ABSTRACT
Cross-kingdom gene regulation by microRNAs (miRNAs) initiated a hot debate on the effective role of orally acquired plant miRNAs on human gene expression. It resulted in the expansion of gene regulation theories and role of plant miRNAs in cross-kingdom regulation of gene expression. This opened up the discussion that ‘Whether we really get what we eat?’ and ‘Whether the orally acquired miRNAs really have a biologically important consequences after entering our digestive and circulatory system?’ The reports of orally acquired plant miRNAs inside human alimentary canal have been a topic of discussion in the scientific community. The cross-kingdom gene regulations have raised our hopes to explore the exciting world of plant miRNAs as therapeutic potential and dietary supplements. However, there are reports which have raised concerns over any such cross-kingdom regulation and argued that technical flaws in the experiments might have led to such hypothesis. This review will give the complete understanding of exogenous application and cross-kingdom regulation of plant miRNAs on human health. Here, we provide update and discuss the consequences of plant miRNA mediated cross-kingdom gene regulation and possibilities for this exciting regulatory mechanism as an augmented therapy against various diseases.
KEYWORDS: miRNA, dietary intake, therapeutics, cross-kingdom, gene expression regulation, exogenous
Introduction
MicroRNAs (miRNAs) are global regulators of gene expression at both transcriptional and post-transcriptional level resulting into pre- and post-transcriptional gene silencing (PTGS) [1]. miRNAs are 21–24 nt long RNA molecules which bind to their targets like RNA and DNA. miRNAs guide miRISC (miRNA-induced silencing complex) to specifically recognize messenger RNA (mRNA) and up- or down-regulate the expression of genes by one of the two post-transcriptional regulatory mechanisms: mRNA cleavage and translational repression [2]. It is reported that about 60% of protein coding genes are the targets and modulated by miRNAs [3]. The endogenous role of plant miRNAs are varied from plant development, signal transduction, stress responses, secondary plant product biosynthesis to plant diseases [4,5]. These miRNAs regulate primary and secondary metabolic pathways through cascade of genetic crosstalk. Most of the miRNAs have regulatory roles at endogenous level. The development of fruits and their ripening is also reported to be regulated by endogenous miRNAs in tomato [6] and banana [7].
Recent reports have presented the roles of miRNAs at another aspect i.e. exogenous regulation. For exogenous path, the miRNAs are reported to be stably encapsulated into P bodies (cytoplasmic bodies), multi vesicular bodies (MVBs) and endosomes for intracellular and intercellular transport [8,9]. The P bodies are formed for the purpose of storage of translationally repressed mRNAs and their disassembly has shown to trigger the activation/inhibition of cellular processes by the released miRNAs in animals. The MVBs and endosomes carry miRNAs and then transport these to other cellular compartments. This provides an opportunity to look into the cellular localization of orally acquired plant miRNAs and to study the stochastic interactions in the complex cellular environment. The plant exogenous miRNA and host mRNA interactions might result into the alterations in genetic regulation of host cellular machinery where these orally acquired plant miRNAs find their complementary targets and modulate the transcriptional or post-transcriptional processes. At the structural level, 2ʹ-O-methylation at 3ʹend of plant miRNAs enhances the stability of plant miRNAs for the survival in various environments due to its exogenous activity [10]. The structural stability of plant miRNAs facilitates to withstand in adverse conditions inside the gastrointestinal (GI) tract such as RNases, phagocytosis and low pH [11].
According to WHO, more than 70% population of developing countries rely upon the traditional medicinal sources for the treatment of their ailments due to paucity of modern medical facilities. Even today, we are relying on the traditional medicinal databases in our quest to counter the diseases like AIDS, cancer, dengue, malaria, psoriasis as well as chronic musculoskeletal disorders. At the same time, some forms of complementary medicine (CM) such as chiropractic, homeopathy, naturopathy, osteopathy and anthroposophic medicine are also in extensive use. According to the WHO Traditional Medicine Strategy 2014–2023, the traditional and complementary medicines have been globally accepted as an alternative form of therapy to various chronic disorders and diseases. The very success of Mongolian TM family medicinal kit project in treatment and control of various diseases is one of the attempts by WHO to promote the use of traditional medicine as augmented therapy. Recent reports on use of mango and pomegranate in prevention of cancer and anti-inflammatory activity have paved the path for probe into the implementation of traditional fruit plants to support augmented therapy [12,13]. Most of the medicinal properties of plant or plant parts have been linked to secondary plant products synthesized and accumulated in specific plants [14–18]. Surprisingly, role of other biomolecules such as miRNAs has not been considered in plant-specific medicinal properties or augmented therapy. Although role of human’s own miRNAs in various diseases have been reported earlier [19,20]. Numerous updations of plant miRNAs from different families of plant kingdom in miRBase [21] have provided a firm ground to the proposition of speculating the role of plant miRNAs as a potential strategy in augmented therapy.
