Abstract
Background
There are concerns that multigene panel testing compared with BRCA1/2-only testing after diagnosis of breast cancer may lead to unnecessary patient worry about cancer because of more ambiguous results.
Methods
Patients with breast cancer diagnosed from 2013 to 2015 and accrued from SEER registries in Georgia and Los Angeles were surveyed (n = 5,080; response rate, 70%), and responses were merged with SEER data and germline genetic testing and results. We examined patient reports of cancer worry by test type and results in 1,063 women who linked to a genetic test and reported undergoing testing.
Results
More than half of the sample (n = 640; 60.2%) received BRCA1/2-only testing versus 423 patients (39.8%) who had a multigene panel. A minority of tested patients reported substantial cancer worry after treatment: 11.1% (n = 130) reported higher impact of cancer worry, and 15.1% (n = 162) reported a high frequency of cancer worry (worrying often or almost always) in the past month. Impact of cancer worry did not substantively differ by test type, test result outcomes, or clinical or treatment factors. The odds ratio for higher impact of cancer worry was 0.81 (95% CI, 0.51 to 1.28) for multigene versus BRCA1/2-only testing. In a separate model, the odds ratios were 1.21 (95% CI, 0.54 to 2.68) and 0.90 (95% CI, 0.50 to 1.62) for pathogenic variant and variant of uncertain significance, respectively, versus a negative test (the reference group).
Conclusion
Compared with BRCA1/2 testing alone, multigene panel testing was not associated with increased cancer worry after diagnosis of breast cancer.
INTRODUCTION
Multiple-gene panel (MGP) testing has rapidly replaced BRCA1/2-only testing after diagnosis of breast cancer.1 However, genetic testing use in patients who need it seems to be low.2,3 One barrier to more robust uptake of MGP testing is uncertainty about its clinical utility. MGP testing yields a higher rate of pathogenic variants than BRCA1/2-only testing but with much wider variability in associated cancer risks, or penetrance. In addition, MGP testing generates higher rates of variants of unknown significance (VUSs). Taken together, there are concerns that more ambiguous findings from MGP testing may lead to unnecessary patient worry and unwarranted interventions. Indeed, some have argued that MGP testing should not replace BRCA1/2-only testing, even in patients with higher pretest risk of a pathogenic mutation, because of these concerns about patient reactions,4 whereas others strongly disagree.5 Current clinical guidelines that recommend genetic testing in women at elevated pretest risk of carrying a pathogenic variant do not specify how many genes should be tested. In this study, we examined the association of patient report of cancer-related worry with genetic testing type (MGP v BRCA1/2-only testing) and results (negative, VUS only, or pathogenic variant) in a large, contemporary population-based sample of patients diagnosed with breast cancer and accrued during the period immediately after introduction of MGP testing into the community.
METHODS
Study Sample and Data Collection
The iCanCare study identified women with who were 20 to 79 years of age, diagnosed with stages 0 to II breast cancer and reported to the Georgia or Los Angeles County SEER registries.1,6,7 We excluded women with prior breast cancer, tumors greater than 5 cm, or more than three involved lymph nodes. We mailed survey materials and a $20 cash gift between July 2013 and August 2015. We used a modified Dillman method8 to encourage response (median time from diagnosis to survey completion, 6 months; standard deviation [SD], 2.8 months). We sent surveys to 7,810 patients: 507 patients were ineligible because of the exclusions noted previously or were deceased, institutionalized, too ill, or unable to complete a survey in Spanish or English, leaving 7,303 patients. The survey was completed by 5,080 eligible patients (response rate, 70%) and was published previously.2 Median time from diagnosis to survey completion was 5.8 months (SD, 2.4 months). Virtually all patients (5,026) had complete information for the test linkage phase, of whom 1,272 (25.3%) linked to a genetic test result, 1,063 of whom (21.2%) reported receipt of genetic testing in the survey.
