Abstract
Supramolecular hydrogels are a class of self-assembled network structures formed via non-covalent interactions of the hydrogelators. These hydrogels capable of responding to external stimuli are considered to be smart materials due to their ability to undergo sol–gel and/or gel–sol transition upon subtle changes in their surroundings. Such stimuli-responsive hydrogels are intriguing biomaterials with applications in tissue engineering, delivery of cells and drugs, modulating tissue environment to promote innate tissue repair, and imaging for medical diagnostics among others. This review summarizes the recent developments in stimuli-responsive supramolecular hydrogels and their potential applications in regenerative medicine. Specifically, various structural aspects of supramolecular hydrogelators involved in self-assembly, the role of external stimuli in tuning/controlling their phase transitions, and how these functions could be harnessed to advance applications in regenerative medicine are focused on. Finally, the key challenges and future prospects for these versatile materials are briefly described.
Keywords: hydrogelators, regenerative medicine, self-assembly, stimuli, supramolecular hydrogels
1. Introduction
Employing molecular conformations to form higher-order structures and undergo changes responding to subtle perturbations in their environmental cues is ubiquitous in biopolymers such as collagen, nucleic acids. Given the unique functionalities such systems offer, extensive research has been dedicated to developing man-made macromolecules with similar functions.[1–5] Polymer hydrogels, cross-linked network of macromolecules, that undergo reversible phase transitions in response to subtle changes in their environment are one of the early examples of such systems.[6,7] The stimuli-responsive hydrogels exhibit volume/phase transitions responding to external and internal triggers such as temperature, pH, electric field, magnetic field, light, and chemical triggers.[8–13] The stimuli-responsive volume/phase transitions are mainly controlled by polymer–polymer and polymer–solvent interactions. The chemical structure of the constituents of the network plays a key role in influencing the nature, extent, and dynamics of these intermolecular interactions and thus stimuli responsiveness. For instance, hydrogels that manifest thermoreversible properties are often composed of molecular moieties exhibiting a balance of hydrophilic and hydrophobic interactions.[8,14,15] Beyond volume/phase transitions, molecules with optimal hydrophilic and hydrophobic interactions have been used to endow chemically cross-linked hydrogels with unique functions such as macroscopic self-organization (solid to hollow interior and sheet to 3D structures), shape memory, and self-healing.[16–20]
Similar to polymer hydrogels, supramolecular hydrogels formed by the assembly of molecular building blocks termed as hydrogelators also show stimuli-responsive properties (gel–sol or sol–gel transitions).[21,22] Although supramolecular hydrogels bear many functional similarities with conventional polymer hydrogels, the network formation in supramolecular hydrogels is different from that of conventional polymer-based hydrogels. In the former case, the network formation is entirely via non-covalent interactions while polymeric hydrogels are formed often through chemical cross-linking. In the case of supramolecular hydrogels, the low-molecular-weight hydrogelators assemble via non-covalent interactions—such as hydrogen bonding (H bonding), electrostatic interactions, hydrophobic interactions, van der Waals interactions, and π–π interactions—into structures like nanofibers, helices, leading to entangled networks imbibed with water.[23,24] In addition to the small molecular hydrogelators, polymeric hydrogelators are also used to form supramolecular hydrogels. For polymeric hydrogelators, functional groups present on the macromolecular chains mediate molecular interactions between the complimentary pairs to form supramolecular networks.[21,25] Several review articles exist covering the structural aspects and potential applications of supramolecular hydrogels based on either small molecular hydrogelators and/or polymeric hydrogelators.[26–29]
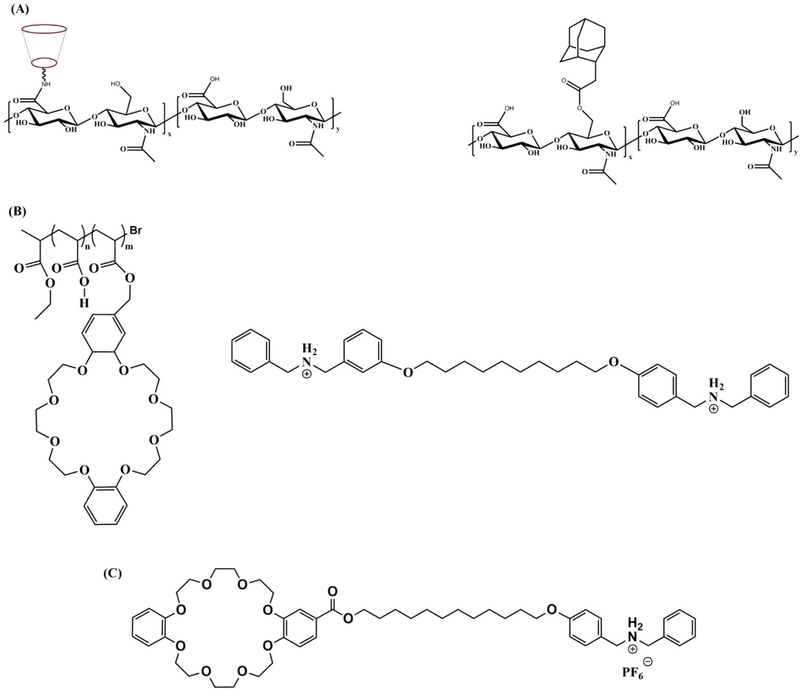
Among synthetic supramolecular hydrogelators, host– guest inclusion complex that allows integration of two or more chemical moieties together in a reversible manner has been used extensively to construct supramolecular hydrogels. A variety of macrocyclic molecules and their derivatives such as cyclodextrins (CDs), cucurbit[n]urils (CBs), calixarenes (CAs), crown ethers, and pillar[n]arenes have been utilized as hosts to create supramolecular structures in the presence of a guest molecule through molecular recognition.[30–43] These macrocyclic moieties, regarded as the hosts, possess cavities to incorporate the guest molecules through non-covalent interactions. In general, while the exterior surface of the host molecules favors interaction with its surrounding solvent, the cavities facilitate inclusion of the guest molecules having an appropriate molecular shape or size via non-covalent interactions such as hydrogen bonding, hydrophobic interactions, electrostatic interaction.[35,44,45] A supramolecular hydrogel by the host–guest interaction can be formed in multiple ways. One such method includes conjugation of both host and guest molecules onto polymer chains.[31,46,47] The complexation between the host and guest molecules via non-covalent inclusion can induce cross-linking between the two polymer chains (Figure 1A). Another method includes conjugation of guest molecules onto a polymer chain which is then reacted with macrocyclic host molecules to achieve inclusion complexation (Figure 1B).[30,48,49] Finally, supramolecular hydrogels have also been developed via direct complexation of host and guest molecules after chemical modification of either host or guest or both host and guest molecules (Figure 1C).[35,50,51]
Figure 1.
Schematic representation of various host–guest hydrogelators. A) Schematic representation and structures of host–guest polymers CD-HA and Ad-HA; B) structures of host polymer poly(methyl methacrylate) with pendent dibenzo[24] crown-8 groups and a guest dibenzylammonium salt; C) structure of host monomer with a guest benzylammonium salt directly attached to it.
Given the strong influence of the chemical structures of the hydrogelators on the macroscopic properties of supramolecular hydrogels, structures with defined/targeted functions can be rationally designed.[21,27,52] For instance, hydrogelators or host/guest molecules encoded with stimuli-responsive chemical moieties can be used toward the development of supramolecular hydrogels with a stimuli-responsive function. To this end, supramolecular hydrogels undergoing phase transition in response to temperature, light, pH, electric field, magnetic field, redox (oxidation–reduction), enzymes, and chemical triggers are developed at a rapid rate.[27,53,54] Together, these stimuli can be roughly classified into three categories— physical, chemical, and biological. The phase transition can be either sol–gel or gel–sol, which is again dictated largely by the chemical structure of the hydrogelators. In the former category, non-covalent molecular association leads to the formation of hierarchical structures while in the case of gel–sol transition, such interactions between the hydrogelators are disrupted to dissemble and solvate the hydrogelators in water or form association with competitive ligands in response to changes in stimuli.[42,55–57]
The past decade has witnessed a rapid progress in the field of stimuli-responsive supramolecular hydrogels, not only due to their aesthetic attributes but also because of their broad applications in bio- and nanotechnology.[23,58,59] Formation of higher-order structures through non-covalent interactions, reversible sol–gel transitions, chemical and structural versatility, and multistimuli responsiveness make supramolecular hydrogels ideal candidates for various biomedical applications like scaffolds for cell/tissue engineering, sensors, bioimaging, and carriers for biomolecules. Multiple reviews discuss various approaches in developing supramolecular hydrogels for these applications.[1,4,23,27,52,53,60–69]
In this review, we aim to discuss the recent developments in the area of stimuli-responsive supramolecular hydrogels and their applications in regenerative medicine. Regenerative medicine is a field which uses endogenous and/or exogenous cells or delivery of biomolecules to repair compromised cell/tissue functions. The transplanted cells can contribute to tissue repair through differentiation or through secreted factors (trophic factors) to rejuvenate the compromised tissue environment to promote tissue repair. Biomaterials as artificial extracellular matrix (ECM) play a key role in regenerative medicine, where they serve as carriers for cells, providing structural integrity and tissue-specific physicochemical cues to facilitate cellular functions like adhesion, proliferation, migration, tissue-specific differentiation when used as a scaffold, and secretome. While decellularized tissues and ECM proteins are known to provide biologically relevant cues for cellular response, considering the complex structure and batch-to-batch variation of natural ECM proteins, focus has been given to using synthetic biomaterials or hybrid biomaterials encompassing both synthetic and biologically derived materials for such applications, with one class of such materials being supramolecular hydrogels.[70–73]
2. Physical Stimuli-Responsive Hydrogels
2.1. Temperature as a Stimulus
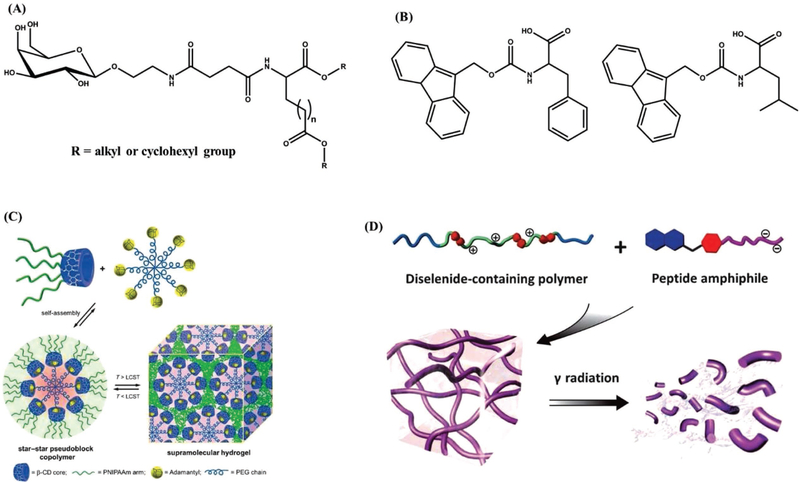
Among various physical stimuli, temperature-responsive supramolecular hydrogels are the most widely studied systems, where the hydrogelators undergo sol–gel or gel–sol transitions in response to subtle changes in their surrounding temperature.[74–80] Akin to thermoreversible hydrogels, the temperature-dependent phase transitions of supramolecular hydrogels are largely driven by hydrophilic and hydrophobic interactions.[81–86] One of the first temperature-responsive supramolecular hydrogels was based on glycosylated amino acid derivatives, for example, N-acetyl-galactosamine-appended amino acid (GalNAc-aa), which self-assembles into a hydrogel above its critical gelation concentration (CGC) (Figure 2A).[87] CGC is the minimum concentration of the hydrogelator at which gelation occurs. The robust hydrogen bonding between the amide groups and the hydrophobic interaction among the cyclic hydrocarbon groups present in the hydrogelators result in network formation at ambient temperature. The phase transition behavior of these molecules is highly sensitive to the molecular structure of the building blocks, wherein the hydrophobic alkyl chain length dictates the self-assembly (Figure 2A). While this study has used hydrophobic interactions involving aliphatic chains, aromatic–aromatic interactions such as π–π stacking have also been employed to create supramolecular hydrogels.[88–90] An example is the use of Fmoc-phenylalanine (Fmoc-F; Fmoc = (fluoren-9-ylmethoxy)carbonyl)) or a combination of Fmoc-protected amino acids (e.g., Fmoc-F and Fmoc-L; L = leucine) as building blocks, where the precursors selfassemble to form 3D network upon cooling to room temperature from a higher temperature (90 °C). Here, the π–π stacking between the Fmoc groups and hydrogen-bonding interactions between the amide groups of the amino acid molecules play an important role in the network formation (Figure 2B).[88] Supramolecular hydrogels formed from thermoreversible poly(N-isopropylacrylamide) (PNIPAM) is an example of using polymer hydrogelators as the building blocks. PNIPAM is widely known for its lower critical solution temperature phenomenon in aqueous solution.[91,92] A star-shaped PNIPAM polymer with a β-cyclodextrin (β-CD) molecule was shown to form supramolecular self-assembled architectures when mixed with adamantylterminated eight-arm poly(ethylene glycol) polymer through inclusion complexation between the β-CD molecules and the adamantyl groups (Figure 2C).[93] Upon heating, this mixture transitioned from a clear solution to a stable hydrogel at around 37 °C, which is mostly driven by the PNIPAM molecules.
