Abstract
With widespread abuse of antibiotics, bacterial resistance has increasingly become a serious threat. Acinetobacter baumannii has emerged as one of the most important hospital-acquired pathogens worldwide. Bacteriophages (also called “phages”) could be used as a potential alternative therapy to meet the challenges posed by such pathogens. Endolysins from phages have also been attracting increasing interest as potential antimicrobial agents. Here, we isolated 14 phages against A. baumannii, determined the lytic spectrum of each phage, and selected one with a relatively broad host range, named vB_AbaP_PD-6A3 (PD-6A3 for short), for its biological characteristics. We over-expressed and purified the endolysin (Ply6A3) from this phage and tested its biological characteristics. The PD-6A3 is a novel phage, which can kill 32.4% (179/552) of clinical multidrug resistant A. baumannii (MDRAB) isolates. Interestingly, in vitro, this endolysin could not only inhibit A. baumannii, but also that of other strains, such as Escherichia coli and methicillin-resistant Staphylococcus aureus (MRSA). We found that lethal A. baumannii sepsis mice could be effectively rescued in vivo by phage PD-6A3 and endolysin Ply6A3 intraperitoneal injection. These characteristics reveal the promising potential of phage PD-6A3 and endolysin Ply6A3 as attractive candidates for the control of A. baumannii-associated nosocomial infections.
Keywords: multidrug resistance Acinetobacter baumannii, endolysin, ESKAPE, sepsis, phage therapy, mice model
Introduction
Acinetobacter baumannii is a non-fermentative, Gram-negative bacillus that can be universally found in nature. It is a conditional pathogen that has been proven to be one of the major pathogens causing hospital-acquired infections, such as bacteremia, urinary tract infections, and ventilator-associated pneumonia (Doi et al., 2015).
An increasing number of A. baumannii strains have now been clinically proven to be multi-drug, and even pan-drug resistant (Lin, 2014; Shivaswamy et al., 2015; Khakhum et al., 2016). According to CHINET surveillance in China, A. baumannii accounts for 10.77% of all clinical isolates, and is ranked the third most prevalent (Hu et al., 2016). The majority of these isolates are multidrug resistant A. baumannii (MDRAB), which is resistant to nearly all accessible antimicrobials. Therefore, the development of new tools to effectively manage A. baumannii infections is of great importance.
Bacteriophages (also called phages), as natural enemies of bacteria, exist abundantly in environments shared by their hosts. Phages were first discovered by Felix d’Hérelle in 1917 (Billiau, 2016; El-Shibiny and El-Sahhar, 2017). The lytic phages can specifically lyse pathogenic bacteria within their host range. In recent years, a huge importance has been given to the phage therapy and phage have been proposed as a valuable approach to control multiple pathogens (Melo et al., 2018).
In addition, phage endolysins, which phages use to digest the host bacterial cell wall to release progeny phages, have been suggested to be potent antibacterial agents in the treatment of Gram-positive pathogens both in vitro and in animal models (Yang et al., 2015; Zhang et al., 2016). However, the Gram-negative bacteria prevent the access of natural endolysins to peptidoglycan layer because of their outer membranes, thus making the exogenously added endolysins limited (Lim et al., 2014). Therefore, endolysins with strong lytic ability for the treatment of Gram-negative bacteria especially A. baumannii are urgently required.
We isolated and characterized a novel A. baumannii phage, PD6A3, and over-expressed its endolysin Ply6A3. Furthermore, the characteristics of the novel endolysin were evaluated. We then applied phage PD6A3 and endolysin Ply6A3 to the treatment of septic infections in mice. The present results support the potential use of phage PD6A3 and endolysin Ply6A3 as alternative antibacterial agents against drug-resistant A. baumannii infections.
Materials and Methods
Bacterial Strains and Culture Conditions
The clinical bacterial strains, human cells, plasmids, and primers used in the present study are listed in Table 1. Altogether, 552 clinical MDRAB isolates (Supplementary Table 1), and Pseudomonas aeruginosa, Escherichia coli, Klebsiella pneumonia, Staphylococcus aureus, and Enterococcus faecium, each comprising 40 isolates were collected between 2014 and 2017. Theses isolates were all collected from different patients at seven different hospitals in Shanghai, China and we have gotten written informed consents from these patients. Isolates were identified using the Vitek 2 compact system (Biomerieux, France). Antimicrobial susceptibility testing was performed using the broth microdilution method, recommended by the Clinical and Laboratory Standards Institute (CLSI, 2017). All isolates were identified by 16S rDNA sequencing analysis. The E. coli DH5α and E. coli BL21 (DE3) cells were purchased from TransGen Biotech, China. The overnight bacterial culture was 1:100 inoculated to fresh Luria-Bertani (LB) broth and cells were grown at 37°C with shaking at 220 rpm (Yang et al., 2015).
Table 1.
Primers, phage, bacterial strains, and plasmids used in this study.
| Name | Characteristics | Source |
|---|---|---|
| Forward primer | CGCTCGAGATGATTCT GACTAAAGACGGATTTAG | Sangon Biotech |
| Reverse primer | GCGAAGCTTCTATA AGCTCCGTAGAGCG | Sangon Biotech |
| pCold-SUMO | Expression vector (kanamycin resistant) | Our Laboratory Collection |
| pCold-SUMO-Ply6A3 | Recombinant vector (kanamycin resistant) | This study |
| DH5α E. coli | Amplification host for plasmid | Transgen Biotech |
| BL21(DE3) E. coli | Expression host for recombinant plasmid | Transgen Biotech |
| PD6A3 | A. baumannii phage | Our Laboratory Collection |
| Ply6A3 | The protein expressed by the endolysin gene | This study |
| AB1-AB200 | MDR-A. baumannii, 200 clinical strains | Our Laboratory Collection |
| AB32 | One of the host bacteria of Phage PD6A3 | Our Laboratory Collection |
| AB01-AB040 | PDR-A. baumannii, 40 clinical strains | Our Laboratory Collection |
| Enterococcus faecium | Vancomycin resistant, 40 clinical strains | Our Laboratory Collection |
| Staphylococcus aureus | Methicillin resistant, 40 clinical strains | Our Laboratory Collection |
| MRSA6 | One of the host bacteria of Phage PD6A3 | Our Laboratory Collection |
| Klebsiella pneumonia | Extended spectrumβ lactamases, 40 clinical strains | Our Laboratory Collection |
| Pseudomonas aeruginosa | Multi-drug Resistance, 40 clinical strains | Our Laboratory Collection |
| Escherichia coli | Extended spectrumβ lactamases, 40 clinical strains | Our Laboratory Collection |
| HEK293T cells | Human renal epithelial cells | Our Laboratory Collection |
| THP-1 cells | Human mononuclear macrophage | Our Laboratory Collection |
PD6A3, a novel Acinetobacter baumannii Phage. Ply6A3, the endolysin encoded by a novel Acinetobacter baumannii Phage PD6A3 gene. AB32, the No. 32 strains of multi-drug resistant Acinetobacter baumannii. MRSA6, the No. 6 strains of methicillin-resistant Staphylococcus aureus. HEK293T cells, human embryonic kidney cells 293. THP-1 cells, a human monocytic cell line derived from an acute monocytic leukemia patient.
