Abstract
Quorum sensing (QS) promotes in situ extracellular enzyme (EE) activity via the exogenous signal N-acylhomoserine lactone (AHL), which facilitates marine particle degradation, but the species that engage in this regulatory mechanism remain unclear. Here, we obtained AHL-producing and AHL-degrading strains from marine particles. The strain Ruegeria mobilis Rm01 of the Roseobacter group (RBG), which was capable of both AHL producing and degrading, was chosen to represent these strains. We demonstrated that Rm01 possessed a complex QS network comprising AHL-based QS and quorum quenching (QQ) systems and autoinducer-2 (AI-2) perception system. Rm01 was able to respond to multiple exogenous QS signals through the QS network. By applying self-generated AHLs and non-self-generated AHLs and AI-2 QS signal molecules, we modulated biofilm formation and lipase production in Rm01, which reflected the coordination of bacterial metabolism with that of other species via eavesdropping on exogenous QS signals. These results suggest that R. mobilis might be one of the participators that could regulate EE activities by responding to QS signals in marine particles.
Keywords: marine particles, quorum sensing, quorum quenching, Ruegeria mobilis, biofilm formation, extracellular enzyme
Introduction
Marine particles are typically derived from organic polymers, phytoplankton, bacteria, fecal pellets, mineral materials, and other suspended matter characteristic of a given water mass (Silver and Alldredge, 1981). They are reservoir for numerous elements and compounds, such as trace metals, C, N, protein, carbohydrate and lipid (Alldredge, 1979), and are responsible for the delivery of the surface organic matter to the seafloor (Alldredge and Silver, 1988). During the sinking process, large numbers of heterotrophic bacteria continuously approach, and colonize these particles (Kiørboe et al., 2002). The heterotrophic bacteria secrete various extracellular enzymes (EEs) to hydrolyze macromolecules into small molecules that are readily absorbed into cells (Cho and Azam, 1988), which promotes marine particle decomposition (Smith et al., 1992; Azam and Long, 2001). Moreover, bacterial abundance and cell-specific EE activity are much higher in marine particles than in ambient seawater (Smith et al., 1992), which indicates the activation of EE production in marine particles. However, studies investigating the regulatory mechanisms of bacterial EE production in marine particles are limited.
Quorum sensing (QS) is the regulation of gene expression in response to high bacterial density. In the QS regulatory process, QS autoinducers (AIs) are key factors delivering messages among bacterial communities, i.e., N-acylhomoserine lactones (AHLs) in Gram-negative bacteria (Williams, 2007; Case et al., 2008), autoinducing peptides (AIPs) in Gram-positive bacteria (Lyon and Novick, 2004) and autoinducer-2 (AI-2) in both Gram-positive and Gram-negative bacteria (Bassler and Losick, 2006). Amendments of QS signal AHLs and AI-2 affected EE production in marine particles (Hmelo et al., 2011; Krupke et al., 2016) and in Trichodesmium consortia (Van Mooy et al., 2012), which demonstrated the regulatory roles of QS on element cycling in oceans. Disruption of QS pathway is another gene regulatory mechanism, termed as quorum quenching (QQ). QQ could be achieved by species producing inhibition molecules (Manefield et al., 2002) or AHL-degrading enzymes AHL lactonases and AHL acylases (Dong and Zhang, 2005). In marine ecosystem, enzymatic QQ activities have been observed in seawater (Hmelo and Van Mooy, 2009) and Trichodesmium colonies (Van Mooy et al., 2012). Although the QS and QQ bacteria are prevalent in marine particles (Gram et al., 2002; Romero et al., 2011), whether they are responsible for the QS-regulated EE production in marine particles is still unclear.
Species belonging to Roseobacter group (RBG) are widespread (Wagner-Dobler and Biebl, 2006; Wietz et al., 2010) and prefer a surface-attached lifestyle (Gram et al., 2002; Zan et al., 2014). They are main forces for AHL producing activities in marine particles (Gram et al., 2002; Doberva et al., 2015). According to a genome analysis, QS in RBG is conserved and widespread thus might participate in multiple metabolisms controlling (Cude and Buchan, 2013). As revealed by several RBG species, QS regulate phenotypic traits related to ecological success especially in particle-attached lifestyle, e.g., flagellar motility and biofilm formation in Ruegeria sp. KLH11 (Zan et al., 2012), antimicrobial indigoidine biosynthesis in Phaeobacter sp. strain Y4I (Cude et al., 2015) and tropodithietic acid (TDA) production in Phaeobacter inhibens DSM 17395 (Berger et al., 2011). Ruegeria mobilis, another common species of the RBG, is capable of biofilm formation and TDA production as well (Porsby et al., 2008; D’Alvise et al., 2014), which may allow them to invade already formed bacterial biofilms and occupy advantageous status on marine particles (Rao et al., 2005). Although the QS circuit in R. mobilis has not been identified, two putative AHL synthases differing from the reported AHL synthases were proposed (Sonnenschein et al., 2016) and the potential role of QS on biofilm formation was suggested by controlling the intracellular signal compound cyclic dimeric guanosinmonophosphate (c-di-GMP) concentration (Srivastava and Waters, 2012; D’Alvise et al., 2014).
In this study, we aimed to extend investigation of QS-regulated marine particle degradation. We collected marine particle samples from the Yellow Sea of China and reported (i) regulations of EE activities in marine particles with exogenous AHL; (ii) the QS and QQ strains isolated from marine particles; (iii) QS networks of a RBG species, R. mobilis Rm01; and (iv) the controls of biofilm formation and EE production in Rm01 by diverse QS signals. This study represents the comprehensive analysis of bacterial assemblage of QS and QQ strains in marine particles; indicates the complex interspecies signaling conducted by Rm01 and provides perspective in revealing mechanisms of QS-regulated marine particle degradation.
Materials and Methods
Marine Particle Collection
The marine particle samples were collected in the Yellow Sea of China on board R/V “Dong Fang Hong 2” in October 2015. Approximately 75 L of seawater at the depth of 2–10 m was obtained via an injection pump at six stations (Supplementary Figure 1). The in situ seawater was filtered through 3-μm and 0.22-μm GTTP Isopore membrane filters (Millipore, Ireland) in succession immediately after sample collection. The marine particles collected on 3-μm filters were rinsed and re-suspended with 10 ml of 0.22-μm-filtered in situ seawater and transferred into a sterile 50-ml polypropylene (PP) tube for temporal storage.
Regulations of EE Activity in Marine Particles by Exogenous AHL
An aliquot of 500 μl of condensed marine particle sample in H33 station was pipetted into a sterile 50-ml PP tube, and then the volume was expanded to 10 ml incubation systems with 0.22-μm-filtered in situ seawater. The marine particle incubations were divided into three treatment groups amended with exogenous N-(3-oxo-octanoyl)-L-homoserine lactone (3OC8-HSL) (a serial concentrations of 50, 100, 500, and 1000 nM), 3OC8-HSL solvent DMSO (negative control) and 0.22-μm-filtered in situ seawater (blank control), respectively. Each treatment included triplicate incubations maintained at 25°C (approximately equivalent to the surface seawater temperature). After incubation of 6 and 24 h, an aliquot of 200 μl sample was centrifuged at 12,000 rpm for 5 min to collect supernatant used for EE assays. Following the methods described by Hoppe and Hmelo (Hoppe, 1993; Hmelo et al., 2011), ten EE activity were assayed using fluorescent substrates, i.e., MUF-α-glucopyranoside (for α-glucosidase activity), MUF-β-glucopyranoside (for β-glucosidase activity), MUF-β-D-xylopyranoside (for β-xylosidase activity), MUF-α-D-mannopyranoside (for mannosidase activity), MUF-β-D-cellobioside (for cellulase activity), MUF-N-acetyl-β-D-galactosaminide (for galactosaminidase activity), MUF-N-acetyl-β-D-glucosamine (for chitobiase activity), MCA-leucine (for aminopeptidase activity), MUF-phosphate (for phosphatase activity) and MUF-butyrate (for lipase activity) (all fluorescent substrates were purchased from Sigma-Aldrich). Released fluorescent signals were detected with a Fluoroskan Ascent FL multi-well plate reader (Thermo) with continuous reading at 2-min intervals during the enzyme activity assay over 1 h. The variations of EE activities were calculated by subtracting the EE activities of blank controls from that of the corresponding treatments. The excitation and emission characteristics of the fluorophores were previously programmed into the instrument (364 and 445 nm, respectively, for MUF; 380 and 440 nm, respectively, for MCA). Standard curves were constructed using the standard fluorophores MUF and MCA (Sigma-Aldrich).
