Abstract
BACKGROUND
Shared traits between prokaryotes and eukaryotes are helpful in the understanding of the tree of life evolution. In bacteria and eukaryotes, it has been shown a particular organisation of tRNA genes as clusters, but this trait has not been explored in the archaea domain.
OBJECTIVE
Explore the occurrence of tRNA gene clusters in archaea.
METHODS
In-silico analyses of complete and draft archaeal genomes based on tRNA gene isotype and synteny, tRNA gene cluster content and mobilome elements.
FINDINGS
We demonstrated the prevalence of tRNA gene clusters in archaea. tRNA gene clusters, composed of archaeal-type tRNAs, were identified in two Archaea class, Halobacteria and Methanobacteria from Euryarchaeota supergroup. Genomic analyses also revealed evidence of the association between tRNA gene clusters to mobile genetic elements and intra-domain horizontal gene transfer.
MAIN CONCLUSIONS
tRNA gene cluster occurs in the three domains of life, suggesting a role of this type of tRNA gene organisation in the biology of the living organisms.
Key words: archaea, tRNA gene cluster, plasmid, haloarchaea, virus, Euryarchaeota
The three domains of life, Archaea, Bacteria and Eukarya are characterised by unique features, even though they share a set of basic characteristics. Archaea and Bacteria share ribosomal genes, transcriptional regulators and overall genome structure, while archaea and eukaryotes are more related about the information transfer system, including replication, transcription and translation processes. 1 , 2 Transfer RNAs (tRNAs) have a major role in the translation machinery and, therefore, belong to the core system of the three domains of life. This molecule is associated with a huge diversity of gene organisation, including canonical and disrupted genes. The latter gene species was revealed to be prevalent among archaea genomes. 3 The tRNA genes are found dispersed in the genomes but occasionally, as in mitogenomes, they are organised in clusters containing two to five tRNA genes. 4 Besides mitogenomes, tRNA gene clusters have been shown to be abundant in Eukarya. 5 In prokaryotes, tRNA gene clusters were characterised in some bacteria, being prevalent in Firmicutes phylum and Mycobacterium genus, 6 , 7 while in archaea, the presence of one tRNA gene cluster was only identified in the Methanobrevibacter M1 strain genome. 6 In addition, tRNA gene clusters have also been particularly observed in mycobacteriophages. 7 , 8 In this case, these tRNA genes could be useful in the late lytic growth overcoming the degradation and/or differences in the codon bias of the host tRNAs. 8 The prevalence of tRNA gene clusters in bacteria, eukaryotes and bacteriophages raise a possible role of this type of gene organisation in the biology of these organisms. The presence of tRNA genes organised in a cluster, some encoding for all the tRNA isotypes, as well as unusual tRNA isotypes (e.g., selenocysteine, pyrrolysine), would provide to the organisms a set of genes basic to protein synthesis in a one-step acquisition way, what may positively impact their fitness. Identifying tRNA gene clusters in the three domains of life is likely to reveal their evolutionary history as well any current function. Therefore, we hypothesised the prevalence of tRNA gene cluster in archaea. In order to test our hypothesis, we analysed 2481 complete and draft archaea genomes overrepresented by Euryarchaeota supergroup.
MATERIALS AND METHODS
Genomes analysed - A total of 2481 complete and draft archaeal genomes (available in May 2018) were retrieved from National Center for Biotechnology Information (NCBI) FTP site (ftp://ftp.ncbi.nlm.nih.gov/genomes/genbank/archaea/).