Phage Isolation and Lytic Spectrum of Single Phages and Phage Cocktail
In this study, 27 non-duplicate clinical strains of extensively drug-resistant (XDR) A. baumannii were used as indicators of phage isolation. All phages were isolated from sewage (Yele et al., 2012). The supernatant (20 mL) from the sewage was added to 10 mL 3 × LB broth and 5 mL of mixed A. baumannii culture, then incubated at 37°C and 100 rpm/min overnight. We used the double-layer agar method to incubate and purify the supernatants which including the phages. Each individual phage was purified using several consecutive rounds of plaque picking until single-plaque morphology was observed. Eventually, 14 virulent phages were obtained. To enrich the phages, each phage was mixed with its host cells by certain proportion then grown in LB soft agar overlays (0.75% agar) overnight. Then the lysate was collected in SM buffer, enriched at 4°C overnight using polyethylene glycol (PEG) 8000, further purified with CsCl density gradient ultracentrifugation and stored at 4°C (Hua et al., 2018).
Then, one drop of phage PD6A3 solution was added onto a copper mesh grid, after natural precipitation about 10 min, 50 μl 2% phosphotungstic acid was added to the grid. After drying, we used a Hitachi transmission electron microscope H-9500 (Japan) to examine the morphology of the phage (Huang et al., 2013).
Phage titers were assessed using double-layer agar method and represented by plaque-forming unit. The lytic spectrum of each phage was tested using the spot assay (Huang et al., 2013; Kitti et al., 2014). We also tested the lytic spectrum of a phage cocktail, which contained all 14 phages.
Biological Characterization and Lytic Effect of Phage PD-6A3
Thermal and pH stability tests were carried out using the double-layer technique (Capra et al., 2004). In the thermal stability tests, aliquots of phage PD6A3 preparations (109 CFU/ml) were treated at pH 7 and at different temperatures (4, 37, 40, 50, 60, and 70°C) in SM Buffer for 1 h. As for the pH stability experiments, phage preparations were treated with various pH buffers at (from pH 2 to pH 11) at 37°C in SM Buffer for 1 h and the phage titers were assessed. An adsorption experiment was performed according to Adams (Adams, 1959). Briefly, exponentially grown AB32 (a clinical pan-drug resistant A. baumannii) were infected with the phage at the MOI of 0.1. Aliquots of 1 ml were taken every minute (from 1 to 10 min) and then the supernatant containing the non-adsorbed phages were titrated using the double-layer method. A one-step growth experiment was determined using the method described by Pajunen et al. (2000). The AB32 was infected by Phage PD6A3 at a MOI of 0.01 and adsorbed for 5 min. During the incubation, samples were taken periodically (10-min intervals, over a period of 50 min) and immediately assayed for plaque titer. In the assay of phage infection curve, AB32 were mixed with the phage PD6A3 at different MOIs (from 0.00001 to 1) at 37°C. The control experiments were performed using equal volume of SM buffer. The OD600 values were measured at 30-min intervals until 7 h post-infection.
Extraction, Sequencing, and Bioinformatics of Phage Genomic DNA
The phage PD6A3 was purified by CsCl gradient ultracentrifugation method as described before. Genome sequence of the phage was obtained using the Illumina HiSeq 3000 Sequencing platform (Illumina, San Diego, CA, United States) and the SOAPdenovo2 software for sequence assembly. Prediction and annotation between genes from phage PD6A3 and other phages were performed using GeneMarkS and Blast1 (Jin et al., 2012). Functional annotation of ORFs and homology assignments between genes from phage PD6A3 and other phages were performed by the BLASTp1. Expasy compute PI/MV tool2 was used to determine isoelectric pH and molecular weight of the predicted ORFs. The Easyfig software was used to conducted the alignment of functional proteins of phage PD6A3 (Hua et al., 2018). An alignment of amino acid sequences between endolysin Ply6A3 and other endolysin gene was created using the Clustal X2 program (Kong and Ryu, 2015). The whole genome of phage PD6A3 was deposited at GenBank under accession number KY388102.1.
Identification and Cloning of the Putative Endolysin From Phage PD-6A3
Gene prediction was performed as described above. The phage PD-6A3 endolysin encoding gene was amplified with P1 and P2 primers (Table 1) and PrimeSTAR HS DNA polymerase kits (TaKaRa, China). The PCR products were purified using a Rapid Mini Plasmid Kit (TIANGEN, China). Furthermore, PCR products were resolved in a 1% agarose gel before purified. The gene was then linked to an expression vector named pCold-SUMO, which comprised a 6× His tag and SUMO tag (Guo et al., 2016). The recombinant vector was transformed into E. coli DH5α cells. After confirmation by restriction enzyme digestion and sequence analysis, the correct plasmids were transformed into the BL21(DE3) cells for over-expression.
Over-Expression and Purification of Endolysin Ply6A3
The BL21(DE3) cells were grown in LB medium containing kanamycin (50 μg/mL, Sangon Biotech, China) at 37°C overnight, diluted 1:100 in 0.5 L LB medium, then incubated until the OD 600 reached 0.6∼0.8. Isopropyl β-D-thiogalactoside (IPTG) (TIANGEN, China) was added until the final concentration was 1 mM, then the mixture was incubated at 16°C for 18 h at 120 rpm. The following procedures were all performed in an ice-cold water bath (Diez-Martinez et al., 2015). The precipitate was resuspended in 25 mL PBS buffer (pH 7.4). The cell lysate was disrupted by sonication (Cole-Parmer, Ultrasonic Processors) (10 s pulse, 20 s rest over 30 min) before centrifuged at 12,000 × g 4°C (Lai et al., 2011). The His-SUMO-Ply6A3 was purified using a Ni-nitrilotriacetic acid column (Novagen, United States), and equilibrated with a binding buffer (20 mM Tris-HCl, 300 mM NaCl, 10 mM imidazole; pH 8.0) at room temperature. The supernatant was filtered using a 0.22-mm membrane, after it was passed through the column at least three times. The His-SUMO- ply6A3 in the supernatant was washed with binding buffer (20 mM Tris-HCl, 300 mM NaCl, 10 mM imidazole; pH 8.0) three times, and the same washing buffer was then eluted with elution buffer (20 mM Tris-HCl, 300 mM NaCl, 50 mM imidazole; pH 8.0). All samples were stored for SDS-PAGE and western blot analyses.
Cleavage of the SUMO Tag and Protein Concentration
The reaction assay was performed in a dialysis bag in a 4°C-refrigerator overnight. The SUMO protease and purified His-SUMO-ply6A3 were placed in dialysis at a ratio of 50:1 (vol/vol). The His-SUMO protein was cleaved with continuous stirring overnight, and the dialysis bag was placed in PBS buffer (pH 7.0) (Guo et al., 2016). The enzyme digested product was then purified using an affinity chromatography column (Novagen. United States). The protein solution was collected into a protein concentration tube with a molecular diameter of 10 kD (Amicon Ultra), and centrifuged at 4,000 × g at 4°C for 30 min. The concentration of the purified protein was then measured using a Micro BCATM protein assay kit (Tiangen, China).
SDS-PAGE and Western Blot Analysis
The samples collected during expression, purification, and cleavage were subjected to 12% SDS-PAGE, and then transferred to polyvinylidene fluoride (PVDF) membranes (Kong and Ryu, 2015). The PVDF membranes were blocked with 5% bovine serum albumin in phosphate buffered saline with Tween 20 (PBST) for 3 h and then incubated with anti-His antibody (1:3000) (Sangon Biotech, China) overnight, followed by peroxidase-affiniPure goat anti-mouse IgG (1:10000) (Sangon Biotech, China) for 1 h.
