Abstract
Chromosomal translocations are associated with several tumor types, including hematopoietic malignancies, sarcomas, and solid tumors of epithelial origin, due to their activation of a proto-oncogene or generation of a novel fusion protein with oncogenic potential. In many cases, the availability of suitable human models has been lacking because of the difficulty in recapitulating precise expression of the fusion protein or other reasons. Further, understanding how translocations form mechanistically has been a goal, as it may suggest ways to prevent their occurrence. Chromosomal translocations arise when DNA ends from double-strand breaks (DSBs) on two heterologous chromosomes are improperly joined. This review provides a summary of DSB repair mechanisms and their contribution to translocation formation, the various programmable nuclease platforms that have been used to generate translocations, and the successes that have been achieved in this area.
Keywords: double-strand break, CRISPR-Cas9, chromosomal translocation, NHEJ
Introduction
Chromosomal translocations join DNA segments derived from two heterologous chromosomes. Translocations influence the evolution of species, but they are mainly considered in the context of disease. In particular, they are prominent features of several types of cancers, from hematopoietic to solid tumors, leading to the expression of a new fusion oncogene or to the mis-regulation of a proto-oncogene. Models for translocation-associated cancers typically rely on ectopic expression of fusion genes in cell lines or on endogenous expression in transformed tumor cells. While valuable to the scientific community for providing insights into mechanisms of oncogenesis, these models may fail to fully recapitulate the human disease. For instance, mouse models overexpressing the NPM1-ALK fusion (implicated in Anaplasic Large Cell Lymphoma, or ALCL) mostly induce B cell lymphomas rather than T cell lymphomas associated with the human disease, therefore failing to provide a robust pre-clinical model (Turner and Alexander, 2005). Moreover, patient-derived tumors cells will invariably have a number of tumor-acquired mutations.
Chromosomal translocations appear to arise from improper repair of DNA double-strand breaks (DSBs), which are highly toxic lesions. The “guardians” of genome integrity mostly ensure reliable repair of DSBs; also, unrepaired DSBs can lead to apoptosis or senescence. However, imprecise repair of DSBs has the potential to be highly deleterious, as it can lead to genome instability, including the formation of chromosomal rearrangements. In particular, chromosomal translocations can arise when DNA ends from DSBs on two heterologous chromosomes are improperly joined (Scott et al., 2000). Given this, researchers have been taking advantage of various nucleases, especially the recently developed programmable nucleases, to deliberately induce DSBs at loci of interest to generate translocations. The goal is to ultimately generate faithful tumor models and also to understand the DSB “misrepair” mechanisms that lead to translocations.
1. Multiple DSB repair pathways: Repairing a dangerous lesion
Given that DSBs can compromise the integrity of the genome, it is perhaps not surprising that multiple pathways exist to repair DSBs (Chapman et al., 2013; Jasin and Haber, 2016; Jasin and Rothstein, 2013) (Figure 1). The two major DSB repair pathways in mammalian cells are nonhomologous end-joining (NHEJ) and homologous recombination (HR), also termed homology-directed repair. The relationship of these pathways is complex. In some cases, DSB repair is limited to one pathway, as in programmed DSBs in the immune system (NHEJ) or during meiosis (HR), but the pathways can also compete with each other for the repair of a single DSB and surprisingly even collaborate (Kass and Jasin, 2010), such that DSB repair initiates by HR but is completed by NHEJ (Costantino et al., 2014; Johnson and Jasin, 2000; Richardson and Jasin, 2000a)
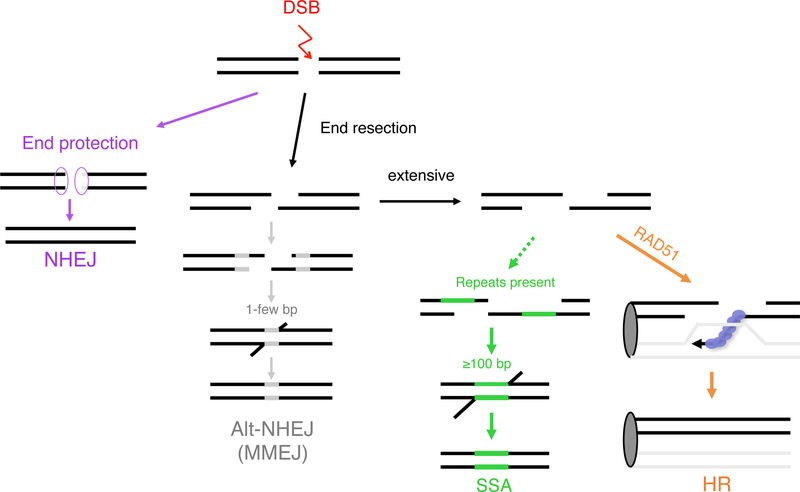
Figure 1 : DSB repair pathways.
The two major DSB repair pathways in mammalian cells are nonhomologous end-joining (NHEJ) and homologous recombination (HR). In addition, end resection provides also single-stranded DNA intermediates for two other pathways: Alternative NHEJ (alt-NHEJ) using microhomology and Single Strand Annealing (SSA).