tRNA gene prediction and tRNA gene cluster identification - The tRNA gene prediction of the data set was primarily performed by tRNAscan-SE 2.0 9 using the archaeal model, and inconsistent predictions were verified using ARAGORN v1.2.38. 10 Here we surveyed tRNA gene clusters with a minimum of 10 tRNA genes and used the approach described in a previous study to identify them. 7 tRNA genes were considered clustered if presented a tRNA gene density ≥ 2 tRNA/kb. 5
Genetic analysis - The genetic relationships of the genomes harboring tRNA gene clusters (Euryarchaeota supergroup) were assayed performing a core genome Multilocus Sequence Analysis (cgMLSA). The orthologous genes were retrieved using GET_HOMOLOGUES v3.0.5 11 considering a coverage of ≥ 70% and identity ≥ 40%. The 66 orthologous genes were concatenated (yielding ~75 kb length) and submitted to Maximum-likelihood analysis using PhyML v3.1. 12 Genetic relationships of the tRNA genes from the tRNA gene clusters were also assessed by concatenation of their nucleotide sequence (~3 kb length), and subsequent Maximum-likelihood analysis. The alignments were performed by MAFFT v7.271, 13 and the tree figures generated by iTOL. 14
Sequence annotation, plasmid, and viral identification - The sequence of tRNA gene cluster regions and their flanking regions (~10 kb) were annotated using Prokka v1.12. 15 A bipartite network of gene content was build using AcCNET v1.2 16 and visualised in Cytoscape v3.6.0. 17 The topology of the contigs harboring tRNA gene clusters was assayed as described by Jørgensen et al. 18 . These contigs were also submitted to blastn and virSorter 19 analysis for plasmid and viral/proviral identification, respectively.
RESULTS
Detection and distribution of tRNA gene clusters among archaea genomes - In order to identify the presence of tRNA gene clusters in archaea genomes, we used the methodology described in a previous study. 7 The analysis identified tRNA gene clusters in 29/2481 archaea genomes from GenBank (Table). The number of tRNA genes in the arrays ranged from 10 to 29, including five clusters with 20 or more tRNA genes. The tRNA clusters encompassed regions from ~1.1 to ~7 kb, representing a density from ~2.14 to ~9.17 tRNA gene/kb (Table). The number of tRNA genes present in these clusters corresponds to ~ 17 to 46% of the total number of tRNA genes in these archaea genomes. Two genomes, Methanobrevibacter sp. RUG344 and Methanobrevibacter sp. RUG833 were the only ones harboring two tRNA gene clusters (Table).
TABLE. Archaeal genomes harboring tRNA gene clusters.
| Genome | Genome (Mb) | # tRNAs (genome) | # tRNAs (cluster) | Length (bp) | BEGIN (bp) | END (bp) | Density (tRNA/kb) | Archaeal type | Isolation location | Access number | # pseudogenes | # introns |
| Haloarcula amylolytica JCM 13557 | 4.3 | 70 | 23 | 6311 | 21449 | 27759 | 3.64 | Halophilic | China | AOLW01000050.1 | 2 | - |
| Haloarcula argentinensis DSM 12282 | 4.2 | 71 | 22 | 6998 | 216765 | 223762 | 3.14 | Halophilic | Argentina | AOLX01000027.1 | 2 | - |
| Haloarcula californiae ATCC 33799 | 4.4 | 60 | 12 | 2825 | 9001 | 11825 | 4.24 | Halophilic | - | AOLS01000072.1 | 2 | - |
| Haloarcula vallismortis ATCC 29715 | 4.2 | 72 | 25 | 3569 | 254165 | 257733 | 7.00 | Halophilic | USA | AOLQ01000004.1 | 3 | 1 |
