Abstract
Background
Immune checkpoint blockade targeting programmed cell death ligand-1 (PD-L1)/programmed death-1 (PD-1) signaling was approved recently for locally advanced and metastatic urothelial bladder carcinoma (UBC). Some patients experience a very rapid tumor progression, rather than clinical benefit, from anti-PD-L1/PD-1 therapy.
Case presentation
A 58-year-old male diagnosed with non-muscle-invasive bladder cancer 3 years ago received transurethral resection of bladder tumor (TURBT) and intravesical chemotherapy. TURBT was repeated a year later for recurrent and progressive UBC. Following further disease progression, he received a radical cystectomy (RC), pathologically staged as T2bN2M0, and adjuvant cisplatin-containing combination chemotherapy. When his disease progressed to metastatic UBC, he was started on anti-PD-L1 monotherapy and experienced ultrarapid disease progression within 2 months; imaging scans ruled out pseudoprogression. We observed a fourfold increase in tumor growth rate, defined as the ratio of post- to pretreatment rates. Next-generation sequencing of formalin-fixed paraffin-embedded RC tissues showed MDM2 amplification without MDM4 amplification, EGFR aberrations, or DNMT3A alterations. Immunohistochemistry showed grade 2+ PD-L1 labeling intensity of the RC tissues, with 15%–25% and 5%–10% PD-LI immunopositive tumor cells and tumor-infiltrating immune cells, respectively.
Conclusion
Even in cases with PD-L1-positive tumors, MDM2 gene amplification may result in failure of anti-PD-L1 immunotherapy and rapid tumor growth. Therefore, genomic profiling may identify patients at risk for hyperprogression before immunotherapy.
Keywords: urothelial bladder carcinoma, programmed cell death ligand-1, immune checkpoint blockade, hyperprogression, MDM2
Introduction
Although platinum-based combination chemotherapy often prolongs the survival of patients with locally advanced or metastatic urothelial bladder carcinoma (UBC), progression remains almost inevitable with a median overall survival of only 14 months in 2014.1 The recent US FDA approval of immune checkpoint inhibitors that target the programmed cell death ligand-1 (PD-L1)/programmed death-1 (PD-1) receptor axis has changed how advanced or metastatic UBC is managed.2 Monoclonal PD-L1 antibodies can revitalize and enhance anticancer immunity by preventing PD-L1 from binding to PD-1 receptors.3
PD-L1 antibody was confirmed to produce durable objective responses and to have good tolerability in patients with inoperable advanced or metastatic UBC,4–7 leading to its approval for use in patients whose disease progressed during or within 12 months following neoadjuvant or adjuvant platinum-based chemotherapy.8 However, immunomodulatory therapies, such as PD-L1 immunotherapy, can produce opposing effects in a subset of patients. Indeed, there have been several recent reports of patients who experienced rapid tumor progression while on immune checkpoint blockade (ICB), consistent with ICB-promoted hyperprogression.9–14 Thus, there is a critical and urgent need to identify the predictors and mechanisms of such hyperprogression to prevent tragic adverse outcomes of ICB. A recent study showed an association between tumor hyperprogression and specific genomic alterations, including MDM2 family amplification and EGFR aberrations.14
Here, we report the case of an adult male patient with recurrent metastatic UBC whose disease progressed following platinum-based chemotherapy and then hyperprogressed shortly after initiation of ICB. UBCs have been reported to have relatively high PD-L1 expression among all cancers, and elevated PD-L1 expression intensity has been related to a higher probability of clinical response.6,7,15–17 Thus, we investigated the genomic profile and PD-L1 protein expression of the patients’ primary tumor following radical cystectomy (RC).
Case presentation
Patient characteristics and history
A 55-year-old man presented with left hip pain in October 2014. An initial workup revealed a left posterior mass in his bladder. Transurethral resection of bladder tumor (TURBT) pathology indicated stage-TaG3 UBC. After the TURBT procedure, he began a 12-month course of intravesical instillation of epirubicin chemotherapy. However, 14 months after the resection surgery, a cystoscope examination revealed bladder tumor recurrence. TURBT pathology indicated that the recurrent tumor was stage T1G3. The patient then received an additional 12-month course of adjuvant intravesical epirubicin chemotherapy instillations.
Disease progression was detected 11 months later, and TURBT pathology indicated that the advancing lesion was a stage T2G3 N0 UBC. He then received a RC, and the removed tumor was pathologically staged as T2bN2M0. Subsequently, he was treated with adjuvant cisplatin-containing combination chemotherapy for 3 months.
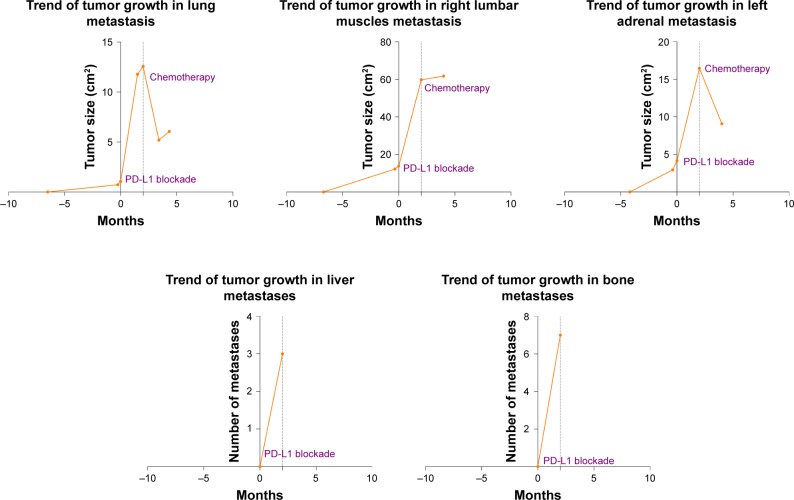
Twelve months after the RC, follow-up chest radiography and computer tomography (CT) revealed metastases in the right lumbar muscles, left adrenal gland, and lungs (Figure 1). In addition to bladder cancer, patient had no other history of cancer. The patient’s right lumbar mass biopsy puncture results indicated urothelial carcinoma. The patient was started on PD-L1 blockade monotherapy on December 19, 2017. Chest radiography and a full-body CT on January 15, 2018 showed pronounced enlargement of a left lung metastasis (1,004% increase from preimmunotherapy size) and progression of the right lumbar muscle and left adrenal gland metastases, as well as new multiple lymph node metastases involving a mediastinal, a left supraclavicular, and two hilar lymph nodes (Figure 1). He had developed a progressively enlarging right back mass with localized swelling and persistent severe pain, and was therefore admitted to our hospital.
