Abstract
Heavy metal pollution has become an increasingly serious problem worldwide. Co-contamination with toxic mercury (Hg) and arsenic (As) presents a particularly difficult bioremediation trouble. By oxidizing the greenhouse gas methane, methanotrophs have been demonstrated to have high denitrification activity in eutrophic waters, indicating their possible potential for use in bioremediation of Hg(II) and As(V) in polluted water. Using metagenomics, a novel Methylocystis species (HL18), which was one of the most prevalent bacteria (9.9% of the relative abundance) in a CH4-based bio-reactor, is described here. The metagenomic-assembled genome (MAG) HL18 had gene products whose average amino acid identity against other known Methylocystis species varied from 69 to 85%, higher than the genus threshold but lower than the species boundary. Genomic analysis indicated that HL18 possessed all the genes necessary for the reduction of Hg(II) and As(V). Phylogenetic investigation of mercuric reductase (MerA) found that the HL18 protein was most closely affiliated with proteins from two Hg(II)-reducing bacteria, Bradyrhizobium sp. strain CCH5-F6 and Paracoccus halophilus. The genomic organization and phylogeny of the genes in the As(V)-reducing operon (arsRCCB) had significant identity with those from a As(V)-reducing bacterium belonging to the Rhodopseudomonas genus, indicating their reduction capability of As(V). Further analysis found that at least eight genera of methanotrophs possess both Hg(II) and As(V) reductases, illustrating the generally overlooked metabolic potential of methanotrophs. These results suggest that methanotrophs have greater bioremediation potential in heavy metal contaminated water than has been appreciated and could play an important role in the mitigation of heavy metal toxicity of contaminated wastewater.
Keywords: heavy metals, reductase, methanotroph, metagenomics, Methylocystis
Introduction
Mercury (Hg), a toxic and bio-accumulative trace metal, has increased remarkably because of anthropogenic activities such as mining and fossil fuel combustion, and as a result seriously threatens the environment and is a growing public health concern (Streets et al., 2011). Inorganic mercury ion (Hg(II)), with high water solubility, can irritate the gut, cause kidney damage, and will bind to thiol group of essential proteins and consequently inactivate them (Langford and Ferner, 1999; Mathema et al., 2011). Industrial activities can also produce arsenic (As), a cancerogenic and mutagenic element, resulting in a co-contaminated waste stream and further polluting natural waters (Rahman et al., 2014). For example, Hg in shallow-water lagoons have concentrations of 0.38–231 ng/L, and As concentration in natural waters, including freshwater and groundwater, ranges from 0.50 to 5000 μg/L (Smedley and Kinniburgh, 2002; Faganeli et al., 2012). This co-occurrence makes the treatment of contaminated waters much more challenging.
The most common forms of Hg in waters are divalent Hg(OH)2 and HgCl2, while the inorganic arsenate (As(V)) and arsenite (As(III)) are the primary types of As in aquatic environments (Lievremont et al., 2009). Considering that (1) the higher valency forms of Hg and As are more soluble and bio-available; (2) Hg(0) vapor can simply volatilize from waters (Munthe and Mcelroy, 1992), and that As(III) can easily precipitate with sulfide or sulfide-containing materials and be removed by centrifugation or filtration (Newman et al., 1997), it is the bio-reduction of Hg(II) and As(V) that is the major focus of Hg/As remediation efforts.
Many Gram-positive and Gram-negative bacteria are able to reduce Hg(II) with mercuric reductase. The operons whose gene products confer mercury-resistance contain several functional genes such as merA, merB, merP, merT, merD, merR, merC et al., depending on the particular strain (Dash and Das, 2012). Typically, MerR and MerD initially detect Hg(II) and regulate the transcription of other genes (Huang et al., 2002; Champier et al., 2004). Then, MerP, MerT, and MerC transport the mercury from the extracellular space into the cytoplasm (Liebert et al., 1997; Powlowski and Sahlman, 1999; Rossy et al., 2004). Finally, MerA and MerB detoxify inorganic and organic mercury compounds by reducing the ionic form thus generating volatile Hg0 which is expelled from cells (Osborn et al., 1997).
As(V) reduction involves two metabolic systems: the arr respiration system or the ars detoxification system (Mukhopadhyay and Rosen, 2002). Saltikov and Newman (2003) reported that the respiratory As(V)-reducing ability of Shewanella sp. strain ANA-3 was carried out by a two-subunit enzyme. The smaller subunit of the dimer, ArrB, contains four 4Fe–4S clusters and transfers electrons through the respiratory chain to the larger catalytic subunit ArrA, which utilizes a molybdenum cofactor and then reduces As(V) (Silver and Phung, 2005). The ars system is cytoplasmic and thus not coupled to respiration. This operon has three core genes (arsRBC), encoding a transcriptional repressor, a transmembrane efflux pump, and an As(V) reductase, respectively (Lievremont et al., 2009). A larger version composed of five genes (arsRDABC) has also been reported (Martin et al., 2001). ArsA is an ATPase which provides energy to the pump for discharge of As(III) while ArsD is an arsenic chaperone which transfers the As(III) to ArsA and activates the ArsAB As(III) excretion pump (Lin et al., 2007).
Microbes with these metal reduction pathways have been utilized to remediate Hg/As contaminated waters. For example, Yu et al. (2014) reported Staphylococcus epidermidis strain Lzu-02 could reduce Hg(II) to less toxic volatile Hg(0) using glucose as electron donor. Bachate et al. (2009) isolated twenty bacteria which contained the arsC gene and were capable of reducing As(V) to As(III) with gluconate as the electron donor. More recently, some studies have reported that methanotrophs possess a Hg(II)-reducing system. Boden and Murrell (2011) demonstrated that Methylococcus capsulatus (Bath) was able to oxidize CH4 in the presence of Hg(II) although neither the production of Hg(0) nor the involvement of MerA was confirmed. Methylosinus trichosporium OB3b and Methylocystis strain SB2 were reported to have the capability to synthesize methanobactin to detoxify Hg through complexation (Vorobev et al., 2013; Baral et al., 2014). However, whether methanotrophs have the ability to reduce heavy metals remains to be directly demonstrated.
