Abstract
Background
Studies have shown strong positive associations between serum urate (SU) levels and chronic kidney disease (CKD) risk; however, whether the relation is causal remains uncertain. We evaluate whether genetic data are consistent with a causal impact of SU level on the risk of CKD and estimated glomerular filtration rate (eGFR).
Methods and findings
We used Mendelian randomization (MR) methods to evaluate the presence of a causal effect. We used aggregated genome-wide association data (N = 110,347 for SU, N = 69,374 for gout, N = 133,413 for eGFR, N = 117,165 for CKD), electronic-medical-record-linked UK Biobank data (N = 335,212), and population-based cohorts (N = 13,425), all in individuals of European ancestry, for SU levels and CKD. Our MR analysis showed that SU has a causal effect on neither eGFR level nor CKD risk across all MR analyses (all P > 0.05). These null associations contrasted with our epidemiological association findings from the 4 population-based cohorts (change in eGFR level per 1-mg/dl [59.48 μmol/l] increase in SU: −1.99 ml/min/1.73 m2; 95% CI −2.86 to −1.11; P = 8.08 × 10−6; odds ratio [OR] for CKD: 1.48; 95% CI 1.32 to 1.65; P = 1.52 × 10−11). In contrast, the same MR approaches showed that SU has a causal effect on the risk of gout (OR estimates ranging from 3.41 to 6.04 per 1-mg/dl increase in SU, all P < 10−3), which served as a positive control of our approach. Overall, our MR analysis had >99% power to detect a causal effect of SU level on the risk of CKD of the same magnitude as the observed epidemiological association between SU and CKD. Limitations of this study include the lifelong effect of a genetic perturbation not being the same as an acute perturbation, the inability to study non-European populations, and some sample overlap between the datasets used in the study.
Conclusions
Evidence from our series of causal inference approaches using genetics does not support a causal effect of SU level on eGFR level or CKD risk. Reducing SU levels is unlikely to reduce the risk of CKD development.
Author summary
Why was this study done?
Epidemiological studies have shown strong correlations between serum urate (SU) levels and chronic kidney disease (CKD) risk.
Elevated SU levels are often found in patients with CKD, but it is not clear whether high serum urate is a cause of kidney disease or just a common co-occurrence.
Previous studies examining whether SU levels had a causal effect on CKD were limited due to not having large enough samples to detect a true causal relationship if it existed and/or had limitations related to the methodology.
Several clinical trials have been started that aim to use urate-lowering medication to prevent CKD.
What did the authors do and find?
To determine whether SU level has a causal effect on CKD, we used a methodology known as Mendelian randomization to test whether genetic variants known to increase SU level also increased the risk of CKD.
We used multiple datasets to perform Mendelian randomization analyses, which included meta-analyses performed across multiple population-based cohorts, 4 individual population-based cohorts, and the large electronic-medical-record-linked UK Biobank.
Across all datasets, we found no significant causal connection between SU level and risk of CKD.
What do these findings mean?
Our findings do not support a causal role of SU level in CKD.
Lower SU levels would be unlikely to translate into reduced risk of CKD.
Introduction
Approximately 10% of the global population has chronic kidney disease (CKD) [1,2], which can result in end-stage renal disease, associated with shortened life expectancy and requirement for dialysis or kidney transplantation [3]. There are limited therapeutic options for CKD, with management predominantly focused on control of blood pressure, diabetes, and complications. Hence, there is an intense search for novel therapeutic targets.
Observational studies have consistently shown strong positive associations between serum urate (SU) levels and the risk of CKD [4,5]; however, whether the relation is causal remains unknown. Speculated mechanisms for the potential impact of SU levels on CKD have included nitric oxide and renin-angiotensin pathways [6], stimulation of the renin-angiotensin system [7], and vascular smooth muscle cell proliferation [8]. Furthermore, findings in induced-hyperuricemia rodent models have suggested a causal role of urate in hypertension and associated renal pathophysiology [9]. However, these findings are difficult to directly translate to humans as rodents have considerably lower urate levels owing to functional uricase [9].
As effective medications to lower SU levels are available, the potential causal role of SU level in CKD has become one of the most investigated targets for a renoprotective agent. As such, clinical trials of xanthine oxidase inhibitors (allopurinol or febuxostat) for the endpoint of CKD progression/development are currently underway [10].
Genetic epidemiology can be used to evaluate the causality of risk factors with respect to potential endpoints of interest. This approach, called Mendelian randomization (MR), posits that causality can be inferred because the alleles of a particular exposure-associated genotype are assigned randomly at conception. This can minimize the bias that can occur by confounding and reverse causation in conventional observational studies [11]. Genotypes can be used as a genetically determined lifetime exposure of interest and can be tested for a causal effect on outcomes of interest. For example, MR studies have found that low-density lipoprotein cholesterol is causally related to the risk of coronary artery disease, whereas high-density lipoprotein cholesterol is not [12].
Our objective was to evaluate whether genetic data are consistent with a causal effect of SU levels on estimated glomerular filtration rate (eGFR) and risk of CKD. We performed a series of MR analyses of SU level and eGFR and CKD using genetic variants influencing SU level as the exposure without the influence of confounders.