Figure 1.
Treatment intervention process and imaging of disease progress after PD-L1 blockade.
Notes: (A) Summary of interventions received by the present patient. Arrowheads indicate time points for each intervention. (B) PET/CT or CT images for metastatic lesions before and after PD-L1 blockade.
Abbreviations: PET, positron emission tomography; CT, computer tomography; PD-L1, programmed cell death ligand-1; IHC, immunohistochemistry; NGS, next-generation sequencing.
In the hospital, while still receiving PD-L1 blockade monotherapy, the patient experienced unusually rapid disease progression demonstrated in repeated CT scans to rule out pseudoprogression. The patient terminated the immunotherapy after receiving two cycles of PD-L1 blockade treatment due to his rapid disease progression. A full-body CT, upper abdomen MRI, and positron emission tomography-CT on January 29, 2018 showed rapid progression of the metastatic lung lesions (1,078% increase from pre-immunotherapy cumulative size) and continued growth of the right lumbar muscle and left adrenal metastases, as well as the emergence of three liver metastases and at least seven bone metastases. Upon discovery of these changes, the patient’s treatment plan was changed to cisplatin/gemcitabine chemotherapy. One month after the patient began cisplatin/gemcitabine chemotherapy, we observed drastic reductions in lesion size (Figure 1).
To evaluate the patient’s treatment responses, we calculated tumor growth rate (TGR) vis-à-vis comparisons of tumor volume over time. TGR ratio was defined as the ratio of tumor volume growth change after, relative to that observed prior to, the treatment of interest. Comparing the TGR for the 8-week period following ICB to that for the 1-week period prior to ICB, we determined that the patient had a TGR ratio of 4.0, reflecting a fourfold increase in growth rate in association with ICB onset, meeting the criteria for hyperprogression (Figure 2). We employed Kato et al’s definition of hyperprogression criteria as follows: time-to-treatment failure (TTF) <2 months; increase in tumor burden >50%; and a >2-fold increase in TGR.14 All procedures performed in studies involving human participants were conducted in accordance with the ethical standards of Shanghai Tenth People’s Hospital (SHSY-IEC-4.0/17-16/01) and with the 1964 Helsinki declaration and its amendments or comparable ethical standards. Written informed consent was obtained from the patient to have the case details and any accompanying images published. The publication of the case details was approved by ethics committee of Shanghai Tenth People’s Hospital.
Figure 2.
Tumor metastasis changes over time. The 0-month time point represents the start of PD-L1 blockade treatment.
Abbreviation: PD-L1, programmed cell death ligand-1.
Assessments
Formalin-fixed paraffin embedded RC tissue samples were obtained from the Department of Pathology, Shanghai Tenth People’s Hospital. The samples were subjected to next-generation sequencing (NGS) and immunohistochemistry (IHC) with the aim of identifying possible predictive factors for immunotherapy-triggered hyperprogression. NGS was performed with a 499-gene panel assay (Table S1). The panel included sequences for multiple gene variants previously suggested to be associated with hyperprogression including MDM2 family amplification, EGFR aberration, and DNMT3A alteration sequences. The mean sequencing coverage depth exceeded 15,000×. The NGS method employed revealed copy number alterations, gene rearrangements, and somatic mutations with 95% specificity and >90% sensitivity. The presence of ≥3 gene copies was considered gene amplification. IHC carried out with monoclonal rabbit anti-PD-L1 antibody (clone MXR003, working solution for 15 hours; Fujian Maixin, Fujian, PR China), goat anti-rabbit and -mouse secondary antibody (PV-6000, working solution for 1 hour; ZSGB-BIO, Beijing, PR China), and horse-radish peroxidase to enhance visualization (ZLI-9017; ZSGB-BIO). IHC-AP cell membrane staining intensity was graded as follows: 0, none; 1+, weak or incomplete; 2+, weak to medium; 3+, medium to strong and complete.
Predictors of hyperprogression
NGS showed that the RC specimen from the present case had several malignancy-related alterations, including MDM2 amplification, a KRAS mutation, and a KMT2D mutation. It was not harboring an MDM4 amplification, EGFR aberrations, or DNMT3A alterations. The genomic alterations found are reported in Table 1 with descriptive information, including abundance, location, base and amino acid changes, and type of mutation.
Table 1.