In cycling cells, an early determinant of DSB repair pathway choice is whether DNA ends undergo resection to generate 3′ single-stranded overhangs, which is promoted during S/G2 phases of the cell cycle but suppressed during G1 (Symington and Gautier, 2011). The resected DNA is then coated with the RAD51 protein to form a nucleoprotein filament, which performs strand invasion of a homologous template to prime repair synthesis (Moynahan and Jasin, 2010). If templated by the sister chromatid, the preferred HR partner, repair synthesis leads to restoration of the original DNA sequence prior to breakage. By contrast, during canonical NHEJ (c-NHEJ), DNA ends are protected from resection and can be precisely joined, if ends do not require modification, or imprecisely joined after processing to make ends ligatable to give rise to a variety of junctions.
End resection also provides single-stranded DNA intermediates for two other pathways. Alternative NHEJ (alt-NHEJ) using microhomology, a major pathway of which is microhomology-mediated NHEJ (MMEJ), involves annealing at short sequence identities present near the DNA ends and thus only requires limited end resection (<100 bp) (Sfeir and Symington, 2015). As with c-NHEJ, this pathway gives rise to a variety of junctions, although deletions may be longer with MMEJ due to resection (Simsek and Jasin, 2010). Because any particular breakpoint junction can likely form by either NHEJ pathway, identifying which pathway is responsible requires statistical analysis to determine if microhomology is over represented or the use of pathway mutants (for example, in ligase IV for canonical NHEJ and ligase III for alt-NHEJ (Simsek et al., 2011; Simsek and Jasin, 2010). Single-strand annealing (SSA) also involves annealing at repeats flanking the DSB, but the repeats are much longer, and thus requires more extensive end resection than alt-NHEJ to uncover complementary single-strands. The physiological role of SSA in cells is unclear, but it has been used to distinguish whether HR mutants are defective in the early end-resection step of HR (defective in both HR and SSA) from those defective only at the strand invasion step (defective in HR but elevated SSA) (Stark et al., 2004).
2. Elucidation of translocation mechanisms in mouse cells using a rare-cutting endonuclease
As the complexity of DSB repair pathways in mammalian cells was uncovered, investigators sought to determine how each pathway participates in translocation formation. In the era before programmable endonucleases, DSBs were introduced into the genome using the yeast homing endonuclease I-SceI (Colleaux et al., 1988), which has an ~18 bp recognition site and thus is suitable for studies in complex mammalian genomes. Several reporters were developed in mammalian cells to determine which DSB repair pathway(s) gives rise to translocations upon DSB formation (Weinstock et al., 2006). I-SceI sites were introduced at specified chromosomal locations by gene targeting in mouse embryonic stem cells, which have a diploid chromosome complement. Translocations were selected by reconstruction of a drug resistance marker and confirmed by fluorescence in situ hybridization.
Initial studies focused on HR between repeats on different chromosomes, given that translocations will form by HR in budding yeast (Harris et al., 1993) and that mammalian genomes are replete with sequence repeats. Introducing a DSB into a repeat on one chromosome did not give rise to a translocation; rather the DSB was repaired by a simple gene conversion event with the other chromosome without exchange of flanking markers (Richardson et al., 1998). This study, as well as subsequent ones (e.g., (LaRocque et al., 2011; Stark and Jasin, 2003)), indicated that HR in mammalian cells is rarely associated with crossing over. A follow up study attempted to drive translocation formation by HR by truncating the repeats, such that restoration of the selectable marker would seem to require HR. However, in this case too, HR did not lead to translocations; rather, HR was coupled to NHEJ, involving a break-induced replication type of HR that was completed by NHEJ (Richardson and Jasin, 2000a). Presumably, the BLM helicase plays a major role in suppressing crossing over that would drive translocation formation, as it does between homologous chromosomes (LaRocque et al., 2011). Thus, these studies indicated that while a DSB on one chromosome was sufficient to induce HR with another chromosome, it was not sufficient to drive translocation formation. Although HR between repeated sequences has been reported in several contexts, it is notable that many or most of these events likely occur during meiosis (Kim et al., 2016), when HR may be under different constraints.
Subsequent studies focused on introducing two DSBs, one on each chromosome. Using this approach, both NHEJ and SSA were found to give rise to reciprocal translocations; in fact, both derivative chromosome could form by NHEJ or SSA or one derivative chromosome could form by NHEJ and the other SSA (Elliott et al., 2005; Richardson and Jasin, 2000b; Weinstock et al., 2007). SSA was highly proficient for translocation formation with identical sequence repeats and presumably because the DSBs occurred close by the repeats and on opposite sides (Elliott et al., 2005). However, when divergent repeats were used, specifically two different Alu elements from the MLL gene, the frequency of SSA-mediated translocations dropped substantially, resulting in more NHEJ-mediated events. Mismatch repair components likely suppress SSA between diverged, but homologous sequences, as it does in other contexts (Elliott et al., 1998). These studies highlight the constraints on SSA for mediating translocation formation, in particular, the degree of sequence identity between the repeats and the positions of the DSBs relative to the repeats.
