Abstract
Previously, we constructed and characterized the vaccine efficacy of attenuated S. agalactiae strain YM001 in tilapia. In this study, the potential impacts of YM001 on the tilapia intestinal microbiota were assessed by qPCR and 16S rRNA sequencing methods. The results showed that YM001 distributed unevenly in different parts of intestine, peaked in the intestine at 12 h after oral administration, and then declined gradually. YM001 caused 0% mortality of fish during the entire experimental period, while the referent strain HN016 caused 100% mortality at 3 d after oral administration. However, the intestinal microbiota could be changed by YM001, the diversity of intestinal microbiota decreased first and gradually recovered after oral administration. The diversity of intestinal microbiota of tilapia was negatively correlated with the content of HN016 in the intestinal tract. The oral YM001 mainly changed the abundance of Streptococcus, Cetobacterium, Akkermansia, Romboutsia, Bacteroides, Brevinema, Lachnospiraceae_NK4A136-group, coprothermobactter, presiomonas, and Roseburia in intestine. The present study indicate that oral administration of YM001 altered the diversity and composition of intestinal microbiota in tilapia, but these change were only temporary, non-lethal, and recoverable. The results provide a more comprehensive experimental basis for the safety of oral YM001 vaccines.
Keywords: gut microbiota, oral vaccine, Streptococcus agalactiae, tilapia, microbiology
Introduction
Streptococcus agalactiae, or Group B streptococcus (GBS) is an important pathogen causing sepsis and meningitis in newborns and mastitis in bovine (Edwards and Baker, 2005; Tazi et al., 2011; Edmond et al., 2012; Lamagni et al., 2013). S. agalactiae is also a common aquaculture pathogen that can harm different fish species, such as tilapia, Barcoo grunter (Scortum barcoo), golden pompano (Trachinotus blochii), ya-fish (Schizothorax potanini), giant queensland grouper (Epinephelus lanceolatus), and silver pomfret (Pampus argenteus) (Jafar et al., 2008; Amal et al., 2012; Li et al., 2015). Tilapia is an important aquaculture species. Outbreaks of S. agalactiae in tilapia have cost more than 40 million dollars every year worldwide (Mian et al., 2009; Chen et al., 2012). Currently, S. agalactiae control in tilapia mainly relies on antibiotics, but the overuse of antibiotics will lead to food safety and problems with drug-resistant strains. Therefore, the use of antibiotics in aquaculture is severely restricted (Depaola et al., 1995; Baquero et al., 2008).
Vaccines are the safest and most reliable method for the prevention of human and animal infectious diseases, but most of them are given by injection. However, in the field of aquatic products, the use of injections for immunization is greatly limited due to the large population of aquatic animals and the greater stress response resulted from the injection of vaccines. Compared to injectable vaccines, oral vaccines are more suitable for aquaculture animals. These vaccines are generally classified as live attenuated vaccines (LAVs), inactivated vaccines and subunit vaccines. Compared with inactivated vaccines or subunit vaccines, oral LAV have great advantages by avoiding the degradation of intestinal enzymes, directly entering the liver, kidney, spleen, and other immune related sites and simultaneously stimulating systemic immunity and local mucosal immunity (Chen et al., 2015; Makesh et al., 2015). Because of the above advantages, three types of LAVs against S. agalactiae in tilapia have been developed recently (Pridgeon and Klesius, 2013; Huang et al., 2014; Li et al., 2015).
Although oral LAVs have many advantages in application, their effects on the intestinal microbiota cannot be ignored. Long-term natural selection and differences in habitat, diet and habits have led to the formation of unique microbial symbiosis among different animals (Rawls et al., 2004; Ley et al., 2008). For fish, the micro-ecosystem composed of gut microorganisms is also formed during long-term historical evolution. These microorganisms are involved in the development of fish epithelial barrier, nutrition, digestion, as well as immunity (Gómez and Balcazar, 2008; Kelly, 2010; Roeselers et al., 2011; Sommer and Backhed, 2013; Xia et al., 2014). However, the balance of intestinal community composition could be altered by many factors (Xia et al., 2014), including stress (O'mahony et al., 2009; Kelly, 2010), antibiotic exposure (Tanaka et al., 2009), nutritional status (Turnbaugh et al., 2009), age (Hopkins et al., 2002), degree of hygiene (Schmidt et al., 2011), and bacterial infection (Dethlefsen et al., 2006). Gut microorganisms are continuously shaping the development of their host's immune system, directly modulating the innate and adaptive immune responses (Sommer and Backhed, 2013; Barroso-Batista et al., 2015). Differences in the host's intestinal microbiota can affect the effectiveness of the vaccine. For instance, intestinal microbiome composition correlates significantly with the immunogenicity of rotavirus vaccine (RVV) and may contribute to the diminished RVV immunogenicity. RVV response is correlated with an increased abundance of Streptococcus bovis and a decreased abundance of Bacteroidetes phylum (Harris et al., 2017). Therefore, it is necessary to analyze the effect of oral vaccines on intestinal microbiota.
In the previous study, we attenuated and domesticated the virulent S. agalactiae strain HN016 and obtained a safe, stable, and highly immunogenic attenuated S. agalactiae strain YM001. Oral immunization of tilapia with this strain produced a good immune protection (Chen et al., 2012; Li et al., 2015; Wang et al., 2015). Despite this, the effect of oral administration of YM001 on tilapia intestinal microbiota is still unknown. In this study, we analyzed the pathogenicity of oral administration of YM001 as well as its distribution in the intestine and influence on intestinal microbiota in tilapia using quantitative PCR and 16S bacterial sequencing with the hope to provide a more comprehensive experimental basis for the safety of YM001 oral vaccines.
Materials and Methods
Ethics Statement
This study was carried out in accordance with the principles of good animal practice as defined by the European Union guidelines for the handling of laboratory animals (http://ec.europa.eu/environment/chemicals/lab_animals/home_en.htm) and the protocol was approved by Guangxi Institute of Fisheries Animal Ethics Committee and Guangxi Medical University Animal Ethics Committee. The institute did not issue a number or ID to this animal study, because the studied fish are not an endangered or protected species.
Bacterial Strains and Fish
S. agalactiae strain HN016 (serotype Ia) was isolated from an outbreak epidemical disease in tilapia from Hainan, China in 2010 (Chen et al., 2012). S. agalactiae strain YM001 was obtained by continuously passaging HN016 in vitro for 840 passages (Li et al., 2015). These two strains were stored at −80°C. Non-infected Nile tilapia was obtained from the National Tilapia Seed Farm (Nanning, Guangxi, China), with average weight of 155.45 ± 20.85 g. Prior to experiments, the fishes were acclimated in plastic tanks (800 L each) with a stocking rate of 4 g/L at 30 ± 4°C for 2 weeks. The experimental fishes were checked randomly to verify pathogen free by bacteriological analysis of the brain and kidney samples.
Oral Administration
The bacteria for oral gavage were recovered and cultured as described previously (Li et al., 2015). In brief, the stored HN016 and YM001 were thawed, streaked onto 5.0% sheep blood agar plates, and incubated at incubator (Jing Hong, China) 28°C for 24 h. Then single colony of every strain was inoculated in sterile tryptone soy broth (TSB) and incubated at 28°C for 24 h under low agitation. The concentration of the bacteria was determined as colony forming unit (CFU) per mL by plating 100 μL of 10-fold serial dilutions onto blood agar plates. The details are as follow: 100 μL culture of HN016 or YM001was added to 900 μL PBS and blended. Then 100 μL diluent was added to another 900 μL PBS and blended. By the same method, the culture was 10-fold serial diluted to different concentration. Hundred micro liter diluent with different concentration were plated onto the blood agar plates and then incubated at 28°C for 24 h. Only the agar plates with 30–100 colony of bacteria was selected to calculated the concentration of primary culture.