Summary of NGS-revealed gene mutations
| Gene | Location | Base mutation | Amino acid change | Abundance | Mutation type | |
|---|---|---|---|---|---|---|
| KRAS | chr12:25398284 | c.35G>A | p.Gly12Asp | 22.82% | Missense | |
| KMT2D | chr12:49426895 | c.11593C>T | p.Gln3865Ter | 21.78% | Nonsense | |
| MDM2 | – | – | – | 11.92 copies | Amplification | |
| SPEN | chr1:16264490 | c.10693C>T | p.Arg3565Ter | 1.26% | Nonsense | |
| NOTCH2 | chr1:120462059 | c.5657G>A | p.Arg1886His | 1.09% | Missense | |
| AR | chrX:66765516 | c.528C>A | p.Ser176Arg | 95.11% | Missense | |
| MUTYH | chr1:45798136 | c.715G>A | p.Val239Ile | 1.52% | Missense | |
| DDR2 | chr1:162748503 | c.2417G>A | p.Arg806Gln | 1.43% | Missense | |
| TCF7L2 | chr10:114910785 | c.904C>T | p.His302Tyr | 13.55% | Missense | |
| PTPN11 | chr12:112926915 | c.1535G>A | p.Arg512Gln | 1.09% | Missense | |
| IDH2 | chr15:90630711 | c.775G>A | p.Asp259Asn | 1.19% | Missense | |
| IGF1R | chr15:99465453 | c.2278G>A | p.Ala760Thr | 1.32% | Missense | |
| PLCG2 | chr16:81902844 | c.505A>G | p.Ile169Val | 46.35% | Missense | |
| AXIN2 | chr17:63554353 | c.386G>A | p.Arg129Gln | 1.08% | Missense | |
| SMARCA4 | chr19:11100064 | c.1190G>A | p.Arg397Gln | 1.16% | Missense | |
| LRP1B | chr2:141283458 | c.7981G>A | p.Gly2661Arg | 1.55% | Missense | |
| CASP8 | chr2:202136289 | c.533C>A | p.Ser178Tyr | 53.30% | Missense | |
| BAP1 | chr3:52442077 | c.272G>T | p.Cys91Phe | 15.58% | Missense | |
| EPHA5 | chr4:66231683 | c.2017T>A | p.Ser673Thr | 46.75% | Missense | |
| TET2 | chr4:106155794 | c.695A>G | p.Gln232Arg | 46.42% | Missense | |
| INPP4B | chr4:143043366 | c.2050G>A | p.Val684Ile | 27.15% | Missense | |
| FAT1 | chr4:187524812 | c.10868C>T | p.Thr3623Met | 44.02% | Missense | |
| FAT1 | chr4:187541475 | c.6265G>A | p.Val2089Ile | 41.88% | Missense | |
| PDGFRB | chr5:149501461 | c.2326G>A | p.Asp776Asn | 50.00% | Missense | |
| ARID1B | chr6:157405827 | c.2069C>T | p.Thr690Met | 37.91% | Missense | |
| ETV1 | chr7:14027789 | c.55G>A | p.Gly19Arg | 45.92% | Missense | |
| MAGI2 | chr7:78150951 | c.550G>A | p.Gly184Ser | 1.45% | Missense | |
| KMT2C | chr7:151860428 | c.10234C>T | p.Arg3412Trp | 1.07% | Missense | |
| KAT6A | chr8:41906155 | c.341G>C | p.Gly114Ala | 4.56% | Missense | |
| PREX2 | chr8:69033224 | c.3664C>A | p.Pro1222Thr | 50.00% | Missense | |
| GID4 | chr17:17942909 | c.131G>C | p.Arg44Pro | 12.48% | Missense | |
| SOX10 | chr22:38370185 | c.718A>C | p.Thr240Pro | 10.47% | Missense | |
Abbreviation: NGS, next-generation sequencing.
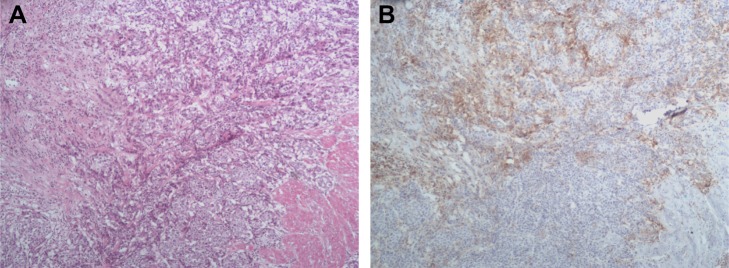
To calibrate PD-L1 expression relative to the proportion of tumor cells present in the RC specimen, alternate sections were subjected to H&E staining and anti-PD-L1 IHC prior to evaluating PD-L1 expression. In the H&E-stained sections (Figure 3A), we observed a 40% tumor cell ratio; >100 PD-L1 immunopositive tumor cells were examined under a light microscope. PD-L1 staining was localized primarily to cell membranes, with some non-specific cytoplasm staining. Tumor-associated immune cells had PD-L1 immunopositive cytoplasm and membranes. Both tumor cells and tumor-infiltrating immune cells had grade 2+ PD-L1 staining intensity. We found that 15%–25% and 5%–10% of tumor cells and tumor-infiltrating immune cells, respectively, showed PD-L1 immunopositivity (Figure 3B).
Figure 3.
Anti-PD-L1 immunohistochemistry of bladder cancer tissues.
Notes: (A) H&E stained tumor section with 40% tumor cell proportion. (B) Image of IHC PD-L1 labeled section subjected to PD-L1 percentage scoring. The percentages of tumor cells and tumor-infiltrating immune cells are 15%–25% and 5%–10%, respectively.
Abbreviations: PD-L1, programmed cell death ligand-1; IHC, immunohistochemistry.
Discussion
Blockade of the PD-1/PD-L1 pathway has produced durable clinical responses for some solid tumors and anti-PD-L1 agents have demonstrated a manageable safety profile and favorable clinical activity in patients with advanced, previously treated UBC.2,5–7 Currently, it is still a challenge to select the patients most likely to respond to treatment with immunotherapeutic agents. Robertson et al reported that clustering by mRNA, lncRNA, and miRNA expression converged to identify subsets with differential epithelial–mesenchymal transition status, carcinoma-in-situ scores, histologic features, and survival in bladder cancer. Their analyses identified five expression subtypes that may stratify response to different treatments. Among these, mRNA luminal-papillary subtype and basal-squamous subtypes show increased expression of CD270 (PD-L1) and PD-1 immune markers, which correspond to lncRNA 1 and miRNA 2 subtypes, lncRNA 4 and miRNA 4 subtypes, respectively. These two subtypes may serve as predictive markers for response to immune checkpoint therapy.18 However, the occurrence of immunotherapy-induced hyperprogression in some patients with various cancer types has drawn attention to a critical potential risk of immunotherapy.13,14 Reports of UBC hyperprogression with anti-PD-1 antibody treatment specifically are rare. To the best of our knowledge, the presently reported circumstance of dramatic growth and metastatic spreading of neoplastic lesions following anti-PD-L1 antibody initiation in an MDM2-amplified patient with UBC is quite rare. The rapid shrinking of multiple metastatic lesions, especially in the lungs, observed during the subsequent cisplatin-gemcitabine treatment indicated that the ICB-associated progression observed in this patient was not pseudoprogression but rather true hyperprogression.