In the present study, we identified a novel bacterium belonging to Methylocystis genus from a CH4-fed bioreactor by metagenomic analysis, and mainly focused on its metals reduction pathways instead of generally basic characterizations such as morphology. Analysis of MAG found it has both Hg- and As-reduction gene operons, suggesting it will be of practical use for heavy metal bioremediation in contaminated waters.
Materials and Methods
Sample Enrichment and DNA Isolation
This microbial community was enriched in a CH4-fed membrane biofilm bio-reactor using the original culture from Lai et al. (2016a). Biofilm samples were collected through cutting off several ∼10-cm sections from fibers (Zhong et al., 2017). We then placed the sections into a centrifuge tube with DNA-free water, and vortexed at the highest speed for 15 min to detach the biofilm from the fibers as described by Lai et al. (2016c). DNA from the biofilm was isolated using the PowerSoil DNA Isolation Kit (MO BIO) following the manufacturer’s instructions (Lai et al., 2016b).
Metagenome Sequencing and Genome Reconstruction
Shotgun metagenomic sequencing (2 × 150 bp) was performed using the Illumina HiSeq X Ten. The Illumina TruSeq DNA Nano Library Prep Kit was used for library preparation. About 205 million paired-end reads were generated. The quality of the raw data was confirmed using FastQC1. We trimmed the partial primers and adapters from the reads using the BBmap package2, and further removed the reads which: (i) contained the spike-in PhiX sequences; (ii) were low-quality (Phred Quality Score < 15) or short (<30 bp); (iii) had low complexity. All reads that passed quality control were assembled into contigs using metaSPAdes (Bankevich et al., 2012), yielding one assembly. Quality-controlled reads were mapped to assembled contigs (>500 bp) through BBmap and contigs were annotated using an in-house pipeline, meta-annotator. Metabat3 and CheckM4 were used to bin the contigs and check the completeness of bins, respectively.
Phylogenetic Analysis
Phylogenetic trees of relevant proteins were constructed using MEGA 6 with a multi-step approach (Tamura et al., 2013; Chen et al., 2016). Briefly, functional amino acid sequences were initially aligned, and then fragmentary and non-overlapping regions were removed. Phylogenetic trees were subsequently generated using the neighbor-joining algorithm based on distance analysis by MEGA 6 program packages. Reference proteins were found in the NCBI database through BLASTp algorithm. The accuracy of inferred topologies was tested by bootstrap re-sampling of 1000 replicates using the same distance model.
Gene Abundance Analysis
Genes for arsenate reductase (arsC) and the house-keeping RNA polymerase β subunit (rpoB) were searched in the six-frame translated metagenomic contigs in HL18 genome, using hmmsearch with e-5 of the cutoff e-value. The ArsC (PF03960, PF00462) and the RpoB (TIGR02013, TIGR03670) hidden Markov models were downloaded from the Pfam and TIGRFAMs database, respectively (Haft et al., 2013; Finn et al., 2016). The metagenomic reads after quality control were mapped against the arsC and rpoB nucleotide gene sequences using bowtie2 (Langmead and Salzberg, 2012). Picard5was used to remove PCR duplicates in the mapping files, and then BEDTools was applied to calculate coverage values for the extracted genes (Quinlan and Hall, 2010). To avoid that longer genes had higher values due to their length, the coverages were normalized to the length of corresponding genes, and further normalized to the total number of the rpoB gene, yielding an approximate number of each extracted arsC gene in HL18 assembled genome.
Data Availability
All raw Illumina sequences were submitted to the Sequence Read Archive (SRA) under Accession No. SRP136696 as described by Ontiveros-Valencia et al. (2014).
Results
Genome Reconstruction and Phylogeny
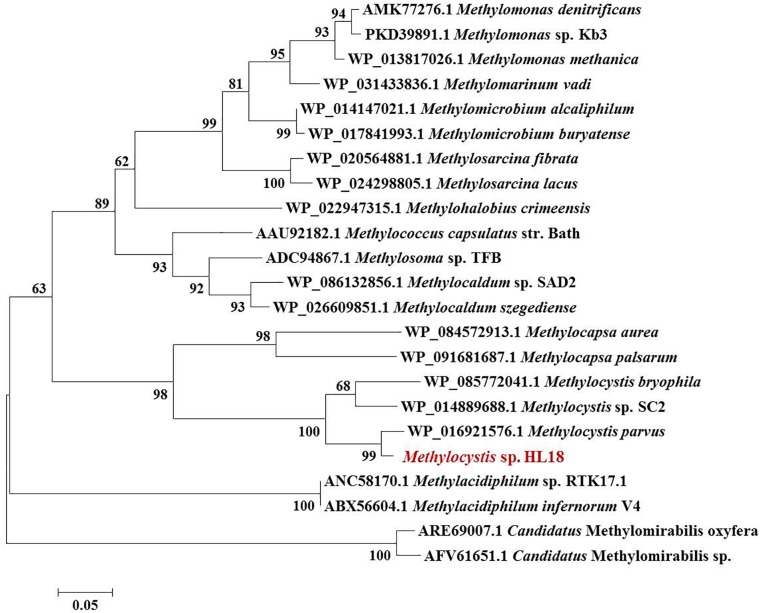
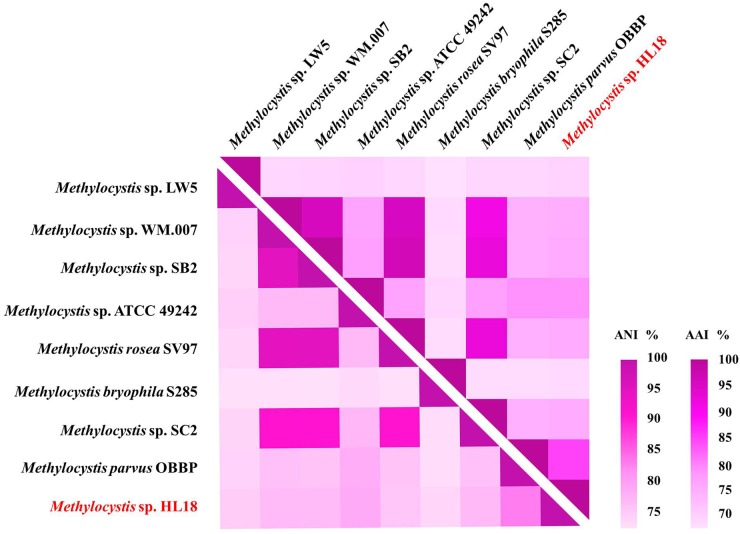
