Abstract
Candida albicans is one of the most common fungal pathogens of humans. Currently, there are limitations in the evaluation of C. albicans infection in existing animal models, especially in terms of understanding the influence of specific infectious stages of the fungal pathogen on the host. We show that C. albicans infects, grows and invades tissues in the planarian flatworm Schmidtea mediterranea, and that the planarian responds to infection by activating components of the host innate immune system to clear and repair host tissues. We study different stages of C. albicans infection and demonstrate that planarian stem cells increase division in response to fungal infection, a process that is likely evolutionarily conserved in metazoans. Our results implicate MORN2 and TAK1/p38 signaling pathways as possible mediators of the host innate immune response to fungal infection. We propose the use of planarians as a model system to investigate host-pathogen interactions during fungal infections.
Keywords: Innate immunity, planarians, Platyhelmithes, invertebrates, Candida albicans, pathogenic fungi, clearance, infection, neoblasts, stem cells
One Sentence Summary:
Planaria: a new model for fungal infections
INTRODUCTION
Fungal infections rank among the top ten causes of human death, with over 1 billion people currently infected with a fungal disease worldwide (WHO, 2018). Together with the increase in antifungal drug resistance emerging in recent years, pathogenic fungi represent a notable global threat to human health (Fisher et al., 2018; Fisher et al., 2012). Candida albicans can lead to infections that range from superficial cutaneous and mucosal infections to severe systemic and invasive infections resulting in over 40% human mortality (Bongomin et al., 2017; Brown et al., 2012). The current gold-standard in the field to study C. albicans virulence is the mouse tail-vein injection model, where C. albicans is injected directly into the host bloodstream (Clancy et al., 2009). This model, however, only captures the late stages of C. albicans infection in humans, once the fungus has breached the host defenses to enter the bloodstream. Therefore, there is a need to understand the earlier stages of infection, such as how C. albicans breaches host barriers and defends against the host innate immune system.
C. albicans is a polymorphic fungal pathogen that has the ability to change morphologies depending on its environment. For example, C. albicans can reversibly switch between round yeast form cells to elongated filamentous cells (hyphae and pseudohyphae) in response to specific environmental cues, which is an important virulence factor for C. albicans host infection (Braun and Johnson, 1997; Saville et al., 2003). The yeast to hyphal transition is important for C. albicans infection, colonization, and evasion of the host immune system; the yeast form cells facilitate dissemination through the blood stream, while the hyphal cells cause tissue damage and invasion as well as aid in the escape out of phagocytic cells. In fact, it is well-established that C. albicans strains genetically modified to remain “locked” in the yeast or filamentous forms are unable to undergo the yeast to hyphal transition, resulting in highly attenuated conditions for virulence in mouse models of infection (Lo et al., 1997; Saville et al., 2003).
Planarians are flatworms (Platyhelminthes) with high rates of cellular turnover and an extraordinary capacity to regenerate, both processes of which are based on adult stem cells (SCs) called neoblasts (Aboobaker, 2011; King and Newmark, 2012; Pellettieri and Sanchez Alvarado, 2007; Reddien and Sanchez Alvarado, 2004; Wagner et al., 2011). The immune system of planaria has a high-degree of evolutionary conservation to the innate immune system of humans (Abnave and Ghigo, 2018; Li et al., 2018; Lu et al., 2017; Maciel and Oviedo, 2018; Peiris et al., 2014; Tsoumtsa et al., 2018). Additionally, similar to humans, planarians are able to distinguish between commensal and pathogenic bacteria (Abnave et al., 2014; Arnold et al., 2016; Keating et al., 2017; Torre et al., 2017b). In fact, it takes only a few days for planaria to eliminate infections of a wide range of pathogenic Gram-negative and Gram-positive bacteria, as well as mycobacteria (e.g. L. pneumophila, S. aureus, and M. tuberculosis, respectively) (Abnave et al., 2014). Thus, previous analyses of the planaria innate immune system during bacterial infection has provided useful information about host response strategies aimed at clearance of bacterial pathogens (Abnave and Ghigo, 2018; Abnave et al., 2014; Arnold et al., 2016; Gao et al., 2017; Hammoudi et al., 2018; Li et al., 2018; Lu et al., 2017; Pang et al., 2016; Torre et al., 2017a; Tsoumtsa et al., 2018; Tsoumtsa et al., 2017). Indeed, the function of the gene encoding the human homolog of Membrane Occupation and Recognition Nexus repeat containing-2 (MORN2) was unknown until a screen in planarians allowed for the identification of its function in phagocytosis and clearance of bacterial infections (Abnave et al., 2014). Interestingly, MORN2 is conserved in planarians and humans, but it is not present in other invertebrate models namely D. melanogaster or C. elegans (Abnave et al., 2014), and thus its identification would have been missed using other invertebrate infection models.
The study of vertebrate and invertebrate in vivo models of candidiasis to date has provided important insights into the pathogenesis of fungal infections (Bergeron et al., 2017; Chamilos et al., 2007; Fuchs et al., 2010; Glittenberg et al., 2011; Gratacap et al., 2017; Mallick et al., 2016; Mylonakis, 2008; Mylonakis et al., 2007; Peterson and Pukkila-Worley, 2018; Pukkila-Worley et al., 2009a; Segal and Frenkel, 2018). However, there are still existing limitations in analyzing host- pathogen interactions in the context of the adult body and the stem cell response. For example, although mammalian models such as the rat, mouse, and rabbit are the closest species used to mimic human infections, their use is limited by sample size, handling, and cost. On the other hand, invertebrate models, including Drosophila melanogaster, Caenorhabditis elegans and Galleria mellonella, although cost-effective, are limited in their use to analyze embryonic stages, infection methodology (e.g. injections cause injury), and evolutionary divergence from humans, among other limitations. Here, we introduce the use of the planarian Schmidtea mediterranea as an alternative invertebrate model to study different stages of fungal infection with C. albicans. The simplified anatomy of adult planarians enables an in-depth analysis of fungal infections and the effects they may have on signaling pathways regulating the crosstalk between stem cells and various differentiated tissues. We developed a fungal infection strategy using C. albicans as the pathogen and the planaria S. mediterranea as the host. We established protocols to visualize C. albicans distribution during infection of planarian tissues. These analyses determined that fungi rapidly infect and grow inside planaria causing damage to superficial and deep tissues. Despite this aggressive pathogen invasion, it only takes a few days for S. mediterranea to halt C. albicans growth and clear the infection while reestablishing form and function of damaged tissues. Furthermore, we find that fungal infection activates neoblast proliferation and increases the expression of highly conserved host molecular cascades such as MORN2 and TAK1/p38.
METHODS
Planarian Culture
The asexual strain CIW4 of planaria species Schmidtea mediterranea was used for all assays. The planaria culture was maintained as previously described (Oviedo et al., 2008).