Isolation and Identification of Bacterial Strains From Marine Particles
The condensed particle sample was serially diluted from 10-1 to 10-6 with 0.22-μm-filtered in situ seawater and 100 μl of each dilution was spread on 2216E marine agar (MA, Difco) plates. Colonies appeared were transferred three to four times on MA plates to obtain pure isolates. Further identification of the isolates relied on PCR using the B8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and B1510 (5′-GGTTACCTTGTTACGACTT-3′) primers (Weisburg et al., 1991) for 16S rRNA gene amplification followed by sequencing at BGI (Qingdao, China). Sequence similarities between isolates and their closest relatives were calculated using the EzTaxon-e server (Kim et al., 2012).
Screening for AHL-Producing and AHL-Degrading Bacterial Species
Screening for AHL-producing and AHL-degrading species was conducted using the reporter strain Agrobacterium tumefaciens (pCF218) (pCF372) A136 (Zhu et al., 2003). AHL-producing activity was confirmed by the cross-feeding method according to Chu et al. (2011). In brief, the reporter strain A136 and the tested strains were streaked adjacently on the same agar plates enriched with modified marine broth medium (MB 2216E; Difco, Detroit, MI, United States) and 0.5% X-Gal. The pH of MB medium was adjusted to 6.7 in avoid of the spontaneous alkaline hydrolysis of AHLs (Decho et al., 2009). After co-incubation at 28°C for 24 h, positive results were confirmed by the presence of indigo spots on A136.
AHL-degrading activity was detected using the high-throughput method described by Tang (Tang et al., 2013). N-hexanoyl-L-homoserine lactone (C6-HSL, representative for AHLs with short acyl chains) and N-dodecanoyl-L-homoserine lactone (C12-HSL, representative for AHLs with long acyl chains) were used as the substrates. Briefly, C6-HSL or C12-HSL was mixed with the bacterial culture and MB medium (negative control) and maintained at 28°C for 24 h. After incubation, the supernatant was obtained, mixed with an A136 X-gal assay solution (an overnight broth culture of A136 inoculated in AT minimal glucose medium and mixed with 0.5% X-Gal) in 96-well plates and incubated at 28°C for another 24 h. Positive results were indicated by reduced indigo color compared to the negative control.
Identification of AHLs Produced and Degraded by Ruegeria mobilis Rm01
A RBG species R. mobilis Rm01 was identified for holding both AHL-producing and AHL-degrading abilities. The initial time for QS and QQ activities were primarily detected for further identification of AHLs produced and degraded by Rm01.
AHLs secreted by Rm01 were extracted using an ethyl acetate protocol (Shaw et al., 1997; Zhu et al., 1998) and further evaluated by gas chromatography-mass spectrometer (GC-MS) system (Wagner-Dobler et al., 2005; Cataldi et al., 2007). Briefly, the GC-MS analysis was performed using a GC system 6890 N connected to an Agilent-5973 mass selective detector (Agilent Technologies, United States). One microliter of sample was injected into a HP-5 MS capillary column (30 m × 0.25 mm ID and 0.25 μm film thickness) and analyzed in split mode. Helium (99.99%) was used as a carrier gas at a flow rate of 0.8 ml min-1. The oven temperature was held at 150°C for 3 min and then increased to 280°C at a rate of 15°C min-1. The mass spectrometer was run in single ion monitoring (SIM) mode at m/z 143. A compassion of mass spectra and retention time of chromatographic peak with AHL standards (at the final concentration of 1 mg/ml) allowed a quick detection of AHLs in sample.
The AHL degrading ability of Rm01 was detected following the procedures described above with a few modifications. The strain Rm01 was inoculated in 2216E liquid and cultured for 8 h on a shaker (170 rpm) at 28°C. The cells were harvested after centrifugation for 10 min at 4°C and 6,000 rpm and re-suspended in HEPES buffer (20 mM Na-HEPES with 0.5 M NaCl, 10% glycerol, and 0.1% Triton X-100, pH 8.5) for sonication. Additional centrifugation for the lysed cells was conducted at 4°C and 12,000 rpm to obtain the crude enzyme supernatant. The supernatant inactivated by proteinase K (final concentration, 200 μg/ml) was used as the negative control. After co-incubation, the QQ activity of Rm01 was indicated with well-diffusion assays (Gram et al., 2002) using the reporter strains A136 and Chromobacterium violaceum VIR24 (Someya et al., 2009). Another acidification test to determine QQ enzymatic mechanisms was conducted according to Yates et al. (2002).
Identification of QS and QQ Enzyme Encoding Genes in Rm01
Rm01 genomic DNA was obtained using the phenol-chloroform method (Murray and Thompson, 1980) and the whole-genome sequencing was performed using Illumina Hiseq 2000 platform and Pacific Biosciences (PacBio) sequencing at BGI. PacBio sequence read data were assembled de novo using Canu version 1.0 (Berlin et al., 2015) and further polished using Illumina sequencing data to resolve single nucleotide errors. The assembled genome sequences were submitted into RAST server for genome annotation (Aziz et al., 2008).
Putative QS (AHL synthase) and QQ enzyme (AHL lactonase or AHL acylase) encoding genes were searched from Rm01 genome through local BLASTP against known QS and QQ enzyme databases. The putative QS and QQ enzyme encoding genes were amplified with primers (Supplementary Table S1) and merged into vector pET-24a. The recombined plasmids were transformed into Escherichia coli BL21 (DE3).
The putative QS and QQ genes were heterogeneously expressed in BL21 (DE3) and examined for QS and QQ activities. The recombinant strains were grown to OD600 0.4–0.6 and induced with 0.1 mM IPTG for 12 h on a shaker (150 rpm) at 16°C. AHLs synthesized by QS enzymes expressed in E. coli (DE3) were extracted and analyzed by the GC-MS approach. The activity of QQ enzyme expressed in E. coli (DE3) was detected by well-diffusion bioassays described above.
The Influence of AIs on Biofilm Formation and EE Production in Rm01
The influence of diverse AIs on physiological metabolisms in Rm01 was investigated by growing bacterium in MB medium supplemented with 10 μM self-generated N-(3-oxo-decanoyl)-L-homoserine lactone (3OC10-HSL), N-decanoyl-L-homoserine lactone (C10-HSL) and C12-HSL; non-self-generated 3OC8-HSL, N-tetradecanoyl-L-homoserine lactone (C14-HSL) and AI-2; and purified AHL lactonase MomL (0.5 U ml-1) (Tang et al., 2015). The reagent solvent DMSO, 1 M PIPES and solution buffer (HEPES buffer supplemented with 25% glycerol) were used as negative controls for AHLs, AI-2 and MomL, respectively. Each treatment included triplicate repeats, and all experiments were repeated at least 3 times.
For biofilm mass quantification, Rm01 was grown in 200 μl of MB medium using 96-well microtiter plate and was maintained at 28°C for 8 h. Biofilm mass quantification was carried out as described previously (Sheikh et al., 2001) with a few modifications. Briefly, the attached cells were stained with 0.2% crystal violet (CV) and washed three times with ddH2O. The remaining CV was re-dissolved in 75% ethanol, and finally, the absorbance at 570 nm was monitored with a Tecan SunriseTM microplate absorbance reader.
Primarily identification of EE production in Rm01 was conducted using fluorescent substrates mentioned above and only lipase was detected in the sterile filtered culture supernatant. For lipase activity assay, Rm01 was grown in 100 ml of MB medium using 500 ml schott flasks and was incubated at 28°C for 24 h. Growth was measured at OD600 and sterile culture supernatants were obtained for lipase activity using fluorescent substrates MUF-butyrate. Additionally, the enzyme activities of exogenous AHLs, AI-2 and MomL were also assayed under the same incubation conditions and subtracted from the corresponding assayed lipase activities.
Results
Exogenous AHL Regulates EE Production in Marine Particles
Since GC-MS analyses of AHLs cannot be conducted on R/V “Dong Fang Hong 2”, the AHL 3OC8-HSL, which was previously identified in marine particles (Hmelo et al., 2011), was applied as the exogenous AHL. After incubation for 6 and 24 h, the activity of ten EEs (seven carbohydrases, one aminopeptidase, one lipase and one phosphatase) was assayed. Of the EEs, four carbohydrases were modulated with the addition of 3OC8-HSL (Figure 1). Galactosaminidase activity was up to 3–4-fold higher than that of the control after 24 h, and the activity of initially undetectable β-xylosidase was elevated at both time points. By contrast, β-glucosidase and mannosidase activities were inhibited, and this effect was sustained from 6 to 24 h. The addition of 3OC8-HSL in higher concentrations (100, 500, and 1000 nM) did not enhance the effects induced by 3OC8-HSL in low concentration (50 nM) (Figure 1), which is suggestive of the inducing threshold of 3OC8-HSL. No significant variations were observed in the activities of chitobiase, aminopeptidase, lipase and phosphatase (Supplementary Figure 2). We propose that the modulations on EE activities might be achieved by species possessing QS networks in marine particles.