Survey responses were merged with SEER clinical data and genetic testing information obtained from four laboratories (Ambry Genetics, Aliso Viejo, CA; GeneDx, Gaithersburg, MD; Invitae, San Francisco, CA; Myriad Genetics, Salt Lake City, UT) that performed nearly all germline cancer genetic testing in the study regions. Test type and results were merged to 5,026 patients with complete information on all variables for SEER genetic testing linkage using a probabilistic matching strategy performed by Information Management Services (Rockville, MD). The Safe Harbor method was used to de-identify the data set before transfer to the University of Michigan for analysis.9
The collaboration was covered under data use agreements between the University of Michigan, genetic laboratories, and Information Management Services. The research was approved by institutional review boards of the University of Michigan, Emory University, the University of Southern California, the Georgia Department of Public Health, the California State Committee for the Protection of Human Subjects, and California Cancer Registry. Signed consent was waived for the survey. Waivers of informed consent and authorization were approved, given the use of a third-party honest broker to conduct the linkage and create a de-identified data set for analyses.
Measures
We used two questions to evaluate patient worry about future cancer. Impact of Cancer Worry was the primary outcome, which was assessed using three survey items: “During the past month, how often has worrying about your cancer coming back…made you feel upset?; made it difficult for you to carry out your usual daily activities at home or work?; made you feel distant from family or friends?” (responses were based on 5-point Likert categories from almost never to almost always). We developed a scale by averaging the responses across the three items to create a continuous range from 1 to 5, with higher values indicating greater impact of cancer worry. The mean score was 1.7 (SD, 0.8), with a Cronbach’s alpha of .87. We then created a binary variable indicating higher impact of cancer worry versus low impact using a cut point of 3.0, which corresponded to responses of sometimes, often, or almost always. We also measured Frequency of Cancer Worry for validation, which was assessed using a single item: “In the past month, how often have you worried about your cancer coming back?” (almost never to almost always; five point categories). We created a binary variable indicating high frequency (often or almost always) versus low frequency (almost never, rarely, or sometimes).
Test measures from laboratories.
Genetic laboratories provided results at the level of the gene tested (eg, BRCA1) and the clinical interpretation sent to the ordering clinician (consisting of pathogenic, likely pathogenic, uncertain significance, likely benign, or benign). We combined interpretations of pathogenic and likely pathogenic together as pathogenic and combined likely benign and benign together as benign. We categorized a test that assessed only BRCA1/2 as BRCA1/2 only and a test that assessed any additional gene (eg, ATM, CHEK2) as a multigene panel. If patients received both tests on separate occasions (eg, a BRCA1/2-only test and later MGP), they were coded as having received MGP. We created mutually exclusive results categories: (1) negative for pathogenic variants or VUS in any gene, (2) one or more VUS but no pathogenic variant in any gene, (3) positive for a pathogenic variant in BRCA1/2 with or without VUS but no pathogenic variant in another gene, or (4) positive for a pathogenic variant in another gene with or without VUS in any gene.
Other independent variables.
We assessed clinical factors related to future cancer risk (tumor behavior, triple-negative disease, nodal status, bilateral disease), treatments received, age (in 10-year groups), education, and family history. On the basis of the National Comprehensive Cancer Network guidelines, family history was defined as having two or more first-degree relatives with breast cancer; any relatives with ovarian cancer, sarcoma, or male breast cancer; Ashkenazi Jewish ancestry; or a family history of a mutation-conferring risk (eg, BRCA1/2).
Statistical Analysis
We first examined cancer worry variables by test type, test results, and selected covariables. We then regressed high Impact of Cancer Worry (binary variable) on test type and selected covariables using a logistic regression model. We then repeated the analysis using test results instead of test type. We confirmed findings by repeating these analyses using high Frequency of Cancer Worry as the dependent variable. SAS version 9.4 (SAS Institute, Cary, NC) was used to compute odds ratios, adjusting for covariables and weighted for survey design and nonresponse.