Detection of Peptidoglycan Degradation by PlyAB3 Using the Diffusion Method
According to the enrichment technique recommended by Sun et al. (2005). The AB32 was incubated in 0.5 L LB medium at 37°C for 24 h then harvested by centrifugation, before washed several times with ddH2O. Then placed in a boiling water bath for 45 min and broken by sonication. Next, centrifuged at 1000 r/min for 10 min, the supernatant was decanted. Centrifuged the supernatant again, the precipitate was incubated in 4% SDS before washed with ddH2O and dehydrated alcohol, digested with 0.02% trypsin in PBS (pH 7.5) for 24 h. The walls were resuspended in 10% trichloroacetic acid at 70°C for 30 min. Then harvested by centrifugation and washed by ddH2O, the peptidoglycan was then mixed in proportion (0.5%) with 15% SDS-polyacrylamide gel. After solidification, the gel was perforated with a sterilizing puncher. A volume of 20 μL endolysin Ply6A3 (2 mg/mL) was added to the hole formed. The negative control was PBS, and the positive control was lysozyme (2 mg/mL). The mixtures were sealed in a wet box and cultured at 37°C overnight, following which the samples were dyed with methylene blue stain for 2 h, and bleached to transparent strips with ddH2O (Blázquez et al., 2016).
Cytotoxicity of Phage PD-6A3 and Endolysin Ply6A3 to HEK293T and THP-1 Cells
The human embryonic kidney cells 293 (HEK293T) and a human monocytic cell THP-1 cells were both supplemented with fetal bovine serum (10%) and incubated in 5% CO2 at 37°C (Shen et al., 2018). Cells were seeded into 96-well plates (104 cells/well), then incubated for 24 h. They were then incubated with different concentrations of phage PD-6A3 or endolysin Ply6A3 for an additional 8 and 24 h (Yin et al., 2018). Cell viability was measured using the Cell Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan) and absorbance was measured at 450 nm.
Biological Characterization and Enzyme Activity of Endolysin Ply6A3
To test the thermal and pH stability of endolysin Ply6A3, host bacteria Ab32, which grew to log phase, was repeatedly suspended three times with PBS. A volume of 100 μL endolysin (1 mg/mL, dissolved in PBS) was combined with 100 μL Ab32 (1 × 109 cfu/mL), and added to a 96-well plate incubated at temperatures of 22, 27, 32, 37, and 42°C, at pH 7 for 0.5 h. For the pH assay, the mixture was incubated at pH 5.5, 6.0, 6.5, 7.0, 7.5, 8.0, and 8.5, at 37°C for 0.5 h (Kong and Ryu, 2015). A mixture of bacteria and PBS was used as the blank control group. Absorbance was measured before and after incubation. Based on the two readings, the percentage drop in bacterial turbidity was calculated.
The enzyme activity unit was defined as the reciprocal of the dilution at 37°C, after 30 min, which could have reduced the turbidity of the bacterium by 50%. The Ab32 was removed the bacterial outer membrane by 10% trichloromethane. The OD600 of Ab32 which suspended in PBS was 1. The enzyme activity unit was determined by turbidimetric regression. The bacterial suspension was added to the 96-well plate, 100 μL per well. A volume of 100 μL of a double dilution of endolysin Ply6A3, with an initial concentration of 3.9 mg/mL, was added to each well. Absorbance was measured at 600 nm before and after incubation. Based on the two readings, the percentage drop in bacterial turbidity was calculated (Oliveira et al., 2016).
Lytic Spectrum of Endolysin Ply6A3 Against Clinical ESKAPE Isolates
The acronym ESKAPE stands for bacterial pathogens and include E. faecium, S. aureus, K. pneumoniae, A. baumannii, P. aeruginosa, and Enterobacter. The lytic spectrum of endolysin Ply6A3 against the 440 clinical isolates were determined by the plate method (Zhang et al., 2016). The clinical strains were suspended in 300 μL PBS (pH 7.4), to yield a final concentration of 1 × 109 cfu/mL. The cells were plated with 4 mL agar (Lood et al., 2015). After solidification, the gel was perforate with a sterilizing puncher. A volume of 20 μL endolysin Ply6A3 (1 mg/mL) was added into a hole. The phage PD-6A3 (1 × 108 PFU/mL), cocktail (1 × 108 PFU/mL) which contain the 14 phages and the PBS were added into three other holes as comparison. The mixtures were cultured at 37°C overnight.
Phage Therapy in the Mouse Sepsis Model
We investigated the ability of the phage PD6A3, phage cocktail (contain all the 14 phages) and endolysin Ply6A3, after removed endotoxin by the EndotoxinOUT kit (G-bioscience, United States), to work systemically to rescue sepsis in mice. Briefly, 80 6∼8 weeks-old female BALB/c mice (Shanghai Laboratory Animal Company, China) were randomly assigned to eight groups (n = 10 per group) (Jun et al., 2014). The first three groups were inoculated intraperitoneally with 1 mL 1 × 109 CFU/mL of AB32 (minimum lethal dose, Supplementary Figure 2) and intraperitoneal administration with either 1 mL of endolysin Ply6A3 at 2 mg/mL (10 mice), 1 mL of phage PD6A3 (109 PFU/mL), or 1 mL of PBS (10 mice), 1 h later. The remaining five groups were only inoculated intraperitoneally with 1 mL PBS, 1 mL of endolysin Ply6A3 at 2 mg/mL, 1 mL of phage PD6A3 (109 PFU/mL), 1 mL of phage Cocktail (109 PFU/mL) or 1ml of the mixture [0.5 mL of endolysin Ply6A3 at 2 mg/mL + 0.5 mL of phage PD6A3 (109 PFU/mL)] respectively. The animals were monitored for 7 days and the survival rate of each group was calculated. The white blood cell (WBC) count, levels of IL-10 (interleukin-10) and PCT (procalcitonin) were evaluated to assess the immune status (Henein et al., 2016). At 2 days post-infection, three mice were selected from each group and subjected to 0.1 mL pentobarbital sodium (10 g/mL) anesthesia by intraperitoneal injection. Blood was collected from each anesthetized mouse then was sent to the clinical laboratory for examination of the WBC. The serum for measurement of IL-10 and PCT (Entenza et al., 2005) were used DuoSet enzyme-linked immunosorbent assay kits (R&D Systems). All the mice were sacrificed by cervical dislocation after experiment.
Statistical Analysis
Statistical calculations for mice survival rate were performed by Kaplan–Meier survival analysis with log-rank test (GraphPad Prism 7.0 software), and the comparison of cytokine levels between groups were analyzed statistically by Student’s t-test (SPSS Statistics 21.0 software). P-values < 0.05 were considered statistically significant.
Ethics Statement
This study was carried out in accordance with the recommendations of “Animal experiment guidelines, the Animal Care Committee of Fudan University.” The protocol was approved by the ‘the Animal Care Committee of Fudan University.’
Results
Isolation and Lytic Spectrum of A. baumannii Phages
About 40 different virulent phages were isolated, and 14 of those were additionally tested host spectra. The 14 phages were designated as PD-Ab1, PD-Ab8, PD-Ab9, PD-Ab11, PD-Ab 15, PD-Ab16, PD-Ab17, PD-Ab18, PD-6A1, PD-6A2, PD-6A3, PD-6A4, PD-7A1, and PD-7Ab3 (Supplementary Table 2). The results showed that all the phages have completely different lytic spectrum. Compared with the other 13 phages, phage PD-6A3 had a broader host range 32.4% (179/552) of the isolates. The cocktail which contains all the 14 phages was capable of lyzing 54.0% (298/552) of the isolates. Thus, we chose phage PD-6A3 for further study.