Predictors of and mechanisms underlying ICB-triggered hyperprogression remain to be elucidated. The limited information available to date has implicated two clinical variables, namely older age and regional recurrence in an irradiated field,13 and a handful of genomic alterations, namely MDM2/4 amplification, EGFR aberrations, and DNMT3A alterations, in hyperprogression.14 In a study of 131 patients, encompassing 21 tumor types, treated with PD-1/PD-L1 pathway blockade, without genomic profiling, Champiat et al observed rapid progression in 12 patients (9%), including 2/8 patients (25%) with bladder cancer.13 In a study of 155 patients with diverse cancers, Kato et al reported that 49 patients (31.6%) had poor clinical outcomes of immunotherapy, defined as a TTF <2 months. Molecular profiling of Kato et al’s patient group showed that those with a poor clinical outcome harbored MDM2/4, EGFR, and/or DNMT3A alterations, each of which emerged as an independent predictor of a poor outcome. Six patients had MDM2 or –4 amplification, and all of them experienced hyperprogression, including one patient with bladder cancer harboring an MDM2 amplification.14
In the presently reported case, this patient was only 58 years old and had not received radiation therapy (RT). Upon starting anti-PD-L1 antibody treatment, the patient experienced rapid clinical deterioration with a marked acceleration in tumor growth (fourfold increase in progression rate and TTF of 1.4 months) accompanied by the emergence of new liver and bone metastases. IHC revealed PD-L1 expression in up to a quarter of RC tumor cells and up to a tenth of tumor-infiltrating immune cells, which suggests that PD-L1 immunopositivity is not a reliable indicator of immunotherapy sensitivity. Retrospective genomic profiling by NGS aimed at identifying hyperprogression predictors and clues regarding its mechanism showed MDM2 amplification without accompanying MDM4 or ERGR alterations. Similarly, Kriegmair et al found that patients with low MDM4 and high MDM2 expression tended to have poor muscle-invasive bladder cancer outcomes.19 These data point to MDM2 amplification as a predictive biomarker candidate for rapid ICB-triggered cancer progression.
Normally, PD-1/PD-L1 pathway activation is associated with anti-tumor immunity evasion that enables immunogenic tolerance. However, unfortunately, in some patients with UBC, the PD-1/PD-L1 pathway appears to have been linked with oncogenic signaling that triggers tumor proliferation and progression. Melanoma cell-intrinsic functions of PD-1/PD-L1 signaling might modulate several alternative signaling networks, including some that favor tumor growth.20 Such an effect may be secondary to an accumulation of oncogenes in tumor cells. Because our patient’s tumor had MDM2 amplification, in the absence of a p53 mutation, it may be that amplification of MDM2 inhibited the wild-type p53 tumor suppressor.21 Indeed, antigen-specific CD4+ T-cell responses have been reported to down-modulate tumor suppressor p53 through T-cell receptor signaling by decreasing expression of p53 while escalating expression of MDM2, the protein product of which mediates posttranscriptional inactivation of p53.22 In addition to T-cell receptor signaling increasing interferon-γ suppression of the PD-1 pathway – which activates JAK-STAT signaling thereby increasing interferon regulatory factor-8 expression – it may also induce MDM2 expression.23–26
Immune checkpoints occupy crucial regulatory pathways for the maintenance of immune homeostasis. Numerous immune cell subsets express PD-1 in tumor microenvironments, including macrophages, T cells, B cells, natural killer cells, and dendritic cells.27 Thus, ICB could trigger compensatory mechanisms and adaptive immune resistance, enabling an acceleration of tumor growth.
If the presently observed hyperprogression phenomenon is specific to anti-PD-1/PD-L1 monotherapy, it might be solved with mechanistically sound combination therapies. In metastatic castration-resistant prostate cancer mouse models, intratumoral myeloid-derived suppressor cells inhibited CD4+ and CD8+ T-cell proliferation, and PD-1/PD-L1 blockade combined with myeloid-derived suppressor cell-targeted therapies yielded excellent synergistic efficacy against ICB resistance.28 Indeed, RT has been reported to enhance T-cell recognition of malignant cells through induction of MHCI expression and neoantigen generation.29 Meanwhile, PD-L1 has been found to be upregulated after RT,12,30 and combining RT with PD-L1 blockade has been found to enhance anti-tumor treatment effects.30,31 Likewise, chemotherapy has been reported to augment intra-tumor CD8+ T-cell infiltration, consistent with the notion that immunogenic chemotherapies could increase the anticancer efficacy of ICB.32–34 These studies support the strategy of developing innovative combination therapies to overcome undesirable tumor responsivity to PD-1/PD-L1 blockade.
In summary, genomic testing of malignant tumors prior to treatment, preferably in an early stage, may reveal which patients harbor genetic alterations associated with hyperprogression. The present case indicates that patients with MDM2 amplification in particular should not receive anti-PD-L1 monotherapy, even in cases where tumor cells or tumor-associated immune cells are found to express PD-L1. Large-cohort studies are needed to confirm this link. ICB-triggered hyperprogression may be avoided with a combined treatment.
Supplementary material
Table S1.
