Summary
Contact-dependent growth inhibition (CDI) entails receptor-mediated delivery of CdiA-derived toxins into Gram-negative target bacteria. Using electron cryotomography, we show that each CdiA effector protein forms a filament extending ~33 nm from the cell surface. Remarkably, the extracellular filament represents only the N-terminal half of the effector. A programmed secretion arrest sequesters the C-terminal half of CdiA, including the toxin domain, in the periplasm prior to target-cell recognition. Upon binding receptor, CdiA secretion resumes, and the periplasmic FHA-2 domain is transferred to the target-cell outer membrane. The C-terminal toxin region of CdiA then penetrates into the target-cell periplasm, where it is cleaved for subsequent translocation into the cytoplasm. Our findings suggest that the FHA-2 domain assembles into a transmembrane conduit for toxin transport into the periplasm of target bacteria. We propose that receptor-triggered secretion ensures that FHA-2 export is closely coordinated with integration into the target-cell outer membrane.
Introduction
Bacteria have long been known to release diffusible antibiotics and bacteriocins that inhibit competitor cells. Recent research has revealed that bacteria also commonly antagonize their neighbors through direct inter-cellular transfer of protein toxins. In Gram-negative bacteria, type I (Garcia-Bayona et al., 2017), type II (Jamet et al., 2015), type IV (Souza et al., 2015), type V (Aoki et al., 2005) and type VI (Hood et al., 2010; MacIntyre et al., 2010) secretion systems have all been shown to deploy toxic antibacterial effectors. Gram-positive species exploit distinct mechanisms, using cell-wall associated YD-repeat proteins (Koskiniemi et al., 2013) and type VII secretion systems (Cao et al., 2016; Whitney et al., 2017) to deliver toxins into target bacteria. Direct inter-bacterial toxin exchange was first discovered in Escherichia coli isolate EC93, which uses the CdiB-CdiA two-partner secretion (TPS) system to inhibit other E. coli strains in a process called contact-dependent growth inhibition (CDI) (Aoki et al., 2005). CdiB is an outer-membrane localized Omp85 β-barrel protein that exports and presents the toxic CdiA effector on the cell surface. CdiA is a very large protein (~320 kDa) that is predicted to form a filament extending several hundred Å from the cell surface (Aoki et al., 2005; Kajava et al., 2001). CdiA recognizes target bacteria through specific binding interactions with a receptor. Upon binding to receptor, CdiA transfers its C-terminal toxin domain (CdiA-CT) into the target cell to inhibit growth (Aoki et al., 2008). To prevent self-inhibition, E. coli EC93 also produces an immunity protein, CdiI, which specifically neutralizes CdiA-CT toxins delivered from neighboring sibling cells. Thus, E. coli EC93 uses CDI to suppress the growth of non-isogenic E. coli strains, presumably to compete for limited environmental resources.
Since its discovery in E. coli EC93, CDI systems have been identified in many other Gram-negative bacteria and are particularly common in pathogens (Aoki et al., 2010; Zhang et al., 2012; Zhang et al., 2011). Though CdiA is not known to intoxicate eukaryotic cells, the effectors contribute to virulence by promoting bacterial auto-aggregation and biofilm formation (Neil and Apicella, 2009; Rojas et al., 2002). CdiA proteins are heterogeneous in sequence and length, ranging from ~180 kDa in Moraxella to over 630 kDa in some Pseudomonas species (Willett et al., 2015b). Despite this diversity, CdiA proteins share the same general domain architecture. The N-terminus carries a signal peptide for Sec-dependent secretion into the periplasm and a conserved TPS transport domain for CdiB-mediated export across the outer membrane (Figure 1A). The N-terminal half of CdiA is dominated by FHA-1 hemagglutinin peptide repeats (Pfam: PF05594) (Figure 1A). These motifs were first identified in the filamentous hemagglutinin (FHA) adhesins produced by Bordetella species (Relman et al., 1989). Structural modeling and electron microscopy of FHA indicate that FHA-1 repeats form an elongated β-helix, with each ~20-residue motif extending the helix ~4.8 Å (Kajava et al., 2001; Makhov et al., 1994). The C-terminal half of CdiA contains a second repeat domain composed of uncharacterized FHA-2 repeats (PF13332) (Figure 1A). FHA-2 repeats are predicted to adopt β secondary structure, but these sequences are distinct from FHA-1, and it is unclear whether this domain is filamentous. The receptor-binding domain (RBD) of CdiA resides between the two FHA repeat regions. RBD sequences can differ between otherwise closely related CdiA proteins, enabling the effectors to recognize unique receptors (Ruhe et al., 2017). In E. coli, three RBD classes have been characterized that bind to BamA (Aoki et al., 2008), heterotrimeric OmpF/OmpC (Beck et al., 2016), and the Tsx nucleoside transporter (Ruhe et al., 2017) (Figure 1A). Lastly, growth inhibition activity resides within the CdiA-CT region, which often varies dramatically in sequence between bacteria (Aoki et al., 2010). E. coli isolates collectively encode at least 18 CdiA-CT sequence types that exhibit distinct toxic activities. For example, CdiA-CTEC93 from E. coli EC93 dissipates the proton gradient in target bacteria (Aoki et al., 2009), whereas CdiA-CTSTECO31 from E. coli STEC_O31 contains an EndoU RNase domain that cleaves tRNAGlu (Michalska et al., 2018) (Figure 1A). CdiA-CT regions are typically demarcated by conserved peptide motifs, such as the ELYN sequence in Burkholderia species or VENN in enterobacteria (Willett et al., 2015b). The latter VENN motif is part of a larger "pre-toxin" (PT) domain annotated as the PT-VENN region (PF04829) (Zhang et al., 2011) (Figure 1A). The function of the PT domain is unknown, but its location suggests it could mediate auto-proteolysis to release the toxin for transport into target bacteria.
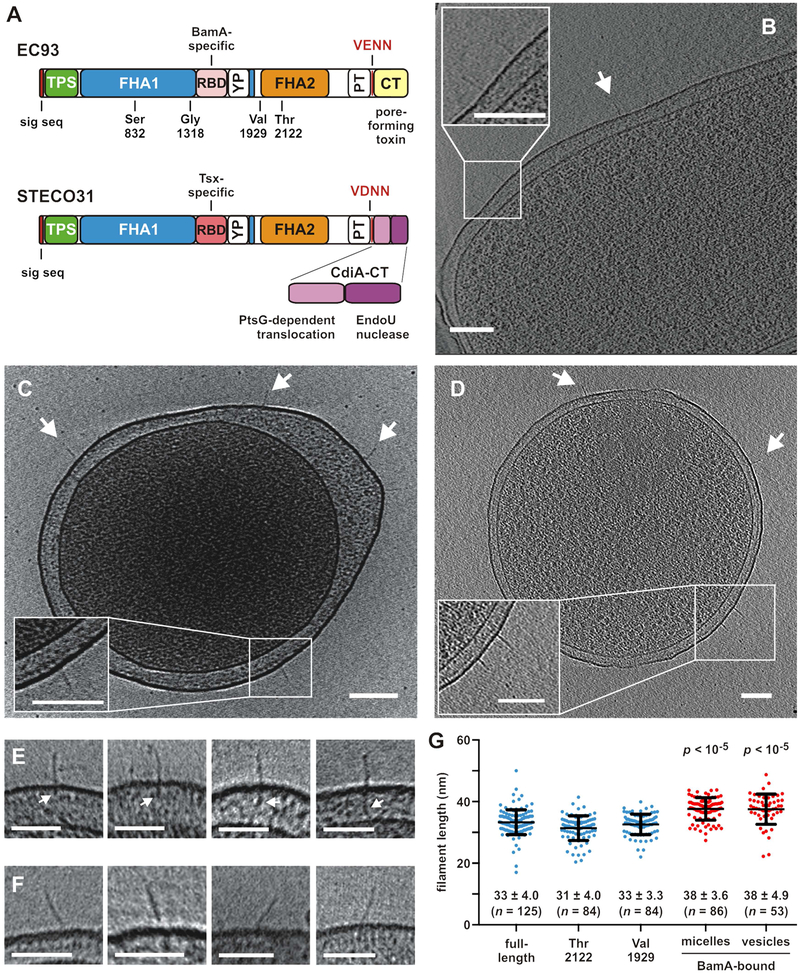
Figure 1. CdiA forms a filament on the cell surface.
A) CdiA domain architecture. VENN/VDNN motifs demarcating the CdiA-CT regions are indicated. The YP domain is not annotated in current databases. See Figure S3. B) CdiAEC93 on the cell surface. See Figure S1B & Movie S1. C) CdiAEC93 on an E. coli minicell. See Figure S1C & Movie S2. D) CdiASTECO31 on an E. coli minicell. See Movie S3. Arrows indicate individual CdiA proteins, and scale bars = 100 nm for A, B & C. E) Periplasmic densities associated with filaments. F) Angled filaments. Scale bars = 50 nm for E & F. G) Lengths of CdiAEC93 filaments. Data are presented with mean ± SD for each condition. The p values are from twotailed t-test comparisons with full-length CdiAEC93.
Although CdiA is thought to form a cell-surface filament, its orientation and architecture have yet to be examined experimentally. The original "toxin-on-a-stick" model proposed that CdiA extends its C-terminus away from the cell to facilitate toxin transfer into target bacteria (Aoki et al., 2011). This model implies that the FHA-1 and FHA-2 domains form a continuous filament, with the C-terminal toxin near the distal tip. This scenario appears to be incongruent with the recent discovery that the RBD is located between the FHA repeat regions (Ruhe et al., 2017), which would place the RBD in the middle of the filament. The mechanisms governing CdiA-CT delivery are also poorly understood. E. coli CdiA proteins recognize various OMPs as receptors, suggesting that these β-barrel proteins could also be exploited as toxin translocation conduits. However, the E. coli BamA β-barrel is not open to the cell surface (Albrecht et al., 2014), and the narrow (~3-5 Å) hydrophobic lumen of Tsx presumably restricts the passage of peptides (Ye and van den Berg, 2004). Further, some CdiA proteins use lipopolysaccharide as a receptor (Koskiniemi et al., 2015), suggesting that OMPs are not necessarily required for toxin translocation. CDI toxin transport across the target-cell cytoplasmic membrane is also dependent on specific receptors. The CdiA-CT region is typically composed of two variable domains, with the extreme C-terminal domain constituting the actual toxin (Willett et al., 2015b) (Figure 1A). Genetic evidence suggests that the N-terminal domain of the CdiA-CT hijacks integral membrane proteins to transport the tethered C-terminal toxin into the target-cell cytoplasm (Willett et al., 2015a). Each N-terminal "translocation" domain recognizes a different membrane protein. For example, the PtsG glucose transporter is required for CdiA-CTSTECO31 delivery (Figure 1A), but CdiA-CTDd3937 from Dickeya dadantiii 3937 uses the ribose ABC transporter RbsC to enter the cytoplasm (Willett et al., 2015a). Though the co-opted membrane proteins are commonly metabolite transporters, their solute-transport activities are not required for toxin import.
Here, we uncover several new and unexpected insights into CdiA secretion and toxin delivery. Electron cryotomography (ECT) reveals that individual CdiA proteins form thin, 33-nm long filaments on the cell surface. Remarkably, CdiA export across the outer membrane is arrested during biogenesis, and the extracellular filament corresponds solely to the N-terminal half of the effector. We identify and delineate a previously unrecognized CdiA domain that is required for this secretion arrest. This Tyr/Pro-enriched region ensures that the C-terminal half of CdiA – including its toxin domain – remains sequestered within the periplasm prior to target-cell recognition. CdiA export resumes upon binding receptor, and the FHA-2 region is deposited directly onto the surface of target bacteria. The FHA-2 domain associates stably with the target-cell outer membrane, and this interaction is required for CdiA-CT transport into the periplasm of target bacteria. We propose that FHA-2 forms a membrane-embedded structure for CdiA-CT translocation. This unprecedented mechanism presumably contributes to CdiA modularity, enabling the effectors to deliver diverse cargoes.
Results
CdiA forms a filament on the cell surface
We used ECT to visualize CdiAEC93 in its native context on the bacterial cell surface. To image effectors in the pre-delivery state, we used E. coli EC93 bamALT2 cells, which express BamALT2 from Salmonella typhimurium LT2. The surface epitopes of BamALT2 differ significantly from E. coli BamA, thus preventing recognition by CdiAEC93 (Ruhe et al., 2015; Ruhe et al., 2013). Tomography revealed poorly resolved filaments on EC93 cells (Figure S1A, Table S1). We reasoned that the surface O-antigen layer of E. coli EC93 could obscure the filaments, and therefore we produced CdiA in E. coli K-12 strains that lack both O-antigen and CdiA-receptors. CdiAEC93 filaments were readily apparent on the surface of these latter cells (Figures 1B, S1B, Table S1, Movie S1), and this approach facilitated further experiments using minicells for higher resolution imaging (Figures 1C, S1C, Movie S2). Similar filaments were also observed on bacteria and minicells that express CdiASTECO31 from E. coli STEC_O31 (Figure 1D, Table S1, Movie S3). Notably, we did not detect filaments on control cells that lack cdi expression constructs (Table S1, Movie S4). Analysis of >100 CdiAEC93 filaments revealed considerable heterogeneity. A few filaments were associated with additional periplasmic density (Figure 1E), and others projected from the cell surface at an angle (Figure 1F). We also observed some bent structures (Figure S1D) and instances of twinned and clustered filaments (Figure S1E).The average length of CdiAEC93 filaments was 33 ± 4 nm (Figure 1G). Though filament width could not be determined at this resolution, it was clearly ≤4 nm, consistent with a parallel β-helix.