Microorganisms
Candida albicans strain SN250 was used as the wild-type control strain, and hyper-filamentous strain TF125 (nrg1/nrg1) (Homann et al., 2009), and non- filamentous strain TF156 (efg1/efg1) (Homann et al., 2009) were used to assess the effects of filamentation on the infection assays. Strains were grown on yeast peptone dextrose (YPD) agar plates for two days at 30°C, and single colonies were selected and cultured in YPD liquid medium overnight at 30°C to obtain cultures for infection assays.
S. mediterranea challenge/infection assay
About ten planarians were kept in plastic wells containing 3 mL of water to which specific concentrations of C. albicans cells were added and the total volume was adjusted to 4mL. The animals were kept in the infected media for three days. After this initial three-day exposure, the planaria were washed daily with fresh water and observed under the microscope to record any behavioral or macroscopic defects for the next seven days. All procedures before, during and after infection with C. albicans were performed in water at room temperature, which is the ideal medium and temperature for planaria. Under these conditions, C. albicans are not actively dividing in the media, and thus the infection period was extended to three days.
CFU Counting
Planaria were collected after three days of infection as well as at other indicated time points. The animals were homogenized in 1mL of 1xPBS. The homogenate was further diluted into 10mL of 1xPBS, and 250uL of the homogenate was then plated onto YPD media agar plates containing ampicillin (1|jg/mL). After an overnight incubation at 30°C, C. albicans colonies were counted.
Whole Mount Immunofluorescence
Planarians were sacrificed by placing them in 5.7% of 12N HCL solution for 5 mins and fixing them in Carnoys solution for 2 hours on ice. After this, planarians were stored in methanol at −20°C for at least an hour and then bleached overnight in 6% H2O2 solution. Animals were rehydrated in dilutions of Methanol:PBSTx and stained as previously described (Thiruvalluvan et al., 2018). Primary antibodies: α-H3p, 1:250 (Millipore Cat# 05–817R) and caspase- 3, 1:500 (Abcam ab13847). Secondary antibodies: Goat anti-rabbit Alexa568, 1:800 (Invitrogen Cat# 11036) for H3P, HRP-conjugated goat anti-rabbit antibody (Millipore Cat# 12–348) with TSA-Alexa568 anti-HRP for caspase-3, 1:2000. For C. albicans staining and p-38 staining in the planaria whole mounts, animals were sacrificed in 10% NAC diluted in PBS. The animals were then fixed in 4% formaldehyde in 0.3% PBSTx and permeabilized with 1% SDS. Animals were then bleached in 6% H2O2 in 1xPBS. Primary antibodies: Anti-Candida, 1:500 (ThermoFisher Cat# PA1–27158) and anti-phospho p38, 1:800 (Cell Signaling Technologies CAT# 9211).
TUNEL Assay
Worms were prepared for TUNEL or immunostaining identically to those prepared for C. albicans staining. TUNEL was then performed as previously described (Pellettieri et al., 2010). During the double staining, the animals were fixed in preparation for TUNEL staining. The double stained always started with TUNEL stain followed by blocking with PSTB for four hours before the caspase stain.
Imaging and data processing
Area measurements were calculated using ImageJ software bundled with Java 1.8.0_172 (Schneider et al., 2012) and the differences in animal sizes were determined as fold change in reference relative to control group at each time point. Digital pictures were collected using a Nikon AZ-100 multizoom microscope and NIS Elements AR 3.2 software (Nikon). Brightness and contrast were adjusted using Adobe Photoshop. Neoblasts were counted and normalized to the area using ImageJ. Caspase-3 signal was quantified by measuring levels of fluorescence using NIS element software (Nikon) as previously described (Thiruvalluvan et al., 2018).
Gene expression analysis
RNA was extracted with Trizol from control and infected animals at all three time points. The qPCR reactions were performed using SYBR Green Master Mix in a 7500 Fast Real Time PCR cycler (Applied Biosystems). Each of the reactions were performed in triplicate using the median cycle threshold value for analysis and normalized to the ubiquitously expressed clone H.55.12e, as described previously (Peiris et al., 2016).
RESULTS
Effects of Candida albicans Infection in Planarians
We developed a fungal infection strategy in which C. albicans was introduced to liquid (water) media containing planaria. After three days of incubation at room temperature (i.e. days post-infection-dpi), the media was replaced daily with fresh water and worms were evaluated under the microscope for seven days to record any macroscopic or behavioral changes (Figure 1A). Three strains of C. albicans were used: wild type/WT (SN250), non-filamentous (TF156, an efg1/efg1 mutant), and hyper-filamentous (TF125, an nrg1/nrg1 mutant). In a mouse model of C. albicans bloodstream infection, the non-filamentous efg1/efg1 mutant strain was found to be attenuated for virulence, and in a C. elegans infection model, the hyper-filamentous nrg1/nrg1 mutant strain was highly invasive and virulent (Lo et al., 1997; Pukkila-Worley et al., 2009b). Overall, three days of exposure to C. albicans was generally lethal to planarians (Figure 1B). However, planarian survival rate was lower when exposed to the hyper-filamentous C. albicans strain, requiring 4–8 times the concentration of wild-type and non-filamentous strains to kill 50% of the worms 3 dpi (Figure 1B). Planarian exposure to C. albicans for three days demonstrated a dual host response characterized by either survival or death of the planarians. To better understand the mechanisms guiding survival in the presence of C. albicans, we focused our analysis on animals that survived after 3 dpi. To that end, we established an effective infection concentration of C. albicans in the planarian model at 5 million cells/mL for the hyper-filamentous and 15 million cells/mL for the WT and non-filamentous strains. Next, we determined the capacity of planaria to clear C. albicans from their body by macerating individual worms at different time points of infection and plating the content on fungal-selective medium to determine CFUs. Strikingly, the hyper-filamentous strain grew fewer colonies than the WT and the non- filamentous strains; and at 10 dpi, all C. albicans strains were eliminated from the planaria (Figure 1C).
Figure 1: Infection protocol with C. albicans leads to tissue damage in S. mediterranea.
A) Graphical illustration of the planaria infection timeline with C. albicans. Planaria were placed in small wells with 4mL of water that was inoculated with C. albicans (0.5 × 107 to 6 × 107 cells/mL) and incubated for three days in the dark at room temperature. After the third day liquid was removed and planarians rinsed with freshwater daily for seven days. B) Planarian survival under different concentrations of three strains of C. albicans at the third day of infection. C) Number of C. albicans cells (three different strains) in planarians during the course of infection expressed as colony forming units (CFUs) over time. Note that C. albicans growth peaks during the first few days of incubation but by day 10 post-infection the presence of fungi is dramatically reduced. The experiment was replicated three times with 10 animals per experiment. The C. albicans concentration is 7.5×106 cells/mL for each condition. Two-Way-ANOVA, ****P<0.0001. D) Representative images and percentage of morphological defects observed after incubating planarians with the different C. albicans strains at 7.5 × 106 cells/mL between 4 and 7 days post-infection. Arrows point toward macroscopic lesions in the dorsal side of planarians. E) Percentage of macroscopic defects associated with the different growth forms at 7.5 × 106 cells/mL between 4–7 days post-infection. Scale bar is 200|jm. Two independent replicates were performed with 20 animals each and statistical analyses were based on comparison against the non-filamentous strain. Two-Way-ANOVA, ***P<0.001; ****P<0.0001, NS= no significant (C, E).