Most translocation breakpoint junctions observed in tumors from patients join at sequences that do not share significant lengths of homology, indicating that they arose by NHEJ (Mani and Chinnaiyan, 2010). The role of canonical NHEJ was investigated using the NHEJ-based translocations reporters. Translocations were found to be suppressed by canonical NHEJ components ligase IV (LIG4) and Ku70 (Simsek and Jasin, 2010; Weinstock et al., 2007), consistent with results in lymphoid systems where oncogenic translocations increased in the absence of these proteins (Ferguson and Alt, 2001). Translocation junctions were biased in the presence of microhomologies, with or without these canonical NHEJ components, suggesting that alt-NHEJ (MMEJ) gave rise to translocations, even in wild-type cells (Simsek and Jasin, 2010). Consistent with this, translocations were found to be largely dependent on the alt-NHEJ components ligase III (LIG3) (Simsek et al., 2011), the end resection factor CtIP (Zhang and Jasin, 2011), and polymerase theta (Mateos-Gomez et al., 2015), as determined using programmable nucleases. Taken together, these results indicate that alt-NHEJ is the major mechanism for translocation formation in mouse embryonic stem cells.
3. When ZFNs then TALENs arrived on the scene: tailored nucleases for tailored translocations
The rare cutting I-SceI endonuclease proved to be a valuable tool to induce chromosomal translocations. The limitation of this method is the necessity to target genomic loci with the recognition site, yet genome modification prior to 2005 was much more laborious in human cells than in mouse cells. The advent of programmable nucleases tailored to cleave any possible locus within genomes has opened tremendous possibilities to create de novo translocations to generate cancer models.
The development of tailored endonucleases originated in 1996 with the report of a fusion of a zinc finger DNA binding domain with the cleavage domain of the FokI restriction enzyme to create a zinc finger nuclease (ZFN) (Kim et al., 1996). Almost a decade later, a ZFN developed by Sangamo Biosciences was shown to cleave an endogenous locus in human cells to lead to its modification (Urnov et al., 2005). Building on the initial results obtained with I-SceI, our group harnessed ZFNs to induce chromosomal translocations at two endogenous loci in human cells (Brunet et al., 2009; Weinstock et al., 2008). Breakpoint junctions were identified by PCR and clones carrying translocations could be recovered from tumor cell lines.
This study provided the first proof of concept of modeling translocations using custom-designed nucleases. Remarkably, translocations were also obtained in multipotential stem cells, both human embryonic stem cells and mesenchymal cells derived from them (Brunet et al., 2009). Following this work, a cancer-relevant translocation was recapitulated, t(11;22)(q24;q12), the most common rearrangement found in Ewing sarcoma, by designing ZFNs to target the most common breakpoints found in patients. Reciprocal translocations were readily recovered in mesenchymal precursor cells, leading to EWSR1-FLI1 fusion gene expression from the endogenous EWSR1 promoter. Notably, the joining characteristics – deletions, insertions, mutations – found in translocations resulting from ZFN cleavage fully recapitulated those from Ewing patient cells and demonstrated that the junctions arose by an NHEJ pathway. Of note, the FokI cleavage domain in a ZFN works as a dimer, with each monomer fused to a different assembly of zinc fingers for DNA recognition. This leaves open the possibility that incorrect ZFNs could form to cleave newly formed translocation junctions or off-target sites. Modified FokI domains that heterodimerize have been developed that strongly promote the use of the correct partner (Doyon et al., 2011); these are particularly valuable for the simultaneous use of pairs of ZFNs as required for translocation formation
The technical complexity of designing and assembling highly specific and active zinc fingers, however, prohibited the wide spread use of ZFNs (“democratization”; (Jasin and Haber, 2016)) by academic researchers. In 2010 the development of TALENs (Transcription Activator-like Effector Nucleases) extended the repertoire of tailored nucleases to one with a much more elementary code of base recognition (Miller et al., 2011). TALENs use the same homo- or heterodimeric FokI cleavage domains as ZFNs, but assembling modules for DNA sequence recognition became much more trivial. As with ZFNs, the use of two TALENs enabled the formation of translocations (Piganeau et al., 2013). Modeling t(2;5)(p23;q35), found in cases of ALCL, our group showed expression of oncogenic NPM1-ALK kinase activation in human cell lines. Conversely, the NPM1-ALK translocation in a patient cell line could be reverted with the same pair of TALENs, restoring the integrity of the two participating chromosomes and potentially permitting the analysis of phenotypic consequences of fusion protein loss once cells are transformed.
4. The CRISPR-Cas9 revolution: when easy is made easier
Soon after TALEN development, CRISPR-Cas9 appeared upon the scene as a highly simplified tailored nuclease, using a guide RNA (gRNA or sgRNA) to recognize the complementary DNA sequence in the genome (Jinek et al., 2012). This new nuclease was quickly used to induce chromosomal translocations, including the previously described models of NPM1-ALK (Ghezraoui et al., 2014) and Ewing sarcoma (Torres et al., 2014) (Renouf et al., 2014) and new models of lung cancer translocations (Choi and Meyerson, 2014) and acute myelogenous leukemia (Renouf et al., 2014; Torres et al., 2014). CRISPR-Cas9 was also used by other teams to create chromosomal translocations in mouse embryonic stem cells (Jiang et al., 2016) and in mouse myoblasts, the latter modeling the human alveolar rhabdyomyosarcoma Pax3-Foxo1 (Lagutina et al., 2015), but also in other organisms, namely C. elegans (Chen et al., 2015) and Leishmania (Zhang et al., 2017).