Metagenome sequencing, assembly, and binning reconstructed a number of high quality draft genomes from the CH4-fed bioreactor. The most abundant microorganism responsible for methane oxidation was a member of the Methylocystis, a Type II methanotroph, which accounted for 9.9% of the community (by calculating the sequence percentage). This MAG, referred to as HL18, contained 69 contigs, with an N50 of 97.898 Kbp. The total genome size was 3.816 Mbp (Table 1). HL18 was estimated to have >99% completeness and <1% contamination by CheckM, indicating a nearly complete genome. Unfortunately, the 16S rRNA gene was not present in the assembly. Skennerton et al. (2015) reported the consistent phylogenetic placement using pmoA gene to replace the 16S rDNA. Thus we adopted the pmoA gene encoding protein as the criterion to analyze the phylogeny of HL18. Figure 1 shows the phylogenetic tree of the PmoA based on amino acid sequences, with organisms from the candidate division NC10 as the outgroup members. HL18 was located within other members of the Methylocystis genus, and most closely affiliated with Methylocystis parvus. However, the average nucleotide identity (ANI) between HL18 and Methylocystis parvus was about 83%, which was only slightly above that (74–80%) of other members of this genus (Figure 2). ANI values for species circumscription are typically 95–96% (Richter and Rosselló-Móra, 2009), while between different species within the same genus these values range from 62 to 100% (Kim et al., 2014). Additionally, the average amino acid identity (AAI) of HL18 and other species varied from 69 to 85% (Figure 2), higher than the genus threshold but lower than the species boundary (Konstantinidis and Tiedje, 2007). These results indicated HL18 should be a novel species belonging to Methylocystis genus.
Table 1.
Overview of the HL18 assembled genome.
| Draft genome information | |
|---|---|
| Genome size | 3.816 Mbp |
| Number of contigs | 69 |
| N50 (contigs) | 97.898 Kbp |
| GC content | 64.89% |
| completeness | 99.37% |
| Contamination | 0.95% |
FIGURE 1.
Phylogenetic tree of PmoA based on amino acid sequences. The microorganism in red font was the one recovered in this study. PmoA of Methylomirabilis genus belonging to NC10 phylum were used as the outgroup members of this tree. Numbers indicated the bootstrap support (1000 replicates).
FIGURE 2.
Heatmap of genome relatedness between HL18 and other methanotrophs belonging to Methylocystis, generated by HemI (Deng et al., 2014). The average nucleotide identity (ANI; lower triangle) and average amino acid identity (AAI; upper triangle) between pairs of genomes were calculated using JSpeciesWS (Richter et al., 2016) and CompareM package (https://github.com/dparks1134/CompareM), respectively.
Potential of Inorganic Mercury Reduction
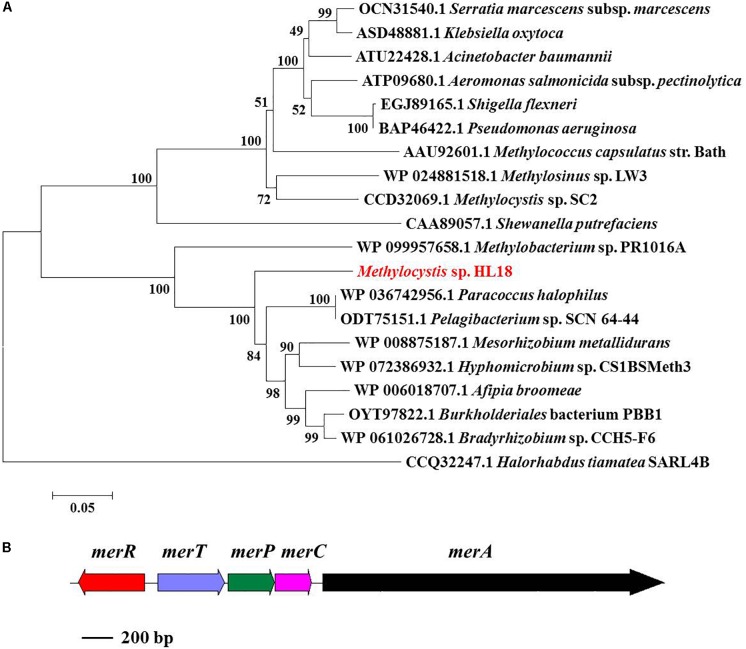
merA, the gene encoding the inorganic Hg(II) reductase, was found in the HL18 MAG. Phylogenetic analysis (Figure 3A) demonstrated that MerA has the same ancestor as similar proteins in Bradyrhizobium sp. strain CCH5-F6 and Paracoccus halophilus. Moreover, merR, merT, merP, and merC (Figure 3B) were found upstream of merA, whose products might contribute to the complete metabolic pathway for Hg(II) reduction. We compared GC content of the mercury-resistant operon, the contig where operon located, and the whole genome (Supplementary Figure S1), to test the possible sources of other microbes resulting from mis-assembly or mis-binning, even though the estimated contamination value was very low (0.95%). The differences between these contents were small (0.01–1.21%), suggesting this region had not been incorrectly assigned to HL18.
FIGURE 3.
(A) Phylogenetic tree of MerA based on amino acid sequences and (B) mer operon containing five genes (merR, merT, merP, merC, and merA). MerA of archaea Halorhabdus tiamatea was used as the outgroup member of this tree. Numbers indicated the bootstrap support (1000 replicates).
