Abstract
Homologous recombination (HR) is a universally conserved DNA repair pathway that can result in the exchange of genetic material. In eukaryotes, HR has evolved into an essential step in meiosis. During meiosis many eukaryotes utilize a two-recombinase pathway. This system consists of Rad51 and the meiosis-specific recombinase Dmc1. Both recombinases have distinct activities during meiotic HR, despite being highly similar in sequence and having closely related biochemical activities, raising the question of how these two proteins can perform separate functions. A likely explanation for their differential regulation involves the meiosis-specific recombination proteins Hop2 and Mnd1, which are part of a highly conserved eukaryotic protein complex that participates in HR, albeit through poorly understood mechanisms. To better understand how Hop2–Mnd1 functions during HR, here we used DNA curtains in conjunction with single-molecule imaging to measure and quantify the binding of the Hop2–Mnd1 complex from Saccharomyces cerevisiae to recombination intermediates comprising Rad51– and Dmc1–ssDNA in real time. We found that yeast Hop2–Mnd1 bound rapidly to Dmc1–ssDNA filaments with high affinity and remained bound for ∼1.3 min before dissociating. We also observed that this binding interaction was highly specific for Dmc1 and found no evidence for an association of Hop2–Mnd1 with Rad51–ssDNA or RPA–ssDNA. Our findings provide new quantitative insights into the binding dynamics of Hop2–Mnd1 with the meiotic presynaptic complex. On the basis of these findings, we propose a model in which recombinase specificities for meiotic accessory proteins enhance separation of the recombinases' functions during meiotic HR.
Keywords: meiosis, homologous recombination, DNA recombination, DNA, DNA enzyme, Dmc1, Hop2–Mnd1, Rad51, DNA curtains, DNA repair, yeast, DNA–protein interaction, recombinase
Introduction
Homologous recombination (HR),3 a DNA repair pathway used to repair double-strand DNA breaks (DSB), is essential for maintaining genome stability. In addition, HR-mediated pairing of homologous chromosomes during the first meiotic division is essential for efficient chromosome segregation, and meiotic recombination also allows for the generation of genetically diverse progeny (1). As such, defects in HR-related proteins can predispose patients to infertility, cancer, and other severe genetic disorders (2, 3).
HR utilizes a homologous DNA as a template to guide break repair in a nearly error-free manner (4–7). During mitotic growth, DSBs can arise from spontaneous DNA damage because of exogenous agents (e.g. ionizing radiation or DNA-damaging chemicals), programmed breaks during developmentally regulated DNA rearrangements (e.g. MAT switching in Saccharomyces cerevisiae), or errors during DNA replication (6–8). Whereas during meiosis, programmed DSBs are initiated through the action of the universally conserved Spo11 enzyme (9, 10), HR proceeds through the resection of broken double-strand DNA (dsDNA), yielding long single-strand DNA (ssDNA) overhangs that are paired with a homologous dsDNA elsewhere in the genome and used as a template for the repair of the damaged DNA (11–13). The 3′-ssDNA overhangs are first bound by the heterotrimeric eukaryotic ssDNA–binding protein, RPA (replication protein A), which protects the ssDNA from nucleases, removes secondary structure, and serves as a signal for initiating the DNA damage response (14–16). RPA is replaced by either Rad51 during mitosis or by both Rad51 and Dmc1 during meiosis (7, 17–21). Rad51 and Dmc1 are closely related ATP-dependent DNA recombinases that form extended right-handed helical filament on the ssDNA (22–25). These nucleoprotein filaments, referred to as presynaptic complexes, are responsible for promoting key DNA transactions during the early stages of HR (7, 21, 26). The presynaptic complex first locates a homologous dsDNA template through a process referred to as the homology search (17, 27–30). The presynaptic complex then pairs the bound ssDNA with the complementary strand from the homologous dsDNA template (7, 17, 21, 24, 31–33). This strand invasion reaction results in displacement of the noncomplementary strand, and the resulting D-loop intermediate can be processed through several different pathways, leading to repair of the damaged DNA (1, 4, 6, 7, 26, 34–38).