A total of 105 fishes were orally administered with HN016 or YM001 at the dose of 1.0 × 109 CFU/fish (35 fishes/tank, with 3 replicates), and the control group was treated with 1.0 mL/fish of TSB. The experimental fishes were cultured in plastic tanks (800 L each) at 30 ± 4°C, then monitored and fed twice a day for 30 days. Five fishes in each group were sampled randomly at 0h, 12h, 24h, 3d, 7d, 15d after oral gavage. Sampled fishes were dissected immediately with sterile scissors. The intestine was aseptically removed from their abdominal cavity, and cut into duodenum, foregut, midgut and hindgut, separately. All the samples then were stored at −80°C immediately until use.
DNA Extraction and Real-Time RT-PCR Assays
Microbial DNA was extracted from fish intestine samples by using the Bacterial DNA Kit (50) (Takara, Japan) following the manufacturer's protocol. For each sample, DNA was extracted in triplicate to avoid bias, and the extracts from the same sample were pooled. The purity of DNA extracts was verified by electrophoresis on ethidium bromide staining 1% agarose gels, and their concentration was analyzed spectrophotometrically. To detect the distribution of YM001 or HN016 in tilapia intestines, specific primers and probe for S. agalactiae were design according to the cfb gene (Gene ID: 3686873). The Primer and probe sequences are listed in Table 1. Real-time PCR assays were performed using the Premix Ex Taq according to the protocol of Probe qPCR Kit (Takara, Japan) in a LightCycler 480 System (Roche, Germany). The PCR assays were performed at a volume of 20.0 μL comprised of 10.0 μL of SYBR Green I Master Mix (Takara, Japan), 0.5 μL of cfb-F, 0.5 μL of cfb-R, 0.5 μL of TaqMan Probe, 4.0 μL of template DNA and 7.5 μL of ddH2O. The cycling parameters were 30 s at 95°C followed by 40 cycles of 5 s at 95°C 5 and 30 s at 60°C. In blank control group, ddH2O was used to replace the template DNA. All Real-time PCR reactions were performed in triplicate.
Table 1.
Sequences of primers and probe.
| Primer | Sequence (5′ to 3′) |
|---|---|
| cfb-F | CGGTTAATGAGGCTATTACTAGTG |
| cfb-R | ATCTGTTAAGGCTTCTACACGAC |
| cfb-P probe | FAM-TTCATTGCGTGCCAACCCTGAGACA-Eclipse |
Illumina HiSeq Sequencing
The extracted genomic DNA samples were diluted to 1 ng/μl with sterile water and used as templates to amplify the selected sequencing region using specific primers with barcodes and DNA polymerase with high efficiency and high fidelity. The PCR products were separated on 2% agarose gels, purified and prepared as libraries using TruSeq® DNA PCR Free Sample Preparation Kit. The constructed libraries were analyzed using Tapestation for quality control and Qubit and qPCR for quantification and sequenced using a Hiseq 2500PE250 platform.
The obtained FASTQ data were demultiplexed. After removal of barcodes and poor quality data, the obtained effective reads were analyzed using Upplese software (Uparse v7.0.1001, http://drive5.com/uparse/) and clustered into operational taxonomic units (OTUs) based on 97% identity. The sequences of each OUT with the highest abundance were selected as the representative sequence and used for annotation against the GreenGene database (http://greengenes.secondgenome.com) using the RDP Classifier (Version 2.2, http://sourceforge.net/projects/rdp-classifier/) at the setting threshold of 0.8–1 to calculate the community composition of each sample at the levels of kingdom, phylum, class, order, family, genus, and species.
The data for each sample was normalized and subjected to Alpha diversity analysis and Beta diversity analysis.
Results
Disease Symptoms of the Experimental Fish
All fish in the YM001 group showed no significant changes except that their appetite dropped at 24–72 h after oral administration of YM001. The appetite returned to normal at 4 d after the administration. No fish in the YM001 group died during the entire experiment period. By contrast, fish in the HN016 group showed typical symptoms of S. agalactiae infection at 24 h after administration of HN016 including lack of energy, abnormal swimming posture and body blackness and died 3 d after administration of HN016.
Distribution of YM001 in Fish Intestine
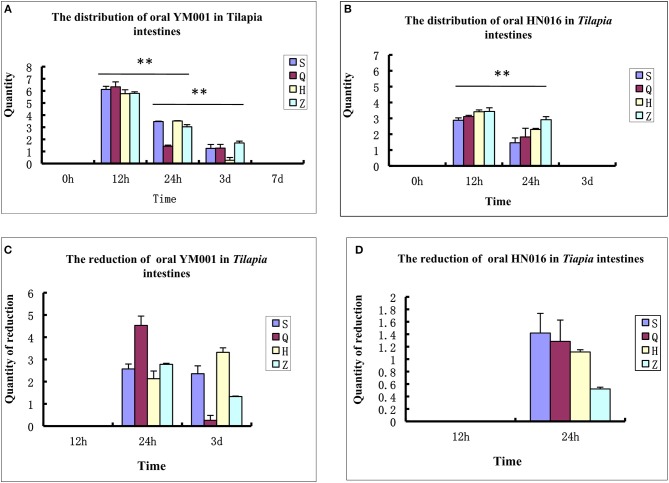
Tilapia intestinal samples were collected at 0 h, 12 h, 24 h, 3 d, 7 d, and 15 d after oral administration of S. agalactiae, respectively. Their DNA were extracted and analyzed using fluorescence quantitative PCR to detect the distribution and colonization of YM001 and HN016 in the intestine of tilapia. The results showed that both YM001 and HN016 can be colonized in the foregut, rectum, duodenum and hindgut, but their number and distribution were significantly different. The contents of YM001 and HN016 were the highest in all intestinal segments at 12 h and gradually decreased with time (Figures 1A,B). The relative reductions of YM001 and HN016 in different segments of the intestine were different. The reduction of YM001 was in the order of foregut> rectum> duodenum> hindgut, and hindgut> duodenum> rectum> foregut at 24 h−3d (Figure 1C) while that of HN016 was duodenum> foregut> hindgut> rectum (Figure 1D).
Figure 1.
The retention time of YM001 and HN016 is different in the intestinal tract. (A) The distribution of YM001 in Tilapia intestines. (B) The distribution of HN016 in Tilapia intestines. (C) The reduction of YM001 in Tilapia intestines. (D) The reduction of HN016 in Tilapia intestines. Real-time RT-PCR was performed in triplicate for each sample. The data were provided as the mean fold changes (means ± S.D., n = 3) relative to the control group. The statistical significance was calculated by the Student's t-test (**p < 0.01).
16S rRNA Gene Sequencing Analysis of Intestinal Microbiota in the Experimental Tilapia
The V3-V4 region of the 16S rRNA gene of each sample was sequenced on the Illumina HiSeq platform. A total of 2,891,942 original sequences and 2201874 effective reads were obtained from 36 samples. The total effective rate was 76.13%. The number of original sequences per sample was 63,842–92,065 and the effective read number was 48,541–70,400. The average length (AvgLen) was 405–426 nt (Table 2). To study the compositional diversity information of the microbiota, the effective sequences of all samples were clustered using the Uparse software into Operational Taxonomic Units (OTUs) according to 97% sequence similarity. A total of 964 OTUs were obtained from 36 tilapia intestinal samples with average OTUs of 339–1,623 per sample.
Table 2.