Methods
Study design overview
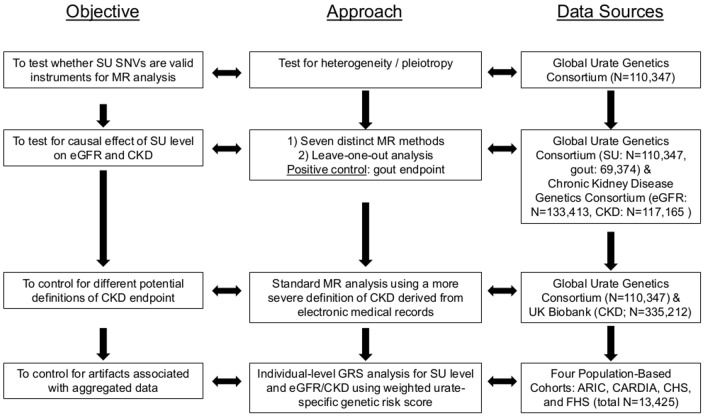
Our study approach was composed of several complementary components (Fig 1). We first performed a test of heterogeneity to detect the presence of pleiotropy [13] in urate-associated single nucleotide variants (SNVs) identified in a large meta-analysis of genome-wide association (GWA) studies of SU (see S1 Text) [14]. We then conducted 7 distinct MR analyses to evaluate the potential causal role of SU level in eGFR and CKD risk. Since we found significant heterogeneity (see Results), indicating potential pleiotropy, these 7 analytic approaches were specifically chosen to be robust to heterogeneity and pleiotropy in our MR analysis (see S1 Text and below for details). As a positive control endpoint, we assessed whether the same genetic instrumental variables showed the known causal effect of SU on gout. To assess the effect of varying disease endpoint definitions and to control for potential artifacts produced by meta-analyses of heterogeneous populations, we conducted MR analyses using the same SNVs in the UK Biobank, as well as individual-level MR analyses based on 4 population-based cohorts (Atherosclerosis Risk in Communities [ARIC] [15], Coronary Artery Risk Development in Young Adults [CARDIA] [16], Cardiovascular Health Study [CHS] [17], and Framingham Heart Study [FHS] [18]) (Fig 1).
Fig 1. Study design overview.
Overview of the study design. ARIC, Atherosclerosis Risk in Communities; CARDIA, Coronary Artery Risk Development in Young Adults; CHS, Cardiovascular Health Study; CKD, chronic kidney disease; eGFR, estimated glomerular filtration rate; FHS, Framingham Heart Study; GRS, genetic risk score; MR, Mendelian randomization; SNV, single nucleotide variant; SU, serum urate.
Data sources and participants
We examined 26 SNVs strongly associated with SU level identified in a GWA meta-analysis study of 110,347 participants of European ancestry conducted by the Global Urate Genetics Consortium (S1 Table) [14].
For MR analyses for eGFR and CKD endpoints, we retrieved GWA study summary statistics for eGFR and CKD from a published meta-analysis conducted by the Chronic Kidney Disease Genetics Consortium (CKDGen) [19]. We used summary statistics for eGFR values calculated from serum creatinine, available in up to 133,413 participants of European ancestry, and for CKD status defined as eGFR < 60 ml/min/1.73 m2, available in up to 12,385 cases and 104,780 controls.
For MR analyses for gout (as a positive control endpoint), we retrieved GWA study summary statistics (effect sizes and standard errors) for gout from a published meta-analysis in 2,115 cases and 67,259 controls of European ancestry from the Global Urate Genetics Consortium [14].
Our additional datasets consisted of electronic-medical-record-linked UK Biobank data (N = 335,212), and 4 population-based cohorts (ARIC, CARDIA, CHS, and FHS; N = 13,425 total). In the UK Biobank, based on recommendations from UK Biobank (http://www.ukbiobank.ac.uk/wp-content/uploads/2018/03/ukb_genetic_data_description_v3.txt), we excluded samples that belonged to any of the following categories: outliers in heterozygosity and missing rates, putative sex chromosome aneuploidy, self-reported non-white British ancestry, and related individuals. Excluded related individuals were defined as 1 individual in each pair with relatedness up to the third degree. In total, 335,212 individuals of white British ancestry remained for analyses. For samples from the UK Biobank, we treated the following ICD-10 codes as indicative of CKD: N03 (chronic nephritic syndrome), N05 (unspecified nephritic syndrome), N17.1 (acute kidney failure with acute cortical necrosis), N17.2 (acute kidney failure with medullary necrosis), N18 (chronic kidney disease), N19 (unspecified kidney failure), N26.9 (renal sclerosis, unspecified), N25.0 (renal osteodystrophy), R80.2 (orthostatic proteinuria, unspecified), I12 (hypertensive chronic kidney disease), I13 (hypertensive heart and chronic kidney disease), Z99.2 (dependence on renal dialysis), Z94.0 (kidney transplant status), and Z49 (encounter for care involving renal dialysis). In total, this produced 5,615 cases. We performed logistic regression for each of the 26 SU-associated SNVs to generate association summary statistics for CKD using this case definition.
In the 4 population-based cohorts (ARIC, CARDIA, CHS, and FHS), we conducted individual-level analyses using a genetic risk score (GRS) of the 26 SNVs associated with SU level. eGFR was recalculated in the 4 prospective cohorts using the Modification of Diet in Renal Disease (MDRD) 4-variable equation. CKD cases were defined as individuals who had an eGFR less than 60 ml/min/1.73 m². A description of the 4 cohorts is provided in the S2 Text.
Statistical analyses
First, we tested our 26 instrumental SNVs for heterogeneity/pleiotropy using a published test for heterogeneity, the MR-PRESSO [13] global test (see S1 Text for details). We noted the presence of significant heterogeneity/pleiotropy. Accordingly, we selected 7 MR analyses designed to be robust to the presence of heterogeneity in instrumental variables: (i) inverse variance weighted least squares (WLS) regression with a random effects model [20], (ii) MR-Egger regression [21,22], (iii) and (iv) weighted and unweighted median tests [22], (v) and (vi) weighted and unweighted mode-based estimates, and (vii) WLS regression after removing outliers identified by the MR-PRESSO outlier test [13]. For all methods, we used the effect size for the outcome variable (eGFR or CKD) as the response variable, the effect size for the exposure variable (SU) as the predictor variable, and, for weighted methods, the inverse square of the standard error for the outcome variable (eGFR or CKD) as the weight. We used the same 7 methods for analysis of the UK Biobank data. For details of all these methods, see S1 Text. To test the robustness of our analysis to the choice of SNVs, we conducted a leave-one-out analysis, removing each urate-specific SNV separately from the WLS regression test. We also conducted an analysis specifically excluding 2 SNVs: rs12498742 in the SLC2A9 gene and rs2231142 in the ABCG2 gene. These are the 2 SNVs with the most significant effects on SU, and are also the only 2 SNVs identified in the SU GWA analysis as having significant sex-specific effects after applying multiple test correction [14].