Dmc1 and Rad51 arose from a gene duplication event during the early history of eukaryotic evolution (39); these proteins still retain ∼46% identical amino acid sequence and share the same basic biochemical activities (22, 33, 40–42). Both recombinases are ATP-dependent DNA–binding proteins that form extended helical filaments on ssDNA (17, 33). However, the catalytic activity of Rad51 is not required for the completion of meiosis; instead the role of Rad51 during meiosis is to facilitate the loading of Dmc1 onto the ssDNA overhangs (18, 20, 43, 44). Dmc1 is then able to promote strand invasion (43, 45, 46). Given the striking similarity between these recombinases, we do not have a complete understanding of how the basic biochemical activities of these two recombinases are differentially regulated or even why both recombinases are necessary.
Recently, several biochemical differences between Rad51 and Dmc1 have been identified. These include the requirement for calcium ions in Dmc1 activity (40, 47–49); the ability of Dmc1 to stabilize strand invasion complexes with partially mismatched homology in vitro (27, 28, 33); and the ability of Rad51 and Dmc1 to self-segregate into isolated filaments both in vitro and in vivo (18, 48). It is also likely that the differential regulation of these two recombinases during mitotic and meiotic growth may be dictated by cofactor proteins that are able to interact exclusively with either Rad51 or Dmc1. For instance, in S. cerevisiae, the meiosis-specific protein Hed1 binds exclusively to Rad51 and inhibits the binding of Rad54 (49–52). Rad54 is an essential cofactor for Rad51 strand invasion activity, and thus the Hed1-mediated inhibition of Rad54 binding prevents Rad51 from performing strand invasion during meiosis (53–55).
Eukaryotic Hop2–Mnd1 is a highly conserved heterodimeric protein complex, which stabilizes the presynaptic complex and facilitates capture of homologous DNA templates during the early stages of HR (40, 56–60). Like Hed1, S. cerevisiae Hop2–Mnd1 is expressed only during meiosis, and Hop2–Mnd1 interacts with Dmc1 but does not stimulate strand exchange by Rad51, indicating that yeast Hop2–Mnd1 is a meiosis-specific recombination accessory factor (40). Accordingly, S. cerevisiae hop2Δ and mnd1Δ mutants exhibit numerous meiotic defects including aberrant synapsis between nonhomologous chromosomes, defects in completion of DSB repair, and developmental arrests at the pachytene stage of meiosis (57, 61, 62). Interestingly, Hop2 and Mnd1 are found in many eukaryotic organisms, but they are absent in organisms such as Sordaria macrospora, Drosophila melanogaster, and Caenorhabditis elegans, all of which also lack Dmc1; this strongly suggests that Hop2–Mnd1 has a specific role in promoting Dmc1-dependent HR during meiosis (63).
Surprisingly, both biochemical and genetic data have revealed a mitotic role for Hop2–Mnd1 in higher eukaryotes. Hop2–Mnd1 is broadly expressed in ALT (alternative lengthening of telomeres) cell lines and plays a role in promoting a Rad51-dependent homology search that allows for interchromosomal recombination between telomeres in these mitotic cells (64). Moreover, mammalian HOP2–MND1 stimulates the DNA strand exchange activities of both RAD51 and DMC1 in vitro (65–67), mutations in human HOP2 have been found in early-onset familial breast and ovarian cancer patients (68), and HOP2 point mutations have also been linked to XX ovarian dysgenesis (65). Together, these studies imply that mammalian HOP2–MND1 may have a broader role in genome maintenance and cancer biology. Thus, studies of yeast Hop2–Mnd1 may yield new insights into HOP2–MND1 functions in vertebrates.
Hop2 and Mnd1 together function as an obligate heterodimer (69), and early structural experiments with Hop2–Mnd1 suggest that the complex forms an elongated structure that exists as an isolated heterodimer in solution (67). Recently, a crystal structure of Giardia lamblia Hop2–Mnd1 confirmed that the complex has an elongated V-like shape and revealed that the interface between Hop2 and Mnd1 is stabilized by three sequential α-helices connected by leucine zipper domains (70). The structure also revealed a winged-helix domain consisting of regions from both Hop2 and Mnd1 that form a unified dsDNA-binding domain (70, 71). Hop2–Mnd1 has high affinity and specificity for dsDNA over ssDNA, and functional analysis of Mus musculus Hop2–Mnd1 has determined that the recombinase-binding domain of Hop2–Mnd1 is located on the opposite end of the complex from the DNA-binding domain (65, 67). Together, these studies suggest a model in which Hop2–Mnd1 may promote HR during meiosis by helping Dmc1 (in yeast) or both Rad51 and Dmc1 (in higher eukaryotes) to capture dsDNA.