Statistics the data of the samples sequencing.
| ID | Raw reads number | Effective reads number | Effective reads ratio (%) | AvgLen (bp) |
|---|---|---|---|---|
| S1 | 67,202 | 51,024 | 75.93 | 407 |
| Q1 | 87,705 | 63,545 | 72.45 | 415 |
| H1 | 88,978 | 63,048 | 70.86 | 418 |
| Z1 | 80,330 | 61,860 | 77.00 | 400 |
| RS1 | 85,012 | 66,061 | 77.71 | 411 |
| RQ1 | 69,790 | 53,911 | 77.25 | 399 |
| RH1 | 77,753 | 60,507 | 77.82 | 411 |
| RZ1 | 87,127 | 68,186 | 78.26 | 414 |
| RS2 | 82,023 | 60,768 | 74.09 | 423 |
| RQ2 | 77,184 | 56,278 | 72.91 | 425 |
| RH2 | 90,328 | 67,106 | 74.29 | 425 |
| RZ2 | 92,065 | 68,313 | 74.20 | 424 |
| RS3 | 79,045 | 60,180 | 76.13 | 414 |
| RQ3 | 80,394 | 62,708 | 78.00 | 408 |
| RH3 | 76,219 | 59,523 | 78.09 | 413 |
| RZ3 | 88,974 | 68,382 | 76.86 | 411 |
| RS4 | 81,896 | 61,065 | 74.56 | 413 |
| RQ4 | 84,224 | 67,639 | 80.31 | 412 |
| RH4 | 75,255 | 58,985 | 78.38 | 410 |
| RZ4 | 80,641 | 64,536 | 80.03 | 413 |
| RS5 | 64,970 | 48,541 | 74.72 | 404 |
| RQ5 | 80,847 | 63,449 | 78.48 | 407 |
| RH5 | 75,724 | 60,644 | 80.09 | 407 |
| RZ5 | 89,025 | 70,400 | 79.08 | 409 |
| RS6 | 83,729 | 60,600 | 72.38 | 409 |
| RQ6 | 63,842 | 50,847 | 79.65 | 403 |
| RH6 | 81,714 | 59,235 | 72.49 | 407 |
| RZ6 | 71,835 | 55,227 | 76.88 | 412 |
| QS1 | 90,604 | 67,067 | 74.02 | 422 |
| QQ1 | 86,118 | 65,332 | 75.86 | 416 |
| QH1 | 69,187 | 52,006 | 75.17 | 415 |
| QZ1 | 76,567 | 58,042 | 75.81 | 419 |
| QS2 | 82,843 | 58,386 | 70.48 | 426 |
| QQ2 | 82,196 | 63,483 | 77.23 | 412 |
| QH2 | 77,184 | 60,891 | 78.89 | 425 |
| QZ2 | 83,312 | 64,101 | 76.94 | 418 |
Distribution of Microorganisms in Various Intestinal Segments of Tilapia Before Oral Administration of S. agalactiae
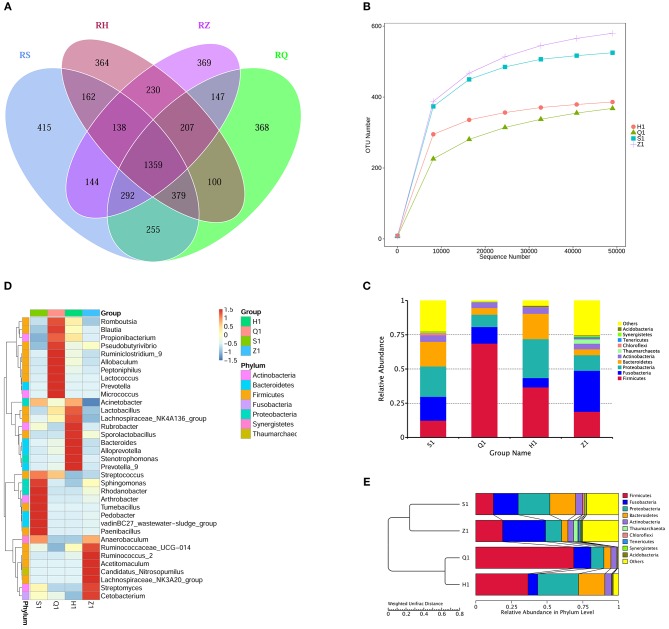
Analysis of the Venn diagram of intestinal microbiota showed that 1,359 OTUs were shared in the intestine of normal tilapia, and the number of OTUs unique to the duodenum, foregut, hindgut, rectum was 415, 365, 364, and 469, respectively (Figure 2A).
Figure 2.
The intestinal flora in different segments of tilapia were different in structure and diversity. (A) Venn diagram (Petals figure) illustrating the Common or characteristic OTUs in duodenum (S), foregut (Q), midgut (Z), hindgut (H). (B) Rarefaction curves of OTUs illustrating the microbial diversity in duodenum (S), foregut (Q), midgut (Z), hindgut (H). When the curve flattens, it shows that the amount of sequencing data is gradually reasonable, and more data will produce only a small number of new species (OTUs). (C) Column diagram illustrating the relative abundance of Intestinal flora at phylum level in duodenum (S), foregut (Q), midgut (Z), hindgut (H). The horizontal axis is the sample name, and the vertical axis represents the relative abundance of a classification. Different colors correspond to different species at the same level. (D) Heat maps illustrating the relative abundance of Intestinal flora at genus level in duodenum (S), foregut (Q), midgut (Z), hindgut (H). The color and intensity of each square represent the value of relative abundance. (E) UPGMA clustering tree illustrating the similarity between samples of duodenum (S), foregut (Q), midgut (Z), hindgut (H).
At the phylum level, the dominant microbial floras in tilapia intestine are mainly firmicutes, fusobacterium, proteobacteria, bacteroidetes, actinobacteria, thaumarchaeota, chloroflexi, tenericutes, verrucomicrobia, synergistetes, and actidobacteria (Figure 2C). The dominant microbiota in tilapia duodenum were proteobacteria (22.14.38%), bacteroidetes (18.02%), fusobacterium (17.41%), firmicutes (12.39%), actinobacteria (4.77%), chloroflexi (1.79%), and Others (23.37%); The dominant microbiota in tilapia foregut were Firmicutes (68.67%), fusobacterium (12.12%), proteobacteria (8.99%), bacteroidetes (4.86%), actinobacteria (4.23%), and others (1.12%); The dominant microbiota in tilapia hindgut were Firmicutes (36.66%), proteobacteria (28.41%), bacteroidetes (18.56%), fusobacterium (6.84%), actinobacteria (4.68%), Chloroflexi (1.79%), Others (23.37%); The dominant microbiota in tilapia rectum were fusobacterium (29.96%), firmicutes (18.86%), proteobacteria (11.35%), bacteroidetes (4.32%), actinobacteria (4.08%), thaumarchaeota (2.99%), tenericutes (1.29%), and others (27.14%).
At genus level, different positions of tilapia intestine also have different dominant bacterial structures (Figure 2D). The dominant microbiota in tilapia duodenum were streptococcus, sphingomonas, rhodanobacter, arthrobacter, tumebacillus, pedobacter, vadinBC27_wastewater-sludge_group; paenibacillus and anaerobaculum; the dominant microbiota in tilapia foregut were romboutsia, blautia, propionibacterium, pseudobutyrivibrio, ruminiclostrid_9, allobaculum, peptoniphilus, lactococcus, prevotella and micrococcus; the dominant microbiota in tilapia hindgut were Acinetobacter, Lactobacillus, Lachnospiraceae_NK4A136_group, Rubrobacter, Sporolactobacillus, Bacteroides Alloprevotella, Stenotrophomonas and Prevotella_9; and the dominant microbiota in tilapia rectum were Anaerobaculum, Ruminococcaceae_UCG-014, Ruminococcus_2, Acetitomaculum, Candidatus_Nitrosopumilus, Lachnospiraceae_NK3A20_group, Streptomyces, Cetobacterium.
The clustering tree analysis showed that the microbiota in tilapia foregut and hindgut were more similar than that in the duodenum and rectum (Figure 2E). The dilution curve can directly reflect the rationality of the sequencing data and indirectly reflect the abundance of species in the sample. Our results in the dilution curves showed that the diversity of microbes in different gut segments of tilapia was different, showing an order of rectum> duodenum> hindgut> foregut in species richness (Figure 2B).