We previously showed that CdiAEC93 is also produced in a truncated form that lacks the FHA-2, PT and CdiA-CT domains (Aoki et al., 2005; Ruhe et al., 2015). Although truncated CdiAEC93 cannot mediate CDI, it retains BamA-specific adhesin activity (Ruhe et al., 2015), indicating that it is presented on the cell surface. To determine whether truncation affects filament structure, we imaged CdiAEC93 truncated after residues Ser832, Gly1318, Val1929 and Thr2122 (Figure 1A). We did not detect surface structures with the two shortest constructs (Table S1), but proteins truncated after Val1929 and Thr2122 form filaments with the same dimensions as CdiAEC93 produced from the full-length construct (Figure 1G). Thus, the FHA-2, PT and CdiA-CT domains are not part of the surface structure, indicating that the extracellular filament corresponds to the N-terminal half of CdiA.
CdiA surface topology
ECT indicates that the C-terminal half of CdiA is either sequestered within the cell or not resolved because it is disordered or flexible. To distinguish between these possibilities, we examined CdiA surface topology using biochemical approaches. CdiASTECO31 was used for these experiments, because antisera to its N- and C-terminal regions are available. Immunoblotting showed that CdiASTECO31 is produced in both full-length and truncated forms when expressed in cells that lack its receptor Tsx (Figure 2A, lanes 1 & 4). The N-terminal TPS domain can be digested with extracellular proteinase K (Figure 2A, lane 2), but a C-terminal fragment of >180 kDa is resistant to proteolysis (Figure 2A, lane 5). Trypsin generates a similar C-terminal fragment, but the TPS domain is resistant to this protease (Figure 2A, lanes 3 & 6). These results suggest that FHA-2 and CdiA-CT are intracellular, but it is also possible that these domains are simply protease resistant. Therefore, we developed a fluorescent dye-labeling approach to differentiate periplasmic and extracellular residues. We first showed that the outer membrane is impermeable to maleimide-conjugated IRDye® 680LT using His6-tagged maltose-binding protein (MBP) engineered to carry a single Cys residue. Periplasmic MBP-Cys-His6 is not labeled when whole cells are treated with dye, but becomes labeled when the outer membrane is permeabilized with polymyxin B (Figure 2B, lanes 1 & 2). The cytoplasmic membrane remains intact during polymyxin treatment, because MBP-Cys-His6 lacking a signal peptide is not labeled under these conditions (Figure 2B, lanes 2 & 5). Cytoplasmic MBP becomes accessible to dye only when cells are completely disrupted (Figure 2B, lane 6). Thus, polymyxin selectively permeabilizes the outer membrane and can be used to probe the periplasm.
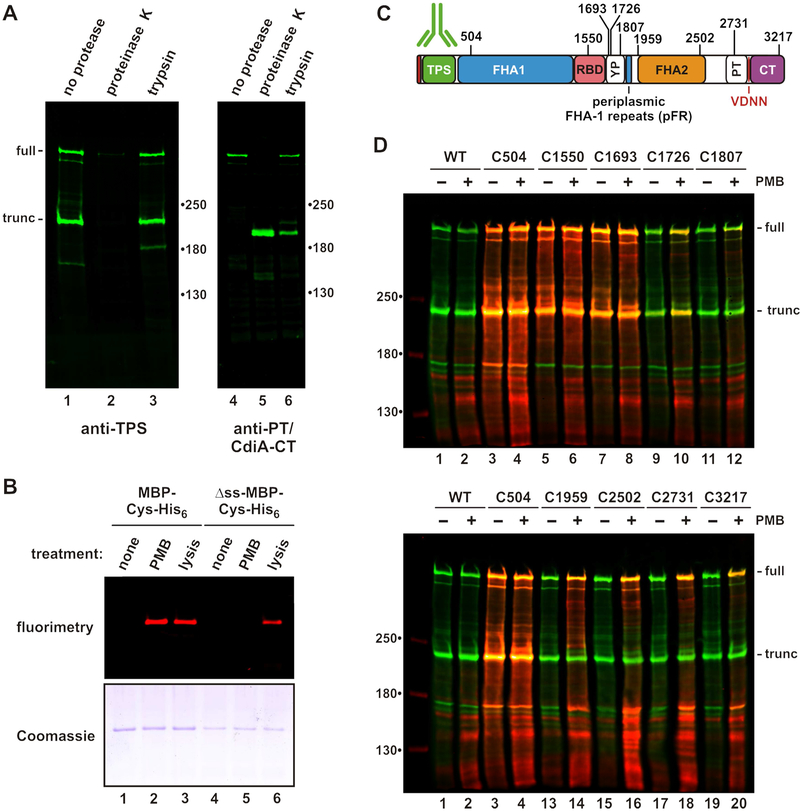
Figure 2. CdiA surface topology.
A) Protease protection assays. Immunoblot analysis with antisera the TPS and PT/CdiA-CT domains of CdiASTECO31. B) Polymyxin B (PMB) permeabilizes the outer membrane. Cells expressing periplasmic or cytoplasmic (Δss) maltose-binding protein (MBP) were incubated with maleimide-dye and permeabilized with PMB or lysed as indicated. Purified MBP was analyzed by fluorimaging (top) and SDS-PAGE (bottom). C) Cys substitutions in CdiASTECO31. The periplasmic FHA-1 repeat (pFR) cluster and major domains are indicated. See Figure S2. D) Immunoblot analysis of dye-labeled CdiA. CdiA expressing cells were incubated with maleimide-dye and permeabilized with PMB where indicated. CdiA was analyzed by immunoblotting with TPS antisera. Dye fluorescence (red) is superimposed onto antibody fluorescence (green).
We then introduced Cys residues along CdiASTECO31 for dye-labeling studies (Figure 2C). Each Cys-substituted effector retains growth inhibition activity against target bacteria (Figure S2A), demonstrating that the mutations do not perturb function. Wild-type CdiASTECO31 contains four Cys residues that do not react with dye (Figure 2D, lanes 1 & 2), suggesting that they form disulfide bonds. Engineered Cys residues at positions 504, 1550 and 1693 label efficiently, with dye fluorescence superimposing with anti-TPS antibody signal (Figure 2D, lanes 3 - 8). These Cys residues are extracellular because they are accessible without polymyxin treatment. By contrast, positions 1726, 2502, 2731 and 3217 of CdiASTECO31 are labeled only when the outer membrane is permeabilized (Figure 2D). Position 1807 is relatively unreactive even with polymyxin treatment (Figure 2D, lanes 11 & 12), suggesting that its side-chain is occluded. These results indicate that the FHA-2, PT and CdiA-CT domains reside in the periplasm prior to target-cell recognition. The periplasmic region also contains a small cluster of FHA-1 motifs, which we have termed periplasmic FHA-1 repeats (pFR) (Figure 2C). The function of these repeats is unclear, but this region is conserved in other CdiA proteins (Figure S3).
CdiA-receptor binding interactions
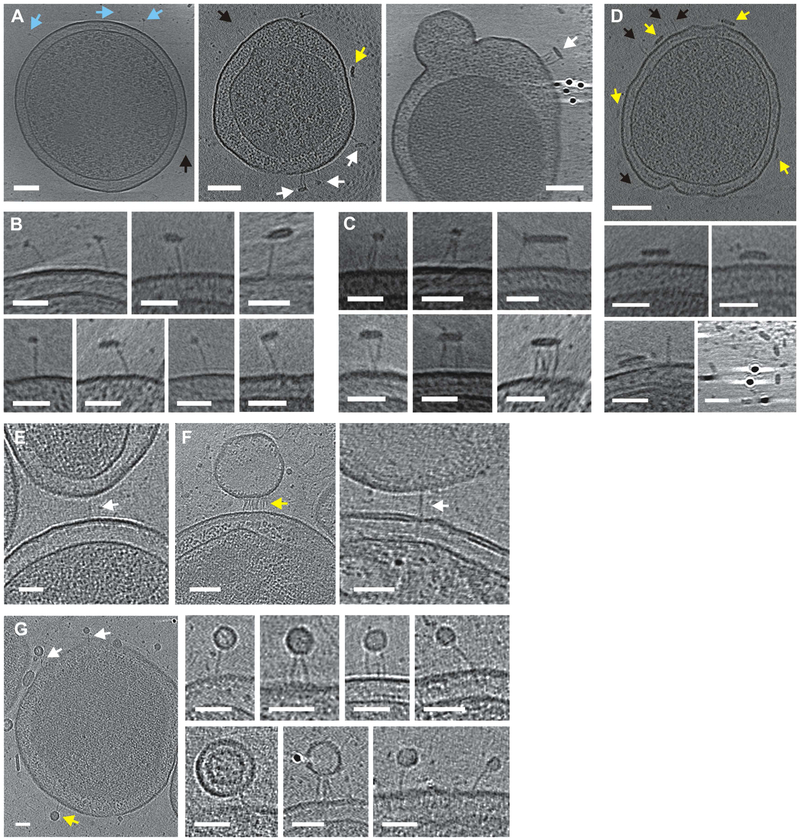
The receptor-binding domain (RBD) of CdiA presumably forms the distal end of the filament. To localize the RBD, we incubated CdiAEC93-expressing minicells with purified E. coli BamA and imaged using ECT. As predicted, BamA-containing micelles bind to the distal tips of the filaments, forming "lollipop"-like structures (Figures 3A & 3B). In some instances, filaments bundle together as they interact with larger micelles that presumably contain multiple receptors (Figures 3A & 3C). These interactions are BamA-dependent, because detergent micelles lacking BamA do not bind CdiAEC93 and instead tend to interact with the cell surface (Figure 3D). Remarkably, BamA-bound filaments are significantly elongated, increasing approximately 5 nm to an average length of 38 ± 3.6 nm (Figure 1G, see Methods for statistical comparisons and p-values). We obtained similar results with target minicells that express E. coli BamA, and captured an image of two minicells bridged by an individual filament (Figure 3E). However, most of the interactions involved vesicles that were presumably derived from the BamA-expressing target cells (Figures 3F & 3G). Given that the periplasmic space is often widened and distorted in minicells, these vesicles may have been stripped from target cells upon binding to CdiAEC93. The vesicle-bound filaments were also elongated to 38 ± 4.9 nm (Figure 1G). These data indicate that the RBD is at the distal end of the filament, suggesting that the TPS domain is associated with CdiB at the cell surface. Therefore, the CdiA chain must make a hairpin-turn at the RBD and return to the cell for the C-terminal domains to remain in the periplasm. Notably, only 176 residues separate position 1550 in the RBD from position 1726 in the periplasm, indicating that some portion of the intervening sequence must be extended (at ~3.5 Å per residue) to span the 33-nm filament. This region is enriched for Tyr (Y) and Pro (P) residues relative to the surrounding FHA repeat domains (Figure S4) and is abruptly demarcated from the variable RBD sequence by a conserved YPLP motif (Figure S3). This previously unrecognized "YP domain" is conserved in other species (Table S2 & Figure S3) and is of particular interest because dye-labeling indicates that this region crosses the outer membrane.
Figure 3. CdiA-receptor binding interactions.
A) CdiAEC93 bound to solubilized BamA in globular (blue arrows) and flattened micelles (white arrows). See Movie S5. Some filaments (black arrows) and micelles (yellow arrows) do not interact. B) Enlarged views of BamA-bound filaments. C) Micelles bound to multiple filaments. D) CdiAEC93 expressing minicell incubated with detergent in buffer. E) CdiAEC93 filament bridging minicells. F) Filaments bound to vesicles from BamA-expressing minicells. G) Filaments bound to small vesicles from BamA minicells. Scale bars 100 nm for minicell views and 50 nm for close-up views.
CdiA export resumes upon binding receptor
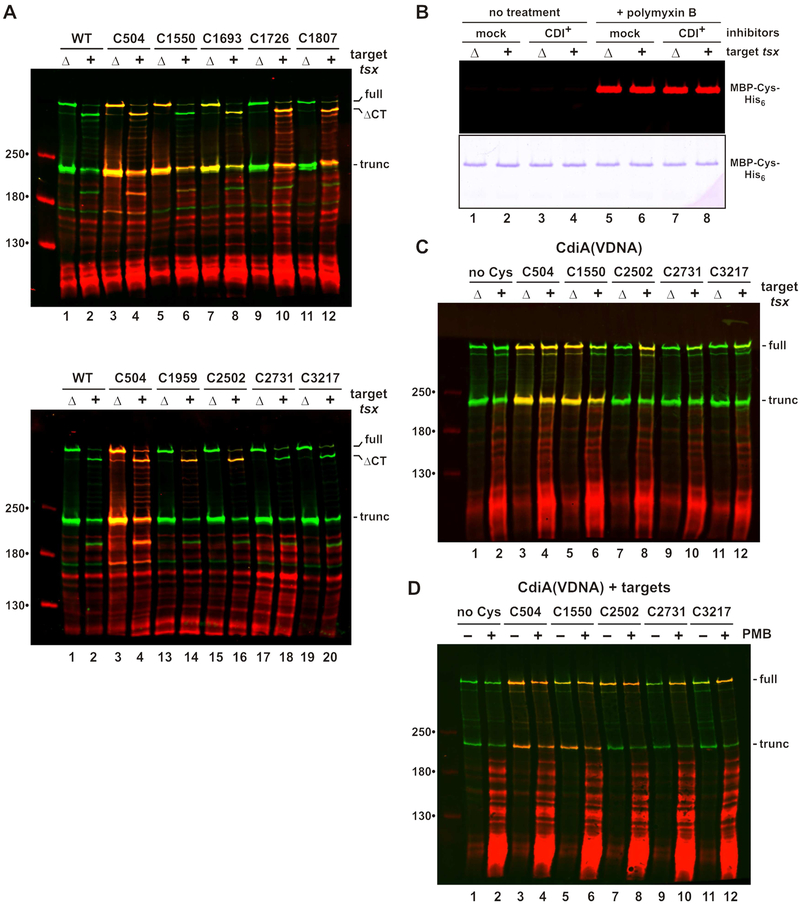
Filament elongation suggests that CdiA undergoes structural reorganization upon binding receptor. Moreover, effector topology must change dramatically for the C-terminal toxin domain to be transferred into target bacteria. Therefore, we used the dye-labeling approach to monitor topological changes in response to the addition of target bacteria. Immunoblotting revealed that the CdiA-CT region of CdiASTECO31 is cleaved upon mixing with tsx+ target cells, but not with Δtsx mock targets (Figure 4A, lanes 1 & 2). Thus, this CdiA-CT processing is indicative of toxin delivery. The reactivity of extracellular positions 504 and 1693 is unaltered by target bacteria, but labeling at position 1550 is diminished in the presence of tsx+ targets (Figure 4A, lanes 5 & 6). Because residue 1550 lies within the RBD, it may become occluded through direct interaction with Tsx. Strikingly, positions 1726, 1807, 1959 and 2502 all become accessible to extracellular dye when tsx+ cells are introduced, but remain periplasmic with mock Δtsx targets (Figure 4A). These data suggest that the C-terminal region is exported upon binding Tsx, but it is also possible that outer membranes become permeable to dye in response CdiA-receptor binding. This is not the case, however, as MBP-Cys-His6 in the inhibitor-cell periplasm remains unreactive when tsx+ targets are introduced (Figure 4B, lanes 3 & 4). In contrast to other Cys substitutions, residue 2731 in the PT domain remains unlabeled with tsx+ targets (Figure 4A, lanes 17 & 18), suggesting that it may be transferred into the target cell. We note that position 3217 cannot be detected because it lies within the cleaved CdiA-CT region. To monitor this latter position, we blocked CdiA-CT processing by mutating the VDNN motif to VDNA (Figure 4C, lanes 1 & 2), and repeated the topology mapping with a subset of Cys substitutions. The VDNA mutation has no effect on Cys reactivity at positions 504, 1550 and 2502 (Figure 4C, lanes 3 - 8). Positions 2731 and 3217 show some labeling in the presence of tsx+ targets, but at lower efficiencies than position 2502 (Figure 4C, compare lanes 8, 10 & 12). Residues 2731 and 3217 appear to be transferred into the periplasm of target cells, because they are labeled more efficiently when the mixed cell suspensions are permeabilized with polymyxin (Figure 4D, lanes 10 & 12). Together, these data indicate that CdiA export resumes upon binding receptor, with the PT and CdiA-CT domains being transferred into target bacteria. Because N-terminal residues remain extracellular, the CdiA chain must pass through the outer membrane of the target cell.