Soaking planarians in media containing the different strains of C. albicans (7.5 × 106 cells/mL) for three days led to morphological defects including epithelial lesions, tissue loss (e.g. head regression), partial body lysing and lethality (Figure 1D, E). Live images were taken after the 3rd day of the infection right after the daily rinse with fresh water. The hyper-filamentous strain was the most virulent leading to over 50% of animal death and the remainder of the animals experienced phenotypes of tissue lesion and lysing that affected ~90% of the animals (Figure 1E). Exposure to the WT strain led to lesions, head regression and death that affected 45% of the animals, while the non-filamentous strain affected only 10% of the planarian exposed (Figure 1E). These results demonstrate that the hyper-filamentous strain renders the most aggressive form of infection in S. mediteranea. Moreover, we noticed that at 10 dpi, lesions were completely healed in the surviving animals and their behavior was indistinguishable from the untreated animals.
Visualizing Candida albicans Infection in Planarians
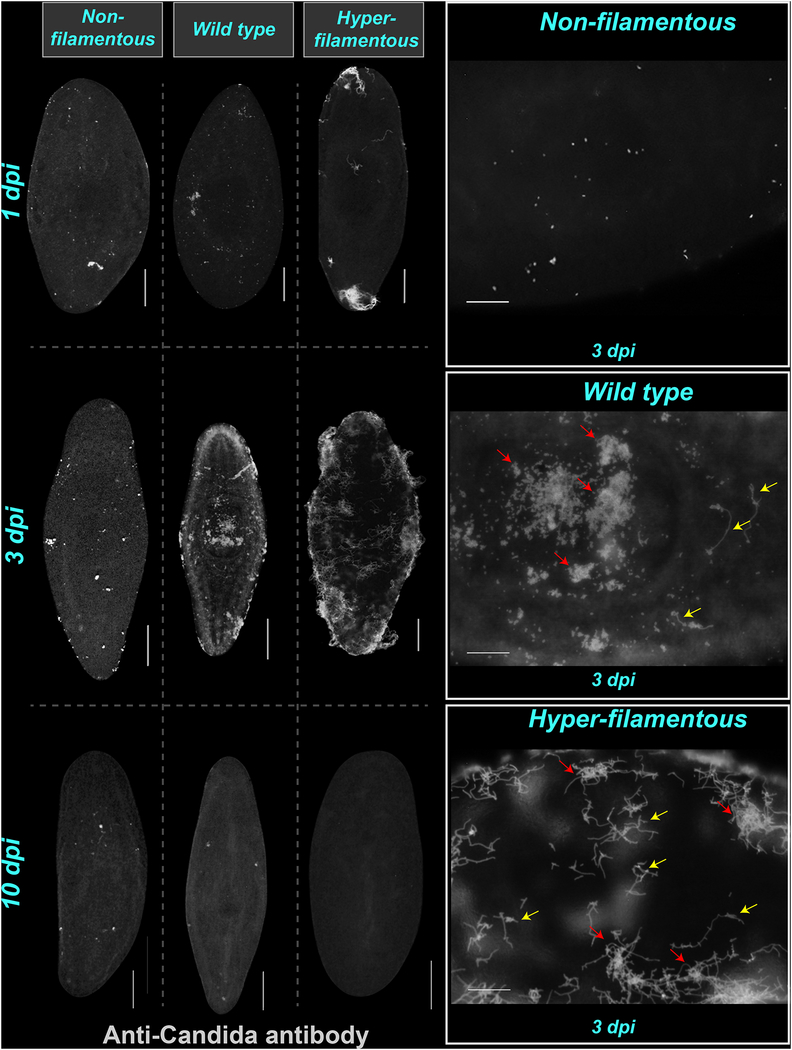
To assess the effects and the spatiotemporal distribution of C. albicans in planaria, we used the anti-Candida antibody (Abcam, #ab53891) at different time points after infection with each C. albicans strain (Figure 2). This approach allowed us to distinguish the distinct morphological forms of C. albicans in whole-mount and in tissue sections, and determine their locations and qualitatively analyze their abundance throughout the planarian body (Figures 2, 3). Generally, C. albicans cells were detected on planarian tissues from day 1 and the concentration gradually increased and peaked by 3 dpi. Interestingly, at 10 dpi C. albicans cells from all strains were dramatically reduced or absent from the planarian body with all three strains (Figure 2). Overall, the infection with the WT and hyper-filamentous strains were more prolific and aggressive by 3 dpi relative to the non-filamentous strain. No obvious tissue tropism was observed as C. albicans cells were observed throughout the planaria body, including within dorsal and ventral surfaces and in deep tissues (Figure 2B).
Figure 2: Visualization of C. albicans during and after infection.
A-C) Representative images of whole-mount anti-Candida antibody stain during infection days 1, 3 and 10. The images show the presence of C. albicans cells on the most superficial layer of planaria tissue. Animals were infected with 2 × 107 cells/mL for the non-filamentous, 1.5 × 107 cells/mL for the wild type and 5 × 106 cells/mL for the hyper-filamentous C. albicans strains. D-F) High-magnification images of planarian tissue with anti-Candida antibody stain at day 3 postinfection with the non-filamentous, wild type and hyper-filamentous strains, respectively. The yellow arrows indicate the presence of hyphae in tissue infected with the wild type strain. The red arrows point towards clusters of C. albicans cells. Scale bar is 200μm. Five independent replicates were performed using 10 animals each.
Figure 3: C. albicans invades epithelium and deeper tissues in S. mediterranea.
A, B) Transverse cross-section images from the middle part of the planarian body exposed to C. albicans strains (red signal) at days 1 and 3 post-infection. The pharynx is the circular structure in the middle of the section. The planaria tissue is counterstained with DAPI (blue). Animals were infected with 2 × 107 cells/mL for the non-filamentous, 1.5 × 107 cells/mL for the wild type and 5 × 106 cells/mL for the hyper-filamentous C. albicans strains. The thickness of the sections was 10 μm and dorsal is shown at the top and ventral at the bottom.
In addition, to the general distribution of C. albicans in the animal, we also noted the tendency of C. albicans to form clusters as the infection progressed (e.g. 3 dpi). The nature of these clusters is unclear but may be suggestive of biofilm development, which is a common C. albicans virulence strategy (Lohse et al., 2018; Nobile and Johnson, 2015). Close up microscopic observations revealed that hyphal cells were present at 3 dpi in planaria infected with the WT strain (Figure 2E), indicating the growth of the tissue invasive morphological form at this time point. We also noted that infection with the hyper-filamentous strain typically started at either the anterior or the posterior end of the animal (head or tail) but by 3 dpi the antibody signal was spread throughout the animal (Figure 2F).