Figure 2.
Schematic representation of hydrogelators and supramolecular structures. A) Structure of GalNAc-aa; B) structure of Fmoc-F and Fmoc-L hydrogelators; C) formation of self-assembled supramolecular architecture via inclusion complexation in aqueous solution and the formation of supramolecular hydrogel at lower critical solution temperature. Adapted with permission.[93] Copyright 2013, John Wiley & Sons. D) Schematic representation of γ-ray responsive supramolecular hydrogel formed from a diselenide-containing polymer and a peptide amphiphile. Adapted with permission.[107]
Biomaterials exhibiting sol–gel transition at physiological temperature (37 °C) are highly attractive in tissue engineering as scaffolds and for delivery of biomolecules. The sol–gel transition at 37 °C enables easy encapsulation of cells and their minimally invasive administration in vivo.[94,95] Conversely, the gel–sol transition at physiological temperature can be used to release biomolecules or cells within the defect site. Incorporation of growth factors along with cells during encapsulation and their subsequent controlled release enables continuous supply of growth factor(s) to control the fate/phenotype of the differentiating cells. Hong et al. have used thermogelling of injectable polyethylene glycol-b-poly (l-alanine) (PEG-b-poly (l-alanine)) supramolecular hydrogelator to encapsulate tonsilderived mesenchymal stem cells with hepatogenic growth factors and found overexpression of hepatic marker mRNA and proteins.[96] Studies have also explored the potential of phase transition of supramolecular structures to mitigate the cell damage encountered during cryopreservation. In this case, the supramolecular structures of Boc-O-dodecyl-l-tyrosine hydrogelator minimized cell damage by confining the ice crystal growth and reducing osmotic shock during cryopreservation.[97] Huang et al. developed a polymeric hydrogelator based on copolymer of PEG and oligo(tyrosine).[98] This copolymeric precursor showed concentration-dependent thermoresponsive sol–gel transition depending upon the length of the tyrosine oligomer. The resulting supramolecular hydrogels showed the controlled delivery of encapsulated proangiogenic drug, desferrioxamine, over 7 days. The sustained delivery of drug coupled with noncytotoxicity of the hydrogelator makes it an ideal candidate as an injectable drug depot.
2.2. Light as a Stimulus
Another widely used physical stimulus is light, which leverages hydrogelators encoded with light-sensitive groups (e.g., photocleavable groups, photoisomerizable groups) to trigger phase transitions.[99–102] One key advantage of light as a trigger is the potential use of coherent light sources such as laser, which allows creation of supramolecular hydrogels with high spatial resolution.[103,104] Polypeptides or small molecules encoded with light-sensitive moieties such as 2-nitrobenzyl groups have been shown to form supramolecular structures when exposed to light of wavelength 260 < λ < 365 nm.[105,106] Upon exposure to light, the 2-nitrobenzyl group is cleaved from the precursors allowing the interactions between the building blocks and subsequent formation of 3D networks. A specific example includes MAX7CNB, a photocaged peptide, which exhibits an unfolded structure in aqueous solution and upon exposure to light undergoes folding due to the exclusion of the photocaged group.[105] This subsequently leads to the formation of ordered amphiphilic β-hairpins resulting in viscoelastic hydrogel structures. Similarly, peptide amphiphile (PA)—hybrid molecule containing a hydrophilic peptide sequence attached to a hydrophobic alkyl chain or aromatic rings—with bioactive epitope Arg-Gly-Asp-Ser (RGDS) bearing a photocleavable 2-nitrobenzyl group undergoes sol–gel transition as a result of light-induced structural change. This small structural change was found to induce a significant change in the architecture of the network from nanospheres to nanofibers.[106] Another example is γ-ray (often used in clinical radiotherapy)-responsive supramolecular hydrogel based on diselenide-containing polymers and peptide amphiphiles Nap–Nitrob–GFFYGE (Figure 2D).[107] The resulting hydrogel undergoes gel–sol transition upon exposure to γ-ray following a disruption of the diselenium bonds present in the network.
Apart from the use of photocleavable moieties, light-induced cis–trans isomerization has also been employed to generate photoresponsive supramolecular hydrogels.[108–111] For example, a supramolecular hydrogel was developed by mixing α-CD conjugated curdlan (β−1,3 glucan, CUR) polymer with pAC12Azo polymer containing an azobenzene group.[112] Upon mixing, the polymers form supramolecular network via inclusion of the trans-azo group into the CD core. This supramolecular hydrogel undergoes gel–sol transition upon photoirradiation at 365 nm, which induces isomerization of the trans-azo group to the cis-azo group (trans/cis = 12:88).
The ability of the hydrogelators to respond to light has been used to achieve spatiotemporal control over cellular response. A study by He et al. has used bi-orthogonal photoclick/photodegradation to control the structural and mechanical properties of the hydrogels surrounding the encapsulated cells.[113] The hydrogelators were prepared by linking N-terminal of peptides to biaryl-substituted tetrazole containing o-allyloxy group on the N-phenyl group. Hydrogelators undergo self-assembly due to hydrophilic interaction in peptide and π–π stacking in aromatic moieties. On exposure to UV light, intramolecular photoclick reaction converts tetrazole moiety to fluorescent pyrazoline cycloadduct, thus disrupting π–π stacking and self-assembly. Human mesenchymal stem cells encapsulated within these hydrogels were stable for 3 weeks while UV light–mediated degradation of the hydrogel structures facilitated change in cell morphology where the cells with spherical shape adopted a spindle morphology over time.
Photomodulation of hydrogel accompanied by fluorescence offered by these hydrogelators can be potentially used to study the cellular behavior using photopatterned channels. In addition to their use as scaffolds for cell encapsulation and culture, light-responsive supramolecular hydrogels are also explored for controlled delivery of different biomolecules such as vitamins, drugs, and phototherapeutic agents.[114–117] Although most of the light-sensitive biomaterials focus on UV light as a trigger, in vivo application of UV-sensitive smart materials is limited due to its low penetrability through the tissues. While both photodegradation- and photoisomerization-induced phase transitions of hydrogelators require the use of high-energy UV or visible light, light with lesser energy such as near-infrared (NIR) light has also been employed to trigger phase transition of supramolecular hydrogels.[118–120] Compared with UV light, NIR light not only deeply penetrates through the tissues but also is less damaging to the cells. A hydrogel was developed via mixing of α-CD and PEG-terminated poly(amino amine) dendrimer bearing NIR-active platinum (Pt) nanoparticles in the core. The inclusion of the PEG chains into the cavities of α-CD and the strong hydrogen bonding between the PEG-terminated poly(amino amine) dendrimers led to network formation. Upon irradiation with NIR light (for about 20 min), self-heating and subsequent disruption of the supramolecular network led to gel–sol transition.[118] NIR as a stimulus could also extend the in vivo application of these hydrogels for chemotherapy, as they can be irradiated even within deep tissues. In another study, a peptide-based hydrogelator containing two-photon absorption dye dimethylaminocoumarin-4-yl-methoxycarbonyl was designed to undergo gel–sol transition on exposure to twophoton NIR light. Spatiotemporal control of fluidity of the said hydrogelator was found to be useful for remote manipulation of bacterial chemotaxis, thus showing its potential for controlled drug release and studying migration and differentiation of cells in deep tissues.[121]
Another example where light was effectively used as a stimulus in biomedical application is in the area of actuators. Stimuli-responsive hydrogel-based actuators, which manifest different kinds of deformation and movements can find various biomedical applications.[122,123] Leveraging the actuating potential of supramolecular hydrogel, artificial muscles have been designed using photoresponsive azo isomerization in azobenzene and its inclusion complex with α-CD. Here the supramolecular hydrogel expands on exposure to UV light at 365 nm due to the formation of inclusion complex with trans form of α-CD, while the exposure to visible light at 430 nm dissociates the complex due to the trans isomerization to cis form of αCD. By controlling the volume of the hydrogel exposed in the direction of incident light, macroscopic deformation can be controlled.[124] Various multistimuli-responsive supramolecular hydrogelators using light as a stimulus in conjunction with other stimuli such as pH, temperature, redox conditions, are also explored for various applications, which are discussed in multiresponsive hydrogels.[125–127]
2.3. Electric and Magnetic Fields as Stimuli
Though only few examples exist, other physical stimuli that have been explored toward supramolecular hydrogels include electric and magnetic fields.[128–131] Development of electric and magnetic field responsive supramolecular hydrogels relies on hydrogelators with the ability to respond to the respective stimulus. Similar to traditional/polymeric hydrogels that are responsive to electric field, supramolecular hydrogels with similar function can exhibit fast response and deform under electric field.[123,128,132] Design of magnetic field–responsive supramolecular hydrogels utilizes hydrogelators with magnetically active centers such as paramagnetic or diamagnetic species (e.g., metal ions, aromatic moieties) that form aligned fibrillar structures in the presence of external magnetic field.[131,133–135] The subsequent hydrogel formation requires retention of these aligned structures and their assembly which is usually achieved by the addition of ions or pH change.[131,136] The switchable nature of the magnetic field coupled with the slow diffusion of ions enables precise control of the anisotropy across the networks, something that is generally very difficult to achieve.
Unlike temperature and light, very few supramolecular hydrogels based on electric or magnetic field have been explored toward biomedical applications. One of the very early works by Kwon et al. on polymeric supramolecular hydrogel has shown the potential of using electrical trigger to release payload of insulin due to disruption of the self-assembly formed between hydrogen-bonded poly(ethyloxazoline) and poly(methacrylic acid).[137] Xue et al. have demonstrated the use of electrically responsive 2-naphthalenyl-glycine-phenylalaninephenylalanine-based hydrogelator for multiple applications such as controlled drug release, artificial muscle, and microfluidic device.[138]
3. Chemical Stimuli
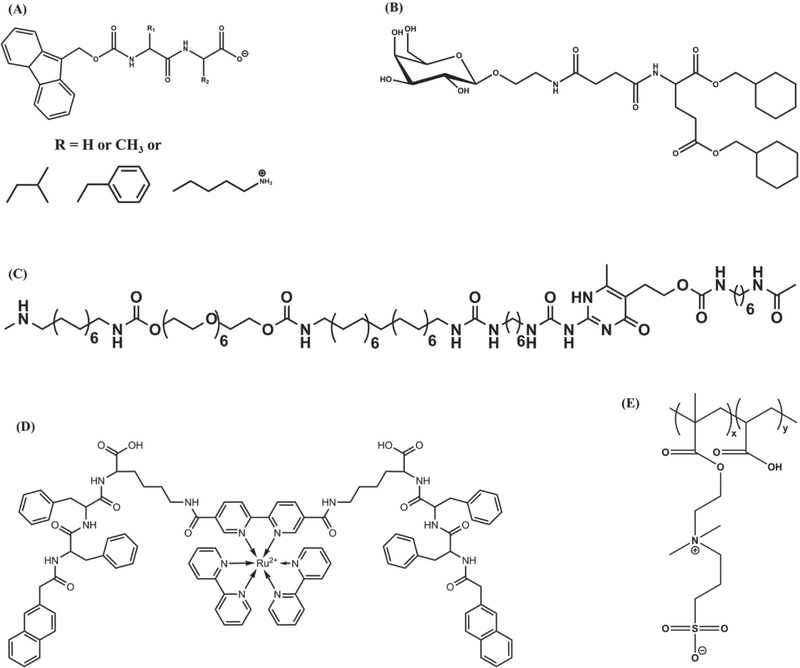
One of the most effective methods to initiate supramolecular hydrogelation is the use of pH as a stimulus since small amount of acid or base can provide large pH-shift through rapid diffusion of protons or hydroxide ions.[139–142] In general, the reversible protonation/deprotonation of an acidic/basic group increases the solubility of the hydrogelators in aqueous medium upon change in pH, which in turn can result in the formation of supramolecular hydrogels above their CGC. Furthermore, change in pH is also known to significantly affect the intensity and strength of hydrogen bonds between the hydrogelators.[75,141,143–145] For example, Patra et al. developed histidinebased hydrogelators that showed varying CGC over a pH range of 2–12.[145] In this case, depending upon the pH, amphiphilic hydrogelators exhibited different ionic forms which influenced their intermolecular interactions, solubilities, and aggregation. Among various hydrogelators, peptide-based molecules are the most commonly used to form pH-sensitive supramolecular structures. As an example, Fmoc-diphenylalanine (Fmoc-FF; Figure 3A) self-assembles into fibrillar- or ribbonlike structures which ultimately form supramolecular hydrogels at pH 4.[143] The self-assembly of such hydrogelators causes two apparent pKa shifts (pKa ≈6.4 and ≈2.2), which are different from the theoretical pKa of the monomer Fmoc-FF (pKa 3.5).[146]
Figure 3.
