Abstract
Introduction:
Multiple myeloma (MM) and light chain monoclonal gammopathy of undetermined significance (LCMGUS) are plasma cell disorders associated with the secretion of monoclonal free light chain (LC) proteins. Due to the high concentrations of LC in circulation, both of these populations are at risk for developing LC-associated amyloidosis (AL) – a protein misfolding disease characterized by the deposition of LC protein fibrils in organs and tissues, leading to dysfunction and significant morbidity. At present, accurate identification of subjects at risk for developing amyloidosis is not possible, but with the advent of novel, amyloid-targeted therapies, identification of pre-symptomatic individuals is of clinical import.
Methods:
To address this, a competition assay has been developed to discern LC proteins with enhanced amyloidogenic potential. Numerous factors that may influence the efficacy of the assay have been evaluated to yield optimal conditions.
Results:
Using a panel of nine patient-derived LC, we have demonstrated that amyloid-associated LC inhibited the recruitment of a biotinyl- λ 6 variable domain by homologous amyloid-like fibrils significantly more than MM LC (p < .01).
Conclusion:
The assay accurately discriminated AL from MM patient populations, suggesting that it may aid in the identification of patients with monoclonal gammopathies who have an increased risk of developing amyloidosis.
Keywords: Light-chain amyloidosis, multiple myeloma, light-chain protein, risk assessment, assay
Introduction
Multiple myeloma (MM) is a plasma cell disorder that accounts for approximately 1% of all cancer cases in the United States and roughly 10% of all blood-related malignancies [1,2]. It is estimated by the American Cancer Society that 30,000 new cases of MM are diagnosed in the United States annually. Myeloma is characterized by the presence of monoclonal plasma cells in the bone marrow, resulting in high concentrations of Ig and, often, free monoclonal immunoglobulin light chain (LC) protein in the blood that can result in pathology, notably in the kidneys [2,3]. Elevated serum free LC proteins are also found in patients with LC monoclonal gammopathy of undetermined significance (LCMGUS) and smoldering MM (SMM). Both conditions may precede, by decades, the appearance of MM, and protein aggregation pathologies are a significant source of morbidity in these patients [4–9].
In approximately 10–15% of MM patients, and a smaller subset of LCMGUS and SMM, the production of LCs can lead to LC-associated amyloidosis (AL) – a condition in which highly ordered protein fibrils composed of LC, or their fragments, form extracellular deposits in various organs and tissues including the liver, heart, kidneys, spleen, intestines, vasculature and nerves [10,11]. Amyloid deposits can be cytotoxic [12,13], but the incessant accumulation that causes architectural damage in affected organs is likely the major cause of dysfunction and morbidity. Amyloid formation is characterized by a two-step process – the initial formation of a nidus, or seed, followed by recruitment of LC proteins, or fragments thereof, sequestered from the circulation [14]. While the life expectancy of MM patients has improved due to advances in treatment regimens and the availability of novel new agents [2,15,16], the prognosis for MM patients with LC amyloidosis remains significantly worse [3,17,18]; renal and cardiac amyloid involvement contribute significantly to the mortality of MM patients with amyloidosis. All of these become more formidable in an aging population where treatment options become limited [19]. Early detection and the development of preemptive measures are desirable.
Currently, diagnosis of pre-symptomatic LC amyloidosis in patients with MM is not commonplace, and methods to accurately identify those at greater risk of developing the pathology have not been developed. Studies have shown, however, that germline gene usage and serum monoclonal free LC (FLC) concentration may be indicators of potential disease progression and disease severity. Indeed, analysis of LC proteins from cohorts of AL and MM patients has indicated that λ6 LC appear at significantly higher frequency in AL patients [10,20,21]. Given this, a prospective, observational clinical trial evaluating the germline gene of patients with λ LC SMM and λ LCMGUS is currently underway. The goal of the SAVE trial (Seeking AL Amyloidosis Very Early; clinicaltrials.gov identifier: NCT02741999) is to determine if germline gene usage in patients with a plasma cell dyscrasia can predict progression to AL amyloidosis. Furthermore, since monoclonal serum FLC is hallmark in patients with MM and LCMGUS, FLC concentration may provide additional insight into progression to LC-associated amyloidosis in these patients. Recently, Weiss et al. described a retrospective case–control study in which patient serum samples collected prior to a confirmed diagnosis of AL amyloidosis were analysed for FLC concentration [22]. The data demonstrated the occurrence of a prolonged prodromal state for patients who developed AL amyloidosis characterized by increasing levels of serum FLC for up to 11 years prior to diagnosis, suggesting that an increase in FLC precedes the development of AL amyloidosis [22], but this value alone is not diagnostic of a patient population likely to develop the disease [23].
The ability to identify patients with the greatest risk of developing amyloidosis might provide more effective and focused clinical management in this subset of patients, particularly in light of the recent development of amyloid-clearing immunotherapeutic agents [24–26]. Patients identified as being at greater risk for developing LC-associated amyloidosis could be monitored more closely for amyloid throughout their treatment for MM, by incorporating routine fat pad biopsy [27], evaluating organ-specific biomarkers [28], or by molecular imaging with amyloid-targeting agents [29–31]. If detected, intervention with amyloid-reactive antibodies, such as NEOD001 [24], 11–1F4 [25] or the SAP reactive antibody [26], all of which are currently being evaluated in clinical trials, could facilitate amyloid clearance or stabilize amyloid load in the pre-symptomatic population. This could minimize amyloid-associated organ damage and improve patient prognosis, quality of life and survival.