We performed cross sections at different time points post-infection to visualize the distribution of C. albicans cells in superficial and deeper planarian tissues. For simplicity, we display representative images of the cross sections taken at the middle part of the body where the pharynx is located (Figure 3). All three strains were observed to localize to the epithelial layer by 1 dpi, and as the infection progressed C. albicans cells were found in deeper tissues (parenchyma) and the amount of pathogen present was strain-dependent (Figure 3). Specifically, the hyper-filamentous and the WT strains had the largest presence in both the epithelia and parenchyma (Figure 3C–F). Despite the massive growth of C. albicans in the planaria, the fungus became undetectable by 10 dpi, consistent with the CFUs results in Figure 1C. These results demonstrate that fungi are able to attach, penetrate and grow inside planarian tissues, and that planaria subsequently respond to the infection by recognizing and clearing the pathogen.
Candida albicans Infection Triggers Neoblast Division and Cell Death
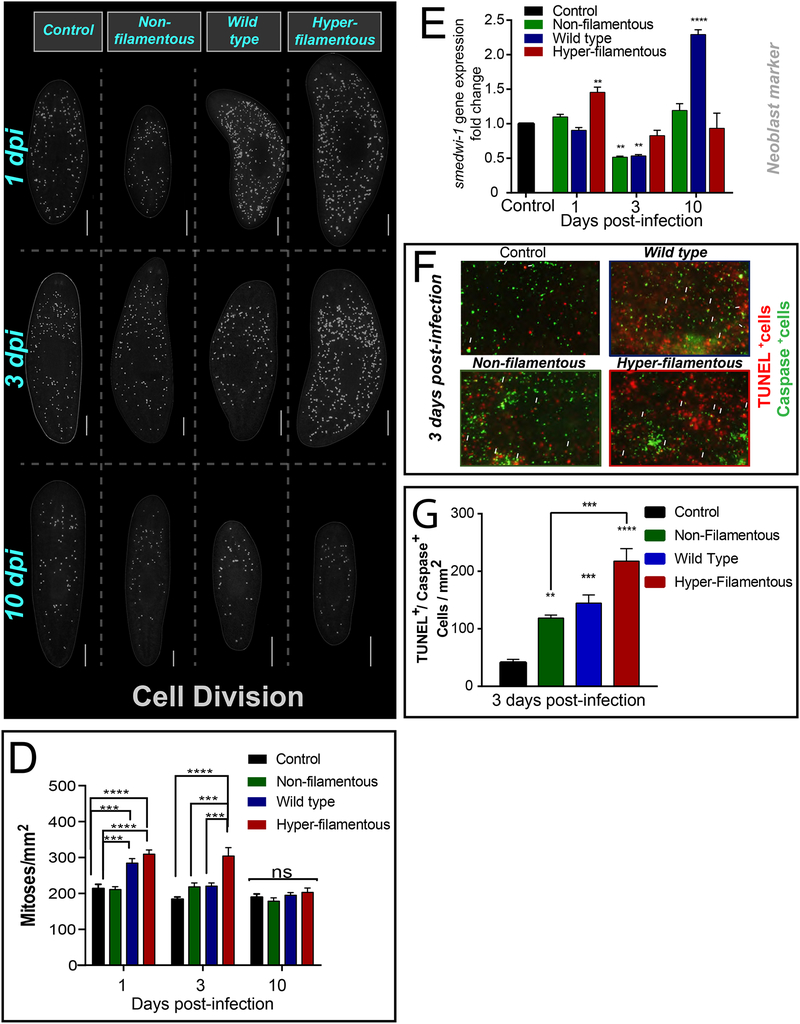
To learn about the planaria host response to C. albicans infection, we first observed the cellular changes taking place. Neoblasts are the only cells with the capacity to divide in planaria (Bardeen and Baetjer, 1904; Newmark and Sanchez Alvarado, 2000; Reddien et al., 2005; van Wolfswinkel et al., 2014; Wagner et al., 2011; Zeng et al., 2018). Thus, neoblast division provides the cellular progeny required to renew and repair all planarian tissues. Mitotic neoblasts are recognized with anti-histone-3 phosphorylated antibody (H3P) (Newmark and Sanchez Alvarado, 2000). We found that infection with the hyper- filamentous strain was accompanied with an increase in neoblast division during the first 3 dpi, while the WT strain only increased mitotic activity on the first day of infection (Figure 4A–D). Remarkably, mitotic levels returned to basal levels by 10 dpi in all groups (Figure 4D). Furthermore, we also found that the expression of the pan-neoblast marker smedwi-1 displayed significant changes during infection. Specifically, smedwi-1 expression levels increased at 1 dpi with the hyper-filamentous strain and unexpectedly also increased at 10 dpi with the WT strain (Figure 4E). Interestingly, there was a dramatic decrease in smedwi-1 expression at 3 dpi with the WT and non-filamentous C. albicans strains, while the group exposed to the hyper-filamentous strain display similar levels of expression as the uninfected control (Figure 4E). Although the changes in gene expression associated with this neoblast marker are not mechanistically clear, these findings suggest that neoblasts may form part of the host response to counteract the effects of C. albicans infection.
Figure 4: C. albicans infection induces hyper-proliferation and cell death in planaria.
A-C) Whole-mount immunostaining with anti-phospho-histone H3 (Ser10) antibody, which labels mitotic cells (yellow dots) at different times of infection with C. albicans strains: non-filamentous (2 × 107 cells/mL), wild type (1.5 × 107 cells/mL) and hyper-filamentous (5 × 106 cells/mL). D) Number of mitoses at different times of infection with C. albicans strains and the non-infected control. E) Levels of smedwi-1 gene expression at different times of infection with C. albicans strains. The expression of smedwi-1 is normalized against a noninfected control. The internal reference is based on the ubiquitously expressed clone H.55.12e. F) Double staining with anti-caspase-3 antibody (green signal) and TUNEL positive cells (red signal) in planarian tissue at day 3 post-infection with C. albicans strains (7.5 × 106 cells/mL). In all cases the control sample corresponds to uninfected animal. G) Levels of double positive cells for caspase+/TUNEL+ in planarian tissue at day 3 post-infection with C. albicans strains (7.5 × 106 cells/mL). Cell division experiments consisted of three biological replicates using 10 animals each. Cell death experiments consisted of two biological replicates using 4 animals each. All graphs represent mean ±SEM. All statistical comparisons are against control unless noted with bars. Scale bar is 200μm. Two-Way-ANOVA, *P<0.01; ***P<0.001; ****P<0.0001 and ns= no significant.
Simultaneous whole-mount immunostaining with the TUNEL (terminal deoxynucletidyl transferase dUTP nick end labeling) assay and anti-caspase-3 antibody were performed to determine whether C. albicans infection affects cell death in the host (Figure 4F, G). Since the TUNEL assay also labels cell death in C. albicans and caspase-3 is specific to the planarian, we co-labeled planarian tissue with both TUNEL and the caspase-3 antibody to quantify double positive cells. These experiments revealed that all three C. albicans strains triggered an increase in cell death, although the infection with the hyper-filamentous strain displayed four-fold the amount of cell death compared to the control (uninfected sample) (Figure 4F, G). Taken together, these data demonstrate that the hyper-filamentous C. albicans strain triggers the most dramatic changes in the host involving cell death and cell division.