| Haloarculaceae archeaon HArcel1 | 2.7 | 66 | 18 | 5757 | 2632007 | 2637763 | 3.12 | Halophilic | Russia | CP028858.1 | 1 | - |
| Halobellus rufus CBA1103 | 3.8 | 60 | 14 | 4602 | 6479 | 11080 | 3.04 | Halophilic | - | BBJO01000043.1 | 1 | - |
| Halobiforma haloterrestris DSM 13078 | 4.4 | 61 | 14 | 5924 | 13582 | 19505 | 2.36 | Halophilic | - | FOKW01000014.1 | 1 | - |
| Halogranum rubrum CGMCC 1.7738 | 4.5 | 70 | 15 | 3838 | 160615 | 164452 | 3.90 | Halophilic | - | FOTC01000006.1 | 1 | - |
| Halogranum salarium B-1 | 4.5 | 70 | 15 | 3838 | 40762 | 44599 | 3.90 | Halophilic | South Korea | ALJD01000014.1 | 3 | - |
| Halomicrobium zhouii CGMCC 1.10457 | 4.2 | 62 | 14 | 5662 | 253014 | 258675 | 2.47 | Halophilic | - | FOZK01000002.1 | - | - |
| Halopenitus persicus CBA1233 | 2.9 | 65 | 16 | 3934 | 1151067 | 1155000 | 4.06 | Halophilic | South Korea | AP017558.1 | 2 | - |
| Halopenitus persicus DC30 | 3.4 | 61 | 14 | 4601 | 55752 | 60352 | 3.04 | Halophilic | - | FNPC01000012.1 | 1 | - |
| Haloplanus natans DSM 17983 | 3.8 | 66 | 14 | 6473 | 401627 | 408099 | 2.16 | Halophilic | Israel | ATYM01000003.1 | 3 | - |
| Halorientalis persicus IBRC-M 10043 | 5 | 81 | 29 | 5739 | 15276 | 21014 | 5.05 | Halophilic | Iran | FOCX01000031.1 | 6 | 2 |
| Halorientalis regularis IBRC-M 10760 | 4 | 66 | 16 | 6676 | 110496 | 117171 | 2.39 | Halophilic | - | FNBK01000012.1 | 2 | 2 |
| Halorientalis sp. IM1011 | 3.8 | 79 | 29 | 5677 | 2239040 | 2244716 | 5.10 | Halophilic | China | CP019067.1 | 5 | 1 |
| Halorubrum chaoviator DSM 19316 | 3.6 | 62 | 15 | 4553 | 11178 | 15730 | 3.29 | Halophilic | Mexico | FZNK01000013.1 | 3 | - |
| Halorubrum ezzemoulense DSM 17463 | 3.6 | 63 | 15 | 4606 | 2376 | 6981 | 3.25 | Halophilic | - | NEDJ01000026.1 | 2 | - |
| Halorubrum ezzemoulense LD3 | 3.7 | 60 | 14 | 4552 | 12826 | 17377 | 3.07 | Halophilic | Iran | NHOW01000026.1 | 1 | - |
| Halorubrum ezzemoulense LG1 | 4.6 | 72 | 14 | 4554 | 12827 | 17380 | 3.07 | Halophilic | Iran | NHOV01000832.1 | 1 | - |
| Halorubrum sp. BV1 | 2.7 | 63 | 14 | 4541 | 180660 | 185200 | 3.08 | Halophilic | - | JQKV01000005.1 | 2 | - |
| Halorubrum sp. SD683 | 3.1 | 62 | 14 | 4607 | 33841 | 38447 | 3.03 | Halophilic | Namibia | NEWJ01000007.1 | 1 | - |
| Halorubrum vacuolatum DSM 8800 | 3.4 | 61 | 12 | 3781 | 10871 | 14651 | 3.17 | Halophilic | Kenya | FZNQ01000022.1 | 1 | - |
| Methanobrevibacter ruminantium M1 | 3 | 57 | 18 | 3608 | 360196 | 363803 | 4.98 | Methanogenic | - | CP001719.1 | 2 | - |
| Methanobrevibacter sp. RUG344 | 2.3 | 63 | 19 | 3477 | 32563 | 36039 | 5.46 | Methanogenic | Scotland | ONJC01000013.1 | 4 | - |
| 10 | 1115 | 27774 | 28888 | 8.96 | ONJC01000014.1 | 2 | - | |||||
| Methanobrevibacter sp. RUG648 | 2.3 | 40 | 13 | 2348 | 6705 | 9052 | 5.53 | Methanogenic | Scotland | ONUN01000043.1 | 4 | - |
| Methanobrevibacter sp. RUG833 | 2.5 | 73 | 16 | 1743 | 187745 | 189487 | 9.17 | Methanogenic | Scotland | OMSG01000001.1 | 6 | - |
| 15 | 6999 | 40007 | 47005 | 2.14 | OMSG01000002.1 | 12 | - | |||||
| Methanosarcina mazei 1.H.A.2.3 | 4 | 68 | 12 | 2884 | 647 | 3530 | 4.16 | Methanogenic | USA | JJQM01000083.1 | - | - |
| Methanosarcina mazei 1.H.A.2.6 | 4 | 69 | 12 | 2759 | 531 | 3289 | 4.34 | Methanogenic | USA | JJQN01000165.1 | - | - |
#: pseudogenes/introns, number of these elements in the tRNA gene clusters.