Figure 4. CdiA export resumes upon binding receptor.
A) Topology of receptor-bound CdiASTECO31. CdiA expressing cells were mixed with tsx+ or Δtsx targets and incubated with maleimide-dye. CdiA was analyzed by immunoblotting with anti-TPS antibodies. Full-length, CdiA-CT processed (ΔCT) and truncated proteins are indicated. B) Inhibitors co-expressing MBP-Cys-His6 were mixed with targets and incubated with maleimide-dye. Purified MBP was analyzed by fluorimetry (top panel) and SDS-PAGE (bottom panel). C) CdiA(VDNA) expressing cells were mixed with targets and analyzed as in panel A. D) CdiA(VDNA) expressing cells were mixed with tsx+ targets and incubated with maleimide-dye. Mixed cell suspensions were treated with PMB where indicated.
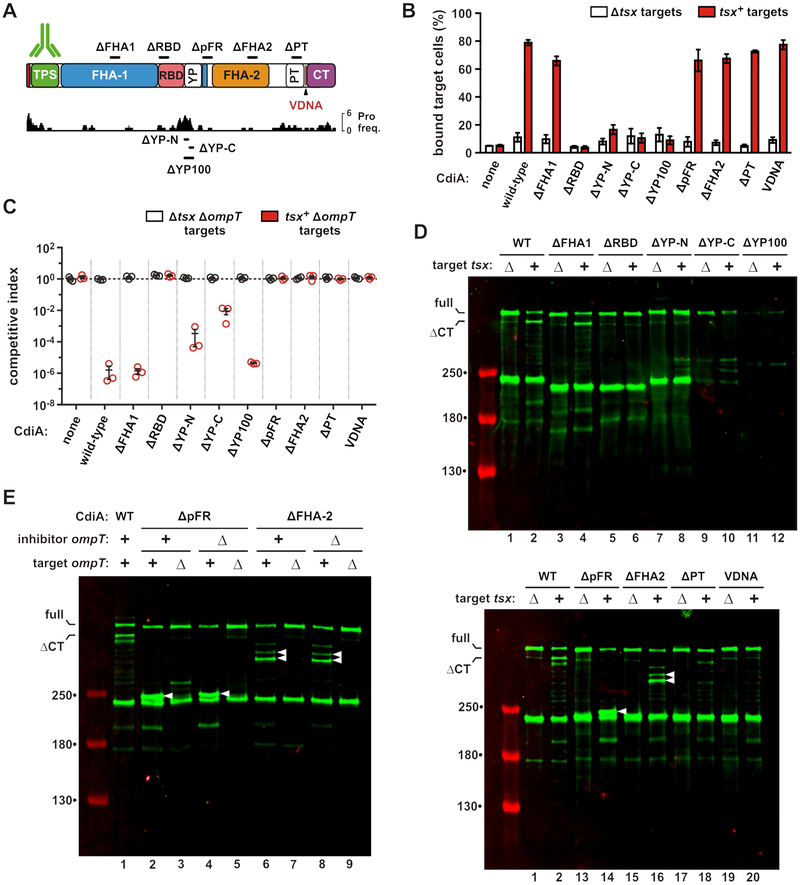
The YP domain is required for cell-surface presentation
We next disrupted the FHA-1, RBD, YP, FHA-2 and PT domains with 100-residue in-frame deletions to examine their functions during CDI (Figure 5A). The YP domain was dissected further with 50-residue deletions to test its extracellular region (ΔYP-N) and membrane-spanning segment (ΔYP-C). We also removed a portion of the periplasmic FHA-1 repeat (ΔpFR) region to probe its function. Though the ΔFHA-1 effector retains nearly wild-type inhibition activity, the other deletion constructs fail to inhibit target cells in shaking-broth co-cultures (Figure S2B). Receptor-binding function was then assessed by flow cytometry to monitor cell-cell adhesion. GFP-labeled inhibitor cells were mixed with DsRed-labeled target cells, and the mixtures analyzed by flow cytometry to quantify aggregates with dual green and red fluorescence. Cells expressing wild-type CdiASTECO31 bind ~80% of tsx+ target cells (Figure 5B). Adhesion is CDI-dependent because Δtsx cells are bound at much lower levels, and mock inhibitors lacking CdiASTECO31 do not bind tsx+ targets (Figure 5B). As expected, the ΔRBD deletion abrogates target-cell binding, but effectors carrying YP domain deletions are also defective for adhesion (Figure 5B). The remaining effectors retain nearly wild-type cell-adhesion activity (Figure 5B). Immunoblotting showed low levels of ΔYP-C and ΔYP-100 relative to the other effectors, and these proteins were released into the culture supernatants (Figure S5A, lanes 5 & 6). This latter finding suggests that their defects in cell-cell adhesion are due to an unstable association with the inhibitor-cell surface. This phenomenon could also interfere with growth inhibition activity, particularly in shaking broth co-cultures. Therefore, we re-examined deletion construct activity on solid media, where inhibitor and target cells are held in close proximity within a structured environment. Strikingly, the YP deletion constructs inhibited target bacteria on solid media, with the ΔYP-100 effector exhibiting nearly wild-type growth-inhibition activity (Figure 5C). Further, this activity is tsx-dependent (Figure 5C), demonstrating that the ΔYP-100 deletion does not disrupt the adjacent RBD. Thus, the YP domain is not necessary for toxin delivery, but is critical when cell-cell interactions are transient. Collectively, these results show that the membrane-spanning segment of the YP domain is required for stable presentation on the inhibitor-cell surface.
Figure 5. The YP domain is required for cell-surface presentation.
A) CdiASTECO31 deletion constructs. See Figures S2. B) Target cell adhesion. GFP-labeled inhibitors were mixed with DsRed-labeled targets, and analyzed by flow cytometry to quantify the fraction of target cells bound to inhibitors. Data are presented as mean ± SEM. C) Competition co-cultures. Inhibitors were mixed at a 1:1 ratio with targets and co-cultured on LB-agar. Competitive indices equal the final ratio of targets to inhibitors divided by the initial ratio. Data are presented as mean ± SEM. D) Inhibitors were mixed with targets and CdiA analyzed by anti-TPS immunoblotting. E) ΔpFR and ΔFHA-2 inhibitors were mixed with target cells, and CdiA analyzed by anti-TPS immunoblotting. Inhibitor and target cells carried ΔompT mutations where indicated. White carets indicate aberrantly processed CdiA. See Figure S5.
The FHA-2 domain is required for toxin delivery
To determine which steps in the CDI pathway are disrupted by the various deletions, we examined how each effector responds to target bacteria. Immunoblotting showed that Tsx-dependent CdiA-CT processing is defective for each effector except the ΔFHA-1 protein (Figure 5D, lanes 3 & 4). Notably, the ΔpFR and ΔFHA-2 proteins are cleaved at novel sites when mixed with tsx+ cells, but not mock targets (Figure 5D, white carets in lanes 14 & 16), suggesting aberrant processing during toxin delivery. We screened protease-deficient E. coli strains and found that OmpT is required for this activity. OmpT protease is embedded in the outer membrane with its active site facing the extracellular milieu (Kramer et al., 2001; Vandeputte-Rutten et al., 2001). In principle, OmpT on either the inhibitor or target cell could catalyze cleavage, but CdiA degradation depends solely on OmpT in the target cell (Figure 5E, lanes 2, 4, 6 and 8). Further, OmpT-mediated cleavage releases C-terminal CdiA fragments into the culture supernatant (Figure S5B, lanes 10 & 12). The masses of the OmpT-dependent fragments indicate that the ΔpFR and ΔFHA-2 chains are each cleaved near their respective deletion sites, suggesting that the entire FHA-2 region is brought into close proximity with the target-cell surface. However, OmpT-mediated proteolysis does not explain the defects in inhibition activity, because tsx+ ΔompT target cells are not inhibited by the ΔpFR and ΔFHA-2 effectors, even when co-cultured on solid media (Figure 5C). Moreover, the CdiA-CT toxin region is not processed when ΔpFR and ΔFHA-2 effectors bind to tsx+ ΔompT target cells (Figure 5E, lanes 3, 5, 7 and 9). Therefore, the pFR and FHA-2 regions are critical for CdiA-CT delivery into the target-cell periplasm.
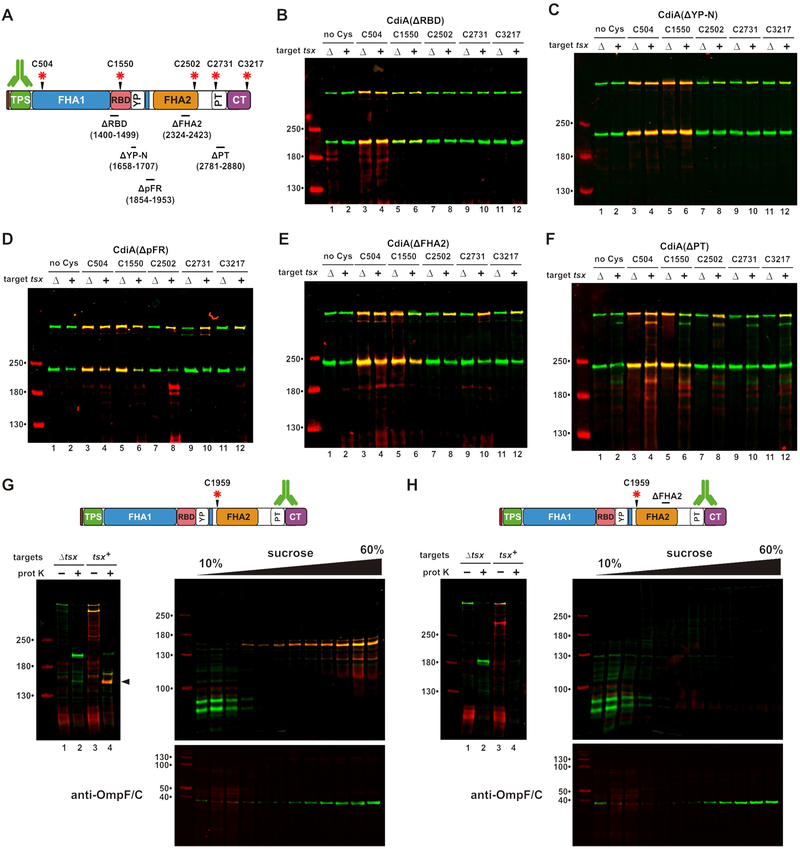
We introduced Cys substitutions into the ΔRBD, ΔYP-N, ΔpFR, ΔFHA-2 and ΔPT effectors (Figure 6A) and monitored surface topology after addition of tsx+ ΔompT target cells. As expected, Cys residues in the ΔRBD protein show no changes in reactivity when target cells are introduced (Figure 6B). Similarly, the ΔYP-N protein shows only modest labeling of periplasmic residues 2502, 2731 and 3217 when mixed with targets (Figure 6C). In contrast, all periplasmic positions become accessible to extracellular dye when the ΔpFR and ΔFHA-2 effectors bind to target bacteria (Figures 6D & 6E), indicating that Tsx-interactions trigger export, but the PT and CdiA-CT domains are not transferred into target bacteria. Disruption of the PT domain also interferes with toxin delivery, because position 3217 in the toxin domain remains extracellular when the ΔPT effector binds target cells (Figure 6F, lane 12). However, labeling at position 2731 is suppressed in the ΔPT effector, similar to what is observed when this residue is probed in the context of full-length CdiASTECO31 (compare Figure 6F, lane 10 with Figure 4A, lane 18). This latter observation suggests that residue 2731, which is N-terminal to the ΔPT deletion, is still transferred into the target-cell periplasm. Therefore, the pFR, FHA-2 and PT disruptions have no effect on receptor-triggered export, but prevent CdiA-CT delivery into target bacteria.