Gene detection list
| ABL1 | ABL2 | ACVR1 | ACVR1B | AGO2 | AKT1 | AKT2 | AKT3 | ALK | ALOX12B |
| AMER1 | AR | APC | ANKRD11 | ARAF | ARFRP1 | ARID1A | ARID1B | ARID2 | ARID5B |
| ASXL1 | ASXL2 | ATM | ATR | ATRX | AURKA | AURKB | AXIN1 | AXIN2 | AXL |
| B2M | BAP1 | BARD1 | BCL10 | BCL2 | BCL2L1 | BCL2L11 | BCL2L2 | BCL6 | BCOR |
| BCORL1 | BIRC3 | BLM | BMPR1A | BRAF | BRCA1 | BRCA2 | BRD3 | BRD4 | BRIP1 |
| BTG1 | BTK | C11orf30 | CARD11 | CALR | CARM1 | CASP8 | CBFB | CBL | CCND1 |
| CCND2 | CCND3 | CCNE1 | CD274 | CD276 | CD74 | CD79A | CD79B | CDC42 | CDC73 |
| CDH1 | CDK12 | CDK4 | CDKN1A | CDK6 | CDKN2A | CDKN1B | CDK8 | CDKN2B | CIC |
| CEBPA | CHD2 | CHD4 | CHEK1 | CHEK2 | CDKN2C | CREBBP | CRKL | CRLF2 | CSDE1 |
| CSF1R | CSF3R | CTCF | CTLA4 | CUL3 | CTNNB1 | CTNNA1 | CXCR4 | CYLD | CYSLTR2 |
| DAXX | DDR2 | DICER1 | DNMT3B | DNAJB1 | DNMT1 | DNMT3A | DIS3 | DOT1L | DROSHA |
| DUSP4 | E2F3 | EED | EGF | EGFR | EIF1AX | EIF4A2 | ELF3 | EML4 | EP300 |
| EPAS1 | EPCAM | EPHA3 | EPHA5 | EPHA7 | EPHB1 | ERBB2 | ERBB3 | ERBB4 | ERCC1 |
| ERCC2 | ERCC3 | ERCC4 | ERCC5 | ERF | ERG | ERRFI1 | ESR1 | ETV1 | ETV6 |
| EZH2 | FAM46C | FAM58A | FAM175A | FANCA | FANCD2 | FANCE | FANCC | FANCF | FANCG |
| FANCL | FAS | FAT1 | FBXW7 | FGF10 | FGF14 | FGF19 | FGF23 | FGF3 | FLT3 |
| FGF4 | FGF6 | FGFR1 | FGFR2 | FGFR3 | FGFR4 | FLCN | FH | FLT1 | FLT4 |
| FOLR3 | FOXA1 | FOXL2 | FOXO1 | FOXP1 | FRS2 | FUBP1 | FYN | GABRA6 | GATA1 |
| GATA2 | GATA3 | GATA4 | GATA6 | GID4 | GLI1 | GNA11 | GNA13 | GNAQ | GNAS |
| GOPC | GPR124 | GREM1 | GRIN2A | GRM3 | GSK3B | GSTA1 | H3F3A | H3F3B | HDAC1 |
| HDAC4 | HIST1H1C | HGF | HIST1H3B | HLA-A | HLA-B | HIST1H2BD | HNF1A | HIST1H3G | HOXB13 |
| HRAS | HSP90AA1 | HSD3B1 | ID3 | IDH1 | IDH2 | IFNGR1 | IGF1 | IGF1R | IGF2 |
| IKBKE | IKZF1 | IL10 | IL7R | INHBA | INPP4A | INPP4B | INPPL1 | INSR | IRF2 |
| IRF4 | IRS1 | IRS2 | JAK1 | JAK2 | JAK3 | JUN | KAT6A | KDM5A | KDM5C |
| KDM6A | KDR | KEAP1 | KEL | KIT | KLF4 | KLHL6 | KMT2A | KMT2B | KMT2C |
| KMT2D | KNSTRN | KRAS | LATS1 | LATS2 | LMO1 | LRP1B | LRRK2 | LYN | LZTR1 |
| MAGI2 | MALT1 | MAP2K1 | MAP2K2 | MAP2K4 | MAP3K1 | MAP3K13 | MAPK1 | MAP3K14 | MAPK3 |
| MAX | MCL1 | MDC1 | MDM2 | MDM4 | MED12 | MEF2B | MEN1 | MET | MGA |
| MITF | MLH1 | MPL | MRE11A | MSH2 | MSH3 | MSH6 | MSI2 | MST1R | MTOR |
| MUTYH | MYC | MYCL | MYCN | MYD88 | MYOD1 | NAT2 | NBN | NCOA3 | NCOR1 |
| NEGR1 | NF1 | NF2 | NFE2L2 | NFKBIA | NKX2-1 | NOTCH1 | NOTCH2 | NOTCH3 | NOTCH4 |
| NPM1 | NRAS | NSD1 | NTRK1 | NTRK2 | NTRK3 | NUF2 | NUP93 | OPRM1 | PAK1 |
| PAK3 | PAK7 | PALB2 | PARK2 | PARP1 | PARP2 | PAX5 | PBRM1 | PDCD1 | PDK1 |
| PDGFRA | PDGFRB | PDPK1 | PGR | PIK3CA | PHOX2B | PDCD1LG2 | PIK3C3 | PIK3C2B | PIK3C2G |
| PIK3CB | PIK3CD | PIK3CG | PIK3R1 | PIK3R2 | PIK3R3 | PPP2R1A | PIM1 | PLCG2 | PMS1 |
| PMS2 | PNRC1 | POLD1 | PTPN11 | POLE | PPARG | PPM1D | PPP6C | PRDM1 | PRDM14 |
| PREX2 | PRKAR1A | PRKCI | PRKD1 | PRKDC | PRSS8 | PTCH1 | PTCH2 | PTEN | PTPRD |
| PTPRS | PTPRT | QKI | RAB35 | RAC1 | RAC2 | RAD21 | RAD50 | RAD51 | RAD51B |
| RAD51C | RAD51D | RAD52 | RAD54L | RAF1 | RANBP2 | RARA | RASA1 | RB1 | RBM10 |
| RECQL | RECQL4 | REL | RET | RFWD2 | RHEB | RHOA | RICTOR | RIT1 | RNF43 |
| ROCK1 | ROS1 | RPTOR | RUNX1T1 | RRAGC | RPS6KB1 | RPS6KA4 | RRAS2 | RRM1 | RTEL1 |
| RUNX1 | RXRA | RYBP | SDHAF2 | SDHA | SDHB | SLC19A1 | SDHC | SDHD | SETD2 |
| SF3B1 | SMARCA4 | SH2B3 | SMARCB1 | SHOC2 | SHQ1 | SMARCD1 | SLIT2 | SLX4 | SMAD2 |
| SMAD3 | SNCAIP | SMAD4 | SOCS1 | SMO | SOS1 | STAT5A | SOX10 | SOX17 | SOX2 |
| SOX9 | STAT5B | SPEN | STK11 | SPOP | SPTA1 | SUFU | SRC | SRSF2 | STAG2 |
| STAT3 | SUZ12 | STAT4 | SYK | TAF1 | TAP1 | TAP2 | TBX3 | TCEB1 | TCF3 |
| TEK | TCF7L2 | TERT | TGFBR1 | TET1 | TET2 | TGFBR2 | TOP1 | TMEM127 | TMPRSS2 |
| TNFAIP3 | TNFRSF14 | TOP2A | TP53 | TP63 | TP53BP1 | TRAF7 | TRAF2 | TSC1 | TSC2 |
| TSHR | TYMS | U2AF1 | VEGFA | VHL | WHSC1 | WHSC1L1 | WISP3 | WT1 | WWTR1 |
| XIAP | XPO1 | XRCC2 | YAP1 | YES1 | ZBTB2 | ZFHX3 | ZNF217 | ZNF703 |
Acknowledgments
This study was funded by the Natural Science Foundation of China (grant number 81472389) and Shanghai Health and Family Planning Commission Key Project (grant number 20124008). The abstract of this paper was presented at the Global Congress on Bladder Cancer 2018 as a poster presentation with interim findings. The poster’s abstract was published in Abstract book (ISBN 9789462210165) and as an e-poster online: https://abstracts.mirrorsmed.org/abstracts/hyperprogression-after-immunotherapy-patient-recurrent-and-metastatic-urothelial-bladder.