Effect of YM001 on Intestinal Microbiota in Tilapia
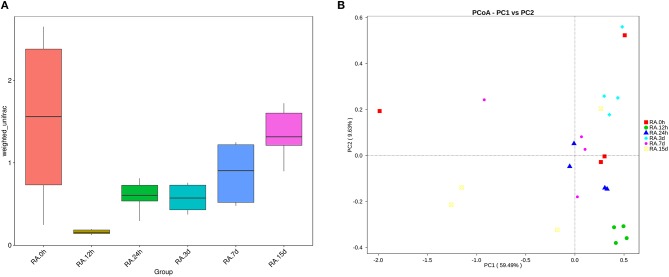
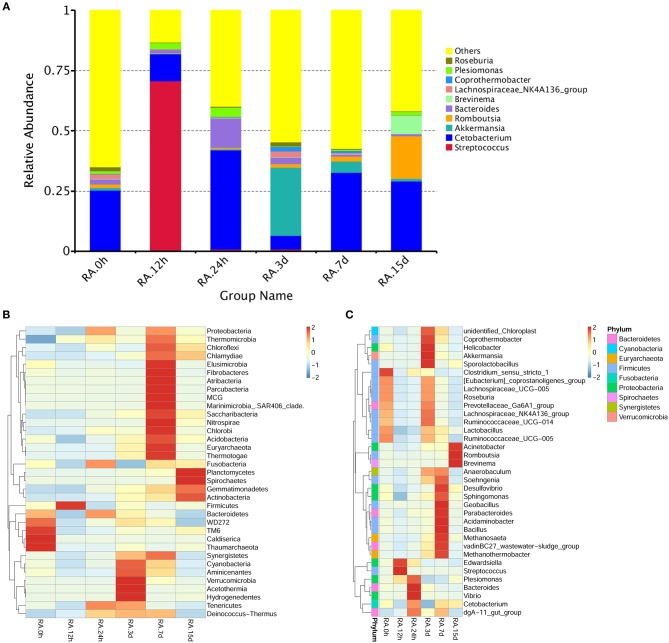
β diversity analysis of intestinal samples of tilapia found that the diversity of gut microbiota lowered after oral administration of YM001, reaching the lowest at 12 h and gradually recovered afterward to the normal level at 15 d (Figure 3). Furthermore, YM001 significantly changed the composition of microbiota in tilapia intestine, and the relative proportion of dominant bacteria fluctuated at different time (Figure 4A). At genus level, the top 10 intestinal microbe were Streptococcus, Cetobacterium, Akkermansia, Romboutsia, Brevinema, Lachnospiraceae_NK4A136-group, Coprothermobactter, Plesiomonas, and Roseburia after the YM001 administration. The abundance of Streptococcus in the intestine reached the highest level (70.8%) at 12 h after YM001 administration, then dramatically decreased at 24 h after YM001 administration, and almost restored to the normal level at 5 d and 7 d after YM001 administration (Table 3). The trend of changes in Streptococcus abundance is consistent with that measured by fluorescence quantitative PCR (Figure 1). Cetobacterium accounted for approximately 25% of the total bacteria before oral administration of YM001 and showed a significant decrease at 12 h after administration. After that, the abundance of Cetobacterium rebounded to 25% at 24 h, decreased again at 3 d after administration and almost returned to the original level at 7 and 15 d after administration (Table 3). The abundance of Bacteroides was about 2.2% before oral administration of YM001 but increased to 12.4% at 24 h after administration and then gradually decreased to the original level later (Table 3). Akkermansia accounted for about 1% of the total bacteria before oral administration of YM001, but the abundance rose to 28.3% at 3 d after oral administration of YM001 and then gradually decreased at 5 and 7 d (Table 3). Romboutsia accounted for about 1.5% of the total bacteria before oral administration of YM001, but the abundance reduced to 0.8% at 12 h after oral administration of YM001 and then gradually increased to 17.7% at 15 d (Table 3). The content of Brevinema increased to 7% at 15 d after oral administration of YM001, and was less than 1% at all other times (Table 3).
Figure 3.
YM001 alters the diversity of intestinal flora of tilapia at different observing times. (A) A box chart based on Weighted Unifrac Beta diversity. (B) Principle coordinate analysis (PCoA) Based on Weighted Unifrac distance. The samples at 0 h, 12 h, 24 h, 3 d, 7d, 15d were represented as color icons with red square, green dot, blue triangle, turquoise square, pink dot, orange box.
Figure 4.
YM001 alters the intestinal flora composition at different observing times. (A) The relative abundances of the top 10 bacteria at the genus level in YM001 groups. (B) The alteration of intestinal bacterial patterns at the genus level. The heat map is color-based on row Z-scores. The highest and lowest bacterial levels are showed in red and blue, respectively. (C) The alteration of intestinal bacterial patterns at the phylum level. The heat map is color-based on row Z-scores. The highest and lowest bacterial levels are showed in red and blue, respectively.
Table 3.
The composition of the top ten intestinal flora in YM001 group.
| Taxonomy | Strep | Cetob | Akker | Romb | Bacte | Brevi | Lachn | Copro | Plesi | Roseb | Others |
|---|---|---|---|---|---|---|---|---|---|---|---|
| RA.0h | 0.005012 | 0.247655 | 0.01045 | 0.015298 | 0.022154 | 0.001102 | 0.019525 | 0.001347 | 0.011202 | 0.017022 | 0.649234 |
| RA.12h | 0.708163 | 0.110283 | 0.002242 | 0.002841 | 0.014807 | 0.000169 | 0.001974 | 0.00042 | 0.024902 | 0.001407 | 0.132791 |
| RA.24h | 0.008764 | 0.411216 | 0.003523 | 0.006643 | 0.124136 | 0.000976 | 0.003 | 0.000807 | 0.039513 | 0.002274 | 0.399147 |
| RA.3d | 0.008623 | 0.056982 | 0.283192 | 0.014076 | 0.028616 | 0.000774 | 0.024422 | 0.018505 | 0.001767 | 0.016662 | 0.54638 |
| RA.7d | 0.004887 | 0.322307 | 0.046462 | 0.020419 | 0.009822 | 0.000862 | 0.005208 | 0.007788 | 0.004058 | 0.004483 | 0.573704 |
| RA.15d | 0.001756 | 0.290315 | 0.009332 | 0.177731 | 0.00895 | 0.076065 | 0.002476 | 0.00018 | 0.013984 | 0.001189 | 0.418023 |
Strep, Streptococcusl; Cetob, Cetobacterium; Akker, Akkermansia; Romb, Romboutsia; Bacte, Bacteroides; Brevi, Brevinema; Lachn, Lachnospiraceae_NK4A136_group; Copro, Coprothermobacter; Plesi, Plesiomonas; Roseb, Roseburia.
The contents of Lachnospiraceae_NK4A136-group, Coprothermobactter, Plesiomonas, and Roseburia in the intestine also fluctuated but to less extent compared with the other genera. Before oral administration of YM001, the content of Lachnospiraceae_NK4A136-group was 7% in duodenum and <1% in other segments, after oral administration of YM001, its content showed a decreasing trend (Table 3). The content of Presiomonas was approximately 0.4% in total intestinal bacteria in the case of normal, but after oral administration of YM001, its content rose to 2.4% at 12 h and 3.9% at 24 h, and then gradually reduced to 1.3% at 15 d, close to the initial level (Table 3). Before oral administration of YM001, the content of Coprothermobactter in each gut segment was <0.1%. However, at 3 d after administration of YM001, its content in the foregut increased to 7% and then returned to the normal level at 7 and 15 d after administration of YM001 (Table 3). Before oral administration of YM001, the content of Roseburia in each gut segment was < 0.1% except that of 6.3% in duodenum. After administration, its content in duodenum gradually decreased, reaching to the normal level at 7 and 15 d after administration of YM001 (Table 3).
In addition to the above-mentioned predominant bacteria, oral administration of YM001 also caused changes in other non-predominant gut bacteria. The heat maps show the changes in the highest 35 gut bacteria at both phylum and genus levels (Figures 4B,C).