RESULTS
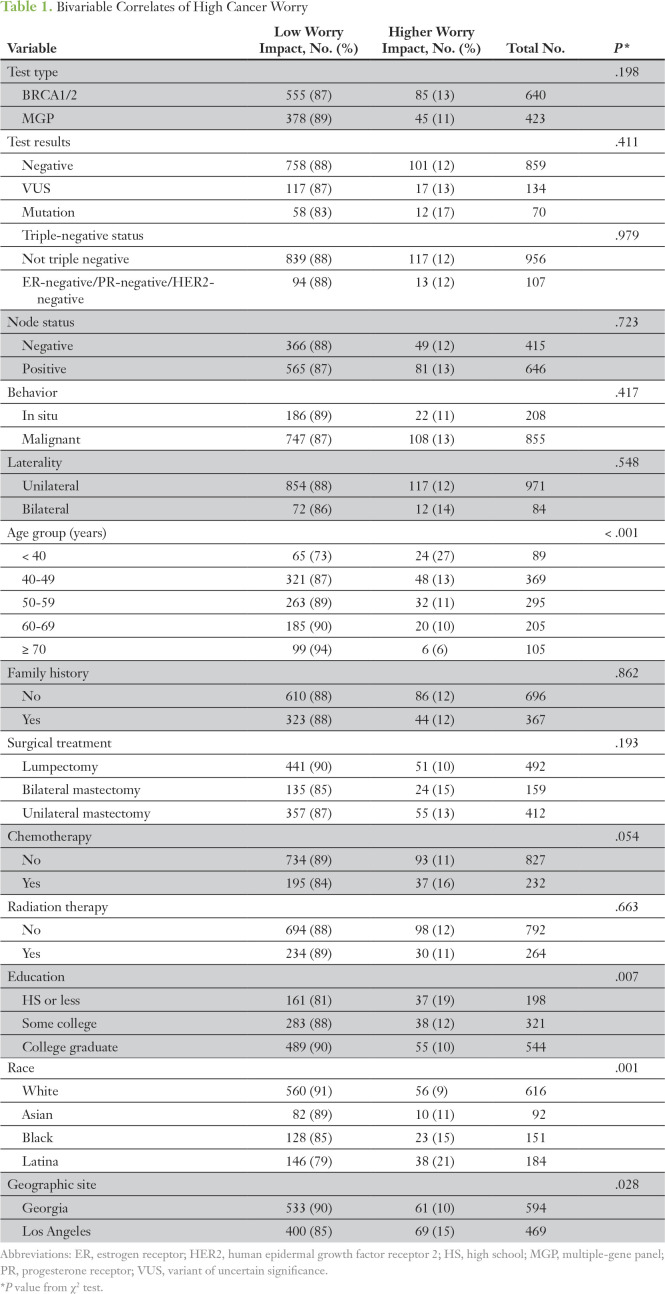
More than half of the sample (n = 640; 60.2%) received BRCA1/2-only testing versus 423 (39.8%) who had multigene panels. A minority of tested patients reported substantial cancer worry after treatment: 11.1% (n = 130) reported high impact of cancer worry, and 15.1% (n = 162) reported high frequency of worry (worrying often or almost always in the past month). Impact of cancer worry did not substantively differ by test type, test result, or clinical or treatment factors in bivariable analyses (Table 1). Impact of cancer worry was higher for patients of younger age and lower education level, and for ethnic minorities. We observed similar findings in a model of frequency of cancer worry (data not shown).
Table 1.
Bivariable Correlates of High Cancer Worry
Figure 1 shows the results of a multivariable model regressing a binary variable indicating higher versus low impact of cancer worry on test type, controlling for selected predisposing, clinical, and treatment factors. The odds ratio for higher cancer worry was 0.81 (95% CI, 0.51 to 1.28) for MGP versus BRCA1/2-only testing. Younger age, black race (ref: white), and lower education were associated with higher cancer worry. We listed the modeling approach in Appendix Figure A1, regressing high cancer worry on outcomes of test results and found no association; the odds ratios were 1.21 (95% CI, 0.54 to 2.68) and 0.90 (95% CI, 0.50 to 1.62) for pathogenic variant and VUS, respectively, versus a negative test (the reference group). In sensitivity analyses, we examined different permutations of the cancer worry variables, including different cut points for higher worry, and using a continuous variable, and results were consistent.