Footnotes
Author contributions
YY and XY designed and guided the present study. SM, JZ, and YG collected the study data. SM, YW, ZZ, and WZ analyzed and interpreted the data. LW, JZ, and YG made figures and tables. SM was a major contributor in writing the manuscript. JG, YY, and XY revised the manuscript. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bellmunt J, Orsola A, Leow JJ, et al. Bladder cancer: ESMO practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25(Suppl 3):iii40–iii48. doi: 10.1093/annonc/mdu223. [DOI] [PubMed] [Google Scholar]
- 2.Balar AV, Galsky MD, Rosenberg JE, et al. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: a single-arm, multicentre, phase 2 trial. Lancet. 2017;389(10064):67–76. doi: 10.1016/S0140-6736(16)32455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stewart R, Morrow M, Hammond SA, et al. Identification and characterization of MEDI4736, an antagonistic anti-PD-L1 monoclonal antibody. Cancer Immunol Res. 2015;3(9):1052–1062. doi: 10.1158/2326-6066.CIR-14-0191. [DOI] [PubMed] [Google Scholar]
- 4.Rosenberg JE, Hoffman-Censits J, Powles T, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387(10031):1909–1920. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Powles T, Eder JP, Fine GD, et al. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature. 2014;515(7528):558–562. doi: 10.1038/nature13904. [DOI] [PubMed] [Google Scholar]
- 6.Powles T, O’Donnell PH, Massard C, et al. Efficacy and safety of durvalumab in locally advanced or metastatic urothelial carcinoma: updated results from a phase 1/2 open-label study. JAMA Oncol. 2017;3(9):e172411. doi: 10.1001/jamaoncol.2017.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Massard C, Gordon MS, Sharma S, et al. Safety and efficacy of durvalumab (MEDI4736), an anti-programmed cell death ligand-1 immune checkpoint inhibitor, in patients with advanced urothelial bladder cancer. J Clin Oncol. 2016;34(26):3119–3125. doi: 10.1200/JCO.2016.67.9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Syed YY. Durvalumab: first global approval. Drugs. 2017;77(12):1369–1376. doi: 10.1007/s40265-017-0782-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saâda-Bouzid E, Defaucheux C, Karabajakian A, et al. Hyperprogression during anti-PD-1/PD-L1 therapy in patients with recurrent and/or metastatic head and neck squamous cell carcinoma. Ann Oncol. 2017;28(7):1605–1611. doi: 10.1093/annonc/mdx178. [DOI] [PubMed] [Google Scholar]
- 10.Chubachi S, Yasuda H, Irie H, et al. A case of non-small cell lung cancer with possible “disease flare” on nivolumab treatment. Case Rep Oncol Med. 2016;2016:1–3. doi: 10.1155/2016/1075641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshida T, Furuta H, Hida T. Risk of tumor flare after nivolumab treatment in patients with irradiated field recurrence. Med Oncol. 2017;34(3):3434. doi: 10.1007/s12032-017-0895-4. [DOI] [PubMed] [Google Scholar]
- 12.Alexander GS, Palmer JD, Tuluc M, et al. Immune biomarkers of treatment failure for a patient on a phase I clinical trial of pembrolizumab plus radiotherapy. J Hematol Oncol. 2016;9(1):96. doi: 10.1186/s13045-016-0328-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Champiat S, Dercle L, Ammari S, et al. Hyperprogressive disease is a new pattern of progression in cancer patients treated by anti-PD-1/PD-L1. Clin Cancer Res. 2017;23(8):1920–1928. doi: 10.1158/1078-0432.CCR-16-1741. [DOI] [PubMed] [Google Scholar]
- 14.Kato S, Goodman A, Walavalkar V, Barkauskas DA, Sharabi A, Kurzrock R. Hyperprogressors after immunotherapy: analysis of genomic alterations associated with accelerated growth rate. Clin Cancer Res. 2017;23(15):4242–4250. doi: 10.1158/1078-0432.CCR-16-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zou W, Chen L. Inhibitory B7-family molecules in the tumour micro-environment. Nat Rev Immunol. 2008;8(6):467–477. doi: 10.1038/nri2326. [DOI] [PubMed] [Google Scholar]