Figure 1.

In situ galactosaminidase (A), β-xylosidase (B), β-glucosidase (C), and mannosidase (D) activities in marine particles treated with 3OC8-HSL (the final concentrations of 3OC8-HSL are shown in different colors). The data are shown as the mean ± standard deviation (SD). The difference between the 3OC8-HSL-treated groups and the un-treated control groups was calculated by Student’s t-test (∗∗P < 0.01; ∗P < 0.05).
AHL-Producing and AHL-Degrading Bacteria in Marine Particles
In order to figure out the species participating in the QS-regulated EE production, we screened for AHL-producing and AHL-degrading strains from marine particles using the reporter strain A136. A total of 122 bacterial species were identified according to 16S rRNA gene sequence analyses and phylogenetically classified into four phyla: Proteobacteria, Bacteroidetes, Actinobacteria, and Firmicutes. Among these species, 20 and 62 species possessed AHL-producing and AHL-degrading abilities, respectively (Table 1 and Supplementary Figure 3). Most AHL-producing isolates belonged to the orders Rhodobacterales (5 species), Sphingomonadales (4 species), Vibrionales (4 species) and Rhizobiales (3 species), while AHL-degrading isolates belonged to the orders Alteromonadales (19 species), Flavobacteriales (9 species), Rhodobacterales (7 species) and Oceanospirillales (7 species) (Figure 2). The results demonstrated high proportions of species possessing QS or QQ activities and suggested a large scale of bacteria possibly involving in QS-regulated EE production in marine particles. Notably, the isolate R. mobilis Rm01 was found possessing both AHL-producing and AHL-degrading abilities thus it was chosen as representative for further investigations.
Table 1.
Summary of AHL-producing and/or AHL-degrading bacterial isolates.
| Phylum or group | Strains | Identification by EZBioCloud alignment | % identity to EZBioCloud sequence | AHL-producing ability ( ) ) |
AHL-degrading ability ( ) ) |
|---|---|---|---|---|---|
| Actinobacteria | H19-20 | Kocuria palustris DSM 11925T | 100 |  |
|
| H10-56 | Microbacterium aquimaris JS54-2T | 98.1 |  |
||
| H19-37 | Nocardioides marinus CL-DD14T | 98.7 |  |
||
| Cytophagia | H18-59 | Algoriphagus boritolerans T-22T | 98 |  |
|
| H01-35 | Croceitalea litorea CBA3205T | 96.4 |  |
||
| Flavobacteria | H33-3 | Dokdonia genika Cos-13T | 99.8 |  |
|
| H33-47 | Empedobacter falsenii NF 993T | 98.3 |  |
||
| H33-97 | Maribacter dokdonensis DSW-8T | 99.9 |  |
||
| H33-75 | Meridianimaribacter vietnamensis KMM 6217T | 99.5 |  |
||
| H19-32 | Muricauda ruestringensis DSM 13258T | 97.2 |  |
||
| H33-69 | Non-labens tegetincola UST030701-324T | 100 |  |
||
| H19-18 | Tenacibaculum mesophilum MBIC1140T | 99.1 |  |
||
| H01-25 | Tenacibaculum xiamenense WJ-1T | 97.6 |  |
||
| Alphaproteobacteria | H33-92 | Aliiroseovarius pelagivivens GYSW-22T | 97.3 |  |
|
| H10-30 | Brevundimonas abyssalis TAR-001T | 100 |  |
||
| H19-55 | Brevundimonas nasdae GTC 1043T | 99.2 |  |
||
| H10-60 | Citromicrobium bathyomarinum JF-1T | 99.8 |  |
||
| H33-63 | Erythrobacter citreus RE35F/1T | 99.4 |  |
 |
|
| H18-45 | Erythrobacter gaetbuli SW-161T | 98.9 |  |
||
| H19-12 | Erythrobacter pelagi UST081027-248T | 100 |  |
||
| H19-15 | Henriciella aquimarina P38T | 98 |  |
||
| H33-86 | Henriciella marina DSM 19595T | 98.3 |  |
||
| H19-22 | Hoeflea suaedae YC6898T | 98.9 |  |
||
| H33-59 | Hyphomonas atlantica22II1-22F38T | 100 |  |
||
| H19-9 | Jiella aquimaris LZB041T | 100 |  |
||
| H10-43 | Labrenzia aggregata IAM 12614T | 100 |  |
||
| H10-55 | Labrenzia alba CECT 5094T | 98.7 |  |
||
| H19-11 | Mesorhizobium tamadayense Ala-3T | 97.4 |  |
||
| H19-35 | Mesorhizobium thiogangeticum SJTT | 97.4 |  |
||
| H33-8 | Nautella italica CCUG 55857T | 97.8 |  |
||
| H33-70-1 | Oceanicaulis stylophorae GISW-4T | 99.8 |  |
||
| H33-105 | Pelagibaca bermudensis HTCC2601T | 99.7 |  |
||
| H10-50 | Phenylobacterium falsum AC-49T | 98 |  |
||
| H19-2 | Ponticaulis koreensis DSM 19734T | 98.2 |  |
||
| H19-23 | Roseovarius mucosus DSM 17069T | 100 |  |
||
| Rm01 | Ruegeria mobilis NBRC 101030T | 100 |  |
 |
|
| H33-41 | Sinorhodobacter ferrireducens SgZ-3T | 100 |  |
||
| H18-57 | Sphingobium abikonense NBRC 16143T | 99.6 |  |
||
| H18-18 | Sphingopyxis alaskensis RB2256T | 98.4 |  |
||
| H18-22 | Sphingopyxis italica SC13E-S73T | 99.9 |  |
||
| H10-48-1 | Thalassobaculum salexigens DSM 19539T | 99.7 |  |
||
| Gammaproteobacteria | H33-18 | Acinetobacter venetianus RAG-1T | 100 |  |
|
| H33-19 | Aestuariibacter aggregatus WH169T | 100 |  |
||
| H19-53 | Alcanivorax borkumensis SK2T | 99.5 |  |
 |
|
| H33-67 | Alcanivorax gelatiniphagus MEBiC08158T | 99.5 |  |
||
| H33-94 | Alcanivorax jadensis T9T | 98.7 |  |
||
| H18-16 | Alcanivorax marinus R8-12T | 99.8 |  |
||
| H19-7 | Alcanivorax venustensis ISO4T | 100 |  |
||
| H33-5 | Alteromonas macleodii ATCC 27126T | 99.8 |  |
||
| H18-4 | Alteromonas marina SW-47T | 99.6 |  |
||
| H33-31 | Alteromonas tagae BCRC 17571T | 99.4 |  |
||
| H18-42 | Cobetia marina DSM 4741T | 100 |  |
||
| H33-11 | Escherichia flexneri ATCC 29903T | 99.5 |  |
||
| H10-4 | Halomonas titanicae BH1T | 100 |  |
||
| H33-82 | Klebsiella pneumoniae ATCC 13884T | 100 |  |
||
| H33-13-1 | Marinobacter adhaerens HP15T | 98.6 |  |
||
| H33-64 | Marinobacter algicola DG893T | 99.9 |  |
||
| H33-50 | Marinobacter goseongensis En6T | 99.8 |  |
||
| H33-20 | Marinobacter hydrocarbonoclasticus ATCC 49840T | 99.7 |  |
||
| H33-13-2 | Marinobacter koreensis DD-M3T | 98.9 |  |
||
| H33-14-2 | Marinobacter litoralis SW45T | 96.8 |  |
||
| H33-14-1 | Marinobacter maritimus CK47T | 97.6 |  |
||
| H33-48 | Marinobacter sediminum R65T | 99.7 |  |
||
| H18-44 | Marinobacter similis A3d10T | 99.7 |  |
||
| H33-70-2 | Mediterranea mediterranea DET | 98.7 |  |
||
| H33-107 | Neptuniibacter caesariensis MED92T | 97.5 |  |
||
| H19-31 | Pseudoalteromonas hodoensis H7T | 100 |  |
||
| H10-48-2 | Pseudoalteromonas shioyasakiensis SE3T | 97.9 |  |
||
| H33-29 | Pseudoalteromonas spongiae UST010723-006T | 99.8 |  |
||
| H33-24 | Pseudomonas pachastrellae KMM 330T | 99.7 |  |
||
| H33-35 | Pseudomonas sabulinigri J64T | 98.6 |  |
||
| H33-32 | Rheinheimera nanhaiensis E407-8T | 99.4 |  |
||
| H10-16 | Serratia arcescens ATCC13880T | 99.7 |  |
 |
|
| H33-54 | Shewanella corallii fav-2-10-05T | 98.9 |  |
||
| H10-24 | Vibrio harveyi ATCC 14126T | 99.9 |  |
||
| H18-1 | Vibrio neocaledonicus NC470T | 100 |  |
||
| H10-21 | Vibrio sinaloensis CAIM797T | 99.1 |  |
||
| H10-39 | Vibrio variabilis R-40492T | 98.3 |  |
The identification of bacterial isolates was based on partial (∼800 bp) 16S rRNA gene sequences. The solid circle ( ) indicates AHL-producing bacterial isolate. The solid triangle (
) indicates AHL-producing bacterial isolate. The solid triangle ( ) indicates AHL-degrading bacterial isolate.