Schematic representation of various hydrogelators. A) Molecular structure of Fmoc-dipeptide; B) structure of GalNAc-appended (GalNAc = N-acetylgalactosamine) glutamate ester; C) structure of PEG-UPy chain-extended (co)polymers; D) structure of a metallohydrogelator with Ru(II) metal ion; E) chemical structure of the polyelectrolyte (PAA-co-DMAPS).
Zhou et al. have alternatively used the intrinsic changes in pH associated with mixing of N-acetylgalactosamine (GalNAc)appended glutamate ester with an amphiphilic carboxylic acid to form supramolecular hydrogels (Figure 3B). Unlike systems that rely on external pH, herein the formation of supramolecular network structures was triggered by in situ changes in pH.[147] Several other hydrogelators with different functional groups have been developed to form supramolecular hydrogels in response to various pH conditions like acidic,[148] neutral,[149–151] and alkaline conditions.[152–154] For instance, segmented amphiphilic macromolecules consisting of hydrophilic poly(ethylene glycol) (PEG) moieties and 2-ureido-4[1H]-pyrimidinone (UPy) units self-assemble into hydrogel structures due to the hydrogen bonding between self-complementary UPy units (Figure 3C).[155] These supramolecular UPy-hydrogels can undergo gel–sol transition at basic pH, with a pH threshold of 8.5, and reversibly transform back into a gel state at neutral pH.[156]
Since biological tissues could exhibit different pH (gastric pH of 1–3 vs intestinal pH of 6.1–7), such pH-responsive supramolecular hydrogels can be used as smart carriers for drug delivery.[157] Moreover, many pathological conditions such as cancer have been characterized to exhibit differential extracellular pH compared with the healthy state.[158] This differential pH within the compromised tissues has often been leveraged toward the delivery of drugs. One such system is the oral delivery of drugs for gastrointestinal treatment. In this case, pH-responsive polymeric supramolecular structures based on a mixture of poly(acryloyl 6-aminocaproic acid) and poly(methacrylic acid-co-ethyl acrylate) have shown potential utility as a gastric retentive device for the delivery of oral drugs, nutritional modulation in bariatric intervention, and ingestible diagnostics.[159] The acidic environment–induced hydrogen bonding between the 6-aminocaproic acid molecules leads to an elastomeric structure formation which dissolves in neutral or alkaline solutions providing the necessary structural and enteric functions. A similar strategy of using abnormal extracellular acidic pH of cancerous tissue has been adopted for the development of delivery vehicle for antitumor drugs.[160]
Besides changes in pH, many other chemical stimuli such as redox reactions, metal ligations, acid–base reactions, and ring-opening metathesis polymerization triggering sol–gel transition have been used toward supramolecular hydrogel formation.[161–166] A self-assembling peptide sequence, AcC(FKFE)2CG-NH2, cyclized via disulfide bonding of the flanking cysteine residues has been shown to self-assemble into fibrillar structures upon reduction of the disulphide bond.[162] In another report, Ac-I3CGK-NH2, a cysteine-containing small peptide, formed hydrogels at low concentrations under an oxidative environment, where their mechanical properties have been tuned via degree of oxidation.[167] Of these chemical stimuli-responsive supramolecular hydrogels, redox-responsive supramolecular hydrogels have been extensively studied, especially for their self-healing property and potential applications as actuators.[168,169] Using redox stimuli-triggered gel–sol transition of olsalazine-containing supramolecular hydrogel, a site-specific delivery of anti-inflammatory drug was shown by Li et al.[170] In another study, Wojciechowski et al. have shown temporal control over transient gelation of redox-triggered supramolecular hydrogelators by tuning the competing kinetics of dissolution and gelation, thus expanding the scope of chemical stimuli-responsive supramolecular hydrogels.[171]
In the case of supramolecular hydrogels harnessing metal ion coordination, the gel–sol transition can be tuned by oxidation of the metal ion center which undergoes geometrical changes following redox reaction, thereby shifting the intermolecular interactions.[172–175] For example, a tripeptide derivative (Nap-FFK), integrated with a ruthenium(II) tris(bipyridine) ([Ru(bipy)3]2+) complex, undergoes spontaneous self-assembly in aqueous solution resulting in supramolecular hydrogels (Figure 3D).[172] Upon oxidation of the metal ions from Ru(II) to Ru(III), the geometry of the metal complex changes from hexagonal to octahedral affecting the intermolecular π–π stacking and hydrogen bonding interactions among the tripeptide molecules eventually causing gel–sol transition. The ability of metal complexation to alter the non-covalent interactions in macromolecules has long been recognized and used to create biomimetic structures.[176,177] Qin et al. developed peptide dendron based hydrogelators which showed sol–gel transition to form higher-order structures in the presence of various multivalent metal ions.[178] Using divalent metal ions, researchers developed a shrinkable supramolecular hydrogel with potential application in controlled drug delivery.
Supramolecular hydrogels formed by host–guest reactions were designed to be redox responsive. For instance, supramolecular hydrogels formed from poly(acrylic acid) modified with cyclodextrins (pAA-CDs) (host) and pAA modified with ferrocene (pAA-Fc) (known for its redox-responsive properties) (guest) underwent gel–sol transition upon addition of oxidant sodium hypochlorite (NaClO). In contrast, the addition of glutathione (GSH) to the solution was shown to induce hydrogel formation via sol–gel transition. Due to the hydrophobic nature of the Fc group, β-CD displayed a high affinity for the Fc group leading to gelation (sol–gel transition), whereas the oxidized state of the Fc group, that is, cationic Fc+ group, exhibited a low affinity for β-CD leading to gel–sol transition.[179]
Employing specific chemical molecules as a trigger to drive self-assembly is another approach to form supramolecular hydrogels. While utilizing specific small molecules as a trigger to drive/disrupt self-assembly could expand the biological application of supramolecular hydrogels, it is much more challenging as it requires strong molecular recognition to ensure efficacy and avoid nontarget effects.[45] Chemical molecules, such as sodium chloride and vancomycin, have also been used as a trigger to form/disrupt the supramolecular network structures.[180,181] N-(fluorenyl-9-methoxycarbonyl)-D-Ala-D-Ala-based supramolecular hydrogels exhibited gel–sol transition upon binding to vancomycin.[180] The interaction of vancomycin with the D-Ala-D-Ala peptide (via hydrogen bonding between amide and/or carboxyl groups and aromatic interactions between the phenyl groups) is thought to disturb the balance between hydrophobic and hydrophilic interactions in N-(fluorenyl9-methoxycarbonyl)-D-Ala-D-Ala hydrogel network resulting in a gel–sol transition. In contrast, the hydrogel formed by N-(fluorenyl-9-methoxycarbonyl)-L-Ala-L-Ala, enantiomer of N-(fluorenyl-9-methoxycarbonyl)-D-Ala-D-Ala, showed no such response to vancomycin, thus demonstrating the ability of supramolecular systems in chiral recognition. In a recent report, a supramolecular polyelectrolyte hydrogel was prepared through random copolymerization of acrylic acid (AA) and 3-dimethyl (methacryloyloxyethyl) ammonium propane sulfonate (DMAPS) followed by swelling in NaCl aqueous solution (Figure 3E).[181] The resulting supramolecular network exhibited robust mechanical properties.
Another variant of creating hierarchical structures is the use of dissipative self-assembly driven by chemical reaction.[182,183] In this case, precursors are converted into self-assembling building blocks by the conversion of a source of energy, normally photon or a reactant. These intrinsically unstable self-assembling building blocks revert to their original precursors, either on removal or depletion of energy source, giving the building blocks a limited lifetime. Here the rate of energy consumption for self-assembly should be higher than the rate of energy dissipation during disintegration to form self-assembled structures. An example is the hydrogel formation by the amide surfactants (e.g., (S)-4-((1-(dodecylamino)-3-methyl-1-oxobutan2-yl)amino)-4-oxobutanoic acid), which was indirectly fuelled by sucrose conversion. In this case, sucrose was converted to ethanol and carbon dioxide using yeast. Produced carbon dioxide in equilibrium with bicarbonate in water subsequently released a proton. This available proton was used to protonate the soluble negatively charged amide surfactant, subsequently leading to surfactant assembly. As the gaseous carbon dioxide gradually left the system, the chemical equilibrium was shifted to the carbon dioxide side over time and the protons were gradually removed from the assembled network, resulting in the collapse of the network.[183] In addition to reactants, enzymes and light energy like UV light have also been used to control dissipative self-assembly of molecules.[182] Although properties of supramolecular hydrogels based on dissipative energy assembly have been studied extensively in recent years, their probable application as self-healing materials, actuators, or adaptive materials is still far from realization.
4. Biological Stimuli
Given the key role played by the enzymatic reactions in the formation of hierarchical structures in nature, enzyme-triggered supramolecular hydrogels have drawn increased attention in the recent years. An excellent article on enzyme-catalyzed supramolecular hydrogelation reviewed the use of enzymes to trigger and control self-assembly of supramolecular hydrogels.[184] A number of enzymes such as phosphatase,[185–189] esterase/lipase,[190] β-lactamase,[191] β-galactosidase,[192] glucose oxidase,[193] peroxidase,[194,195] chymotrypsin,[196] thermolysin,[197,198] thrombin,[199] matrix metalloproteinase (MMP)-9,[200,201] have been utilized in developing supramolecular networks.
The key factor that drives hydrogelation is the enzyme-catalyzed hydrolysis/formation of bonds resulting in active hydrogelator which results in higher-order structures via self-assembly.[185,202] Zelzer et al. in their review discussed various strategies to design enzyme-responsive materials, including supramolecular hydrogels.[203] Enzymes can trigger hydrogelation either by functioning as a trigger to drive the self-assembly or by generating active hydrogelators from the precursors which undergo further self-assembly.[204]
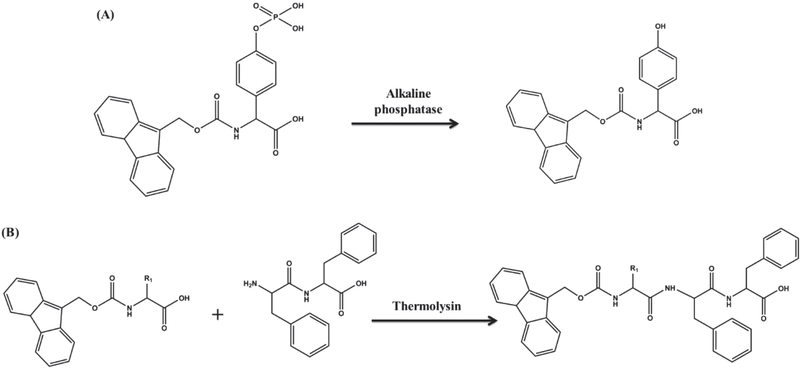
One of the early examples of enzyme-triggered supramolecular hydrogel formation is the use of alkaline phosphatase to induce self-assembly of Fmoc-tyrosine phosphate via catalytic hydrolysis of the phosphate group (Figure 4A).[185] Studies have also harnessed the ability of enzymes such as thermolysin to form peptide bonds for creating supramolecular structures.[198,205] Thermolysin is a thermally stable enzyme with a preference toward hydrophobic or aromatic residues on the N-terminus of the peptide chains to form amide bonds with another molecule. Together, the properties have been used to create a tripeptide hydrogelator by reacting phenylalanine dipeptide (Phe)2 with Fmoc-amino acids, which spontaneously self-assemble to form supramolecular hydrogels (Figure 4B).[198] A major advantage of enzyme-catalyzed peptide bond formation is the absence of by-products except water. Though the promise of enzymatic hydrogelation in understanding biological functions and in developing biomimetic systems are far from fully realized, the use of enzymes has several advantages such as the ability to obtain sophisticated non-covalent structures, flexibility in structural modification, and, most importantly, availability of enzymes both in vitro and in vivo.