Thus, it is now possible to faithfully model the full outcome of these chromosome rearrangements, including the formation of the reciprocal translocation, loss of one intact copy of each participating gene, recapitulates potential haploinsufficiency, and fusion gene expression from the endogenous promoter: basically the holy grail for those in quest of relevant translocation cancer models.
5. Isolating translocation clones
Despite their success, these studies also showed that isolation of translocation clones induced de novo remains tedious irrespective of the type of nuclease used and particularly in primary cells for which long sib-selection cycles are mostly unworkable. Whether it is just a matter of efficiency – translocation formation being much less efficient than intra-chromosomal repair – or whether expression of the fusion gene directly affects proliferation of the cells remains to be elucidated. Attempts to increase the translocation frequency have involved short single-strand oligonucleotides matching the DSB ends formed by CRISPR-Cas9 to “guide” joining of the two chromosome ends for translocation formation (Torres-Ruiz et al., 2017).
A strategy for selecting translocation clones has also recently been developed, using EWSR1-WT1 found in desmoplastic round cell tumors as a model (Vanoli et al., 2017). The approach uses CRISPR-Cas9 to induce integration of a homologous donor fragment containing a selectable marker at DSBs on the translocating chromosomes. The selectable marker is promoterless and contains an upstream splice acceptor to strongly enrich for HR events at the EWSR1 locus. A further refinement is that the selectable marker is flanked by LoxP sites, such that fusion protein expression is conditional and dependent on removal of the selectable marker cassette by expression of Cre recombinase. This strategy has also proved to be effective in a tumor cell line (Spraggon et al., 2017).
6. Modeling oncogenesis using tailored nucleases: first steps towards full transformation in vivo
The first example of oncogenesis obtained in vivo from a nuclease-induced rearrangement was published in 2014 and involved formation of the EML4-ALK fusion through an 11 Mb inversion (Maddalo et al., 2014). Although not a translocation per se involving two different chromosomes, this report provided direct evidence that the formation of the EML4-ALK fusion induced by CRISPR-Cas9 activity in the lungs of mice leads to lung adenocarcinomas. In parallel, another group obtained similar results (Blasco et al., 2014). While the high efficiency of transformation obtained in these studies is likely related to a high rate of intra-chromosomal rearrangement, this study suggests translocations between two chromosomes could be tested in a similar manner.
While TALENs have been used to generate an MLL-AF9 fusion by knock-in (Buechele et al., 2015)), faithful de novo translocations giving rise to MLL-AF4 and MLL-AF9 have also been produced by TALENs in CD34+ cells (Breese et al., 2015). Although transformation to leukemia did not occur in vitro, a range of phenotypes was observed, from frequent loss of cells to persistent proliferative advantage of a few cells, as well as some clones showing a transient proliferative advantage. More recently another group reached similar conclusions with the MLLENL translocation in CD34+ cells in vitro, with cells forming normal hematopoietic colonies but eventually ceasing to proliferate, even if some clones shown extended plating capacity (Reimer et al., 2017). However, while the first round recipient mice initiated a “monocytic leukemia-like phenotype” but not immature AML, the second round recipients developed AML but with incomplete penetrance.
Despite the limitations uncovered in these studies, nuclease induced-translocation models reveal new aspects of tumorigenesis and will undoubtedly in a near future provide new insights about the timing between the translocation occurrence and the appearance of the disease, more relevant to progression development found in patients by bypassing limitations of models expressing ectopically fusion genes. The role of the in vivo environment and the undeniably pivotal role of accumulation of secondary mutations certainly remains to be elucidated.