) indicates AHL-degrading bacterial isolate.
Figure 2.
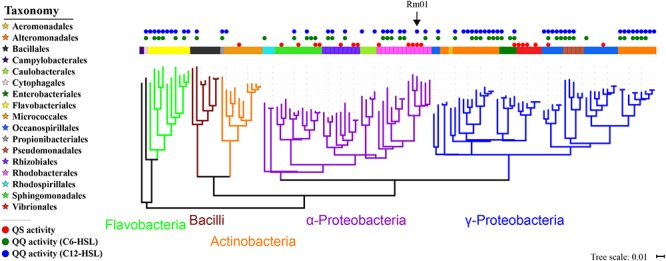
Phylogenetic tree of cultivable bacterial community in marine particles based on the 16S rRNA gene. The arrow indicates the phylogenetic position of Ruegeria mobilis Rm01.
AHL-Producing Profile of R. mobilis Rm01
We applied biosensor strains and GC-MS analysis to detect the AHLs produced by Rm01. As indicated by the responses of the biosensor strains C. violaceum VIR24 (sensitive to AHLs with long acyl chains) and C. violaceum CV026 (sensitive to AHLs with short acyl chains), AHLs with long acyl chains were preliminarily inferred to be synthesized by Rm01 (Supplementary Figure 4). Following the observation of biosensor responses, the supernatant of Rm01 cultured for 24 h (Figure 3A) was extracted and analyzed by GC-MS. Three AHLs, 3OC10-HSL, C10-HSL and C12-HSL, were identified in Rm01 (Figure 3B), consistent with the results initially detected by the biosensor strains VIR24 and CV026.
Figure 3.
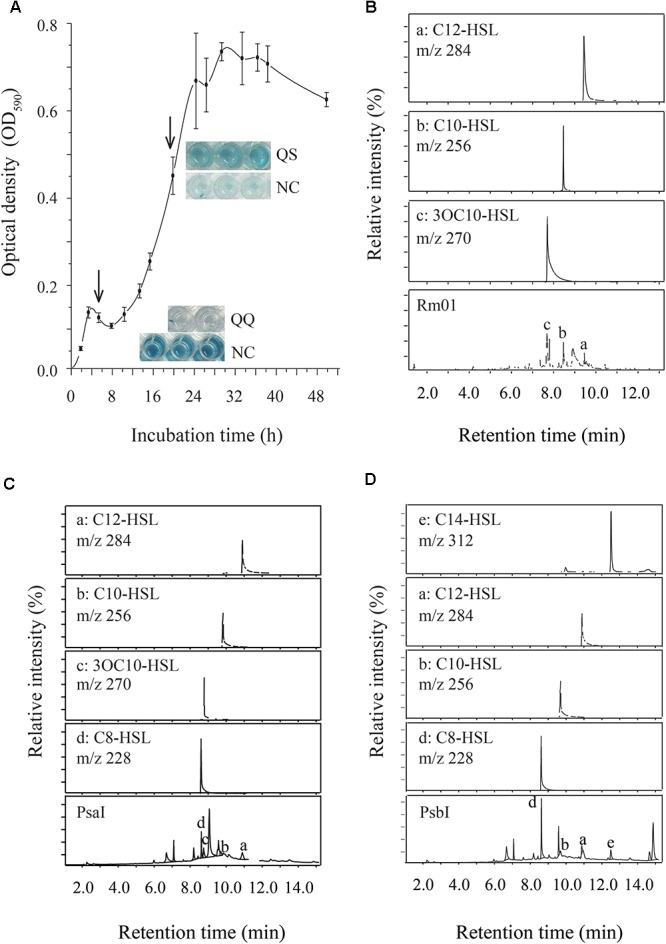
AHL-producing ability of R. mobilis Rm01. (A) The growth curve of Rm01. The arrows indicate the initiation time for QQ and QS activities. (B–D) GC-MS chromatograms in SIM mode at m/z 143 of cell-free supernatant extracts of wild-strain Rm01 and recombinant strains BL21 (DE3)/pET-24a–MG001458 (PsaI) and BL21 (DE3)/pET-24a–MG001460 (PsbI), respectively.
In the genome of Rm01, two putative AHL synthases PsaI (Accession No. MG001458) and PsbI (Accession No. MG001460) (particle-associated symbiont locus A and B luxI homolog, respectively) were found and further verified as functional AHL synthases by heterogeneous expression in E. coli BL21 (DE3). PsbI shared 41% identity at the amino acid level with LuxI1 in Dinoroseobacter shibae DFL 12 (Neumann et al., 2013) while PsaI did not resemble to any reported AHL synthase. According to the BLASTP results, PsaI is conserved in Rhodobacteracea genomes, sharing at least 80% identities with the homologous proteins (Supplementary Figure 6). However, the homologs are automatically annotated as GNAT family N-acetyltransferase and no experimental evidence has been raised to verify their functions. The AHLs produced by PsaI and PsbI were also extracted and analyzed by GC-MS. As a result, N-octanoyl-L-homoserine lactone (C8-HSL), 3OC10-HSL, C10-HSL and C12-HSL were produced by PsaI (Figure 3C), while C8-HSL, C10-HSL, C12-HSL, and C14-HSL were produced by PsbI (Figure 3D). Except of C10-HSL, other AHLs identified in the recombinant strains were not detected in the AHL production background of the E.coli strain BL21 (DE3)/pET-24a (supplementary Figure 5). Most of the AHLs produced by PsaI and PsbI were included in the AHL-producing profile of Rm01. However, C8-HSL produced by both PsaI and PsbI, and C14-HSL produced by PsbI were not identified in the AHL extractions of Rm01, which might be explained by the differential acyl-ACP substrate pools between R. mobilis and E. coli (Zan et al., 2012).
AHL-Degrading Ability of R. mobilis Rm01
In addition to AHL-synthesizing ability, Rm01 also demonstrated AHL-degrading ability, termed as QQ. The QQ activity of Rm01 was detected at the beginning of the exponential growth phase indicated by both reporter strains A136 and VIR24 (Figures 3A, 4A). The AHL molecules C10-HSL, C12-HSL and C14-HSL were partially degraded, whereas 3OC8-HSL and 3OC10-HSL were not attenuated (Figure 4A). Based on these results, saturated AHLs were much more easily degraded by Rm01 than oxo-substituted AHLs.
Figure 4.
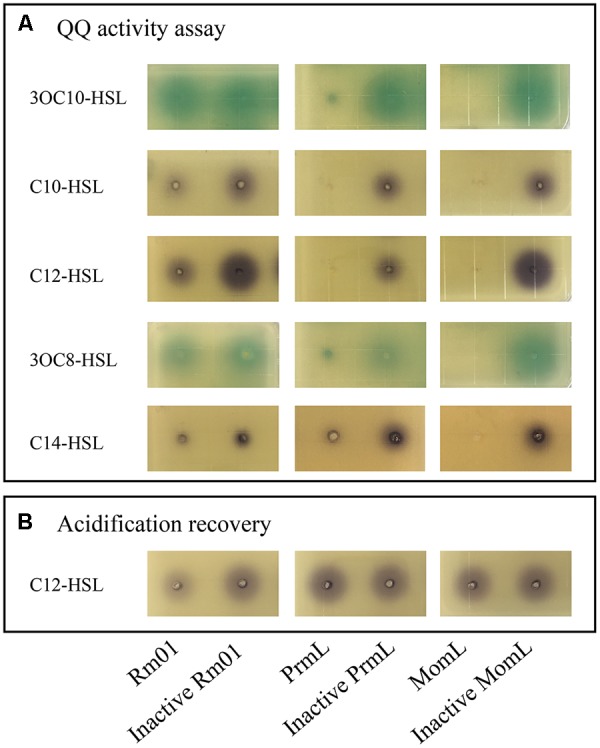
AHL-degrading ability of R. mobilis Rm01. The results are shown via well-diffusion assays supplemented with the AHL reporter strains A136 and VIR24. The degradation results for the self-generated AHLs 3OC10-HSL, C10-HSL and C12-HSL and non-self-generated AHLs 3OC8-HSL and C14-HSL are shown in frame (A). Acidification recovery of hydrolyzed C12-HCL was conducted and is shown in frame (B). Columns 1–6 represent the following samples, in order: Rm01 cell lysate; inactive Rm01 cell lysate; whole cells of BL21 (DE3)/pET-24a-MG001461 (PrmL); inactive whole cells of BL21 (DE3)/pET-24a (PrmL); AHL lactonase MomL (positive control); and inactive AHL lactonase MomL.