Figure 4.
Schematic representation of enzymatic reaction on active precursors. A) Reaction scheme showing the enzymatic hydrolysis of a precursor N-(fluorenylmethyloxycarbonyl) tyrosine phosphate to active hydrogelator N-(fluorenylmethyloxycarbonyl) tyrosine; B) reaction scheme showing the enzymatic bond formation between diphenylalanine dipeptide and Fmoc-phenylalanine to produce active tripeptide hydrogelator.
Akin to use of differential pH of the diseased tissues, use has been also made of elevated levels of extracellular enzymes like MMP at the tumor environment to assist delivery of drugs.[206] For instance, Tanaka et al. used sol–gel transition to form the supramolecular hydrogels to induce cell necrosis.[207] Herein, the authors harnessed the MMP-7-triggered self-assembly of peptide lipids to induce gelation of these molecules inside the cells, which exerted stress leading to cell death. Similarly, Pires et al. have used the high availability of alkaline phosphatase within the osteosarcoma to achieve sol–gel transition of carbohydrate amphiphiles to induce apoptosis of the cells.[208] The selective inhibitory effects of alkaline phosphatase–assisted self-assembly and the extent of dephosphorylation of d-peptides on the effective inhibition of cancer cells have been demonstrated by Zhou et al.[204] Researchers found the antitumor effect of enzymatic self-assembled hydrogels to be dependent upon the level of dephosphorylation of the hydrogelator. Unphosphorylated d-peptides did not have any inhibitory effect while mono- and diphosphorylated d-peptides showed inhibitory effects on cancer cells and not on normal cells. Further, monophosphorylated d-tetrapeptides showed pronounced inhibitory effect as compared with their diphosphorylated counterparts. In another study, 2-cyano-6-aminobenzothiazole (CBT)-based precursor Cys (SEt)-Glu-Tyr-(H2PO3)-Phe-Phe-Gly-CBT was found to undergo sequential gelation in the presence of alkaline phosphatase and glutathione to form nanofiber assemblies at each stage.[209] Excessive extracellular alkaline phosphatase in the tumor environment dephosphorylated the hydrogelator precursors to yield an intermediate structure. Upon endocytosis, this intermediate structure underwent a second transformation inside the cell due to the presence of intracellular glutathione to form the nanofiber assembly. This dual mechanism opens up a possibility of using such precursors to detect the infected cells and deliver the drugs simultaneously. Further studies have demonstrated the application of enzyme-instructed self-assembly of d-peptide in intracellular imaging.[210] Enzyme-triggered transformations have also been used to develop logic gate systems for biosensors.[211] For instance, tripeptide-based supramolecular hydrogels BPmoc-F3 (boronophenylmethoxycarbonyl-phenylalanine) and BNmoc-F3 (borononaphthylmethoxycarbonyl-phenylalanine) showed oxidation-responsive and reduction-responsive gel–sol transitions, respectively.[212] When BFmoc-F3 or BNmoc-F3 precursors were encapsulated with the corresponding two oxidase or reductase enzymes, they responded to the corresponding input by OR logic gate. However, a hybrid gel based on BPmoc-F3-glucose oxidase and BNmoc-F3-nitroreductase responded to glucose and nicotinamide adenine dinucleotide (NADH) only through AND gate to undergo gel–sol transition. This hybrid supramolecular hydrogel enabled sensing of both biomarkers, glucose and NADH, and thereby the release of the encapsulated drug. Development of such intelligent drug-release system based on logic sensing can improve the specificity of the targeted delivery of a drug.
Small molecules and metabolites produced by the cells can also be used as a trigger toward phase transitions of supramolecular hydrogels.[213,214] For instance, higher level of reactive oxygen species (ROS) is associated with pathophysiology of various diseases such as arthritis, diabetes, cancer, and neurodegenerative diseases. Hence, systems that can be responsive to extracellular ROS level can have various applications such as drug delivery.[215] Incorporation of functional groups such as disulfide units can be used to render the supramolecular structures cell responsive, as cell-secreted molecules like cysteine, glutathione, thioredoxin-1, protein disulfide isomerase, can cleave the disulfide bonds.[216–220] Cleavable disulfide linker containing peptide-based hydrogelator precursors with the ability to undergo sol–gel transition in the presence of glutathione or dithiothreitol have also been developed.[221] Such systems can be used toward cell encapsulation and 3D cell culture.
5. Multistimuli
The aforedescribed studies focus on supramolecular hydrogels where the sol–gel or gel–sol transition is triggered by a single stimulus; however, most biological systems are capable of responding to multiple stimuli. To this end, significant efforts have been dedicated to design supramolecular hydrogels with multistimuli responsiveness.[30,222–225] Such multistimuli-responsive hydrogels respond to two or more stimuli such as pH, temperature, light, enzymes, redox potential, electric or magnetic field, or small molecules.[214,223,226–231] Here, the phase transition behavior can be regulated through the use of multiple stimuli in a sequential manner or in concert.[168] Harnessing host–guest interactions to form supramolecular hydrogels offers an ideal system to create multistimuli-responsive systems by using precursors with responsiveness to different stimuli.[34,38,168,179,232–239]
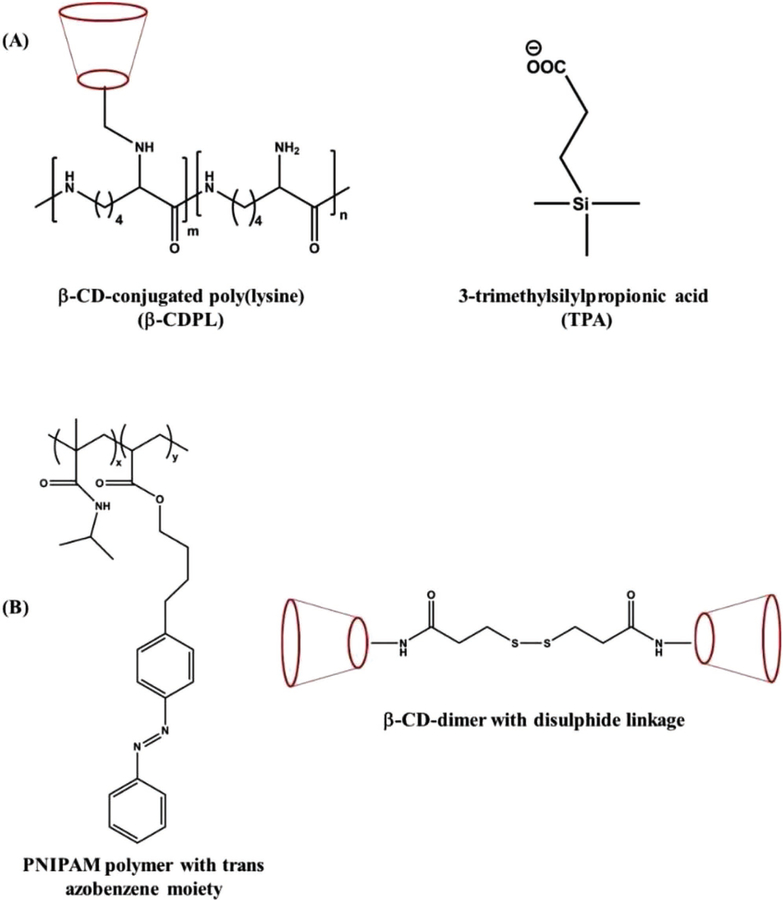
Supramolecular hydrogel consisting of β-CD-conjugated poly(lysine) (β-CDPL) and 3-trimethylsilylpropionic acid (TPA) is one such system manifesting multistimuli responsiveness.[240] The rapid phase transition observed in this system is mostly associated with the temperature- and PH-driven intermolecular associations via inclusion complexation between β-CD and TPA as well as ionic complexation between β-CDPL and TPA, respectively (Figure 5A). Another such example is a supramolecular hydrogel made by mixing PNIPAM polymers containing azo groups (where PNIPAM and azo groups are temperature and light sensitive, respectively) and CD dimers connected through disulphide bonds (which can function as redox-responsive switch), where the former functions as the guest and the CD molecules as the host (Figure 5B).[228] The said supramolecular hydrogel exhibited reversible sol–gel transition upon exposure to temperature and light, while showing the ability to undergo gel–sol transition responding to reducing agents. Similarly, a temperature- and pH-responsive host–guest supramolecular hydrogel was developed by mixing poly(l-glutamic acid)block-poly(ethylene oxide) (PLG-b-PEO) and α-CD in aqueous solution at room temperature. It was proposed that normal micelle formation mediated by the H bonding among PLG within PLG-b-PEO copolymer was formed in the first stage, and then the supramolecular inclusion of the PEO into the α-CD core formed the physical cross-linking, resulting in the hydrogelation. The hydrogel was shown to undergo reversible sol–gel transition upon changes in temperature and pH.[237] The resulting supramolecular hydrogel showed sustained release of anticancer drug, doxorubicin hydrochloride, over 45 days. The versatility of host–guest inclusion complexes based supramolecular hydrogels in terms of their synthetic chemistry and multiple stimuli responsiveness has led to such multiple studies for their potential applications in controlled delivery of drugs, biomolecules, etc.[45,241]
Figure 5.
Schematic representation of various multistimuli-responsive hydrogelators. A) Structures β-CD-conjugated poly(lysine) (β-CDPL) and 3-trimethylsilylpropionic acid (TPA); B) structures of PNIPAM polymers containing azo groups and cyclodextrin (CD) dimers connected through disulphide bonds.
A hydrogelator based on phenylalanine derivative and azobenzene derivative and responding to temperature, pH, and photoirradiation was designed for cell encapsulation. Here, cell encapsulation was controlled by using temperature- and pH-driven sol–gel transition, while the encapsulated cells were subsequently released by using photoirradiation.[242]
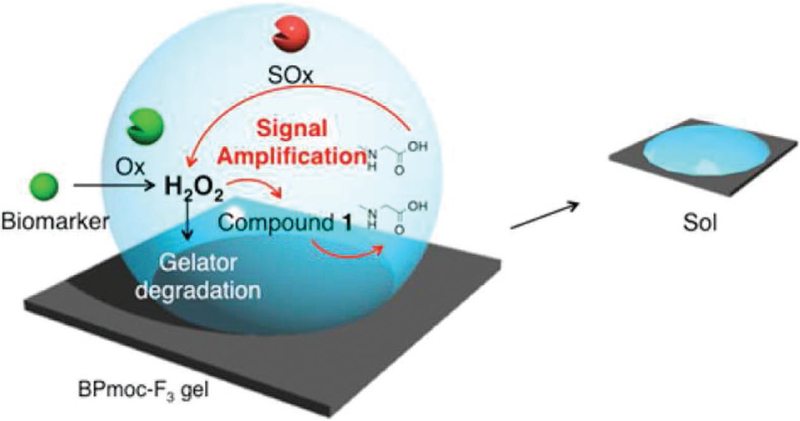
Multistimuli-responsive systems offer advantages such as incorporation of a feedback loop to develop diagnostic tools with improved sensitivity or using multiple stimuli for specificity. Such a feedback system was developed and used to amplify the chemosensor signals by a cascade of enzyme-responsive gelation of BPmoc-F-based hydrogelator. This study utilized reactive BPmoc-F-based supramolecular hydrogel and self-immolating dendron to achieve seamless detection of multiple analytes in human plasma.[243] Herein, the reaction of BPmoc-F with hydrogen peroxide releases boronic acid moiety, which leads to disassembly of peptide hydrogel. It was observed that the self-assembly of supramolecular hydrogel was not disturbed by the embedded sarcosine oxidase (SOx) and self-immolating dendron in it. H2O2 (biomarker of hyperuricemia) can react with the boronic acid moiety of dendron resulting in sarcosine generation. The released sarcosine molecule gets oxidized by sarcosine oxidase to generate two molecules of H2O2 as a reaction by-product (Figure 6). The released H2O2 then rapidly dissembles BPmoc-F hydrogel. This cascade of reactions increases the sensitivity toward H2O2 (analyte) detection by gel–sol transition of hydrogels. Similar to H2O2 detection, the authors also demonstrated the use of such signal amplification to detect glucose (biomarker of diabetes) and uric acid (biomarker of gout) in human plasma with higher sensitivity.
Figure 6.
Schematic representation of the use of enzyme-responsive supramolecular hydrogelator (BPmoc-F3) for signal amplification in biomarker detection. Reprinted with permission.[243] Copyright 2015, American Chemical Society.