Phage PD-6A3 formed clear, round, significant halos, 2–3 mm in diameter (Figure 1A). TEM showed that phage PD-6A3 possessed an isometric head (50 ± 10 nm in diameter) and a very short tail (9 ± 2 nm in length), which was nearly invisible (Figure 1B). The phage was assigned to the order Caudovirales and family Podoviridae.
Figure 1.
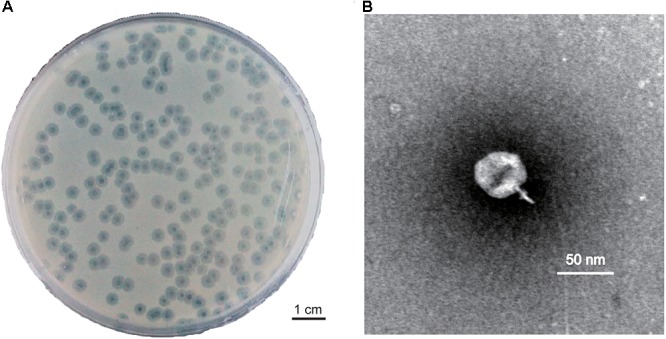
Plaque and TEM morphology of phages. (A) Plaque morphologies of phage PD-6A3. Scale bar, 1 cm. (B) TEM morphology of phage PD-6A3. Scale bar, 50 nm. PD-6A3, a novel Acinetobacter baumannii Phage. TEM, transmission electron micrograph.
Biological Characterization and Lytic Effect of Phage PD-6A3
The stability of phage PD-6A3 was investigated at different temperatures and pH conditions. No significant loss in phage activity was observed after heating to temperatures between 4 and 50°C. In addition, it remained relatively stable after incubation at pH levels ranging from 5 to 10 (Supplementary Figures 1A,B). The adsorption experiment showed that more than 90% of the phage particles were adsorbed within 5 min incubation (Supplementary Figure 1C). One-step growth experiments showed that the latent and eclipse periods were both 20 min and the burst size was 129 PFU per infected cell (Supplementary Figure 1D). The infection assay of phage PD-6A3 against AB32 was evaluated in vitro. The results showed that phage PD-6A3 could effectively reduce growth of the host bacteria, with OD values declining more quickly at MOI 1 than at MOI 0. 1, 0.01, or 0.001 (Figure 2).
Figure 2.
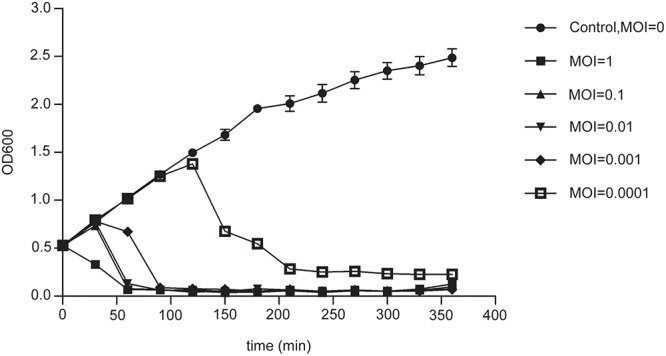
The infection assay of phage PD-6A3 against A. baumannii in vitro. AB32 was infected by phage PD6A3 at MOI of 0, 1, 0.1, 0.01, 0.001 or 0.0001 and cultured for 7 h. The control experiments were performed using equal volume of SM buffer. This experiment was repeated three times, and the data were shown in the mean ± SD. PD-6A3, a novel Acinetobacter baumannii Phage.
Sequencing and Bioinformatics Analysis of Phage PD-6A3 Genomic DNA
The phage PD-6A3 is a linear double-stranded DNA with a length of 41,563 bp and GC content of 39.48% (GenBank accession No. KT388102.1). A total of 48 putative genes were identified by GeneMarks, on the positive strand (Supplementary Table 3). The average length of a gene was 776 bp. Genome analysis also reviewed that this phage carried no virulence and antibiotic resistance genes which assure its safety to be applied for phage therapy in the future.
The proteins encoded by phage PD-6A3 could be divided into five categories morphogenesis, DNA packing protein, host lysis, DNA replication/regulation, and hypothetical proteins (Figure 3). According to the NCBI database, phage PD-6A3 has similarities with Acinetobacter phage SH-Ab15519 (GenBank accession number: KY082667.1; 99% identity, 93% coverage). The relatedness of these two phages was also determined using the Easy fig software show that phage PD-6A3 shares only 22 completely identical proteins with Acinetobacter phage SH-Ab15519, which is relatively limited (Figure 3). These all showed that Phage PD6A3 is a novel phiKMV-like phage.
Figure 3.
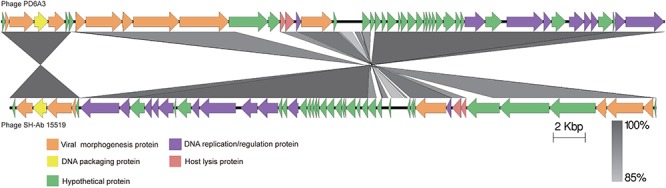
The comparison of the complete genome sequence of the Phage PD6A3 using Easy fig against the closet homolog phage SH-AB15519. The colored arrows indicate ORFs according to their predicted function. The homologous regions between phages are indicated by gray shading.
The endolysin gene of the phage PD6A3 is 558 bp and contains 185 amino acid residues (22.4 kDa protein), which belongs to glycoside hydrolase family 19. Conserved domain analysis has revealed the presence of alysozyme-like (N-acetyl-b-D-muramidase) catalytic domain between residues 75 and 128. A ClustalX2 alignment of endolysin Ply6A3 with two phage endolysins showed similarity in the domain region, however, 8 amino acid mutations were found (Figure 4).
Figure 4.

Amino acid sequence alignment of the endolysin of Phage PD6A3. The sequences used for alignment analysis were the endolysin gene of Phage 6A3, Phage SH-Ab15519 and Phage Abp1. Eight mutation residues are indicated by different color.
Cloning, Expression, Purification, and Assembly of Endolysin Ply6A3
The molecular weight of recombinant endolysin Ply6A3 is approximately 40 kDa (Figure 5A). A specific band (His-SUMO tag) of approximately 23 kDa was identified using specific His-tag antibodies (Figure 5B). The concentration of purified endolysin reached about 3.9 mg/mL.
Figure 5.
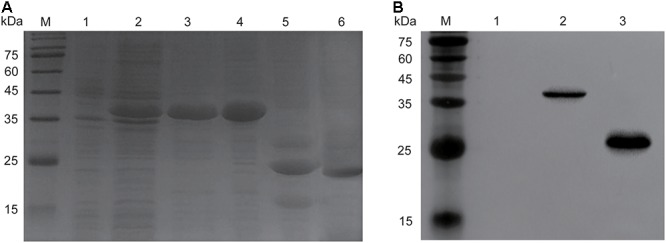
Over expression and purification of endolysin Ply6A3. (A) Lane M, molecular weight marker; Lane 1, un-induced bacterial lysate; Lane 2, IPTG-induced bacterial lysate; Lane 3, purified His-SUMO-Ply6A3 protein; Lane 4, dialyzed His-SUMO-Ply6A3 protein; Lane 5, Ply6A3 protein after cleavage of the His-SUMO tag; Lane 6, purified endolysin Ply6A3 protein after cleavage and dialyze of the His-SUMO tag. (B) Lane M, molecular weight marker; Lane 1 un-induced bacterial lysate; Lane 2-4, IPTG-induced bacterial lysate; Lane 5, the His-SUMO tag cleaved from endolysin Ply6A3. Ply6A3, the endolysin encoded by a novel Acinetobacter baumannii Phage PD6A3 gene. IPTG, Isopropyl β-D-Thiogalactoside.