Potential of Arsenate Reduction
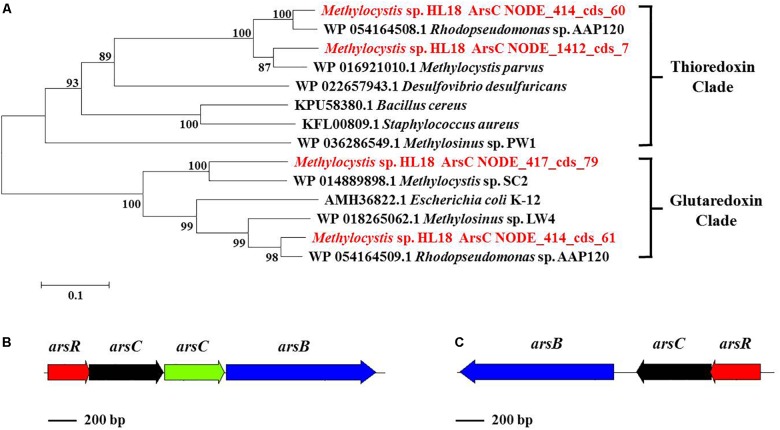
In addition to Hg(II) reductase, HL18 also possessed the As(V) reductase (ArsC) (Figure 4). There were four arsC genes distributed on three contigs (Figure 4A). Two reductases belonged to the glutaredoxin (Grx) clade, whose prototype is the enzyme of Escherichia coli plasmid R773 (Gladysheva et al., 1994; Martin et al., 2001); the other two were associated with the thioredoxin (Trx) clade having ArsC of Staphylococcus aureus plasmid pI258 as prototype (Ji et al., 1994). We compared the GC content of the arsC operons, corresponding contigs, and the whole genome, to preliminarily determine whether the affiliation was rational. Supplementary Figure S1 reveals that one arsC gene, labeled as NODE_417_cds_79, has quite different GC content with those of both the relevant contig and the whole genome. This deviation (∼9%) implied the possible contamination of this arsC gene. Besides, no other associated genes (e.g., arsB, the gene encoding the As(III) transporter) could be found nearby this gene, thus it was discarded in the subsequent analysis. For the remaining three genes, two arsC were adjacent to one another on the same contig, with each having closest affiliations to the proteins in Rhodopseudomonas sp. AAP120, while the other was on a separate contig and its closest affiliation was Methylocystis parvus. Genes encoding the regulator protein ArsR and the As(III) transporter ArsB could also be found in the vicinity of arsC genes (Figures 4B,C), forming two operons encoding proteins whose products are involved in As(V) reduction.
FIGURE 4.
(A) Phylogenetic tree of ArsC based on amino acid sequences, (B) ars operon containing four genes (arsR, arsC, arsC, and arsB) and (C) another ars operon containing three genes (arsR, arsC, and arsB). ArsC sequences recovered in this study from the metagenome were in red font and suffixed with their contig names. Numbers indicated the bootstrap support (1000 replicates).
Furthermore, the arsC gene number was calculated based on the single-copy gene rpoB, the house-keeping gene for the RNA polymerase β subunit. Average abundances of arsC ranged from 3.6 to 3.9 fold more than the number of rpoB, leading to total ∼11 fold higher in HL18 MAG. The effect of the ars operon on As(V) resistance has been reported to be cumulative (Diorio et al., 1995; Cai et al., 1998). Two operons responsible for As(V) reduction and high abundances of arsC are consistent with the increased As(V)-resistance of HL18, suggesting it may have better efficiency to bio-remediate As(V) contaminated wastewater.
Potential for Metal Resistance in Other Methanotrophs
The presence of Hg(II) and As(V) reductases in other methanotrophs has not been reported. Analysis of available genomes found that members of at least eight genera that had the capacity to oxidize methane possessed both Hg(II) and As(V) reductases. Identities of relevant reductases between HL18 and 10 type species were calculated using BLASTp. Primary sequence identity of the Hg(II) reductases ranged from 39–97% while As(V) reductases were 33–97% similar (Supplementary Figures S2–S4). Phylogenetic analysis demonstrated significant diversity of both reductases within methanotrophs as shown by the fact that many reductase clades occur (Supplementary Figures S5–S7). The MerA of HL18, along with that of Methylosarcina lacus, did not form clades with any of the other proteins, revealing the divergent sequences of these two reductases. Similarly, a variety of methanotrophic genera belonging to seven taxonomic families contained different clades of As(V) reductases. Two thioredoxin-dependent enzymes from HL18 were found clustered with proteins from Methylocystaceae and Beijerinkiaceae clusters (Supplementary Figure S6). This result suggests that the gene encoding ArsC (NODE_414_cds_60) was derived from horizontal gene transfer rather than the duplication of another ArsC (NODE_1412_cds_7) gene. Interestingly, the glutaredoxin-clade ArsC (NODE_414_cds_61), which is directly adjacent to NODE_414_cds_60 also clustered with enzymes in the Beijerinkiaceae cluster, lending further support to the possibility that these genes were acquired by lateral gene transfer (Supplementary Figure S7).
Discussion
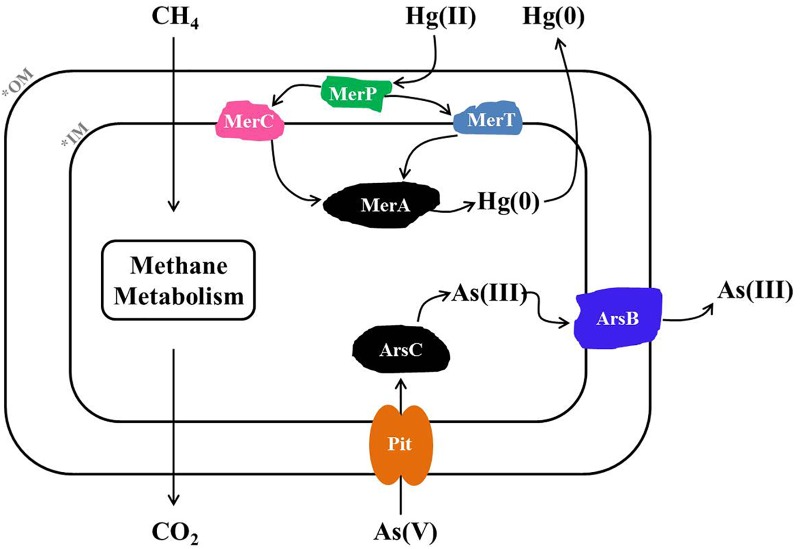
Based on metagenome sequencing, we generated a MAG from a methanotroph (HL18) found in a CH4-fed bioreactor that is a new member of the genus Methylocystis. HL18 was the highest occurring methanotroph in the community and contained both Hg(II) and As(V) reductases, revealing a multiplicity of heavy metal reduction capacities. The assembled genome of HL18 contained the genes necessary for detoxication of Hg(II) including the mercuric reductase MerA. The MerA in HL18 shares significant primary sequence identity (80%) with that from Bradyrhizobium sp. strain CCH5-F6 which has been reported capable of Hg(II) reduction, implying that HL18 also had the potential to reduce Hg(II) (Wang et al., 2013). Moreover, the protein with the high sequence identity (77%) to the MerA in HL18 is found in Paracoccus halophilus, a bacterium able to tolerate Hg(II), also providing evidence for its Hg(II)-reducing potential (Lapanje et al., 2010). Based on previous work and analysis of the HL18 MAG, when HL18 is exposed to mercury, MerR would detect the presence of Hg(II) and activate the transcription of downstream genes (Huang et al., 2002). This would enable the transport of Hg(II) into the cytoplasm via MerP where it would be subsequently transferred to the inner membrane bound protein MerT and MerC (Hamlett et al., 1992). Hg(II) would then be transferred to MerA and further reduced to elemental Hg(0) by an NAD(P)H-dependent reaction (Dash and Das, 2012). The proposed metabolic pathway based on the genes in HL18 is shown in Figure 5.