MORN2 and TAK1/p38 Signaling Pathways May Facilitate Efficient Clearance of C. albicans Infection
To further understand the mechanisms of the innate immune system activated during planarian clearance of C. albicans infection, we focused on pattern recognition receptors (PRRs) that are frequently involved in the recognition of fungi during an infection (Drummond and Brown, 2013; Goyal et al., 2018; Tang et al., 2018). Specifically, we performed gene expression analyses of components associated with C-type lectin receptors (CTLs) and Toll like receptors (TLRs) upon infection with different strains of C. albicans (Gao et al., 2017; Tsoumtsa et al., 2018). Components of the CTL and TLR pathways are depicted in Figure 5A. To assess the involvement of the CTL pathway postinfection, we measured the expression of the upstream component, SYK, which encodes an adaptor protein, and the downstream component, TAK1, which encodes a signaling kinase. To evaluate the TLR pathway, we measured MyD88, which encodes an adaptor protein of TLRs that is known to play important roles during C. albicans infection and is present in the planarian genome (Roeder et al., 2004; Tsoumtsa et al., 2018). Two other genes MORN2 and Tyrosinase were also selected for expression measurements due to their known roles in clearing other microbial pathogens (Abnave et al., 2014; Cerenius et al., 2008). The gene expression measurements during the infection time course revealed that MYD88 was not upregulated during any of the infections or time points while TRAF6 only showed increased expression for the non-filamentous strain in day 1 (Figure 5B). Conversely, SYK and TAK1 were upregulated in the planaria at different days depending on the strain of C. albicans. The non-filamentous and hyper- filamentous strains upregulated most of the selected genes during day 1 and 3, which was different from what was observed for the WT strain, where WT upregulated most genes at day 10 (Figure 5B). Since the overall upregulation of the CTL effector genes was observed during the infection for all three C. albicans strains, we proceeded to look further into the CTL pathway. Recent studies revealed that bacterial infection triggers phosphorylation of p38 MAPK through activation of TAK1, which in turn leads to responses mediated by NF-kB (Arnold et al., 2016). To test whether planarians clear C. albicans infection through activation of phospho-p38 MAPK we used the P-p38 human antibody at different time points of infection. The results showed that by day 3, the signal for P-p38 was increased compared to the control (uninfected planaria) and compared to planaria infected with the other two strains of C. albicans (Figure 5C, D). These results suggest that MORN2 and TAK1/p38 signaling pathways may facilitate efficient clearance of C. albicans infection.
Figure 5: Downstream effectors of C Type Lectins are upregulated during C. albicans infection.
A) Graphical representation of the canonical signaling cascade activated by two particular pattern recognition receptors, C-type lectins (CTLs) and Toll like receptors (TLRs). The left side of the illustration depicts three of the most commonly known C type lectin pattern recognition receptors in response to C. albicans infection in mammals. The right side of the illustration shows two of the common toll like receptors that recognize C. albicans along with its downstream effectors. B) Gene expression levels of different components associated with CTL and TLR pathways during time course of infection with C. albicans strains. Levels are shown in a heat map and the scale bar displayed indicates red is upregulation and blue is downregulation. Gene expression values represent mean ± SEM of triplicate samples; each condition was generated by extracting RNA from 10 animals. The internal control is the ubiquitously expressed clone H.55.12e. C, D) Whole-mount immunostaining with anti-phosphorylated p38 antibody (P-p38, white dots) at different time points of infection with C. albicans strains: non-filamentous (2 × 107 cells/mL), wild type (1.5 × 107 cells/mL) and hyper-filamentous (5 × 106 cells/mL). E) Number of P-p38+cells at different time points of infection with C. albicans strains and the non-infected control. Experiments consisted of two biological replicates with 10 animals each. All graphs represents mean ± SEM. All statistical comparisons are against control unless noted with bars. Scale bar is 200μm. Two-Way-ANOVA, *P<0.01; ***P<0.001; ****P<0.0001 and ns= no significant.
DISCUSSION
Our results demonstrate that the fungal pathogen C. albicans can infect planarians, and that this flatworm is capable of recognizing and effectively eliminating C. albicans in a relatively short period of time. Here, we introduce a new invertebrate and low-cost model system to enable studies of host-pathogen interactions at different stages of fungal infection. Our findings also provide unique opportunities to analyze the activation of the host innate immune response independently of the adaptive immune response and underscore the privileged evolutionary position of flatworms and their biology that closely relate to human health.
We developed a highly reproducible infection protocol based on soaking planarians in a solution containing C. albicans. This cost-effective strategy allows for the visualization of the initial C. albicans contact with host tissues, as well as its successive interactions to penetrate and invade deeper within tissues and organs throughout the host. We noticed that although some planaria succumb to fungal infection, some survive, and additional experimentation involving the use of different methods of exposure (e.g. injection and feeding), will be required to understand the underlying basis leading to this differential behavior. Although infecting planarians by feeding has been used before for bacterial infections (Abnave et al., 2014; Arnold et al., 2016), our method of infection through soaking offers several advantages. First, it allows for the control of both the concentration of pathogens exposed to the host as well as for the easy and rapid removal of those pathogens in the media through washes. Second, this novel model is also useful to study mechanisms of host-pathogen epithelial infections by fungi. Third, our protocol is time-effective, taking only a few days from initial exposure to clearance of the infection. Finally, our methods can be easily scaled up to perform high-throughput screens to identify mutants with virulence defects and or antimicrobial compounds that are effective against C. albicans infection.
Exposure to three different morphological strains of C. albicans (WT, non- filamentous (efg1/efg1) and hyper-filamentous (nrg1/nrg1)) revealed that filamentation is an important virulent factor that affects planarian tissue. These findings also demonstrate that, similar to vertebrate animal models, the amount of tissue damage and the number of C. albicans cells penetrating deeper organs is dependent on the ability of C. albicans to undergo the yeast to hyphal transition. Interestingly, all tested C. albicans strains were able to adhere to the planarian epithelial cells, but the non-filamentous strain was less efficient at disseminating to internal tissues. This dissemination defect of the non- filamentous strain is likely due to the inability of this strain to cause tissue damage, and thus preventing penetration into the deeper tissues of the planarians. This non-filamentous strain is also likely to be eliminated more easily through phagocytosis. Wild-type C. albicans cells can adhere and transition to invasive forms in planarian tissue, and thus this new model can be used to gain mechanistic insights into virulence and possibly biofilm formation during fungal infection. Future studies will address the specific mechanisms used by pathogenic fungi to penetrate and invade host planarian tissues, and will provide deeper mechanistic insights into the host response and whether phagocytosis plays a central role in C. albicans clearance. Nonetheless, the current results provide opportunities to analyze direct early interactions between C. albicans and epithelial surfaces in the host. We propose that this infection strategy has some resemblance to mucosal infections in vertebrate models, and is consistent with the ability of fungal filamentation to facilitate tissue invasion, biofilm formation, and evasion of host macrophages (Cleary et al., 2016; Fuchs et al., 2010; Kadosh and Lopez-Ribot, 2013; Lo et al., 1997; Mitchell, 1998).