Despite the wealth of biochemical understanding of Hop2–Mnd1 function, the relationship among Dmc1, Rad51, and Hop2–Mnd1 is not fully understood. For instance, to date there are no quantitative data available describing the interactions between Hop2–Mnd1 and the presynaptic complex. As part of a continuing effort to understand the structural properties and dynamics of the meiotic presynaptic complex, here we have used ssDNA curtains with single-molecule imaging to directly visualize the binding properties of S. cerevisiae Hop2–Mnd1 with presynaptic complexes composed of yeast Rad51–ssDNA, Dmc1–ssDNA, or both recombinases in real time. In this study we showed that yeast Hop2–Mnd1 binds specifically to Dmc1–ssDNA and does not interact with Rad51–ssDNA. Furthermore, we found that although Hop2–Mnd1 has high affinity for Dmc1–ssDNA, the binding is dynamic and Hop2–Mnd1 readily exchanges between the free and bound states. Finally, we show that Hop2–Mnd1 retains its binding specificity for Dmc1 even in mixed recombinase filaments containing both Rad51 and Dmc1. We discuss the possible implications of these findings for Hop2–Mnd1 function during meiotic recombination.
Results
Hop2–Mnd1 binding is specific for Dmc1
Hop2–Mnd1 is essential during meiotic HR (59, 60, 62). However, many of the basic biophysical properties of Hop2–Mnd1 binding to Dmc1 presynaptic filaments remain unknown. To address these questions, we generated a Mnd1–mCherry fusion protein. We chose to fuse the mCherry fluorescent protein to the C terminus of Mnd1 because Mnd1–GFP compliments the mnd1Δ phenotype in yeast cells, suggesting that this fusion construct retains biological function in vivo (57). This fusion protein forms a stable complex with His6–Hop2 when co-expressed in Escherichia coli. Hop2 and Mnd1–mCherry can then be co-purified though the His6 tag on Hop2 (see “Experimental procedures”). For brevity, the protein complex will be referred to as Hop2–Mnd1–mCherry. We then visualized the binding of Hop2–Mnd1–mCherry to Dmc1–ssDNA presynaptic complexes using ssDNA curtains with a total internal reflection fluorescence microscope (TIRFM) (Fig. 1) (reviewed in Refs. 72 and 73).
Figure 1.
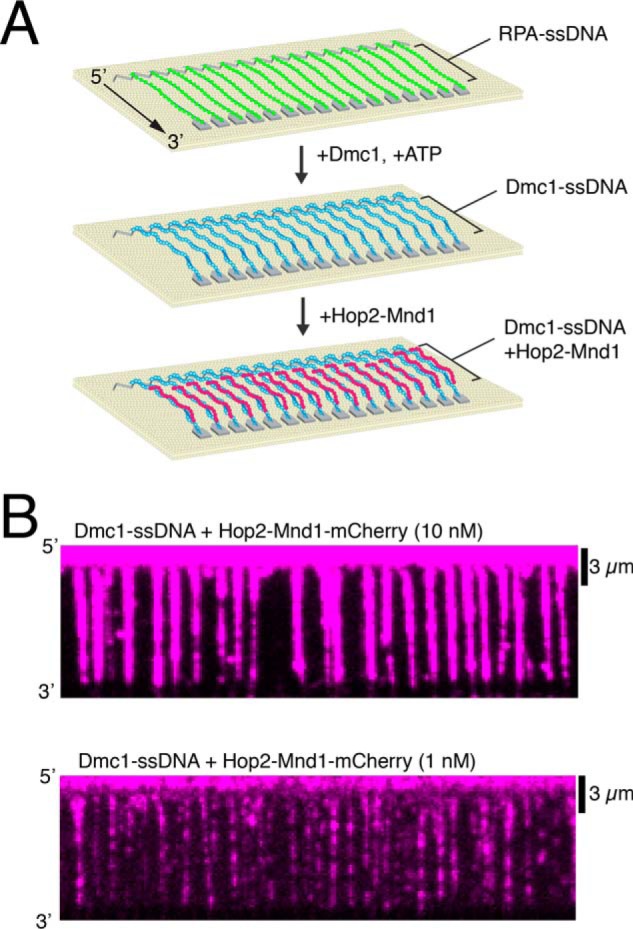
Using ssDNA curtains to measure the association of Hop2–Mnd1 with Dmc1–ssDNA filaments. A, schematic diagram of DNA curtains experiments that follows the assembly of Dmc1–ssDNA filaments from RPA-ssDNA, as described (27, 48), followed by the binding of Hop2–Mnd1–mCherry. B, wide-field TIRFM images of Dmc1–ssDNA curtains (unlabeled) bound with 10 nm (top) and 1 nm (bottom) Hop2–Mnd1–mCherry (magenta).