The use of multistimuli-responsive hydrogel has also been exploited in creating logic gate function for various applications such as controlled release of biomolecules and intelligent soft materials for diagnostics and therapeutics.[212,244] Such systems offer sol–gel or gel–sol transition as a response to different stimuli depending upon the input patterns and also provide complex control over the phase transition of the resulting hydrogels. Similar to the use of hybrid supramolecular hydrogels responding to two-enzyme inputs in AND logic gate for chemosensors, there are possibilities of using multiple types and combination of stimuli with logic gates. For example, when there are two or more input stimuli, the hydrogelator can respond to simultaneous addition of inputs (AND gate), either input (OR), all inputs (NAND), and at least one input (NOR), or a combination of these patterns such as AND-OR.[212,245] Logic gate–based phase transitions of supramolecular hydrogels are especially useful for improving the selectivity of controlled release of biomolecules.
6. Conclusion and Perspective
While most of the potential applications of stimuli-responsive supramolecular hydrogels are in their infancy and under development, the ability of these materials to undergo sol–gel transition at physiological temperature has led to a number of commercially available ECM mimetics for 3D cell culture. Prominent examples of these include Biogelx (by Biogelx Inc.) Fmocmodified peptides, Curodont (by Credentis)—peptide based on anionic groups P11–4, and PuraMatrix (by 3D Matrix)—RADA 16.[246–248] Most of these materials are used as 3D culture or implants for in vivo filling of voids for tissue regeneration. In addition to temperature and metal ligation (as with Biogelx) triggered gelation, the physicochemical properties of the supramolecular hydrogels can be tuned to achieve various desirable properties. For instance, the ability of PuraMatrix and Curodont is under consideration as fillers for dental lesions due to their ability to undergo biomineralization, which can be influenced by both chemistry and hydrophobicity of the precursors.[249–251] In essence, temperature-responsive supramolecular hydrogels that can undergo biomineralization could guide enamel regeneration through de novo mineralization and growth.
Most hydrogels as ECM mimetics provide static cues (based on chemical and mechanical properties of the hydrogel). However, employing a hydrogelator with sensitivity toward multiple stimuli and/or different kinetics will enable the development of synthetic matrices emulating dynamic features of the native ECM. The dynamic nature of the ECM is pivotal to its function, and the reciprocal interactions of the cells with their ECM play a key role in tissue development, tissue homeostasis, and diseases.[252] For example, stimuli-responsive supramolecular hydrogels functioning as actuators can be used to provide multiple cues to the encapsulated cells. Previously, we have used electric field–responsive hydrogels with dynamic bending/ extension as multifunctional (providing electrical, chemical, and mechanical cues) scaffolds for stem cell culture.[123] Multistimuli-responsive supramolecular hydrogel actuators with the ability to exhibit reversible “on–off” swelling or deformations can also contribute to soft robots and actuators with applications in surgical tools, smart carriers for biomolecules, and smart implants for soft tissue replacements such as heart valves.
One of the unique features of stimuli-responsive supramolecular structures is their reversible interactions. These stimuli-specific reversible interactions (formation and disruption of interactions to exhibit gel–sol or sol–gel behavior) enable reverse modulation of matrix properties. Sequential exposure of the hydrogel structures to various stimuli can be used to erode or increase the overall cross-link density of the hydrogel structures. Such spatiotemporal control over matrix properties can be used to provide spatially varying chemical (e.g., growth factors’ availability) and physical cues (mechanical and space available for the cells to spread or migrate). Supramolecular hydrogels that can change the material properties such as matrix degradation responding to cell secreted molecules in a controlled and predetermined fashion can be an ideal scaffold for both in vitro and in vivo tissue engineering. The degradation of the scaffolds as a function of tissue formation will lead to tissues with optimal structure and function, making tissue engineering a vital approach to treat critical defects.
Another major area to which stimuli-responsive supramolecular hydrogels can contribute is immunotherapy. To this end, supramolecular hydrogels have been tested to modulate both adaptive and innate immunity. One of the most widely exploited applications of supramolecular hydrogels is as vaccine adjuvants. Vaccines are widely used to protect against or treat diseases through immune response. While proteins and peptides are useful antigens to induce both humoral and cellular immune response, they often fall short in function due to their poor immunogenicity. To circumvent the problem, various immunological adjuvants are developed. Supramolecular hydrogels have been considered as a good candidate for immune adjuvants due to their biocompatibility, low self-immunogenicity, and control over design. Various stimuli such as temperature and presence of enzymes have been used to create supramolecular hydrogel-based vaccine adjuvants.[253–255] Besides as vaccine adjuvants, sol–gel transition of supramolecular hydrogelators has been used to create de novo chemoattractant depots to attract the immune cells to the desired location and modulate their functions.[256,257] Given the power of immunotherapy to treat various debilitating diseases such as cancer, the use of supramolecular hydrogels in immunotherapy will experience further advancements.
Moving forward, there are several opportunities to use stimuli-responsive supramolecular hydrogels to advance regenerative medicine. These include the design of multifunctional, adaptive scaffolds to engineer tissues from stem cells as carriers for on-demand delivery of cells and biomolecules and as vehicles to modulate the immune system to accelerate the innate tissue repair and for combating the diseases.
Although rational design of supramolecular hydrogelators can be used toward artificial ECM emulating various functions of native ECM, there still remains a gap between synthetic supramolecular systems and living molecules. While molecular engineering of hydrogelators and supramolecular hydrogels with multistimuli responsiveness is moving toward this goal, design of molecular structures to achieve coordinated spatiotemporal dynamics and precise molecular recognitions is still challenging. Another class of stimuli-responsive hydrogels with tremendous biomedical applications is enzyme- or biomolecule-responsive supramolecular hydrogels. The expression and concentration levels of these molecules are closely related to the health (or pathology), and hence, biomolecule-responsive hydrogels have great promise for developing therapies and diagnostics. However, the development of such systems with the necessary sensitivity and selectivity requires mitigating many challenges such as overcoming thermodynamic equilibrium of reactants and products of reversible reaction. Nevertheless, further advances in molecular engineering of building blocks to control the function of the resulting structures will truly expand their biomedical applications. These advances not only create new supramolecular hydrogelators and hydrogels but will also lead to new frontiers in science.
Acknowledgements
J.H. and N.S. contributed equally to this work. The authors acknowledge the financial support from National Institutes of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number NIH R01 AR063184 and NIH R01 AR071552.
Biographies

Jiaul Hoque is a postdoctoral associate at the Department of Orthopaedic Surgery at Duke University. He received his M.Sc. degree in organic chemistry from the University of North Bengal, India, in 2009 and his Ph.D. degree in polymer and materials science from Jawaharlal Nehru Centre for Advanced Scientific Research, India, in 2017. His current research focuses on developing polymer-based biomaterials for bone tissue engineering and regenerative medicine.

Nivedita Sangaj is a senior research associate at the Department of Orthopaedic Surgery at Duke University. She received her doctorate in technology of plastics from the Institute of Chemical Technology, India, and postdoctoral training at the University of California, San Diego, in biomaterials for tissue engineering. She has worked as a research scientist at GE India Technology Centre, India, and DuPont Knowledge Center, India. She also worked as chief technology officer at IPUA Technical Center, India. She is currently working on biomaterials for regenerative medicine.

Shyni Varghese is a professor of biomedical engineering, mechanical engineering and materials science, and orthopedic surgery at Duke University. She is the inaugural MEDx investigator at Duke University. Prior to moving to Duke, she was a professor of bioengineering at the University of California, San Diego. Her research covers a broad range of topics including stem cells, biomaterials, biologically inspired systems, and regenerative medicine.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Jiaul Hoque, Department of Orthopaedic Surgery, Duke University, Durham 27710, NC, shyni.varghese@duke.edu.
Nivedita Sangaj, Department of Orthopaedic Surgery, Duke University, Durham 27710, NC.
Shyni Varghese, Department of Orthopaedic Surgery, Department of Biomedical Engineering, Department of Mechanical Engineering and Materials Science, Duke University, Durham 27710, NC.