Fig 1.
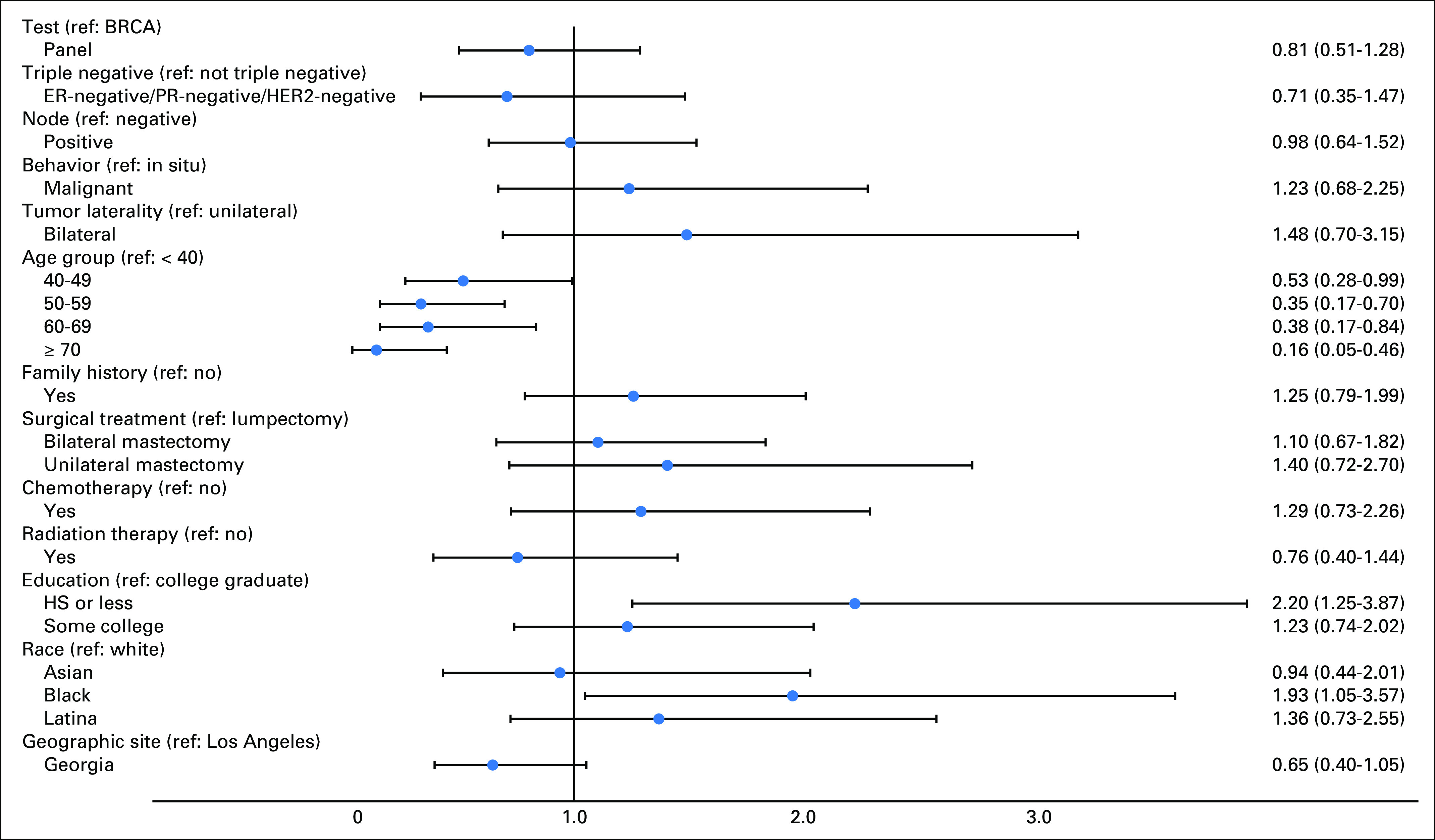
Factors associated with higher impact of cancer worry. Forest plot (adjusted odds ratios and 95% CIs) for results of a multivariable model regressing a binary variable indicating higher versus low impact of cancer worry on test type, controlling for selected predisposing, clinical, and treatment factors and weighted for survey design and nonresponse. ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; HS, high school; MGP, multigene panel; PR, progesterone receptor; ref, reference.