Fibrillogenesis is characterized by an initial seeding event followed by an exponential fibril growth phase [32,33]. Previous studies have demonstrated efficient in vitro formation of amyloid-like fibrils by patient-related recombinant λ6 light chain variable domains (rVλ6Wil) as well as κ1- and κ4-derived amyloidogenic proteins. Recently, we demonstrated that rVλ6Wil amyloid-like fibrils can serve as an effective template for the recruitment of heterologous (non-λ6) patient-derived LC proteins isolated from urine [34]. Furthermore, it was shown that LCs from AL patients bound the amyloid fibrils significantly more effectively than did MM LCs, and the two patient populations could be readily discerned based upon the amount of LC bound. Moreover, a LC protein derived from an MM patient who was later diagnosed with severe hepatosplenic LC amyloidosis was recruited with the efficiency of other AL-associated LC proteins [34].
Based on these preliminary findings, we hypothesized that the binding of AL-associated LC proteins to rVλ6Wil fibrils would inhibit the further recruitment of monomeric rVλ6Wil and, thereby, prevent fibril growth more effectively than MM-derived LC. Herein, we describe a microplate based competition assay designed to quantitatively assess the ability of patient-derived LCs to inhibit recruitment of biotinylated rVλ6Wil by homologous, surface-adsorbed fibrils (Figure 1). We report studies conducted for the optimization of assay conditions as well as an initial assessment of previously well-characterized LC proteins [34,35] where accurate discrimination of AL-associated and MM LC proteins was possible. In conjunction with other clinical measurements, the assay may assist in the identification of patients with monoclonal plasma cell disorders, for example, MM and LCMGUS, who are at increased risk of developing amyloidosis.
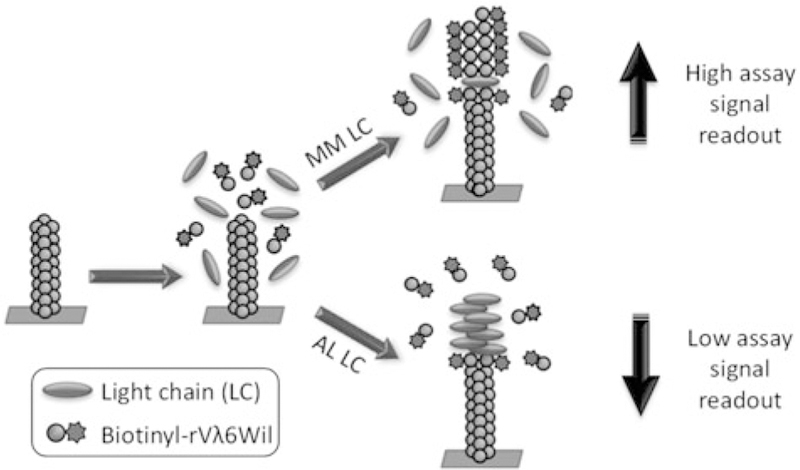
Figure 1.
Schematic representation of the biotinyl-rVλ6Wil competition assay. Light chain proteins mixed with biotinyl-rVλ6Wil are incubated in the presence of surface adsorbed fibrils composed of rVλ6Wil. In the presence of a benign (MM) LC, little interaction with the fibril occurs and biotinyl-rVλ6Wil is efficiently recruited by the fibrils leading to a high fluorescent read out. Conversely, an AL LC protein binds to the fibrils and hinders recruitment leading to a decrease in fluorescence intensity following incubation.
Materials and methods
Preparation of patient-derived light-chain proteins
Light chain proteins were isolated from the urine of patients with a diagnosis of MM (n =4; 2 κ and 2 #x03BB;) or AL amyloidosis (n = 4; 2 κ and 2 λ). Additionally, one κ LC protein (Hig) from and later developed LC amyloidosis, was used; thus, a total of 9 LCs were analyzed in our studies. These proteins were isolated from urine specimens as previously described [36]. Subgroup classification of the LC proteins had been previously determined either serologically using monoclonal VL-subgroup-specific antisera [36] or by amino acid sequence analysis [37]. Isolated LC proteins were lyophilized and stored at room temperature (RT) until use. Prior to use, lyophilized material was dissolved in phosphate buffered saline (PBS; 150 mM NaCl, pH 7.2) and the concentrations determined using a micro-bicinchoninic acid kit (Pierce, Rockford, IL).
Production of rVλ6Wil protein monomer and fibrils
The rVλ6Wil protein was isolated from the periplasmic space of recombinant E. coli and purified by reverse phase high-pressure liquid chromatography, as previously described [33]. Synthetic, amyloid-like fibrils of rVλ6Wil were generated by shaking monomeric protein in PBS at 37 ○C and 225 revolutions per min (C24 Incubator Shaker, New Brunswick Scientific, Edison, NJ) for ~72 h. Generally, fibrils were prepared using 1 mg/mL monomer unless otherwise stated. All preparations were centrifuged (10,000 × g; 5 min) and assayed to ensure fibril formation. For this, 5 µg of fibril sample was mixed with thioflavin T (50 µM) and the fluorescence emission measured at 490 nm using an excitation wavelength of 450 nm. For use in the fibril extension assay, rVλ6Wil protein monomer (2 mg) was biotinylated using a N-hydroxy-succinimidyl linker, and free reagent was removed per manufacturer’s instructions (Pierce).
rVλ6Wil fibril extension assays
Unless otherwise stated, the rVλ6Wil recruitment assay was performed as follows. Amyloid-like fibrils composed of rVλ6Wil suspended in up to 100 µL PBS (1 mg/mL) were sonicated at 60% power (Tekmar Sonic Disruptor with microprobe) for 10 s, and a 96-well microplate was coated with 0.5µg of the protein per well (50 µL/well of a 0.83 µM fibril suspension) and dried at 37 ○C for 17 h. Microplates were washed twice with PBS 0.05% tween-20 (PBST) and blocked for 1 h at 37 ○C with 100 µl of 1% bovine serum albumin (BSA) in PBS. Biotinyl-rVλ6Wil monomer (5 nM) diluted in PBS with 1% BSA 0.05% tween-20 (BSAT) was added at 100 µL/well, and the microplate was again incubated for 1 h at 37 ○C prior to another 2 washes with PBST. A 1:1000 dilution of europium/streptavidin in BSAT was added and the microplate incubated at 37 ○C for another hour. After three washes with PBST, 100 µL/well enhancement solution (Perkin Elmer) was added, and the time- resolved fluorescence emission measured (Wallac Victor3, Perkin Elmer, Shelton, CT).