Figure 6. The FHA-2 domain is required for toxin delivery.
A) CdiA deletions and Cys substitutions. Deleted residues are indicated below domain identifiers. B - F) Cells expressing the indicated CdiA effectors were mixed with targets and incubated with maleimide-dye. CdiA proteins were analyzed by anti-TPS immunoblotting. G) FHA-2 localizes to the outer membrane after export. Cells expressing Cys1959 CdiA were mixed with tsx+ targets, then treated with dye and proteinase K. The sample from lane 4 was run on a sucrose gradient, and fractions analyzed by anti-TPS and anti-OmpF/C immunoblotting. H) Cells expressing Cys1959 ΔFHA2 were treated and analyzed as in panel G.
The ΔpFR and ΔFHA-2 deletions both prevent CdiA-CT transfer into the target-cell periplasm, and these effectors are susceptible to OmpT-mediated cleavage at the target-cell surface. These observations raise the possibility that FHA-2 integrates into the target-cell outer membrane to form a translocon for toxin delivery. To explore this hypothesis, we incubated tsx+ target cells with inhibitors that express Cys1959-substituted CdiASTECO31, then incubated the suspension with maleimide-dye and proteinase K. Analysis of CdiA from this experiment revealed a large (>130 kDa) dye-labeled fragment that is resistant to proteolysis (Figure 6G, lane 4). Dye-labeling demonstrates that this fragment was derived from CdiA that was exported in response to target bacteria (Figure 6G, compare lanes 2 & 4). The dye-labeled fragment also reacted with PT/CdiA-CT antisera, indicating that it encompasses the entire FHA-2 region. Finally, we found that this fragment migrates in the outer-membrane fraction on sucrose gradients (Figure 6G), consistent with a stable interaction between FHA-2 and target-cell outer membranes. By contrast, a protease-resistant fragment was not detected with ΔFHA-2 effectors carrying the Cys1959 substitution (Figure 6H, lane 4). These results strongly suggest that an intact FHA-2 region is required for stable association with the target-cell outer membrane.
Discussion
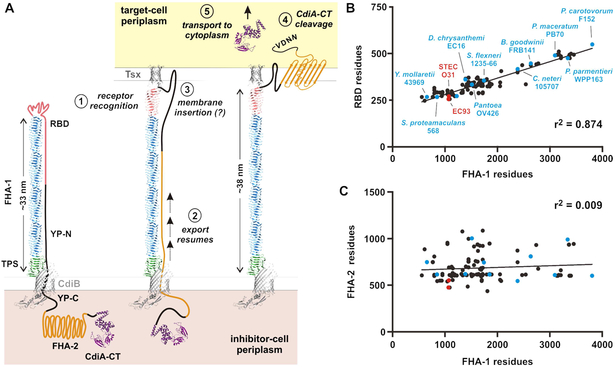
Here, we present the first ultrastructural images of CdiA in its near-native state. ECT revealed that each CdiA effector forms a thin filament extending ~33 nm from the cell surface. These dimensions are consistent with the β-helix model (Kajava et al., 2001), which predicts that the FHA-1 region of CdiAEC93 should form a ~27 nm filament. The TPS transport domain could account for another ~3 nm based on the crystal structures of homologous domains in Bordetella FhaB and Haemophilus HWM1 (Clantin et al., 2004; Yeo et al., 2007). Given our finding that the RBD forms the filament tip, the TPS domain must be at the other end of the filament, likely associating with CdiB at the cell surface (Figure 7A). We also discovered that CdiA is subject to secretion arrest, and the entire C-terminal half of the effector is retained within the periplasm. Therefore, the extracellular CdiA filament must follow a hairpin-like trajectory, first extending outward to form the RBD, then returning back to the cell to enter the periplasm (Figure 7A). This topology imposes constraints on the sequence linking the distal RBD to the periplasmic FHA-2 domain. The peptide chain must be extended in this region to span the length of the extracellular filament. Because the YP domain traverses the outer membrane and is required for secretion arrest, we predict that it passes through the central lumen of CdiB to halt export and stabilize CdiA on the cell surface. Further, this export arrest appears to be universal, because similar YP domains are found in all recognizable CdiA proteins. Given that FHA-1 varies considerably in size between species (Willett et al., 2015b), our model predicts that the extended return region should exhibit commensurate changes to accommodate different filament lengths. Indeed, analysis of 142 CdiA proteins from enterobacteria revealed that the size of the variable RBD is directly proportional to FHA-1 length (Figure 7B & Table S2). This correlation is remarkably robust (r2 = 0.87) for filaments ranging from ~14 to 100 nm in predicted length. Therefore, the RBD must contribute significantly to the extended region, because the length of the YP domain is essentially invariant (Table S2). These observations suggest that the RBD, which is defined as the variable region between the FHA-1 and YP domains, may in fact contain two domains with distinct functions. We hypothesize that the N-terminal portion of the RBD makes direct contact with receptors based on sequence alignments showing limited homology in this region across species (Figure S3). This model predicts that the additional residues found in longer CdiA proteins do not contribute to receptor-binding per se, but are required to span the filament as the chain returns to the inhibitor-cell surface (Figure 7A). We also note that FHA-2 does not scale with filament length (Figures 7C & Table S2), demonstrating that size correlation is limited to the RBD and FHA-1. Together, these observations argue that all CdiA proteins adopt the same general surface topology.
Figure 7. Model for CdiA surface topology and receptor-triggered toxin delivery.
A) Model of CdiA surface topology. Receptor recognition (1) triggers export of the C-terminal half of CdiA (2). The FHA-2 domain interacts with the target-cell outer membrane (3), perhaps forming a β-barrel translocon. Once transferred into the target-cell periplasm, the CdiA-CT region is cleaved (4) for transport into the cytoplasm (5). B) RBD size is proportional to filament length. RBD residues are plotted as a function of FHA-1 residues. See Figure S3 & Table S2. C) Plot of FHA-2 versus FHA-1 residues for the effectors in panel B. See Table S2.
Though CdiA undergoes export arrest, it remains poised to deliver toxin upon encountering target bacteria. Presumably, receptor-recognition generates a signal that causes CdiA export to resume. One possible mechanism involves receptor-induced folding of the RBD, which would exert a force on the extended region and release the YP domain from its "locked" state within CdiB (Figure 7A, step 2). This model may also explain why receptor-bound filaments become elongated. Alternatively, the additional length could correspond to the newly exported FHA-2 domain, which we showed makes contact with target bacteria and becomes stably associated with their outer membranes (see Figures 5E & 7G). Our results indicate that the FHA-2 domain likely passes through the target-cell outer membrane to deliver the CdiA-CT into the periplasm. The structure of FHA-2 is unknown, but the I-TASSER server (Yang et al., 2015) predicts that it could resemble the β-rich RsaA S-layer protein from Caulobacter, or the LptD lipopolysaccharide transporter from Shigella. The latter model is appealing because LptD forms a 26-stranded β-barrel in the outer membrane, which would be ideal for toxin translocation. Irrespective of its precise structure, we propose that FHA-2 interaction with the target-cell outer membrane is closely coupled with its export from the inhibitor cell (Figure 7A, step 3). This would allow FHA-2 to integrate as an unfolded chain emerging from the inhibitor cell. This hypothesis could explain why the constitutively released ΔYP-100 effector is inactive in shaking broth co-culture (see Figure S2B), yet retains nearly wild-type growth inhibition activity on solid media (see Figure 5C). Following release into the medium, the FHA-2 domain is presumably free to adopt a conformation that precludes membrane integration. However, when released in a crowded sessile community, some of the CdiA will likely be deposited directly onto target bacteria before FHA-2 has the opportunity to fold. Thus, secretion arrest is not strictly required for CDI, but this feature is critical to ensure that the FHA-2 translocon is deployed only when a suitable target membrane is nearby.
The outer-membrane translocation mechanism proposed here is novel and distinct from other inter-bacterial competition systems. Type VI secretion systems rely on mechanical force to deliver toxic payloads. Type VI systems utilize a phage-related contractile apparatus to eject a toxin-laden spear that perforates the outer membranes of neighboring bacteria (Basler et al., 2012; Russell et al., 2014). This mechanism is sufficient to deliver lytic enzymes that degrade the bacterial cell wall and membrane lipids, but it remains unclear how these systems transfer nuclease toxins into the cytoplasm. Myxobacteria have the unique ability to merge outer membranes, thereby allowing lipids and OMPs to be shared freely between cells (Pathak et al., 2013; Vassallo et al., 2015). Wall and coworkers have recently shown that polymorphic lipoprotein toxins are also transferred during this process, enabling cells to discriminate kin when engaging in this social behavior (Vassallo et al., 2017). Because the lipoprotein toxins are tethered to the inner leaflet of the outer membrane, they are able to flow between cells without physical translocation across a lipid bilayer. Toxin translocation has been studied most extensively with colicins. These diffusible protein toxins bind specific surface receptors, then recruit porins as portals for entry into the target-cell periplasm (Cascales et al., 2007; Housden and Kleanthous, 2012). Transport through the porin is powered by Tol and Ton system proteins, which harness energy from the proton gradient across the bacterial cytoplasmic membrane. Thus, colicins cannot penetrate the outer membrane when bacteria are treated with uncoupling agents that dissipate the proton gradient. Intriguingly, Δtol Δton mutants are completely resistant to colicin-mediated killing, but are fully sensitive to CDI (Ruhe et al., 2014). Moreover, CdiA-CT toxins are readily delivered into the periplasm of de-energized cells, though the proton gradient is required for subsequent transport into the cytoplasm (Ruhe et al., 2014). These observations underscore fundamental differences between colicins and CDI, but also raise important unresolved questions about the thermodynamics of CdiA-CT transport across the outer membrane.
Finally, our results could have implications for the function of FHA adhesins in Bordetella. FHA is derived from the FhaB pre-protein, which is processed during biogenesis to remove its large C-terminal "prodomain" (Scheller and Cotter, 2015). The domain architecture of FhaB is quite similar to CdiA, and the prodomain contains an FHA-2 repeat region (Aoki et al., 2005; Willett et al., 2015b). In another parallel, Cotter and coworkers have shown that the FhaB prodomain resides in the periplasm during biogenesis (Noel et al., 2012). This phenomenon is thought to promote folding of the exported adhesin domain, after which the prodomain is degraded by periplasmic proteases to release mature FHA from the cell (Noel et al., 2012; Scheller and Cotter, 2015). Our work suggests an alternative function for the prodomain. We hypothesize that the FHA-2 repeats in FhaB are used to transport cargo across target-cell membranes. Instead of an antibacterial toxin, the prodomain of FhaB carries an unusual prolyl-rich domain adjacent to the FHA-2 region. The function of this prolyl-rich region is enigmatic, but recent work from the Cotter lab shows that it is important for bacterial persistence in the lower respiratory tract (Melvin et al., 2015). Collectively, these observations raise the possibility that the prolyl-rich domain is delivered into host cells to modulate immune responses. Thus, Bordetella species may have repurposed the primordial antibacterial CDI mechanism to play a new role in pathogenesis.
STAR Methods
Contact for reagent and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Christopher Hayes (chayes@lifesci.ucsb.edu).
Experimental model and subject details
Bacterial growth conditions.
Bacterial strains were derivatives of E. coli K-12 strains MG1655 and EPI100. Bacteria were cultured in lysogeny broth (LB) or on LB agar at 37 °C. Unless indicated otherwise, media were supplemented with antibiotics at the following concentrations: 150 μg/mL ampicillin (Amp), 100 μg/mL chloramphenicol (Cm), 50 μg/mL kanamycin (Kan), 200 μg/mL spectinomycin (Spc), and 25 μg/mL tetracycline (Tet).
Method details
Strain constructions.
