between these aromatic ring pairs, and the exact stacking orientation depends on their centroid distance. Interestingly, we found that the interpeptide ZZ, ZF, and FF ring pairs have an almost identical free energy surface, indicating that all interaromatic ring pairs have similar stacking patterns. The parallel/T-shaped stacking pattern between two inter/intra-molecular FF aromatic rings is shown in a parallel z-FF dimer (bottom right of Figure 6d) from the z-FF spheres/fibrils. We also investigated the self-assembly dynamics by calculating the normalized solvent accessible surface area (SASA) of the z-FF assemblies in ethanol and in an ethanol−water mixture as a function of simulation time. The time evolution of normalized SASA indicated that hydrophobic interactions played an important role in the early stage of z-FF peptide self-assembly (Figure 6e,f), as the hydrophobic SASA decreased rapidly during the assembly process. Notably, the slopes of the SASA curves in the ethanol−water mixture were steeper than those in ethanol, indicating that the self-assembly in the ethanol−water mixture is faster than in ethanol. A possible explanation for this effect is that the ethanol−water mixture is more polar than ethanol, resulting in stronger aromatic and hydrophobic interactions between the z-FF molecules.
Conclusions
In this work, we explored the self-assembly process of the z-FF peptide, an FF derivative and a minimal member of the triaromatic peptide family, as a building block for nanostructure fabrication. By altering the solvent composition and peptide concentration, we revealed that z-FF assembles into various architectures, including nanowires, nanospheres, fibers, and noncanonical toroids. Toroid formation is especially intriguing as toroids made of short aromatic peptides were previously obtained only by co-assembly of FF with FFF.34
The z-FF building block was also found to form hydrogels, regardless of stock solution type and peptide concentration, as long as the water ratio in the solution was 75% or higher. The z-FF hydrogels were found to be slightly stiffer than those formed by Fmoc-FF. Additionally, self-healing properties were observed, even after several cycles of breakage and relaxation.
The secondary structure of the various nanostructures, including the hydrogels, is predominantly β-sheet stabilized by π−π interactions between the aromatic rings in the peptides. The crystal structure supports the secondary structure studies, as indicated by plotting the torsion angles in a Ramachandran plot. All assembly architectures are governed by the same thermodynamic forces, namely π−π stacking and hydrogen bonds, resulting in a similar secondary structure. However, due to the different initial conditions, the various assembly architectures are formed as different products, kinetically trapped in different local minima of the potential energy profile.
The crystal structure is bridged by two methanol molecules, forming hydrogen bonds between two z-FF molecules, in different conformations and with their carboxylic termini pointing toward each other. The peptide conformations alternate along the c-axis so that opposing z-FF molecules inhabit the opposite conformation, resulting in a “wishbone” structure. To the best of our knowledge, this is the first report of such a lattice organization of short peptide crystals.
All-atom and CG-MD simulations revealed that the z-FF molecules occupy conformations similar to those observed in the crystal structure. The z-FF peptide assembles into spheres in ethanol and into fibers in a 1:1 ethanol:water mixture, regardless of the peptide concentration. The assembly mechanism includes initial aggregation into amorphous clusters, which then form the larger, ordered spheres and fibers. Toroid formation was not observed in the simulations, which can be explained by the relatively short time scales and number of molecules simulated, compared to the experimental system.
This work extends the scope of bioinspired building blocks for self-assembly and nanostructure fabrication, gaining insight into the mechanism and theory behind nanostructure formation.
The study implies that simple triaromatic systems contain all the necessary information to form noncanonical supramolecular assemblies, such as toroids. It will be interesting to explore other triaromatic systems, as well as tetra-aromatic systems, including z-FFF, to learn about the role of aromatic moieties and to increase the possible structural space for nano-assemblies.
Materials and Methods
All solvents were purchased from Bio-Lab Ltd. (Israel), unless otherwise stated. The z-FF peptide was purchased from Bachem (Switzerland).