FIGURE 5.
Metabolic pathway of Hg(II) and As(V) reduction inferred from the HL18 genome sequence. OM, outer membrane; IM, inner membrane; MerA, mercuric reductase; MerT, mercuric transport protein MerT; MerC, mercuric transport protein MerC; MerP, mercuric transport protein periplasmic component; Pit, phosphate transporter; ArsC, arsenate reductase; ArsB, arsenic efflux pump protein.
Likewise, HL18 also contains As(V) reduction potential conferred by two distinct As(V)-reducing operons, arsRCCB and arsRCB. The difference of arsC number in two operons is not unreasonable since a variety of arrangements of As(V)-resistance genes have been discovered in other microorganisms (Martin et al., 2001; Li and Krumholz, 2007; Lievremont et al., 2009). The structure and phylogeny of arsRCCB operon in HL18 were very similar with those in Rhodopseudomonas sp. AAP120 (BioProject Accession No. PRJNA255569). While the arsenate reductase activity of R. sp. AAP120 has not been reported, its relative R. palustris C1 was found to reduce As(V), with a specific activity of 12.07 U mg-1 protein, which was comparable to that of the As(V)-reducing bacterium Pseudomonas sp. (12.15 U mg-1 protein) (Srivastava et al., 2010; Nookongbut et al., 2017). Based on the similarity of the proteins in HL18 to proteins with known As(V)-reducing activity, it is very likely that these proteins in HL18 are active as well. The gene organization of the other ars operon in HL18 was very similar other than the absence of one copy of arsC. The closest affiliation of the arsC in this operon was with a protein from Methylocystis parvus. To date, there have been no articles reporting As(V) reducing capacity in methanotrophs. However, all genes required for As(V) reduction could be found flanking the arsC in both operons (Figures 4B,C). Furthermore, we inoculated the consortium which contained HL18 (Supplementary Table S1) in serum bottles along with CH4 and As(V), and observed significant reduction of As(V) that did not happen without CH4 in presence or with killed inoculum (Supplementary Figure S8). Among the consortium, the highest abundant bacterium was related to genus Pseudoxanthomonas, of which the closest member was P. suwonensis with the ANI of 76.7%. Although an isolate from this genus has been reported able to reduce As(V), it was phylogenetically distant from P. suwonensis (Mohapatra et al., 2018), implying the different metabolisms between species in the same genus. Additionally, arsB whose gene product could expel As(III) and thus confer As-resistance (Fu et al., 2009), was not found in the assembled genome of putative Pseudoxanthomonas sp., indicating this species had no capacibility for As(V) reduction in our experiment. Likewise, arsC, arsR, arsB in the genomes of putative Meiothermus sp. distributed dispersely and did not constitute an operon, excluding its possibility for As(V) reduction. Inversely, other four bacteria, including HL18, possessed different forms of As(V)-reducing operons, implying their active role in CH4-based As(V) reduction. While it is possible that one or several alternate genera were the primary As(V)-reducing strains, we could not exclude the circumstance that Methylocystis sp. HL18 may have been responsible for some of the As(V) reduction. Based on the available genes, we propose that when As(V) is present, inactivation of arsR will result in the expression of the ars system (Rosenstein et al., 1992). As(V) would then be transported from outside to the cytoplasm through the phosphate transporter Pit and then reduced by ArsC (Willsky and Malamy, 1980; Martin et al., 2001). The product As(III) was then be transported out of the cytoplasm via ArsB (Rosen, 2002).
In addition to HL18, multiple methanotrophic bacteria also contain Hg(II) and As(V) reductases. However, the Hg(II) reductase of HL18 was notably different with proteins of other Methylocystis species, only sharing ∼47% identity (Supplementary Figure S2). These identities were much smaller than those (>80%) between other species within the same genus (Supplementary Figure S2), and that (80%) with MerA from Bradyrhizobium sp. CCH5-F6. Lateral gene transfer may explain why the MerA from HL18 is more related to the Bradyrhizobium protein instead of proteins from other Methylocystis. Unlike the MerA, As(V) reductases from the same genus, Methylocystis, have the highest sequence relatedness with the ArsC from HL18 (Supplementary Figures S3, S4). These arsenate reductase genes are widespread in methanotrophic bacteria, with at least seven families and more than 10 genera (Supplementary Figures S6, S7). This distribution implies the common occurrence of As(V) reduction in methanotrophs.