E. coli strains are presented in the Key Resources Table. The mreB(A125V) allele (linked to ΔyhdE::cat) from E. coli DS612 (Shiomi et al., 2013) was transferred into E. coli CH9591 and CH9604 using phage Pi-mediated transduction. The ΔyhdE::cat Cm resistance cassette was subsequently removed using FLP recombinase expressed from pCP20 (Cherepanov and Wackernagel, 1995) to generate strains ZR82 and ZR83 (respectively). ZR82 was transformed with pZR108 (minE overexpression) and CdiAEC93 expressing plasmids to generate minicell producing strains for ECT. The resulting strains were cultured in media supplemented with 0.4% L-arabinose to generate minicells. The Δwzb::kan, Δtsx::kan and ΔompT::kan disruptions were introduced into E. coli MG1655 from the Keio collection (Baba et al., 2006) by phage P1-mediated transduction. For strains that carry multiple deletions, Kan resistance cassettes were removed with FLP recombinase.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rabbit polyclonal antisera to the CdiAEC93 TPS transport domain (residues Val33 – Gly285) | (Ruhe et al., 2015) | N/A |
| rabbit polyclonal antisera to the CdiASTECO31 pretoxin and CdiA-CT domains (residues Gly2648 – Lys3253) | This paper | N/A |
| rabbit polyclonal antisera to E. coli OmpC/OmpF | Thomas Silhavy | N/A |
| IRDye® 800CW goat anti-rabbit IgG | LI-COR | Cat# P/N 925-32211 |
| Bacterial and Virus Strains | ||
| E. coli K-12 sub-strain MG1655 | E. coli Genetic Stock Center | Strain #7740 |
| E. coli K-12 sub-strain EPI100 | Epicentre/Lucigen | Cat# EC10010 |
| E. coli: DS612: BW25113 ΔyhdE::cat mreB-A125V | (Shiomi et al., 2013) | N/A |
| E. coli: CH2016: X90 (DE3) Δrna ΔslyD::kan | (Garza-Sánchez et al., 2011) | N/A |
| E. coli: CH7286: MG1655 Δwzb::kan | This paper | N/A |
| E. coli: CH7367: MG1655 Δwzb | This paper | N/A |
| E. coli: CH9591: EPI100 ΔbamA::cat pZS21-bamAECL | (Ruhe et al., 2013) | |
| E. coli: CH9604: EPI100 ΔbamA::cat pZS21-bamA+ | (Ruhe et al., 2013) | |
| E. coli: CH10060: EC93 bamALT2 | (Ruhe et al., 2015) | N/A |
| E. coli: CH10093: EC93 ΔcdiA | (Ruhe et al., 2015) | N/A |
| E. coli: CH14016: MG1655 Δwzb Δtsx | This paper | N/A |
| E. coli: CH14017: MG1655 Δwzb Δtsx ΔompT | This paper | N/A |
| E. coli: ZR82: EPI100 ΔbamA::cat ΔyhdE mreB-A125V pZS21-bamAECL | This paper | N/A |
| E. coli: ZR83: EPI100 ΔbamA::cat ΔyhdE mreB-A125V pZS21-bamA+ | This paper | N/A |
| E. coli: DL4259: MC4100 λ640-13 PpapIB-gfp-mut3 | (Webb et al., 2013) | N/A |
| E. coli: ZR373: EPI100 Δwzb Δtsx | This paper | N/A |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| IRDye® 680LT maleimide | LI-COR | Cat# P/N 929-71008 |
| polymyxin B sulfate | Sigma-Aldrich | Cat# P4932 |
| Benzonase® nuclease | Sigma-Aldrich | Cat# E1014 |
| Halt™ Protease Inhibitor Cocktail | ThermoFisher | Cat# 78430 |
| Critical Commercial Assays | ||
| Deposited Data | ||
| I-TASSER output for FHA-2 domain structure prediction | This paper; Mendeley data | http://dx.doi:10.17632/hrbmtfv754.1 |
| Individual fluorescence channels for immunoblotting data | This paper; Mendeley data | http://dx.doi:10.17632/hrbmtfv754.1 |
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| Oligonucleotides | ||
| Complete list of PCR primers | This paper | see Table S3 |
| Recombinant DNA | ||
| pBluescript II SK+ | Agilent/Stratagene | Cat# 212205 |
| pDsRedExpress2 | Clontech | Cat# 632535 |
| pET21b | Novagen | Cat# 69741-3 |
| pCP20 | (Cherepanov and Wackernagel, 1995) | N/A |
| pZS21-bamA+ | (Ruhe et al., 2013) | N/A |
| pZS21-bamAECL | (Ruhe et al., 2013) | N/A |
| pDAL660 | (Aoki et al., 2005) | N/A |
| pCH253 | This paper | N/A |
| pCH450 | (Hayes and Sauer, 2003) | N/A |
| pCH649 | This paper | N/A |
| pCH7277 | (Garza-Sánchez et al., 2011) | N/A |
| pCH9674 | (Ruhe et al., 2017) | N/A |
| pCH9216 | (Ruhe et al., 2017) | N/A |
| pCH9231 | (Ruhe et al., 2017) | N/A |
| pCH10145 | (Beck et al., 2016) | N/A |
| pCH12705 | (Aoki et al., 2005) | N/A |
| pCH12706 | (Aoki et al., 2005) | N/A |
| pCH12707 | (Aoki et al., 2005) | N/A |
| pCH13602 | (Ruhe et al., 2017) | N/A |
| pCH13603 | (Ruhe et al., 2017) | N/A |
| pCH13604 | (Ruhe et al., 2017) | N/A |
| pCH13658 | This paper | N/A |
| pCH13709 | This paper | N/A |
| pCH14028 | This paper | N/A |
| pCH14029 | This paper | N/A |
| pCH14030 | This paper | N/A |
| pCH14031 | This paper | N/A |
| pCH14032 | This paper | N/A |
| pCH14033 | This paper | N/A |
| pCH14034 | This paper | N/A |
| pCH14035 | This paper | N/A |
| pCH14036 | This paper | N/A |
| pCH14047 | This paper | N/A |
| pCH14092 | This paper | N/A |
| pCH14093 | This paper | N/A |
| pCH14094 | This paper | N/A |
| pCH14095 | This paper | N/A |
| pCH14096 | This paper | N/A |
| pCH14102 | This paper | N/A |
| pCH14103 | This paper | N/A |
| pCH14104 | This paper | N/A |
| pCH14105 | This paper | N/A |
| pCH14106 | This paper | N/A |
| pCH14107 | This paper | N/A |
| pCH14108 | This paper | N/A |
| pCH14109 | This paper | N/A |
| pCH14110 | This paper | N/A |
| pCH14111 | This paper | N/A |
| pCH14112 | This paper | N/A |
| pCH14113 | This paper | N/A |
| pCH14114 | This paper | N/A |
| pCH14115 | This paper | N/A |
| pCH14116 | This paper | N/A |
| pCH14117 | This paper | N/A |
| pCH14118 | This paper | N/A |
| pCH14119 | This paper | N/A |
| pCH14120 | This paper | N/A |
| pCH14121 | This paper | N/A |
| pCH14202 | This paper | N/A |
| pCH14203 | This paper | N/A |
| pCH14204 | This paper | N/A |
| pCH14205 | This paper | N/A |
| pCH14206 | This paper | N/A |
| pCH14213 | This paper | N/A |
| pCH14214 | This paper | N/A |
| pCH14215 | This paper | N/A |
| pCH14216 | This paper | N/A |
| pCH14217 | This paper | N/A |
| pCH14218 | This paper | N/A |
| pCH14219 | This paper | N/A |
| pCH14220 | This paper | N/A |
| pCH14221 | This paper | N/A |
| pZR108 | This paper | N/A |
| pZR113 | (Aoki et al., 2005) | N/A |
| pZR420 | This paper | N/A |
| pZR432 | This paper | N/A |
| pZR433 | This paper | N/A |
| pZR434 | This paper | N/A |
| pZR435 | This paper | N/A |
| pZR437 | This paper | N/A |
| pZR439 | This paper | N/A |
| pZR441 | This paper | N/A |
| pZR442 | This paper | N/A |
| pZR444 | This paper | N/A |
| pZR462 | This paper | N/A |
| pZR463 | This paper | N/A |
| Software and Algorithms | ||
| UCSF Tomo | (Zheng et al., 2007) | http://www.msg.ucsf.edu/Tomography/tomography_main.html |
| IMOD | (Kremer et al., 1996) | http://bio3d.colorado.edu/imod/ |
| Other | ||
Plasmid constructions.
Plasmids are presented in the Key Resources Table and oligonucleotide primers are listed in Table S3.
Cys-substitution mutations
Cys substitutions were generated by PCR and the resulting fragments introduced into plasmid pET21b::cdiBAISTECO31 (pCH13604) using restriction endonuclease based cloning. The product from primers ZR260/ZR267 was ligated via NotI/KpnI to generate pZR432 (Ser504Cys). The product from CH403/ZR271 was ligated via SacI to generate pZR433 (Ser1550Cys). PCR fragments from primer pairs ZR268/ZR274 and ZR275/ZR287 were cloned sequentially into pBluescript II SK+ using KpnI/SalI and SalI/NotI, respectively. The KpnI/NcoI fragment was then subcloned to generate pZR434 (Ser1693Cys). PCR fragments from primer pairs ZR268/ZR277 and ZR281/ZR287 were cloned sequentially into pBluescript II SK+ using KpnI/XbaI and XbaI/NotI, respectively. The KpnI/NcoI fragment was then subcloned to generate pZR435 (Gly1726Cys). The product from ZR280/ZR261 was ligated via SexAI/Xho I to generate pZR437 (Ala1807Cys). The product from ZR286/ZR261 was ligated via NcoI/Xho I to generate pZR439 (Asp1959Cys). PCR fragments from primer pairs ZR347/ZR288 and ZR290/ZR261 were cloned sequentially into pBluescript II SK+ using NotI/EcoRI and EcoRI/XhoI, respectively. The NcoI/XhoI fragment was then subcloned to generate pZR441 (Thr2502Cys). PCR fragments from primer pairs ZR347/ZR348 and ZR349/ZR261 were cloned sequentially into pBluescript II SK+ using NotI/BamHI and BamHI/Xho I, respectively. The NcoI/XhoI fragment was then subcloned to generate pZR442 (Ser2731Cys). The product from ZR294/ZR261 was ligated via MfeI/XhoI to generate pZR444 (Ser3217Cys).
In-frame deletion constructs
In-frame deletions within cdiASTECO31 were generated by PCR and introduced into plasmid pCH13604. The fragments from primer pairs ZR268/CH4240 and CH4241/ZR287 were cloned sequentially into pBluescript II SK+ using KpnI/XhoI and XhoI/NotI, respectively. The KpnI/NcoI fragment was then subcloned to generate pCH14028 (ΔFHA-1: ΔAsp900-Ala999). The fragments from primer pairs ZR268/CH4243 and CH4244/ZR287 were cloned sequentially into pBluescript II SK+ via KpnI/XhoI and XhoI/NotI, respectively. The KpnI/NcoI fragment was then subcloned to generate pCH14029 (ΔRBD: ΔAsn1400-Ser1499). The fragments from primer pairs ZR268/CH4381 and CH4382/ZR287 were cloned sequentially into pBluescript II SK+ via KpnI/XhoI and XhoI/NotI, respectively. The KpnI/NcoI fragment was then subcloned to generate pCH14032 (ΔYP-100: Δpro1658-Asn1757). The fragments from primer pairs ZR268/CH4383 and CH4382/ZR287 were cloned sequentially into pBluescript II SK+ via KpnI/XhoI and XhoI/NotI, respectively. The KpnI/NcoI fragment was then subcloned to generate pCH14030 (ΔYP-N: ΔPro1658-Pro1707). The fragments from primer pairs ZR268/CH4383 and CH4384/ZR287 were cloned sequentially into pBluescript II SK+ via KpnI/XhoI and XhoI/NotI, respectively. The KpnI/NcoI fragment was then subcloned to generate pCH14031 (ΔYP-C: ΔGly1708-Asn1757). The product from ZR268/CH4245 was ligated via KpnI/NcoI to generate pCH14033 (ΔpFR: ΔGly1854-Ser1953). The product from ZR347/CH4247 was ligated via NcoI/AflII to generate pCH14034 (ΔFHA-2: ΔThr2324-Gln2423). The fragments from primer pairs CH4371/CH4372 and CH4373/ZR261 were cloned sequentially into pET21b via NheI/SacI and SacI/XhoI, respectively. The AflII/XhoI fragment was then subcloned to generate pCH14035 (ΔPT: ΔPro2781-Met2880).
The Asn2934Ala mutation in the VDNN motif was generated in three steps. Plasmid pCH13604 was digested with XbaI/SalI, end-filled with T4 DNA polymerase and re-ligated to remove restriction sites upstream of the cdiBAISdTECO31 gene cluster. An NheI restriction site was introduced into the resulting pCH13658 construct by ligating the PCR product of primers CH4282/CH4283 via AflII/XhoI to generate plasmid pCH13709. Finally, the product from CH4412/CH4374 amplification was used as a template for a second PCR with CH4413/CH4374. The final product was ligated to pCH13709 via NheI/XhoI to generate pCH14036 (VDNA).
Arabinose-inducible constructs for cell-cell adhesion
For cell-cell adhesion assays, the cdiBAISTECO31 gene cluster was over-expressed under an L-arabinose inducible promoter from plasmid pCH13602. Deletion constructs were introduced into pCH13602 through subcloning. The NotI/SacI fragment from pCH14028 was ligated to generate pCH14213 (ΔFHA-1: ΔAsp900-Ala999). The NotI/PmlI fragment from pCH14029 ligated to generate pCH14214 (ΔRBD: ΔAsn1400-Ser1499). SacI/XcmI fragments from pCH14030, pCH14031 and pCH14032 were ligated to generate plasmids pCH14215 (ΔYP-N: ΔPro1658-Pro1707), pCH14216 (ΔYP-C: ΔGly1708-Asn1757) and pCH14217 (ΔYP-100: Δpro-1658-Asn1757), respectively. NotI/XhoI fragments from pCH14033, pCH14034, pCH14035 and pCH14036 were ligated to generate pCH14218 (ΔpFR: ΔGly1854-Ser1953), pCH14219 (ΔFHA-2: ΔThr2324-Gln2423), pCH14220 (ΔPT: ΔPro2781-Met2880), and pCH14221 (VDNA: Asn2934Ala), respectively.
Combination of Cys substitutions with in-frame deletions
Cys substitutions were combined with the ΔAsn1400-Ser1499 (ΔRBD) deletion as follows. The NheI/KpnI fragment from pZR432 was ligated into pCH14029 to generate pCH14092 (S504C/ΔN1400-S1499). Plasmid pCH14029 was amplified with CH4239/CH4243 and pZR433 amplified with CH4244/ZR287, and the products ligated sequentially into pBluescript II SK+ with KpnI/XhoI and XhoI/NotI, respectively. The KpnI/NcoI fragment was then ligated to pCH13604 to generate pCH14093 (S1550C/ΔN1400-S1499). The KpnI/NcoI fragment from pCH14029 was ligated to plasmids pZR441, pZR442 and pZR444 to generate pCH14094 (T2502C/ΔN1400-S1499), pCH14095 (S2731C/ΔN1400-S1499) and pCH14096 (S3217C/ΔN1400-S1499), respectively.
Cys substitutions were combined with the ΔPro1658-Pro1707 (ΔYP-N) deletion as follows. The NheI/KpnI fragment from pZR432 was ligated into pCH14030 to generate pCH14202 (S504C/ΔP1658-P1707). Plasmid pZR433 was amplified with CH4239/CH4381 and with CH4382/ZR287, and the products ligated sequentially into pBluescript II SK+ via KpnI/XhoI and XhoI/NotI, respectively. The KpnI/NcoI fragment was then ligated to pCH13604 to generate pCH14203 (S1550C/ΔP1658-P1707). The KpnI/NcoI fragment from pCH14030 was ligated to plasmids pZR441, pZR442 and pZR444 to generate pCH14204 (T2502C/ΔP1658-P1707), pCH14205 (S2731C/ΔP1658-P1707), pCH14206 (S3217C/ΔP1658-P1707), respectively.