Nanostructure Formation
Stock solutions of z-FF were prepared by diluting the lyophilized form of the peptide in ethanol, HFIP (Sigma-Aldrich, Israel), and glacial acetic acid, at a concentration of 50 mg/mL (112 mM). The solutions were sonicated to solubilize the peptide and to break preformed aggregates. To avoid pre-aggregation of the peptide in the stock solutions, fresh stock solutions were prepared for each experiment. The stock solutions were then diluted in ethanol−water solutions with ethanol ratios of 100, 75, and 50% to final concentrations of 0.5, 1.0, 5.0, and 10 mg/mL. Hydrogels were prepared similarly to nanostructures, with ethanol contents of 25 and 0% in the ethanol−water solution.
High-Resolution Scanning Electron Microscopy (HR-SEM)
The samples were prepared by drop casting on siliconized glass and drying in room temperature overnight. The samples were sputtered with chrome and imaged using a JSM-6700F high-resolution field emission SEM (Jeol, Japan), operating at an acceleration voltage of 10 kV.
High-Resolution Transmission Electron Microscopy (HR-TEM)
Samples were prepared on 400-mesh copper grids, stained with 2% uranyl acetate, and imaged using a FEI F20 field emission gun transmission electron microscope (Tecnai, USA), operating at an acceleration voltage of 80 kV.
Circular Dichroism Spectroscopy (CD)
Samples were prepared in a 0.1 mm optical pathway cuvette by ×10 dilution of the examined peptide nanostructure solution. CD spectra were recorded using a Chirascan CD spectrometer (Applied Photophysics, UK), equipped with a temperature-controlled cell. The spectra were obtained with a resolution of 1 nm and were averaged from 3 separate measurements. Spectra were subtracted and smoothed using the Pro-Data software (Applied Photophysics, Leatherhead, UK).
Fourier-Transform Infrared Spectroscopy (FTIR)
Samples of 30 μL were deposited onto polyethylene IR sample cards (Sigma-Aldrich, Israel). The samples were dried overnight in vacuo. IR spectra were collected using a Nexus 470 FTIR spectrometer (Nicolet, Offenbach, Germany) with a deuterated triglycine sulfate (DTGS) detector. Measurements were made using the atmospheric suppression mode, averaging 64 scans at 2 cm−1 resolution. Subtraction and smoothing were performed using the OMNIC software (Nicolet, Offenbach, Germany). A second derivative calculation and curve fitting were then performed using the Peakfit software, version 4.12 (SYSTAT, Richmond, CA, USA).
Hydrogel Rheology
Hydrogels were prepared in situ and characterized using a Discovery HR-3 rheometer (TA Instruments, New Castle, DE, USA), operating with a 40 mm parallel plate geometry at a 600 μm measuring gap. Time-sweep oscillation tests were first performed at 0.5% strain and 5.0 Hz frequency. The linear viscoelastic region, at which the time-sweep tests were conducted, was examined by frequency-sweep (1.0−100 Hz) and strain amplitude-sweep tests (0.01−16.67%) after a 1200 s soak time to ensure gelation. Examination of thixotropic properties of the gels was done by periodically alternating strain from 0.1 to 100% in a time-sweep test.
X-ray Crystallography
Crystals suitable for X-ray diffraction were obtained by slowly evaporating a 25 mg/mL solution of z-FF in methanol at room temperature. A crystal was transferred to Hampton Paratone oil (Hampton Research) as a cryoprotectant, mounted on a MiTeGen loop, and flash frozen in liquid nitrogen.
X-ray diffraction data were collected at 100(2)K on a Rigaku XtaLabPro diffractometer equipped with a microfocus sealed tube and Max-Flux optics, Cu Kα1 (λ = 1.54184 Å) and a Dectris PILATUS S200 detector. The data were processed with CrysAlisPro 1.171.39.4c (Rigaku OD, 2015). The structure was solved by direct methods, using SHELXT-16. Full matrix least-squares refinement based on F2 with SHELXL-13 on 637 parameters gave a final R1= 0.0750 (based on F2) for data with I > 2σ(I) and R1= 0.2198 on 10182 reflections/Å−3. The crystallographic data of z-FF are given in Table S1 and the cif file in the Supporting Information.
The crystal structure was illustrated using Mercury 3.9 (Cambridge Crystallographic Data Centre, Cambridge, UK).