Optimization of rVλ6Wil competition assay
For competition studies using patient-derived LCs, the fibril extension assay described above was used with the following modifications. In addition to the 5 nM biotinyl-rVλ6Wil monomer, increasing concentrations of AL or MM-patient derived LC protein (4 nM–8 µM) were premixed and added to the fibril-coated wells and incubated for 1 h at 37 ○C. In some assays, 5 µM of LC was added to 5 nM rVλ6Wil monomer using fibril substrates generated from various concentrations of rVλ6Wil monomeric protein in PBS (0.2 mg/mL, 0.8 mg/mL or 1.35 mg/mL) and using 1 mg/mL rVλ6Wil fibril with a 1-h, 2-h or 3-h reaction incubation time.
Final competition assay conditions
The nine AL or MM-patient derived LCs were assessed as described above with the following specifications: rVλ6Wil fibrils were prepared at 1 mg/mL and sonicated for 10 s. A 50 mL aliquot of fibril suspension (diluted 100-fold in PBS), 0.83 µM, was added to the microplate and dried at 37 ○C for ~17 h. A 100 µL volume of 5 nM biotinyl-rVλ6Wil monomer containing 0.05 µM, 0.1 µM, 0.5 µM or 2.0 µM LC (n=6 per concentration) was added to the well and incubated at 37 ○C for 1 h.
Data analysis and statistics
Recruitment inhibition data with increasing concentrations of LC were plotted using a log10 x-axis and the data fit using a semilog linear equation (y= m logx + c). The concentration-dependent gradient of the increase in inhibition (given by the gradient, m) was calculated. For statistical analyses, comparisons were made between sets of data using ANOVA with Bonferroni or Tukey’s correction for multiple comparisons, as noted, with α= 0.05. Pairwise comparisons were made using a two-tailed, unpaired t-test with α= 0.05. Receiver operating characteristic (ROC) curve cut-offs were established with an area under the curve of 100%, p =.01. Inter-assay repeatability was assessed using and intra-class correlation coefficient (ICC), two-way random model, performed in SPSS, v0.22 (Armonk, NY; IBM Corp.). In all cases, *p < .05, **p < .01, ***p < .001, ****p < .0001.
Results
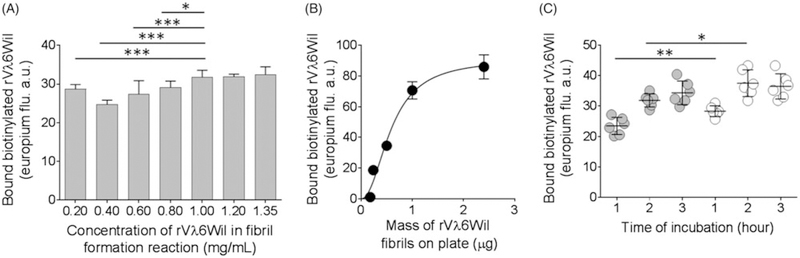
A series of experimental conditions were evaluated to optimize the efficacy of the rVλ6Wil microplate-based fibril extension assay. To assess the effect of initial monomer concentration used in the rVλ6Wil fibril formation step, fibrils were prepared using monomer concentrations from 0.2 mg/mL to 1.35 mg/mL. All fibrils were shown to be ThT positive (>7.5 105 fluorescence units per 5 µg), and each preparation supported the recruitment of biotinyl-rVλ6Wil monomer (Figure 2(A)). When compared to the 1 mg/mL preparation (the standard method for fibril formation), there was significantly less biotinyl-rVλ6Wil monomer bound to fibrils generated using lower concentrations of monomer (Figure 2(A)). There was no significant difference in the recruitment capacity of fibrils made from >1.0 mg/mL monomer; thus, we used fibrils prepared from a 1 mg/mL rVλ6Wil monomer preparation in future assays. We next examined recruitment of biotinyl-rVλ6Wil by increasing amounts of microplate-well coated fibrils (Figure 2(B)). There was a linear increase in the recruitment of biotinylrVλ6Wil as the mass of plated fibrils increased from 0.18 µg to 1.0 µg, and at higher concentrations, recruitment plateaued (Figure 2(B)). The assay signal achieved with 0.5 µg fibrils (equivalent to 50 mL of a 10 µg/mL [0.83 µM] stock added per well) was significantly above background and was in the linear range of response (Figure 2(B)). To assess the effect of the presence of salt in the fibril coating milieu, we compared recruitment of biotnyl-rVλ6Wil by fibrils coated onto the microplate wells, in water or PBS, after 1 h, 2 h, and 3 h of incubation (Figure 2(C)). At 1 h and 2 h of incubation, there was a small, but significant, increase in recruitment when the fibrils were coated onto microplate wells using PBS; however, after a 3-h incubation, this benefit was lost (Figure 2(C)). Additionally, there was a significant increase in the recruitment of biotnyl-rVλ6Wil by fibrils as the reaction incubation time increased, independent of the use of PBS or water as the coating solution (Figure 2(C)).
Figure 2.