Cys substitutions were combined with the ΔGly1854-Ser1953 (ΔpFR) deletion as follows. The NheI/KpnI fragment from pZR432 was ligated into pCH14033 to generate pCH14102 (S504C/ΔG1854-S1953). The KpnI/PmlI fragment from pZR433 was ligated to pCH14033 to generate pCH14103 (S1550C/ΔG1854-S1953). AflII/XhoI fragments from plasmids pZR441, pZR442 and pZR444 were ligated to pCH14033 to generate pCH14104 (T2502C/ΔG1854-S1953), pCH14105 (S2731C/ΔG1854-S1953) and pCH14106 (S3217C/ΔG1854-S1953), respectively.
Cys substitutions were combined with the ΔThr2324-Gln2423 (ΔFHA-2) deletion as follows. The NheI/KpnI fragment from pZR432 was ligated to pCH14034 to generate pCH14107 (S504C/ΔT2324-Q2423). The KpnI/PmlI fragment from pZR433 was ligated to pCH14034 to generate pCH14108 (S1550C/ΔT2324-Q2423). AflII/XhoI fragments from plasmids pZR441, pZR442 and pZR444 were ligated to pCH14034 to generate pCH14109 (T2502C/ΔT2324-Q2423), pCH14110 (S2731C/ΔT2324-Q2423) and pCH14111 (S3217C/ΔT2324-Q2423). Plasmid pZR439 was amplified with CH4384/CH4247 and the product ligated into pCH13604 via NcoI/AflII to generate pCH649 (D1959C/ΔT2324-Q2423).
Cys substitutions were combined with the ΔPro2781-Met2880 (ΔPT) deletion as follows. The NheI/KpnI fragment from pZR432 was ligated to pCH14035 to generate pCH14112 (S504C/ΔP2781-M2880). The KpnI/PmlI fragment from pZR433 was ligated to pCH14035 to generate pCH14113 (S1550C/ΔP2781-M2880). Plasmid pZR441 was amplified with CH4371/CH4372 and pZR432 with CH4373/ZR261, and the products ligated sequentially into pET21b via NheI/SacI and SacI/XhoI, respectively. The KpnI/NcoI fragment was then subcloned into pCH13604 to generate pCH14114 (T2502C/ΔP2781-M2880). Plasmid pZR442 was amplified with CH4371/CH4372 and pCH14035 with CH4373/ZR261, and the products ligated sequentially into pET21b via NheI/SacI and SacI/XhoI, respectively. The KpnI/NcoI fragment was then subcloned into pCH13604 to generate pCH14115 (S2731C/ΔP2781-M2880). The MfeI/XhoI fragment from pZR444 was ligated to pCH14035 to generate pCH14116 (S3217C/ΔP2781-M2880).
Cys substitutions were combined with the Asn2934Ala (VDNA) mutation as follows. The AflII/XhoI fragment from pCH14036 was ligated to plasmids pZR432 and pZR433 to generate pCH14117 (S504C/N2934A) and pCH14118 (S1550C/N2934A), respectively. Plasmid pZR441 was amplified with CH4371/CH4283 and the product ligated to pCH14036 via AflII/NheI to generate pCH14119 (T2502C/N2934A). Plasmid pZR442 was amplified with CH4371/CH4283 and the product ligated to pCH14036 via AflII/NheI to generate pCH14120 (S2731C/N2934A). The AflII/MfeI fragment from pCH14036 was ligated to pZR444 to generate pCH14121 (S3217C/N2934A).
Other plasmid constructs
The E. coli minE gene was amplified with CH2770/CH2771 and ligated to plasmid pTrc99a via NcoI/BamHI to generate pCH253. The NcoI/SbfI fragment from pCH253 was subcloned into pCH450 (Hayes and Sauer, 2003) to generate pZR108. The E. coli malE gene was amplified with ZR295/ZR297 and ligated to pCH450 via NotI/XhoI to generate pZR462. The malE signal sequence was removed by cloning the product of ZR296/ZR297 into pCH450 via NotI/XhoI to generate pZR463. The E. coli tsx gene was amplified with ZR256/ZR257 and ligated to pZS21-MCS(Kan) (Beck et al., 2016) via EcoRI/XbaI to generate pCH14047. A fragment encoding the PT and CdiA-CT domains (Gly2648 – Lys3253) of cd/ASTECO31 and cdilSTECO31 was amplified with primer pair ZR293/ZR261 and ligated to plasmid pCH7277 (Garza-Sánchez et al., 2011) via SpeI/Xhoi to generate pZR420. This latter plasmid was used to over-produce and purify His6-tagged PT/CdiA-CT domains to raise polyclonal antisera in rabbits (Cocalico Biologicals, Stevens, PA).
Electron cryotomography.
E. coli ZR82 cells carrying pCH253 and pDAL660 (full-length CdiAEC93) were grown in LB media supplemented with Amp and Cm (100 μg/mL) at 37 °C, and minicell production induced with 1.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). E. coli ZR82 strains carrying pZR108 and the various CdiAEC93 truncation constructs were grown in LB media supplemented with Tet and Amp and induced with 0.2% L-arabinose to produce minicells. E. coli CH14017 cells carrying pZR108 and pCH13604 were used to visualize CdiASTECO31 filaments. Lysed E. coli cells were prepared in 2 mg/mL lysozyme, 10 mM Tris-HCl (pH 8.0) at 37 °C for 30 min. The lysis mixture was adjusted to 2 mM MgCl2, 250 μM CaCl2 and treated with 1 mg/mL DNase I for 15 min at 37 °C. Cells were then collected by centrifugation and re-suspended in 10 mM Tris-HCl (pH 8.0) for imaging. All cell suspensions (16 μL) were mixed with bovine serum albumin (BSA)-treated 10-nm-diameter colloidal gold fiducial markers (Iancu et al., 2006; Mastronarde, 2006). This mixture (3 μL) was applied to a glow-discharged, X-thick carbon-coated, R2/2 200 mesh copper Quantifoil holey grid (Quantifoil Microtools) in a Vitrobot Mark III (FEI Company, Hillsboro, OR). The Vitrobot chamber was maintained at 22 °C and 80% humidity. Excess liquid was blotte d from the grid with a blot time of 3 - 4 s and a drain offset of −3.5 or −4. The grid was then plunge-frozen in a liquid ethane-propane mixture and imaged by ECT (Tivol et al., 2008). Imaging was performed on an FEI Polara G2 (FEI Company, Hillsboro, OR) 300-keV field emission gun electron microscope equipped with a Gatan image filter (Gatan, Pleasanton, CA) and K2 Summit counting electron detector camera (Gatan, Pleasanton, CA). Data were collected using the UCSF Tomo software (Zheng et al., 2007), with each tilt series ranging from −60° to 60° (or −65° to 65°) in 1° increments, an under-focus of ~4-15 μm, and a cum ulative electron dose of ~190 e/Å2 or less for each tilt series. The IMOD software package was used to calculate three-dimensional (3D) reconstructions (Kremer et al., 1996).
To image BamA binding to CdiAEC93 by ECT, E. coli ZR82 strains carrying pCH253 and pDAL660 were grown and minicells produced as described above. Cells were collected by centrifugation, washed three times in 1× phosphate buffered saline (PBS) and re-suspended in 100 μg/mL BSA for 30 min at 4 °C. Purified E. coli BamA [50 nM in 20 mM Tris-HCl (pH 8.0), 0.5% Triton X-100] was also incubated with 100 μg/mL BSA for 30 min at 4 ° to prevent non-specific interactions with the cell surface. Cells were incubated with BamA-BSA for 1 h, collected by centrifugation at 3,000 × g for 5 min and re-suspended in PBS. For the negative control, the same procedure was conducted using a buffer-BSA solution that lacks purified BamA. Inhibitor cells (E. coli ZR82 carrying pCH253 and pDAL660) and BamA-expressing target cells (E. coli ZR83 carrying pCH253) were grown separately in LB media supplemented with the appropriate antibiotics as described above. Minicell production was induced with IPTG for 3.5 h, then cultures were mixed and incubated with shaking at 240 rpm for 10 min and 75 rpm for 10 min. The suspension was incubated at 37 °C without shaking for 10 min prior to flash-freezing. Mixtures (4 μL) were applied to a glow-discharged, X-thick carbon-coated, R2/2 200 mesh copper Quantifoil holey grid in a Vitrobot Mark IV chamber maintained at 22 °C and 70% humidity. Excess liquid was blotted off the grid with a blot force of 6 or 3, a blot time of 3 or 2.5 s, and a drain time of 1 s. The grid was then plunge-frozen in a liquid ethane-propane mixture and imaged by ECT. Images were collected using either an FEI G2 300-keV field emission gun microscope or a FEI TITAN Krios 300-keV field emission gun microscope equipped with correction for lens aberration. Both microscopes were equipped with Gatan imaging filters and K2 summit counting electron-detector cameras (Gatan). Data were collected using the UCSF Tomo software with each tilt series ranging from −65° to 65° or (−60° to 60°) in 1 or 2° increments, an under-focus of ~10 μm, and a cumulative electron dose of ~180 e/A2 or less for each tilt series.
Length measurements.
The IMOD software package was used to calculate three-dimensional (3D) reconstructions of tilt series (Kremer et al., 1996). Alternatively, the images were aligned and contrast transfer function corrected using IMOD software package before producing SIRT reconstructions using the TOMO3D program (Agulleiro and Fernandez, 2011). CdiA structures on cell envelopes were located by visual inspection. Using the IMOD software package, a stack of 10 slices displaying the best view for each structure was used to measure lengths (by eye) from the center of the outer membrane to the tip of the filament. All filaments were measured separately by three individuals. The mean lengths within each particular dataset (as determined by the three individuals) did not differ with statistical significance (p-values > 0.05 in all instances). In contrast, the mean lengths of unbound CdiA versus BamA(micelle)-bound CdiA, and unbound versus BamA(vesicle)-bound measurements were significantly different (p-values < 0.00001 in both case for all three individuals).
Protease protection, SDS-PAGE and immunoblotting.
E. coli CH14016 cells carrying plasmid pCH13604 were diluted to OD600 ~ 0.05 in LB medium supplemented with Amp and cultured with shaking at 37 °C. Once in mid-log phase, cultures were treated with Spc for 20 min to block protein synthesis. Cells were harvested by centrifugation and re-suspended in 1× phosphate buffered saline (PBS) supplemented with 1 mM MgSO4 (PBS-Mg). Proteases were added to 20 μg/mL, and the cell suspensions incubated at ambient temperature for 20 min. Cells were collected by centrifugation and washed four times with PBS-Mg supplemented with 6 mM 2-mercaptoethanol (2-ME), 2 mM phenylmethanesulfonyl fluoride. Cell pellets were re-suspended in urea-lysis buffer [50% urea, 150 mM NaCl, 20 mM Tris-HCl (pH 8.0)] and subjected a freeze-thaw cycle to extract proteins for SDS-PAGE and immunoblotting.
Urea-soluble protein samples (5 μL) were analyzed by SDS-PAGE on Tris-tricine 6% polyacrylamide gels run at 100 V (constant) for 3 h. Gels were soaked for 15 min in 25 mM Tris, 192 mM glycine (pH 8.6), 10% methanol, then electroblotted to low-fluorescence PVDF membranes using a semi-dry transfer apparatus at 17 V (constant) for 1 h. Membranes were blocked with 4% non-fat milk in PBS for 1 h at room temperature, and incubated with primary antibodies in 0.1% non-fat milk, PBS overnight at 4 °C. Rabbit polyclonal antisera (Coca lico Biologicals, Stevens, PA) to the N-terminal TPS domain was used at a 1:10,000 dilution and anti-PT/CdiA-CT antisera was used at a 1:3,000 dilution. Blots were incubated with 800CW-conjugated goat anti-rabbit IgG (1:40,000 dilution, LICOR) in 0.1% non-fat milk in PBS. Immunoblots were visualized with a LI-COR Odyssey infrared imager.
To analyze culture supernatants for released CdiASTECO31 chains, cells were grown to mid-log phase in LB media supplemented with Amp. Protein synthesis was then blocked with Spc and the culture incubated with shaking for 20 min at 37 °C. Cells were collected by centrifugation at 3,000 × g for 5 min, and 900 μL of the supernatant was added to 100 μL of trichloroacetic acid. Proteins were collected by centrifugation at 18,000 × gfor 30 min at 4 °C. Precipitates were washed thrice with 1.0 ml of cold acetone. Air-dried pellets were dissolved in 45 μL of urea-lysis buffer, and 5 μL of each sample analyzed by SDS-PAGE and immunoblotting.
Maleimide-dye labeling to map surface topology.
E. coli CH14016 cells carrying CdiASTECO31 expression plasmids were diluted to OD600 ~ 0.05 in Amp supplemented LB media and grown to mid-log phase at 37 °C. Cultures were then treated with Spc for 20 m in to block protein synthesis. Cells were harvested by centrifugation and re-suspended at OD600 ~ 0.3 in 1.0 mL of: i) PBS-Mg to probe intact cells; ii) PBS-Mg with 100 μg/mL polymyxin B to probe permeabilized cells; or iii) PBS-Mg supplemented with 100 μg/mL polymyxin B, 100 μg/mL lysozyme, 250 U/mL benzonase to probe lysed cells. IRDye680LT-maleimide (LI-COR) was added to a final concentration of 40 μM for intact cells and 120 μM for permeabilized/lysed cells. Labeling reactions were incubated in the dark at room temperature for 15 min, then quenched with 6 mM 2-ME. Cells were collected by centrifugation and washed with PBS-Mg supplemented with 6 mM 2-ME. Cell pellets were re-suspended in 50 μL of urea-lysis buffer and subjected to one freeze-thaw cycle to extract proteins. Urea-soluble proteins (5 μL) were resolved by SDS-PAGE and analyzed by immunoblotting using anti-TPS domain antisera. 680LT (dye-maleimide) and 800CW (secondary antibody) fluorescence was visualized using a LI-COR Odyssey infrared imager. His6-tagged MBP was purified from urea-solubilized lysates by Ni2+-affinity chromatography prior to analysis by SDS-PAGE and blotting.