Differences of Gut Microbial Changes in Tilapia Subjected to Oral Administration of YM001 and HN016
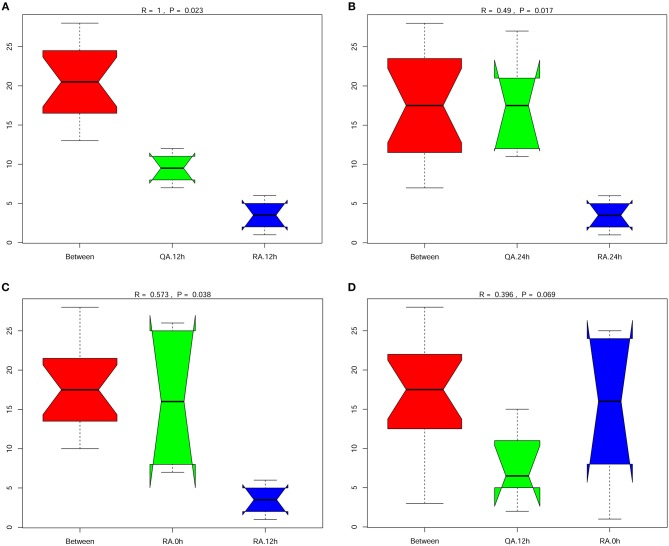
Anosim analysis is a non-parametric test used to examine whether the differences between groups are significantly greater than the differences within groups. Comparison of intestinal microbiota composition between any two group, YM001 and HN016 or normal control group (0 h), showed that all the R-values of Anosim analysis were >0 (Figures 5A–D), indicating that the composition of intestinal microbiota between any two group was significantly different and the difference between the groups were greater than that within the group.
Figure 5.
Significant difference between the group oral with YM001 and the group fed with HN016. (A) The differences between group R (YM001) and group Q (HN016) at 12 h by Anosim analysis. (B) The differences between group R (YM001) and group Q (HN016) at 24 h. (C) The differences of group R (YM001) between 12 h and 24 h. (D) The differences of group Q (HN016) between 12 and 24 h. R-value is between (−1, 1) and R-value is >0, indicating significant difference between groups. R-value is < 0, indicating that the difference in the group is greater than that between groups. The credibility of statistical analysis is expressed in p-value, and P < 0.05 indicates that the statistics are significant.
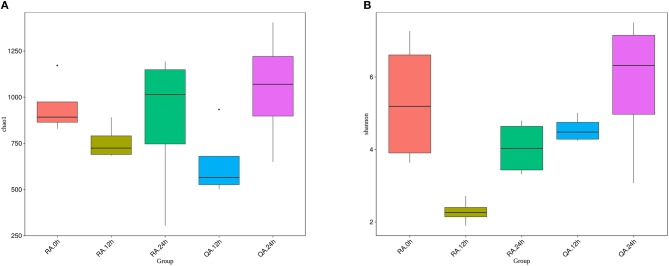
To determine whether two different S. agalactiae strain altered the composition of the gut microbiota in a similar pattern, we compared the diversity and richness indices of intestinal microbiota of tilapia at different periods after administration of YM001 and HN016. The results showed that both Chao1 index and Shannon index dropped at 12 h after administration of YM001 or HN016, then increased again at 24 h. Especially, the Chao1 index of YM001 or HN016 groups at 24 h was higher than that of the control groups. Except the Chao1 index at 12 h, the Chao1 index and the Shannon index of YM001 groups at all other time points were lower than those of HN016 groups (Figures 6A,B). These data indicated that the richness of the intestinal microbiota of tilapia at 12 h after administration of YM001 or HN016 were attenuated, especially in tilapia after 12 h after oral administration of YM001.
Figure 6.
The diversity of intestinal flora of tilapia, which fed with YM001 or HN016, changed differently. (A) Diversity of bacterial species as indicated by Chao1 rarefaction measure. The larger the number of OTU in the sample is, the larger the number of species in the sample. (B) Shannon index. The richness and evenness of the species composition in the sample were evaluated. The larger the value was, the more abundant the species was, and the more homogeneous the species were. Evenness based on species-level operational taxonomic units (OTUs). Error bars indicate SD,* indicates significant association (0.005 < P < 0.05).
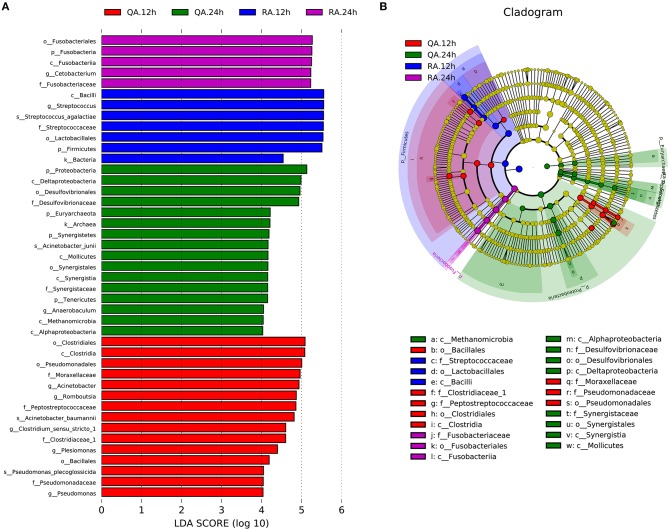
LDA effect size (LEfSe) analysis is used to identify biomarkers or the species that have statistically significant differences between two groups. Comparing with other groups, the following biomarkers, including k__Bacteria, p__Firmicutes, c__Bacilli, o__Lactobacillales, f__Streptococcaceae, g__Streptococcus and s__Streptococcus_agalactiae, of YM001 group at 12 h were most different (Figures 7A,B). On other hand, the following biomarkers, including p__Fusobacteria, c__Fusobacteriia, o__Fusobacteriales, f__Fusobacteriaceae, g__Cetobacterium, of YM001 group at 24 h were most different compared with other groups. In addition, the biomarkers c__Clostridia, o__Clostridiales, o__Pseudomonadales, o__Bacillales, f__Clostridiaceae_1, f__Pseudomonadaceae, f__Peptostreptococcaceae, f__Moraxellaceae, g__Pseudomonas, g__Plesiomonas, g__Romboutsia, g__Acinetobacter, g__Clostridium_sensu_stricto,s__Acinetobacter_baumannii, s__Pseudomonas_plecoglossicida were significantly different in the intestine of tilapia at 12 h after administration of subjected to virulent S. agalactiae strain HN016 compared with other groups; and the biomarkers k__Archaea, p__Tenericutes, p__Synergistetes, p__Euryarchaeota, p__Proteobacteria, c__Alphaproteobacteria, c__Methanomicrobia, c__Mollicutes, c__Synergistia c__Deltaproteobacteria, o__Synergistales, o__Desulfovibrionales, f__Desulfovibrionaceae, f__Synergistaceae, g__Anaerobaculum, and s__Acinetobacter_junii were significantly different in the intestine of tilapia at 24 h after administration of subjected to HN016 compared with other groups (Figures 7A,B).
Figure 7.
YM001 and HN016 alter the intestinal flora composition differently. (A) The taxa whose abundance differed between the RA samples (blue and violet bars) and QA samples (red and green bars). The cutoff value of ≥4.0 used for the linear discriminant analysis (LDA) is shown. (B) Taxonomic cladogram obtained from LEfSe sequence analysis (relative bundance ≥ 0.5%). Biomarker taxa are highlighted by colored circles and shaded areas (RA samples are shown in blue or violet and QA are shown in red or green). Each circles diameter reflects the abundance of that taxa in the community.