One putative QQ lactonase gene, prmL (Accession No. MG001461) (particle-associated R. mobilis lactonase), was found in the genome of Rm01. PrmL expressed in heterologous system, degraded C10-HSL, C12-HSL and C14-HSL (Figure 4A) as well as 3OC8-HSL and 3OC10-HSL, which were not quenched by Rm01. Moreover, the enzymatic mechanisms of Rm01 and PrmL were revealed with an acidification experiment. Under acidic conditions, hydrolyzed C12-HSL by PrmL was completely recovered (Figure 4B), which indicated that PrmL was an AHL lactonase (Yates et al., 2002). The unrecovered C12-HSL might own to other unidentified QQ enzymes in Rm01 that can further hydrolyze ring-opened AHLs, e.g., AHL acylase.
Exogenous AIs Regulate Biofilm Formation and Lipase Production in Rm01
Biofilm formation is essential for bacteria to colonize on marine particles and get access to resources (Dang and Lovell, 2016). To investigate the roles of different AIs play on biofilm formation in Rm01, we conducted feedback experiments using a variety of QS AI molecules (including self-generated AHLs and non-self-generated AHLs and AI-2). We demonstrated that the biofilm formation of Rm01 was reduced with 3OC10-HSL, 3OC8-HSL and AI-2, whereas increased with C10-HSL, C12-HSL, and C14-HSL (Figure 5A). Additionally, biofilm formation was also increased with an effective AHL lactonase MomL, which completely eliminated the intrinsic AHLs secreted by Rm01. The modest variations of biofilm mass may own to the counteraction by the intrinsic QS systems of Rm01. But according to the statistical analyses, the modulations of biofilm formation truly happened in Rm01 by sensing and responding to diverse AIs. The results might indicate bacterial adjustment during habitat colonization when encountering different bacterial species.
Figure 5.

Changes in biofilm formation ability (A) and production of extracellular lipase (B) in R. mobilis Rm01 treated with self-generated 3OC10-HSL, C10-HSL and C12-HSL and non-self-generated 3OC8-HSL, C14-HSL, AI-2 and AHL lactonase MomL. The y-axis presents the ratios of experimental groups relative to control groups, and a value of 1.0 indicates no difference. The data are shown as the mean ± SD, and the difference between the amended groups and the control groups was calculated by Student’s t-test (∗∗P < 0.01; ∗P < 0.05).
Bacterial EE production is important for the acquisition of nutrition, which is also the key factor regulating the marine particle degradation process. With amendment of above AIs, the growth of Rm01 was not affected whereas the lipase production in Rm01 was modulated. As is shown in Figure 5B, lipase activity in Rm01 was stimulated by all applied AHLs and AI-2, while inhibited by the AHL lactonase MomL. Above results indicated cooperations of Rm01 with other QS and QQ strains in nutrient acquisition, which ultimately affect the marine particle degradation process.
Discussion
In this study, we extended our investigation of QS-regulated marine particle degradation by applying amendment experiments in situ and in vitro. Compared with previous findings (Hmelo et al., 2011; Krupke et al., 2016), the regulations of EE production in marine particles with exogenous AHLs were further observed on carbohydrases, xylosidase, galactosaminidase, β-glucosidase and mannosidase. RBG species are frequently observed on marine particles (Gram et al., 2002; Slightom and Buchan, 2009; Thiele et al., 2015; Table 1) and have been shown to possess AHL networks (Gram et al., 2002; Zan et al., 2014; Table 1), which indicates the significance of RBG species in QS-regulated metabolic mechanisms in niches. To obtain more concrete information, we identified the AHL-based QS network in an RBG species, R. mobilis Rm01, which possesses the additional QQ and AI-2 pathways. Further amendment experiments clearly demonstrated that biofilm formation and lipase production by Rm01 were tightly associated with its multiple QS networks described above. Our results here suggest that QS networks in Rm01 enable interspecies communications and coordinations of metabolisms in R. mobilis, which might be one of factors affecting marine particle degradation.
QQ- and AI-2-Related Processes Are Important Components of the QS Regulatory Network of R. mobilis Rm01
Ruegeria mobilis Rm01 has a typical AHL-based QS system. In RBG species, many studies have demonstrated that QS systems participate in TDA production (Berger et al., 2011), flagellar motility and biofilm formation (Zan et al., 2012), which facilitate their high abundance (20%-40% of the 16S rRNA genes) in ocean waters (Moran et al., 2003; Buchan et al., 2005; Wagner-Dobler and Biebl, 2006). In addition, RBG species have been found more dominant in particle-associated fractions than in free-living fractions (Zan et al., 2014; Thiele et al., 2015). Though we did not measure the abundance of R. mobilis in this study, previous study has demonstrated that R. mobilis, one of the typical RBG species, was more abundant in particle-associated fractions than free-living fractions, with 40% and 6% occurrences respectively (Sonnenschein et al., 2016). However, the R. mobilis QS system has not been studied in detail. In this study, two AHL synthase genes, psaI and psbI, were detected in Rm01. According to genomic analysis, psaI is located downstream of a luxR transcriptional regulator psaR (Accession No. MG001459), whereas psbI is an “orphan” AHL synthase gene without a cognate transcriptional regulator. PsaI and PsbI fell into two new distinct clusters (Supplementary Figure 6) and exhibited few conserved residues with other identified AHL synthases (Supplementary Figure 7) according to neighbor-joining phylogenetic tree and multiple-sequence alignment analyses, respectively. We propose that the QS system in Rm01 represents a new, previously undiscovered QS system in RBG species.
QQ might be another important process for the adaptation of Rm01 to marine particles. Previous studies demonstrated that many strains capable of AHL interference have been isolated from sediments, biofilms, the surface of the alga Fucus vesiculosus (Romero et al., 2012) and the Mediterranean seagrass angiosperm Posidonia oceanica (Blanchet et al., 2017), and marine particles (Table 1) in which QS activity is easily concentrated. The QQ processes in dense microbial niches limit the coordination of antagonistic bacteria and even affect vital activities of their phytoplankton host (Tait et al., 2009; Rolland et al., 2016). Coordinated mechanism achieved by QS and QQ systems is essential for bacterial survival. QS systems in Burkholderia pseudomallei (Lumjiaktase et al., 2006), Vibrio cholera (Joelsson et al., 2007) and Pseudomonas aeruginosa (García-Contreras et al., 2015) could enhance their stress tolerance and even affect bacterial ecology by promoting their resistance to QQ strains (García-Contreras et al., 2015). In the extremophilic bacterium Deinococcus radiodurans, the QS and QQ systems cooperate in regulating gene to resist oxidative stress (Lin et al., 2016). Combined QS and QQ systems have not been reported in RBG species yet. Our work on the cooperation of QS and QQ processes in Rm01 extends the existing evidence regarding successful regulatory mechanisms in marine particles.
An AI-2-based QS network enables cross-species communications on a wider scale. AI-2 synthase LuxS homologs have been found in 537 out of the 1402 sequenced bacterial genomes (Pereira et al., 2013). However, in Rm01 or even other RBG species, neither the signal molecule AI-2 (Supplementary Figure 8) nor its synthase LuxS homolog has been identified. In our experiments, lipase production and biofilm formation in Rm01 were regulated by AI-2 (Figure 4), which suggested the existence of AI-2 perception pathway. Rm01 genome annotation results conducted on the RAST server (Aziz et al., 2008) identified homologs of periplasmic AI-2 binding protein LsrB (Accession No. MG00148), proteins involved in an AI-2 internalization system (LsrA, Accession No. MG00145; LsrC, Accession No. MG00146; LsrD, Accession No. MG00147), the AI-2 phosphokinase LsrK (Accession No. MG001463) and the lsr repressor LsrR (Accession No. MG001464), which illustrates the genetic capabilities of Rm01 to respond to exogenously supplied AI-2. Coincidently, a similar pathway was observed in a plant symbiont, Sinorhizobium meliloti, which is unable to produce but nonetheless responds to AI-2 in the surrounding environment (Pereira et al., 2008). By “eavesdropping” on AI-2 produced by other species, the strain was capable of interfering with AI-2-regulated behaviors such as virulence. Although AI-2 has not been detected in marine particles, it is very likely to occur due to the prevalence of AI-2-producing Vibrio species (Bassler et al., 1994) in marine particles (Hmelo et al., 2011; Jatt et al., 2015; Table 1). The AI-2 pathway might facilitate Rm01 competing against antagonistic species in the microflora. Until now, research on AI-2-regulated mechanisms in marine bacteria has been rare, with the exception of studies investigating Vibrio species. Thus, our investigations of AI-2-regulated mechanisms in Rm01 might provide more hints that reveal the roles of AI-2 systems in marine bacteria and interspecies communications in marine particles.