Very recently, Ettwig et al. (2016) and Cai et al. (2018) described that archaea related to Candidatus Methanoperedens nitroreducens were able to couple the reduction of iron/manganese ions to the oxidation of methane, demonstrating additional possibilities of high valent heavy metals reduction of methanotrophs. Although there is yet no direct biochemical evidence illustrating Hg(II) and As(V) reduction of methanotrophs, genome information strongly supports the conclusion that a variety members of this group have this physiological capacity. In addition, some members of Methylomonas genus contain both metals reductases as well as denitrification enzymes, and one species in this genus (Methylomonas denitrificans sp. strain FJG1) has been reported to reduce nitrate (Kits et al., 2015), though the capacity for Hg(II) or As(V) reduction across members of the genus are unknown. Meanwhile, pure culture of Methylococcus capsulatus (Bath) has been demonstrated possessing the mercuric reduction potential (Boden and Murrell, 2011). Our supplemented experimental result preliminarily confirmed arsenate reduction ability of the consortium which contains high abundance of HL18. These results together illustrate that many methanotrophs, including HL18, have an unappreciated metabolic potential and versatile capabilities for bio-remediation. Furthermore, Hg and As are globally dispersed (Wedepohl, 1995; Mukhopadhyay et al., 2002) and very likely co-occur with methane, indicating that the links between the carbon, arsenic, and mercury cycles may be more common than previously recognized. However, further ecophysiological experiments are required to confirm the function of these methanotrophs both in lab reactors and natural ecology.
Author Contributions
L-DS and H-PZ constructed the experiments, drafted, and revised the manuscript. JS helped to analyze the data and revise the manuscript. Y-SC, J-JD, and Y-QH helped to revise the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors greatly appreciate for the collaboration with Marc Strous and Xiaoli Dong (Department of Geoscience, University of Calgary, Calgary, AB, Canada) for metagenomic analysis.
Funding. The authors greatly thank the Natural Science Funds for Distinguished Young Scholar of Zhejiang Province (Grant No. LR17B070001), the National Natural Science Foundation of China (Grant No. 21577123), and the Fundamental Research Funds for the Central Universities (Grant Nos. 2016QNA6007 and 2017XZZX010-03) for their financial support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.03297/full#supplementary-material.
References
- Bachate S. P., Cavalca L., Andreoni V. (2009). Arsenic-resistant bacteria isolated from agricultural soils of Bangladesh and characterization of arsenate-reducing strains. J. Appl. Microbiol. 107 145–156. 10.1111/j.1365-2672.2009.04188.x [DOI] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baral B. S., Bandow N. L., Vorobev A., Freemeier B. C., Bergman B. H., Herdendorf T. J., et al. (2014). Mercury binding by methanobactin from Methylocystis strain SB2. J. Inorgan. Biochem. 141 161–169. 10.1016/j.jinorgbio.2014.09.004 [DOI] [PubMed] [Google Scholar]
- Boden R., Murrell J. C. (2011). Response to mercury (II) ions in Methylococcus capsulatus (bath). Fems Microbiol. Lett. 324 106–110. 10.1111/j.1574-6968.2011.02395.x [DOI] [PubMed] [Google Scholar]
- Cai C., Leu A. O., Xie G. J., Guo J., Feng Y., Zhao J. X., et al. (2018). A methanotrophic archaeon couples anaerobic oxidation of methane to Fe(III) reduction. ISME J. 12 1929–1939. 10.1038/s41396-018-0109-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J., Salmon K., Dubow M. S. (1998). A chromosomal ars operon homologue of Pseudomonas aeruginosa confers increased resistance to arsenic and antimony in Escherichia coli. Microbiology 144 2705–2713. 10.1099/00221287-144-10-2705 [DOI] [PubMed] [Google Scholar]
- Champier L., Duarte V., Michaud-Soret I., Coves J. (2004). Characterization of the MerD protein from Ralstonia metallidurans CH34: a possible role in bacterial mercury resistance by switching off the induction of the mer operon. Mol. Microbiol. 52 1475–1485. 10.1111/j.1365-2958.2004.04071.x [DOI] [PubMed] [Google Scholar]
- Chen R., Luo Y. H., Chen J. X., Zhang Y., Wen L. L., Shi L. D., et al. (2016). Evolution of the microbial community of the biofilm in a methane-based membrane biofilm reactor reducing multiple electron acceptors. Environ. Sci. Pollut. Res. 23 9540–9548. 10.1007/s11356-016-6146-y [DOI] [PubMed] [Google Scholar]
- Dash H. R., Das S. (2012). Bioremediation of mercury and the importance of bacterial mer genes. Int. Biodeterior. Biodegradation 75 207–213. 10.1016/j.ibiod.2012.07.023 [DOI] [Google Scholar]
- Deng W., Wang Y., Liu Z., Cheng H., Xue Y. (2014). HemI: a toolkit for illustrating heatmaps. PLoS One 9:e111988. 10.1371/journal.pone.0111988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diorio C., Cai J., Marmor J., Shinder R., Dubow M. S. (1995). An Escherichia coli chromosomal ars operon homolog is functional in arsenic detoxification and is conserved in gram-negative bacteria. J. Bacteriol. 177 2050–2056. 10.1128/jb.177.8.2050-2056.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettwig K. F., Zhu B. L., Speth D., Keltjens J. T., Jetten M. S. M., Kartal B. (2016). Archaea catalyze iron-dependent anaerobic oxidation of methane. Proc. Natl. Acad. Sci. U.S.A. 113 12792–12796. 10.1073/pnas.1609534113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faganeli J., Hines M. E., Covelli S., Emili A., Giani M. (2012). Mercury in lagoons: an overview of the importance of the link between geochemistry and biology. Estuar. Coast. Shelf Sci. 113 126–132. 10.1016/j.ecss.2012.08.021 [DOI] [Google Scholar]