Plant and animal miRNAs biogenesis and target specificity: similarities and dissimilarities
The miRNAs found extensively in both plants and animals and regulate the gene expression through interaction in a sequence specific manner with their corresponding target mRNAs. Despite the similarities, there are differences in plant and animal miRNAs with respect to their biogenesis, function and evolution [22]. In both the systems, miRNAs are first transcribed as primary transcripts (pri-miRNA) with 5ʹ capping and polyadenylation at 3ʹ end by RNApolymerase II and III from the gene. The precursor miRNAs (pre-miRNAs) are transcribed from pri-miRNAs and form a short 70-nucleotide stable step-loop structure. The processing of pre-miRNA from pri-miRNAs in animals is carried out by Drosha protein whereas in plants RNase-III-like protein, Dicer-like 1 (DCL 1). DCL 1 protein further catalyzes pre-miRNA to miRNA:miRNA* duplex in nucleus of plants [23]. In contrary, the same step occurs in cytoplasm of animals but catalyzed by Dicer. In plants miRNA-miRNA* duplexes are transported to cytoplasm from nucleus with the help of exportin protein [24]. Further, the duplex get separated and the active (mature) strand of miRNA incorporates with RISC (RNA-induced silencing), protein complex. RICS protein acts as a guide for mature miRNA to recognize its complementary site of target gene (Fig 1).
Figure 1.
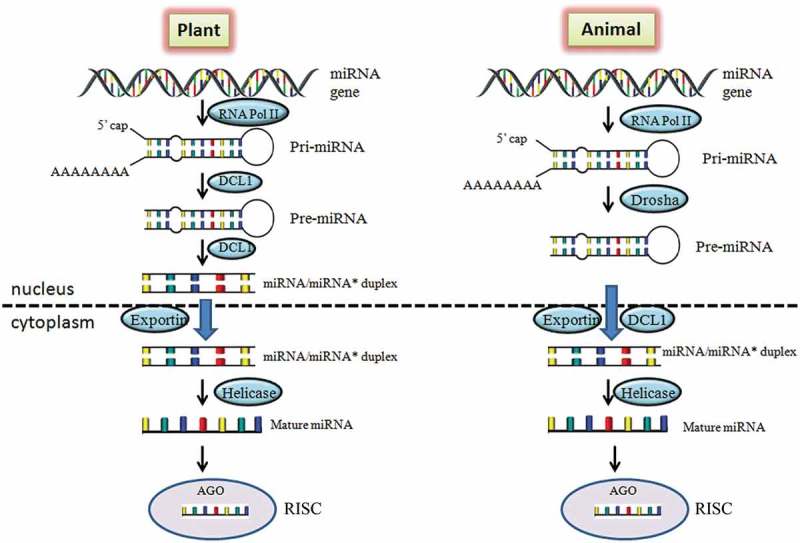
Comparison of miRNA biogenesis and function in plant and animal. In plants, the pri-miRNAs are synthesized from MIR genes through RNA Pol II and processed to pre-miRNA by DCL-1 protein which further leads to miRNA/miRNA* duplex formation. The duplex is transported from nucleus to cytoplasm and forms mature miRNAs. The mature miRNA binds with AGO protein to regulate the corresponding gene. While in animals, pre-miRNAs is transported to cytoplasm from nucleus and processes further same as in plants. DCL1, Dicer like 1 protein; AGO, Argonaute protein; RISC, RNA-induced silencing complex.
The location specificity for mature miRNA binding sites within target genes in plant and animals has been reviewed earlier [25]. In animals, miRNA binds at 3ʹ UTR and in plants at protein coding region of gene. However, later several reports came into existence for explaining the binding of miRNA in almost every sites of gene in both the kingdoms. In animals, the miRNA binding sites occur in 3ʹ UTR [26,27], 5ʹ UTR and coding region [28,29] of target gene. The miRNAs regulate the expression of their corresponding mRNAs through cleavage or translational repression mechanisms. Earlier, it was reported that cleavage occurs in plant miRNA and translational repression in animals [25]. However, in the present scenario both the mechanisms are reported in plants as well as animals [30]. The ultimate goal of miRNA interaction is to inhibit the formation of encoded proteins.