7. Elucidation of translocation mechanisms in human cells using programmable nucleases
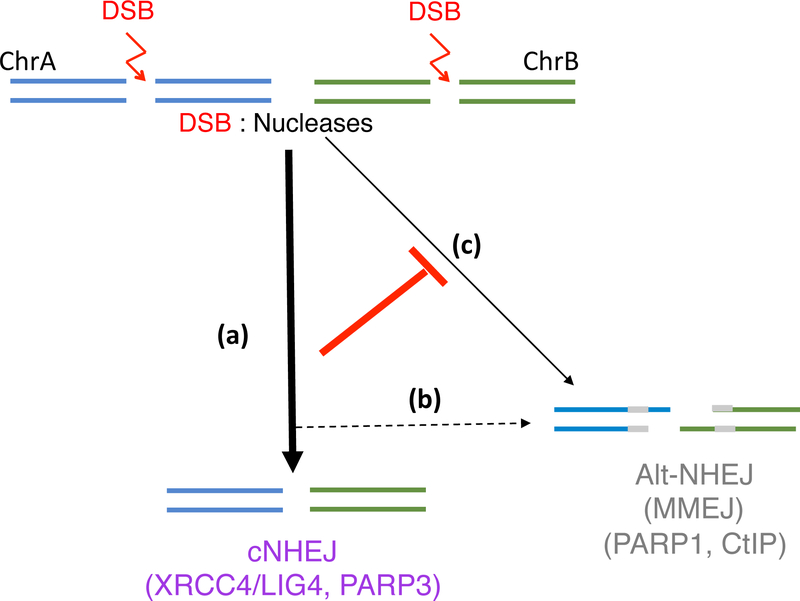
While translocations induced in mouse cells primarily arise by alt-NHEJ and are suppressed by c-NHEJ components, translocations induced by programmable nucleases (ZFNs, TALENs, CRISPR-Cas9) in human cells have breakpoint junctions characterized by little or no end processing, suggestive of c-NHEJ (Brunet et al., 2009; Ghezraoui et al., 2014). In one study, half of the breakpoints in c-NHEJ-proficient cells demonstrated almost perfect joining of the ends (≤1bp deletion) with few microhomologies (Ghezraoui et al., 2014). Confirming the involvement of c-NHEJ, loss of LIG4 (or its partner XRCC4) reduced translocations in multiple cell lines. More recently, PARP3, which cooperates in the recruitment of c-NHEJ factors, has also been shown to participate to translocation formation (Day et al., 2017). Conversely, translocation frequency in human cells is not affected by loss of alt-NHEJ components LIG3 or CtIP (Ghezraoui et al., 2014). In the absence of active c-NHEJ, the repertoire of breakpoint junctions is substantially modified with the appearance of numerous long deletions and the presence of longer microhomologies, consistent with a switch from c-NHEJ to alt-NHEJ (MMEJ). This conclusion is supported by the drastic reduction in translocation frequency when both c-NHEJ and alt-NHEJ components are lost.
Paired Cas9 nickases (nCas9) have also been used to induce translocations (Ghezraoui et al., 2014). In this case, two gRNAs directed to opposite strands of each chromosome are used to generate two nicks which can be converted to DSBs with 5’ overhangs of ~40 bp that join to form translocations. At breakpoint junctions, deletions are substantially longer than found with unmodified Cas9, involving loss of sequences from the overhangs. The portions of the overhangs that are preserved are filled by DNA synthesis, which leads to duplications of sequences at breakpoint junctions. Microhomology is also increased, consistent with annealing of bases within the overhangs. Paired nickase-generated translocations are largely dependent on LIG4, demonstrating that c-NHEJ can give rise to junctions with microhomology.
Other groups have implicated PARP1, a reported alt-NHEJ component (Audebert et al., 2004), in translocation formation in human cells. PARP1 knockdown and PARP1 inhibition (olaparib) have both been shown to reduce translocations (Soni et al., 2014; Wray et al., 2013), while another report has shown that PARP1 overexpression can increase translocations in some, but not all, cell lines (Torres-Ruiz et al., 2017). Translocations induced by irradiation have also been studied. Interestingly, CtIP has been shown to affect translocation formation in G1-irradiated cells at late time points after irradiation, but not in G2-irradiated cells (Barton et al., 2014; Biehs et al., 2017). The authors suggest that these events arise from a subtype of c-NHEJ they called resection-dependent c-NHEJ, which is dependent on Artemis, DNA-PK and the exonuclease activity of MRE11. As IR induced complex DSBs which probably need a step of maturation before joining, we can wonder what is the exact contribution of resection-dependent c-NHEJ in the formation of nuclease-induced translocations where “cleaner” DSBs are induced. In conclusion, c-NHEJ is directly implicated in translocation formation in human cells, although a small contribution of alt-NHEJ cannot be excluded (Figure 2). However, when c-NHEJ is impaired alt-NHEJ become critical. It should be noted that the absence of NHEJ components can leave spontaneously arising DSBs unrepaired; the increased frequency of breakage genome wide may then promote translocations even in the absence of induced DSBs.
Figure 2 : Translocation mechanisms in human with programmable nucleases.
Chromosomal translocations in human cells are principally formed by c-NHEJ (a) although a small contribution of alt-NHEJ cannot be excluded (b). In absence of active c-NHEJ, the contribution of alt-NHEJ become critical (c).
8. How model systems recapitulate patient breakpoints
Numerous studies have reported breakpoint junctions for various translocations found in tumors. Oftentimes, only the junction sequences for the oncogenic translocation have been reported, although several publications include both junctions from the reciprocal (balanced) translocation. Concerning deletions, while most of the breakpoints are accompanied by deletions, the median deletion length remains short (e.g., 1 bp, (Nilsson et al., 2017); 5 bp, (Reiter et al., 2003); 14 bp, (Gillert et al., 1999)). Larger deletions (>1000 bp) are also observed at lower frequency, arising either by resection or possibly as the result of several breaks. Notably, perfect joining of ends has also been reported (e.g., 37% of junctions (Nilsson et al., 2017)). Microhomologies have been observed in 20–40% of translocation breakpoint junctions from tumors (Mattarucchi et al., 2008; Nilsson et al., 2017; Weckselblatt et al., 2015). The definition of microhomology can differ, however, as authors have sometimes considered there to be microhomologies when short repeated sequences are found after short (templated or not) insertions but not exactly corresponding to the 2 original breakpoint sequences (Mattarucchi et al., 2008; Nilsson et al., 2017), making it difficult to compare various studies. Particularly, a mechanism of template switching potentially use microhomologies to facilitate ligation of templated insertions with the partner chromosome, not reflecting altNHEJ activity.