QS-Regulated Biofilm Formation and EE Production Promote the Adaption of Rm01 to Marine Particles
Biofilms are essential for heterotrophic bacteria colonizing marine particles, facilitating their access to resources (Dang and Lovell, 2016) and maintaining the integrity and activity of secreted bacterial EEs (Flemming and Wingender, 2010). Moreover, the enrichment of bacteria in a biofilm stimulates bacterial QS, which in turn affects biofilm formation and EE production in microflora (Dang and Lovell, 2016). Bacterial biofilm formation, EE production and bacterial QS are undoubtedly highly interactive and inseparable in marine particles.
Previous studies demonstrated that biofilm formation is characteristic feature of RBG species in transition between motile and sessile life stages, which is regulated by a second messenger c-di-GMP (Hengge, 2009; Mcdougald et al., 2011; D’Alvise et al., 2014). Here, we suggest that the transition between motile and sessile states in R. mobilis Rm01 is also regulated with self-generated and non-self-generated AIs by modifying biofilm formation. Notably, biofilm formation was inhibited by self-generated AHL 3OC10-HSL, which might accumulate faster than C10-HSL and C12-HSL in early proliferation period due to its QQ activities. Reduced biofilm mass enables Rm01 searching for a substrate or a host more effectively. Once settle down, self-generated C10-HSL and C12-HSL accumulated faster and subsequently up-regulated the biofilm formation in Rm01, facilitating its persistence in high organic substrate (e.g., marine particles).
Additionally, the transition between sessile and motile life of Rm01 is also affected by non-self-generated AIs produced by bacteria in marine particles. Bacteria secreting 3OC8-HSL and AI-2 (e.g., Vibrio species) (Bassler et al., 1997; Surette et al., 1999; Rajput et al., 2016) might resist the colonization of Rm01 by inhibiting its biofilm formation, while C14-HSL synthesizing species (e.g., RBG species) (Ziesche et al., 2015) might assist the colonization of Rm01 by promoting its biofilm formation. Relying on the eavesdropping behaviors, the strain Rm01 is expected to colonize marine particles more selectively by developing a biofilm matrix with commensal rather than antagonistic bacteria.
All tested AIs in this study increased lipase production in Rm01. We suggest that bacteria in marine particles secreting AIs could impact lipase production in Rm01. Thus they may together affect marine particle degradation process by QS systems. In this study, we also found that bacterial lipase was prominent in tested EEs from marine particles as well as the bacterial isolates (Supplementary Table S2; Martinez et al., 1996). Lipids in marine environment are derived from cell membranes of dead organisms and a variety of other biological sources (Martinez et al., 1996), which lets them to be one of the inevitable components in marine particles (Lee et al., 2004). The hydrolyzed products of lipids, glycerol and fatty acids, are readily usable sources of energy for heterotrophic microbial communities (Zoppini et al., 2005). Therefore, the QS-regulated production of bacterial lipase is clearly meaningful to bacterial survival in dense microbial habitats and further affects the cycling of organic matter in the oceans.
Future Perspectives
In this study, R. mobilis Rm01 was proposed as the model strain to reveal the mechanisms underlying QS-regulated degradation processes of marine particles. Rm01 was capable of perceiving and interfering with diverse exogenous AIs to regulate its biofilm formation and lipase production. Our results demonstrate the molecular basis how Rm01 intercommunicate with other species and affects the marine particle degradation process to a certain extent. We suggest that R. mobilis play significant roles in marine particle degradation through bacterial interactions realized by QS networks. However, details of the pathways need to be better probed. In future investigations, transcriptome analysis is required to reveal the differential gene expression in detail. Metagenomic and metatranscriptomic analyses are also expected to characterize microbial structure and unravel QS pathways in marine particle microflora in avoid of culturability bias.
Author Contributions
X-HZ, YS, and KT contributed to the conception and design of the study. YS and YZ performed the molecular biological and biochemical experiments. YS, JL, and YW performed the statistical analysis. YS and YZ wrote the first draft of the manuscript. All authors contributed to manuscript revision, read and approved the submitted version.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Robert J. C. McLean (Texas State University, United States), Tomohiro Morohoshi (Utusnomiya University, Japan), and Bonnie L. Bassler (Princeton University, Princeton) for providing us the reporter strains C. violaceum CV026 and A136, C. violaceum VIR24 and V. harveyi TL88.
Footnotes
Funding. This work was supported by projects from the National Natural Science Foundation of China (Nos. 41476112, 41730530, and 91751202) and the National Key Research and Development Program of China (No. 2016YFA0601303).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.03304/full#supplementary-material
References
- Alldredge A. L. (1979). The chemical composition of macroscopic aggregates in two neretic seas1. Limnol. Oceanogr. 24 855–866. 10.4319/lo.1979.24.5.0855 [DOI] [Google Scholar]
- Alldredge A. L., Silver M. W. (1988). Characteristics, dynamics and significance of marine snow. Prog. Oceanogr. 20 41–82. 10.1016/0079-6611(88)90053-5 [DOI] [Google Scholar]
- Azam F., Long R. A. (2001). Oceanography: sea snow microcosms. Nature 414 495–498. 10.1038/35107174 [DOI] [PubMed] [Google Scholar]
- Aziz R. K., Bartels D., Best A. A., DeJongh M., Disz T., Edwards R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler B. L., Greenberg E. P., Stevens A. M. (1997). Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179 4043–4045. 10.1128/jb.179.12.4043-4045.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler B. L., Losick R. (2006). Bacterially speaking. Cell 125 237–246. 10.1016/j.cell.2006.04.001 [DOI] [PubMed] [Google Scholar]
- Bassler B. L., Wright M., Silverman M. R. (1994). Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13 273–286. 10.1111/j.1365-2958.1994.tb00422.x [DOI] [PubMed] [Google Scholar]
- Berger M., Neumann A., Schulz S., Simon M., Brinkhoff T. (2011). Tropodithietic acid production in Phaeobacter gallaeciensis is regulated by N-acyl homoserine lactone-mediated quorum sensing. J. Bacteriol. 193 6576–6585. 10.1128/JB.05818-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin K., Koren S., Chin C. S., Drake J. P., Landolin J. M., Phillippy A. M. (2015). Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat. Biotechnol. 33 623–630. 10.1038/nbt.3238 [DOI] [PubMed] [Google Scholar]
- Blanchet E., Prado S., Stien D., Oliveira da Silva J., Ferandin Y., Batailler N., et al. (2017). Quorum sensing and quorum quenching in the mediterranean seagrass Posidonia oceanica microbiota. Front. Mar. Sci. 4:218 10.3389/fmars.2017.00218 [DOI] [Google Scholar]
- Buchan A., Gonzalez J. M., Moran M. A. (2005). Overview of the marine Roseobacter lineage. Appl. Environ. Microbiol. 71 5666–5677. 10.1128/AEM.71.10.5665-5677.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case R. J., Labbate M., Kjelleberg S. (2008). AHL-driven quorum-sensing circuits: their frequency and function among the Proteobacteria. ISME J. 2 345–349. 10.1038/ismej.2008.13 [DOI] [PubMed] [Google Scholar]
- Cataldi T. R., Bianco G., Palazzo L., Quaranta V. (2007). Occurrence of N-acyl-L-homoserine lactones in extracts of some Gram-negative bacteria evaluated by gas chromatography-mass spectrometry. Anal. Biochem. 361 226–235. 10.1016/j.ab.2006.11.037 [DOI] [PubMed] [Google Scholar]
- Cho B. C., Azam F. (1988). Major role of bacteria in biogeochemical fluxes in the ocean’s interior. Nature 332 441–443. 10.1038/332441a0 19759822 [DOI] [Google Scholar]