Plant miRNAs in cross-kingdom gene regulation
Among all organism derived miRNAs, the plant miRNAs are majorly reported to regulate cross-kingdom gene expression and cellular processes through dietary intake. This fact came into existence after the detection of plant miRNAs in mammalian specimens (human, mouse, mice, calf, rat, horse, sheep etc.) [31–33]. Studies demonstrated that plant miRNAs could survive exogenously in mature form in animals. No other forms such as double-stranded miRNA (dsmiR), precursor miRNA (pre-miR), single-stranded DNA miRNA (ssDNA-miR) and DNA precursor miRNA (pre-DNA-miR) are reported to be detected in cross-kingdom level [31]. As mature miRNAs are detected in mammalian species, importance of the plant miRNAs in traditional medicinal system as well as dietary food supplement can be a topic of interest. However, investigations have been started in this area very recently and limited information is available in this area. Recent studies suggested that the exogenous application of plant miRNAs could be taken up through food intake. Interestingly, plant miRNAs could be consumed from raw as well as cooked form [34] as these are stable even in the form of cooked food [31].
Exogenous role of plant miRNAs in mammals
The exogenous miRNAs of rice origin were demonstrated for the first time not only to be present in serum and plasma but also down-regulating the host genes [31]. The availability of rice miR156a and miR168a is reported from Chinese human serum and other tissues suggest the exogenous application of plant miRNAs. The isolated miRNAs were from rice which is the main diet of Chinese population [31]. No metabolic dysfunction was reported due to the presence of these rice miRNAs. It was also demonstrated that miR168a down-regulated the LDLRAP1 (low-density lipoprotein receptor adapter protein 1) gene through near-perfect complementarity results in the increase of plasma LDL cholesterol level in mammals. This supports the theory about the role of plant miRNAs in cross-kingdom gene regulation. In this study, miR168a miR156a showed higher levels whereas miR166a with moderate in animal serum and tissues revealing its higher stability as compared to other forms of nucleotides. These miRNAs were also investigated in other animals like mouse, rat, calf, horse and sheep serum [31]. In another report, the mice were ingested with miR172 through feeding of total RNA of Brassica oleracea mixed in gavage. It was demonstrated that miR172 survived in GI tract and entered to various organs passing through blood stream [32]. After examination, miR172 was found in stomach, intestine, spleen, liver kidney, serum, blood and feces of mice. During the experimentation, no phenotypic changes were observed in the mice when fed with these small RNAs.
The survival and persistence of plant miRNAs in animal GI tract was the topic of debate till 2014 [32]. However in the present scenario, there are a number of reports available suggesting the survival and persistence of plant miRNAs with in exogenous environment. In one report, the mice were fed with rapeseed (Brassica campestris) bee pollen and the plant miRNAs were detected its blood [33]. miR166a was identified to be highly enriched exogenous plant miRNA followed by miR159 which are absorbed by mice during feeding. It is also reported that the rapeseed bee pollen could also be used as supplement of plant miRNAs in addition to the form of food and healthy products for the prevention and treatment of diseases.
The antiviral activity of miRNA from honeysuckle (Lonicera periclymenum) plant has also been reported for miR2911 [35]. This miRNA is effective against different Influenza A viruses such as H1N1, H5N1 and H7N9 through accumulation in mouse lungs. The accumulation of miR2911 was detected through bright fluorescent dots in mouse lung sections. It was observed that in the presence of miR2911, the viral replication was inhibited in mouse influencing the prevention of weight loss and mortality due infection of viruses. Another research group has also studied the dietary role of miR29111 from Lonicera japonica in mice. After feeding the herb based diet to mice, the level of miR2911 was detected to be enhanced in sera and urine [36].
Transcription factor 7 (TCF7) is a transcription factor gene of Wnt signaling pathway and up-regulated in breast cancer. Recently, it was demonstrated that miR159 acts as breast cancer suppressor through targeting TCF7 gene. It is reported that the level of TCF7 genes is high in cancerous cell lines as compared to non-cancerous. miRNA159 was reported to be present in human serum and transported to breast tissues and able to reduce the transcriptional activity of TCF7 gene in breast cancer cell lines but not in non-cancerous [34]. The suppression of growth of xenograft breast tumors in mice was observed. This miRNA is present in all plants including Arabidopsis thaliana, Glycine max and Broccoli. It was the first report that plant miRNAs can inhibit the cancer growth in humans. Thus, plant miR159 could be a potential therapeutic agent for the treatment of breast cancer.