For our individual-level analyses in the 4 population-based cohorts (ARIC, CARDIA, FHS, and CHS), we calculated the weighted GRS per individual based on the number of risk alleles for the SNVs and the effect size of these SNVs on SU level based on summary results from the Global Urate Genetics Consortium [14]. Individual-level analyses in these cohorts were performed using linear regression when the outcome was a continuous trait or logistic regression when the outcome was a dichotomous trait, after adjustment for age and sex. An inverse variance weighted meta-analysis was performed across the 4 cohorts. To account for relatedness in the FHS cohort, a linear mixed-effects kinship model implemented in the coxme package [23] was used. To account for relatedness in the ARIC cohort, 66 individuals with known family relationships were removed. The other cohorts are not known to have kinship between individuals. To account for possible nonlinearity in the causal relationship between SU and eGFR, the weighted GRS per individual was stratified into 100 strata to calculate localized average causal effect (LACE), and a fractional polynomial fit was performed [24]. The fractional polynomial fit was performed using the mfp package [25].
Epidemiological associations between SU level and eGFR were assessed using linear regression, and between SU level and CKD using logistic regression, adjusting for age and sex in the 4 population-based cohort studies.
We performed analytic power calculations using mRnd [26] based on the strength of observed epidemiological associations of SU with CKD in the 4 population-based cohorts (odds ratio [OR] = 1.34–1.74 per 1 mg/dl [59.48 μmol/l] SU), the fraction of variance in SU explained by each SNV in the Global Urate Genetics Consortium meta-analysis (0.5% to 3%), and the numbers of cases and controls in the CKDGen meta-analysis (12,385 cases and 104,780 controls).
All statistical analyses were performed using R (R Project for Statistical Computing, Vienna, Austria) or Python 2.7 (Python Software Foundation, Wilmington, DE, US).
Results
Primary MR analysis for eGFR level and CKD endpoints
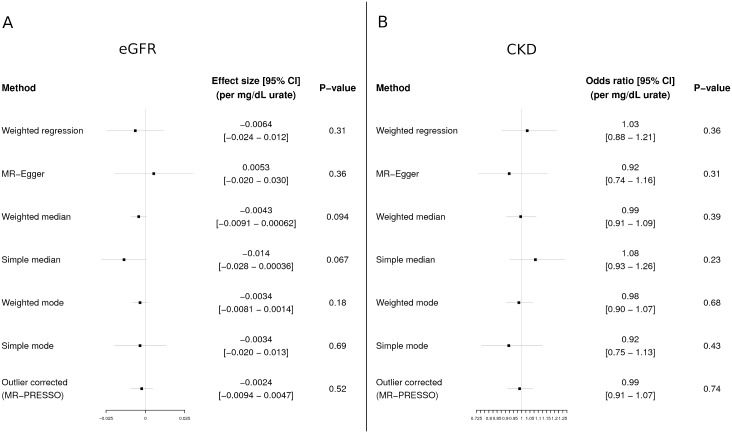
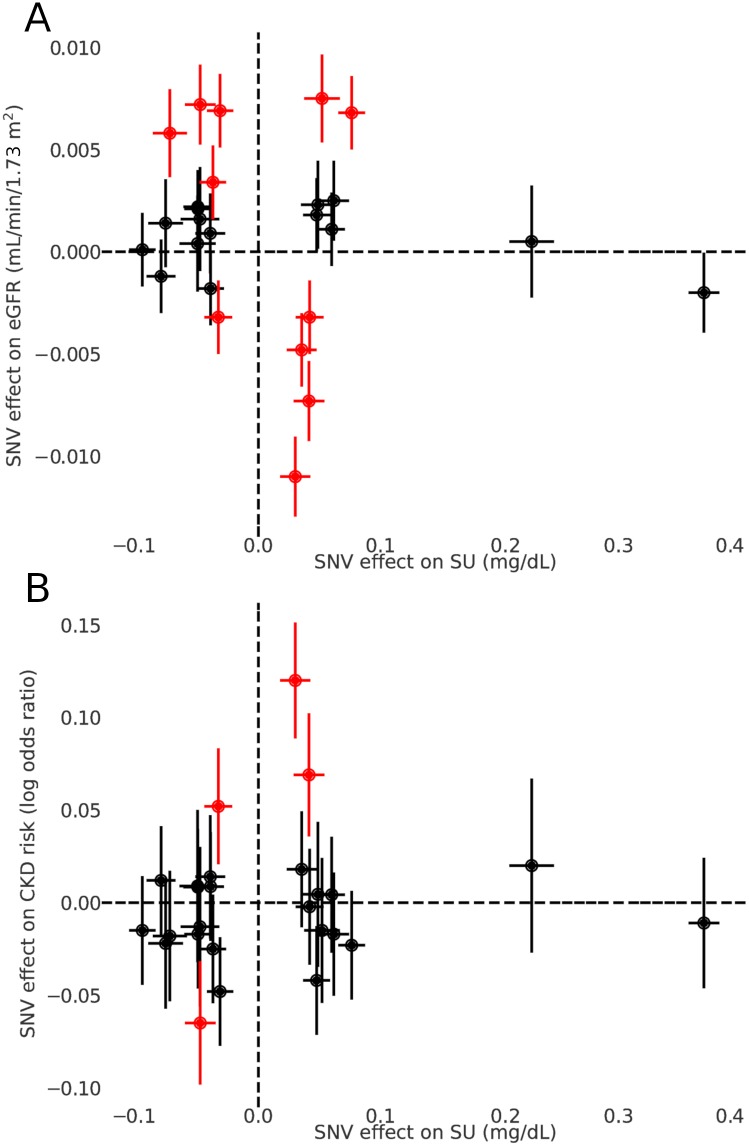
We utilized 26 SNVs associated with SU level identified from the GWA study for SU [14] for our MR analyses (S1 Table). We detected significant heterogeneity when testing for a causal effect of SU on both CKD and eGFR (both P < 10−6 for MR-PRESSO global test), which may indicate the presence of pleiotropy. Accordingly, we selected 7 MR approaches considered to be appropriate for instrumental variables displaying potential pleiotropy: a standard WLS regression analysis with a random effects model, 5 alternative MR methods designed to be robust to heterogeneity, and a WLS regression analysis after removing 4 SNVs identified as outliers for the effect of SU on CKD and 11 SNVs identified as outliers for the effect of SU on eGFR (S2 Table). All 7 approaches detected no causal relationship between SU level and eGFR level (Figs 2A and 3A). Similarly, there was no causal effect detected on CKD risk (Figs 2B and 3B). We observed no heterogeneity/pleiotropy after removal of outliers in the MR test of the effect of SU on CKD (P = 0.06); heterogeneity/pleiotropy in the eGFR analysis was still present but reduced (P = 0.004). We also performed leave-one-out analysis with the WLS regression method (S1 Fig), as well as an analysis excluding the 2 SNVs with the most significant effects on SU (SLC2A9 rs12498742 and ABCG2 rs2231142), which are also known to have sex-specific effects (S2 Fig). Both analyses showed no causal effect.