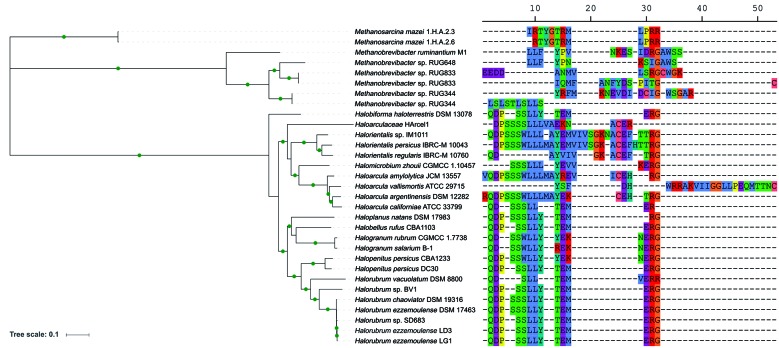
The 29 genomes harboring the tRNA gene clusters correspond to 18 species and seven spp. belonging to 11 archaea genera, all from Euryarchaeota supergroup, including Haloarcula, Halobellus, Halobiforma, Halogranum, Halomicrobium, Halopenitus, Haloplanus, Halorientalis, Halorubrum, Methanobrevibacter, and Methanosarcina. The latter two genera are from methanogenic archaea, while the others, halophilic. Analysing the sequences of the tRNA genes in the clusters, we observed that all but Methanosarcina are archaeal-type tRNAs. The phylogenetic analysis revealed that all genomes from Halorubrum genus but Halorubrum vacuolatum DSM 8800 and Halorubrum sp. BV1, belong to a single lineage as well as the two genomes from Methanosarcina mazei (Fig. 1). Strains harboring tRNA gene clusters with a same tRNA amino acid isotype pattern were isolated from different geographic locations, even considering species from the same lineage (Table). Despite the evolutionary relationship among some strains, their tRNA isotype organisation and genomic context differ substantially (Figs 1-2). Interestingly, 23/29 and 6/29 tRNA gene clusters were identified among 239 halophilic and 618 methanogenic archaea genomes, respectively, revealing a bias to the prevalence of tRNA gene clusters in halophilic archaea. On the other hand, tRNA gene clusters were not identified in any other archaea supergroup.
Fig. 1: maximum likelihood tree based on 66 concatenated orthologous genes. In the right side, the tRNA isotype organisation (using the single-letter amino acid code) of the tRNA gene clusters from each genome is related to the tree branches. The gaps (- symbol) may not represent the actual distance between two adjacent tRNA genes, but the distance from the reference tRNA gene cluster. The green circles in the branches indicate bootstrap values ≥ 70.
tRNA gene cluster organisation and composition - Overall, the halophilic archaea share tRNA gene clusters presenting similar tRNA amino acid isotype composition and synteny. The exception is the Haloarcula vallismortis ATCC 29715 which has a distinct tRNA amino acid isotype composition (Fig. 1). In contrast, the methanogenic archaea harboring tRNA gene clusters present a quite distinct tRNA isotype composition and synteny. This scenario is also observed when considering the nucleotide sequences of the tRNA genes from the clusters [Supplementary data (2.4MB, pdf) (Fig. 1)]. The most represented isotypes in the tRNA gene clusters were tRNA-Leu and tRNA-Ser, with up to four gene copies per cluster, while the less frequent being tRNA-His and tRNA-Phe, absent in 25 and 21 tRNA gene clusters, respectively [Supplementary data (2.4MB, pdf) (Table I)]. Only H. vallismortis ATCC 29715 tRNA gene cluster presented all universal 20 isotypes, while Halorientalis sp. IM1011 cluster only has tRNA-His absent. In 26/29 archaea genomes, the tRNA-Tyr and tRNA-Glu from tRNA gene clusters represent a significant mean increment (~45%) to the strains. In general, all tRNA isotypes present in the tRNA gene clusters are redundant with the tRNA isotype repertoire from archaea genomes, the exception is tRNA-Gly in Methanobrevibacter sp. RUG833, which is only present in its tRNA gene cluster [Supplementary data (2.4MB, pdf) (Table I)]. However, we observed that for 9/29 genomes there was an increase of isoacceptors provided by some tRNA gene clusters, which increased from one to eight species, depending on the strain [Supplementary data (2.4MB, pdf) (Table II)]. Almost all clusters presented tRNA genes annotated as pseudogenes. In Methanobrevibacter sp. RUG833, which harbors two tRNA gene clusters summing 31 tRNA genes, 17 tRNA genes were annotated as pseudogenes (Table). This was also observed among the Halorientalis genomes, which also showed some tRNA genes with introns.