Recently, one more study supported cross-kingdom gene regulation by plant miRNAs. The human plasma small RNA sequencing data were analyzed for the identification of plant origin miRNAs. One miRNA, miR2910 conserved in fruits and vegetables was identified in relatively higher amount in human plasma samples supports the evidence that this miRNA reached through food ingestion. miR2910 from Populus euphratica was detected to be more abundant than most of the human miRNAs in the samples. miR2910 was identified in human plasma and targets Sprouty RTK Signaling Antagonist 4 (SPRY4) gene of the Janus kinase/signal transducers and activators of transcription (JAK-STAT) signaling pathway and transcription regulation gene indicating its role in human diseases [37]. Earlier reported miR168 [31] and miR159 [34] were also detected in the considered human samples even in small amount and given supporting evidence of the reports (Table 1). In a recent report on the presence of miR156a from green vegetables in human serum and blood has demonstrated their role in cardiovascular diseases (CVD). Through targeting the junction adhesive molecule-A (JAM-A), it was demonstrated that miR156a reduces cytokine-induced monocytes adhesion in endothelial cells and act as a vasoprotective molecule [38].
Table 1.
Plant miRNAs identified for their involvement in cross-kingdom gene regulation.
| Sr. No. | miRNAs | Host plant | Targeted organisms | Disease/target gene | References |
|---|---|---|---|---|---|
| 1. | miR168a | Oryza sativa | Human, Mouse, Rat, Calf, Horse, Sheep | low-density lipoprotein receptor adapter protein 1 (LDLRAP1) | (Zhang et al., 2012) |
| 2. | miR156a | Oryza sativa | Human, Mouse, Rat, Calf, Horse, Sheep | low-density lipoprotein receptor adapter protein 1 (LDLRAP1) | (Zhang et al., 2012) |
| 3. | miR166a | Oryza sativa | Human, Mouse, Rat, Calf, Horse, Sheep | low-density lipoprotein receptor adapter protein 1 (LDLRAP1) | (Zhang et al., 2012) |
| 4. | miR172 | Brassica oleracea | Mice | - | (Liang et al., 2014) |
| 5. | miR166a | Brassica campestris | Mice | - | (Chen et al., 2016) |
| 6. | miR 159 | Brassica campestris | Mice | - | (Chen et al., 2016) |
| 7. | miR2911 | Lonicera japonica | Mice | Influenza A virus (H1N1, H5N1, H7N9) | (Zhou et al., 2015) |
| 8. | miR2911 | Lonicera japonica | Mice | - | (Yang et al., 2015) |
| 9. | miR159 | Arabidopsis thaliana | Mice | Breast cancer/Transcription factor 7 (TCF7) | (Chin et al., 2016) |
| 10. | miR159 | Glycine max | Mice | Breast cancer/Transcription factor 7 (TCF7 | (Chin et al., 2016) |
| 11. | miR159 | Broccoli | Mice | Breast cancer/Transcription factor 7 (TCF7 | (Chin et al., 2016) |
| 12. | miR2910 | Populus euphratica | Human | JAK-STAT pathway | (Liu et al., 2017) |
| 13. | miRNAs (multiple) | Camptotheca acuminata | Human | Cancer | (Kumar et al., 2017) |
| 14. | miR14 | Curcuma longa | Human | Rheumatoid arthritis | (Sharma et al., 2017) |
| 15. | miR156a | cabbage, spinach and lettuce | Human | Cardiovascular disease | (Hou et al., 2018) |
In silico analysis of exogenous plant miRNAs
Different justifications have been provided in relation to the stability of miRNAs even after the food is cooked [31] and the medicinal plants boiled for decoction [35]. The stability of miRNAs is due to unique sequence and high GC content in addition to its methylation [35]. Besides various in vitro and in vivo studies, two recent in silico analyses also strengthened cross-kingdom gene regulation [39,40]. In these reports, the miRNAs were predicted from the available EST datasets of important medicinal plants happy tree (Camptotheca acuminata) [39] and Curcuma longa [40]. In happy tree, the identified miRNAs were predicted to be targeting the cancer associated genes of human and their regulation of expression. In Curcuma longa, the identified miR14 was analyzed for their stability in mammalian serum implicating their involvement in the regulation of inflammation-related proteins leading to Rheumatoid arthritis. Liang et al. (2013) also reviewed the cross-kingdom roles of miRNAs of different origin [11]. Here, other than plant-human, human-parasite and virus-human miRNA communication is explained (Table 1).