References
- [1].Stupp SI, Nano Lett 2010, 10, 4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kumar VA, Wang BK, Kanahara SM, Exp. Biol. Med 2016, 241, 899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Goor OJGM, Hendrikse SIS, Dankers PYW, Meijer EW, Chem. Soc. Rev 2017, 46, 6621. [DOI] [PubMed] [Google Scholar]
- [4].Tu Y, Peng F, Adawy A, Men Y, Abdelmohsen LK, Wilson DA, Chem. Rev 2016, 116, 2023. [DOI] [PubMed] [Google Scholar]
- [5].Huang G, Li F, Zhao X, Ma Y, Li Y, Lin M, Jin G, Lu TJ, Genin GM, Xu F, Chem. Rev 2017, 117, 12764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hoffman AS, Adv. Drug Delivery Rev 2012, 64, 18. [Google Scholar]
- [7].Koetting MC, Peters JT, Steichen SD, Peppas NA, Mater. Sci. Eng. R Rep 2015, 93, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lim HL, Hwang Y, Kar M, Varghese S, Biomater Sci 2014, 2, 603. [DOI] [PubMed] [Google Scholar]
- [9].Tsitsilianis C, Soft Matt 2010, 6, 2372. [Google Scholar]
- [10].Lin CC, RSC Adv 2015, 5, 39844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Appel EA, del Barrio J, Loh XJ, Scherman OA, Chem. Soc. Rev 2012, 41, 6195. [DOI] [PubMed] [Google Scholar]
- [12].Liow SS, Dou Q, Kai D, Karim AA, Zhang K, Xu F, Loh XJ, ACS Biomater. Sci. Eng 2016, 2, 295. [DOI] [PubMed] [Google Scholar]
- [13].Liow SS, Karim AA, Loh XJ, MRS Bull 2016, 41, 557. [Google Scholar]
- [14].Varghese S, Lele AK, Mashelkar RA, J. Chem. Phys 2000, 112, 3063. [Google Scholar]
- [15].Tamai Y, Tanaka H, Nakanishi K, Macromolecules 1996, 29, 6750. [Google Scholar]
- [16].Varghese S, Lele AK, Srinivas D, Sastry M, Mashelkar RA, Adv. Mater 2001, 13, 1544. [Google Scholar]
- [17].Zhang YM, Ionov L, Langmuir 2015, 31, 4552. [DOI] [PubMed] [Google Scholar]
- [18].Phadke A, Zhang C, Arman B, Hsu CC, Mashelkar RA, Lele AK, Tauber MJ, Arya G, Varghese S, Proc. Natl. Acad. Sci 2012, 109, 4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gulyuz U, Okay O, Macromolecules 2014, 47, 6889. [Google Scholar]
- [20].Lowenberg C, Balk M, Wischke C, Behl M, Lendlein A, Acc. Chem. Res 2017, 50, 723. [DOI] [PubMed] [Google Scholar]
- [21].Appel EA, del Barrio J, Loh XJ, Scherman OA, Chem. Soc. Rev 2012, 41, 6195. [DOI] [PubMed] [Google Scholar]
- [22].Raeburn J, Zamith Cardoso A, Adams DJ, Chem. Soc. Rev 2013, 42, 5143. [DOI] [PubMed] [Google Scholar]
- [23].Webber MJ, Appel EA, Meijer EW, Langer R, Nat. Mater 2016, 15, 13. [DOI] [PubMed] [Google Scholar]
- [24].Estroff LA, Hamilton AD, Chem. Rev 2004, 104, 1201. [DOI] [PubMed] [Google Scholar]
- [25].Jia YG, Zhu XX, Chem. Mater 2015, 27, 387. [Google Scholar]
- [26].Krieg E, Bastings MMC, Besenius P, Rybtchinski B, Chem. Rev 2016, 116, 2414. [DOI] [PubMed] [Google Scholar]
- [27].Du X, Zhou J, Shi J, Xu B, Chem. Rev 2015, 115, 13165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Dong RJ, Pang Y, Su Y, Zhu XY, Biomater. Sci 2015, 3, 937. [DOI] [PubMed] [Google Scholar]
- [29].Mann JL, Yu AC, Agmon G, Appel EA, Biomater. Sci 2017, 6, 10. [DOI] [PubMed] [Google Scholar]
- [30].Ren LX, He LH, Sun TC, Dong X, Chen YM, Huang J, Wang C, Macromol. Biosci 2009, 9, 902. [DOI] [PubMed] [Google Scholar]
- [31].Rodell CB, Kaminski AL, Burdick JA, Biomacromolecules 2013, 14, 4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wu DQ, Wang T, Lu B, Xu XD, Cheng SX, Jiang XJ, Zhang XZ, Zhuo RX, Langmuir 2008, 24, 10306. [DOI] [PubMed] [Google Scholar]
- [33].Park KM, Yang JA, Jung H, Yeom J, Park JS, Park KH, Hoffman AS, Hahn SK, Kim K, ACS Nano 2012, 6, 2960. [DOI] [PubMed] [Google Scholar]
- [34].Appel EA, Biedermann F, Rauwald U, Jones ST, Zayed JM, Scherman OA, J. Am. Chem. Soc 2010, 132, 14251. [DOI] [PubMed] [Google Scholar]
- [35].Zhang J, Guo DS, Wang LH, Wang Z, Liu Y, Soft Matter 2011, 7, 1756. [Google Scholar]
- [36].Ni MF, Zhang N, Xia W, Wu X, Yao CH, Liu X, Hu XY, Lin C, Wang LY, J. Am. Chem. Soc 2016, 138, 6643. [DOI] [PubMed] [Google Scholar]
- [37].Barrow SJ, Kasera S, Rowland MJ, del Barrio J, Scherman OA, Chem. Rev 2015, 115, 12320. [DOI] [PubMed] [Google Scholar]
- [38].Hwang I, Jeon WS, Kim HJ, Kim D, Kim H, Selvapalam N, Fujita N, Shinkai S, Kim K, Angew. Chem. Int. Ed 2007, 46, 210. [DOI] [PubMed] [Google Scholar]
- [39].Kim K, Selvapalam N, Ko YH, Park KM, Kim D, Kim J, Chem. Soc. Rev 2007, 36, 267. [DOI] [PubMed] [Google Scholar]
- [40].Cheng H, Fan X, Wang X, Ye E, Loh XJ, Li Z, Wu YL, Biomacromolecules 2018, 19, 1926. [DOI] [PubMed] [Google Scholar]
- [41].Park KM, Yang JA, Jung H, Yeom J, Park JS, Park KH, Hoffman AS, Hahn SK, Kim K, ACS Nano 2012, 6, 2960. [DOI] [PubMed] [Google Scholar]
- [42].Boekhoven J, Rubert Perez CM, Sur S, Worthy A, Stupp SI, Angew. Chem. Int. Ed 2013, 52, 12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Qi Z, Schalley CA, Acc. Chem. Res 2014, 47, 2222. [DOI] [PubMed] [Google Scholar]
- [44].Murray J, Kim K, Ogoshi T, Yao W, Gibb BC, Chem. Soc. Rev 2017, 46, 2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ma X, Zhao YL, Chem. Rev 2015, 115, 7794. [DOI] [PubMed] [Google Scholar]
- [46].Appel EA, Loh XJ, Jones ST, Dreiss CA, Scherman OA, Biomaterials 2012, 33, 4646. [DOI] [PubMed] [Google Scholar]
- [47].Lin N, Dufresne A, Biomacromolecules 2013, 14, 871. [DOI] [PubMed] [Google Scholar]
- [48].Zhang MM, Xu DH, Yan XZ, Chen JZ, Dong SY, Zheng B, Huang FH, Angew. Chem. Int. Ed 2012, 51, 7011. [DOI] [PubMed] [Google Scholar]
- [49].Li J, Li X, Ni X, Wang X, Li H, Leong KW, Biomaterials 2006, 27, 4132. [DOI] [PubMed] [Google Scholar]
- [50].Dong SY, Luo Y, Yan XZ, Zheng B, Ding X, Yu YH, Ma Z, Zhao QL, Huang FH, Angew. Chem. Int. Ed 2011, 50, 1905. [DOI] [PubMed] [Google Scholar]
- [51].Deng W, Yamaguchi H, Takashima Y, Harada A, Angew. Chem. Int. Ed 2007, 46, 5144. [DOI] [PubMed] [Google Scholar]
- [52].Shigemitsu H, Hamachi I, Acc. Chem. Res 2017, 50, 740. [DOI] [PubMed] [Google Scholar]
- [53].Shigemitsu H, Hamachi I, Chem. Asian J 2015, 10, 2026. [DOI] [PubMed] [Google Scholar]
- [54].Segarra-Maset MD, Nebot VJ, Miravet JF, Escuder B, Chem. Soc. Rev 2013, 42, 7086. [DOI] [PubMed] [Google Scholar]
- [55].Sangeetha NM, Maitra U, Chem. Soc. Rev 2005, 34, 821. [DOI] [PubMed] [Google Scholar]
- [56].Meazza L, Foster JA, Fucke K, Metrangolo P, Resnati G, Steed JW, Nat. Chem 2013, 5, 42. [DOI] [PubMed] [Google Scholar]
- [57].Xu W, Song Q, Xu JF, Serpe MJ, Zhang X, ACS Appl. Mater. Interfaces 2017, 9, 11368. [DOI] [PubMed] [Google Scholar]
- [58].Dong R, Zhou Y, Huang X, Zhu X, Lu Y, Shen J, Adv. Mater 2015, 27, 498. [DOI] [PubMed] [Google Scholar]
- [59].Ma X, Zhao Y, Chem. Rev 2015, 115, 7794. [DOI] [PubMed] [Google Scholar]
- [60].Yang H, Yuan B, Zhang X, Scherman OA, Acc. Chem. Res 2014, 47, 2106. [DOI] [PubMed] [Google Scholar]
- [61].Uhlenheuer DA, Petkau K, Brunsveld L, Chem. Soc. Rev 2010, 39, 2817. [DOI] [PubMed] [Google Scholar]
- [62].Zhang SG, Biotechnol. Adv 2002, 20, 321. [DOI] [PubMed] [Google Scholar]
- [63].Brinkmann J, Cavatorta E, Sankaran S, Schmidt B, van Weerd J, Jonkheijm P, Chem. Soc. Rev 2014, 43, 4449. [DOI] [PubMed] [Google Scholar]
- [64].Pape ACH, Dankers PYW, Adv. Polym. Sci 2015, 268, 253. [Google Scholar]
- [65].Shao Y, Jia H, Cao T, Liu D, Acc. Chem. Res 2017, 50, 659. [DOI] [PubMed] [Google Scholar]
- [66].Dong R, Pang Y, Su Y, Zhu X, Biomater. Sci 2015, 3, 937. [DOI] [PubMed] [Google Scholar]
- [67].Wang D, Hu Y, Liu PF, Luo D, Acc. Chem. Res 2017, 50, 733. [DOI] [PubMed] [Google Scholar]
- [68].Trausel F, Versluis F, Maity C, Poolman JM, Lovrak M, van Esch JH, Eelkema R, Acc. Chem. Res 2016, 49, 1440. [DOI] [PubMed] [Google Scholar]
- [69].Kahn JS, Hu YW, Willner I, Acc. Chem. Res 2017, 50, 680. [DOI] [PubMed] [Google Scholar]
- [70].Toh WS, Spector M, Lee EH, Cao T, Mol. Pharmaceutics 2011, 8, 994. [DOI] [PubMed] [Google Scholar]
- [71].Ko IK, Lee SJ, Atala A, Yoo JJ, Exp. Mol. Med 2013, 45, e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Boekhoven J, Stupp SI, Adv. Mater 2014, 26, 1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Phadke A, Shih YRV, Varghese S, Macromol. Biosci 2012, 12, 1022. [DOI] [PubMed] [Google Scholar]
- [74].Bhattacharjee S, Datta S, Bhattacharya S, Chem. Eur. J 2013, 19, 16672. [DOI] [PubMed] [Google Scholar]
- [75].Shome A, Debnath S, Das PK, Langmuir 2008, 24, 4280. [DOI] [PubMed] [Google Scholar]
- [76].Suzuki M, Yumoto M, Shirai H, Hanabusa K, Chem. Eur. J 2008, 14, 2133. [DOI] [PubMed] [Google Scholar]
- [77].Wang H, Zhang W, Dong X, Yang Y, Talanta 2009, 77, 1864. [DOI] [PubMed] [Google Scholar]
- [78].Zhang ZX, Liu X, Xu FJ, Loh XJ, Kang ET, Neoh KG, Li J, Macromolecules 2008, 41, 5967. [Google Scholar]
- [79].Ochi R, Nishida T, Ikeda M, Hamachi I, J. Mater. Chem. B 2014, 2, 1464. [DOI] [PubMed] [Google Scholar]
- [80].Latxague L, Ramin MA, Appavoo A, Berto P, Maisani M, Ehret C, Chassande O, Barthelemy P, Angew. Chem. Int. Ed 2015, 54, 4517. [DOI] [PubMed] [Google Scholar]
- [81].Yan X, Wang F, Zheng B, Huang F, Chem. Soc. Rev 2012, 41, 6042. [DOI] [PubMed] [Google Scholar]
- [82].Wang L, Shi X, Wang J, Soft Matt 2018, 14, 3090. [DOI] [PubMed] [Google Scholar]
- [83].Zhang ZX, Liu KL, Li J, Macromolecules 2011, 44, 1182. [Google Scholar]
- [84].Ramin MA, Latxague L, Sindhu KR, Chassande O, Barthelemy P, Biomaterials 2017, 145, 72. [DOI] [PubMed] [Google Scholar]
- [85].Ci TY, Chen L, Yu L, Ding JD, Sci. Rep 2014, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Zhang L, Shen WJ, Luan JB, Yang DX, Wei G, Yu L, Lu WY, Ding JD, Acta Biomater 2015, 23, 271. [DOI] [PubMed] [Google Scholar]
- [87].Kiyonaka S, Sugiyasu K, Shinkai S, Hamachi I, J. Am. Chem. Soc 2002, 124, 10954. [DOI] [PubMed] [Google Scholar]
- [88].Irwansyah I, Li YQ, Shi W, Qi D, Leow WR, Tang MB, Li S, Chen X, Adv. Mater 2015, 27, 648. [DOI] [PubMed] [Google Scholar]
- [89].Zhang Y, Gu H, Yang Z, Xu B, J. Am. Chem. Soc 2003, 125, 13680. [DOI] [PubMed] [Google Scholar]