All-Atom Molecular Dynamics (MD) Simulations
All-atom MD simulations were carried out using the GROMACS 4.5.4 software package.58 The z-FF monomer was structurally optimized by Spartan’1059 and then energy minimized by the GAMESS software.60 Atomic partial charges were derived using the R.E.D III package.61 Other z-FF parameters were assigned based on the AMBER99SB force field.62 The z-FF molecules were randomly placed in a cubic 5 nm3 box and fully solvated with pre-equilibrated methanol,63 using the GAFF64 force field. z-FF systems (20-, 27-, and 50-mer) were simulated containing 1436, 1321, and 1037 methanol molecules, respectively. The concentrations used here were high compared to those of the experimental procedure in order to accelerate the assembly process and reduce the computational cost. Simulation length was 20 ns. Bond lengths within z-FF and methanol molecules were constrained by the LINCS65 and SETTLE algorithms,66 allowing an integration step of 2 fs. Electrostatic interactions were calculated using the particle mesh Ewald (PME) method with a real space cutoff of 1.0 nm. vdW interactions were calculated using a cutoff of 1.4 nm. The solute and solvent were separately coupled to an external temperature bath using a velocity rescaling method67 and a pressure bath using the Parrinello−Rahman method.68 The temperature and pressure were maintained at 298 K and 1 bar using coupling constants of 0.1 and 1.0 ps, respectively.
Coarse-Grained MD (CG-MD) Simulations
All MD simulations were performed using the GROMACS 4.5.3 software69 in combination with the MARTINI coarse-grained model, version 2.1.70,71 One phenylalanine residue was represented by four beads: one bead for the main chain (SC4 interaction) and three beads for the side chain ring (P5 interaction). The Cbz protecting group at the N-terminus of a z-FF molecule was represented by four beads: one bead for the C−O−C═O (the interaction type is Na according to Zheng et al.72) and three beads for the benzene ring. One ethanol molecule was represented by one bead (Figure S5). Using a P3 interaction type for the ethanol bead, we performed multiple microsecond time scale CG-MD simulations. Systems (300-, 600-, and 900-mer) were simulated in ethanol and a 1:1 ethanol−water mixture corresponding to concentrations of ~40, ~80, and ~120 mg/mL, respectively. The z-FF peptides were randomly placed in a 17.7 nm3 cubic box. Four independent microsecond MD simulations were carried out for each peptide concentration and solvent system, using similar temperature and pressure parameters as those used for the all-atom MD simulations, with integration steps of 20 fs. Electrostatic and vdW interactions were treated in their shifted forms with a cutoff of 1.2 nm. The neighbor-list was updated every 10 steps with a cutoff distance of 1.4 nm.
MD Simulations Analysis
The trajectory analyses were performed using in-house-developed codes and tools implemented in the Gromacs software package.58,69 The angle between two benzene rings was obtained by calculating the angle between the normal vectors of the two rings. If the angle was larger than 90°, the supplementary angle was used as the angle between the two benzene rings. Two benzene rings were considered only if their centroid distance was within 1.0 nm. The two-dimensional potential mean force (PMF) was constructed using the relation
where H(x,y) is the probability of a conformation having a certain value of two selected reaction coordinates, x and y. In this work, the angle and centroid distance between two benzene rings were chosen as x and y, respectively. The data in the last 10/700 ns of all-atom/CG MD trajectories, respectively, were used to construct the PMF. The end-to-end distance of a z-FF molecule was defined as the distance between the alkyl carbon of Cbz and the carbonyl carbon of the C-terminal phenylalanine residue. Trajectory visualization and graphical structure analysis were performed using the VMD software suite.73
Supplementary Material
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.7b07723.
Conditions matrices, HR-TEM images, Ramachandran plot analysis, additional rheological data, and coarse-graining scheme (PDF)
Acknowledgments
This project was funded by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (Grant agreement BISON-694426 to E.G.). We thank S. Gilead for her insight and the members of the Gazit lab for helpful discussions. G.W. gives thanks for the financial support from National Key R&D Program of China (Grant 2016YFA0501702) and the NSF of China (Grant 11674065-91227102). All simulations were performed using the high-performance computational facilities at the National High-Performance Computing Center of Fudan University and Guangzhou.