Gene content of archaea tRNA gene cluster - The number of protein-coding genes accompanying the tRNA gene clusters is variable, most of them encoding hypothetical proteins. A gene encoding for a TROVE domain-containing protein is presented in 20/29 clusters [Supplementary data (2.4MB, pdf) (Fig. 2)]. The majority of the genes shared by the clusters belong to the halophilic archaea (except for H. vallismortis and Halogranum genomes), therefore this group represents the basis of the tRNA gene content network in archaea [Supplementary data (2.4MB, pdf) (Fig. 3)].
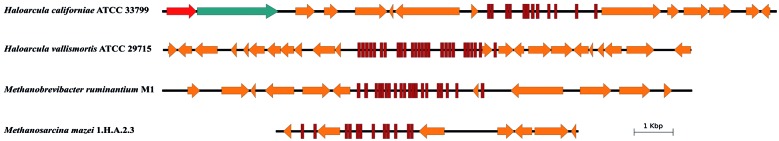
Fig. 2: linear representation of the tRNA gene cluster genomic regions of some representative genomes. The orange arrows and red bars represent CDS and tRNA genes, respectively. In Haloarcula californiae representation the red and blue arrows indicate the genes encoding for Relaxase and conjugative transfer protein TraD, respectively.
Several genes associated with DNA transfer mechanisms were identified in the genomic context of the tRNA gene clusters, including transposases, integrases, HNH endonucleases, relaxase, toxin-antitoxin systems and secretion systems related genes [Supplementary data (2.4MB, pdf) (Table III)]. Besides these, there were also resistance genes to antibiotics/metals (teicoplanin, bleomycin, chloramphenicol, tetracycline, arsenic). Interestingly, Halogranum salarium and Halogranum rubrum genomes presented a gene encoding for a conjugal transfer protein (HerA). In the methanogenic archaea genomes, it was identified a gene encoding for a putative phage holin family protein (Methanobrevibacter ruminantium M1), and in M. mazei there were protein-coding genes with high similarity to bacteria, bacteriophages, and plasmids from Proteobacteria phylum [Supplementary data (2.4MB, pdf) (Table IV)], including chloramphenicol O-acetyltransferase gene sequences [Supplementary data (2.4MB, pdf) (Fig. 4)].
tRNA gene clusters in archaea mobilome - Since many genes surrounding the tRNA gene clusters are related to lateral gene transfer and the GC content of some tRNA gene cluster regions contrast with the genomic GC content [Supplementary data (2.4MB, pdf) (Table V)], we surveyed the contigs in order to find sequences related to mobile genetic elements. Among the methanogens, a quite discrepancy intra and inter tRNA gene cluster region GC content could be observed, but none evidence of mobile genetic elements presence was obtained.
By the other hand, in the Haloarcula californiae ATCC 33799 strain there is a ~51 kb length contig (access number AOLS01000072.1) harboring the tRNA gene cluster and protein-coding genes with amino acid sequence similarity to Relaxase (identity 94% and coverage 100%) and conjugative TraD proteins (identity 95% and coverage 100%) present in the pNYT1 plasmid from Haloarcula taiwanensis strain Taiwanensis. In addition, it also presents a type II toxin-antitoxin system from hicA/B family. In Halorubrum sp. BV1, a contig with ~188 kb (access number JQKV01000005.1) also presented similarity with sequences from many archaea plasmids, showing coverage of 30% and identity of 96% with Halorubrum trapanicum plasmid pCBA1232-02. Besides that, the in-silico analysis determined the circular topology to this contig, characterising it as a replicon. Considering all these evidences, it seems that some archaea plasmids can be carrying tRNA gene clusters. Interestingly, some contigs harboring tRNA gene clusters also presented features common to archaea plasmids that could represent an ancestral presence of a plasmid.