ZFN-induction of the common Ewing translocation has been shown to give rise to breakpoint junctions that fully recapitulate those found in patient tumor cells with a comparable proportion of each type of junction (deletions, insertions, microhomology) (Piganeau et al., 2013) (Zucman-Rossi et al., 1998). More complex junctions, albeit happening to a lesser extent, can also be recovered in these this model and are likely to arise by similar repair mechanisms as in tumor cells from patients. In some cases, a plausible mechanism for their formation is replication primed by one of the DNA ends using microhomology reminiscence of the template switching mechanism described in patient cells (Mattarucchi et al., 2008; Nilsson et al., 2017). Of note the use of nCas9 provides more flexibility in DNA end structures potentially leading to more complex rearrangements found in certain type of tumors, i.e., duplications (Ghezraoui et al., 2014; Renouf et al., 2014)
In summary, the induction of chromosomal translocations in human cells with programmable nucleases provides a relevant model for deciphering repair mechanisms leading to this genome rearrangement and holds promise for deciphering the early events leading to oncogenesis.
Acknowledgments
MSK research is supported by NIH/NCI Cancer Center support grant P30 CA008748. This work was supported in part by an Alex’s Lemonade Stand Innovation Award and NIH R01CA185660 and R35GM118175 (M.J.). E.B. research is supported by ANR-12-JSV6–0005, the Canceropole IdF, the Institut National du Cancer and la Ligue Contre le Cancer (Equipe de Villartay, Labelisée LA LIGUE).
References
- Audebert M, Salles B, and Calsou P (2004). Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem 279, 55117–55126. [DOI] [PubMed] [Google Scholar]
- Barton O, Naumann SC, Diemer-Biehs R, Kunzel J, Steinlage M, Conrad S, Makharashvili N, Wang J, Feng L, Lopez BS, et al. (2014). Polo-like kinase 3 regulates CtIP during DNA double-strand break repair in G1. J Cell Biol 206, 877–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biehs R, Steinlage M, Barton O, Juhasz S, Kunzel J, Spies J, Shibata A, Jeggo PA, and Lobrich M (2017). DNA Double-Strand Break Resection Occurs during Non-homologous End Joining in G1 but Is Distinct from Resection during Homologous Recombination. Mol Cell 65, 671–684 e675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco RB, Karaca E, Ambrogio C, Cheong TC, Karayol E, Minero VG, Voena C, and Chiarle R (2014). Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell Rep 9, 1219–1227. [DOI] [PubMed] [Google Scholar]
- Breese EH, Buechele C, Dawson C, Cleary ML, and Porteus MH (2015). Use of Genome Engineering to Create Patient Specific MLL Translocations in Primary Human Hematopoietic Stem and Progenitor Cells. PLoS One 10, e0136644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet E, Simsek D, Tomishima M, DeKelver R, Choi VM, Gregory P, Urnov F, Weinstock DM, and Jasin M (2009). Chromosomal translocations induced at specified loci in human stem cells. Proc Natl Acad Sci U S A 106, 10620–10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buechele C, Breese EH, Schneidawind D, Lin CH, Jeong J, Duque-Afonso J, Wong SH, Smith KS, Negrin RS, Porteus M, et al. (2015). MLL leukemia induction by genome editing of human CD34+ hematopoietic cells. Blood 126, 1683–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Barral P, Vannier JB, Borel V, Steger M, Tomas-Loba A, Sartori AA, Adams IR, Batista FD, and Boulton SJ (2013). RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol Cell 49, 858–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Li M, Feng X, and Guang S (2015). Targeted Chromosomal Translocations and Essential Gene Knockout Using CRISPR/Cas9 Technology in Caenorhabditis elegans. Genetics 201, 1295–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi PS, and Meyerson M (2014). Targeted genomic rearrangements using CRISPR/Cas technology. Nat Commun 5, 3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleaux L, d’Auriol L, Gailbert F, and Dujon B (1988). Recognition and cleavage site of the intron-encoded omega transposase. Proc Natl Acad Sci USA 85, 6022–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino L, Sotiriou SK, Rantala JK, Magin S, Mladenov E, Helleday T, Haber JE, Iliakis G, Kallioniemi OP, and Halazonetis TD (2014). Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science 343, 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day TA, Layer JV, Cleary JP, Guha S, Stevenson KE, Tivey T, Kim S, Schinzel AC, Izzo F, Doench J, et al. (2017). PARP3 is a promoter of chromosomal rearrangements and limits G4 DNA. Nat Commun 8, 15110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J, Xia DF, Miller JC, Urnov FD, Gregory PD, and Holmes MC (2011). Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods 8, 74–79. [DOI] [PubMed] [Google Scholar]
- Elliott B, Richardson C, and Jasin M (2005). Chromosomal translocation mechanisms at intronic alu elements in mammalian cells. Molecular cell 17, 885–894. [DOI] [PubMed] [Google Scholar]
- Elliott B, Richardson C, Winderbaum J, Nickoloff JA, and Jasin M (1998). Gene conversion tracts from double-strand break repair in mammalian cells. Mol Cell Biol 18, 93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson DO, and Alt FW (2001). DNA double strand break repair and chromosomal translocation: lessons from animal models. Oncogene 20, 5572–5579. [DOI] [PubMed] [Google Scholar]