- Chu W., Vattem D. A., Maitin V., Barnes M. B., McLean R. J. (2011). Bioassays of quorum sensing compounds using Agrobacterium tumefaciens and Chromobacterium violaceum. Methods Mol. Biol. 692 3–19. 10.1007/978-1-60761-971-0_1 [DOI] [PubMed] [Google Scholar]
- Cude W. N., Buchan A. (2013). Acyl-homoserine lactone-based quorum sensing in the Roseobacter clade: complex cell-to-cell communication controls multiple physiologies. Front. Microbiol. 4:336. 10.3389/fmicb.2013.00336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cude W. N., Prevatte C. W., Hadden M. K., May A. L., Smith R. T., Swain C. L., et al. (2015). Phaeobacter sp. strain Y4I utilizes two separate cell-to-cell communication systems to regulate production of the antimicrobial indigoidine. Appl. Environ. Microbiol. 81 1417–1425. 10.1128/AEM.02551-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alvise P. W., Magdenoska O., Melchiorsen J., Nielsen K. F., Gram L. (2014). Biofilm formation and antibiotic production in Ruegeria mobilis are influenced by intracellular concentrations of cyclic dimeric guanosinmonophosphate. Environ. Microbiol. 16 1252–1266. 10.1111/1462-2920.12265 [DOI] [PubMed] [Google Scholar]
- Dang H., Lovell C. R. (2016). Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 80 91–138. 10.1128/MMBR.00037-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decho A. W., Visscher P. T., Ferry J., Kawaguchi T., He L., Przekop K. M., et al. (2009). Autoinducers extracted from microbial mats reveal a surprising diversity of N-acylhomoserine lactones (AHLs) and abundance changes that may relate to diel pH. Environ. Microbiol. 11 409–420. 10.1111/j.1462-2920.2008.01780.x [DOI] [PubMed] [Google Scholar]
- Doberva M., Sanchez-Ferandin S., Toulza E., Lebaron P., Lami R. (2015). Diversity of quorum sensing autoinducer synthases in the Global ocean sampling metagenomic database. Aquat. Microb. Ecol. 74 107–119. 10.3354/ame01734 [DOI] [Google Scholar]
- Dong Y.-H., Zhang L.-H. (2005). Quorum sensing and quorum-quenching enzymes. J. Microbiol. 43 101–109. [PubMed] [Google Scholar]
- Flemming H. C., Wingender J. (2010). The biofilm matrix. Nat. Rev. Microbiol. 8 623–633. 10.1038/nrmicro2415 [DOI] [PubMed] [Google Scholar]
- García-Contreras R., Nunez-Lopez L., Jasso-Chávez R., Kwan B. W., Belmont J. A., Rangel-Vega A., et al. (2015). Quorum sensing enhancement of the stress response promotes resistance to quorum quenching and prevents social cheating. ISME J. 9:115. 10.1038/ismej.2014.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gram L., Grossart H. P., Schlingloff A., Kiørboe T. (2002). Possible quorum sensing in marine snow bacteria: production of acylated homoserine lactones by Roseobacter strains isolated from marine snow. Appl. Environ. Microbiol. 68 4111–4116. 10.1128/AEM.68.8.4111-4116.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge R. (2009). Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7 263–273. 10.1038/nrmicro2109 [DOI] [PubMed] [Google Scholar]
- Hmelo L., Van Mooy B. A. S. (2009). Kinetic constraints on acylated homoserine lactone-based quorum sensing in marine environments. Aquat. Microb. Ecol. 54 127–133. 10.3354/ame01261 [DOI] [Google Scholar]
- Hmelo L. R., Mincer T. J., Van Mooy B. A. S. (2011). Possible influence of bacterial quorum sensing on the hydrolysis of sinking particulate organic carbon in marine environments. Environ. Microbiol. Rep. 3 682–688. 10.1111/j.1758-2229.2011.00281.x [DOI] [PubMed] [Google Scholar]
- Hoppe H.-G. (1993). “Use of fluorogenic model substrates for extracellular enzyme activity (EEA) measurement of bacteria,” in Handbook of Methods in Aquatic Microbial Ecology, eds Kemp P. F., Cole J. J., Sherr B. F., Sherr E. B. (Boca Raton, FL: CRC Press; ), 423–431. [Google Scholar]
- Jatt A. N., Tang K., Liu J., Zhang Z., Zhang X. H. (2015). Quorum sensing in marine snow and its possible influence on production of extracellular hydrolytic enzymes in marine snow bacterium Pantoea ananatis B9. FEMS Microbiol. Ecol. 91 1–13. 10.1093/femsec/fiu030 [DOI] [PubMed] [Google Scholar]
- Joelsson A., Kan B., Zhu J. (2007). Quorum sensing enhances the stress response in Vibrio cholerae. Appl. Environ. Microbiol. 73 3742–3746. 10.1128/AEM.02804-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim O.-S., Cho Y.-J., Lee K., Yoon S.-H., Kim M., Na H., et al. (2012). Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62 716–721. 10.1099/ijs.0.038075-0 [DOI] [PubMed] [Google Scholar]
- Kiørboe T., Grossart H. P., Ploug H., Tang K. (2002). Mechanisms and rates of bacterial colonization of sinking aggregates. Appl. Environ. Microbiol. 68 3996–4006. 10.1128/AEM.68.8.3996-4006.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupke A., Hmelo L. R., Ossolinski J. E., Mincer T. J., Van Mooy B. A. S. (2016). Quorum sensing plays a complex role in regulating the enzyme hydrolysis activity of microbes associated with sinking particles in the ocean. Front. Mar. Sci. 3:55 10.3389/fmars.2016.00055 [DOI] [Google Scholar]
- Lee C., Wakeham S., Arnosti C. (2004). Particulate organic matter in the sea: the composition conundrum. Ambio 33 565–575. 10.1579/0044-7447-33.8.565 [DOI] [PubMed] [Google Scholar]
- Lin L., Dai S., Tian B., Li T., Yu J., Liu C., et al. (2016). DqsIR quorum sensing-mediated gene regulation of the extremophilic bacterium Deinococcus radiodurans in response to oxidative stress. Mol. Microbiol. 100 527–541. 10.1111/mmi.13331 [DOI] [PubMed] [Google Scholar]
- Lumjiaktase P., Diggle S. P., Loprasert S., Tungpradabkul S., Daykin M., Camara M., et al. (2006). Quorum sensing regulates dpsA and the oxidative stress response in Burkholderia pseudomallei. Microbiology 152 3651–3659. 10.1099/mic.0.29226-0 [DOI] [PubMed] [Google Scholar]
- Lyon G. J., Novick R. P. (2004). Peptide signaling in Staphylococcus aureus and other gram-positive bacteria. Peptides 25 1389–1403. 10.1016/j.peptides.2003.11.026 [DOI] [PubMed] [Google Scholar]
- Manefield M., Rasmussen T. B., Henzter M., Andersen J. B., Steinberg P., Kjelleberg S., et al. (2002). Halogenated furanones inhibit quorum sensing through accelerated LuxR turnover. Microbiology 148 1119–1127. 10.1099/00221287-148-4-1119 [DOI] [PubMed] [Google Scholar]
- Martinez J., Smith D. C., Steward G. F., Azam F. (1996). Variability in ectohydrolytic enzyme activities of pelagic marine bacteria and its significance for substrate processing in the sea. Aquat. Microb. Ecol. 10 223–230. 10.3354/ame010223 [DOI] [Google Scholar]
- Mcdougald D., Rice S. A., Barraud N., Steinberg P. D., Kjelleberg S. (2011). Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Microbiol. 10:39. 10.1038/nrmicro2695 [DOI] [PubMed] [Google Scholar]
- Moran M. A., González J. M., Kiene R. P. (2003). Linking a bacterial taxon to sulfur cycling in the sea: studies of the marine Roseobacter group. Geomicrobioloy 20 375–388. 10.1080/01490450303901 [DOI] [Google Scholar]
- Murray M. G., Thompson W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8 4321–4326. 10.1093/nar/8.19.4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann A., Patzelt D., Wagner-Dobler I., Schulz S. (2013). Identification of new N-acylhomoserine lactone signalling compounds of Dinoroseobacter shibae DFL-12(T) by overexpression of luxI genes. ChemBioChem 14 2355–2361. 10.1002/cbic.201300424 [DOI] [PubMed] [Google Scholar]
- Pereira C. S., McAuley J. R., Taga M. E., Xavier K. B., Miller S. T. (2008). Sinorhizobium meliloti, a bacterium lacking the autoinducer-2 (AI-2) synthase, responds to AI-2 supplied by other bacteria. Mol. Microbiol. 70 1223–1235. 10.1111/j.1365-2958.2008.06477.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira C. S., Thompson J. A., Xavier K. B. (2013). AI-2-mediated signalling in bacteria. FEMS Microbiol. Rev. 37 156–181. 10.1111/j.1574-6976.2012.00345.x [DOI] [PubMed] [Google Scholar]