Besides the plasmids, we performed a screening for provirus. VirSorter analysis detected putative viral sequences encompassing the regions of the tRNA gene clusters in two genomes, H. vallismortis ATCC 29715 and Halorientalis sp. IM101 [Supplementary data (2.4MB, pdf) (Table VI)].
DISCUSSION
Shared traits between prokaryotes and eukaryotes are helpful in the understanding of the tree of life evolution. tRNA gene cluster, a particular feature in RNA gene organisation, has been demonstrated to be prevalent in bacteria and eukaryotes, but there is a gap concerning its occurrence in archaea. Despite the significant presence in the Bacteria and Eukarya domains, the role of the tRNA gene cluster is under debate. In Bacteria, it has been implicated with a faster cell growing and in the modulation of the tRNA transcription and translation process. 20 In contrast, some studies have not observed any improvement in the fitness of the organism by tRNA gene clusters. In addition, some of the genes from these clusters would be inactivated. 21 In eukaryotes, there is a negative correlation between clusters of tRNA genes and chromosomal stability, since they can act as barriers to DNA replication and the consequent formation of genomic fragile sites. 5 Besides that, the tRNA-derived fragments (tsRNAs or tRFs), identified in the three domains of life, 22 could be generated from the tRNA gene clusters, being another common trait between prokaryotes and eukaryotes. The presence of tRNA gene clusters in the three domains of life reveals a common type of tRNA gene organisation, and as the function of these tRNA genes seems not be canonical, these clusters may be presenting a different functionality.
In this study, based on analyses of complete and draft archaeal genomes we demonstrated the prevalence of tRNA gene clusters in archaea. Considering the four archaea supergroups (Asgard, DPANN, TACK, and Euryarchaeota), 23 tRNA gene cluster occurs at least in two class (Halobacteria and Methanobacteria) from Euryarchaeota. In fact, there is a bias concerning the number of genomes and drafts from Euryarchaeota, and therefore it is not possible to assume that this trait is exclusive of this supergroup.
Similarly to what was found in bacteria, 6 , 7 archaea tRNA gene clusters seem to favor tRNA isotype redundancy, just increasing the number of tRNA gene copies. However, there was a slight increase in the diversity of isoacceptors for few tRNA species and genomes. For most genomes, the tRNA isoacceptors identified in the clusters were redundant. Since the tRNA isoacceptor abundance determine the codon translation efficiency, 24 this result suggests that there is a selective pressure against the diversification of the isoacceptors encoded in tRNA gene clusters, favoring the translation efficiency of certain codons in the archaea. Indeed, it was observed in Halobacteria class a biased codon usage which was related to translational speed. 2 Another common trait between archaea and bacteria is the presence of protein-coding genes within some tRNA gene clusters. In archaea, most of tRNA gene clusters harbor less than 20 tRNA genes, while in bacteria there is a considerable number of tRNA gene clusters with more than 20 tRNA genes. Besides tRNA genes, there are also tRNA pseudogenes in the tRNA gene clusters from prokaryotes and eukaryotes, 5 , 21 suggesting a compromised canonical function, or may be related to other functions.