Detection of Peptidoglycan Degradation by Endolysin PlyAB3 Using the Diffusion Method
This assay was performed to determine whether the endolysin Ply6A3 indeed with lytic activities toward cell walls (Lai et al., 2013). According to the principle of methylene blue staining, peptidoglycan stains blue (Figure 6A). Endolysin Ply6A3 (2 mg/mL) and lysozyme (2 mg/mL) can degrade peptidoglycan to form a transparent ring; whereas the negative control, PBS, showed no degradation and formed no transparent rings.
Figure 6.

(A) Detection of peptidoglycan degradation by endolysin Ply6A3 by diffusion method. Hole 1: 20 μl PBS, Hole 2: 20 μl Lysozyme (2 mg/ml), Hole 3: 20 μl endolysin (2 mg/ml). (B) Lytic activity of endolysin against clinical AB32 isolates. (C) Lytic activity of endolysin Ply6A3 against clinical MRSA6 isolates. Hole 1: 20 μl endolysin (2 mg/ml), Hole 2: 20 μl Phage PD6A3 (108cuf/ml), Hole 3: 20 μl Phage Cocktail (108cuf/ml) Hole 4: 20 μl PBS. Scale bar, 1 cm. CFU, colony-forming units. PD6A3, a novel Acinetobacter baumannii Phage. Ply6A3, the endolysin encoded by a novel Acinetobacter baumannii Phage PD6A3 gene. AB32, the No. 32 strains of Multidrug Resistance Acinetobacter baumannii. MRSA6, the No. 6 strains of methicillin-resistant Staphylococcus aureus.
Cytotoxicity of Phage PD6A3 and Endolysin Ply6A3 to HEK293T and THP-1 Cells
The potential cytotoxicity of phage PD6A3 were evaluated using the HEK293T and THP-1cells by the CCK-8 assay. The results showed that the percentage of viable HEK293T cells were around 100%, treated with Phage PD6A3 in 8 h and 24 h. We changed the cells to THP-1, and the result is the same (Figures 7A,B). And no significant cytotoxicity was observed in either HEK293T or THP-1 cells, treated with endolysin Ply6A3 (Figures 7C,D) (Shen et al., 2018; Yin et al., 2018).
Figure 7.
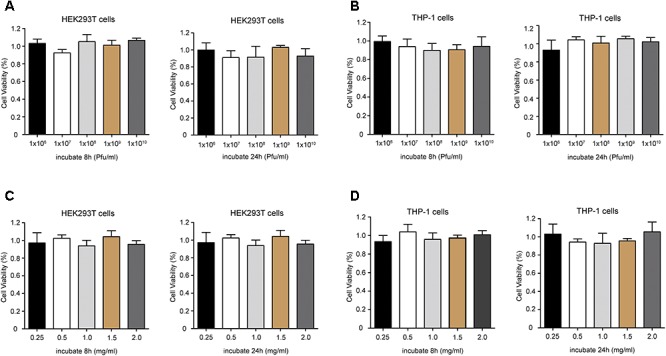
Cell viability of HEK293T and THP-1 cells after incubation with different concentrations of Phage PD6A3 (A,B) and endolysin Ply6A3 (C,D). Cells were incubated with different concentrations of phage PD-6A3 (106–1010 Pfu/mL) and endolysin Ply6A3 (0.25–2 mg/mL) for 8 h then followed by fresh complete media for 16 h. Or only incubated with different concentrations of phage PD-6A3 (106–1010 Pfu/mL) and endolysin Ply6A3 (0.25–2 mg/mL) for 24 h. Cell viability was measured using the CCK-8. Data are mean ± SD (n = 6). All the result shows that cell lines remained highly viable after incubation with endolysin Ply6A3. Ply6A3, the endolysin encoded by a novel Acinetobacter baumannii Phage PD6A3 gene. HEK293T cells, human embryonic kidney cells 293. THP-1 cells, a human monocytic cell line derived from an acute monocytic leukemia patient. CCK-8, Cell Counting Kit-8.
Biological Characterization and Lytic Effect of Endolysin Ply6A3
The temperature and pH stability test revealed that endolysin Ply6A3 remained high activity at temperature between 22 and 42°C and at pH range from 5.5 to 8.5; however, its activity was significantly reduced at pH levels above 7.5 (Figures 8A,B). The broad range of thermal and pH stability facilitates the application of endolysin in phage therapy.
Figure 8.
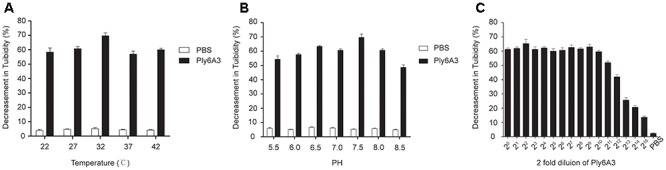
(A) The thermal stability of endolysin Ply6A3. Endolysin was incubated at various temperatures for 30 min. (B) The pH stability of endolysin Ply6A3. Endolysin was incubated at the indicated pH conditions for 30 min. (C) Activity unit of endolysin Ply6A3. The initial concentration was 3.9 mg/mL, then double dilution. Data were obtained from three independent experiments and are shown as mean ± SD. Ply6A3, the endolysin encoded by a novel Acinetobacter baumannii Phage PD6A3 gene. pH, hydrogen ion concentration. AB32, the No. 32 strains of Multidrug Resistance Acinetobacter baumannii.
The lytic effect shown that, when the original concentration of endolysin Ply6A3 was 3.9 mg/mL, its dilution ratio was 1:211, and the turbidity of AB32 could have been reduced by nearly 50% (Figure 8C). The results showed that the activity unit of endolysin Ply6A3 was 2048 unit/mL or 525 unit/mg. The amount of endolysin protein contained in each active unit was 1.9 μg. After fitting to a function curve, the IC50 of endolysin was calculated to be 1.6 μg.
Lytic Spectrum of Endolysin Ply6A3
Lytic spectrum was determined by spot test, the results show that 141 of the 200 MDRAB strains could have been lysed by endolysin Ply6A3 (Table 2). Moreover, 21 of the 40 PDR-AB strains could have been lysed. In addition, the protein also showed lytic activity for other septic bacterial strains as follows: E. faecium, S. aureus, K. pneumonia, and E. coli strains. The lytic spectrum of endolysin Ply6A3 is broader than that of phage PD6A3 and phage Cocktail which contain 14 phages (Table 3). None of the 40 P. aeruginosa strains could have been lysed. Only one hole (added endolysin Ply6A3) formed a bright, clear inhibition zone (Figures 6B,C).
Table 2.
Lysis spectrum of endolysin Ply-6A3.
| Bacteria | Positive | Total | Positive rate (%) |
|---|---|---|---|
| Enterococcus faecium | 5 | 40 | 12.5 |
| Staphylococcus aureus | 9 | 40 | 22.5 |
| Klebsiella pneumonia | 3 | 40 | 7.5 |
| Pseudomonas aeruginosa | 0 | 40 | 0 |
| Escherichia coli | 8 | 40 | 20.0 |
| MDR-A. baumannii | 141 | 200 | 70.5 |
| PDR-A. baumannii | 21 | 40 | 52.5 |
MDR-A. baumannii, multi-drug resistant Acinetobacter baumannii. PDR-A. baumannii, pan-drug resistant Acinetobacter baumannii.
Table 3.