DISCUSSION
We found that the type of genetic testing patients received (MGP v BRCA1/2 only) was not associated with their report of cancer-related worry (impact or frequency). We further observed no association between patient cancer worry and test result (pathogenic variant, VUS, negative). As noted in our prior work, cancer-related worry was associated with younger age and lower educational level, but not clinical factors.10,11
Strengths of this study include a large population-based sample of patients recently diagnosed with breast cancer and accrued shortly after the advent of multigene panel testing in the community, genetic testing results from companies linked to SEER clinical data, and granular information about patients, including their report of cancer-related worry. But there are several limitations. First, we had limited information about patient perspectives and appraisal specifically related to genetic testing experiences. Second, some patients who linked to a genetic test did not recall it at the time of the survey. We excluded them from the analytic sample because we did not think it was valid to assess the association of a test type with attitudes about health risk among patients who did not recall that they had the test. Finally, generalizability of the findings are limited to two large geographic regions of the United States.
The results from this study and our prior work suggest that testing more genes versus BRCA1/2 alone does not seem to foment more negative patient reactions. Our prior research in this cohort suggested that MGP testing (v BRCA1/2-only testing) does not lead to inappropriate rates of contralateral prophylactic mastectomy.1 Virtually all testers in this study received some form of genetic counseling,6 and our results suggest these efforts may sufficiently frame results in a manner that did not alarm patients. Uncertainty remains about the clinical utility of MGP versus BRCA1/2-only testing after diagnosis of breast cancer. However, our results in this study and in our prior work suggest that testing more genes than BRCA1/2 does not seem to increase cancer-related worry or unwarranted interventions.1 More research will be needed to examine the impact of genetic testing on patient outcomes in this rapidly evolving clinical context.