Concerning the origin of the tRNA gene clusters, in bacteria, it was assumed that they originated from Firmicutes phylum through a horizontal gene transfer event. 6 Indeed, there is evidence showing the association of the tRNA gene clusters with mobile genetic elements. In several bacteria species there are plasmids carrying tRNA gene clusters, 6 , 7 and in archaea, we have evidence that some contigs harboring tRNA gene clusters are associated with homologous sequences of archaea plasmids (this work). Interestingly, one of these contigs presented a relaxase gene, which is rare trait in archaeal plasmids, 25 but a common trait of conjugative bacterial plasmids. In fact, the BLAST analyses of the nucleotide/protein sequence of this relaxase revealed high similarity with archaeal relaxase, but not with bacterial relaxase. Moreover, tRNA gene clusters are abundant in mycobacteriophages, 7 , 8 , 26 also occurring in some bacteriophages. 27 So far, the tRNA gene cluster had been only identified in one haloarchaeal virus genome, being composed of 36 tRNA genes. 28 Here, using VirSorter, it was raised an evidence of two archaea genomes presenting tRNA gene clusters in regions predicted as provirus sequences. This result suggests that, in archaea, as in bacteria, viruses play a role in the transfer of the tRNA gene clusters.
While most of the tRNA gene clusters were composed of archaeal-type tRNAs, those from M. mazei genomes were bacterial-type and presented associated genes with high similarity to bacteria and bacteriophages. One of these genes encodes for chloramphenicol O-acetyltransferase, which has already been reported in other methanogenic archaea species. 29 These results would suggest an inter-domain horizontal gene transfer, however, we could not discard the possibility of contamination, since the contigs that harbor these tRNA gene clusters are small (~7/8 kb) and contain only bacterial sequences, preventing the discrimination of the insertion sites in the archaea genome. Although we could not state the occurrence of this inter-domain transfer, there have already been reports of horizontal gene transfer between M. mazei species and bacteria. 30 , 31
Altogether, it is clear that the tRNA gene cluster is a common feature in the three domains of life, which has independently evolved in each domain. In addition, it appears to be associated with mobile elements in bacteria and archaea.
Financial support:
CAPES (Finance Code 001; PNPD-CAPES fellowship), CNPq and Instituto Oswaldo Cruz (grants).
REFERENCES
- 1.Capes MD, Coker JA, Gessler R, Grinblat-Huse V, DasSarma SL, Jacob CG. The information transfer system of halophilic archaea. Plasmid. 2011;65(2):77–101. doi: 10.1016/j.plasmid.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 2.Iriarte A, Jara E, Leytón L, Diana L, Musto H. General trends in selectively driven codon usage biases in the domain archaea. J Mol Evol. 2014;79(3-4):105–110. doi: 10.1007/s00239-014-9647-7. [DOI] [PubMed] [Google Scholar]
- 3.Fujishima K. Kanai A tRNA gene diversity in the three domains of life Front. Genet. 2014;5:142–142. doi: 10.3389/fgene.2014.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li E, Li X, Wu X, Feng G, Zhang M, Shi H. Complete nucleotide sequence and gene rearrangement of the mitochondrial genome of Occidozyga martensii. J Genet. 2014;93(3):631–641. doi: 10.1007/s12041-014-0418-4. [DOI] [PubMed] [Google Scholar]
- 5.Bermudez-Santana C, Attolini CS-O , Kirsten T, Engelhardt J, Prohaska SJ, Steigele S. Genomic organization of eukaryotic tRNAs. BMC Genomics. 2010;11:270–270. doi: 10.1186/1471-2164-11-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tran TTT, Belahbib H, Bonnefoy V, Talla EA. Comprehensive tRNA genomic survey unravels the evolutionary history of tRNA arrays in prokaryotes. Genome Biol Evol. 2016;8(1):282–295. doi: 10.1093/gbe/evv254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morgado SM, Vicente ACP. Beyond the limits tRNA array units in Mycobacterium genomes. Front Microbiol. 2018;9:1042–1042. doi: 10.3389/fmicb.2018.01042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pope WH, Anders KR, Baird M, Bowman CA, Boyle MM, Broussard GW. Cluster M Mycobacteriophages Bongo, PegLeg, and Rey with unusually large repertoires of tRNA isotypes. J Virol. 