- Porsby C. H., Nielsen K. F., Gram L. (2008). Phaeobacter and Ruegeria species of the Roseobacter clade colonize separate niches in a Danish Turbot (Scophthalmus maximus)-rearing farm and antagonize Vibrio anguillarum under different growth conditions. Appl. Environ. Microbiol. 74 7356–7364. 10.1128/AEM.01738-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajput A., Kaur K., Kumar M. (2016). SigMol: repertoire of quorum sensing signaling molecules in prokaryotes. Nucleic Acids Res. 44 D634–D639. 10.1093/nar/gkv1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao D., Webb J. S., Kjelleberg S. (2005). Competitive interactions in mixed-species biofilms containing the marine bacterium Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 71 1729–1736. 10.1128/AEM.71.4.1729-1736.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland J. L., Stien D., Sanchez-Ferandin S., Lami R. (2016). Quorum sensing and quorum quenching in the phycosphere of phytoplankton: a case of chemical interactions in ecology. J. Chem. Ecol. 42 1201–1211. 10.1007/s10886-016-0791-y [DOI] [PubMed] [Google Scholar]
- Romero M., Martin-Cuadrado A. B., Otero A. (2012). Determination of whether quorum quenching is a common activity in marine bacteria by analysis of cultivable bacteria and metagenomic sequences. Appl. Environ. Microbiol. 78 6345–6348. 10.1128/AEM.01266-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero M., Martin-Cuadrado A. B., Roca-Rivada A., Cabello A. M., Otero A. (2011). Quorum quenching in cultivable bacteria from dense marine coastal microbial communities. FEMS Microbiol. Ecol. 75 205–217. 10.1111/j.1574-6941.2010.01011.x [DOI] [PubMed] [Google Scholar]
- Shaw P. D., Ping G., Daly S. L., Cha C., Cronan J. E., Rinehart K. L., et al. (1997). Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. U.S.A. 94 6036–6041. 10.1073/pnas.94.12.6036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh J., Hicks S., Dall’Agnol M., Phillips A. D., Nataro J. P. (2001). Roles for Fis and YafK in biofilm formation by enteroaggregative Escherichia coli. Mol. Microbiol. 41 983–997. 10.1046/j.1365-2958.2001.02512.x [DOI] [PubMed] [Google Scholar]
- Silver M. W., Alldredge A. L. (1981). Bathypelagic marine snow: deep-sea algal and detrital community. J. Mar. Res. 39 501–530. [Google Scholar]
- Slightom R. N., Buchan A. (2009). Surface colonization by marine Roseobacters: integrating genotype and phenotype. Appl. Environ. Microbiol. 75 6027–6037. 10.1128/AEM.01508-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. C., Simon M., Alldredge A. L., Azam F. (1992). Intense hydrolytic enzyme activity on marine aggregates and implications for rapid particle dissolution. Nature 359 139–142. 10.1038/359139a0 [DOI] [Google Scholar]
- Someya N., Morohoshi T., Okano N., Otsu E., Usuki K., Sayama M., et al. (2009). Distribution of N-acylhomoserine lactone-producing fluorescent Pseudomonads in the phyllosphere and rhizosphere of potato (Solanum tuberosum L.). Microbes Environ. 24 305–314. 10.1264/jsme2.ME09155 [DOI] [PubMed] [Google Scholar]
- Sonnenschein E. C., Nielsen K. F., D’Alvise P., Porsby C. H., Melchiorsen J., Heilmann J., et al. (2016). Global occurrence and heterogeneity of the Roseobacter-clade species Ruegeria mobilis. ISME J. 11 569–583. 10.1038/ismej.2016.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D., Waters C. M. (2012). A tangled web: regulatory connections between quorum sensing and cyclic Di-GMP. J. Bacteriol. 194 4485–4493. 10.1128/JB.00379-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surette M. G., Miller M. B., Bassler B. L. (1999). Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. U.S.A. 96 1639–1644. 10.1073/pnas.96.4.1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait K., Williamson H., Atkinson S., Williams P., Camara M., Joint I. (2009). Turnover of quorum sensing signal molecules modulates cross-kingdom signalling. Environ. Microbiol. 11 1792–1802. 10.1111/j.1462-2920.2009.01904.x [DOI] [PubMed] [Google Scholar]
- Tang K., Su Y., Brackman G., Cui F., Zhang Y., Shi X., et al. (2015). MomL, a novel marine-derived-N-acyl homoserine lactonase from Muricauda olearia. Appl. Environ. Microbiol. 81 774–782. 10.1128/AEM.02805-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang K., Zhang Y., Yu M., Shi X., Coenye T., Bossier P., et al. (2013). Evaluation of a new high-throughput method for identifying quorum quenching bacteria. Sci. Rep. 3:2935. 10.1038/srep02935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele S., Fuchs B. M., Amann R., Iversen M. H., Wommack K. E. (2015). Colonization in the photic zone and subsequent changes during sinking determine bacterial community composition in marine snow. Appl. Environ. Microbiol. 81 1463–1471. 10.1128/AEM.02570-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Mooy B. A., Hmelo L. R., Sofen L. E., Campagna S. R., May A. L., Dyhrman S. T., et al. (2012). Quorum sensing control of phosphorus acquisition in Trichodesmium consortia. ISME J. 6 422–429. 10.1038/ismej.2011.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner-Dobler I., Biebl H. (2006). Environmental biology of the marine Roseobacter lineage. Annu. Rev. Microbiol. 60 255–280. 10.1146/annurev.micro.60.080805.142115 [DOI] [PubMed] [Google Scholar]
- Wagner-Dobler I., Thiel V., Eberl L., Allgaier M., Bodor A., Meyer S., et al. (2005). Discovery of complex mixtures of novel long-chain quorum sensing signals in free-living and host-associated marine Alphaproteobacteria. ChemBioChem 6 2195–2206. 10.1002/cbic.200500189 [DOI] [PubMed] [Google Scholar]
- Weisburg W. G., Barns S. M., Pelletier D. A., Lane D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173 697–703. 10.1128/jb.173.2.697-703.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wietz M., Gram L., Jørgensen B., Schramm A. (2010). Latitudinal patterns in the abundance of major marine bacterioplankton groups. Aquat. Microb. Ecol. 61 179–189. 10.3354/ame01443 17284217 [DOI] [Google Scholar]
- Williams P. (2007). Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology 153 3923–3938. 10.1099/mic.0.2007/012856-0 [DOI] [PubMed] [Google Scholar]
- Yates E. A., Philipp B., Buckley C., Atkinson S., Chhabra S. R., Sockett R. E., et al. (2002). N-acylhomoserine lactones undergo lactonolysis in a pH-, temperature-, and acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa. Infect. Immun. 70 5635–5646. 10.1128/IAI.70.10.5635-5646.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan J., Cicirelli E. M., Mohamed N. M., Sibhatu H., Kroll S., Choi O., et al. (2012). A complex LuxR-LuxI type quorum sensing network in a roseobacterial marine sponge symbiont activates flagellar motility and inhibits biofilm formation. Mol. Microbiol. 85 916–933. 10.1111/j.1365-2958.2012.08149.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan J., Liu Y., Fuqua C., Hill R. (2014). Acyl-homoserine lactone quorum sensing in the Roseobacter clade. Int. J. Mol. Sci. 15 654–669. 10.3390/ijms15010654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Beaber J. W., Moré M. I., Fuqua C., Eberhard A., Winans S. C. (1998). Analogs of the autoinducer 3-oxooctanoyl-homoserine lactone strongly inhibit activity of the TraR protein of Agrobacterium tumefaciens. J. Bacteriol. 180 5398–5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Chai Y., Zhong Z., Li S., Winans S. C. (2003). Agrobacterium bioassay strain for ultrasensitive detection of N-acylhomoserine lactone-type quorum-sensing molecules: detection of autoinducers in Mesorhizobium huakuii. Appl. Environ. Microbiol. 69 6949–6953. 10.1128/AEM.69.11.6949-6953.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziesche L., Bruns H., Dogs M., Wolter L., Mann F., Wagner-Dobler I., et al. (2015). Homoserine lactones, methyl oligohydroxybutyrates, and other extracellular metabolites of macroalgae-associated bacteria of the Roseobacter clade: identification and functions. ChemBioChem 16 2094–2107. 10.1002/cbic.201500189 [DOI] [PubMed] [Google Scholar]
- Zoppini A., Puddu A., Fazi S., Rosati M., Sist P. (2005). Extracellular enzyme activity and dynamics of bacterial community in mucilaginous aggregates of the northern Adriatic Sea. Sci. Total Environ. 353270–286. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