Discussion
For oral attenuated vaccines, safety and efficacy are critical. In this study, each tilapia was given 109 YM001 or 109 HN016 via oral gavage, respectively. Compared with 100% mortality of fish given HN016 within 3 d, the mortality of fish given YM001 was 0% within 15 d, indicating that YM001 has higher safety. The decrease of virulence of YM001 was related to the change of its genetic structure. Our previous data have shown that compared to HN016, YM001 lost two 5832bp, and 11,116 bp DNA fragments and possesses more than 40 genes with deletions or point mutations (Wang et al., 2015).
Live vaccines administrated via gavage in the intestine could stimulate immunity through two routes. On the one hand, live bacteria will colonize on the mucosa of intestine, stimulating mucosal immunity. On the other hand, these bacteria will break through the intestinal barrier and enter other immune organs, stimulating systemic immunity. Therefore, the number of colonization in the intestine, retention time, and intestinal barrier breakthrough efficiency of YM001 are related to the immune efficacy of the YM001 oral vaccine. In this study, real-time quantitative PCR was used to detect the retention of YM001 and HN016 in the intestine of tilapia. It was found that the content of both was the highest at 12 h after administration and then greatly reduced to almost undetectable at 3d (HN016) or 7d (YM001) after administration (Figure 1A). The above results show that the intestinal environment of tilapia is not suitable for long-term colonization of YM001. The intestine as well as its intrinsic microbiota has a strong repulsive effect on YM001.
The symbiotic bacteria colonize in the intestinal mucosa of fish to form a bacterial plexus, which acts as a barrier against the colonization and proliferation of pathogens and to a certain extent protects the host against pathogens (Gómez and Balcazar, 2008; Kelly, 2010). Our previous studies showed that YM001 could be detected in the brain, liver, spleen, and kidney of tilapia at 6 h after administration of YIM001, and the levels of YM001 in the tissues gradually decreased after 24 h of YM001 administration (Li et al., 2015), indicating that although YM001 is repulsed by the intestine or intrinsic microbiota of tilapia, it can still quickly cross the intestinal barrier to reach other parts of the body. The content of YM001 in the intestine and other organs began to decrease simultaneously at 24 h after oral administration, indicating that the defense responses of intestine in tilapia are highly synchronized with the internal defense responses of the body. The responses of tilapia intestine to YM001 and HN016 may be different. Comparison of the contents of YM001 and HN016 in various segments of the intestine showed that there was still a large amount of YM001 in tilapia intestine at 3 d after oral administration, but HN016 was almost undetectable, indicating that YM001 was likely to colonize on the intestine much longer that than HN016 (Figures 1A,B). In addition, comparison of the content of YM001 and HN016 in each segment of the intestine also showed that the concentration of YM001 was higher than that of HN016 at each time point except that the concentration of YM001 was lower than that of HN016 in foregut at 24 h after administration (Figures 1A–D). There may be two reasons for this situation. One is that the virulent strain HN016 enters the intestinal barrier faster than the attenuated strain YM001, resulting in a lower content of HN016 in the intestine than that of the attenuated strain YM001. The other one is because HN016 could produce toxins, resulting in strong intestine responses, which accelerate the discharge of HN016, and eventually result in lower content in the intestine than that of the attenuated strain YM001. Regardless of the situation, the longer colonization time of the weak strain YM001 in the intestine is obviously beneficial to stimulate the body to produce long-lasting immunity.
To analyze the influence of YM001 on intestinal microbiota of tilapia, it is necessary to understand the structure of the intestinal microbiota of healthy tilapia. 16S RNA sequencing analysis showed that proteobacteria, firmicutes, fusobacterium, bacteroidetes, verrucomicrobia, spirochaetes, Euryarchaeota, actinobacteria, synergistetes, and tenericutes are the top abundant bacteria genera in the intestine of healthy tilapia (Figure 2C). Swapnil Sopan Gaikwad et al. used 16S rRNA gene sequencing on the Illumina Miseq platform and found that Fusobacteria and Proteobacteria are the dominated genera in the digestive tract of tilapia collected from rivers while Actinobacteria, Cyanobacteria, Planctomyctes, and Proteobacteria are the dominant genera in in the digestive tract of tilapia collected from lakes (Gaikwad et al., 2017). Our results are similar to those of Swapnil Sopan Gaikwad et al., but there are also some bacterial species that are inconsistent with their research, such as Verrucomicrobia, Spirochaetes, Euryarchaeota, Synergistetes, and Tenericutes. These discrepancies may be related to the experimental fish species, different living environments and diet, and may also be related to different sequencing methods and sequencing depths. Microbial colonies are widely distributed in the digestive tract and are found in almost every segment of the digestive tract. At the same time, unique colonies are formed in various segments of the digestive tract. To our best knowledge, there are no reports about the bacterial structures in different segments of the digestive tract in tilapia. We found that there are significant differences in the microbial composition at different segments of the digestive tract of tilapia (Figures 2A,B). The differences in microbiota at different parts of segments of the digestive tract are closely related to their physiological characteristics.
The diversity and abundance of intestinal microbiota in animals may change due to factors such as age, dietary changes, medication, and infection (Xia et al., 2014). One instance is that canine distemper virus infection distorted the intestinal microbiota composition by reducing the prevalence of the dominant genera, Escherichia and Clostridium, and increasing microbial diversity. In this study, we performed high-throughput 16S RNA gene sequencing of the intestinal microbiota of tilapia after oral administration of S. agalactiae YM001. Through β diversity analysis, it was found that early oral YM001 immunization could decrease intestinal microbiota diversity in tilapia (Figure 3A). Changes in intestinal microbiota may have an impact on the health of the host. Previous studies have shown that changes in intestinal microbiota may have an impact on the health of the host. For human, the decrease in bacterial richness and diversity is consistent with other disease states that have alterations in the intestinal microbiota such as inflammatory bowel disease and diabetes (Jandhyala et al., 2017; Nishino et al., 2018). Despite this, administration of YM001 only temporarily reduced the diversity of intestinal microbiota in tilapia (Figures 3A,B) and changes in intestinal microbial diversity in tilapia appear to be closely related to YM001 concentration in the intestine. By analyzing the relationship between the retention of oral streptococcus and the diversity of intestinal microbiota, we found that when the concentration of YM001 was highest, the diversity of intestinal microbiota in tilapia was the lowest, and with the concentration of YM001 decreasing, the diversity was gradually recovered (Figures 3A,B). Oral administration of virulent strain HN016 also decreased the intestinal microbiota diversity of tilapia, but as the concentration of HN016 decreasing, the intestinal microbiota diversity also recovered or even exceeded the normal level (Figures 6A,B). More and more scholars started to explore the effects of oral vaccines on intestinal microbiota in recent years. It has been reported that oral administration of live-attenuated typhoid vaccine Ty21a could considerably affect inter- and intra-individual variability, yet not discernibly perturb the bacterial assemblage related to vaccine administration (Eloe-Fadrosh et al., 2013). Oral administration of different S. Typhimurium vaccine strains can differentially influence the presence, but not the relative abundance of microbiota in the ceca (Park et al., 2017). Similar studies have also been reported in aquatic animals. When bath-immunizing grass carp using recombinant Aeromonas hydrophila vaccine (Aera), there was no significant difference in the abundance of genera Acinetobacter, Cetobacterium, and Gemmobacter compared to the Control (Liu et al., 2015). All the above examples indicate that oral vaccines can affect the composition of the intestinal microbiota to varying degrees.