2014;88(5):2461–2480. doi: 10.1128/JVI.03363-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lowe TM. Chan PP tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes Nucleic Acids. Res. 2016;44(W1):W54–W57. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32(1):11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Contreras-Moreira B, Vinuesa P. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol. 2013;79(24):7696–7701. doi: 10.1128/AEM.02411-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 13.Katoh K, Standley DM. MAFFT Multiple Sequence Alignment Software Version 7 improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Letunic I, Bork P. Interactive tree of life (iTOL) v3 an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44(W1):W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seemann T. Prokka rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 16.Lanza VF, Baquero F. de la Cruz F.Coque TM AcCNET (Accessory Genome Constellation Network) comparative genomics software for accessory genome analysis using bipartite networks. Bioinformatics. 2017;33(2):283–285. doi: 10.1093/bioinformatics/btw601. [DOI] [PubMed] [Google Scholar]
- 17.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D. Cytoscape a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jørgensen TS, Xu Z, Hansen MA, Sørensen SJ, Hansen LH. Hundreds of circular novel plasmids and DNA elements identified in a rat cecum metamobilome. PLoS One. 2014;9(2):e87924. doi: 10.1371/journal.pone.0087924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roux S, Enault F, Hurwitz BL, Sullivan MB. VirSorter mining viral signal from microbial genomic data. PeerJ. 2015;3:e985. doi: 10.7717/peerj.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ardell DH, Kirsebom LA. The genomic pattern of tDNA operon expression in E Coli. PLoS Comput Biol. 2005;1(1):e12. doi: 10.1371/journal.pcbi.0010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puerto-Galan L, Vioque A. Expression and processing of an unusual tRNA gene cluster in the syanobacterium Anabaena sp PCC 7120. FEMS Microbiol Lett. 2012;337:10–17. doi: 10.1111/j.1574-6968.2012.02664.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhu L, Ow DW, Dong Z. Transfer RNA-derived small RNAs in plants. Sci China Life Sci. 2018;61(2):155–161. doi: 10.1007/s11427-017-9167-5. [DOI] [PubMed] [Google Scholar]
- 23.Spang A, Caceres EF, Ettema TJG. Genomic exploration of the diversity, ecology, and evolution of the archaeal domain of life. Science. 2017;357(6351) doi: 10.1126/science.aaf3883. [DOI] [PubMed] [Google Scholar]
- 24.Novoa EM, Pavon-Eternod M, Pan T, de Pouplana LR. A role for tRNA modifications in genome structure and codon usage. Cell. 2012;149(1):202–213. doi: 10.1016/j.cell.2012.01.050. [DOI] [PubMed] [Google Scholar]
- 25.Guglielmini J. de la Cruz F.Rocha EP Evolution of conjugation and type IV secretion systems. Mol Biol Evol. 2013;30(2):315–331. doi: 10.1093/molbev/mss221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delesalle VA, Tanke NT, Vill AC, Krukonis GP. Testing hypotheses for the presence of tRNA genes in mycobacteriophage genomes. Bacteriophage. 2016;6(3):e1219441. doi: 10.1080/21597081.2016.1219441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bailly-Bechet M, Vergassola M, Rocha E. Causes for the intriguing presence of tRNAs in phages. Genome Res. 2007;17(10):1486–1495. doi: 10.1101/gr.6649807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sencilo A, Jacobs-Sera D, Russell DA, Ko CC, Bowman CA, Atanasova NS. Snapshot of haloarchaeal tailed virus genomes. RNA Biol. 2013;10(5):803–816. doi: 10.4161/rna.24045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dridi B, Fardeau ML, Ollivier B, Raoult D, Drancourt M. The antimicrobial resistance pattern of cultured human methanogens reflects the unique phylogenetic position of archaea. J Antimicrob Chemother. 2011;66(9):2038–2044. doi: 10.1093/jac/dkr251. [DOI] [PubMed] [Google Scholar]
- 30.Fournier GP, Gogarten JP. Evolution of acetoclastic methanogenesis in methanosarcina via horizontal gene transfer from cellulolytic clostridia. J Bacteriol. 2008;190(3):1124–1127. doi: 10.1128/JB.01382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Youngblut ND, Wirth JS, Henriksen JR, Smith M, Simon H, Metcalf WW. Genomic and phenotypic differentiation among Methanosarcina mazei populations from Columbia River sediment. ISME J. 2015;9(10):2191–2205. doi: 10.1038/ismej.2015.31. [DOI] [PMC free article] [PubMed] [Google Scholar]