- 16.Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med. 2015;373(17):1627–1639. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferris RL, Blumenschein G, Fayette J, et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med. 2016;375(19):1856–1867. doi: 10.1056/NEJMoa1602252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robertson AG, Kim J, Al-Ahmadie H, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell. 2018;174(4):1033. doi: 10.1016/j.cell.2018.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kriegmair MC, Balk M, Wirtz R, et al. Expression of the p53 inhibitors MDM2 and MDM4 as outcome predictor in muscle-invasive bladder cancer. Anticancer Res. 2016;36(10):5205–5214. doi: 10.21873/anticanres.11091. [DOI] [PubMed] [Google Scholar]
- 20.Kleffel S, Posch C, Barthel SR, et al. Melanoma cell-intrinsic PD-1 receptor functions promote tumor growth. Cell. 2015;162(6):1242–1256. doi: 10.1016/j.cell.2015.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13(2):83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watanabe M, Moon KD, Vacchio MS, Hathcock KS, Hodes RJ. Downmodulation of tumor suppressor p53 by T cell receptor signaling is critical for antigen-specific CD4(+) T cell responses. Immunity. 2014;40(5):681–691. doi: 10.1016/j.immuni.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng W, Liu C, Xu C, et al. PD-1 blockade enhances T-cell migration to tumors by elevating IFN-γ inducible chemokines. Cancer Res. 2012;72(20):5209–5218. doi: 10.1158/0008-5472.CAN-12-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schindler C, Levy DE, Decker T. JAK-STAT signaling: from interferons to cytokines. J Biol Chem. 2007;282(28):20059–20063. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- 25.Waight JD, Netherby C, Hensen ML, et al. Myeloid-derived suppressor cell development is regulated by a STAT/IRF-8 axis. J Clin Invest. 2013;123(10):4464–4478. doi: 10.1172/JCI68189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Y, Yu H, Hu W. The regulation of MDM2 oncogene and its impact on human cancers. Acta Biochim Biophys Sin (Shanghai) 2014;46(3):180–189. doi: 10.1093/abbs/gmt147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gordon SR, Maute RL, Dulken BW, et al. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature. 2017;545(7655):495–499. doi: 10.1038/nature22396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu X, Horner JW, Paul E, et al. Effective combinatorial immunotherapy for castration-resistant prostate cancer. Nature. 2017;543(7647):728–732. doi: 10.1038/nature21676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newcomb EW, Demaria S, Lukyanov Y, et al. The combination of ionizing radiation and peripheral vaccination produces long-term survival of mice bearing established invasive GL261 gliomas. Clin Cancer Res. 2006;12(15):4730–4737. doi: 10.1158/1078-0432.CCR-06-0593. [DOI] [PubMed] [Google Scholar]
- 30.Dovedi SJ, Adlard AL, Lipowska-Bhalla G, et al. Acquired resistance to fractionated radiotherapy can be overcome by concurrent PD-L1 blockade. Cancer Res. 2014;74(19):5458–5468. doi: 10.1158/0008-5472.CAN-14-1258. [DOI] [PubMed] [Google Scholar]
- 31.Walshaw RC, Honeychurch J, Illidge TM, Choudhury A. The anti-PD-1 era – an opportunity to enhance radiotherapy for patients with bladder cancer. Nat Rev Urol. 2018;15(4):251–259. doi: 10.1038/nrurol.2017.172. [DOI] [PubMed] [Google Scholar]
- 32.Galluzzi L, Buqué A, Kepp O, Zitvogel L, Kroemer G. Immunological effects of conventional chemotherapy and targeted anticancer agents. Cancer Cell. 2015;28(6):690–714. doi: 10.1016/j.ccell.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 33.Dosset M, Vargas TR, Lagrange A, et al. PD-1/PD-L1 pathway: an adaptive immune resistance mechanism to immunogenic chemotherapy in colorectal cancer. Oncoimmunology. 2018;7(6):e1433981. doi: 10.1080/2162402X.2018.1433981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song W, Shen L, Wang Y, et al. Synergistic and low adverse effect cancer immunotherapy by immunogenic chemotherapy and locally expressed PD-L1 trap. Nat Commun. 2018;9(1):2237. doi: 10.1038/s41467-018-04605-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Gene detection list