ACKNOWLEDGMENT
We thank Jill S. Dolinsky, CGC, and Melissa Pronold at Ambry Genetics, Delores Bowman and Benjamin Solomon at GeneDx, Edward Esplin and Stephen Lincoln at Invitae, and Johnathan Lancaster and Brian Dechairo at Myriad Genetics for their collaboration on the genetic test data linkage to SEER data. Written permission was obtained to include the names of all acknowledged individuals.
Appendix
Fig A1.
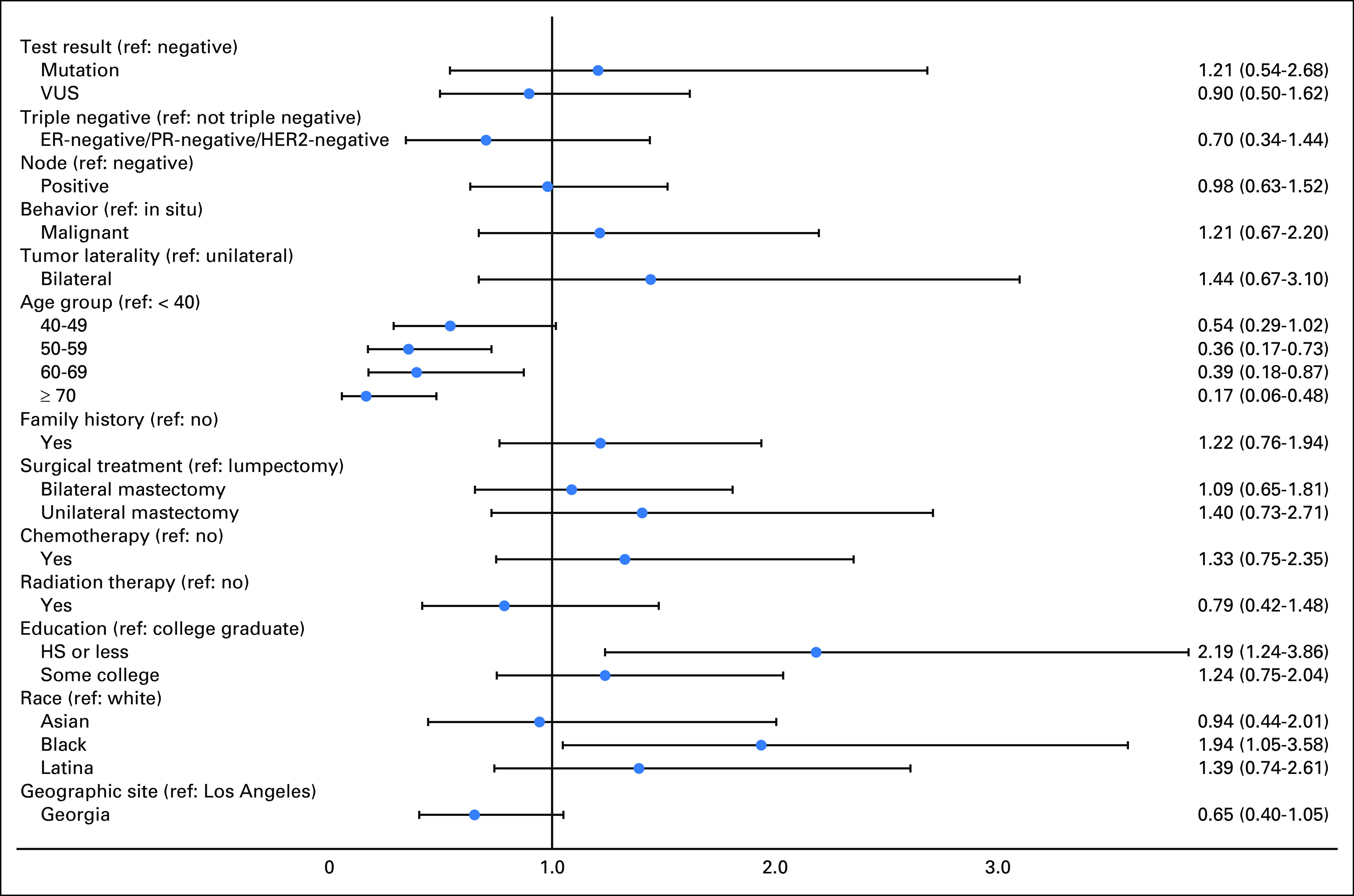
Association of genetic test result with high impact of cancer worry (odds ratios and 95% CIs). Forest plot (adjusted odds ratios and 95% CIs) showing results of multivariable logistic regression for higher impact of cancer worry (complete case n = 1,044). ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; HS, high school; MGP, multiple-gene panel; PR, progesterone receptor; VUS, variant of uncertain significance.
Footnotes
Supported by Grant No. P01 CA163233 to the University of Michigan from the National Cancer Institute. Conducted work was also supported by the University of Michigan Cancer Center Biostatistics, Analytics and Bioinformatics shared resource (P30CA46592). The collection of Los Angeles County cancer incidence data used in this study was supported by the California Department of Public Health pursuant to California Health and Safety Code Section 103885, Centers for Disease Control and Prevention’s (CDC’s) National Program of Cancer Registries, under cooperative agreement 5NU58DP003862-04/DP003862, the National Cancer Institute’s SEER Program under contract HHSN261201000140C awarded to the Cancer Prevention Institute of California, contract HHSN261201000035C awarded to the University of Southern California, and contract HHSN261201000034C awarded to the Public Health Institute. The collection of cancer incidence data in Georgia was supported by contract HHSN261201300015I, Task Order HHSN26100006 from the National Cancer Institute and cooperative agreement 5NU58DP003875-04-00 from the CDC. The ideas and opinions expressed herein are those of the authors, and endorsement by the States of California and Georgia, Department of Public Health, the National Cancer Institute, and the CDC or their contractors and subcontractors is not intended nor should be inferred.