Lysis spectrum of endolysin Ply-6A3 and Phage PD6A3 toward 200 MDR-AB strains.
| Positive | Total | Positive rate (%) | |
|---|---|---|---|
| Ply6A3 | 141 | 200 | 70.5 |
| Phage PD6A3 | 58 | 200 | 29.0 |
| Cocktail | 90 | 200 | 45.0 |
PD6A3, a novel Acinetobacter baumannii Phage. Ply6A3, the endolysin encoded by a novel Acinetobacter baumannii Phage PD6A3 gene. Cocktail, the cocktail which contain all the 14 phages mentioned in this article.
Phage Therapy in the Mouse Sepsis Model
To ascertain the effect of phage and endolysin against the sepsis mouse model, mouse was developed by intraperitoneal administration of A. baumannii. The survival rate of endolysin therapy group, endolysin + phage therapy group, phage therapy group and phage cocktail therapy group were 70, 70, 60, and 50%, respectively, higher than the bacterial group (Figure 9). Mice in the endolysin therapy group had milder clinical signs (slow reactions, lethargy, and activity reduction) than the mice in other three therapy groups. Furthermore, all mice in the negative control group (injected with PBS, phage or endolysin respectively) remained alive after 7 days.
Figure 9.
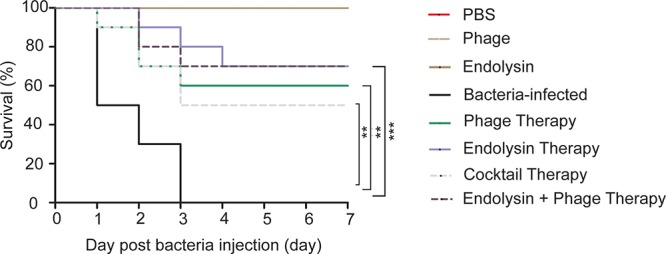
Therapeutic efficacy of Phage PD6A3 and endolysin Ply6A3 against mice A. baumannii sepsis infections. Survival of mice that were sepsis infected with AB32 and then treated with endolysin Ply6A3, phage PD6A3; or only intraperitoneal injected with PBS, Phage, endolysin. Using log-rank (Mantel-Cox) test. Each group harbors 10 mice. ∗∗P < 0.01, ∗∗∗P < 0.001 compared with the Bacteria group. PD6A3, a novel Acinetobacter baumannii Phage. Ply6A3, the endolysin encoded by a novel Acinetobacter baumannii Phage PD6A3 gene. AB32, the No. 32 strains of Multidrug Resistance Acinetobacter baumannii.
On the second day of the experiment, WBC counts in all the 4 therapy groups were significantly lower than those in the sepsis control group (P < 0.05). Furthermore, WBC counts in the endolysin therapy group and endolysin + phage therapy group were significantly lower than those in the phage therapy group and phage cocktail group (P < 0.05). The levels of IL-10 and PCT were significantly elevated in sepsis group, while all the 4 therapy groups effectively reduced the level. The expression of WBC, IL-10 and PCT remained at basal levels in the mice from control, phage group and endolysin group (Figure 10).
Figure 10.
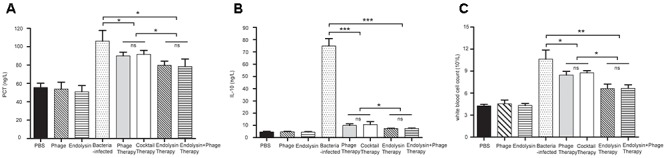
Levels of WBC and cytokines in the blood of the different groups of mice. The blood of the mice in the groups of PBS, phage, endolysin, bacteria-infected, Phage therapy, Phage cocktail therapy, Endolysin therapy and Endolysin + phage therapy were collected at 2 days. (A) PCT and (B) IL-10 concentrations of serum were measured by ELISA. (C) Blood was sent to the clinical laboratory to examine WBC. The data were shown in the mean ± SD (n = 3). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. WBC, white blood cell count. IL-10, interleukin-10. PCT, procalcitonin.
Discussion
Acinetobacter baumannii is an important nosocomial pathogen worldwide. This bacterium can be easily spread throughout healthcare facilities (Kusradze et al., 2016). The treatment of A. baumannii infection becomes increasingly difficult because of its resistance to many antimicrobial agents. Moreover, phages and their endolysins have drawn the attention of researchers to explore their potential as antimicrobial agents, because of the low probability of developing resistance, effectiveness against bacteria both in vitro and vivo, and ease of access (Oliveira et al., 2016).
The use of phages in animals has been reported in various applications, such as in the treatment of lung infections and skin wounds, and the results of such therapy have been significant (Lood et al., 2015; Shivaswamy et al., 2015; Henein et al., 2016). Hua and colleagues used an A. baumannii phage to rescue carbapenem-resistant A. baumannii (CRAB) lung infections (Hua et al., 2018). The use of endolysin treatments in animals is less common than that of phages. Zhang et al. (2016) proved the safety and efficacy of Streptococcus suis phage endolysin against Streptococcus suis infection in mice (Zhang et al., 2016). These experiments demonstrate that the phage and its endolysin could provide a promising future in the quest for treatments against bacterial infections. However, these studies have some limitations as follows: only a few resistant strains were used to test the lytic spectrum; all strains were collected from one hospital; the lack of a specific, effective concentration of the endolysin; and the lack of quantitative analysis with reliance on qualitative analysis. These limitations all render the reported lytic rates less convincing (Huang et al., 2014; Defraine et al., 2016; Larpin et al., 2018).
We isolated 40 A. baumannii phages, among which phage PD-6A3 had a relatively broad host range of 32.4% (179/552) among clinical MDR A. baumannii isolates, and was selected for further study. Phage PD-6A3 showed outstanding characteristics compared to the other phages such as remained stable over a wide range of temperatures and pH conditions. Genome analysis also reviewed that this phage carried no virulence and antibiotic resistance genes. Considering these advantages, phage PD-6A3 proves to be promising for potential applications in A. baumannii infections.
We over-expressed and purified the endolysin. The final concentration of endolysin Ply6A3 was 3.9 mg/mL. Previous studies have rarely included quantitative descriptions. The endolysin remained stable over a wide range of temperatures and pH conditions. Our results demonstrate that as a murein hydrolase, endolysin Ply6A3 was able to degrade 141/200 clinical MDRAB isolates and 21/40 PDR-AB isolates. In addition, it was also able to degrade Enterococci, MRSA, K. pneumonia, MDR- P. aeruginosa, and ESBL-E. coli, but Phage PD6A3 could not lysis any of them. Interestingly, we also over-expressed the endolysin of Phage Ab9 (GenBank accession number KY388103.), but it only could degrade 18/200 clinical MDRAB isolates which was very narrow compared to endolysin Ply6A3. In our future study, we will analysis this phenomenon in depth. Compared with previous articles, our research group tested the greatest number of bacterial strains, including A. baumannii and other common pathogens (Lai et al., 2013). Interestingly, when endolysin Ply6A3, phage PD6A3 and phage cocktail acted on 200 MDRAB strains, the lytic spectrum of the endolysin was much broader than the other groups. This might be attributed to different features of the outer membrane structure, like a different composition in the lipopolysaccharides, which may prevent or facilitate the access to exogenous lysins (Huang et al., 2014). We are further studying this phenomenon.
We extracted the cell wall of A. baumannii (Endersen et al., 2015). The results demonstrated that the peptidoglycan can be degraded by endolysin Ply6A3 and lysozyme. However, only endolysin could lyse A. baumannii strains, whereas lysozyme could not. These findings indicate that endolysin Ply6A3 is exceptional in its ability to directly destroy the cell wall of Gram-positive bacteria. These findings are also consistent with previous reports of the ability of endolysins from Gram-negative phages to lyse bacterial cells through enzymatic activity.