- Ghezraoui H, Piganeau M, Renouf B, Renaud JB, Sallmyr A, Ruis B, Oh S, Tomkinson AE, Hendrickson EA, Giovannangeli C, et al. (2014). Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol Cell 55, 829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillert E, Leis T, Repp R, Reichel M, Hosch A, Breitenlohner I, Angermuller S, Borkhardt A, Harbott J, Lampert F, et al. (1999). A DNA damage repair mechanism is involved in the origin of chromosomal translocations t(4;11) in primary leukemic cells. Oncogene 18, 4663–4671. [DOI] [PubMed] [Google Scholar]
- Harris S, Rudnicki KS, and Haber JE (1993). Gene conversions and crossing over during homologous and homeologous ectopic recombination in Saccharomyces cerevisiae. Genetics 135, 5–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M, and Haber JE (2016). The democratization of gene editing: Insights from site-specific cleavage and double-strand break repair. DNA Repair (Amst) 44, 6–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M, and Rothstein R (2013). Repair of strand breaks by homologous recombination. Cold Spring Harbor perspectives in biology 5, a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Zhang L, Zhou X, Chen X, Huang G, Li F, Wang R, Wu N, Yan Y, Tong C, et al. (2016). Induction of site-specific chromosomal translocations in embryonic stem cells by CRISPR/Cas9. Sci Rep 6, 21918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, and Charpentier E (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RD, and Jasin M (2000). Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J 19, 3398–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass EM, and Jasin M (2010). Collaboration and competition between DNA double-strand break repair pathways. FEBS letters 584, 3703–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Peterson SE, Jasin M, and Keeney S (2016). Mechanisms of germ line genome instability. Semin Cell Dev Biol 54, 177–187. [DOI] [PubMed] [Google Scholar]
- Kim YG, Cha J, and Chandrasegaran S (1996). Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci U S A 93, 1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagutina IV, Valentine V, Picchione F, Harwood F, Valentine MB, Villarejo-Balcells B, Carvajal JJ, and Grosveld GC (2015). Modeling of the human alveolar rhabdomyosarcoma Pax3-Foxo1 chromosome translocation in mouse myoblasts using CRISPR-Cas9 nuclease. PLoS Genet 11, e1004951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRocque JR, Stark JM, Oh J, Bojilova E, Yusa K, Horie K, Takeda J, and Jasin M (2011). Interhomolog recombination and loss of heterozygosity in wild-type and Bloom syndrome helicase (BLM)-deficient mammalian cells. Proc Natl Acad Sci U S A 108, 11971–11976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddalo D, Manchado E, Concepcion CP, Bonetti C, Vidigal JA, Han YC, Ogrodowski P, Crippa A, Rekhtman N, de Stanchina E, et al. (2014). In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature 516, 423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani RS, and Chinnaiyan AM (2010). Triggers for genomic rearrangements: insights into genomic, cellular and environmental influences. Nat Rev Genet 11, 819–829. [DOI] [PubMed] [Google Scholar]
- Mateos-Gomez PA, Gong F, Nair N, Miller KM, Lazzerini-Denchi E, and Sfeir A (2015). Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature 518, 254–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattarucchi E, Guerini V, Rambaldi A, Campiotti L, Venco A, Pasquali F, Lo Curto F, and Porta G (2008). Microhomologies and interspersed repeat elements at genomic breakpoints in chronic myeloid leukemia. Genes Chromosomes Cancer 47, 625–632. [DOI] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. (2011). A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29, 143–148. [DOI] [PubMed] [Google Scholar]
- Moynahan ME, and Jasin M (2010). Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol 11, 196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson D, Pettersson M, Gustavsson P, Forster A, Hofmeister W, Wincent J, Zachariadis V, Anderlid BM, Nordgren A, Makitie O, et al. (2017). Whole-Genome Sequencing of Cytogenetically Balanced Chromosome Translocations Identifies Potentially Pathological Gene Disruptions and Highlights the Importance of Microhomology in the Mechanism of Formation. Hum Mutat 38, 180–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piganeau M, Ghezraoui H, De Cian A, Guittat L, Tomishima M, Perrouault L, Rene O, Katibah GE, Zhang L, Holmes MC, et al. (2013). Cancer translocations in human cells induced by zinc finger and TALE nucleases. Genome Res 23, 1182–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer J, Knoess S, Labuhn M, Charpentier EM, Gohring G, Schlegelberger B, Klusmann JH, and Heckl D (2017). CRISPR-Cas9-induced t(11;19)/MLL-ENL translocations initiate leukemia in human hematopoietic progenitor cells in vivo. Haematologica. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter A, Saussele S, Grimwade D, Wiemels JL, Segal MR, Lafage-Pochitaloff M, Walz C, Weisser A, Hochhaus A, Willer A, et al. (2003). Genomic anatomy of the specific reciprocal translocation t(15;17) in acute promyelocytic leukemia. Genes Chromosomes Cancer 36, 175–188. [DOI] [PubMed] [Google Scholar]