- [90].Das D, Dasgupta A, Roy S, Mitra RN, Debnath S, Das PK, Chem. Eur. J 2006, 12, 5068. [DOI] [PubMed] [Google Scholar]
- [91].Badiger MV, Lele AK, Bhalerao VS, Varghese S, Mashelkar RA, J. Chem. Phys 1998, 109, 1175. [Google Scholar]
- [92].Bhalerao VS, Varghese S, Lele AK, Badiger MV, Polymer 1998, 39, 2255. [Google Scholar]
- [93].Zhang ZX, Liu KL, Li J, Angew. Chem. Int. Ed 2013, 52, 6180. [DOI] [PubMed] [Google Scholar]
- [94].Wu EC, Zhang SG, Hauser CAE, Adv. Funct. Mater 2012, 22, 456. [Google Scholar]
- [95].Webber MJ, Bioeng. Transl. Med 2016, 1, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Hong JH, Lee HJ, Jeong B, ACS Appl. Mater. Interfaces 2017, 9, 11568. [DOI] [PubMed] [Google Scholar]
- [97].Zeng J, Yin YX, Zhang L, Hu WH, Zhang CC, Chen WY, Macromol. Biosci 2016, 16, 363. [DOI] [PubMed] [Google Scholar]
- [98].Huang J, Hastings CL, Duffy GP, Kelly HM, Raeburn J, Adams DJ, Heise A, Biomacromolecules 2013, 14, 200. [DOI] [PubMed] [Google Scholar]
- [99].Tomatsu I, Peng K, Kros A, Adv. Drug Delivery Rev 2011, 63, 1257. [DOI] [PubMed] [Google Scholar]
- [100].Frkanec L, Jokic M, Makarevic J, Wolsperger K, Zinic M, J. Am. Chem. Soc 2002, 124, 9716. [DOI] [PubMed] [Google Scholar]
- [101].Ramamurthy V, Gupta S, Chem. Soc. Rev 2015, 44, 119. [DOI] [PubMed] [Google Scholar]
- [102].Ji W, Qin MG, Feng CL, Macromol. Chem. Phys 2018, 219, 1700398. [Google Scholar]
- [103].Wilson RM, Schnapp KA, Chem. Rev 1993, 93, 223. [Google Scholar]
- [104].Oberg KI, Chem. Rev 2016, 116, 9631. [DOI] [PubMed] [Google Scholar]
- [105].Haines LA, Rajagopal K, Ozbas B, Salick DA, Pochan DJ, Schneider JP, J. Am. Chem. Soc 2005, 127, 17025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Muraoka T, Koh CY, Cui HG, Stupp SI, Angew. Chem. Int. Ed 2009, 48, 5946. [DOI] [PubMed] [Google Scholar]
- [107].Cao W, Zhang XL, Miao XM, Yang ZM, Xu HP, Angew. Chem. Int. Ed 2013, 52, 6233. [DOI] [PubMed] [Google Scholar]
- [108].Liao XJ, Chen GS, Liu XX, Chen WX, Chen F, Jiang M, Angew. Chem. Int. Ed 2010, 49, 4409. [DOI] [PubMed] [Google Scholar]
- [109].Peng K, Tomatsu I, Kros A, Chem. Commun 2010, 46, 4094. [DOI] [PubMed] [Google Scholar]
- [110].Pianowski ZL, Karcher J, Schneider K, Chem. Commun 2016, 52, 3143. [DOI] [PubMed] [Google Scholar]
- [111].Matsumoto S, Yamaguchi S, Ueno S, Komatsu H, Ikeda M, Ishizuka K, Iko Y, Tabata KV, Aoki H, Ito S, Noji H, Hamachi, Chem. Eur. J 2008, 14, 3977. [DOI] [PubMed] [Google Scholar]
- [112].Tamesue S, Takashima Y, Yamaguchi H, Shinkai S, Harada A, Angew. Chem. Int. Ed 2010, 49, 7461. [DOI] [PubMed] [Google Scholar]
- [113].He M, Li J, Tan S, Wang R, Zhang Y, J. Am. Chem. Soc 2013, 135, 18718. [DOI] [PubMed] [Google Scholar]
- [114].Huang Y, Qiu Z, Xu Y, Shi J, Lin H, Zhang Y, Org. Biomol. Chem 2011, 9, 2149. [DOI] [PubMed] [Google Scholar]
- [115].Tomatsu I, Peng K, Kros A, Adv. Drug Delivery Rev 2011, 63, 1257. [DOI] [PubMed] [Google Scholar]
- [116].Swaminathan S, Garcia-Amoros J, Fraix A, Kandoth N, Sortino S, Raymo FM, Chem. Soc. Rev 2014, 43, 4167. [DOI] [PubMed] [Google Scholar]
- [117].Cheng TY, Wu HC, Huang MY, Chang WH, Lee CH, Wang TW, Nanoscale 2013, 5, 2734. [DOI] [PubMed] [Google Scholar]
- [118].Wang X, Wang C, Zhang Q, Cheng Y, Chem. Commun 2016, 52, 978. [DOI] [PubMed] [Google Scholar]
- [119].Zheng Z, Hu JJ, Wang H, Huang JL, Yu YH, Zhang Q, Cheng YY, ACS Appl. Mater. Interfaces 2017, 9, 24511. [DOI] [PubMed] [Google Scholar]
- [120].Tong X, Xiang J, Shi F, Zhao Y, Adv. Opt. Mater 2016, 4, 1392. [Google Scholar]
- [121].Yoshii T, Ikeda M, Hamachi I, Angew. Chem. Int. Ed 2014, 53, 7264. [DOI] [PubMed] [Google Scholar]
- [122].Wang E, Desai MS, Lee SW, Nano Lett 2013, 13, 2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Lim HL, Chuang JC, Tuan T, Aung A, Arya G, Varghese S, Adv. Funct. Mater 2011, 21, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Takashima Y, Hatanaka S, Otsubo M, Nakahata M, Kakuta T, Hashidzume A, Yamaguchi H, Harada A, Nat. Commun 2012, 3, 1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Jones CD, Steed JW, Chem. Soc. Rev 2016, 45, 6546. [DOI] [PubMed] [Google Scholar]
- [126].Sun Z, Li Z, He Y, Shen R, Deng L, Yang M, Liang Y, Zhang Y, J. Am. Chem. Soc 2013, 135, 13379. [DOI] [PubMed] [Google Scholar]
- [127].Sun Z, Huang Q, He T, Li Z, Zhang Y, Yi L, ChemPhysChem 2014, 15, 2421. [DOI] [PubMed] [Google Scholar]
- [128].Samanta SK, Bhattacharya S, J. Mater. Chem 2012, 22, 25277. [Google Scholar]
- [129].Sardone L, Palermo V, Devaux E, Credgington D, De Loos M, Marletta G, Cacialli F, Van Esch J, Samori P, Adv. Mater 2006, 18, 1276. [Google Scholar]
- [130].Alsberg E, Feinstein E, Joy MP, Prentiss M, Ingber DE, Tissue Eng 2006, 12, 3247. [DOI] [PubMed] [Google Scholar]
- [131].Wallace M, Cardoso AZ, Frith WJ, Iggo JA, Adams DJ, Chem. Eur. J 2014, 20, 16484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Yoshio M, Shoji Y, Tochigi Y, Nishikawa Y, Kato T, J. Am. Chem. Soc 2009, 131, 6763. [DOI] [PubMed] [Google Scholar]
- [133].Zhou J, Du XW, Gao Y, Shi JF, Xu B, J. Am. Chem. Soc 2014, 136, 2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Piepenbrock MOM, Lloyd GO, Clarke N, Steed JW, Chem. Rev 2010, 110, 1960. [DOI] [PubMed] [Google Scholar]
- [135].Ma D, Zhang LM, J. Phys. Chem. B 2008, 112, 6315. [DOI] [PubMed] [Google Scholar]
- [136].Hua YQ, Pu GJ, Ou CW, Zhang XL, Wang L, Sun JT, Yang ZM, Chen MS, Sci. Rep 2017, 7, 40172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Kwon IC, Bae YH, Kim SW, Nature 1991, 354, 291. [DOI] [PubMed] [Google Scholar]
- [138].Xue B, Qin M, Wang T, Wu J, Luo D, Jiang Q, Li Y, Cao Y, Wang W, Adv. Funct. Mater 2016, 26, 9053. [Google Scholar]
- [139].Hassanali A, Prakash MK, Eshet H, Parrinello M, Proc. Natl. Acad. Sci 2011, 108, 20410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Nanda J, Banerjee A, Soft Matt 2012, 8, 3380. [Google Scholar]
- [141].Aufderhorst-Roberts A, Frith WJ, Kirkland M, Donald AM, Langmuir 2014, 30, 4483. [DOI] [PubMed] [Google Scholar]
- [142].Spitzer D, Marichez V, Formon GJM, Besenius P, Hermans TM, Angew. Chem. Int. Ed 2018, 57, 11349. [DOI] [PubMed] [Google Scholar]
- [143].Jayawarna V, Ali M, Jowitt TA, Miller AE, Saiani A, Gough JE, Ulijn RV, Adv. Mater 2006, 18, 611. [Google Scholar]
- [144].Zhao F, Gao YA, Shi JF, Browdy HM, Xu B, Langmuir 2011, 27, 1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Patra T, Pal A, Dey J, Langmuir 2010, 26, 7761. [DOI] [PubMed] [Google Scholar]
- [146].Tang C, Smith AM, Collins RF, Ulijn RV, Saiani A, Langmuir 2009, 25, 9447. [DOI] [PubMed] [Google Scholar]
- [147].Zhou SL, Matsumoto S, Tian HD, Yamane H, Ojida A, Kiyonaka S, Hamachi I, Chem. Eur. J 2005, 11, 1130. [DOI] [PubMed] [Google Scholar]
- [148].Grigoriou S, Johnson EK, Chen L, Adams DJ, James TD, Cameron PJ, Soft Matt 2012, 8, 6788. [Google Scholar]
- [149].Marchesan S, Waddington L, Easton CD, Winkler DA, Goodall L, Forsythe J, Hartley PG, Nanoscale 2012, 4, 6752. [DOI] [PubMed] [Google Scholar]
- [150].Nonoyama T, Ogasawara H, Tanaka M, Higuchi M, Kinoshita T, Soft Matt 2012, 8, 11531. [Google Scholar]
- [151].Xu XD, Liang L, Cheng H, Wang XH, Jiang FG, Zhuo RX, Zhang XZ, J. Mater. Chem 2012, 22, 18164. [Google Scholar]
- [152].Liu YF, Yang YL, Wang C, Zhao XJ, Nanoscale 2013, 5, 6413. [DOI] [PubMed] [Google Scholar]
- [153].Fletcher NL, Lockett CV, Dexter AF, Soft Matt 2011, 7, 10210. [Google Scholar]
- [154].Imura Y, Matsue K, Sugimoto H, Ito R, Kondo T, Kawai T, Chem. Lett 2009, 38, 778. [Google Scholar]
- [155].Guo M, Pitet LM, Wyss HM, Vos M, Dankers PY, Meijer EW, J. Am. Chem. Soc 2014, 136, 6969. [DOI] [PubMed] [Google Scholar]
- [156].Bastings MMC, Koudstaal S, Kieltyka RE, Nakano Y, Pape ACH, Feyen DAM, van Slochteren FJ, Doevendans PA, Sluijter JPG, Meijer EW, Chamuleau SAJ, Dankers PYW, Adv. Healthcare Mater 2014, 3, 70. [DOI] [PubMed] [Google Scholar]
- [157].Cook MT, Tzortzis G, Charalampopoulos D, Khutoryanskiy VV, J. Controlled Release 2012, 162, 56. [DOI] [PubMed] [Google Scholar]
- [158].Webb BA, Chimenti M, Jacobson MP, Barber DL, Nat. Rev. Cancer 2011, 11, 671. [DOI] [PubMed] [Google Scholar]
- [159].Zhang SY, Bellinger AM, Glettig DL, Barman R, Lee YAL, Zhu JH, Cleveland C, Montgomery VA, Gu L, Nash LD, Maitland DJ, Langer R, Traverso G, Nat. Mater 2015, 14, 1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Li F, He J, Zhang M, Tam KC, Ni P, RSC Adv 2015, 5, 54658. [Google Scholar]
- [161].Wang M, Bao YL, Wu Y, Yu CL, Meng XY, Huang YX, Sun Y, Zheng LH, Li YX, J. Cell. Biochem 2010, 111, 75. [DOI] [PubMed] [Google Scholar]
- [162].Bowerman CJ, Nilsson BL, J. Am. Chem. Soc 2010, 132, 9526. [DOI] [PubMed] [Google Scholar]
- [163].Nakahata M, Takashima Y, Yamaguchi H, Harada A, Nat. Commun 2011, 2, 511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Sun ZF, Lv FC, Cao LJ, Liu L, Zhang Y, Lu ZG, Angew. Chem. Int. Ed 2015, 54, 7944. [DOI] [PubMed] [Google Scholar]
- [165].Takashima Y, Uramatsu K, Jomori D, Harima A, Otsubo M, Yamaguchi H, Harada A, ACS Macro Lett 2013, 2, 384. [DOI] [PubMed] [Google Scholar]
- [166].Zhang JY, Yan J, Pageni P, Yan Y, Wirth A, Chen YP, Qiao YL, Wang Q, Decho AW, Tang CB, Sci. Rep 2015, 5, 11914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Cao CH, Cao MW, Fan HM, Xia DH, Xu H, Lu JR, Chin. Sci. Bull 2012, 57, 4296. [Google Scholar]
- [168].Lu W, Le X, Zhang J, Huang Y, Chen T, Chem. Soc. Rev 2017, 46, 1284. [DOI] [PubMed] [Google Scholar]
- [169].Nakahata M, Takashima Y, Hashidzume A, Harada A, Angew. Chem. Int. Ed 2013, 52, 5731. [DOI] [PubMed] [Google Scholar]
- [170].Li X, Li J, Gao Y, Kuang Y, Shi J, Xu B, J. Am. Chem. Soc 2010, 132, 17707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Wojciechowski JP, Martin AD, Thordarson P, J. Am. Chem. Soc 2018, 140, 2869. [DOI] [PubMed] [Google Scholar]