Fig 2. MR analysis of the effect of SU on eGFR and CKD.
Estimates of causal effects of serum urate on eGFR (A) and CKD (B) by 7 MR analyses. Effects are shown per 1-mg/dl increase of serum urate. The effect sizes/odds ratios and P values were calculated using all single nucleotide variants (SNVs), except for the outlier-corrected MR, where pleiotropic SNVs (detected by MR-PRESSO) were excluded. In the outlier-corrected analysis, 11 outliers were removed for eGFR and 4 outliers were removed for CKD (see S2 Table). CKD, chronic kidney disease; MR, Mendelian randomization; SNV, single nucleotide variant; SU, serum urate.
Fig 3. Effects of individual SNVs on SU, eGFR, and CKD.
Effects of individual SNVs on SU (x-axis, both panels), eGFR (y-axis, A), and CKD (y-axis, B), as estimated by the respective genome-wide association meta-analyses. Error bars indicate 95% confidence intervals. SNVs identified as outliers by the MR-PRESSO outlier test are highlighted in red. CKD, chronic kidney disease; MR, Mendelian randomization; SNV, single nucleotide variant; SU, serum urate.
MR analysis for gout endpoint
As a positive control, we repeated the same procedure using the same 26 SNVs associated with SU to test for a causal effect of SU on gout. Similarly to the analyses of eGFR and CKD, we observed significant heterogeneity among the 26 SNVs (P < 10−6 for MR-PRESSO global test). However, unlike for eGFR and CKD, all 7 approaches showed a highly significant causal effect of SU on gout (P < 10−3 for all tests; S3 Fig). We observed no heterogeneity/pleiotropy after removal of outliers in the MR test of SU’s effect on gout (P = 0.09). This result serves as a positive control of our approach as it is consistent with the known causal role of SU in gout [27].
MR analysis in the UK Biobank
To account for varying definitions of the CKD endpoint, we performed an additional analysis using a clinical definition of kidney disease based on electronic medical records in the UK Biobank. In total, we identified 5,615 CKD cases and 329,597 controls using this definition. We applied the same 7 MR analyses to the 26 SU-associated SNVs. We observed no significant causal relationship of SU level with kidney disease (P > 0.05 for all analyses; S4 Fig).
Individual-level analysis in 4 population-based cohorts
To account for limitations of using aggregated GWA data, such as heterogeneity among study populations in the meta-analyses or unaccounted for population stratification within the sample, we performed an individual-level analysis on 4 population-based cohorts. Within these cohorts, we observed a highly significant association of a urate-specific GRS composed of the 26 urate-associated SNVs with SU (beta = 1.06; 95% CI = 1.00 to 1.13; P = 7.06 × 10−211). However, the GRS was not associated with either eGFR (beta = −0.42; 95% CI = −1.05 to 0.20; P = 0.18) or CKD risk (OR = 1.05; 95% CI = 0.89 to 1.23; P = 0.59) (Table 1). We repeated the same analysis after stratifying by sex and age, and found similar results (S3 Table). We also tested for a nonlinear causal relationship using a fractional polynomial fit [24]. The nonlinear model does not appear to fit the data better than the linear model (P = 0.49), and still produces a null result for a causal effect of SU on CKD (P = 0.34).
Table 1. Association of a genetic risk score of serum urate single nucleotide variants with serum urate level, eGFR, and CKD in 4 population-based cohorts.
| Group | Serum urate level (mg/dl) | eGFR (ml/min/1.73 m2) | CKD | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Beta | 95% CI | P value | Beta | 95% CI | P value | OR | 95% CI | P value | |
| ARIC | 1.08 | 0.98 to 1.19 | 1.86 × 10−91 | −0.40 | −1.16 to 0.35 | 0.30 | 0.98 | 0.83 to 1.16 | 0.82 |
| CARDIA | 1.05 | 0.87 to 1.24 | 7.28 × 10−29 | −1.16 | −3.09 to 0.77 | 0.24 | 1.62 | 0.78 to 3.46 | 0.20 |
| CHS | 1.10 | 0.93 to 1.26 | 5.35 × 10−38 | −0.47 | −2.18 to 1.25 | 0.59 | 1.14 | 0.85 to 1.52 | 0.38 |
| FHS | 1.01 | 0.88 to 1.14 | 4.82 × 10−53 | 0.33 | −1.76 to 2.41 | 0.76 | N/A | N/A | N/A |
| Meta-analysis | 1.06 | 1.00 to 1.13 | 7.06 × 10−211 | −0.42 | −1.05 to 0.20 | 0.18 | 1.05 | 0.89 to 1.23 | 0.59 |
ARIC, Atherosclerosis Risk in Communities; CARDIA, Coronary Artery Risk Development in Young Adults; CHS, Cardiovascular Health Study; CKD, chronic kidney disease; eGFR, estimated glomerular filtration rate; FHS, Framingham Heart Study; N/A, not applicable; OR, odds ratio.
Association of SNV in the SLC2A9 gene with SU, eGFR, and CKD
We also specifically examined rs12498742 in the SLC2A9 gene, which encodes the GLUT9 transporter (for glucose and urate) in the renal proximal tubule and is the largest contributor to genetic control of SU level, explaining 3% of variance [14]. This SNV was strongly associated with SU (beta = 0.37, P < 10−700) but neither with eGFR (beta = 0.002, P = 0.06) nor with CKD (OR = 0.99, P = 0.53).