Footnotes
ORCID
Noam Brown: 0000-0003-3186-2706
Lihi Adler-Abramovich: 0000-0003-3433-0625
Guanghong Wei: 0000-0001-5814-3328
Notes
The authors declare no competing financial interest.
References
- (1).Mcgaughey GB, Gagné M, Rappé AK. π-Stacking Interactions. J Biol Chem. 1998;273:15458–15463. doi: 10.1074/jbc.273.25.15458. [DOI] [PubMed] [Google Scholar]
- (2).Whitesides GM, Mathias JP, Seto CT. Molecular Self-assembly and Nanochemistry: a Chemical Strategy for the Synthesis of Nanostructures. Science. 1991;254:1312–1319. doi: 10.1126/science.1962191. [DOI] [PubMed] [Google Scholar]
- (3).Hauser CAE, Zhang S. Peptides As Biological Semi-conductors. Nature. 2010;468:516–517. doi: 10.1038/468516a. [DOI] [PubMed] [Google Scholar]
- (4).Fleming S, Ulijn RV. Design of Nanostructures Based On Aromatic Peptide Amphiphiles. Chem Soc Rev. 2014;43:8150–8177. doi: 10.1039/c4cs00247d. [DOI] [PubMed] [Google Scholar]
- (5).Knowles TPJ, Buehler MJ. Nanomechanics of Functional and Pathological Amyloid Materials. Nat Nanotechnol. 2011;6:469–479. doi: 10.1038/nnano.2011.102. [DOI] [PubMed] [Google Scholar]
- (6).Ashkenasy N, Horne WS, Ghadiri MR. Design of Self-assembling Peptide Nanotubes with Delocalized Electronic States. Small. 2006;2:99–102. doi: 10.1002/smll.200500252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Zhang S. Fabrication of Novel Biomaterials Through Molecular Self-assembly. Nat Biotechnol. 2003;21:1171–1178. doi: 10.1038/nbt874. [DOI] [PubMed] [Google Scholar]
- (8).Ikezoe Y, Washino G, Uemura T, Kitagawa S, Matsui H. Autonomous Motors of a Metal–Organic Framework Powered By Reorganization of Self-assembled Peptides at Interfaces. Nat Mater. 2012;11:1081–1085. doi: 10.1038/nmat3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Herbst F, Döhler D, Michael P, Binder WH. Self-Healing Polymers Via Supramolecular Forces. Macromol Rapid Commun. 2013;34:203–220. doi: 10.1002/marc.201200675. [DOI] [PubMed] [Google Scholar]
- (10).Rissanou AN, Georgilis E, Kasotakis E, Mitraki A, Harmandaris V. Effect of Solvent on the Self-Assembly of Dialanine and Diphenylalanine Peptides. J Phys Chem B. 2013;117:3962–3975. doi: 10.1021/jp311795b. [DOI] [PubMed] [Google Scholar]
- (11).Keates RAB, Hall RH. Tubulin Requires an Accessory Protein for Self Assembly Into Microtubules. Nature. 1975;257:418–421. doi: 10.1038/257418a0. [DOI] [PubMed] [Google Scholar]
- (12).Gosline JM, Guerette PA, Ortlepp CS, Savage KN. The Mechanical Design of Spider Silks: from Fibroin Sequence to Mechanical Function. J Exp Biol. 1999;202:3295–3303. doi: 10.1242/jeb.202.23.3295. [DOI] [PubMed] [Google Scholar]
- (13).Vollrath F, Knight DP. Liquid Crystalline Spinning of Spider Silk. Nature. 2001;410:541–548. doi: 10.1038/35069000. [DOI] [PubMed] [Google Scholar]
- (14).Paine ML, Snead ML. Protein Interactions During Assembly of the Enamel Organic Extracellular Matrix. J Bone Miner Res. 1997;12:221–227. doi: 10.1359/jbmr.1997.12.2.221. [DOI] [PubMed] [Google Scholar]
- (15).Kikuchi M, Itoh S, Ichinose S, Shinomiya K, Tanaka J. Self-Organization Mechanism in a Bone-Like Hydroxyapatite/Collagen Nanocomposite Synthesized In Vitro and Its Biological Reaction. Biomaterials. 2001;22:1705–1711. doi: 10.1016/s0142-9612(00)00305-7. [DOI] [PubMed] [Google Scholar]