Stem cells in mammals constantly renew tissues that are targeted by microbial infections (e.g. epithelial surfaces). C. albicans infection is known to activate the proliferation of mesenchymal and hematopoietic SCs leading to an increase in lineage-restricted cells necessary to fight pathogenic fungi (Megias et al., 2016; Megias et al., 2012; Yanez et al., 2011; Yanez et al., 2009; Yang et al., 2012). Planarians constantly renew tissues from stem cell division and we show here that neoblast proliferation is increased when planarians are exposed to invasive forms of C. albicans, which potentially suggests that the stem cell response to fight fungal infection is evolutionarily conserved in metazoans. This claim is also supported by recent findings that associate neoblasts with the ability of the host to combat Staphylococcus aureus infection (Keating et al., 2017; Torre et al., 2017b). Future experiments will determine whether the increase in neoblast division upon C. albicans infection is directed toward a specific lineage such as the reticular cells, which are known to mediate the planarian innate immune response (Morita, 1991, 1995). Our focus at the organismal level also enables access to the host response involving crosstalk between SCs, differentiated tissues and the innate immune system responding to fungal infection. This comprehensive approach has the potential to identify novel molecular players involved in the rapid clearance of pathogenic fungi.
Our results demonstrating an increase in the gene expression of TAK1 and SYK imply that the planarian response to C. albicans infection is mediated by the CTL receptors, which are also activated to eliminate pathogenic bacteria (Abnave et al., 2014; Arnold et al., 2016; Gao et al., 2017). The findings presented here, lead us to propose that C. albicans infection elicits innate immune responses in the host involving multiple mechanisms encompassing traceable cellular (i.e. neoblast proliferation) and humoral (e.g. MORN2, TAK1/p38 signaling) activity to efficiently clear infection by pathogenic fungi. This cellular and humoral strategy is also effective to combat bacterial infection and allow for the innate immune system to adapt to recurrent infection (Abnave et al., 2014; Arnold et al., 2016; Torre et al., 2017a). Our results are consistent with the possibilities that other signaling pathways may also become activated during infection and/or that their mechanistic activation may take place at different times. For example, TAK1 and SYK are expressed at different time points when planaria are infected with the different C. albicans strains. One possible explanation for this observation is that the WT strain may be capable of masking specific fungal cell wall components that are recognized by the PRRs (McKenzie et al., 2010). Most CTL receptors detect either β-glucans or α-mannans, which are two major components of the C. albicans cell wall (Netea et al., 2006). We speculate that the predicted pattern recognition receptor mediating the innate immune response against C. albicans is likely to be a CTL receptor, which will activate the SYK protein, and proceed to activate a complex that contains BCL and CARD proteins (Li et al., 2018; Maciel and Oviedo, 2018). This is also consistent with the idea that TAK1 is also active during this process, and interacts with P38 and other MAP kinase cascades that eventually end in the activation of the transcription factor NF-kβ (Arnold et al., 2016; Torre et al., 2017a; Tsoumtsa et al., 2017). Additional insights about the genetic network activated in the host will be obtained in future studies using genome-wide transcriptomic analyses during a time course of infection with C. albicans. Nonetheless, our work extends previous analyses of host-pathogen interactions in planaria (Abnave and Ghigo, 2018; Abnave et al., 2014; Arnold et al., 2016; Gao et al., 2017; Hammoudi et al., 2018; Li et al., 2018; Lu et al., 2017; Pang et al., 2016; Torre et al., 2017a; Tsoumtsa et al., 2018; Tsoumtsa et al., 2017) and introduces a simplified platform to study evolutionarily conserved mechanisms to overcome infections by pathogenic fungi.
CONCLUSION
Collectively, our work introduces planarian flatworms as a low-cost and time efficient model organism to dissect basic mechanisms of host-pathogen responses during fungal infection.
HIGHLIGHTS.
S. mediterranea is a new animal model to study fungal infections
Host innate immune responses rapidly eliminate C. albicans infection
C. albicans infection activates stem cell proliferation in planarians
C-type lectin and MORN2 signaling pathways are likely involved in the clearance of C. albicans infection
Acknowledgements:
We thank Edelweiss Pfister for lab managing and planarian maintenance, and members of the Oviedo and Nobile labs for insightful discussions and comments on the manuscript.
Funding:
This work was supported by the National Science Foundation (NSF) graduate fellowship award 1744620 to EIM, and the University of California Cancer Research Coordinating Committee (Award# CRR-18–525108) and the National Institutes of Health (NIH) National Cancer Institute (NCI) and National Institute of General Medical Sciences (NIGMS) awards R21CA176114 and R15GM109372 to NJO, and the NIH National Institute of Allergy and Infectious Diseases (NIAID) and NIGMS awards R21AI125801 and R35GM124594 to CJN.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interest:
The authors declare no competing or financial interest.