| ABL1 | ABL2 | ACVR1 | ACVR1B | AGO2 | AKT1 | AKT2 | AKT3 | ALK | ALOX12B |
| AMER1 | AR | APC | ANKRD11 | ARAF | ARFRP1 | ARID1A | ARID1B | ARID2 | ARID5B |
| ASXL1 | ASXL2 | ATM | ATR | ATRX | AURKA | AURKB | AXIN1 | AXIN2 | AXL |
| B2M | BAP1 | BARD1 | BCL10 | BCL2 | BCL2L1 | BCL2L11 | BCL2L2 | BCL6 | BCOR |
| BCORL1 | BIRC3 | BLM | BMPR1A | BRAF | BRCA1 | BRCA2 | BRD3 | BRD4 | BRIP1 |
| BTG1 | BTK | C11orf30 | CARD11 | CALR | CARM1 | CASP8 | CBFB | CBL | CCND1 |
| CCND2 | CCND3 | CCNE1 | CD274 | CD276 | CD74 | CD79A | CD79B | CDC42 | CDC73 |
| CDH1 | CDK12 | CDK4 | CDKN1A | CDK6 | CDKN2A | CDKN1B | CDK8 | CDKN2B | CIC |
| CEBPA | CHD2 | CHD4 | CHEK1 | CHEK2 | CDKN2C | CREBBP | CRKL | CRLF2 | CSDE1 |
| CSF1R | CSF3R | CTCF | CTLA4 | CUL3 | CTNNB1 | CTNNA1 | CXCR4 | CYLD | CYSLTR2 |
| DAXX | DDR2 | DICER1 | DNMT3B | DNAJB1 | DNMT1 | DNMT3A | DIS3 | DOT1L | DROSHA |
| DUSP4 | E2F3 | EED | EGF | EGFR | EIF1AX | EIF4A2 | ELF3 | EML4 | EP300 |
| EPAS1 | EPCAM | EPHA3 | EPHA5 | EPHA7 | EPHB1 | ERBB2 | ERBB3 | ERBB4 | ERCC1 |
| ERCC2 | ERCC3 | ERCC4 | ERCC5 | ERF | ERG | ERRFI1 | ESR1 | ETV1 | ETV6 |
| EZH2 | FAM46C | FAM58A | FAM175A | FANCA | FANCD2 | FANCE | FANCC | FANCF | FANCG |
| FANCL | FAS | FAT1 | FBXW7 | FGF10 | FGF14 | FGF19 | FGF23 | FGF3 | FLT3 |
| FGF4 | FGF6 | FGFR1 | FGFR2 | FGFR3 | FGFR4 | FLCN | FH | FLT1 | FLT4 |
| FOLR3 | FOXA1 | FOXL2 | FOXO1 | FOXP1 | FRS2 | FUBP1 | FYN | GABRA6 | GATA1 |
| GATA2 | GATA3 | GATA4 | GATA6 | GID4 | GLI1 | GNA11 | GNA13 | GNAQ | GNAS |
| GOPC | GPR124 | GREM1 | GRIN2A | GRM3 | GSK3B | GSTA1 | H3F3A | H3F3B | HDAC1 |
| HDAC4 | HIST1H1C | HGF | HIST1H3B | HLA-A | HLA-B | HIST1H2BD | HNF1A | HIST1H3G | HOXB13 |
| HRAS | HSP90AA1 | HSD3B1 | ID3 | IDH1 | IDH2 | IFNGR1 | IGF1 | IGF1R | IGF2 |
| IKBKE | IKZF1 | IL10 | IL7R | INHBA | INPP4A | INPP4B | INPPL1 | INSR | IRF2 |
| IRF4 | IRS1 | IRS2 | JAK1 | JAK2 | JAK3 | JUN | KAT6A | KDM5A | KDM5C |
| KDM6A | KDR | KEAP1 | KEL | KIT | KLF4 | KLHL6 | KMT2A | KMT2B | KMT2C |
| KMT2D | KNSTRN | KRAS | LATS1 | LATS2 | LMO1 | LRP1B | LRRK2 | LYN | LZTR1 |
| MAGI2 | MALT1 | MAP2K1 | MAP2K2 | MAP2K4 | MAP3K1 | MAP3K13 | MAPK1 | MAP3K14 | MAPK3 |
| MAX | MCL1 | MDC1 | MDM2 | MDM4 | MED12 | MEF2B | MEN1 | MET | MGA |
| MITF | MLH1 | MPL | MRE11A | MSH2 | MSH3 | MSH6 | MSI2 | MST1R | MTOR |
| MUTYH | MYC | MYCL | MYCN | MYD88 | MYOD1 | NAT2 | NBN | NCOA3 | NCOR1 |
| NEGR1 | NF1 | NF2 | NFE2L2 | NFKBIA | NKX2-1 | NOTCH1 | NOTCH2 | NOTCH3 | NOTCH4 |
| NPM1 | NRAS | NSD1 | NTRK1 | NTRK2 | NTRK3 | NUF2 | NUP93 | OPRM1 | PAK1 |
| PAK3 | PAK7 | PALB2 | PARK2 | PARP1 | PARP2 | PAX5 | PBRM1 | PDCD1 | PDK1 |
| PDGFRA | PDGFRB | PDPK1 | PGR | PIK3CA | PHOX2B | PDCD1LG2 | PIK3C3 | PIK3C2B | PIK3C2G |
| PIK3CB | PIK3CD | PIK3CG | PIK3R1 | PIK3R2 | PIK3R3 | PPP2R1A | PIM1 | PLCG2 | PMS1 |
| PMS2 | PNRC1 | POLD1 | PTPN11 | POLE | PPARG | PPM1D | PPP6C | PRDM1 | PRDM14 |
| PREX2 | PRKAR1A | PRKCI | PRKD1 | PRKDC | PRSS8 | PTCH1 | PTCH2 | PTEN | PTPRD |
| PTPRS | PTPRT | QKI | RAB35 | RAC1 | RAC2 | RAD21 | RAD50 | RAD51 | RAD51B |
| RAD51C | RAD51D | RAD52 | RAD54L | RAF1 | RANBP2 | RARA | RASA1 | RB1 | RBM10 |
| RECQL | RECQL4 | REL | RET | RFWD2 | RHEB | RHOA | RICTOR | RIT1 | RNF43 |
| ROCK1 | ROS1 | RPTOR | RUNX1T1 | RRAGC | RPS6KB1 | RPS6KA4 | RRAS2 | RRM1 | RTEL1 |
| RUNX1 | RXRA | RYBP | SDHAF2 | SDHA | SDHB | SLC19A1 | SDHC | SDHD | SETD2 |
| SF3B1 | SMARCA4 | SH2B3 | SMARCB1 | SHOC2 | SHQ1 | SMARCD1 | SLIT2 | SLX4 | SMAD2 |
| SMAD3 | SNCAIP | SMAD4 | SOCS1 | SMO | SOS1 | STAT5A | SOX10 | SOX17 | SOX2 |
| SOX9 | STAT5B | SPEN | STK11 | SPOP | SPTA1 | SUFU | SRC | SRSF2 | STAG2 |
| STAT3 | SUZ12 | STAT4 | SYK | TAF1 | TAP1 | TAP2 | TBX3 | TCEB1 | TCF3 |
| TEK | TCF7L2 | TERT | TGFBR1 | TET1 | TET2 | TGFBR2 | TOP1 | TMEM127 | TMPRSS2 |
| TNFAIP3 | TNFRSF14 | TOP2A | TP53 | TP63 | TP53BP1 | TRAF7 | TRAF2 | TSC1 | TSC2 |
| TSHR | TYMS | U2AF1 | VEGFA | VHL | WHSC1 | WHSC1L1 | WISP3 | WT1 | WWTR1 |
| XIAP | XPO1 | XRCC2 | YAP1 | YES1 | ZBTB2 | ZFHX3 | ZNF217 | ZNF703 |