The same general procedure was used to monitor changes in CdiASTECO31 topology in response to target bacteria. E. coli CH14016 inhibitors that express CdiASTECO31 variants were mixed with E. coli CH14016 target cells that carry pCH450 (Δtsx) or pCH13603 (tsx+) at a 1:2 ratio in fresh, pre-warmed LB media supplemented with Spc to block new protein synthesis. After 20 min with shaking, the cell mixtures were collected by centrifugation and re-suspended at OD600 ~ 0.3 in 1.0 mL of PBS-Mg for labeling with 40 μM IRDye680LT-maleimide. Labeling reactions were quenched and proteins extracted for analysis as described above.
Cell mixtures were washed once in PBS-Mg and resuspended in 0.5 mL PBS-Mg. Maleimide dye was added to 40 μM, followed by the addition of proteinase K at 20 μg/mL. Suspensions were incubated at ambient temperature in the dark for 20 min, then quenched with 15 mM 2-ME and 2 mM PMSF. Cells were washed thrice with PBS-Mg supplemented with 15 mM 2-ME, 2 mM PMSF, followed by one wash with PBS-Mg. Cells pellets were resuspended in urea lysis buffer for SDS-PAGE and immunoblot analyses. For sucrose-gradient fractionation, the proteinase K treated cells were resuspended in 1.0 mL PBS-Mg supplemented with 200 μg/mL lysozyme, 50 U/mL benzonase and HALT protease inhibitor cocktail (Thermo-Fisher). Suspensions were subjected to one freeze-thaw, then broken with three French press passages. Lysates were incubated at ambient temperature in the dark for 10 min, then 400 μL was layered onto a 3.6 mL linear 10 – 60% sucrose gradient. Samples were centrifuged at ~150,000 × g for 1 h, and 12 fractions were collected by drip from a puncture in the bottom of the centrifuge tube. Proteins were precipitated by adjusting each fraction to 90% (vol/vol) ethanol. Precipitates were dissolved in urea lysis buffer for SDS-PAGE and immunoblot analyses.
Competition co-cultures.
E. coli CH14016 (Δwzb Δtsx) and CH14017 (Δwzb Δtsx ΔompT) that harbor CdiASTECO31 expression plasmids were used as inhibitor cells in shaking broth co-cultures. E. coli CH14016 and CH14017 strains that carry pCH10145 (Δtsx) or pZR428 (tsx+) were used as target cells. Cells were grown in LB media at 37 °C to mid-log ph ase, adjusted to OD600 ~ 0.3 in fresh pre-warmed LB media without antibiotics, mixed at a 1:1 ratio (5 mL total volume) and incubated with shaking for 3 h at 37 °C. Culture aliquots were taken at the beginn ing of co-culture and after 3 h to quantify viable inhibitor and target cells as colony forming units per milliliter (CFU mL−1). For solid media growth competitions, E. coli CH14017 cells (Δwzb Δtsx ΔompT) that carry pCH10145 (Δtsx) or pZR428 (tsx+) were used as target bacteria. Inhibitor and target cells were grown in LB media at 37 °C to mid-log phase, adjusted to OD600 ~ 1 in 1× M9 salts, and mixed at a 1:1 ratio (100 μL total volume). Samples of the mixed-cell suspensions (10 μL) were spotted onto LB-agar and incubated at 37 °C. After 3 h, cells were harvested using polyester-tipped applicators and resuspended in 500 μL of 1× M9 salts. For all competitions, cell suspensions were serially diluted into 1× M9 salts and plated onto LB-agar supplemented with Amp to enumerate inhibitor cells, and LB-agar supplemented with Kan to enumerate target cells. Competitive indices were calculated as the ratio of target to inhibitor cells at 3 h divided by the initial target to inhibitor cell ratio.
BamA purification.
BamA from E. coli was over-produced in E. coli strain CH2016 carrying plasmid pCH9216 (Ruhe et al., 2017). Cells were grown at 37 °C in LB media supplemented with Amp and BamA production induces with 1.5 mM IPTG for 3 h. Cells were harvested by centrifugation at 6,000 × g for 10 min, then re-suspended in 5 mL of BugBuster reagent and broken by three passages through a French press. The lysate was diluted with 25 mL deionized water and vortexed vigorously. Inclusion bodies were collected by centrifugation at 15,000 × g for 20 min and washed thrice with 5 mL of 0.1 × BugBuster solution. The washed inclusion bodies were dissolved in 0.5 mL of urea-lysis buffer supplemented with 0.05% Triton X-100, then diluted into 50 mL 10 mM Tris-HCl (pH 8.0), 0.5% Triton X-100 to a final concentration of 140 μM. Diluted protein stocks were incubated on a rotisserie for three days at ambient temperature to refold. The refolding reaction was then stored at 4 °C. Greater than 95% of BamA was refolded as determined by heat-modifiable gel mobility as previously described (Robert et al., 2006).
Cell-cell adhesion by flow cytometry.
Tsx-binding studies were conducted using E. coli strain DL4259, which expresses gfp-mut3 from the papBA promoter (Webb et al., 2013). E. coli DL4259 cells were transformed with pCH450-derived CdiASTECO31 expression plasmids and the resulting strains grown at 37 °C in LB media supplemented with Tet until the cells became fluorescent. E. coli ZR373 cells carrying plasmid pDsRedExpress2 were used as targets. Targets were also provided with plasmid pCH450 (Δtsx) or pCH13603 (tsx+). Target cells were grown overnight in LB supplemented with Tet, Amp, 0.4% L-arabinose and 1 mM IPTG to induce expression of Tsx and DsRed prior to mixing with inhibitors. GFP-labeled inhibitor cells were mixed at a 5:1 ratio with DsRed-labeled target bacteria at a final OD600 ~ 0.2. Cell suspensions were shaken at 30 °C for 2 0 min, diluted into 1× PBS, vortexed briefly, then analyzed on an Accuri C6 flow cytometer using FL1 (533/30nm, GFP) and FL2 (585/40nm, DsRed) fluorophore filters. The fraction of target bacteria bound to inhibitor cells was calculated as the number of dual green/red fluorescent events divided by the total number of red fluorescent events. Three independent experiments were performed on separate days, and >2,000 red fluorescent events were scored for each cell mixture per experiment.
CdiA domain analyses.
CdiASTECO31 homologs were identified by BLAST using residues Asn1637 – Asn1902 as a query to search enterobacterial genera. The query sequence includes the FHA-1 peptide immediately adjacent to the RBD and extends through the periplasmic FHA-1 repeats (pFR). 142 full-length CdiA proteins ranging from 2,963 to 6,442 residues were identified for analysis (Table S2). The number of residues comprising each RBD was plotted as a function of the length of the FHA-1 region. The number of residues comprising each FHA-2 domain was determined from automated annotations on the NCBI website (Table S2) and plotted as a function of FHA-1 length. The FHA-2 domain of CdiASTECO31 (residues Leu1941 to Met2560) was submitted to the I-TASSER server for protein structure and function predictions (Yang et al., 2015).
Quantification and statistical analysis
Statistical parameters for each experiment are reported in the figure legends and Method Details.
Supplementary Material
A) Filaments (white arrows) on the surface of E. coli EC93 cells. B) CdiAEC93 filaments on the surface of an E. coli K-12 cell. C) CdiAEC93 filaments on the surface of an E. coli K-12 minicell. Scale bars = 100 nm for A, B & C. D) Bent CdiAEC93 filaments. E) Twinned and clustered filaments. Scale bars = 50 nm for D & E.
A & B) Inhibitors were mixed at a 1:1 ratio with tsx+ or Δtsx targets and incubated in shaking LB media. Competitive indices equal the final ratio of targets to inhibitors divided by the initial ratio. Data are presented as mean ± SEM for three independent experiments.
CdiA sequences were aligned using Clustal Omega. FHA-1 sequences are in blue bold-face. CdiASTECO31 and CdiAEC93 RBD sequences are shown in salmon and pink, respectively. The YP domain is shown in black bold-face for CdiASTECO31 and CdiAEC93, beginning with the YPLP peptide motif and extending to the periplasmic FHA-1 repeats (pFR). Cys-substituted positions (Ser1550, Ser1693, Gly1726 and Ala1807) in CdiASTECO31 are underlined and highlighted in yellow. CdiA sequences are from Pectobacterium carotovorum F152 (NCBI accession: PPE64673.1), Pectobacterium parmentieri WPP163 (ACX88282.1), Pectobacterium maceratum PB70 (POY59994.1), Brenneria goodwinii RBB141 (WP_095834043.1), Cedecea neteri NBRC 105707 (WP_061277518.1), Enterobacter cloacae MGH 54 (WP_084832630.1), Shigella flexneri 1235-66 (EIQ74285.1), Pantoea sp. OV426 (SFN23123.1), Dickeya chrysanthemi EC16 (AAN38708.1), Escherichia coli STEC_O31 (WP_001385946.1), Escherichia coli EC93 (AAZ57198.1), Serratia proteamaculans 568 (WP_012147097.1), and Yersinia mollaretii ATCC 43969 (WP_004876812.1).
Amino acid residues were counted within a sliding 40-residue window along the length of CdiASTECO31. Domains are color-coded: TPS transport (green), FHA-1 (blue), RBD (maroon), FHA-2 (orange), and CdiA-CT (purple).The PT domain corresponds to the region between the dotted line and the CdiA-CT.
CdiA expressing cells were mixed with tsx+ ompT+ or tsx+ ΔompT targets. Cell pellets and culture supernatants were analyzed by immunoblotting with antibodies to the TPS domain and the PT/CdiA-CT region of CdiASTECO31. White carets indicate C-terminal CdiASTECO31 fragments.
Movie S1. 3D reconstruction of E. coli cell expressing CdiAEC93, Related to Figure 1B. Scale bar = 100 nm.
Movie S2. 3D reconstruction of E. coli minicell expressing CdiAEC93, Related to Figure 1C. Scale bar = 100 nm.
Movie S3. 3D reconstruction of E. coli minicell expressing CdiASTECO31, Related to Figure 1D. Scale bar = 100 nm.
Movie S4. 3D reconstruction of E. coli minicell lacking CdiA expression construct, Related to Figure 1. Scale bar = 100 nm.
Movie S5. 3D reconstruction of E. coli minicell with CdiAEC93 bound to detergent solubilized BamA, Related to Figure 3A. Scale bar = 100 nm.
Table S1. Tomograms collected, Related to Figures 1 and 3.
Table S2. CdiA domain analysis, Related to Figures 7B and 7C.
Table S3. Oligonucleotides, Related to Figure STAR Methods.
Acknowledgments
We thank Chuan Hong, Rick Huang and Zhiheng Yu at the HHMI Janelia CryoEM Facility for help with the Titan Krios microscope operation and data collection, and Thomas Silhavy for providing antisera. This work was supported by grants GM117930 (C.S.H.) and GM122588 (G.J.J.) from National Institutes of Health and grant MCB 1545720 (C.S.H. & D.A.L) from the National Science Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
Data and software availability
Individual fluorescent channel data for all immunoblots and the I-TASSER server output for FHA-2 structure predictions are available at http://dx.doi:10.17632/hrbmtfv754.1.