Intestinal microbiota imbalance could cause host health problems, which may be related to reduced levels of beneficial bacteria and increased levels of pathogenic bacteria. By analyzing the microbiota changes in the different gut segments, it was found that at the genus level, the contents of Streptococcus, Cetobacterium, Akkermansia, Romboutsia, Bacteroides, Brevinema and Lachnospiraceae_NK4A136 are changed the most after administration of YM001 (Figure 4A). Ecept the Streptococcus which are directly related to YM001, the function of the other above bacteria are as follow. Cetobacterium is bile resistant and could produce acetic acid as the major end product of metabolism of peptides and carbohydrates (Finegold et al., 2003). A. muciniphila is a mucin-degrading bacterium and inversely associated with obesity, diabetes, inflammation, and metabolic disorders (Everard et al., 2013; Caesar et al., 2015). Romboutsia can utilize different relatively simple carbohydrates and synthesize limited amino acids and vitamins (Gerritsen et al., 2017). Bacteroides play a fundamental role in processing complex molecules to simpler ones in the host intestine (Wexler, 2007). Bacteroides species also benefit their host by excluding potential pathogens from colonizing the gut (Wexler, 2007). Some species (B. fragilis, for example) are opportunistic human pathogens (Wexler, 2007). Brevinema belongs to spirochaetes, which have been extensively studied in termite guts and have been found to be involved in the breakdown of lignocellulose and nitrogen fixation. Lachnospiraceae are a family of bacteria in the order of Clostridiales. Members of Lachnospiraceae family could produce butyric acid and are important for host epithelial cell growth (Meehan and Beiko, 2014). Among these bacteria, the change of Cetobacterium is particularly noteworthy. As the predominant bacteria in the intestine of tilapia, Cetobacterium accounted for 24.87% of the total intestinal bacteria, and its content increased to 41.1% at 24 h after oral administration of YM001, and gradually returned to 32.2 and 29% at 7 d and 15 d after oral administration of YM001. Streptococcus accounted for 0.5% of the total intestinal bacteria in healthy tilapia. At 12 h after oral administration of YM001, its content increased to up to 70.8% and then gradually returned to 0.8% at 24 h after oral administration of YM001. The results showed that oral YM001 changed the composition of intestinal microbiota in a short time; but with the decrease of YM001 content, the content of the predominant bacteria was gradually recovered.
The effects of attenuated strain YM001 and virulent strain HN016 on tilapia intestinal microbiota were very different. The differences in gut bacteria between these two groups were greater than the differenced within each group (Figure 5). Although oral administration of HN016 and YM001 both decreased the intestinal flora diversity, the effect of YM001 was more significant (Figure 6). The difference in intestinal flora caused by virulent strains and attenuated strains may be related to the secretion or metabolites of different strains. The specific underlying mechanisms need to be further explored.
In conclusion, after oral administration, YM001 could colonize in the digestive tract of tilapia for a certain period of time, and alter the structure and diversity of the intestinal microbiota. However, these changes do not have fatal consequences. Tilapia intestinal microbiota has a repulsive effect against YM001. With the content of YM001 in the digestive decreasing, the structure and diversity of intestinal microbiota gradually recovered.
Author Contributions
ML, LL, and TH contributed equally to this work. LL, TH, YL, AL, and FC performed experiments and analyzed data. ML and CM provided software and bioinformatics expertise. MC and ML designed the experiments, analyzed data, and wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by Guangxi innovation-driven development special funds (Grant no. AA17204081-3), National Natural Science Foundation of China (31460695), Guangxi Natural Science Foundation (2016GXNSFDA380020), the funds of Guangxi Key Laboratory for Aquatic Genetic Breeding and Healthy Aquaculture (2016–2018).
References
- Amal M. N., Zamri-Saad M., Iftikhar A. R., Siti-Zahrah A., Aziel S., Fahmi S. (2012). An outbreak of Streptococcus agalactiae infection in cage-cultured golden pompano, Trachinotus blochii (Lacepede), in Malaysia. J. Fish Dis. 35, 849–852. 10.1111/j.1365-2761.2012.01443.x [DOI] [PubMed] [Google Scholar]
- Baquero F., Martinez J. L., Canton R. (2008). Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 19, 260–265. 10.1016/j.copbio.2008.05.006 [DOI] [PubMed] [Google Scholar]
- Barroso-Batista J., Demengeot J., Gordo I. (2015). Adaptive immunity increases the pace and predictability of evolutionary change in commensal gut bacteria. Nat. Commun. 6:8945. 10.1038/ncomms9945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caesar R., Tremaroli V., Kovatcheva-Datchary P., Cani P. D., Backhed F. (2015). Crosstalk between gut microbiota and dietary lipids aggravates WAT inflammation through TLR signaling. Cell Metab. 22, 658–668. 10.1016/j.cmet.2015.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Wang R., Li L. P., Liang W. W., Li J., Huang Y., et al. (2012). Screening vaccine candidate strains against Streptococcus agalactiae of tilapia based on PFGE genotype. Vaccine 30, 6088–6092. 10.1016/j.vaccine.2012.07.044 [DOI] [PubMed] [Google Scholar]
- Chen W. H., Garza J., Choquette M., Hawkins J., Hoeper A., Bernstein D. I., et al. (2015). Safety and immunogenicity of escalating dosages of a single oral administration of peru-15 pCTB, a candidate live, attenuated vaccine against enterotoxigenic Escherichia coli and Vibrio cholerae. Clin. Vaccine Immunol. 22, 129–135. 10.1128/CVI.00560-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depaola A., Peeler J. T., Rodrick G. E. (1995). Effect of oxytetracycline-medicated feed on antibiotic resistance of gram-negative bacteria in catfish ponds. Appl. Environ. Microbiol. 61, 2335–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L., Eckburg P. B., Bik E. M., Relman D. A. (2006). Assembly of the human intestinal microbiota. Trends Ecol. Evol. 21, 517–523. 10.1016/j.tree.2006.06.013 [DOI] [PubMed] [Google Scholar]
- Edmond K. M., Kortsalioudaki C., Scott S., Schrag S. J., Zaidi A. K., Cousens S., et al. (2012). Group B streptococcal disease in infants aged younger than 3 months: systematic review and meta-analysis. Lancet 379, 547–556. 10.1016/S0140-6736(11)61651-6 [DOI] [PubMed] [Google Scholar]
- Edwards M. S., Baker C. J. (2005). Group B streptococcal infections in elderly adults. Clin. Infect. Dis. 41, 839–847. 10.1086/432804 [DOI] [PubMed] [Google Scholar]
- Eloe-Fadrosh E. A., Mcarthur M. A., Seekatz A. M., Drabek E. F., Rasko D. A., Sztein M. B., et al. (2013). Impact of oral typhoid vaccination on the human gut microbiota and correlations with s. Typhi-specific immunological responses. PLoS ONE 8:e62026. 10.1371/journal.pone.0062026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A., Belzer C., Geurts L., Ouwerkerk J. P., Druart C., Bindels L. B., et al. (2013). Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. U.S.A. 110, 9066–9071. 10.1073/pnas.1219451110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegold S. M., Vaisanen M. L., Molitoris D. R., Tomzynski T. J., Song Y., Liu C., et al. (2003). Cetobacterium somerae sp. nov. from human feces and emended description of the genus Cetobacterium. Syst. Appl. Microbiol. 26, 177–181. 10.1078/072320203322346010 [DOI] [PubMed] [Google Scholar]