We first measured the binding properties of Hop2–Mnd1–mCherry under equilibrium conditions. For these measurements, we titrated 0.1 to 30 nm Hop2–Mnd1–mCherry into reactions containing preassembled Dmc1–ssDNA (Figs. 1B and Fig. 2, A and B). At the lower end of this titration, Hop2–Mnd1 association with the Dmc1–ssDNA was observed as individual binding events (Fig. 2A) randomly distributed along the Dmc1–ssDNA (Fig. 2C). As the concentration increased, Hop2–Mnd1 began to saturate the Dmc1–ssDNA presynaptic complexes, resulting in a more uniform distribution of mCherry fluorescence signal along the length of the Dmc1–ssDNA complexes (Fig. 2A). To quantify the binding of Hop2–Mnd1, we measured the integrated mCherry signal intensity along each ssDNA molecule at each protein concentration tested. Background from a region lacking DNA was subtracted from the integrated signal intensity. The measured intensities at each concentration were plotted, and the data were fit using the Hill equation (Fig. 2B). The resulting analysis revealed that the Kd of Hop2–Mnd1 for binding to the Dmc1–ssDNA was 11.6 ± 8 nm with an h value (Hill coefficient) of 0.75 ± 0.11 and a mean maximum signal intensity (Bmax) of 39,800 ± 8,552 arbitrary units. These values illustrate the high affinity of Hop2–Mnd1 for the Dmc1–ssDNA filament and reveal a weakly anti-cooperative binding behavior. The weakly anti-cooperative behavior means that individual Hop2–Mnd1 binding events do not promote further binding of Hop2–Mnd1 in the same region of the Dmc1 filament. In contrast, no Hop2–Mnd1 binding to Rad51–ssDNA filaments was observed under similar conditions (Fig. 2D); we also observed no binding to RPA–ssDNA (not shown). This observation is in agreement with previous reports that yeast Hop2–Mnd1 functions exclusively with Dmc1 (40).
Figure 2.