Conventional epidemiological association of SU, eGFR, and CKD
These null associations contrasted with conventional epidemiological association analyses. In a meta-analysis of the same 4 population-based cohorts (ARIC, CARDIA, CHS, and FHS), we observed that an equivalent (1 mg/dl) increase in SU level was associated with reduced eGFR level (beta = −1.99; 95% CI −2.86 to −1.11; P = 8.08 × 10−6) and increased risk of CKD (OR = 1.48; 95% CI 1.32 to 1.65; P = 1.52 × 10−11), after adjustment for age and sex (Table 2).
Table 2. Observational association between serum urate level and eGFR and risk of CKD in 4 population-based cohorts.
| Group | eGFR (ml/min/1.73 m2) | CKD | ||||
|---|---|---|---|---|---|---|
| Beta | 95% CI | P value | OR | 95% CI | P value | |
| ARIC | −1.82 | −1.99 to −1.65 | 3.56 × 10−94 | 1.40 | 1.34 to 1.46 | 1.17 × 10−51 |
| CARDIA | −1.01 | −1.52 to −0.49 | 1.40 × 10−4 | 1.39 | 1.14 to 1.68 | 9.51 × 10−4 |
| CHS | −3.31 | −3.67 to −2.95 | 1.75 × 10−68 | 1.61 | 1.50 to 1.74 | 1.68 × 10−37 |
| FHS | −1.76 | −2.32 to −1.21 | 3.85 × 10−10 | N/A | N/A | N/A |
| Meta-analysis | −1.99 | −2.86 to −1.11 | 8.08 × 10−6 | 1.48 | 1.32 to 1.65 | 1.52 × 10−11 |
ARIC, Atherosclerosis Risk in Communities; CARDIA, Coronary Artery Risk Development in Young Adults; CHS, Cardiovascular Health Study; CKD, chronic kidney disease; eGFR, estimated glomerular filtration rate; FHS, Framingham Heart Study; OR, odds ratio.
Statistical power and the probability of missing a causal effect
We assessed the statistical power of our MR study given the sample size and the variance in SU explained by the 26 SNVs we used as instrumental variables. We calculated that our MR analyses would have greater than 99% power to detect a statistically significant effect at an alpha rate of 5%, if causality between SU and CKD were present at the strength indicated by observational epidemiology (OR = 1.5 per 1 mg/dl of SU) (Table 3).
Table 3. Power calculations for MR analyses of the effect of SU on CKD.
| OR of CKD per 1 mg/dl of SU | Fraction of variance in SU explained by SNV | Power of MR analysis |
|---|---|---|
| 1.3 | 0.5% | 94% |
| 1.3 | 3% | >99% |
| 1.5 | 0.5% | >99% |
| 1.5 | 3% | >99% |
| 1.7 | 0.5% | >99% |
| 1.7 | 3% | >99% |
Power calculations performed with mRnd. OR 1.3–1.7 is the 95% confidence interval of the observational epidemiological association between SU and CKD (Table 2); 0.5%–3% is the range of variance explained reported for single SNVs in the SU genome-wide association study.
CKD, chronic kidney disease; MR, Mendelian randomization; OR, odds ratio; SNV, single nucleotide variant; SU, serum urate.
Discussion
In this study, we investigated a potential causal role for SU level in the development of CKD using a series of complementary MR analyses. Despite previous observational study findings that SU levels were strongly associated with the risk of incident CKD [4,5], our MR analyses found no evidence for a causal role of SU level for eGFR level or incident CKD. In contrast, our positive control MR analysis demonstrated that SU level was causal for the risk of gout, which is consistent with a previous study that showed similar results [28].
Unlike previous studies based on smaller sample sizes, our power calculations show that our study is sufficiently powered to assess a causal relationship between SU level and CKD. One study that examined only eGFR (not CKD) reported that increased SU levels due to SNVs in urate transporter genes were associated with increased eGFR only in men (which is opposite to the expected epidemiological association) [29]. However, this study did not account for pleiotropy, which may explain these unexpected findings. The second MR analysis reported null findings for both eGFR and CKD; however, the study was not sufficiently powered to assess causal relationships for CKD and also did not account for pleiotropy [30]. A third study observed no causal effect of SU levels on renal function in 3,734 Chinese individuals but observed significant effects in some subpopulations: females, individuals under 65 years, individuals with normal eGFR levels, current smokers, and individuals with high fasting glucose levels [31]. This study similarly did not account for pleiotropy and did not directly test for CKD. Importantly, the sample size of the current study is much larger (N > 400,000) and therefore has higher power than the previous studies above. Our failure to detect by MR the expected epidemiological association of SU with CKD/eGFR is therefore not due to lack of power. We suggest instead that it is due to our MR analyses being robust to unknown confounders and reverse causation, which can create positive correlations in observational epidemiological studies.
Clinical trial data on lowering SU levels for preventing CKD progression to date have been conflicting [32,33]. Moreover, no randomized controlled trials have been conducted using targeted interventions to lower SU levels for the prevention of incident CKD, although trials evaluating the role of xanthine oxidase inhibitors in disease populations such as patients with early CKD or type 1 diabetes are ongoing. Furthermore, to date, 2 randomized controlled trials among adolescents with hyperuricemic pre-hypertension or stage-1 hypertension found that urate-lowering therapy lowered blood pressure, a strong risk factor for CKD [34,35], whereas a similarly designed trial among adults did not find such a benefit [36]. Nevertheless, these studies did not address the causal role of urate reduction in the prevention of incident CKD in population-based participants, which would be the relevant context of our study. Furthermore, the potential benefit of xanthine oxidase inhibitors could exclusively come from reducing oxidative stress by inhibiting superoxide generation, rather than from SU reduction [37]. Our findings suggest that SU reduction alone would not result in prevention of incident CKD, consistent with a recent study that showed that the initiation of allopurinol in patients with gout was not associated with a change in CKD risk [38]. Finally, our results are relevant to the impact of lowering urate and do not rule out the potential benefit of superoxide reduction resulting from xanthine oxidase inhibition [37].