- Gaikwad S. S., Shouche Y. S., Gade W. N. (2017). Deep sequencing reveals highly variable gut microbial composition of invasive fish mossambicus tilapia (Oreochromis mossambicus) collected from two different habitats. Indian J. Microbiol. 57, 235–240. 10.1007/s12088-017-0641-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerritsen J., Hornung B., Renckens B., Van Hijum S., Martins Dos Santos V. A. P., Smidt H. (2017). Genomic and functional analysis of Romboutsia ilealis CRIB(T) reveals adaptation to the small intestine. PeerJ. 5:e3698 10.7717/peerj.3698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez G. D., Balcazar J. L. (2008). A review on the interactions between gut microbiota and innate immunity of fish. FEMS Immunol. Med. Microbiol. 52, 145–154. 10.1111/j.1574-695X.2007.00343.x [DOI] [PubMed] [Google Scholar]
- Harris V. C., Armah G., Fuentes S., Korpela K. E., Parashar U., Victor J. C., et al. (2017). Significant correlation between the infant gut microbiome and rotavirus vaccine response in rural ghana. J. Infect. Dis. 215, 34–41. 10.1093/infdis/jiw518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins M. J., Sharp R., Macfarlane G. T. (2002). Variation in human intestinal microbiota with age. Dig. Liver Dis. 34(Suppl. 2):S12–18. 10.1016/S1590-8658(02)80157-8 [DOI] [PubMed] [Google Scholar]
- Huang L. Y., Wang K. Y., Xiao D., Chen D. F., Geng Y., Wang J., et al. (2014). Safety and immunogenicity of an oral DNA vaccine encoding Sip of Streptococcus agalactiae from Nile tilapia Oreochromis niloticus delivered by live attenuated Salmonella typhimurium. Fish Shellfish Immunol. 38, 34–41. 10.1016/j.fsi.2014.02.017 [DOI] [PubMed] [Google Scholar]
- Jafar Q. A., Sameer A. Z., Salwa A. M., Samee A. A., Ahmed A. M., Al-Sharifi F. (2008). Molecular investigation of Streptococcus agalactiae isolates from environmental samples and fish specimens during a massive fish kill in Kuwait Bay. Pak. J. Biol. Sci. 11, 2500–2504. 10.3923/pjbs.2008.2500.2504 [DOI] [PubMed] [Google Scholar]
- Jandhyala S. M., Madhulika A., Deepika G., Rao G. V., Reddy D. N., Subramanyam C., et al. (2017). Altered intestinal microbiota in patients with chronic pancreatitis: implications in diabetes and metabolic abnormalities. Sci. Rep. 7:43640. 10.1038/srep43640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly P. (2010). Nutrition, intestinal defence and the microbiome. Proc. Nutr. Soc. 69, 261–268. 10.1017/S0029665110000108 [DOI] [PubMed] [Google Scholar]
- Lamagni T. L., Keshishian C., Efstratiou A., Guy R., Henderson K. L., Broughton K., et al. (2013). Emerging trends in the epidemiology of invasive group B streptococcal disease in England and Wales, 1991-2010. Clin. Infect. Dis. 57, 682–688. 10.1093/cid/cit337 [DOI] [PubMed] [Google Scholar]
- Ley R. E., Lozupone C. A., Hamady M., Knight R., Gordon J. I. (2008). Worlds within worlds: evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 6, 776–788. 10.1038/nrmicro1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. P., Wang R., Liang W. W., Huang T., Huang Y., Luo F. G., et al. (2015). Development of live attenuated Streptococcus agalactiae vaccine for tilapia via continuous passage in vitro. Fish Shellfish Immunol. 45, 955–963. 10.1016/j.fsi.2015.06.014 [DOI] [PubMed] [Google Scholar]
- Liu L., Gong Y. X., Zhu B., Liu G. L., Wang G. X., Ling F. (2015). Effect of a new recombinant Aeromonas hydrophila vaccine on the grass carp intestinal microbiota and correlations with immunological responses. Fish Shellfish Immunol. 45, 175–183. 10.1016/j.fsi.2015.03.043 [DOI] [PubMed] [Google Scholar]
- Makesh M., Sudheesh P. S., Cain K. D. (2015). Systemic and mucosal immune response of rainbow trout to immunization with an attenuated Flavobacterium psychrophilum vaccine strain by different routes. Fish Shellfish Immunol. 44, 156–163. 10.1016/j.fsi.2015.02.003 [DOI] [PubMed] [Google Scholar]
- Meehan C. J., Beiko R. G. (2014). A phylogenomic view of ecological specialization in the Lachnospiraceae, a family of digestive tract-associated bacteria. Genome Biol. Evol. 6, 703–713. 10.1093/gbe/evu050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mian G. F., Godoy D. T., Leal C. A., Yuhara T. Y., Costa G. M., Figueiredo H. C. (2009). Aspects of the natural history and virulence of S. agalactiae infection in Nile tilapia. Vet. Microbiol. 136, 180–183. 10.1016/j.vetmic.2008.10.016 [DOI] [PubMed] [Google Scholar]
- Nishino K., Nishida A., Inoue R., Kawada Y., Ohno M., Sakai S., et al. (2018). Analysis of endoscopic brush samples identified mucosa-associated dysbiosis in inflammatory bowel disease. J. Gastroenterol. 53, 95–106. 10.1007/s00535-017-1384-4 [DOI] [PubMed] [Google Scholar]
- O'mahony S. M., Marchesi J. R., Scully P., Codling C., Ceolho A. M., Quigley E. M., et al. (2009). Early life stress alters behavior, immunity, and microbiota in rats: implications for irritable bowel syndrome and psychiatric illnesses. Biol. Psychiatry 65, 263–267. 10.1016/j.biopsych.2008.06.026 [DOI] [PubMed] [Google Scholar]
- Park S. H., Kim S. A., Rubinelli P. M., Roto S. M., Ricke S. C. (2017). Microbial compositional changes in broiler chicken cecal contents from birds challenged with different Salmonella vaccine candidate strains. Vaccine 35, 3204–3208. 10.1016/j.vaccine.2017.04.073 [DOI] [PubMed] [Google Scholar]
- Pridgeon J. W., Klesius P. H. (2013). Development of live attenuated Streptococcus agalactiae as potential vaccines by selecting for resistance to sparfloxacin. Vaccine 31, 2705–2712. 10.1016/j.vaccine.2013.03.066 [DOI] [PubMed] [Google Scholar]
- Rawls J. F., Samuel B. S., Gordon J. I. (2004). Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc. Natl. Acad. Sci. U.S.A. 101, 4596–4601. 10.1073/pnas.0400706101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeselers G., Mittge E. K., Stephens W. Z., Parichy D. M., Cavanaugh C. M., Guillemin K., et al. (2011). Evidence for a core gut microbiota in the zebrafish. ISME J. 5, 1595–1608. 10.1038/ismej.2011.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt B., Mulder I. E., Musk C. C., Aminov R. I., Lewis M., Stokes C. R., et al. (2011). Establishment of normal gut microbiota is compromised under excessive hygiene conditions. PLoS ONE 6:e28284. 10.1371/journal.pone.0028284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F., Backhed F. (2013). The gut microbiota–masters of host development and physiology. Nat. Rev. Microbiol. 11, 227–238. 10.1038/nrmicro2974 [DOI] [PubMed] [Google Scholar]
- Tanaka S., Kobayashi T., Songjinda P., Tateyama A., Tsubouchi M., Kiyohara C., et al. (2009). Influence of antibiotic exposure in the early postnatal period on the development of intestinal microbiota. FEMS Immunol. Med. Microbiol. 56, 80–87. 10.1111/j.1574-695X.2009.00553.x [DOI] [PubMed] [Google Scholar]
- Tazi A., Morand P. C., Reglier-Poupet H., Dmytruk N., Billoet A., Antona D., et al. (2011). Invasive group B streptococcal infections in adults, France (2007-2010). Clin. Microbiol. Infect. 17, 1587–1589. 10.1111/j.1469-0691.2011.03628.x [DOI] [PubMed] [Google Scholar]
- Turnbaugh P. J., Hamady M., Yatsunenko T., Cantarel B. L., Duncan A., Ley R. E., et al. (2009). A core gut microbiome in obese and lean twins. Nature 457, 480–484. 10.1038/nature07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Li L., Huang Y., Luo F., Liang W., Gan X., et al. (2015). Comparative genome analysis identifies two large deletions in the genome of highly-passaged attenuated Streptococcus agalactiae strain YM001 compared to the parental pathogenic strain HN016. BMC Genom. 16:897. 10.1186/s12864-015-2026-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler H. M. (2007). Bacteroides: the good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 20, 593–621. 10.1128/CMR.00008-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J. H., Lin G., Fu G. H., Wan Z. Y., Lee M., Wang L., et al. (2014). The intestinal microbiome of fish under starvation. BMC Genom. 15:266. 10.1186/1471-2164-15-266 [DOI] [PMC free article] [PubMed] [Google Scholar]