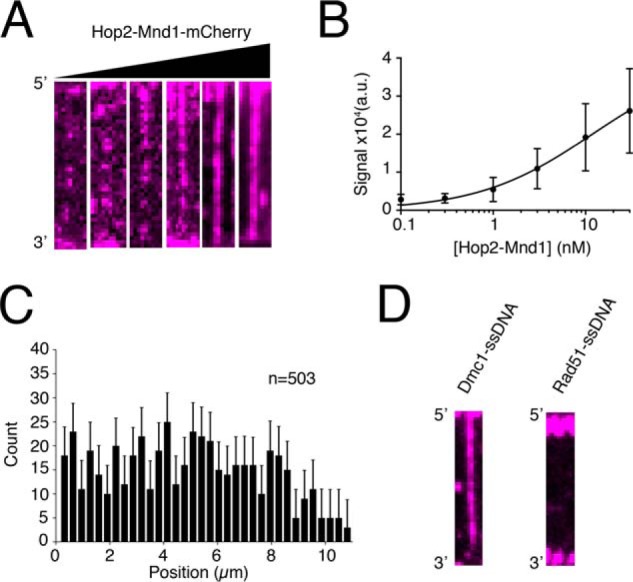
Equilibrium binding properties and specificity of Hop2–Mnd1. A, wide-field TIRFM images of Dmc1–ssDNA filaments with 0.1, 0.3, 1.0, 3.0, 10, of 30 nm Hop2–Mnd1–mCherry (magenta), as indicated. B, quantification of integrated fluorescence signal intensity from Hop2–Mnd1–mCherry binding to Dmc1–ssDNA filaments. The graph was generated by measuring the integrated signal intensity (a.u., arbitrary units) for at least 30 ssDNA molecules at each titration point. The data were fit by a Hill equation, and the error bars represent S.D. of individual molecules. C, position distribution histogram of individual Hop2–Mnd1 molecules bound along Dmc1–ssDNA molecules (n = 503). Error bars were generated by bootstrapping the data using a custom Python script (48). D, wide-field TIRFM images of Dmc1–ssDNA molecules or Rad51-ssDNA molecules in the presence of 10 nm Hop2–Mnd1–mCherry (magenta).
Hop2–Mnd1 binds as isolated heterodimeric units
Previous analysis by size exclusion chromatography revealed that Hop2–Mnd1 behaves primarily as an elongated, heterodimeric 50-kDa complex in free solution (67, 69). We next tested whether the minimal binding unit of Hop2–Mnd1 to Dmc1–ssDNA filaments was in fact a single heterodimer or whether Hop2–Mnd1 might form high-order oligomers prior to interacting with the Dmc1–ssDNA. To measure this quantity, we bound Hop2–Mnd1 to Dmc1–ssDNA filaments at low concentrations (0.1 nm Hop2–Mnd1) and allowed the reaction to reach equilibrium. These measurements were intentionally made at low protein concentrations to ensure that the observed photobleaching steps could be attributed to single rather than multiple binding events. We then monitored the Hop2–Mnd1–mCherry signal without laser shuttering while measuring the rate at which the individual fluorescent signals photobleached. Under these illumination conditions, the bound Hop2–Mnd1–mCherry complexes rapidly photobleached, as revealed by the sudden loss of signal intensity and the number of photobleaching steps, signifying the number of mCherry molecules/binding event (Fig. 3, A and B). We found that a majority (∼75%) of the Hop2–Mnd1 molecules underwent single-step photobleaching with fewer complexes (∼25%) exhibiting two or more photobleaching steps (Fig. 3, B and C). We can state that the protein complex binds as a single heterodimer at 0.1 nm Hop2–Mnd1, but we cannot unequivocally state that it binds as a single heterodimer at 10–30 nm Hop2–Mnd1. Despite this caveat, given that the binding behavior was not positively cooperative, it is not unreasonable to conclude that there was no change in the associating units (i.e. single heterodimers) for the higher concentration regimes. We conclude that under the conditions used for our experiments, Hop2–Mnd1 binds to the Dmc1–ssDNA filaments primarily as a monomeric complex.
Figure 3.