- Renouf B, Piganeau M, Ghezraoui H, Jasin M, and Brunet E (2014). Creating cancer translocations in human cells using Cas9 DSBs and nCas9 paired nicks. Methods Enzymol 546, 251–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C, and Jasin M (2000a). Coupled homologous and nonhomologous repair of a double-strand break preserves genomic integrity in mammalian cells. Mol Cell Biol 20, 9068–9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C, and Jasin M (2000b). Frequent chromosomal translocations induced by DNA double-strand breaks. Nature 405, 697–700. [DOI] [PubMed] [Google Scholar]
- Richardson C, Moynahan ME, and Jasin M (1998). Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev 12, 3831–3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KP, Mercer DK, Richardson AJ, Melville CM, Glover LA, and Flint HJ (2000). Chromosomal integration of the green fluorescent protein gene in lactic acid bacteria and the survival of marked strains in human gut simulations. FEMS Microbiol Lett 182, 23–27. [DOI] [PubMed] [Google Scholar]
- Sfeir A, and Symington LS (2015). Microhomology-Mediated End Joining: A Back-up Survival Mechanism or Dedicated Pathway? Trends Biochem Sci 40, 701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Brunet E, Wong SY, Katyal S, Gao Y, McKinnon PJ, Lou J, Zhang L, Li J, Rebar EJ, et al. (2011). DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet 7, e1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, and Jasin M (2010). Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nature structural & molecular biology 17, 410–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni A, Siemann M, Grabos M, Murmann T, Pantelias GE, and Iliakis G (2014). Requirement for Parp-1 and DNA ligases 1 or 3 but not of Xrcc1 in chromosomal translocation formation by backup end joining. Nucleic Acids Res 42, 6380–6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spraggon L, Martelotto LG, Hmeljak J, Hitchman TD, Wang J, Wang L, Slotkin EK, Fan PD, Reis-Filho JS, and Ladanyi M (2017). Generation of conditional oncogenic chromosomal translocations using CRISPR-Cas9 genomic editing and homology-directed repair. J Pathol 242, 102–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark JM, and Jasin M (2003). Extensive loss of heterozygosity is suppressed during homologous repair of chromosomal breaks. Mol Cell Biol 23, 733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark JM, Pierce AJ, Oh J, Pastink A, and Jasin M (2004). Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol 24, 9305–9316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS, and Gautier J (2011). Double-strand break end resection and repair pathway choice. Annual review of genetics 45, 247–271. [DOI] [PubMed] [Google Scholar]
- Torres R, Martin MC, Garcia A, Cigudosa JC, Ramirez JC, and Rodriguez-Perales S (2014). Engineering human tumour-associated chromosomal translocations with the RNA-guided CRISPR-Cas9 system. Nat Commun 5, 3964. [DOI] [PubMed] [Google Scholar]
- Torres-Ruiz R, Martinez-Lage M, Martin MC, Garcia A, Bueno C, Castano J, Ramirez JC, Menendez P, Cigudosa JC, and Rodriguez-Perales S (2017). Efficient Recreation of t(11;22) EWSR1-FLI1+ in Human Stem Cells Using CRISPR/Cas9. Stem Cell Reports 8, 1408–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SD, and Alexander DR (2005). What have we learnt from mouse models of NPMALK-induced lymphomagenesis? Leukemia 19, 1128–1134. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, and Holmes MC (2005). Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435, 646–651. [DOI] [PubMed] [Google Scholar]
- Vanoli F, Tomishima M, Feng W, Lamribet K, Babin L, Brunet E, and Jasin M (2017). CRISPR-Cas9-guided oncogenic chromosomal translocations with conditional fusion protein expression in human mesenchymal cells. Proc Natl Acad Sci U S A 114, 3696–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckselblatt B, Hermetz KE, and Rudd MK (2015). Unbalanced translocations arise from diverse mutational mechanisms including chromothripsis. Genome Res 25, 937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock DM, Brunet E, and Jasin M (2007). Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat Cell Biol 9, 978–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock DM, Brunet E, and Jasin M (2008). Induction of chromosomal translocations in mouse and human cells using site-specific endonucleases. J Natl Cancer Inst Monogr, 20–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock DM, Richardson CA, Elliott B, and Jasin M (2006). Modeling oncogenic translocations: distinct roles for double-strand break repair pathways in translocation formation in mammalian cells. DNA Repair (Amst) 5, 1065–1074. [DOI] [PubMed] [Google Scholar]
- Wray J, Williamson EA, Singh SB, Wu Y, Cogle CR, Weinstock DM, Zhang Y, Lee SH, Zhou D, Shao L, et al. (2013). PARP1 is required for chromosomal translocations. Blood 121, 4359–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WW, Lypaczewski P, and Matlashewski G (2017). Optimized CRISPR-Cas9 Genome Editing for Leishmania and Its Use To Target a Multigene Family, Induce Chromosomal Translocation, and Study DNA Break Repair Mechanisms. mSphere 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, and Jasin M (2011). An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat Struct Mol Biol 18, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucman-Rossi J, Legoix P, Victor JM, Lopez B, and Thomas G (1998). Chromosome translocation based on illegitimate recombination in human tumors. Proc Natl Acad Sci U S A 95, 11786–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]