AUTHOR CONTRIBUTIONS
Conception and design: Steven J. Katz, Kevin C. Ward, Allison W. Kurian
Financial support: Steven J. Katz
Administrative support: Steven J. Katz
Provision of study materials or patients: Ann S. Hamilton
Collection and assembly of data: Steven J. Katz, Kevin C. Ward, Ann S. Hamilton, Paul Abrahamse, Sarah T. Hawley
Data analysis and interpretation: Steven J. Katz, Kevin C. Ward, Paul Abrahamse, Allison W. Kurian
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Steven J. Katz
No relationship to disclose
Kevin C. Ward
No relationship to disclose
Ann S. Hamilton
No relationship to disclose
Paul Abrahamse
No relationship to disclose
Sarah T. Hawley
No relationship to disclose
Allison W. Kurian
Research Funding: Myriad Genetics (Inst)
REFERENCES
- 1.Kurian AW, Ward KC, Hamilton AS, et al. Uptake, results, and outcomes of germline multiple-gene sequencing after diagnosis of breast cancer. JAMA. 2018;4:1066–1072. doi: 10.1001/jamaoncol.2018.0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kurian AW, Griffith KA, Hamilton AS, et al. Genetic testing and counseling among patients with newly diagnosed breast cancer. JAMA. 2017;317:531–534. doi: 10.1001/jama.2016.16918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurian AW, Li Y, Hamilton AS, et al. Gaps in incorporating germline genetic testing into treatment decision-making for early-stage breast cancer. J Clin Oncol. 2017;35:2232–2239. doi: 10.1200/JCO.2016.71.6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narod S, Sopik V, Cybulski C. Testing Ashkenazi Jewish women for mutations predisposing to breast cancer in genes other than BRCA1 and BRCA2. JAMA Oncol. 2018;4:1012. doi: 10.1001/jamaoncol.2018.0595. [DOI] [PubMed] [Google Scholar]
- 5.King M, Walsh T. Testing Ashkenazi Jewish women for mutations predisposing to breast cancer in genes other than BRCA1 and BRCA2—Reply. JAMA Oncology. 2018;4:1012–1013. doi: 10.1001/jamaoncol.2018.0616. [DOI] [PubMed] [Google Scholar]
- 6.Katz SJ, Ward KC, Hamilton AS, et al. Gaps in receipt of clinically indicated genetic counseling after diagnosis of breast cancer. J Clin Oncol. 2018;36:1218–1224. doi: 10.1200/JCO.2017.76.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurian AW, Bondarenko I, Jagsi R, et al. Recent trends in chemotherapy use and oncologists’ treatment recommendations for early-stage breast cancer. J Natl Cancer Inst. 2018;110:493–500. doi: 10.1093/jnci/djx239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dillman DA, Smyth JD, Christian LM. Internet, Mail, and Mixed-Mode Surveys: The Tailored Design Method. ed 3. Hoboken, NJ: John Wiley & Sons; 2009. [Google Scholar]
- 9.US Department of Health & Human Services Guidance regarding methods for de-identification of protected health information in accordance with the Health Insurance Portability and Accountability Act (HIPAA) privacy rule. https://www.hhs.gov/hipaa/for-professionals/privacy/special-topics/de-identification/index.html#
- 10.Janz NK, Li Y, Zikmund-Fisher BJ, et al. The impact of doctor-patient communication on patients’ perceptions of their risk of breast cancer recurrence. Breast Cancer Res Treat. 2017;161:525–535. doi: 10.1007/s10549-016-4076-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hawley ST, Janz NK, Griffith KA, et al. Recurrence risk perception and quality of life following treatment of breast cancer. Breast Cancer Res Treat. 2017;161:557–565. doi: 10.1007/s10549-016-4082-7. [DOI] [PMC free article] [PubMed] [Google Scholar]