Our research also indicated the safety of phage PD6A3 and endolysin Ply6A3 applied intraperitoneal injection for mice. No obvious side effect was observed after injection during the observation period. And no statistical significance was observed when compared cytokine levels in serum, WBC in blood of the phage group and endolysin group to the PBS group, which provided a further safety evidence for the phage therapy and endolysin therapy by intraperitoneal injection. If in combination, phage PD-6A3 and endolysin Ply6A3 had more activity than the cocktail of 14 phages.
Compared to the sepsis group, all the therapy groups showed significantly higher survival rates and with evident reductions in the WBC count, serum PCT, and serum IL-10. However, the difference in survival between the 4 therapy groups were not statistically significant (P > 0.05). This might because the relatively small sample size could have affected the accuracy of statistical analysis. On the second day of the experiment showed that the WBC count, serum IL-10 and PCT of the endolysin therapy group and endolysin + phage therapy group were lower than that of the phage therapy group and cocktail therapy group (P < 0.05), which implies that endolysins work faster than phages. However, these are just experimental proofs that not very close to clinical situation (such short an interval time in clinical practice is unlikely). And the outcome of endolysin Ply6A3, as a class of enzymes, after entering the body is not clear. These conclusions need to be verified through further studies especially supported by clinical trials (Takemura-Uchiyama et al., 2014; Regeimbal et al., 2016).
Taken together, phage PD-6A3 is a novel phiKMV-like phage with strong lytic activity. Its endolysin Ply6A3 is an effective antibiotic against Gram-negative and Gram-positive bacteria. In addition, we evaluated the safety, effectiveness, and routes of administration for phage PD6A3 and its endolysin Ply6A3, in the treatment of A. baumannii infections, both in vivo and in vitro. Both agents have potentially therapeutic against infections caused by MDRAB strains in the future.
Author Contributions
YS and DC designed and supervised the study. MW, YL, DM, HG, and ZZ performed the experiments, analyzed the data, and drafted the manuscript. KH and YZ performed the experiments and analyzed the data. YX provided advice and suggestions.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Yinxin Shen, Lijun Ming, and Shuai Tao for their efforts in support of this work and Yanfeng Liu for helpful discussions.
Funding. This study was funded by construction of important weak subjects of Shanghai Pudong New Area (Grant No. PWZbr2017-02) and supported by Shanghai Municipal Commission of Health and Family Planning research project (Grant No. 201840261).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.03302/full#supplementary-material
References
- Adams M. H. (1959). Bacteriophages. New York, NY: Interscience Publishers. [Google Scholar]
- Billiau A. (2016). At the centennial of the Bacteriophage: reviving the overlooked contribution of a forgotten pioneer. Richard Bruynoghe (1881–1957). J. Hist. Biol. 49 559–580. 10.1007/s10739-015-9429-0 [DOI] [PubMed] [Google Scholar]
- Blázquez B., Fresco-Taboada A., Iglesias-Bexiga M., Menéndez M., García P. (2016). PL3 amidase, a tailor-made lysin constructed by domain shuffling with potent killing activity against Pneumococci and related species. Front. Microbiol. 7:1156. 10.3389/fmicb.2016.01156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra M. L., Quiberoni A., Reinheimer J. A. (2004). Thermal and chemical resistance of Lactobacillus casei and Lactobacillus paracasei bacteriophages. Lett. Appl. Microbiol. 38 499–504. 10.1111/j.1472-765X.2004.01525.x [DOI] [PubMed] [Google Scholar]
- CLSI (2017). “Performance standards for antimicrobial susceptibility testing,” in Proceedings of the 27th Informational Supplement M100-S20 (Wayne, PA: Clinical and Laboratory Standards Institute; ). [Google Scholar]
- Defraine V., Schuermans J., Grymonprez B., Govers S. K., Aertsen A., Fauvart M., et al. (2016). Efficacy of artilysin Art-175 against resistant and persistent Acinetobacter baumannii. Antimicrob. Agents Chemother. 60 3480–3488. 10.1128/AAC.00285-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Martinez R., De Paz H. D., Garcia-Fernandez E., Bustamante N., Euler C. W., Fischetti V. A., et al. (2015). A novel chimeric phage lysin with high in vitro and in vivo bactericidal activity against Streptococcus pneumoniae. J. Antimicrob. Chemother. 70 1763–1773. 10.1093/jac/dkv038 [DOI] [PubMed] [Google Scholar]
- Doi Y., Murray G., Peleg A. (2015). Acinetobacter baumannii: evolution of antimicrobial resistance—treatment options. Semin. Res. Crit. Care 36 085–098. 10.1055/s-0034-1398388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Shibiny A., El-Sahhar S. (2017). Bacteriophages: the possible solution to treat infections caused by pathogenic bacteria. Can. J. Microbiol. 63 865–879. 10.1139/cjm-2017-0030 [DOI] [PubMed] [Google Scholar]
- Endersen L., Coffey A., Ross R. P., McAuliffe O., Hill C., O’Mahony J. (2015). Characterisation of the antibacterial properties of a bacterial derived peptidoglycan hydrolase (LysCs4), active against C. sakazakii and other Gram-negative food-related pathogens. Int. J. Food Microbiol. 215 79–85. 10.1016/j.ijfoodmicro.2015.08.007 [DOI] [PubMed] [Google Scholar]
- Entenza J. M., Loeffler J. M., Grandgirard D., Fischetti V. A., Moreillon P. (2005). Therapeutic effects of bacteriophage Cpl-1 lysin against Streptococcus pneumoniae endocarditis in rats. Antimicrob. Agents Chemother. 49 4789–4792. 10.1128/AAC.49.11.4789-4792.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Zhu J., Tan Y., Li C., Chen Z., Sun S., et al. (2016). Self-assembly of virus-like particles of rabbit hemorrhagic disease virus capsid protein expressed in Escherichia coli and their immunogenicity in rabbits. Antiviral Res. 131 85–91. 10.1016/j.antiviral.2016.04.011 [DOI] [PubMed] [Google Scholar]
- Henein A. E., Hanlon G. W., Cooper C. J., Denyer S. P., Maillard J. (2016). A partially purified Acinetobacter baumannii phage preparation exhibits no cytotoxicity in 3T3 mouse fibroblast cells. Front. Microbiol. 7:1198. 10.3389/fmicb.2016.01198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F. P., Guo Y., Zhu D. M., Wang F., Jiang X. F., Xu Y. C., et al. (2016). Resistance trends among clinical isolates in China reported from CHINET surveillance of bacterial resistance, 2005–2014. Clin. Microbiol. Infect. 22 S9–S14. 10.1016/j.cmi.2016.01.001 [DOI] [PubMed] [Google Scholar]
- Hua Y., Luo T., Yang Y., Dong D., Wang R., Wang Y., et al. (2018). Phage therapy as a promising new treatment for lung infection caused by carbapenem-resistant Acinetobacter baumannii in mice. Front. Microbiol. 8:2659. 10.3389/fmicb.2017.02659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G., Le S., Peng Y., Zhao Y., Yin S., Zhang L., et al. (2013). Characterization and genome sequencing of phage Abp1, a new phiKMV-like virus infecting multidrug-resistant Acinetobacter baumannii. Curr. Microbiol. 66 535–543. 10.1007/s00284-013-0308-7 [DOI] [PubMed] [Google Scholar]