Potential limitations of this study and, in particular, MR deserve comment. First, MR analysis requires suitable SNVs to act as instrumental variables, and selection of inappropriate or unrepresentative instrumental variables may undermine the validity of the study. Here, we used the SNVs significantly associated with SU in a previously published GWA study. We also performed an additional analysis using a single SNV with a strong effect on SU and a well-understood mechanism (rs12498742 in the SLC2A9 gene), as well as an analysis excluding both this SNV and a second large-effect SNV (rs2231142 in the ABCG2 gene). Second, an assumption of MR is that the SNVs used as instrumental variables are not subject to “horizontal pleiotropy,” meaning that the SNVs should not have pleiotropic effects on the outcome outside of the target biomarker or risk factor. We have used MR approaches designed to be robust to horizontal pleiotropy to account for this. Third, our MR findings cannot predict with absolute certainty that therapeutics lowering SU levels will not result in a lowering of CKD risk, since the effects of genetic variation may not be exactly the same as the effects of a therapeutic intervention. Nevertheless, studies have suggested that genetic findings can predict the effect of drugs on disease outcomes [39]. Fourth, the study is focused on individuals of European ancestry; therefore, it is unclear whether our results can be generalized to non-European populations. Fifth, there are some overlapping samples between the SU and CKD GWA datasets (61% of samples in the SU GWA dataset, 50% in the eGFR GWA dataset, and 43% in the CKD GWA dataset), which can affect the accuracy of MR tests [40]. However, this would only affect our study by producing a false positive result, and we observe only negative results. Furthermore, we also performed an MR analysis with non-overlapping datasets (the SU GWA dataset and the UK Biobank dataset for CKD), and found the same negative result. Sixth, the SU and CKD GWA studies both aggregated heterogeneous populations with differing distributions of age, sex, and other potentially important features, and it is possible that the differences between the populations mask the effect of SU on CKD. However, these studies did account for age and sex as covariates, and we performed individual-level analyses that found a negative result when stratifying by age and sex. Seventh, some individuals included in the GWA datasets for SU and CKD may have been taking urate-lowering medication, which may distort the relationship between SU and CKD. We were not able to remove these individuals from our sample for the GWA summary statistic analysis. However, we did remove individuals taking urate-lowering medication from our individual-level analyses in the population-based cohorts, and these analyses found a similar result to the GWA summary statistic analysis.
In conclusion, our MR analyses do not support a causal effect of SU level on eGFR or CKD. Our results suggest that lowering SU levels would be unlikely to translate into risk reduction for incident CKD.
Supporting information
Lines show 95% confidence intervals; odds ratio estimates are per 1 mg/dl of SU.
(TIFF)
Analyses were performed with 24 SNVs, excluding rs12498742 in the SLC2A9 gene and rs2231142 in the ABCG2 gene. Lines show 95% confidence intervals; odds ratio estimates are per 1 mg/dl of SU. Four outliers were removed in the outlier-corrected (MR-PRESSO) analysis (see S2 Table).
(TIFF)
Lines show 95% confidence intervals; odds ratio estimates are per 1 mg/dl of SU. One outlier was removed in the outlier-corrected (MR-PRESSO) analysis (see S2 Table).
(TIFF)
Lines show 95% confidence intervals; odds ratio estimates are per 1 mg/dl of SU. Four outliers were removed in the outlier-corrected (MR-PRESSO) analysis (see S2 Table).
(TIFF)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
(DOCX)
Acknowledgments
We thank Ha My Vy for assistance with retrieving data and data analysis. The authors thank the staff and participants of the FHS, CHS, ARIC, and CARDIA studies, as well as the UK Biobank, for their important contributions. This research has been conducted using the UK Biobank Resource under project ID 16218.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This manuscript was not prepared in collaboration with, nor approved by, investigators of the FHS, CHS, ARIC, or CARDIA studies and does not necessarily reflect the opinions or views of the FHS, CHS, ARIC, or CARDIA studies; Boston University; or the National Heart, Lung, and Blood Institute.
Abbreviations
- ARIC
Atherosclerosis Risk in Communities
- CARDIA
Coronary Artery Risk Development in Young Adults
- CHS
Cardiovascular Health Study
- CKD
chronic kidney disease
- CKDGen
Chronic Kidney Disease Genetics Consortium
- eGFR
estimated glomerular filtration rate
- FHS
Framingham Heart Study
- GRS
genetic risk score
- GWA
genome-wide association
- MR
Mendelian randomization
- OR
odds ratio
- SNV
single nucleotide variant
- SU
serum urate
- WLS
weighted least squares
Data Availability
All relevant GWAS summary statistic data are included in S1 Table. The data underlying the results for the Atherosclerosis Risk In Communities (ARIC), Coronary Artery Risk Development in Young Adults Study (CARDIA), Cardiovascular Health Study (CHS), and Framingham Heart Study (FHS) cohorts presented in the study are available from the database of Genotypes and Phenotypes (dbGap): https://www.ncbi.nlm.nih.gov/gap, for researchers who meet the criteria for access to the data. The data underlying the results for the UK Biobank presented in the study are available from the UK Biobank: https://www.ukbiobank.ac.uk/, for researchers who meet the criteria for access to the data.