Exogenous role of human and viral miRNAs
In addition to the regulatory roles of plant miRNAs in animal system, there are a few reports suggesting the cross-kingdom role of animal miRNAs. The human miR451 is reported to regulate protozoan gene expression during infection of malarial parasite [41]. Although no endogenous miRNAs are reported in Plasmodium [42], miR451 is translocated to repress mRNA translation of malarial parasite. After infection, viruses take over the miRNA machinery of host and generate their own miRNAs and establish an environment suitable for viral growth. Kaposi sarcoma-associated herpesvirus miRNA, KSHV-miR-K12-11 is reported to take the advantage and regulate the gene expression of its hosts [43,44].
Modes of action of plant miRNAs in human
There are different views related to the modes of action of plant miRNAs as a regulatory element in human and other animal body. The exogenous plant miRNAs are taken up orally as a food intake and pass through GI tract which consists of a number of organs mouth, esophagus, stomach and intestine. From GI tract, the miRNAs are transferred and packaged into Extracellular Vesicles (EVs). The packaging of miRNAs in EVs helps them not to be degraded and leads to its stability. The plant miRNAs are secreted through different EVs such as exomes, micro vesicles, shedding vesicles and apoptotic bodies or in complexes with protein or lipid based carriers [45]. Other than vesicular transportation, the RISC is also involved to carry miRNAs to target genes [46]. From these EVs, miRNAs are transferred to other tissues such as lungs, liver, etc. via blood stream, plasma or serum. After reaching to different tissues, these plant miRNAs modulate the expression of their target genes (Fig 2). The plant miRNAs has also been detected in all biofluids such as serum [31,32] and plasma [37] through secretion. MiR2911 from honeysuckle has also been detected in mouse micro vesicles (MVs) of peripheral blood in associated form with AGO2 complex after passing through GI tract [35].
Figure 2.
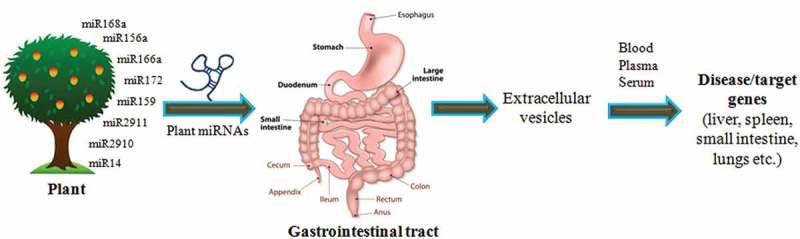
Cross-kingdom regulatory mechanisms of plant miRNAs. A number of miRNAs regulate the genes in other organisms are shown in the figure. These plant miRNAs have been shown to be present in animal tissues to target a number of genes regulating several processes involved in disease control.
Cross-kingdom gene regulation or contamination of plant miRNAs in animal samples
Zhang et al., (2012) [31] has opened up a groundbreaking hopes for plant miRNAs as an important therapeutic agents against various diseases however, plant miRNAs in animals through dietary intake is not globally accepted. A few groups have contradicted different aspects of cross-kingdom gene regulation through various arguments with their scientific viewpoints. There are some reports available explaining against the regulatory effect of exogenous miRNAs as they did not get the effective role of plant miRNAs in human, lack of bioavailability in human tissues and blood or cross-contamination during sequencing studies [47,48]. It was argued that the occurrences of low quantity of plant miRNAs in publicly available animal small RNA sequencing datasets might be due to contamination in sequencing methodologies [49,50] It has been recently reviewed that plant miRNAs present in human sera are due to contamination (technical artifacts) rather than dietary intake (regulating the endogenous genes) [48]. This review was based on various experiments from different groups concluding that the exogenous miRNAs are found in humans are due to contamination [49,51,52]. These findings were based on non-detection of plant miRNAs after proper handling to exclude cross-sample contamination in human sperm cells [49]. However, Hou at al. (2018) [53] responded that groups [49–52] utilized conventional sequencing methods leading inaccuracy of resulting data with biasness for methylated plant miRNAs. Further, suggestions were made to exclude three methods (high-throughput sequencing, quantitative reverse transcriptase PCR and northern blot analysis) for the possibility of contamination or technical artefacts.