- (16).Reches M, Gazit E. Casting Metal Nanowires Within Discrete self-assembled peptide nanotubes. Science. 2003;300:625–627. doi: 10.1126/science.1082387. [DOI] [PubMed] [Google Scholar]
- (17).Görbitz CH. The Structure of Nanotubes Formed by Diphenylalanine, the Core Recognition Motif of Alzheimer’s β-Amyloid Polypeptide. Chem Commun. 2006:2332–2334. doi: 10.1039/b603080g. [DOI] [PubMed] [Google Scholar]
- (18).Kol N, Adler-Abramovich L, Barlam D, Shneck RZ, Gazit E, Rousso I. Self-assembled Peptide Nanotubes are Uniquely Rigid Bioinspired Supramolecular Structures. Nano Lett. 2005;5:1343–1346. doi: 10.1021/nl0505896. [DOI] [PubMed] [Google Scholar]
- (19).Kholkin A, Amdursky N, Bdikin I, Gazit E, Rosenman G. Strong Piezoelectricity in Bioinspired Peptide Nanotubes. ACS Nano. 2010;4:610–614. doi: 10.1021/nn901327v. [DOI] [PubMed] [Google Scholar]
- (20).Amdursky N, Molotskii M, Gazit E, Rosenman G. Self-assembled Bioinspired Quantum Dots: Optical Properties. Appl Phys Lett. 2009;94 261907. [Google Scholar]
- (21).Azuri I, Adler-Abramovich L, Gazit E, Hod O, Kronik L. Why Are Diphenylalanine-based Peptide Nanostructures So Rigid? Insights from First Principles Calculations. J Am Chem Soc. 2014;136:963–969. doi: 10.1021/ja408713x. [DOI] [PubMed] [Google Scholar]
- (22).Adler-Abramovich L, Reches M, Sedman VL, Allen S, Tendler SJB, Gazit E. Thermal and Chemical Stability of Diphenylalanine Peptide Nanotubes: Implications for Nanotechnological Applications. Langmuir. 2006;22:1313–1320. doi: 10.1021/la052409d. [DOI] [PubMed] [Google Scholar]
- (23).Mahler A, Reches M, Rechter M, Cohen S, Gazit E. Rigid, Self-assembled Hydrogel Composed of a Modified Aromatic Dipeptide. Adv Mater. 2006;18:1365–1370. [Google Scholar]
- (24).Rosenman G, Amdursky N, Molotskii M, Aronov D, Adler-Abramovich L, Gazit E. Blue Luminescence Based on Quantum Confinement at Peptide Nanotubes. Nano Lett. 2009;9:3111–3115. doi: 10.1021/nl9008265. [DOI] [PubMed] [Google Scholar]
- (25).Esin A, Baturin I, Nikitin T, Vasilev S, Salehli F, Shur VY, Kholkin AL. Pyroelectric Effect and Polarization Instability in Self-assembled Diphenylalanine Microtubes. Appl Phys Lett. 2016;109 142902. [Google Scholar]
- (26).Kim S, Kim JH, Lee JS, Park CB. β-Sheet-forming, Self-assembled Peptide Nanomaterials Towards Optical, Energy, and Healthcare Applications. Small. 2015;11:3623–3640. doi: 10.1002/smll.201500169. [DOI] [PubMed] [Google Scholar]
- (27).Rosenman G, Beker P, Koren I, Yevnin M, Bank-Srour B, Mishina E, Semin S. Bioinspired Peptide Nanotubes: Deposition Technology, Basic Physics and Nanotechnology Applications. J Pept Sci. 2011;17:75–87. doi: 10.1002/psc.1326. [DOI] [PubMed] [Google Scholar]
- (28).Li Q, Jia Y, Dai L, Yang Y, Li J. Controlled Rod Nanostructured Assembly of Diphenylalanine and Their Optical Waveguide Properties. ACS Nano. 2015;9:2689–2695. doi: 10.1021/acsnano.5b00623. [DOI] [PubMed] [Google Scholar]
- (29).Reches M, Gazit E. Designed Aromatic Homo-Dipeptides: Formation of Ordered Nanostructures and Potential Nanotechnological Applications. Phys Biol. 2006;3:S10–S19. doi: 10.1088/1478-3975/3/1/S02. [DOI] [PubMed] [Google Scholar]