REFERENCES
- Abnave P, Ghigo E, 2018. Role of the immune system in regeneration and its dynamic interplay with adult stem cells. Seminars in cell & developmental biology [DOI] [PubMed] [Google Scholar]
- Abnave P, Mottola G, Gimenez G, Boucherit N, Trouplin V, Torre C, Conti F, Ben Amara A, Lepolard C, Djian B, Hamaoui D, Mettouchi A, Kumar A, Pagnotta S, Bonatti S, Lepidi H, Salvetti A, Abi-Rached L, Lemichez E, Mege J-L, Ghigo E, 2014. Screening in Planarians Identifies MORN2 as a Key Component in LC3- Associated Phagocytosis and Resistance to Bacterial Infection. Cell Host & Microbe 16, 338–350. [DOI] [PubMed] [Google Scholar]
- Aboobaker AA, 2011. Planarian stem cells: a simple paradigm for regeneration. Trends Cell Biol 21, 304–311. [DOI] [PubMed] [Google Scholar]
- Arnold CP, Merryman MS, Harris-Arnold A, McKinney SA, Seidel CW, Loethen S, Proctor KN, Guo L, Sánchez Alvarado A, 2016. Pathogenic shifts in endogenous microbiota impede tissue regeneration via distinct activation of TAK1/MKK/p38. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardeen CR, Baetjer FH, 1904. The inhibitive action of the Roentgen rays on regeneration in planarians. J. Exp. Zool 1, 191–195. [Google Scholar]
- Bergeron AC, Barker SE, Brothers KM, Prasad BC, Wheeler RT, 2017. Polyclonal anti-Candida antibody improves phagocytosis and overall outcome in zebrafish model of disseminated candidiasis. Developmental and comparative immunology 68, 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongomin F, Gago S, Oladele R, Denning D, 2017. Global and Multi-National Prevalence of Fungal Diseases—Estimate Precision. Journal of Fungi 3, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun BR, Johnson AD, 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277, 105–109. [DOI] [PubMed] [Google Scholar]
- Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, White TC, 2012. Hidden killers: human fungal infections. Science translational medicine 4. [DOI] [PubMed] [Google Scholar]
- Cerenius L, Lee BL, Soderhall K, 2008. The proPO-system: pros and cons for its role in invertebrate immunity. Trends in immunology 29, 263–271. [DOI] [PubMed] [Google Scholar]
- Chamilos G, Lewis RE, Albert N, Kontoyiannis DP, 2007. Paradoxical effect of Echinocandins across Candida species in vitro: evidence for echinocandin-specific and candida species-related differences. Antimicrobial agents and chemotherapy 51, 2257–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy CJ, Cheng S, Nguyen M, 2009. Methods in Molecular Biology springer; 499, 65–76. [DOI] [PubMed] [Google Scholar]
- Cleary IA, Reinhard SM, Lazzell AL, Monteagudo C, Thomas DP, Lopez-Ribot JL, Saville SP, 2016. Examination of the pathogenic potential of Candida albicans filamentous cells in an animal model of haematogenously disseminated candidiasis. FEMS Yeast Research 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond RA, Brown GD, 2013. Signalling C-type lectins in antimicrobial immunity. PLoS pathogens 9, e1003417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher MC, Hawkins NJ, Sanglard D, Gurr SJ, 2018. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science 360, 739–742. [DOI] [PubMed] [Google Scholar]
- Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, Gurr SJ, 2012. Emerging fungal threats to animal, plant and ecosystem health. Nature 484, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs B, Eby J, Nobile CJ, Khoury JB, Mitchell AP, Mylonakis E, 2010. Role of filamentation in Galleria mellonella killing by Candida albicans. Microbes and Infection 12, 488–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Han Y, Deng H, Hu W, Zhen H, Li N, Qin N, Yan M, Wu W, Liu B, Zhao B, Pang Q, 2017. The role of a novel C-type lectin-like protein from planarian in innate immunity and regeneration. Developmental and comparative immunology 67, 413–426. [DOI] [PubMed] [Google Scholar]
- Glittenberg MT, Silas S, MacCallum DM, Gow NA, Ligoxygakis P, 2011. Wild-type Drosophila melanogaster as an alternative model system for investigating the pathogenicity of Candida albicans. Disease models & mechanisms 4, 504–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal S, Castrillon-Betancur JC, Klaile E, Slevogt H, 2018. The Interaction of Human Pathogenic Fungi With C-Type Lectin Receptors. Front Immunol 9, 1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratacap RL, Scherer AK, Seman BG, Wheeler RT, 2017. Control of mucosal candidiasis in the zebrafish swimbladder depends on neutrophils that block filament invasion and drive extracellular trap production. Infection and immunity [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoudi N, Torre C, Ghigo E, Drancourt M, 2018. Temperature affects the biology of Schmidtea mediterranea. Scientific reports 8, 14934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann OR, Dea J, Noble SM, Johnson AD, 2009. A Phenotypic Profile of the Candida albicans Regulatory Network. PLoS Genetics 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D, Lopez-Ribot JL, 2013. Candida albicans: Adapting to Succeed. Cell Host & Microbe 14, 483–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating ST, Riksen NP, Netea MG, 2017. Planarians SET New Paths for Innate Immune Memory. EBioMedicine 20, 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RS, Newmark PA, 2012. The cell biology of regeneration. The Journal of cell biology 196, 553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Li A, Zheng K, Liu X, Gao L, Liu D, Deng H, Wu W, Liu B, Zhao B, Pang Q, 2018. Identification and characterization of an atypical RIG-I encoded by planarian Dugesia japonica and its essential role in the immune response. Developmental and comparative immunology 91, 72–84. [DOI] [PubMed] [Google Scholar]
- Lo H-J, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR, 1997. Nonfilamentous C. albicans Mutants Are Avirulent. Cell 90, 939–949. [DOI] [PubMed] [Google Scholar]
- Lohse MB, Gulati M, Johnson AD, Nobile CJ, 2018. Development and regulation of single- and multi-species Candida albicans biofilms. Nature reviews. Microbiology 16, 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Wu S, Zhen H, Deng H, Song Q, Ma K, Cao Z, Pang Q, Zhao B, 2017. 143-3 alpha and 14-3-3 zeta contribute to immune responses in planarian Dugesia japonica. Gene 615, 25–34. [DOI] [PubMed] [Google Scholar]
- Maciel EI, Oviedo NJ, 2018. Platyhelminthes: Molecular dissection of the planarian innate immune system, in: Cooper EL (Ed.), Advances in Comparative Immunology Springer International Publishing, pp. X, 1107. [Google Scholar]
- Mallick EM, Bergeron AC, Jones SK Jr., Newman ZR, Brothers KM, Creton R, Wheeler RT, Bennett RJ, 2016. Phenotypic Plasticity Regulates Candida albicans Interactions and Virulence in the Vertebrate Host. Front Microbiol 7, 780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie CG, Koser U, Lewis LE, Bain JM, Mora-Montes HM, Barker RN, Gow NA, Erwig LP, 2010. Contribution of Candida albicans cell wall components to recognition by and escape from murine macrophages. Infection and immunity 78, 1650–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megías J, Martínez A, Yáñez A, Goodridge HS, Gozalbo D, Gil LM, 2016. TLR2, TLR4 and Dectin-1 signalling in hematopoietic stem and progenitor cells determines the antifungal phenotype of the macrophages they produce. Microbes and Infection 18, 354–363. [DOI] [PubMed] [Google Scholar]
- Megías J, Yáñez A, Moriano S, O’Connor JE, Gozalbo D, Gil ML, 2012. Direct Toll - Like Receptor - Mediated Stimulation of Hematopoietic Stem and Progenitor Cells Occurs In Vivo and Promotes Differentiation Toward Macrophages. STEM CELLS 30, 1486–1495. [DOI] [PubMed] [Google Scholar]
- Mitchell AP, 1998. Dimorphism and virulence in Candida albicans. Current Opinion in Microbiology 1, 687–692. [DOI] [PubMed] [Google Scholar]