In another report, frequent delivery of plant miRNAs through dietary intake has been doubted. Study demonstrated that after ingestion, the plant miRNAs (miR156a, miR159a and miR169a) are not detected or present in negligible amount in plasma of healthy human, mice and honey bee [54]. The limited amount of plant miRNAs in mammalian blood has also been shown through qPCR and droplet digital PCR analyses in Macaca nemestrina, a non-human primate model results inconsistent responses [55]. Dickinson et al. (2013) [47] have reported no measurable uptake of rice miRNAs especially miR168a in plasma and liver of mice through feeding different rice diets. The group also claimed that the level of plasma LDL [unlike Zhang et al. (2012) [31] report] changes due to nutritional imbalances rather than osa-miR168a. Reduction in LDLRAP1 protein level due to presence of rice diet was also argued. It was demonstrated that level of LDLRAP1 was not reduced even in the rice chow diet. This discrepancy was supported by the justification of differences in methods of evaluation adapted for LDLRAP1 expression analyses.
Zhang et al. (2013) [56] disagreed with the claim of Dickinson et al (2013) [47] and argued that improved experimental designs, careful controls and large sequencing data are necessary to extend their findings. The specific sequence interaction between plant based miRNAs and human disease related genes is also required for new era of therapeutics. The transfer and regulation of dietary miRNAs in human have also been represented by other groups [32,34,38] alongwith Zhang et al., (2012) [31]. Hirschi et al., (2015) also argued that no detection of plant derived miRNAs in animals may be due to failure in their metabolism, distribution and elimination rather than absorption through GI tract [57].
Conclusions and future remarks
In addition to several nutrients present in fruits and vegetables, the miRNAs have also shown the beneficiary effects on human health. This suggests that cross-kingdom miRNA delivery could be an herbal mode of gene regulation. The evidences of inhibition of viral infection and other diseases like cancer, rheumatoid arthritis were reported for exogenous application of plant based miRNAs. However, for the effective action of exogenous plant miRNAs, its concentration in human tissues/cells should be equal or greater than endogenous miRNAs. Keeping in mind the possible errors and artifacts during experimentation, this strategy could be taken up further for the effective therapeutic applications against deadly human diseases. The exogenous application of food derived miRNAs could be successful due to its target specificity as well as natural origin. Further, the experimental validation with animal model or human cell lines is also necessary to examine the protective or ameliorative effects of food borne miRNAs interacting with host genome. Keeping in view the physiological and pathophysiological role of miRNAs, the food-derived exogenous miRNAs may be considered as a novel nutrient supplements, like vitamins and minerals as well as therapeutic measure.
Funding Statement
This work was supported by the Council of Scientific and Industrial Research [BSC-107]; Department of Science and Technology, Ministry of Science and Technology [N-PDF].
Acknowledgments
The financial grant given by SERB, DST, New Delhi in the form of National-Post Doctoral Fellowship to Sanchita is highly acknowledged. Authors also acknowledge financial support from Council of Scientific and Industrial Research, New Delhi in the form of Network Project (BSC-0107).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- [1].Catalanotto C, Cogoni C, Zardo G.. MicroRNA in control of gene expression: an overview of nuclear functions. Int J Mol Sci. 2016;17:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wahid F, Shehzad A, Khan T, et al. MicroRNAs: synthesis, mechanism, function, and recent clinical trials. Biochim Biophys Acta. 2010;1803:1231–1243. [DOI] [PubMed] [Google Scholar]
- [3].Friedman RC, Farh KK, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zhang BH, Pan XP, Cobb GP, et al. Plant microRNA: a small regulatory molecule with big impact. Dev Biol. 2006;289:3–16. [DOI] [PubMed] [Google Scholar]
- [5].Sharma D, Tiwari M, Pandey A, et al. MicroRNA858 is a potential regulator of phenylpropanoid pathway and plant development. Plant Physiol. 2016;171:944–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gao C, Ju Z, Cao D, et al. MicroRNA profiling analysis throughout tomato fruit development and ripening reveals potential regulatory role of RIN on microRNAs accumulation. Plant Biotechnol J. 2015;13:370–382. [DOI] [PubMed] [Google Scholar]