- Huang G., Shen X., Gong Y., Dong Z., Zhao X., Shen W., et al. (2014). Antibacterial properties of Acinetobacter baumannii phage Abp1 endolysin (PlyAB1). BMC Infect. Dis. 14:681. 10.1186/s12879-014-0681-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Li Z. J., Wang S. W., Wang S. M., Huang D. H., Li Y. H., et al. (2012). Isolation and characterization of ZZ1, a novel lytic phage that infects Acinetobacter baumannii clinical isolates. BMC Microbiol. 12:156. 10.1186/1471-2180-12-156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun S. Y., Jung G. M., Yoon S. J., Choi Y., Koh W. S., Moon K. S., et al. (2014). Preclinical safety evaluation of intravenously administered SAL200 containing the recombinant phage endolysin SAL-1 as a pharmaceutical ingredient. Antimicrob. Agents Chemother. 58 2084–2088. 10.1128/AAC.02232-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakhum N., Yordpratum U., Boonmee A., Tattawasart U., Rodrigues J. L. M., Sermswan R. W. (2016). Cloning, expression, and characterization of a peptidoglycan hydrolase from the Burkholderia pseudomallei phage ST79. AMB Express 6:77. 10.1186/s13568-016-0251-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitti T., Thummeepak R., Thanwisai A., Boonyodying K., Kunthalert D., Ritvirool P., et al. (2014). Characterization and detection of endolysin gene from three Acinetobacter baumannii bacteriophages isolated from sewage water. Indian J. Microbiol. 54 383–388. 10.1007/s12088-014-0472-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong M., Ryu S. (2015). Bacteriophage PBC1 and its endolysin as an antimicrobial agent against Bacillus cereus. Appl. Environ. Microbiol. 81 2274–2283. 10.1128/AEM.03485-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusradze I., Karumidze N., Rigvava S., Dvalidze T., Katsitadze M., Amiranashvili I., et al. (2016). Characterization and testing the efficiency of Acinetobacter baumannii phage vB-GEC_Ab-M-G7 as an antibacterial agent. Front. Microbiol. 7:1590. 10.3389/fmicb.2016.01590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M., Lin N., Hu A., Soo P., Chen L., Chen L., et al. (2011). Antibacterial activity of Acinetobacter baumannii phage ϕAB2 endolysin (LysAB2) against both Gram-positive and Gram-negative bacteria. Appl. Microbiol. Biotechnol. 90 529–539. 10.1007/s00253-011-3104-y [DOI] [PubMed] [Google Scholar]
- Lai M., Soo P., Lin N., Hu A., Chen Y., Chen L., et al. (2013). Identification and characterisation of the putative phage-related endolysins through full genome sequence analysis in Acinetobacter baumannii ATCC 17978. Int. J. Antimicrob. Agents 42 141–148. 10.1016/j.ijantimicag.2013.04.022 [DOI] [PubMed] [Google Scholar]
- Larpin Y., Oechslin F., Moreillon P., Resch G., Entenza J. M., Mancini S. (2018). In vitro characterization of PlyE146, a novel phage lysin that targets Gram-negative bacteria. PLoS One 13:e0192507. 10.1371/journal.pone.0192507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J., Shin H., Heu S., Ryu S. (2014). Exogenous lytic activity of SPN9CC endolysin against gram-negative bacteria. J. Microbiol. Biotechnol. 24 803–811. 10.4014/jmb.1403.03035 [DOI] [PubMed] [Google Scholar]
- Lin M. (2014). Antimicrobial resistance inAcinetobacter baumannii : from bench to bedside. World J. Clin. Cases 2:787. 10.12998/wjcc.v2.i12.787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lood R., Winer B. Y., Pelzek A. J., Diez-Martinez R., Thandar M., Euler C. W., et al. (2015). Novel phage lysin capable of killing the multidrug-resistant gram-negative bacterium Acinetobacter baumannii in a mouse Bacteremia model. Antimicrob. Agents Chemother. 59 1983–1991. 10.1128/AAC.04641-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo L. D. R., Brandao A., Akturk E., Santos S. B., Azeredo J. (2018). Characterization of a new Staphylococcus aureus kayvirus harboring a lysin active against biofilms. Viruses 10:182. 10.3390/v10040182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira H., Vilas Boas D., Mesnage S., Kluskens L. D., Lavigne R., Sillankorva S., et al. (2016). Structural and enzymatic characterization of ABgp46, a novel phage endolysin with broad anti-gram-negative bacterial activity. Front. Microbiol. 7:208. 10.3389/fmicb.2016.00208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajunen M., Kiljunen S., Skurnik M. (2000). Bacteriophage phiYeO3-12, specific for Yersinia enterocolitica serotype O:3, is related to coliphages T3 and T7. J. Bacteriol. 182 5114–5120. 10.1128/JB.182.18.5114-5120.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regeimbal J. M., Jacobs A. C., Corey B. W., Henry M. S., Thompson M. G., Pavlicek R. L., et al. (2016). Personalized therapeutic cocktail of wild environmental phages rescues mice from Acinetobacter baumannii wound infections. Antimicrob. Agents Chemother. 60 5806–5816. 10.1128/AAC.02877-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Zhang J., Hao W., Wang T., Liu J., Xie Y., et al. (2018). Copolymer micelles function as pH-responsive nanocarriers to enhance the cytotoxicity of a HER2 aptamer in HER2-positive breast cancer cells. Int. J. Nanomed. 13 537–553. 10.2147/IJN.S149942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaswamy V. C., Kalasuramath S. B., Sadanand C. K., Basavaraju A. K., Ginnavaram V., Bille S., et al. (2015). Ability of bacteriophage in resolving wound infection caused by multidrug-resistant Acinetobacter baumannii in uncontrolled diabetic rats. Microb. Drug Resist. 21 171–177. 10.1089/mdr.2014.0120 [DOI] [PubMed] [Google Scholar]
- Sun J., Shi Y., Le G., Ma X. (2005). Distinct immune response induced by peptidoglycan derived from Lactobacillus sp. World J. Gastroenterool. 11 6330–6337. 10.3748/wjg.v11.i40.6330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura-Uchiyama I., Uchiyama J., Osanai M., Morimoto N., Asagiri T., Ujihara T., et al. (2014). Experimental phage therapy against lethal lung-derived septicemia caused by Staphylococcus aureus in mice. Microbes Infect. 16 512–517. 10.1016/j.micinf.2014.02.011 [DOI] [PubMed] [Google Scholar]
- Yang H., Wang M., Yu J., Wei H. (2015). Antibacterial activity of a novel peptide-modified lysin against Acinetobacter baumannii and Pseudomonas aeruginosa. Front. Microbiol. 6:1471. 10.3389/fmicb.2015.01471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yele A. B., Thawal N. D., Sahu P. K., Chopade B. A. (2012). Novel lytic bacteriophage AB7-IBB1 of Acinetobacter baumannii: isolation, characterization and its effect on biofilm. Arch. Virol. 157 1441–1450. 10.1007/s00705-012-1320-0 [DOI] [PubMed] [Google Scholar]
- Yin S., Huang G., Zhang Y., Jiang B., Yang Z., Dong Z., et al. (2018). Phage abp1 rescues human cells and mice from infection by pan-drug resistant Acinetobacter Baumannii. Cell. Physiol. Biochem. 44 2337–2345. 10.1159/000486117 [DOI] [PubMed] [Google Scholar]
- Zhang H., Zhang C., Wang H., Yan Y. X., Sun J. (2016). A novel prophage lysin Ply5218 with extended lytic activity and stability against Streptococcus suis infection. FEMS Microbiol. Lett. 363:fnw186. 10.1093/femsle/fnw186 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