Funding Statement
DMJ was supported by a T32 molecular cardiology training grant (HL007824) by the National Institutes of Health. HKC was supported by R01 AR056291, R01 AR065944, P50 AR060772 from the National Institutes of Health and a research grant from AstraZeneca. RT and TRM were supported by the Health Research Council of New Zealand. HHW was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2016R1C1B2007920). GN was supported by the National Institute Of Diabetes And Digestive And Kidney Diseases of the National Institutes of Health under Award Number K23DK107908. RD was supported by R35GM124836 from the National Institute Of General Medical Sciences of the National Institutes of Health, R01HL139865 from the National Heart, Lung, Blood Institute of the National Institutes of Health, an American Heart Association Cardiovascular Genome-Phenome Discovery grant (15CVGPSD27130014) and research grants from AstraZeneca and Goldfinch Bio. The ARIC study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, N01-HC-55022, R01HL087641, R01HL59367, and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. The FHS and the Framingham SHARe project are conducted and supported by the National Heart, Lung, and Blood Institute in collaboration with Boston University. The Framingham SHARe data used for the analyses described in this manuscript were obtained through dbGaP. The CHS research reported in this article was supported by contract numbers N01-HC-85079, N01-HC-85080, N01-HC-85081, N01-HC-85082, N01-HC-85083, N01-HC-85084, N01-HC-85085, N01-HC-85086, N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133, N01-HC-85239 and HHSN268201200036C; grant numbers U01 HL080295 from the National Heart, Lung, and Blood Institute and R01 AG-023629 from the National Institute on Aging, with additional contribution from the National Institute of Neurological Disorders and Stroke. A full list of principal CHS investigators and institutions can be found at http://www.chs-nhlbi.org/pi.htm. The Coronary Artery Risk Development in Young Adults Study (CARDIA) is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with the University of Alabama at Birmingham (N01-HC95095 & N01-HC48047), University of Minnesota (N01-HC48048), Northwestern University (N01-HC48049), and Kaiser Foundation Research Institute (N01-HC48050). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Xie Y, Bowe B, Mokdad AH, Xian H, Yan Y, Li T, et al. Analysis of the Global Burden of Disease study highlights the global, regional, and national trends of chronic kidney disease epidemiology from 1990 to 2016. Kidney Int. 2018;94(3):567–81. 10.1016/j.kint.2018.04.011 [DOI] [PubMed] [Google Scholar]
- 2.National Kidney Foundation. Global facts: about kidney disease. New York: National Kidney Foundation; 2017. [cited 2018 Dec 13]. https://www.kidney.org/kidneydisease/global-facts-about-kidney-disease. [Google Scholar]
- 3.Levey AS, Stevens LA, Coresh J. Conceptual model of CKD: applications and implications. Am J Kidney Dis. 2009;53(3 Suppl 3):S4–16. 10.1053/j.ajkd.2008.07.048 [DOI] [PubMed] [Google Scholar]
- 4.Weiner DE, Tighiouart H, Elsayed EF, Griffith JL, Salem DN, Levey AS. Uric acid and incident kidney disease in the community. J Am Soc Nephrol. 2008;19(6):1204–11. 10.1681/ASN.2007101075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumagai T, Ota T, Tamura Y, Chang WX, Shibata S, Uchida S. Time to target uric acid to retard CKD progression. Clin Exp Nephrol. 2016;21:182 10.1007/s10157-016-1288-2 [DOI] [PubMed] [Google Scholar]
- 6.Johnson RJ, Kang DH, Feig D, Kivlighn S, Kanellis J, Watanabe S, et al. Is there a pathogenetic role for uric acid in hypertension and cardiovascular and renal disease? Hypertension. 2003;41(6):1183–90. 10.1161/01.HYP.0000069700.62727.C5 [DOI] [PubMed] [Google Scholar]
- 7.Perlstein TS, Gumieniak O, Hopkins PN, Murphey LJ, Brown NJ, Williams GH, et al. Uric acid and the state of the intrarenal renin-angiotensin system in humans. Kidney Int. 2004;66(4):1465–70. 10.1111/j.1523-1755.2004.00909.x [DOI] [PubMed] [Google Scholar]
- 8.Rao GN, Corson MA, Berk BC. Uric acid stimulates vascular smooth muscle cell proliferation by increasing platelet-derived growth factor A-chain expression. J Biol Chem. 1991;266(13):8604–8. [PubMed] [Google Scholar]
- 9.Kang DH, Nakagawa T, Feng L, Watanabe S, Han L, Mazzali M, et al. A role for uric acid in the progression of renal disease. J Am Soc Nephrol. 2002;13(12):2888–97. [DOI] [PubMed] [Google Scholar]
- 10.Hosoya T, Kimura K, Itoh S, Inaba M, Uchida S, Tomino Y, et al. The effect of febuxostat to prevent a further reduction in renal function of patients with hyperuricemia who have never had gout and are complicated by chronic kidney disease stage 3: study protocol for a multicenter randomized controlled study. Trials. 2014;15:26 10.1186/1745-6215-15-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson PC, Choi HK, Do R, Merriman TR. Insight into rheumatological cause and effect through the use of Mendelian randomization. Nat Rev Rheumatol. 2016;12(8):486–96. 10.1038/nrrheum.2016.102 [DOI] [PubMed] [Google Scholar]
- 12.Voight BF, Peloso GM, Orho-Melander M, Frikke-Schmidt R, Barbalic M, Jensen MK, et al. Plasma HDL cholesterol and risk of myocardial infarction: a Mendelian randomisation study. Lancet. 2012;380(9841):572–80. 10.1016/S0140-6736(12)60312-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–8. 10.1038/s41588-018-0099-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kottgen A, Albrecht E, Teumer A, Vitart V, Krumsiek J, Hundertmark C, et al. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet. 2013;45(2):145–54. 10.1038/ng.2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The ARIC investigators. The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. Am J Epidemiol. 1989;129(4):687–702. [PubMed] [Google Scholar]
- 16.Friedman GD, Cutter GR, Donahue RP, Hughes GH, Hulley SB, Jacobs DR Jr, et al. CARDIA: study design, recruitment, and some characteristics of the examined subjects. J Clin Epidemiol. 1988;41(11):1105–16. [DOI] [PubMed] [Google Scholar]
- 17.Fried LP, Borhani NO, Enright P, Furberg CD, Gardin JM, Kronmal RA, et al. The Cardiovascular Health Study: design and rationale. Ann Epidemiol. 1991;1(3):263–76. [DOI] [PubMed] [Google Scholar]