Preparation and fibril coating procedure affects the recruitment of biotinyl-rVλ6Wil. (A) rVλ6Wil fibrils were prepared using a starting monomer peptide concentration ranging from 0.2 mg/mL – 1.35 mg/mL. A 0.5 µg aliquot of each sample (n= 6) was used and the recruitment efficacy for biotinyl-rVλ6Wil (5 nM) determined at 1 h incubation. Data were analysed using ANOVA with Tukey’s correction for multiple comparisons, α= 0.05. (B) Increasing the mass of rVλ6Wil fibrils (prepared from 1 mg/mL monomer) coated onto the microplate well increased recruitment of biotinyl-rVλ6Wil (5 nM) up to 1 µg at 1-h incubation. Mean ± SD, n = 6. (C) Increasing the incubation time of the assay, using rVλ6Wil fibrils coated in water (grey) and in PBS (white) enhanced recruitment of the biotinyl-rVλ6Wil.
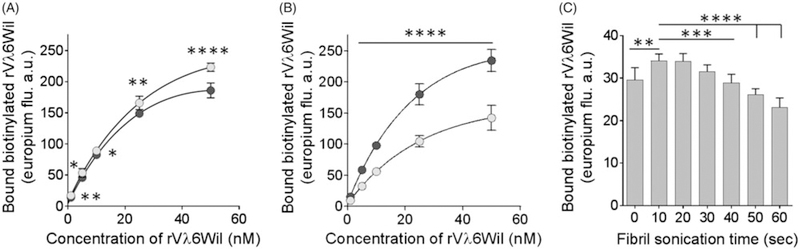
The storage conditions of the rVλ6Wil fibrils, refrigerated or frozen, may contribute to their gross morphology and aggregation state which may, in turn, affect recruitment of biotinyl-rVλ6Wil monomer. We, therefore, tested the recruitment efficacy of fibrils that had been stored at either 4 ○C or –20 ○C prior to use, and each sample was analysed with or without a 10 s sonication step (Figure 3(A,B)). There was a significant decrease in the recruitment of biotinyl-rVλ6Wil when fibrils stored at 4 ○C were sonicated for 10 s prior to coating (Figure 3(A)). In contrast, fibrils stored at 20 ○C recruited monomeric protein significantly better following brief sonication (Figure 3(B)), and unsonicated fibrils stored at 20 ○C recruited significantly less efficiently than all other fibril preparations (Figure 3(A,B)).
Figure 3.
Storage conditions and sonication of rVλ6Wil fibrils affect concentration-dependent recruitment of biotinyl-rVλ6Wil monomer. (A) Sonication (10 s; black) of fibrils stored at 4 ○C were less efficient at recruiting biotinyl-rVλ6Wil than unsonicated fibrils (white). Mean ± SD, n =6. (B) Conversely, sonication (black) enhanced binding of biotinyl-rVλ6Wil by fibrils stored at −20 ○C, as compared to unsonicated fibrils (white). Mean ± SD, n= 6 (C) Sonication of fibrils stored at 4 ○C for more than 10–20 s significantly reduced the recruitment of biotinyl-rVλ6Wil (5 nM). Data in (A) and (B) were analysed using an unpaired t-test, and in (C) were analysed by ANOVA using a Bonferroni correction for multiple comparisons, α = 0.05.
Given the importance of sonication to recruitment efficacy by the rVλ6Wil fibrils, we assessed the effect of increasing sonication times using a fibril preparation stored at 4 ○C (Figure 3(C)). No significant difference was observed in recruitment efficacy between samples sonicated for 10 s, –20 s and 30 s; however, if fibrils were not sonicated, or were treated for 40 s or more, the recruitment efficacy declined significantly (Figure 3(C)). Further studies were performed using rVλ6Wil fibrils prepared using 1 mg/mL monomer, stored at 4 ○C and sonicated for 10 s before being coated on the microplate wells.
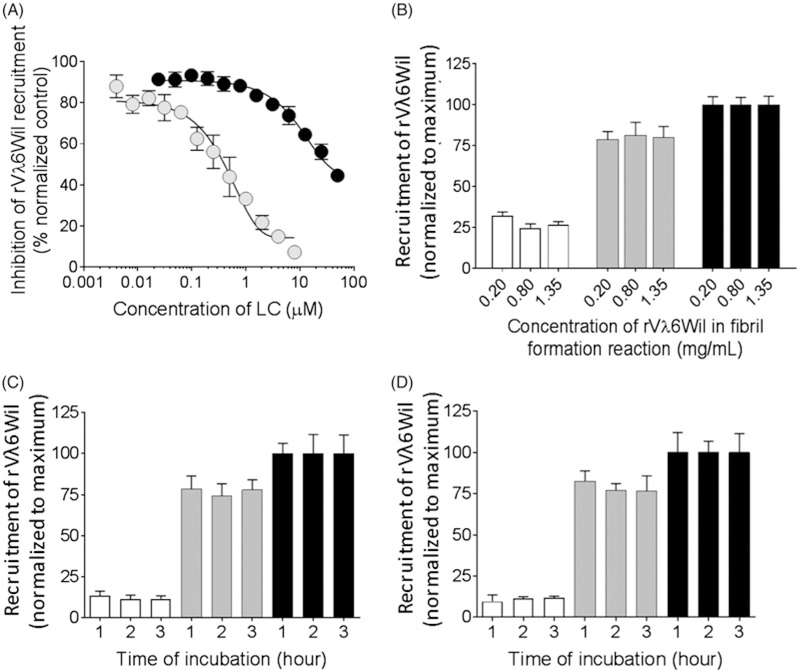
The competition microplate assay was validated using two exemplar κ1 LC proteins isolated from the urine of patients with either AL (protein Cro) or MM (protein Gal) as competitors of the recruitment of biotinyl-rVλ6Wil by fibrils (Figure 4). Addition of increasing concentrations of Cro resulted in inhibition of the recruitment of biotinylrVλ6Wil monomer with 50% inhibition occurring at ~0.5 mM (~ 12 µg/mL) (Figure 4(A)). In contrast, the non- amyloidogenic protein Gal was less potent and inhibited biotinyl-rVλ6Wil recruitment by 50% at ~25 µM (Figure 4(A)). At a concentration of 5 µM (~ 120 µg/mL) LC protein Cro inhibited recruitment by ~85%, whereas Gal inhibited by only ~15%. Therefore, 5 µM was chosen as a concentration of LC that readily discerned AL- from MM-associated protein in the assay. Inhibition of recruitment efficacy by Cro and Gal (at 5 µM) was not dependent upon the concentration of rVλ6Wil monomer used to generate the fibrils (Figure 4(B)). Additionally, neither the fibril coating condition in PBS (Figure 4(C)) or water (Figure 4(D)) nor the time of the recruitment reaction affected the relative inhibition of recruitment by Cro and Gal (Figure 4(C,D)). Thus, a 1-h incubation time was deemed suitable for the competition assay.