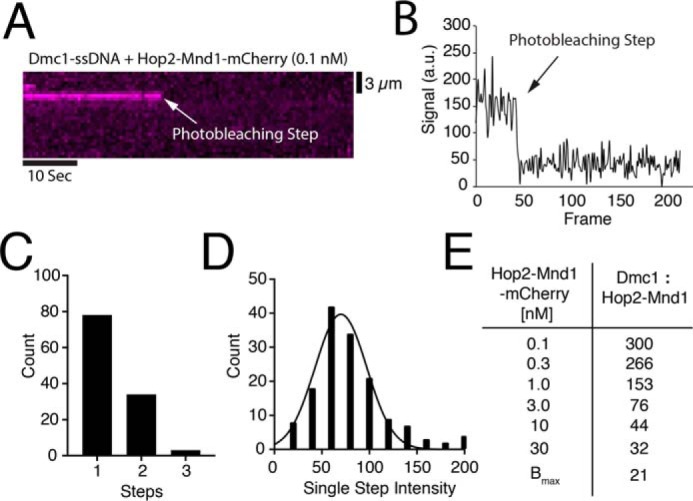
Analysis of individual Hop2–Mnd1–mCherry binding events. A, representative kymograph of single Hop2–Mnd1-mCherry complex (magenta) bound to a Dmc1–ssDNA molecule and visualized with constant laser illumination. Abrupt loss of the signal reflects photobleaching of mCherry as highlighted by the white arrow. B, representative trace of the normalized signal intensity of an individual Hop2–Mnd1–mCherry molecule measured over time (a.u., arbitrary units). The photobleaching step is highlighted with an arrow. C, histogram representing the number of photobleaching steps observed for each observed foci of Hop2–Mnd1–mCherry (n = 115) bound to Dmc1–ssDNA. D, histogram showing the magnitude of the individual mCherry photobleaching steps (n = 155). The change in mean signal intensity during a photobleaching step was determined from a Gaussian fit to the data. E, table displaying estimates for ratio of Dmc1 to Hop2–Mnd1 at each concentration of Hop2–Mnd1–mCherry tested. The Bmax value was determined from the data presented in Fig. 2B and represents the saturation of the Dmc1–ssDNA filaments with Hop2–Mnd1–mCherry.
Substoichiometric association of Hop2–Mnd1 with Dmc1–ssDNA
We also utilized photobleaching data to estimate the ratio of Hop2–Mnd1–mCherry to Dmc1 at various concentrations within the titration curve. For this analysis we quantified the absolute mCherry signal intensity value of each photobleaching step and fit the resulting data to a Gaussian distribution to define the mean signal intensity per bound molecule of Hop2–Mnd1–mCherry. This quantitation revealed a mean mCherry signal intensity value of 70 ± 5 arbitrary units/bound complex of Hop2–Mnd1–mCherry (Fig. 3D). We then estimated the total number of bound Hop2–Mnd1–mCherry complexes by dividing the mean integrated signal intensity values for Hop2–Mnd1–mCherry binding at each concentration point within the titration curve (Fig. 2A) by the mean signal intensity value calculated for a single Hop2–Mnd1–mCherry complex (Fig. 3D). From this analysis, we estimated that at 0.1, 0.3, 1.0, 3.0, 10, and 30 nm Hop2–Mnd1–mCherry there were ∼40, 44, 77, 156, 273, and 373 molecules of Hop2–Mnd1–mCherry bound per molecule of Dmc1–ssDNA, respectively. We estimated that each ssDNA molecule was bound by ∼12,000 Dmc1 monomers; this estimate is based on an ssDNA length of 36,000 nucleotides (measured from the barriers to the pedestals (27)) and bound at a ratio of 1 Dmc1 monomer/3 nucleotides. These values correspond to ratios of Dmc1 to Hop2–Mnd1 of ∼300:1, 266:1, 153:1, 76:1, 44:1, 32:1, and 21:1 for experiments conducted at 0.1, 0.3, 1.0, 3.0, 10 and 30 nm Hop2–Mnd1–mCherry, respectively (Fig. 3E). The maximum amount of Hop2–Mnd1 that could bind to the Dmc1–ssDNA filaments at saturation was estimated from the Bmax value determined when the Hop2–Mnd1 binding data were fit by the Hill equation (Fig. 2B). We concluded that under the conditions of our experiments, Hop2–Mnd1 binds to Dmc1–ssDNA filaments in substoichiometric ratios, which is also consistent with the substoichiometric ratios of Hop2–Mnd1 required to stimulate Dmc1-mediated strand invasion using in vitro assays (40).