References
- Agulleiro JI, and Fernandez JJ (2011). Fast tomographic reconstruction on multicore computers. Bioinformatics 27, 582–583. [DOI] [PubMed] [Google Scholar]
- Albrecht R, Schutz M, Oberhettinger P, Faulstich M, Bermejo I, Rudel T, Diederichs K, and Zeth K (2014). Structure of BamA, an essential factor in outer membrane protein biogenesis. Acta Crystallogr D Biol Crystallogr 70, 1779–1789. [DOI] [PubMed] [Google Scholar]
- Aoki SK, Diner EJ, de Roodenbeke CT, Burgess BR, Poole SJ, Braaten BA, Jones AM, Webb JS, Hayes CS, Cotter PA, et al. (2010). A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature 468, 439–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki SK, Malinverni JC, Jacoby K, Thomas B, Pamma R, Trinh BN, Remers S, Webb J, Braaten BA, Silhavy TJ, et al. (2008). Contact-dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) as the receptor and the inner membrane transport protein AcrB. Mol Microbiol 70, 323–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki SK, Pamma R, Hernday AD, Bickham JE, Braaten BA, and Low DA (2005). Contact-dependent inhibition of growth in Escherichia coli. Science 309, 1245–1248. [DOI] [PubMed] [Google Scholar]
- Aoki SK, Poole SJ, Hayes CS, and Low DA (2011). Toxin on a stick: modular CDI toxin delivery systems play roles in bacterial competition. Virulence 2, 356–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki SK, Webb JS, Braaten BA, and Low DA (2009). Contact-dependent growth inhibition causes reversible metabolic downregulation in Escherichia coli. J Bacteriol 191, 1777–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, and Mori H (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2, 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler M, Pilhofer M, Henderson GP, Jensen GJ, and Mekalanos JJ (2012). Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483, 182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CM, Willett JL, Cunningham DA, Kim JJ, Low DA, and Hayes CS (2016). CdiA Effectors from Uropathogenic Escherichia coli Use Heterotrimeric Osmoporins as Receptors to Recognize Target Bacteria. PLoS Pathog 12, e1005925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Casabona MG, Kneuper H, Chalmers JD, and Palmer T (2016). The type VII secretion system of Staphylococcus aureus secretes a nuclease toxin that targets competitor bacteria. Nat Microbiol 2, 16183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K, Riley M, Slatin S, and Cavard D (2007). Colicin biology. Microbiol Mol Biol Rev 71, 158–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov PP, and Wackernagel W (1995). Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158, 9–14. [DOI] [PubMed] [Google Scholar]
- Clantin B, Hodak H, Willery E, Locht C, Jacob-Dubuisson F, and Villeret V (2004). The crystal structure of filamentous hemagglutinin secretion domain and its implications for the two-partner secretion pathway. Proc Natl Acad Sci U S A 101, 6194–6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bayona L, Guo MS, and Laub MT (2017). Contact-dependent killing by Caulobacter crescentus via cell surface-associated, glycine zipper proteins. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza-Sanchez F, Schaub RE, Janssen BD, and Hayes CS (2011). tmRNA regulates synthesis of the ArfA ribosome rescue factor. Mol Microbiol 80, 1204–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes CS, and Sauer RT (2003). Cleavage of the A site mRNA codon during ribosome pausing provides a mechanism for translational quality control. Mol Cell 12, 903–911. [DOI] [PubMed] [Google Scholar]
- Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, et al. (2010). A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housden NG, and Kleanthous C (2012). Colicin translocation across the Escherichia coli outer membrane. Biochem Soc Trans 40, 1475–1479. [DOI] [PubMed] [Google Scholar]
- Iancu CV, Tivol WF, Schooler JB, Dias DP, Henderson GP, Murphy GE, Wright ER, Li Z, Yu Z, Briegel A, et al. (2006). Electron cryotomography sample preparation using the Vitrobot. Nat Protoc 1, 2813–2819. [DOI] [PubMed] [Google Scholar]
- Jamet A, Jousset AB, Euphrasie D, Mukorako P, Boucharlat A, Ducousso A, Charbit A, and Nassif X (2015). A new family of secreted toxins in pathogenic Neisseria species. PLoS Pathog 11, e1004592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajava AV, Cheng N, Cleaver R, Kessel M, Simon MN, Willery E, Jacob-Dubuisson F, Locht C, and Steven AC (2001). Beta-helix model for the filamentous haemagglutinin adhesin of Bordetella pertussis and related bacterial secretory proteins. Mol Microbiol 42, 279–292. [DOI] [PubMed] [Google Scholar]
- Koskiniemi S, Garza-Sanchez F, Edman N, Chaudhuri S, Poole SJ, Manoil C, Hayes CS, and Low DA (2015). Genetic analysis of the CDI pathway from Burkholderia pseudomallei 1026b. PLoS One 10, e0120265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskiniemi S, Lamoureux JG, Nikolakakis KC, t'Kint de Roodenbeke C, Kaplan MD, Low DA, and Hayes CS (2013). Rhs proteins from diverse bacteria mediate intercellular competition. Proc Natl Acad Sci U S A 110, 7032–7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer RA, Vandeputte-Rutten L, de Roon GJ, Gros P, Dekker N, and Egmond MR (2001). Identification of essential acidic residues of outer membrane protease OmpT supports a novel active site. FEBS Lett 505, 426–430. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, and McIntosh JR (1996). Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116, 71–76. [DOI] [PubMed] [Google Scholar]
- MacIntyre DL, Miyata ST, Kitaoka M, and Pukatzki S (2010). The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc Natl Acad Sci U S A 107, 19520–19524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhov AM, Hannah JH, Brennan MJ, Trus BL, Kocsis E, Conway JF, Wingfield PT, Simon MN, and Steven AC (1994). Filamentous hemagglutinin of Bordetella pertussis. A bacterial adhesin formed as a 50-nm monomeric rigid rod based on a 19-residue repeat motif rich in beta strands and turns. J Mol Biol 241, 110–124. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN (2006). Fiducial marker and hybrid alignment methods for single- and double-axis tomography In Electron Tomography Frank J, ed. (New York, NY: Springer; ), pp. 163–185. [Google Scholar]
- Melvin JA, Scheller EV, Noel CR, and Cotter PA (2015). New Insight into Filamentous Hemagglutinin Secretion Reveals a Role for Full-Length FhaB in Bordetella Virulence. MBio 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalska K, Nhan DQ, Willett JLE, Stols LM, Eschenfeldt WH, Jones AM, Nguyen JY, Koskiniemi S, Low DA, Goulding CW, et al. (2018). Functional plasticity of antibacterial EndoU toxins. Mol Microbiol June 20. doi: 10.1111/mmi.14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil RB, and Apicella MA (2009). Role of HrpA in biofilm formation of Neisseria meningitidis and regulation of the hrpBAS transcripts. Infect Immun 77, 2285–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel CR, Mazar J, Melvin JA, Sexton JA, and Cotter PA (2012). The prodomain of the Bordetella two-partner secretion pathway protein FhaB remains intracellular yet affects the conformation of the mature C-terminal domain. Mol Microbiol 86, 988–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak DT, Wei X, Dey A, and Wall D (2013). Molecular recognition by a polymorphic cell surface receptor governs cooperative behaviors in bacteria. PLoS Genet 9, e1003891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relman DA, Domenighini M, Tuomanen E, Rappuoli R, and Falkow S (1989). Filamentous hemagglutinin of Bordetella pertussis: nucleotide sequence and crucial role in adherence. Proc Natl Acad Sci U S A 86, 2637–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert V, Volokhina EB, Senf F, Bos MP, Van Gelder P, and Tommassen J (2006). Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol 4, e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas CM, Ham JH, Deng WL, Doyle JJ, and Collmer A (2002). HecA, a member of a class of adhesins produced by diverse pathogenic bacteria, contributes to the attachment, aggregation, epidermal cell killing, and virulence phenotypes of Erwinia chrysanthemi EC16 on Nicotiana clevelandii seedlings. Proc Natl Acad Sci U S A 99, 13142–13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhe ZC, Nguyen JY, Beck CM, Low DA, and Hayes CS (2014). The proton-motive force is required for translocation of CDI toxins across the inner membrane of target bacteria. Mol Microbiol 94, 466–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhe ZC, Nguyen JY, Xiong J, Koskiniemi S, Beck CM, Perkins BR, Low DA, and Hayes CS (2017). CdiA effectors use modular receptor-binding domains to recognize target bacteria. MBio 8, e00290–00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhe ZC, Townsley L, Wallace AB, King A, Van der Woude MW, Low DA, Yildiz FH, and Hayes CS (2015). CdiA promotes receptor-independent intercellular adhesion. Mol Microbiol 98, 175–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhe ZC, Wallace AB, Low DA, and Hayes CS (2013). Receptor polymorphism restricts contact-dependent growth inhibition to members of the same species. MBio 4, e00480–00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, Peterson SB, and Mougous JD (2014). Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol 12, 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller EV, and Cotter PA (2015). Bordetella filamentous hemagglutinin and fimbriae: critical adhesins with unrealized vaccine potential. Pathog Dis 73, ftv079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi D, Toyoda A, Aizu T, Ejima F, Fujiyama A, Shini T, Kohara Y, and Niki H (2013). Mutations in cell elongation genes mreB, mrdA and mrdB suppress the shape defect of RodZ-deficient cells. Mol Microbiol 87, 1029–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza DP, Oka GU, Alvarez-Martinez CE, Bisson-Filho AW, Dunger G, Hobeika L, Cavalcante NS, Alegria MC, Barbosa LR, Salinas RK, et al. (2015). Bacterial killing via a type IV secretion system. Nat Commun 6, 6453. [DOI] [PubMed] [Google Scholar]
- Tivol WF, Briegel A, and Jensen GJ (2008). An improved cryogen for plunge freezing. Microsc Microanal 14, 375–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeputte-Rutten L, Kramer RA, Kroon J, Dekker N, Egmond MR, and Gros P (2001). Crystal structure of the outer membrane protease OmpT from Escherichia coli suggests a novel catalytic site. EMBO J 20, 5033–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassallo C, Pathak DT, Cao P, Zuckerman DM, Hoiczyk E, and Wall D (2015). Cell rejuvenation and social behaviors promoted by LPS exchange in myxobacteria. Proc Natl Acad Sci U S A 112, E2939–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassallo CN, Cao P, Conklin A, Finkelstein H, Hayes CS, and Wall D (2017). Infectious polymorphic toxins delivered by outer membrane exchange discriminate kin in myxobacteria. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb JS, Nikolakakis KC, Willett JLE, Aoki SK, Hayes CS, and Low DA (2013). Delivery of CdiA nuclease toxins into target cells during contact-dependent growth inhibition. PLoS oNe 8, e57609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney JC, Peterson SB, Kim J, Pazos M, Verster AJ, Radey MC, Kulasekara HD, Ching MQ, Bullen NP, Bryant D, et al. (2017). A broadly distributed toxin family mediates contact-dependent antagonism between gram-positive bacteria. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett JL, Gucinski GC, Fatherree JP, Low DA, and Hayes CS (2015a). Contact-dependent growth inhibition toxins exploit multiple independent cell-entry pathways. Proc Natl Acad Sci U S A 112, 11341–11346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett JL, Ruhe ZC, Goulding CW, Low DA, and Hayes CS (2015b). Contact-Dependent Growth Inhibition (CDI) and CdiB/CdiA Two-Partner Secretion Proteins. J Mol Biol 427, 3754–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, and Zhang Y (2015). The I-TASSER Suite: protein structure and function prediction. Nat Methods 12, 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, and van den Berg B (2004). Crystal structure of the bacterial nucleoside transporter Tsx. EMBO J 23, 3187–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo HJ, Yokoyama T, Walkiewicz K, Kim Y, Grass S, and Geme JW 3rd (2007). The structure of the Haemophilus influenzae HMW1 pro-piece reveals a structural domain essential for bacterial two-partner secretion. J Biol Chem 282, 31076–31084. [DOI] [PubMed] [Google Scholar]
- Zhang D, de Souza RF, Anantharaman V, Iyer LM, and Aravind L (2012). Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct 7, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Iyer LM, and Aravind L (2011). A novel immunity system for bacterial nucleic acid degrading toxins and its recruitment in various eukaryotic and DNA viral systems. Nucleic Acids Res 39, 4532–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Keszthelyi B, Branlund E, Lyle JM, Braunfeld MB, Sedat JW, and Agard DA (2007). UCSF tomography: an integrated software suite for real-time electron microscopic tomographic data collection, alignment, and reconstruction. J Struct Biol 157, 138–147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Filaments (white arrows) on the surface of E. coli EC93 cells. B) CdiAEC93 filaments on the surface of an E. coli K-12 cell. C) CdiAEC93 filaments on the surface of an E. coli K-12 minicell. Scale bars = 100 nm for A, B & C. D) Bent CdiAEC93 filaments. E) Twinned and clustered filaments. Scale bars = 50 nm for D & E.
A & B) Inhibitors were mixed at a 1:1 ratio with tsx+ or Δtsx targets and incubated in shaking LB media. Competitive indices equal the final ratio of targets to inhibitors divided by the initial ratio. Data are presented as mean ± SEM for three independent experiments.
CdiA sequences were aligned using Clustal Omega. FHA-1 sequences are in blue bold-face. CdiASTECO31 and CdiAEC93 RBD sequences are shown in salmon and pink, respectively. The YP domain is shown in black bold-face for CdiASTECO31 and CdiAEC93, beginning with the YPLP peptide motif and extending to the periplasmic FHA-1 repeats (pFR). Cys-substituted positions (Ser1550, Ser1693, Gly1726 and Ala1807) in CdiASTECO31 are underlined and highlighted in yellow. CdiA sequences are from Pectobacterium carotovorum F152 (NCBI accession: PPE64673.1), Pectobacterium parmentieri WPP163 (ACX88282.1), Pectobacterium maceratum PB70 (POY59994.1), Brenneria goodwinii RBB141 (WP_095834043.1), Cedecea neteri NBRC 105707 (WP_061277518.1), Enterobacter cloacae MGH 54 (WP_084832630.1), Shigella flexneri 1235-66 (EIQ74285.1), Pantoea sp. OV426 (SFN23123.1), Dickeya chrysanthemi EC16 (AAN38708.1), Escherichia coli STEC_O31 (WP_001385946.1), Escherichia coli EC93 (AAZ57198.1), Serratia proteamaculans 568 (WP_012147097.1), and Yersinia mollaretii ATCC 43969 (WP_004876812.1).
Amino acid residues were counted within a sliding 40-residue window along the length of CdiASTECO31. Domains are color-coded: TPS transport (green), FHA-1 (blue), RBD (maroon), FHA-2 (orange), and CdiA-CT (purple).The PT domain corresponds to the region between the dotted line and the CdiA-CT.
CdiA expressing cells were mixed with tsx+ ompT+ or tsx+ ΔompT targets. Cell pellets and culture supernatants were analyzed by immunoblotting with antibodies to the TPS domain and the PT/CdiA-CT region of CdiASTECO31. White carets indicate C-terminal CdiASTECO31 fragments.
Movie S1. 3D reconstruction of E. coli cell expressing CdiAEC93, Related to Figure 1B. Scale bar = 100 nm.
Movie S2. 3D reconstruction of E. coli minicell expressing CdiAEC93, Related to Figure 1C. Scale bar = 100 nm.
Movie S3. 3D reconstruction of E. coli minicell expressing CdiASTECO31, Related to Figure 1D. Scale bar = 100 nm.
Movie S4. 3D reconstruction of E. coli minicell lacking CdiA expression construct, Related to Figure 1. Scale bar = 100 nm.
Movie S5. 3D reconstruction of E. coli minicell with CdiAEC93 bound to detergent solubilized BamA, Related to Figure 3A. Scale bar = 100 nm.
Table S1. Tomograms collected, Related to Figures 1 and 3.
Table S2. CdiA domain analysis, Related to Figures 7B and 7C.
Table S3. Oligonucleotides, Related to Figure STAR Methods.