Figure 4.
Recruitment of biotinyl-rVλ6Wil by rVλ6Wil fibrils is inhibited by AL-associated LC proteins. (A) Concentration-dependent inhibition of biotinyl-rVλ6Wil recruitment by AL- (Cro; grey) and MM-associated LC (Gal; black). (B) Recruitment of biotinyl-rVλ6Wil alone (black), or inhibition in the presence of Cro (white) or Gal (grey) was not affected by the concentration of rVλ6Wil monomer used to generate the fibril substrate (mean ± SD; n= 6). Recruitment of biotinyl-rVλ6Wil alone (black), or inhibition in the presence of Cro (white) or Gal (grey) was not affected by PBS (C) or water (D) as the fibril coating milieu. Data in (C) and (D) were analysed by ANOVA using a Bonferroni correction for multiple comparisons, α = 0.05.
Intra- and inter-assay repeatability determinations were made, for assays performed by a single operator. The raw fluorescence data were analysed for the recruitment of biotinyl-rVλ6Wil in the absence of competitor and in the presence of 5 µM Cro or Gal protein, on one test microplate (intra-assay; Table 1). The coefficient of variation was <7% for all three reaction conditions in an intra-plate repeatability study. Inter-assay repeatability was assessed using Cro and Gal data which had been background-corrected and normalized to the mean value for biotinyl-rVλ6Wil alone on that plate. Using an intra-class correlation coefficient (ICC) for Cro biotinyl-rVλ6Wil assay a value of 0.33 was obtained denoting a poor level of repeatability. Conversely, for biotinyl-rVλ6Wil in the presence Gal, an ICC of 0.59 was found, indicating acceptable levels of repeatability across the observations (Table 2).
Table 1.
Summary of data for evaluation of intra-plate repeatability.
| rVλ6Wil | rVλ6Wil + Gal LC (5µM) | rVλ6Wil + Cro LC (5µM) | |
|---|---|---|---|
| No. replicates | 24 | 12 | 12 |
| Minimum (flu. au)a | 182366 | 152293 | 58493 |
| Maximum (flu. au) | 223117 | 180805 | 72484 |
| Mean (flu. au) | 197814 | 165995 | 63170 |
| Median (flu. au) | 195609 | 166082 | 62034 |
| Lower 95% CI of the mean (flu. au) | 193255 | 160611 | 60490 |
| Upper 95% CI of the mean (flu. au) | 202372 | 171379 | 65849 |
| SD (flu. au) | 10796 | 8474 | 4217 |
| SEM (flu. au) | 2204 | 2446 | 1217 |
| %Coefficient of Variation (%CV) | 5.46% | 5.11% | 6.68% |
(flu. au.): fluorescence emission, arbitrary units.
Table 2.
Summary of data for evaluation of inter-plate repeatability using normalized data.
| rVλ6Wil + Cro LC (5µM) |
rVλ6Wil + Gal LC (5µM) |
|||||
|---|---|---|---|---|---|---|
| Day 1 | Day 2 | Day 3 | Day 1 | Day 2 | Day 3 | |
| No. replicates | 12 | 12 | 12 | 12 | 12 | 12 |
| Minimum (flu. au) | 13.47 | 10.41 | 11.93 | 71.73 | 71.62 | 75.10 |
| Maximum (flu. au) | 22.16 | 19.73 | 18.37 | 89.44 | 85.37 | 92.39 |
| Mean (flu. au) | 16.37 | 14.39 | 13.85 | 80.24 | 76.90 | 79.78 |
| Median (flu. au) | 15.67 | 14.06 | 13.57 | 80.30 | 76.52 | 79.63 |
| Lower 95% CI of the mean (flu. au) | 14.71 | 12.91 | 12.84 | 76.89 | 73.42 | 76.26 |
| Upper 95% CI of the mean (flu. au) | 18.04 | 15.86 | 14.87 | 83.58 | 80.38 | 83.29 |
| SD (flu. au) | 2.62 | 2.33 | 1.60 | 5.27 | 5.48 | 5.53 |
| SEM (flu. au) | 0.76 | 0.67 | 0.47 | 1.52 | 1.58 | 1.60 |
| %Coefficient of Variation (%CV) | 16.00 | 16.18 | 11.53 | 6.56 | 7.12 | 6.93 |
| ICC | 0.33 | 0.59 | ||||
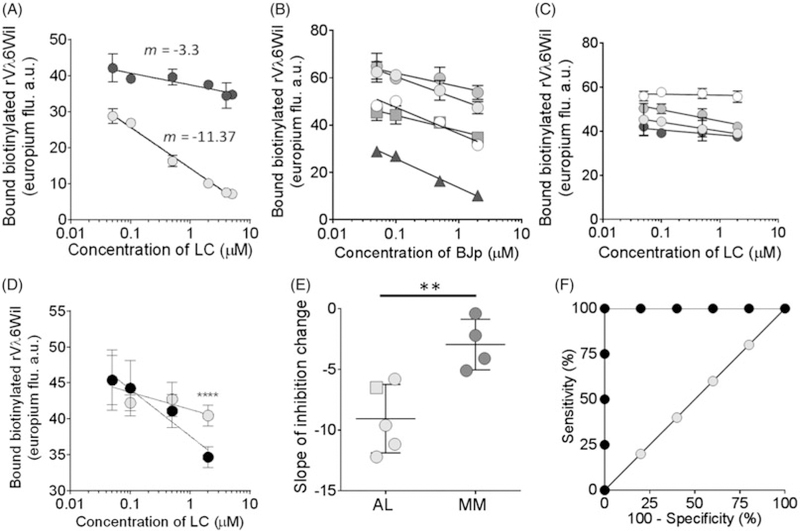
We next assessed the LC concentration-dependent decrease in biotinyl-rVλ6Wil-recruitment as a method for discerning AL from MM patient-derived LC proteins (Figure 5(A)). Using Cro and Gal LC at increasing concentrations from 0.05 µM to 5 µM, a significant difference in the concentration-dependent inhibition of recruitment was observed with estimated gradients (m, where y=m logx+c) of ‒3.3. and ‒11.37 for Gal and Cro, respectively (Figure 5(A)). This approach was then used to analyse cohorts of well-characterized AL (n=4; Figure 5(B)) and MM (n=4; Figure 5(C)) LC as well as LC protein from an MM patient who was later diagnosed with LC amyloidosis (Figure 5(B)). Each LC protein in this analysis was tested at 0.05 µM to 2 µM (Figure 5(B,C)). The concentration-dependent change in recruitment of biotinyl-rVλ6Wil was calculated, and the mean value for AL LC (m = −9.05 ± 1.26) was shown to be significantly different as compared to that for MM LC (m = −2.95 ± 1.04) (Figure 5(D)), indicating that the assay as designed can discriminate between the groups.