- [7].Bi F, Meng X, Ma C, et al. Identification of miRNAs involved in fruit ripening in Cavendish bananas by deep sequencing. BMC Genomics. 2015;16:776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hunter MP, Ismail N, Zhang X, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One. 2008;3:e3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43–51. [DOI] [PubMed] [Google Scholar]
- [10].Ji L, Chen X. Regulation of small RNA stability: methylation and beyond. Cell Res. 2012;22:624–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Liang H, Zen K, Zhang J, et al. New roles for microRNAs in cross-species communication. RNA Biol. 2013;10:367–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Arbizu S, Krenek K, Mertens-Talcott S. Mango polyphenols reduce inflammation in MDA-MB231 breast cancer cell and targets microRNA-21. FASEB J. 2013;27:862–963. [Google Scholar]
- [13].Kim H, Banerjee N, Ivanov I, et al. Comparison of anti-inflammatory mechanisms of mango (Mangifera Indica L.) and pomegranate (Punica Granatum L.) in a preclinical model of colitis. Mol Nutr Food Res. 2016;60:1912–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Choudhary D, Pandey A, Adhikary S, et al. Genetically engineered flavonol enriched tomato fruit modulates chondrogenesis to increase bone length in growing animals. Sci Rep. 2016;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hartmann T. Diversity and variability of plant secondary metabolism: a mechanistic view. Entomol Exp Appl. 1996;80:177–188. [Google Scholar]
- [16].Hussain MS, Fareed S, Ansari S, et al. Current approaches toward production of secondary plant metabolites. J Pharm Bioallied Sci. 2012;4:10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Pandey A, Misra P, Khan MP, et al. Co-expression of Arabidopsis transcription factor, AtMYB12, and soybean isoflavone synthase, GmIFS1, genes in tobacco leads to enhanced biosynthesis of isoflavones and flavonols resulting in osteoprotective activity. Plant Biotechnol J. 2014;12:69–80. [DOI] [PubMed] [Google Scholar]
- [18].Khedgikar V, Ahmad N, Kushwaha P, et al. Preventive effects of withaferin A isolated from the leaves of an Indian medicinal plant Withania somnifera (L.): comparisons with 17-β-estradiol and alendronate. Nutrition. 2015;31:205–213. [DOI] [PubMed] [Google Scholar]
- [19].Cruz KJC, de Oliveira ARS, Morais JBS, et al. Role of microRNAs on adipogenesis, chronic low-grade inflammation, and insulin resistance in obesity. Nutrition. 2017;35:28–35. [DOI] [PubMed] [Google Scholar]
- [20].Marques-Rocha JL, Milagro FI, Mansego ML, et al. Expression of inflammation-related miRNAs in white blood cells from subjects with metabolic syndrome after 8 wk of following a mediterranean diet-based weight loss program. Nutrition. 2016;32:48–55. [DOI] [PubMed] [Google Scholar]
- [21].Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol Biol. 2006;342:129–138. [DOI] [PubMed] [Google Scholar]
- [22].Axtell MJ, Westholm JO, Lai EC. Vive la difference: biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 2011;12:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Tiwari M, Sharma D, Trivedi PK. Artificial microRNA mediated gene silencing in plants: progress and perspectives. Plant Mol Biol. 2014;86:1–18. [DOI] [PubMed] [Google Scholar]
- [24].Wang ZH, Xu CJ. Research progress of microRNA in early detection of ovarian cancer. Chin Med J. 2015;128:3363–3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Millar AA, Waterhouse PM Plant and animal microRNAs: similarities and differences. Funct Integr Genomics. 2005;5:129–135. [DOI] [PubMed] [Google Scholar]
- [26].Felekkis K, Touvana E, Stefanou C, et al. microRNAs: a newly described class of encoded molecules that play a role in health and disease. Hippokratia. 2010;14:236–240. [PMC free article] [PubMed] [Google Scholar]
- [27].Didiano D, Hobert O. Molecular architecture of a miRNA-regulated 3 ‘ UTR. RNA. 2008;14:1297–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Marin RM, Sulc M, Vanicek J. Searching the coding region for microRNA targets. RNA. 2013;19:467–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Brummer A, Hausser J. MicroRNA binding sites in the coding region of mRNAs: extending the repertoire of post-transcriptional gene regulation. Bioessays. 2014;36:617–626. [DOI] [PubMed] [Google Scholar]
- [30].Iwakawa H, Tomari Y. Molecular insights into microRNA-mediated translational repression in plants. Mol Cell. 2013;52:591–601. [DOI] [PubMed] [Google Scholar]