- (30).Santhanamoorthi N, Kolandaivel P, Adler-Abramovich L, Gazit E, Filipek S, Viswanathan S. Diphenylalanine Peptide Nanotube: Charge Transport, Band Gap and Its Relevance to Potential Biomedical Applications. Adv Mater Lett. 2011;2:100–105. [Google Scholar]
- (31).Wang M, Xiong S, Wu X, Chu PK. Effects of Water Molecules On Photoluminescence from Hierarchical Peptide Nanotubes and Water Probing Capability. Small. 2011;7:2801–2807. doi: 10.1002/smll.201100353. [DOI] [PubMed] [Google Scholar]
- (32).Yan X, Zhu P, Li J. Self-Assembly and Application of Diphenylalanine-based Nanostructures. Chem Soc Rev. 2010;39:1877–1890. doi: 10.1039/b915765b. [DOI] [PubMed] [Google Scholar]
- (33).Bosne ED, Heredia A, Kopyl S, Karpinsky DV, Pinto AG, Kholkin AL. Piezoelectric Resonators Based on Self-assembled Diphenylalanine Microtubes. Appl Phys Lett. 2013;102 073504. [Google Scholar]
- (34).Gazit E. The ‘Correctly Folded’ State of Proteins: Is It a Metastable State? Angew Chem, Int Ed. 2002;41:257–259. doi: 10.1002/1521-3773(20020118)41:2<257::aid-anie257>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- (35).Reches M, Gazit E. Self-Assembly of Peptide Nanotubes and Amyloid-Like Structures By Charged-Termini-capped Diphenylalanine Peptide Analogues. Isr J Chem. 2005;45:363–371. [Google Scholar]
- (36).Levin A, Mason TO, Adler-Abramovich L, Buell AK, Meisl G, Galvagnion C, Bram Y, Stratford Sa, Dobson CM, Knowles TPJ, Gazit E. Ostwald’s Rule of Stages Governs Structural Transitions and Morphology of Dipeptide Supramolecular Polymers. Nat Commun. 2014;5 doi: 10.1038/ncomms6219. 5219. [DOI] [PubMed] [Google Scholar]
- (37).Guo C, Arnon ZA, Qi R, Zhang Q, Adler-Abramovich L, Gazit E, Wei G. Expanding the Nanoarchitectural Diversity Through Aromatic Di- and Tri-Peptide Coassembly: Nanostructures and Molecular Mechanisms. ACS Nano. 2016;10:8316–8324. doi: 10.1021/acsnano.6b02739. [DOI] [PubMed] [Google Scholar]
- (38).Mason TO, Chirgadze DY, Levin A, Adler-Abramovich L, Gazit E, Knowles TPJ, Buell AK. Expanding the Solvent Chemical Space for Self-assembly of Dipeptide Nanostructures. ACS Nano. 2014;8:1243–1253. doi: 10.1021/nn404237f. [DOI] [PubMed] [Google Scholar]
- (39).Frederix PWJM, Scott GG, Abul-Haija YM, Kalafatovic D, Pappas CG, Javid N, Hunt NT, Ulijn RV, Tuttle T. Exploring the Sequence Space for (Tri-)Peptide Self-assembly to Design and Discover New Hydrogels. Nat Chem. 2015;7:30–37. doi: 10.1038/nchem.2122. [DOI] [PubMed] [Google Scholar]
- (40).Görbitz CH. Nanotubes from Hydrophobic Dipeptides: Pore Size Regulation Through Side Chain Substitution. New J Chem. 2003;27:1789–1793. [Google Scholar]
- (41).Orbach R, Adler-Abramovich L, Zigerson S, Mironi-Harpaz I, Seliktar D, Gazit E. Self-Assembled Fmoc-Peptides As a Platform for the Formation of Nanostructures and Hydrogels. Biomacromolecules. 2009;10:2646–2651. doi: 10.1021/bm900584m. [DOI] [PubMed] [Google Scholar]
- (42).German HW, Uyaver S, Hansmann UHE. Self-Assembly of Phenylalanine-Based Molecules. J Phys Chem A. 2015;119:1609–1615. doi: 10.1021/jp5077388. [DOI] [PubMed] [Google Scholar]
- (43).Do TD, Kincannon WM, Bowers MT. Phenylalanine Oligomers and Fibrils: The Mechanism of Assembly and the Importance of Tetramers and Counterions. J Am Chem Soc. 2015;137:10080–10083. doi: 10.1021/jacs.5b05482. [DOI] [PubMed] [Google Scholar]