Figure 5.
Concentration-dependent inhibition of biotinyl-rVλ6Wil recruitment by AL-associated LC is significantly greater than for MM-derived LC proteins.(A) AL-associated LC, Cro (grey) yielded a greater concentration-dependent decrease (m, is the gradient) in recruitment inhibition, as compared to MM protein Gal (black). Mean ± SD, n= 6. Concentration-dependent gradient of inhibition of biotinyl-rVλ6Wil recruitment by a panel of 5 amyloid-associated LC (B; Cro, black triangle; Hig, gray square) or 4 MM LC proteins (C). Mean ± SD, n= 6. (D) Comparison of concentration-dependent inhibition of fibrillogenesis by MM protein Gal (grey) and transition protein Hig (black). Data were normalized to the lowest LC concentration. (E) The cohort of AL-associated LC (grey), including transition protein Hig (square), inhibited recruitment significantly more than MM-derived LC proteins (black). Mean ± SD, n = 6. (F) ROC analysis of the data presented in (E). A 100% sensitivity and 100% specificity was achieved at m = −5.45. An unpaired, two-tailed t-test was used to analyze the data in (D and E).
Discussion
In a recent survey detailing more than 500 amyloid patient experiences, it was reported that, for at least one-third of the patients, it took ≥1 year and at least five different physician consultations to obtain a definitive amyloid diagnosis [38]. In 10% of the cases, 3 years were passed before a diagnosis was reached, and in all cases, the diagnosis followed the onset of clinical manifestations of amyloid- related pathology [38]. The development of methods to assist in early detection of amyloidosis, when patients are pre-symptomatic, and accurately predict development of amyloid pathology in at-risk populations is challenging but addresses an unmet clinical need.
The primary goal of this pilot study is to establish a convenient assay capable of identifying amyloidogenic LC proteins. We have previously demonstrated that LCs derived from MM patients, with no known progression to LC amyloidosis, were recruited less efficiently by synthetic amyloid-fibrils composed of rVλ6Wil, as compared to AL patient-derived LC [34]. Notably, the LC protein from transition patient Hig behaved functionally, in our assay, as an AL LC. This observation indicated that the LC protein was predisposed to form amyloid, despite the fact that amyloidosis was not discerned clinically in this patient until three years following a diagnosis of MM. Although this study provided clear discrimination between AL and MM-derived LCs, it relied on urine-derived LC proteins that were radiolabeled, which is an impractical method for translation into the clinical setting.
Herein, we have described a new iteration of this assay based on concentration-dependent inhibition of biotinyl-rVλ6Wil recruitment by amyloid-like fibrils in a 96-well microplate format, which no longer relies on radioactive proteins. The assay is based on our premise that pro-amyloidogenic LC will bind to the surface-adsorbed fibrils at sites required for fibril growth, thus inhibiting the recruitment of biotinyl-rVλ6Wil monomer, and that AL-derived LC will compete more effectively than MM-derived LC for binding sites. We have evaluated optimization of various parameters related to the fibril seed, since amyloid fibrils are known to be heterogeneous structures even when composed of the same precursor protein [39]. In this assay, recruitment of biotinyl-rVλ6Wil by the surface adsorbed fibrils was readily observed. Using exemplar κ1 LC proteins, Cro (AL) and Gal (MM), we demonstrated that Cro was a more effective inhibitor of biotinyl-rVλ6Wil recruitment than Gal; however, at higher concentrations (>5 µM), Gal also effectively inhibited recruitment. This may have resulted from non-specific association of Gal LC with the fibrils, or it may indicate that, at certain concentrations, all LC can be productively recruited by preformed amyloid fibrils serving as a seed. Using the recruitment assay, we effectively discriminated between the two patient populations based on their concentration-dependent inhibition of recruitment, which was significantly greater for AL LC. We recognize the small sample size used in this proof of principle study, but our preliminary findings are encouraging. This assay may also be readily adapted to use patient serum or urine samples containing LC protein. However, the effect of other constituents, not- ably the presence of serum albumin, which hinders amyloid fibril formation [40–42], needs to be assessed and appropriately controlled for in the assay. Recruitment of rVλ6Wil by fibrils in the presence of 10% normal human serum is greatly reduced, likely due to interactions of albumin with the soluble monomeric species. Nonetheless, initial data from a microplate-based competition assay using rVλ6Wil fibrils and serum samples from patients with LCMGUS and MM has demonstrated the feasibility of this approach (EBM and JSW unpublished data).