- 18.Mahmood SS, Levy D, Vasan RS, Wang TJ. The Framingham Heart Study and the epidemiology of cardiovascular disease: a historical perspective. Lancet. 2014;383(9921):999–1008. 10.1016/S0140-6736(13)61752-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pattaro C, Teumer A, Gorski M, Chu AY, Li M, Mijatovic V, et al. Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat Commun. 2016;7:10023 10.1038/ncomms10023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658–65. 10.1002/gepi.21758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–25. 10.1093/ije/dyv080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burgess S, Bowden J, Fall T, Ingelsson E, Thompson SG. Sensitivity analyses for robust causal inference from Mendelian randomization analyses with multiple genetic variants. Epidemiology. 2017;28(1):30–42. 10.1097/EDE.0000000000000559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Therneau TM. coxme: mixed effects Cox models R package version 2.2–10. Vienna: R Project for Statistical Computing; 2018. [Google Scholar]
- 24.Staley JR, Burgess S. Semiparametric methods for estimation of a nonlinear exposure-outcome relationship using instrumental variables with application to Mendelian randomization. Genet Epidemiol. 2017;41(4):341–52. 10.1002/gepi.22041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ambler G, Benner A. mfp: multivariable fractional polynomials R package version 1.5.2. Vienna: R Project for Statistical Computing; 2015. [Google Scholar]
- 26.Brion M-JA, Shakhbazov K, Visscher PM. Calculating statistical power in Mendelian randomization studies. Int J Epidemiol. 2013;42(5):1497–501. 10.1093/ije/dyt179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi HK, Mount DB, Reginato AM. Pathogenesis of gout. Ann Intern Med. 2005;143:499–516. [DOI] [PubMed] [Google Scholar]
- 28.Keenan T, Zhao W, Rasheed A, Ho WK, Malik R, Felix JF, et al. Causal assessment of serum urate levels in cardiometabolic diseases through a Mendelian randomization study. J Am Coll Cardiol. 2016;67(4):407–16. 10.1016/j.jacc.2015.10.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hughes K, Flynn T, de Zoysa J, Dalbeth N, Merriman TR. Mendelian randomization analysis associates increased serum urate, due to genetic variation in uric acid transporters, with improved renal function. Kidney Int. 2014;85(2):344–51. 10.1038/ki.2013.353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Q, Kottgen A, Dehghan A, Smith AV, Glazer NL, Chen MH, et al. Multiple genetic loci influence serum urate levels and their relationship with gout and cardiovascular disease risk factors. Circ Cardiovasc Genet. 2010;3(6):523–30. 10.1161/CIRCGENETICS.109.934455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J, Zhang H, Dong Z, Zhou J, Ma Y, Li Y, et al. Mendelian randomization analysis indicates serum urate has a causal effect on renal function in Chinese women. Int Urol Nephrol. 2017;49(11):2035–42. 10.1007/s11255-017-1686-8 [DOI] [PubMed] [Google Scholar]
- 32.Fleeman N, Pilkington G, Dundar Y, Dwan K, Boland A, Dickson R, et al. Allopurinol for the treatment of chronic kidney disease: a systematic review. Health Technol Assess. 2014;18(40):1–77, v–vi. 10.3310/hta18400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanji T, Gandhi M, Clase CM, Yang R. Urate lowering therapy to improve renal outcomes in patients with chronic kidney disease: systematic review and meta-analysis. BMC Nephrol. 2015;16:58 10.1186/s12882-015-0047-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feig DI, Soletsky B, Johnson RJ. Effect of allopurinol on blood pressure of adolescents with newly diagnosed essential hypertension: a randomized trial. JAMA. 2008;300(8):924–32. 10.1001/jama.300.8.924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soletsky B, Feig DI. Uric acid reduction rectifies prehypertension in obese adolescents. Hypertension. 2012;60(5):1148–56. 10.1161/HYPERTENSIONAHA.112.196980 [DOI] [PubMed] [Google Scholar]
- 36.McMullan CJ, Borgi L, Fisher N, Curhan G, Forman J. Effect of uric acid lowering on renin-angiotensin-system activation and ambulatory BP: a randomized controlled trial. Clin J Am Soc Nephrol. 2017;12(5):807–16. 10.2215/CJN.10771016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.George J, Carr E, Davies J, Belch JJ, Struthers A. High-dose allopurinol improves endothelial function by profoundly reducing vascular oxidative stress and not by lowering uric acid. Circulation. 2006;114(23):2508–16. 10.1161/CIRCULATIONAHA.106.651117 [DOI] [PubMed] [Google Scholar]
- 38.Vargas-Santos AB, Peloquin CE, Zhang Y, Neogi T. Association of chronic kidney disease with allopurinol use in gout treatment. JAMA Int Med. 2018;178(11):1526–33. 10.1001/jamainternmed.2018.4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson MR, Johnson T, Warren L, Hughes AR, Chissoe SL, Xu CF, et al. The genetics of drug efficacy: opportunities and challenges. Nat Rev Genet. 2016;17(4):197–206. 10.1038/nrg.2016.12 [DOI] [PubMed] [Google Scholar]
- 40.Burgess S, Davies NM, Thompson SG. Bias due to participant overlap in two-sample Mendelian randomization. Genet Epidemiol. 2016;40(7):597–608. 10.1002/gepi.21998 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lines show 95% confidence intervals; odds ratio estimates are per 1 mg/dl of SU.
(TIFF)
Analyses were performed with 24 SNVs, excluding rs12498742 in the SLC2A9 gene and rs2231142 in the ABCG2 gene. Lines show 95% confidence intervals; odds ratio estimates are per 1 mg/dl of SU. Four outliers were removed in the outlier-corrected (MR-PRESSO) analysis (see S2 Table).
(TIFF)
Lines show 95% confidence intervals; odds ratio estimates are per 1 mg/dl of SU. One outlier was removed in the outlier-corrected (MR-PRESSO) analysis (see S2 Table).
(TIFF)
Lines show 95% confidence intervals; odds ratio estimates are per 1 mg/dl of SU. Four outliers were removed in the outlier-corrected (MR-PRESSO) analysis (see S2 Table).
(TIFF)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant GWAS summary statistic data are included in S1 Table. The data underlying the results for the Atherosclerosis Risk In Communities (ARIC), Coronary Artery Risk Development in Young Adults Study (CARDIA), Cardiovascular Health Study (CHS), and Framingham Heart Study (FHS) cohorts presented in the study are available from the database of Genotypes and Phenotypes (dbGap): https://www.ncbi.nlm.nih.gov/gap, for researchers who meet the criteria for access to the data. The data underlying the results for the UK Biobank presented in the study are available from the UK Biobank: https://www.ukbiobank.ac.uk/, for researchers who meet the criteria for access to the data.