- [172].Zhang Y, Zhang B, Kuang Y, Gao Y, Shi JF, Zhang XX, Xu B, J. Am. Chem. Soc 2013, 135, 5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Menyo MS, Hawker CJ, Waite JH, Soft Matt 2013, 9, 10314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Yan XZ, Wang F, Zheng B, Huang FH, Chem. Soc. Rev 2012, 41, 6042. [DOI] [PubMed] [Google Scholar]
- [175].Liang H, Zhang ZJ, Yuan QP, Liu JW, Chem. Commun 2015, 51, 15196. [DOI] [PubMed] [Google Scholar]
- [176].Varghese S, Lele AK, Srinivas D, Mashelkar RA, J. Phys. Chem. B 1999, 103, 9530. [Google Scholar]
- [177].Varghese S, Lele AK, Srinivas D, Mashelkar RA, J. Phys. Chem. B 2001, 105, 5368. [Google Scholar]
- [178].Qin L, Duan P, Xie F, Zhang L, Liu M, Chem. Commun 2013, 49, 10823. [DOI] [PubMed] [Google Scholar]
- [179].Nakahata M, Takashima Y, Yamaguchi H, Harada A, Nat. Commun 2011, 2, 511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Zhang Y, Gu HW, Yang ZM, Xu B, J. Am. Chem. Soc 2003, 125, 13680. [DOI] [PubMed] [Google Scholar]
- [181].Lei ZY, Wu PY, Nat. Commun 2018, 9, 1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].van Rossum SAP, Tena-Solsona M, van Esch JH, Eelkema R, Boekhoven J, Chem. Soc. Rev 2017, 46, 5519. [DOI] [PubMed] [Google Scholar]
- [183].Angulo-Pachon CA, Miravet JF, Chem. Commun 2016, 52, 5398. [DOI] [PubMed] [Google Scholar]
- [184].Yang Z, Liang G, Xu B, Acc. Chem. Res 2008, 41, 315. [DOI] [PubMed] [Google Scholar]
- [185].Yang ZM, Gu HW, Fu DG, Gao P, Lam JK, Xu B, Adv. Mater 2004, 16, 1440. [Google Scholar]
- [186].Li JY, Gao Y, Kuang Y, Shi JF, Du XW, Zhou J, Wang HM, Yang ZM, Xu B, J. Am. Chem. Soc 2013, 135, 9907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Gao J, Zheng WT, Kong DL, Yang ZM, Soft Matt 2011, 7, 10443. [Google Scholar]
- [188].Gao J, Wang HM, Wang L, Wang JY, Kong DL, Yang ZM, J. Am. Chem. Soc 2009, 131, 11286. [DOI] [PubMed] [Google Scholar]
- [189].Thornton K, Smith AM, Merry CLR, Ulijn RV, Biochem. Soc. Trans 2009, 37, 660. [DOI] [PubMed] [Google Scholar]
- [190].Yang ZM, Xu KM, Guo ZF, Guo ZH, Xu B, Adv. Mater 2007, 19, 3152. [Google Scholar]
- [191].Yang ZM, Ho PL, Liang GL, Chow KH, Wang QG, Cao Y, Guo ZH, Xu B, J. Am. Chem. Soc 2007, 129, 266. [DOI] [PubMed] [Google Scholar]
- [192].Zhao F, Weitzel CS, Gao Y, Browdy HM, Shi JF, Lin HC, Lovett ST, Xu B, Nanoscale 2011, 3, 2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Liu Y, Javvaji V, Raghavan SR, Bentley WE, Payne GF, Agric J, Food Chem 2012, 60, 8963. [DOI] [PubMed] [Google Scholar]
- [194].Sakai S, Komatani K, Taya M, RSC Adv 2012, 2, 1502. [Google Scholar]
- [195].Sakai S, Ogushi Y, Kawakami K, Acta Biomater 2009, 5, 554. [DOI] [PubMed] [Google Scholar]
- [196].Bremmer SC, Chen J, McNeil AJ, Soellner MB, Chem. Commun 2012, 48, 5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Das AK, Collins R, Ulijn RV, Small 2008, 4, 279. [DOI] [PubMed] [Google Scholar]
- [198].Toledano S, Williams RJ, Jayawarna V, Ulijn RV, J. Am. Chem. Soc 2006, 128, 1070. [DOI] [PubMed] [Google Scholar]
- [199].Bremmer SC, Chen J, McNeil AJ, Soellner MB, Chem. Commun 2012, 48, 5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Bremmer SC, McNeil AJ, Soellner MB, Chem. Commun 2014, 50, 1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Yang ZM, Ma ML, Xu B, Soft Matt 2009, 5, 2546. [Google Scholar]
- [202].Postma SG, Vialshin IN, Gerritsen CY, Bao M, Huck WT, Angew. Chem. Int. Ed 2017, 56, 1794. [DOI] [PubMed] [Google Scholar]
- [203].Zelzer M, Todd S, Hirst A, McDonald T, Ulijn R, Biomater. Sci 2013, 1, 11. [DOI] [PubMed] [Google Scholar]
- [204].Zhou J, Du X, Yamagata N, Xu B, J. Am. Chem. Soc 2016, 138, 3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Guilbaud JB, Vey E, Boothroyd S, Smith AM, Ulijn RV, Saiani A, Miller AF, Langmuir 2010, 26, 11297. [DOI] [PubMed] [Google Scholar]
- [206].Zhou J, Xu B, Bioconjugate Chem 2015, 26, 987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Tanaka A, Fukuoka Y, Morimoto Y, Honjo T, Koda D, Goto M, Maruyama T, J. Am. Chem. Soc 2015, 137, 770. [DOI] [PubMed] [Google Scholar]
- [208].Pires RA, Abul-Haija YM, Costa DS, Novoa-Carballal R, Reis RL, Ulijn RV, Pashkuleva I, J. Am. Chem. Soc 2015, 137, 576. [DOI] [PubMed] [Google Scholar]
- [209].Zheng Z, Chen P, Xie M, Wu C, Luo Y, Wang W, Jiang J, Liang G, J. Am. Chem. Soc 2016, 138, 11128. [DOI] [PubMed] [Google Scholar]
- [210].Li J, Gao Y, Kuang Y, Shi J, Du X, Zhou J, Wang H, Yang Z, Xu B, J. Am. Chem. Soc 2013, 135, 9907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Katz E, Poghossian A, Schoning MJ, Anal. Bioanal. Chem 2017, 409, 81. [DOI] [PubMed] [Google Scholar]
- [212].Ikeda M, Tanida T, Yoshii T, Kurotani K, Onogi S, Urayama K, Hamachi I, Nat. Chem 2014, 6, 511. [DOI] [PubMed] [Google Scholar]
- [213].Liu S, Luo Y, Liang G, Nanoscale 2016, 8, 766. [DOI] [PubMed] [Google Scholar]
- [214].Ooi HW, Hafeez S, van Blitterswijk CA, Moroni L, Baker MB, Mater Horiz 2017, 4, 1020. [Google Scholar]
- [215].Xu Q, He C, Xiao C, Chen X, Macromol. Biosci 2016, 16, 635. [DOI] [PubMed] [Google Scholar]
- [216].Brulisauer L, Gauthier MA, Leroux JC, J. Controlled Release 2014, 195, 147. [DOI] [PubMed] [Google Scholar]
- [217].Kar M, Shih YRV, Velez DO, Cabrales P, Varghese S, Biomaterials 2016, 77, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Holmgren A, Lu J, Biochem. Biophys. Res. Commun 2010, 396, 120. [DOI] [PubMed] [Google Scholar]
- [219].Hatahet F, Ruddock LW, Antioxid. Redox Signaling 2009, 11, 2807. [DOI] [PubMed] [Google Scholar]
- [220].Kar M, Vernon Shih YR, Velez DO, Cabrales P, Varghese S, Biomaterials 2016, 77, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Lv L, Liu H, Chen X, Yang Z, Colloids Surf. B Biointerfaces 2013, 108, 352. [DOI] [PubMed] [Google Scholar]
- [222].Shigemitsu H, Fujisaku T, Tanaka W, Kubota R, Minami S, Urayama K, Hamachi I, Nat. Nanotechnol 2018, 13, 165. [DOI] [PubMed] [Google Scholar]
- [223].Chen Y, Pang XH, Dong CM, Adv. Funct. Mater 2010, 20, 579. [Google Scholar]
- [224].Peng L, Zhang HJ, Feng AC, Huo M, Wang ZL, Hu J, Gao WP, Yuan JY, Polym. Chem 2015, 6, 3652. [Google Scholar]
- [225].Tan L, Liu Y, Ha W, Ding LS, Peng SL, Zhang S, Li BJ, Soft Matt 2012, 8, 5746. [Google Scholar]
- [226].Niu YL, Yuan XY, Zhao YH, Zhang WY, Ren LX, Macromol. Chem. Phys 2017, 218, 1600540. [Google Scholar]
- [227].Vatankhah-Varnoosfaderani M, Hashmi S, GhavamiNejad A, Stadler FJ, Polym. Chem 2014, 5, 512. [Google Scholar]
- [228].Guan Y, Zhao HB, Yu LX, Chen SC, Wang YZ, RSC Adv 2014, 4, 4955. [Google Scholar]
- [229].Gao YF, Dong CM, J. Polym. Sci. A Polym. Chem 2018, 56, 1067. [Google Scholar]
- [230].Sun ZF, Li ZY, He YH, Shen RJ, Deng L, Yang MH, Liang YZ, Zhang Y, J. Am. Chem. Soc 2013, 135, 13379. [DOI] [PubMed] [Google Scholar]
- [231].Zhuang J, Gordon MR, Ventura J, Li L, Thayumanavan S, Chem. Soc. Rev 2013, 42, 7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Li C, Rowland MJ, Shao Y, Cao T, Chen C, Jia H, Zhou X, Yang Z, Scherman OA, Liu D, Adv. Mater 2015, 27, 3298. [DOI] [PubMed] [Google Scholar]
- [233].Cheng W, Zhao D, Qiu Y, Hu H, Wang H, Wang Q, Liao Y, Peng H, Xie X, Soft Matt 2018, 14, 5213. [DOI] [PubMed] [Google Scholar]
- [234].Qi MD, Wang J, Ma XJ, Zhang Q, Wang FF, Kang Y, Liu Y, Lin HX, Zhongguo Zhong Yao Za Zhi 2017, 42, 3901. [DOI] [PubMed] [Google Scholar]
- [235].Abdul Karim A, Chee PL, Chan MF, Loh XJ, ACS Biomater. Sci. Eng 2016, 2, 2185. [DOI] [PubMed] [Google Scholar]
- [236].Miyamae K, Nakahata M, Takashima Y, Harada A, Angew. Chem. Int. Ed 2015, 54, 8984. [DOI] [PubMed] [Google Scholar]
- [237].Chen Y, Pang X-H, Dong C-M, Adv. Funct. Mater 2010, 20, 579. [Google Scholar]
- [238].Deng Z, Guo Y, Zhao X, Ma PX, Guo B, Chem. Mater 2018, 30, 1729. [Google Scholar]
- [239].Wang X, Wang J, Yang YY, Yang F, Wu DC, Polym. Chem 2017, 8, 3901. [Google Scholar]
- [240].Choi HS, Huh KM, Ooya T, Yui N, J. Am. Chem. Soc 2003, 125, 6350. [DOI] [PubMed] [Google Scholar]
- [241].Li J, Drug Deliv J, Sci. Technol 2010, 20, 399. [Google Scholar]
- [242].Liu GF, Ji W, Wang WL, Feng CL, ACS Appl. Mater. Interfaces 2015, 7, 301. [DOI] [PubMed] [Google Scholar]
- [243].Yoshii T, Onogi S, Shigemitsu H, Hamachi I, J. Am. Chem. Soc 2015, 137, 3360. [DOI] [PubMed] [Google Scholar]
- [244].Liu GF, Ji W, Feng CL, Langmuir 2015, 31, 7122. [DOI] [PubMed] [Google Scholar]
- [245].Qi Z, Malo de Molina P, Jiang W, Wang Q, Nowosinski K, Schulz A, Gradzielski M, Schalley CA, Chem. Sci 2012, 3, 2073. [Google Scholar]
- [246].Li Y, Wang F, Cui H, Bioeng. Transl. Med 2016, 1, 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Zhang S, Interface Focus 2017, 7, 20170028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Harper MM, Connolly ML, Goldie L, Irvine EJ, Shaw JE, Jayawarna V, Richardson SM, Dalby MJ, Lightbody D, Ulijn RV, Methods Mol. Biol 2018, 1777, 283. [DOI] [PubMed] [Google Scholar]
- [249].Phadke A, Zhang C, Hwang Y, Vecchio K, Varghese S, Biomacromolecules 2010, 11, 2060. [DOI] [PubMed] [Google Scholar]
- [250].Mari-Buye N, Luque T, Navajas D, Semino CE, Tissue Eng. Part A 2013, 19, 870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [251].Kind L, Stevanovic S, Wuttig S, Wimberger S, Hofer J, Muller B, Pieles U, J. Dent. Res 2017, 96, 790. [DOI] [PubMed] [Google Scholar]
- [252].Rosales AM, Anseth KS, Nat. Rev. Mater 2016, 1, 15012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [253].Rudra JS, Tripathi PK, Hildeman DA, Jung JP, Collier JH, Biomaterials 2010, 31, 8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].Wang HM, Luo Z, Wang YCZ, He T, Yang CB, Ren CH, Ma LS, Gong CY, Li XY, Yang ZM, Adv. Funct. Mater 2016, 26, 1822. [Google Scholar]
- [255].Zhou J, Li J, Du XW, Xu B, Biomaterials 2017, 129, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [256].Zhao F, Li JY, Zhou N, Sakai J, Gao Y, Shi JF, Goldman B, Browdy HM, Luo HBR, Xu B, Bioconjugate Chem 2014, 25, 2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].Moore AN, Silva TLL, Carrejo NC, Marmolejo CAO, Li IC, Hartgerink JD, Biomaterials 2018, 161, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]