Accurate identification of individuals in the MM and LCMGUS population with a greater risk for developing LC amyloidosis may lead to enhanced clinical vigilance. With the advent of novel AL amyloid-reactive monoclonal antibodies, that are showing clinical benefit in ongoing trials [24–26], identification of LC amyloid before the onset of organ dysfunction might aid in effective treatment of these patients using passive immunotherapeutics, thereby enhancing patient survival. An effective approach to assess risk of amyloidosis would likely involve a number of determinants, in addition to the biotinyl-rVλ6Wil assay, including the germline gene usage and the serum FLC concentration. Such a cumulative amyloid risk evaluation algorithm may provide an accurate prediction of amyloidosis in patients, and our assay may play a critical role in the early identification of these patients such that therapeutic intervention enhances survival.
Acknowledgements
We appreciate the thoughts and comments provided by Dr. Raymond Comenzo. We thank Matthew Little for assistance in the production of rVλ6Wil protein.
Funding
This study was supported by a Research Grant from the Amyloidosis Foundation (AF). Additional support was provided by the Department of Medicine at the University of Tennessee Medical Center.
Abbreviations:
- AL
light chain-associated amyloid protein
- BSA(T)
bovine serum albumin (+ tween)
- FLC
serum monoclonal free light chain
- ICC
intra-class correlation coefficient
- LC
immunoglobulin light chain protein
- LCMGUS
light chain monoclonal gammopathy of undetermined significance
- MM
multiple myeloma
- PBS(T)
phosphate buffered saline (+ tween)
- ROC
receiver operating characteristic
- RT
room temperature
- SAP
serum amyloid P-component
- SMM
smoldering multiple myeloma
- ThT
thioflavinT
Footnotes
Disclosure statement
EBM and JSW are co-inventors named on a PCT patent application (claiming priority from U.S. Provisional Patent Application No. 62/ 326,671, filed April 22, 2016, which is titled “Methods & Systems for Identifying Amyloidogenic Proteins”) related to the development of an assay to identify subjects with a high risk of developing amyloidosis in susceptible populations. The remaining authors report no conflicts of interest.
References
- [1].Mateos M-VSM, San Miguel JF. How should we treat newly diagnosed multiple myeloma patients? Hematology Am Soc Hematol Educ Program 2013;2013:488–495. [DOI] [PubMed] [Google Scholar]
- [2].Rajkumar SV Multiple myeloma: 2016 update on diagnosis, risk-stratification, and management. Am j Hematol 2016; 91:719–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rios-Tamayo R, Sanchez MJ, Puerta JM, et al. Trends in survival of multiple myeloma: a thirty-year population-based study in a single institution. Cancer Epidemiol 2015;39:693–699. [DOI] [PubMed] [Google Scholar]
- [4].Comenzo RL Plasma cell neoplasms, their precursor states, and their prediction of organ damage. JCO 2014;32:2679–2682. [DOI] [PubMed] [Google Scholar]
- [5].Dispenzieri A, Katzmann JA, Kyle RA, et al. Prevalence and risk of progression of light-chain monoclonal gammopathy of undetermined significance: a retrospective population-based cohort study. Lancet 2010;375:1721–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Eisele L, Durig J, Huttmann A, et al. Prevalence and progression of monoclonal gammopathy of undetermined significance and light-chain MGUS in Germany. Ann Hematol 2012; 91:243–248. [DOI] [PubMed] [Google Scholar]
- [7].Merlini G, Palladini G Differential diagnosis of monoclonal gammopathy of undetermined significance. Hematology Am Soc Hematol Educ Program 2012;2012:595–603. [DOI] [PubMed] [Google Scholar]
- [8].Rajkumar SV, Kyle RA, Plevak M, et al. Prevalence of light chain monoclonal gammopathy of undetermined significance (LC-MGUS) among Olmsted County, Minnesota residents aged 50 years or greater. Blood 2006;108:5060. [Google Scholar]
- [9].van Rhee F Light chain MGUS: implications for clinical practice. Lancet 2010;375:1670–1671. [DOI] [PubMed] [Google Scholar]
- [10].Dispenzieri A, Gertz MA, Buadi F What do I need to know about immunoglobulin light chain (AL) amyloidosis?. Blood Rev 2012;26:137–154. [DOI] [PubMed] [Google Scholar]
- [11].Wechalekar AD, Gillmore JD, Hawkins PN Systemic amyloidosis. Lancet 2016;387:2641–2654. [DOI] [PubMed] [Google Scholar]
- [12].Lin Y, Marin-Argany M, Dick CJ, et al. Mesenchymal stromal cells protect human cardiomyocytes from amyloid fibril damage. Cytotherapy 2017;19:1426–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].McWilliams-Koeppen HP, Foster JS, Hackenbrack N, et al. Light chain amyloid fibrils cause metabolic dysfunction in human cardiomyocytes. PLoS One 2015;10:e0137716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Blancas-Mejia LM, Ramirez-Alvarado M Recruitment of light chains by homologous and heterologous fibrils shows distinctive kinetic and conformational specificity. Biochemistry 2016;55: 2967–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lakshman A, Abeykoon JP, Kumar SK, et al. Efficacy of daratumumab-based therapies in patients with relapsed, refractory multiple myeloma treated outside of clinical trials. Am J Hematol 2017;92:1146–1155. [DOI] [PubMed] [Google Scholar]