- (44).Tamamis P, Adler-Abramovich L, Reches M, Marshall K, Sikorski P, Serpell L, Gazit E, Archontis G. Self-Assembly of Phenylalanine Oligopeptides: Insights from Experiments and Simulations. Biophys J. 2009;96:5020–5029. doi: 10.1016/j.bpj.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Guo C, Luo Y, Zhou R, Wei G. Probing the Self-Assembly Mechanism of Diphenylalanine-Based Peptide. ACS Nano. 2012;6:3907–3918. doi: 10.1021/nn300015g. [DOI] [PubMed] [Google Scholar]
- (46).Jeon J, Mills CE, Shell MS. Molecular Insights into Diphenylalanine Nanotube assembly: All-atom simulations of oligomerization. J Phys Chem B. 2013;117:3935–3943. doi: 10.1021/jp308280d. [DOI] [PubMed] [Google Scholar]
- (47).Guo C, Luo Y, Zhou R, Wei G. Triphenylalanine peptides self-assemble into Nanospheres and Nanorods That Are Different from the Nanovesicles and Nanotubes Formed by Diphenylalanine Peptides. Nanoscale. 2014;6:2800–2811. doi: 10.1039/c3nr02505e. [DOI] [PubMed] [Google Scholar]
- (48).Frederix PWJM, Ulijn RV, Hunt NT, Tuttle T. Virtual Screening for Dipeptide Aggregation: Toward Predictive Tools for Peptide Self-Assembly. J Phys Chem Lett. 2011;2:2380–2384. doi: 10.1021/jz2010573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Haris PI, Chapman D. The Conformational Analysis of Peptides Using Fourier Transform IR Spectroscopy. Biopolymers. 1995;37:251–263. doi: 10.1002/bip.360370404. [DOI] [PubMed] [Google Scholar]
- (50).Kong J, Yu S. Fourier Transform Infrared Spectroscopic Analysis of Protein Secondary Structures. Acta Biochim Biophys Sin. 2007;39:549–559. doi: 10.1111/j.1745-7270.2007.00320.x. [DOI] [PubMed] [Google Scholar]
- (51).Strickland EH, Beychok S. Aromatic Contributions to Circular Dichroism Spectra of Protein. Crit Rev Biochem. 1974;2:113–175. doi: 10.3109/10409237409105445. [DOI] [PubMed] [Google Scholar]
- (52).Mahalakshmi R, Shanmugam G, Polavarapu PL, Balaram P. Circular Dichroism of Designed Peptide Helices and β-Hairpins: Analysis of Trp- and Tyr-Rich Peptides. ChemBioChem. 2005;6:2152–2158. doi: 10.1002/cbic.200500152. [DOI] [PubMed] [Google Scholar]
- (53).Shlomo Z, Vinod TP, Jelinek R, Rapaport H. Stacking Interactions by Two Phe Side Chains Stabilize and Orient Assemblies of Even the Minimal Amphiphilic β-Sheet Motif. Chem Commun. 2015;51:3154–3157. doi: 10.1039/c4cc09673h. [DOI] [PubMed] [Google Scholar]
- (54).Barnes HA. Thixotropy—a Review. J Non-Newtonian Fluid Mech. 1997;70:1–33. [Google Scholar]
- (55).Gazit E. A Possible Role for π-stacking in the Self-Assembly of Amyloid Fibrils. FASEB J. 2002;16:77–83. doi: 10.1096/fj.01-0442hyp. [DOI] [PubMed] [Google Scholar]
- (56).Xie L, Luo Y, Wei G. Aβ(16–22) Peptides Can Assemble into Ordered β-Barrels and Bilayer β-Sheets, While Substitution of Phenylalanine 19 by Tryptophan Increases the Population of Disordered Aggregates. J Phys Chem B. 2013;117:10149–10160. doi: 10.1021/jp405869a. [DOI] [PubMed] [Google Scholar]