- Finn R. D., Coggill P., Eberhardt R. Y., Eddy S. R., Mistry J., Mitchell A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44 D279–D285. 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H. L., Meng Y. L., Ordonez E., Villadangos A. F., Bhattacharjee H., Gil J. A., et al. (2009). Properties of arsenite efflux permeases (Acr3) from Alkaliphilus metalliredigens and Corynebacterium glutamicum. J. Biol. Chem. 284 19887–19895. 10.1074/jbc.M109.011882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladysheva T. B., Oden K. L., Rosen B. P. (1994). Properties of the arsenate reductase of plasmid R773. Biochemistry 33 7288–7293. 10.1021/bi00189a033 [DOI] [PubMed] [Google Scholar]
- Haft D. H., Selengut J. D., Richter R. A., Harkins D., Basu M. K., Beck E. (2013). TIGRFAMs and genome properties in 2013. Nucleic Acids Res. 41 D387–D395. 10.1093/nar/gks1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlett N. V., Landale E. C., Davis B. H., Summers A. O. (1992). Roles of the Tn21 Mert, Merp, and Merc gene-products in mercury resistance and mercury binding. J. Bacteriol. 174 6377–6385. 10.1128/jb.174.20.6377-6385.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C. C., Narita M., Yamagata T., Endo G., Silver S. (2002). Characterization of two regulatory genes of the mercury resistance determinants from TnMERI1 by luciferase-based examination. Gene 301 13–20. 10.1016/S0378-1119(02)01086-7 [DOI] [PubMed] [Google Scholar]
- Ji G. Y., Garber E. A. E., Armes L. G., Chen C. M., Fuchs J. A., Silver S. (1994). Arsenate reductase of staphylococcus-aureus plasmid Pi258. Biochemistry 33 7294–7299. 10.1021/bi00189a034 [DOI] [PubMed] [Google Scholar]
- Kim M., Oh H. S., Park S. C., Chun J. (2014). Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 64 346–351. 10.1099/ijs.0.059774-0 [DOI] [PubMed] [Google Scholar]
- Kits K. D., Klotz M. G., Stein L. Y. (2015). Methane oxidation coupled to nitrate reduction under hypoxia by the gammaproteobacterium Methylomonas denitrificans, sp nov type strain FJG1. Environ. Microbiol. 17 3219–3232. 10.1111/1462-2920.12772 [DOI] [PubMed] [Google Scholar]
- Konstantinidis K. T., Tiedje J. M. (2007). Prokaryotic taxonomy and phylogeny in the genomic era: advancements and challenges ahead. Curr. Opin. Microbiol. 10 504–509. 10.1016/j.mib.2007.08.006 [DOI] [PubMed] [Google Scholar]
- Lai C. Y., Wen L. L., Shi L. D., Zhao K. K., Wang Y. Q., Yang X. E., et al. (2016a). Selenate and nitrate bioreductions using methane as the electron donor in a membrane biofilm reactor. Environ. Sci. Technol. 50 10179–10186. 10.1021/acs.est.6b02807 [DOI] [PubMed] [Google Scholar]
- Lai C. Y., Wen L. L., Zhan Y., Luo S. S., Wang Q. Y., Luo Y. H., et al. (2016b). Autotrophic antimonate bio-reduction using hydrogen as the electron donor. Water Res. 88 467–474. 10.1016/j.watres.2015.10.042 [DOI] [PubMed] [Google Scholar]
- Lai C. Y., Zhong L., Zhang Y., Chen J. X., Wen L. L., Shi L. D., et al. (2016c). Bioreduction of chromate in a methane-based membrane biofilm reactor. Environ. Sci. Technol. 50 5832–5839. 10.1021/acs.est.5b06177 [DOI] [PubMed] [Google Scholar]
- Langford N. J., Ferner R. E. (1999). Toxicity of mercury. J. Hum. Hypertens. 13:651 10.1038/sj.jhh.1000896 [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9 U357–U354. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapanje A., Zrimec A., Drobne D., Rupnik M. (2010). Long-term Hg pollution-induced structural shifts of bacterial community in the terrestrial isopod (Porcellio scaber) gut. Environ. Pollut. 158 3186–3193. 10.1016/j.envpol.2010.07.001 [DOI] [PubMed] [Google Scholar]
- Li X. K., Krumholz L. R. (2007). Regulation of arsenate resistance in Desulfovibrio desulfuricans G20 by an arsRBCC operon and an arsC gene. J. Bacteriol. 189 3705–3711. 10.1128/JB.01913-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebert C. A., Wireman J., Smith T., Summers A. O. (1997). Phylogeny of mercury resistance (mer) operons of gram-negative bacteria isolated from the fecal flora of primates. Appl. Environ. Microbiol. 63 1066–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lievremont D., Bertin P. N., Lett M. C. (2009). Arsenic in contaminated waters: biogeochemical cycle, microbial metabolism and biotreatment processes. Biochimie 91 1229–1237. 10.1016/j.biochi.2009.06.016 [DOI] [PubMed] [Google Scholar]
- Lin Y. F., Yang J. B., Rosen B. P. (2007). ArsD: an As(III) metallochaperone for the ArsAB As(III)-translocating ATPase. J. Bioenerg. Biomembr. 39 453–458. 10.1007/s10863-007-9113-y [DOI] [PubMed] [Google Scholar]
- Martin P., Demel S., Shi J., Gladysheva T., Gatti D. L., Rosen B. P., et al. (2001). Insights into the structure, solvation, and mechanism of ArsC arsenate reductase, a novel arsenic detoxification enzyme. Structure 9 1071–1081. 10.1016/S0969-2126(01)00672-4 [DOI] [PubMed] [Google Scholar]
- Mathema V. B., Thakuri B. C., Sillanpaa M. (2011). Bacterial mer operon-mediated detoxification of mercurial compounds: a short review. Arch. Microbiol. 193 837–844. 10.1007/s00203-011-0751-4 [DOI] [PubMed] [Google Scholar]
- Mohapatra B., Sar P., Kazy S. K., Maiti M. K., Satyanarayana T. (2018). Taxonomy and physiology of Pseudoxanthomonas arseniciresistens sp nov., an arsenate and nitrate-reducing novel gammaproteobacterium from arsenic contaminated groundwater, India. PLoS One 13:e0193718. 10.1371/journal.pone.0193718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R., Rosen B. P. (2002). Arsenate reductases in prokaryotes and eukaryotes. Environ. Health Pers. 110 745–748. 10.1289/ehp.02110s5745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R., Rosen B. P., Pung L. T., Silver S. (2002). Microbial arsenic: from geocycles to genes and enzymes. Fems Microbiol. Rev. 26 311–325. 10.1111/j.1574-6976.2002.tb00617.x [DOI] [PubMed] [Google Scholar]
- Munthe J., Mcelroy W. J. (1992). Some aqueous reactions of potential importance in the atmospheric chemistry of mercury. Atmos. Environ. A Gen. Topics 26 553–557. 10.1016/0960-1686(92)90168-K [DOI] [Google Scholar]