- [31].Zhang L, Hou D, Chen X, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 2012;22:107–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Liang G, Zhu Y, Sun B, et al. Assessing the survival of exogenous plant microRNA in mice. Food Sci Nutr. 2014;2:380–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Chen X, Dai GH, Ren ZM, et al. Identification of dietetically absorbed rapeseed (Brassica campestris L.) bee pollen microRNAs in serum of mice. Biomed Res Int. 2016;2016:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Chin AR, Fong MY, Somlo G, et al. Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Res. 2016;26:217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhou Z, Li X, Liu J, et al. Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res. 2015;25:39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Yang J, Farmer LM, Agyekum AA, et al. Detection of an abundant plant-based small RNA in healthy consumers. PLoS One. 2015;10:e0137516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Liu YC, Chen WL, Kung WH, et al. Plant miRNAs found in human circulating system provide evidences of cross kingdom RNAi. BMC Genomics. 2017;18:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hou DX, He FF, Ma LN, et al. The potential atheroprotective role of plant MIR156a as a repressor of monocyte recruitment on inflamed human endothelial cells. J Nutr Biochem. 2018;57:197–205. [DOI] [PubMed] [Google Scholar]
- [39].Kumar D, Kumar S, Ayachit G, et al. Cross-kingdom regulation of putative miRNAs drived from happy tree in cancer pathway: a systems biology approach. Int J Mol Sci. 2017;18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Sharma A, Sahu S, Kumari P, et al. Genome-wide identification and functional annotation of miRNAs in anti-inflammatory plant and their cross-kingdom regulation in homo sapiens. J Biomol Struct Dyn. 2017;35:1389–1400. [DOI] [PubMed] [Google Scholar]
- [41].LaMonte G, Philip N, Reardon J, et al. Translocation of sickle cell erythrocyte microRNAs into Plasmodium falciparum inhibits parasite translation and contributes to malaria resistance. Cell Host Microbe. 2012;12:187–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Xue X, Zhang Q, Huang Y, et al. No miRNA were found in Plasmodium and the ones identified in erythrocytes could not be correlated with infection. Malar J. 2008;7:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Hansen A, Henderson S, Lagos D, et al. KSHV-encoded miRNAs target MAF to induce endothelial cell reprogramming. Genes Dev. 2010;24:195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Samols MA, Skalsky RL, Maldonado AM, et al. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 2007;3:e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Turchinovich A, Weiz L, Langheinz A, et al. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Dickinson B, Zhang YJ, Petrick JS, et al. Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat Biotechnol. 2013;31:965–967. [DOI] [PubMed] [Google Scholar]
- [48].Fromm B, Kang W, Rovira C, et al. Plant microRNAs in human sera are likely contaminants. J Nutr Biochem. 2018;S0955-2863:30535–30537. [DOI] [PubMed] [Google Scholar]
- [49].Tosar JP, Rovira C, Naya H, et al. Mining of public sequencing databases supports a non-dietary origin for putative foreign miRNAs: underestimated effects of contamination in NGS. RNA. 2014;20:754–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zhang Y, Wiggins BE, Lawrence C, et al. Analysis of plant-derived miRNAs in animal small RNA datasets. BMC Genomics. 2012;13:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Heintz-Buschart A, Yusuf D, Kaysen A, et al. Small RNA profiling of low biomass samples: identification and removal of contaminants. BMC Biol. 2018;16:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kang W, Bang-Berthelsen CH, Holm A, et al. Survey of 800+ data sets from human tissue and body fluid reveals xenomiRs are likely artifacts. RNA. 2017;23:433–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Hou D, Zhou Z, Chen X, et al. Reply to Fromm et al. J Nutr Biochem. 2018;148:1508. [DOI] [PubMed] [Google Scholar]
- [54].Snow JW, Hale AE, Isaacs SK, et al. Ineffective delivery of diet-derived microRNAs to recipient animal organisms. RNA Biol. 2013;10:1107–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Witwer KW, McAlexander MA, Queen SE, et al. Real-time quantitative PCR and droplet digital PCR for plant miRNAs in mammalian blood provide little evidence for general uptake of dietary miRNAs: limited evidence for general uptake of dietary plant xenomiRs. RNA Biol. 2013;10:1080–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Chen X, Zen K, Zhang CY. Lack of detectable oral bioavailability of plant microRNAs after feeding in mice reply. Nat Biotechnol. 2013;31:967–969. [DOI] [PubMed] [Google Scholar]
- [57].Hirschi KD, Pruss GJ, Vance V. Dietary delivery: a new avenue for microRNA therapeutics? Trends Biotechnol. 2015;33:431–432. [DOI] [PubMed] [Google Scholar]