- [16].Usmani SZ, Weiss BM, Plesner T, et al. Clinical efficacy of daratumumab monotherapy in patients with heavily pretreated relapsed or refractory multiple myeloma. Blood 2016;128: 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Badar T, Srour S, Bashir Q, et al. Predictors of inferior clinical outcome in patients with standard-risk multiple myeloma. Eur J Haematol 2017;98:263–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dinner S, Witteles W, Witteles R, et al. The prognostic value of diagnosing concurrent multiple myeloma in immunoglobulin light chain amyloidosis. Br J Haematol 2013;161:367–372. [DOI] [PubMed] [Google Scholar]
- [19].Sachchithanantham S, Offer M, Venner C, et al. Clinical profile and treatment outcome of older (>75 years) patients with systemic AL amyloidosis. Haematologica 2015;100:1469–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Comenzo RL, Zhang Y, Martinez C, et al. The tropism of organ involvement in primary systemic amyloidosis: contributions of Ig V(L) germ line gene use and clonal plasma cell burden. Blood 2001;98:714–720. [DOI] [PubMed] [Google Scholar]
- [21].Kourelis TV, Dasari S, Theis JD, et al. Clarifying immunoglobulin gene usage in systemic and localized immunoglobulin light- chain amyloidosis by mass spectrometry. Blood 2017; 129:299–306. [DOI] [PubMed] [Google Scholar]
- [22].Weiss BM, Hebreo J, Cordaro DV, et al. Increased serum free light chains precede the presentation of immunoglobulin light chain amyloidosis. JCO 2014;32:2699–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Usnarska-Zubkiewicz L, Holojda J, Debski J, et al. Analysis of free serum light chains in patients suffering from multiple myeloma complicated by light chain amyloidosis. Adv Clin Exp Med 2014;23:531–538. [DOI] [PubMed] [Google Scholar]
- [24].Gertz MA, Landau H, Comenzo RL, et al. First-in-human Phase i/ii study of NEOD001 in patients with light chain amyloidosis and persistent organ dysfunction. JCO 2016;34: 1097–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Langer AL, Miao S, Mapara M, et al. Results of phase 1 study of chimeric fibril-reactive monoclonal antibody 11–1F4 in patients with AL amyloidosis. Blood 2015;126:188. [Google Scholar]
- [26].Richards DB, Cookson LM, Berges AC, et al. Therapeutic clearance of amyloid by antibodies to serum amyloid P component. N Engl J Med 2015;373:1106–1114. [DOI] [PubMed] [Google Scholar]
- [27].Mollee P, Renaut P, Gottlieb D, et al. How to diagnose amyloidosis. Intern Med J 2014;44:7–17. [DOI] [PubMed] [Google Scholar]
- [28].Palladini G, Basset M, Milani P, et al. Biomarker-based screening of organ dysfunction in patients with MGUS allows early diagnosis of AL amyloidosis. Blood 2017;130(Suppl 1):1760.28784598 [Google Scholar]
- [29].Dorbala S, Vangala D, Semer J, et al. Imaging cardiac amyloidosis: a pilot study using (1)(8)F-florbetapir positron emission tomography. Eur J Nucl Med Mol Imaging 2014;41:1652–1662. [DOI] [PubMed] [Google Scholar]
- [30].Hazenberg BP, van Rijswijk MH, Piers DA, et al. Diagnostic performance of 123I-labeled serum amyloid P component scintigraphy in patients with amyloidosis. Am J Med 2006;119:355.e15–355.e24. [DOI] [PubMed] [Google Scholar]
- [31].Law WP, Wang WY, Moore PT, et al. Cardiac amyloid imaging with 18F-florbetaben PET: a pilot study. J Nucl Med 2016; 57:1733–1739. [DOI] [PubMed] [Google Scholar]
- [32].Harper JD, Lansbury PT Jr. Models of amyloid seeding in Alzheimer’s disease and scrapie: mechanistic truths and physiological consequences of the time-dependent solubility of amyloid proteins. Annu Rev Biochem 1997;66:385–407. [DOI] [PubMed] [Google Scholar]
- [33].Wall J, Schell M, Murphy C, et al. Thermodynamic instability of human lambda 6 light chains: correlation with fibrillogenicity. Biochemistry 1999;38:14101–14108. [DOI] [PubMed] [Google Scholar]
- [34].Martin EB, Williams A, Wooliver C, et al. Differential recruitment efficacy of patient-derived amyloidogenic and myeloma light chain proteins by synthetic fibrils-A metric for predicting amyloid propensity. PLoS One 2017;12:e0174152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Blancas-Mejia LM, Martin EB, Williams A, et al. Kinetic stability and sequence/structure studies of urine-derived Bence-Jones proteins from multiple myeloma and light chain amyloidosis patients. Biophys Chem 2017;230:89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Solomon A Light chains of human immunoglobulins. Meth Enzymol 1985;116:101–121. [DOI] [PubMed] [Google Scholar]
- [37].Murphy C, Wang S, Williams T, et al. Characterization of systemic amyloid deposits by mass spectrometry. Meth Enzymol 2006;412:48–62. [DOI] [PubMed] [Google Scholar]
- [38].Lousada I, Comenzo RL, Landau H, et al. Light chain amyloidosis: patient experience survey from the amyloidosis research consortium. Adv Ther 2015;32:920–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Fandrich M, Meinhardt J, Grigorieff N Structural polymorphism of alzheimer abeta and other amyloid fibrils. Prion 2009;3:89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Finn TE, Nunez AC, Sunde M, et al. Serum albumin prevents protein aggregation and amyloid formation and retains chaper- one-like activity in the presence of physiological ligands. J Biol Chem 2012;287:21530–21540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Milojevic J, Raditsis A, Melacini G Human serum albumin inhibits abeta fibrillization through a “monomer-competitor” mechanism. Biophys J 2009;97:2585–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zhou BR, Zhou Z, Hu QL, et al. Mixed macromolecular crowding inhibits amyloid formation of hen egg white lysozyme. Biochim Biophys Acta 2008;1784:472–480. [DOI] [PubMed] [Google Scholar]