- (57).Makin OS, Atkins E, Sikorski P, Johansson J, Serpell LC. Molecular Basis for Amyloid Fibril Formation and Stability. Proc Natl Acad Sci U S A. 2005;102:315–20. doi: 10.1073/pnas.0406847102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Hess B, Kutzner C, Van Der Spoel D, Lindahl E. GROMACS 4: Algorithms for Highly Efficient, Load-balanced, and Scalable Molecular Simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- (59).Shao Y, Molnar LF, Jung Y, Kussmann J, Ochsenfeld C, Brown ST, Gilbert ATB, Slipchenko LV, Levchenko SV, O’neill DP, Distasio RA, et al. Advances in Methods and Algorithms in a Modern Quantum Chemistry Program Package. Phys Chem Chem Phys. 2006;8:3172–3191. doi: 10.1039/b517914a. [DOI] [PubMed] [Google Scholar]
- (60).Schmidt MW, Baldridge KK, Boatz JA, Elbert ST, Gordon MS, Jensen JH, Koseki S, Matsunaga N, Nguyen KA, Su S, Windus TL, et al. General Atomic and Molecular Electronic Structure System. J Comput Chem. 1993;14:1347–1363. [Google Scholar]
- (61).Dupradeau F-Y, Pigache A, Zaffran T, Savineau C, Lelong R, Grivel N, Lelong D, Rosanski W, Cieplak P. The R.E.D. Tools: Advances in RESP and ESP Charge Derivation and Force Field Library Building. Phys Chem Chem Phys. 2010;12:7821–7839. doi: 10.1039/c0cp00111b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of Multiple Amber Force Fields and Development of Improved Protein Backbone Parameters. Proteins: Struct, Funct, Genet. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Caleman C, van Maaren PJ, Hong M, Hub JS, Costa LT, van der Spoel D. Force Field Benchmark of Organic Liquids: Density, Enthalpy of Vaporization, Heat Capacities, Surface Tension, Isothermal Compressibility, Volumetric Expansion Coefficient, and Dielectric Constant. J Chem Theory Comput. 2012;8:61–74. doi: 10.1021/ct200731v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and Testing of a General Amber Force Field. J Comput Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- (65).Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: A Linear Constraint Solver for Molecular Simulations. J Comput Chem. 1997;18:1463–1472. [Google Scholar]
- (66).Miyamoto S, Kollman PA. Settle: An Analytical Version of the SHAKE and RATTLE Algorithm for Rigid Water Models. J Comput Chem. 1992;13:952–962. [Google Scholar]
- (67).Bussi G, Donadio D, Parrinello M. Canonical Sampling through Velocity Rescaling. J Chem Phys. 2007;126 doi: 10.1063/1.2408420. 014101. [DOI] [PubMed] [Google Scholar]
- (68).Parrinello M, Rahman A. Polymorphic Transitions in Single Crystals: A New Molecular Dynamics Method. J Appl Phys. 1981;52:7182–7190. [Google Scholar]
- (69).Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: Fast, Flexible, and Free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- (70).Monticelli L, Kandasamy SK, Periole X, Larson RG, Tieleman DP, Marrink S-J. The MARTINI Coarse-grained Force Field: Extension to Proteins. J Chem Theory Comput. 2008;4:819–834. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- (71).Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. The MARTINI Force Field: Coarse Grained Model for Biomolecular Simulations. J Phys Chem B. 2007;111:7812. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- (72).Zheng X, Zhu L, Zeng X, Meng L, Zhang L, Wang D, Huang X. Kinetics-Controlled Amphiphile Self-Assembly Processes. J Phys Chem Lett. 2017;8:1798–1803. doi: 10.1021/acs.jpclett.7b00160. [DOI] [PubMed] [Google Scholar]
- (73).Humphrey W, Dalke A, Schulten K. VMD: Visual Molecular Dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.