- Newman D. K., Kennedy E. K., Coates J. D., Ahmann D., Ellis D. J., Lovley D. R., et al. (1997). Dissimilatory arsenate and sulfate reduction in Desulfotomaculum auripigmentum sp. nov. Arch. Microbiol. 168 380–388. 10.1007/s002030050512 [DOI] [PubMed] [Google Scholar]
- Nookongbut P., Kantachote D., Krishnan K., Megharaj M. (2017). Arsenic resistance genes of As-resistant purple nonsulfur bacteria isolated from As-contaminated sites for bioremediation application. J. Basic Microbiol. 57 316–324. 10.1002/jobm.201600584 [DOI] [PubMed] [Google Scholar]
- Ontiveros-Valencia A., Tang Y., Zhao H. P., Friese D., Overstreet R., Smith J., et al. (2014). Pyrosequencing analysis yields comprehensive assessment of microbial communities in pilot-scale two-stage membrane biofilm reactors. Environ. Sci. Technol. 48 7511–7518. 10.1021/es5012466 [DOI] [PubMed] [Google Scholar]
- Osborn A. M., Bruce K. D., Strike P., Ritchie D. A. (1997). Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. Fems Microbiol. Rev. 19 239–262. 10.1111/j.1574-6976.1997.tb00300.x [DOI] [PubMed] [Google Scholar]
- Powlowski J., Sahlman L. (1999). Reactivity of the two essential cysteine residues of the periplasmic mercuric ion-binding protein, MerP. J. Biol. Chem. 274 33320–33326. 10.1074/jbc.274.47.33320 [DOI] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S., Kim K. H., Saha S. K., Swaraz A. M., Paul D. K. (2014). Review of remediation techniques for arsenic (As) contamination: a novel approach utilizing bio-organisms. J. Environ. Manag. 134 175–185. 10.1016/j.jenvman.2013.12.027 [DOI] [PubMed] [Google Scholar]
- Richter M., Rosselló-Móra R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106 19126–19131. 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M., Rosselló-Móra R., Oliver Glöckner F., Peplies J. (2016). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32 929–931. 10.1093/bioinformatics/btv681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen B. P. (2002). Biochemistry of arsenic detoxification. Febs Lett. 529 86–92. 10.1016/S0014-5793(02)03186-1 [DOI] [PubMed] [Google Scholar]
- Rosenstein R., Peschel A., Wieland B., Gotz F. (1992). Expression and regulation of the antimonite, arsenite, and arsenate resistance operon of Staphylococcus xylosus plasmid-Psx267. J. Bacteriol. 174 3676–3683. 10.1128/jb.174.11.3676-3683.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossy E., Seneque O., Lascoux D., Lemaire D., Crouzy S., Delangle P., et al. (2004). Is the cytoplasmic loop of MerT, the mercuric ion transport protein, involved in mercury transfer to the mercuric reductase? Febs Lett. 575 86–90. [DOI] [PubMed] [Google Scholar]
- Saltikov C. W., Newman D. K. (2003). Genetic identification of a respiratory arsenate reductase. Proc. Natl. Acad. Sci. U.S.A. 100 10983–10988. 10.1073/pnas.1834303100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver S., Phung L. T. (2005). Genes and enzymes involved in bacterial oxidation and reduction of inorganic arsenic. Appl. Environ. Microbiol. 71 599–608. 10.1128/AEM.71.2.599-608.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skennerton C. T., Ward L. M., Michel A., Metcalfe K., Valiente C., Mullin S., et al. (2015). Genomic reconstruction of an uncultured hydrothermal vent gammaproteobacterial methanotroph (family methylothermaceae) indicates multiple adaptations to oxygen limitation. Front. Microbiol. 6:1425. 10.3389/fmicb.2015.01425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley P. L., Kinniburgh D. G. (2002). A review of the source, behaviour and distribution of arsenic in natural waters. Appl. Geochem. 17 517–568. 10.1016/S0883-2927(02)00018-5 [DOI] [Google Scholar]
- Srivastava D., Madamwar D., Subramanian R. B. (2010). Pentavalent arsenate reductase activity in cytosolic fractions of Pseudomonas sp., isolated from arsenic-contaminated sites of Tezpur, Assam. Appl. Biochem. Biotechnol. 162 766–779. 10.1007/s12010-009-8852-0 [DOI] [PubMed] [Google Scholar]
- Streets D. G., Devane M. K., Lu Z. F., Bond T. C., Sunderland E. M., Jacob D. J. (2011). All-time releases of mercury to the atmosphere from human activities. Environ. Sci. Technol. 45 10485–10491. 10.1021/es202765m [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobev A., Jagadevan S., Baral B. S., Dispirito A. A., Freemeier B. C., Bergman B. H., et al. (2013). Detoxification of mercury by methanobactin from Methylosinus trichosporium OB3b. Appl. Environ. Microbiol. 79 5918–5926. 10.1128/AEM.01673-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. P., Wiatrowski H. A., John R., Lin C. C., Young L. Y., Kerkhof L. J., et al. (2013). Impact of mercury on denitrification and denitrifying microbial communities in nitrate enrichments of subsurface sediments. Biodegradation 24 33–46. 10.1007/s10532-012-9555-8 [DOI] [PubMed] [Google Scholar]
- Wedepohl K. H. (1995). The composition of the continental-crust. Geochim. Cosmochim. Acta 59 1217–1232. 10.1016/0016-7037(95)00038-2 [DOI] [Google Scholar]
- Willsky G. R., Malamy M. H. (1980). Characterization of 2 genetically separable inorganic-phosphate transport-systems in Escherichia Coli. J. Bacteriol. 144 356–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z. S., Li J., Li Y. B., Wang Q., Zhai X. P., Wu G. F., et al. (2014). A mer operon confers mercury reduction in a Staphylococcus epidermidis strain isolated from Lanzhou reach of the Yellow River. Int. Biodeterior. Biodegradation 90 57–63. 10.1016/j.ibiod.2014.02.002 [DOI] [Google Scholar]
- Zhong L., Lai C. Y., Shi L. D., Wang K. D., Dai Y. J., Liu Y. W., et al. (2017). The nitrate effect on chromate bio-reduction in a methane-based MBfR. Water Res. 115 130–137. 10.1016/j.watres.2017.03.003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw Illumina sequences were submitted to the Sequence Read Archive (SRA) under Accession No. SRP136696 as described by Ontiveros-Valencia et al. (2014).