- Morita M, 1991. Phagocytic response of planarian reticular cells to heat-killed bacteria. Hydrobiologia 227, 193–199. [Google Scholar]
- Morita M, 1995. Structure and function of the reticular cell in the planarian Dugesia dorotocephala, in: Cannon L (Ed.), Biology of turbellaria and some related flatworms Springer, Abo/Turku, Finland, pp. 189–196. [Google Scholar]
- Mylonakis E, 2008. Galleria mellonella and the study of fungal pathogenesis: making the case for another genetically tractable model host. Mycopathologia 165, 1–3. [DOI] [PubMed] [Google Scholar]
- Mylonakis E, Casadevall A, Ausubel FM, 2007. Exploiting amoeboid and nonvertebrate animal model systems to study the virulence of human pathogenic fungi. PLoS pathogens 3, e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Gow NA, Munro CA, Bates S, Collins C, Ferwerda G, Hobson RP, Bertram G, Hughes HB, Jansen T, Jacobs L, Buurman ET, Gijzen K, Williams DL, Torensma R, McKinnon A, MacCallum DM, Odds FC, Van der Meer JW, Brown AJ, Kullberg BJ, 2006. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and Toll-like receptors. The Journal of clinical investigation 116, 1642–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmark P, Sanchez Alvarado A, 2000. Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev. Biol 220, 142–153. [DOI] [PubMed] [Google Scholar]
- Nobile CJ, Johnson AD, 2015. Candida albicans Biofilms and Human Disease. Annual review of microbiology 69, 71–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oviedo NJ, Nicolas CL, Adams DS, Levin M, 2008. Establishing and maintaining a colony of planarians. CSH protocols 2008, pdb prot5053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Q, Gao L, Hu W, An Y, Deng H, Zhang Y, Sun X, Zhu G, Liu B, Zhao B, 2016. De Novo Transcriptome Analysis Provides Insights into Immune Related Genes and the RIG-I-Like Receptor Signaling Pathway in the Freshwater Planarian (Dugesia japonica). PLoS One 11, e0151597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris TH, Hoyer KK, Oviedo NJ, 2014. Innate immune system and tissue regeneration in planarians: an area ripe for exploration. Semin Immunol 26, 295302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris TH, Ramirez D, Barghouth PG, Ofoha U, Davidian D, Weckerle F, Oviedo NJ, 2016. Regional signals in the planarian body guide stem cell fate in the presence of genomic instability. Development 143, 1697–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellettieri J, Fitzgerald P, Watanabe S, Mancuso J, Green DR, Sanchez Alvarado A, 2010. Cell death and tissue remodeling in planarian regeneration. Dev Biol 338, 76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellettieri J, Sanchez Alvarado A, 2007. Cell turnover and adult tissue homeostasis: from humans to planarians. Annu Rev Genet 41, 83–105. [DOI] [PubMed] [Google Scholar]
- Peterson ND, Pukkila-Worley R, 2018. Caenorhabditis elegans in high-throughput screens for anti-infective compounds. Current opinion in immunology 54, 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila-Worley R, Holson E, Wagner F, Mylonakis E, 2009a. Antifungal drug discovery through the study of invertebrate model hosts. Current medicinal chemistry 16, 1588–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila-Worley R, Peleg AY, Tampakakis E, Mylonakis E, 2009b. Candida albicans Hyphal Formation and Virulence Assessed Using a Caenorhabditis elegans Infection Model. Eukaryotic Cell 8, 1750–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sánchez Alvarado A, 2005. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science 310, 1327–1330. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Sánchez Alvarado A, 2004. Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol 20, 725–757. [DOI] [PubMed] [Google Scholar]
- Roeder A, Kirschning CJ, Rupec RA, Schaller M, Korting HC, 2004. Toll-like receptors and innate antifungal responses. Trends in microbiology 12, 44–49. [DOI] [PubMed] [Google Scholar]
- Saville SP, Lazzell AL, Monteagudo C, Lopez-Ribot JL, 2003. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot Cell 2, 1053–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW, 2012. NIH Image to ImageJ: 25 years of image analysis. Nature methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Frenkel M, 2018. Experimental in Vivo Models of Candidiasis. Journal of fungi 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Lin G, Langdon WY, Tao L, Zhang J, 2018. Regulation of C-Type Lectin Receptor-Mediated Antifungal Immunity. Front Immunol 9, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiruvalluvan M, Barghouth PG, Tsur A, Broday L, Oviedo NJ, 2018. SUMOylation controls stem cell proliferation and regional cell death through Hedgehog signaling in planarians. Cellular and molecular life sciences: CMLS 75, 1285–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre C, Abnave P, Tsoumtsa L, Mottola G, Lepolard C, Trouplin V, Gimenez G, Desrousseaux J, Gempp S, Levasseur A, Padovani L, Lemichez E, Ghigo E, 2017a. Staphylococcus aureus Promotes Smed-PGRP-2/Smed-setd8–1 Methyltransferase Signalling in Planarian Neoblasts to Sensitize Anti-bacterial Gene Responses During Re-infection. EBioMedicine 20, 150–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre C, Abnave P, Tsoumtsa LL, Mottola G, Lepolard C, Trouplin V, Gimenez G, Desrousseaux J, Gempp S, Levasseur A, Padovani L, Lemichez E, Ghigo E, 2017b. Staphylococcus aureus Promotes Smed-PGRP-2/Smed-setd8–1 Methyltransferase Signalling in Planarian Neoblasts to Sensitize Anti-bacterial Gene Responses During Re-infection. EBioMedicine 20, 150–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoumtsa LL, Sougoufara S, Torre C, Lemichez E, Pontarotti P, Ghigo E, 2018. In silico analysis of Schmidtea mediterranea TIR domain-containing proteins. Developmental and comparative immunology 86, 214–218. [DOI] [PubMed] [Google Scholar]
- Tsoumtsa LL, Torre C, Trouplin V, Coiffard B, Gimenez G, Mege J-L, Ghigo E, 2017. Antimicrobial capacity of the freshwater planarians against S. aureus is under the control of Timeless. Virulence 8, 1160–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wolfswinkel JC, Wagner DE, Reddien PW, 2014. Single-cell analysis reveals functionally distinct classes within the planarian stem cell compartment. Cell stem cell 15, 326–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DE, Wang IE, Reddien PW, 2011. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 332, 811–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO, 2018. The top ten causes of death World Health Organization. [Google Scholar]
- Yáñez A, Megías J, O’Connor J-E, Gozalbo D, Gil LM, 2011. Candida albicans Induces Selective Development of Macrophages and Monocyte Derived Dendritic Cells by a TLR2 Dependent Signalling. PLoS ONE 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez A, Murciano C, O’Connor J-E, Gozalbo D, Gil LM, 2009. Candida albicans triggers proliferation and differentiation of hematopoietic stem and progenitor cells by a MyD88-dependent signaling. Microbes and infection 11, 531–535. [DOI] [PubMed] [Google Scholar]
- Yang R, Liu Y, Kelk P, Qu C, Akiyama K, Chen C, Atsuta I, Chen W, Zhou Y, Shi S, 2012. A subset of IL-17+ mesenchymal stem cells possesses anti-Candida albicans effect. Cell Research 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng A, Li H, Guo L, Gao X, McKinney S, Wang Y, Yu Z, Park J, Semerad C, Ross E, Cheng L-C, Davies E, Lei K, Wang W, Perera A, Hall K, Peak A, Box A, Alvarado A, 2018. Prospectively Isolated Tetraspanin+ Neoblasts Are Adult Pluripotent Stem Cells Underlying Planaria Regeneration. Cell 173, 15931006632960. [DOI] [PMC free article] [PubMed] [Google Scholar]