Hop2–Mnd1 binding kinetics
Our data revealed a ∼12 nm Kd for Hop2–Mnd1–mCherry association with the Dmc1–ssDNA presynaptic complex. This tight binding affinity is in good agreement with Hop2–Mnd1 serving as a component of the meiotic presynaptic complex. To further understand the implications of the observed Kd value, we sought to more fully understand the association and dissociation kinetics of Hop2–Mnd1 during its interactions with the Dmc1–ssDNA complexes. We first tested how quickly Hop2–Mnd1 dissociated from Dmc1–ssDNA by measuring the loss of the Hop2–Mnd1–mCherry signal over time. For these measurements we prebound Hop2–Mnd1–mCherry (10 nm) to Dmc1–ssDNA presynaptic complexes. Unbound Hop2–Mnd1–mCherry was then quickly flushed from the sample chamber while monitoring the loss of mCherry fluorescence signal from the tethered Dmc1–ssDNA molecules over time (Fig. 4A). The integrated fluorescence signal was then plotted over time and used to determine the dissociation rate constant (koff) and binding half-life (t½) for the Hop2–Mnd1–mCherry interaction with Dmc1–ssDNA (Fig. 4, B–D). This analysis revealed a koff value of 0.009 ± 0.0004 s−1, corresponding to a binding half-life of 77.9 ± 5 s (Fig. 4, B–D).
Figure 4.
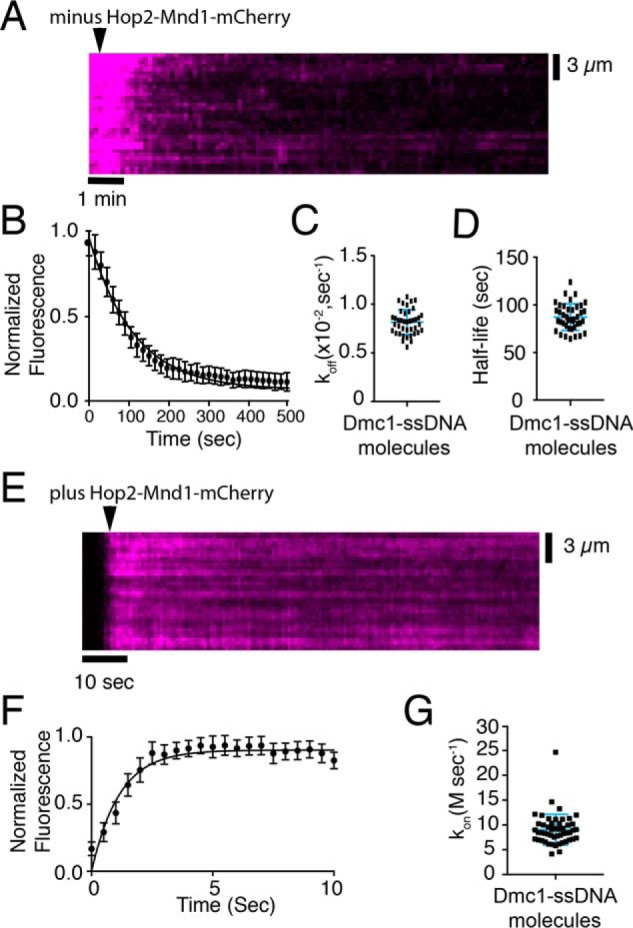
Hop2–Mnd1 dissociation and association kinetics. A, representative kymograph measuring Hop2–Mnd1 dissociation from Dmc1–ssDNA molecules. The arrowhead highlights the time point at which free Hop2–Mnd1–mCherry was flushed from the sample chamber, and the dissociation of Hop2–Mnd1–mCherry (magenta) is readily observed as the loss of mCherry signal. B, quantification of the normalized fluorescent intensities during dissociation of Hop2–Mnd1–mCherry from Dmc1–ssDNA. The graph represents the mean of all DNA molecules tested (n = 40), and the error bars are S.D. between individual molecules. C, measured dissociation rates (koff) of Hop2–Mnd1–mCherry for each individual Dmc1–ssDNA molecule tested (n = 40); error bars represent S.D. of the data. D, observed Hop2–Mnd1–mCherry half-lives for each individual Dmc1–ssDNA molecule that was measured. The half-life was determined from fitting the data with a single exponential decay function (n = 40). The error bars represent S.D. of the data. E, representative kymograph showing the association of Hop2–Mnd1–mCherry (magenta) with the Dmc1–ssDNA molecules (unlabeled). The arrowhead highlights the time point of the Hop2–Mnd1 injection. F, graph representing the normalized fluorescent intensity increase as Hop2–Mnd1–mCherry (10 nm) binds to Dmc1–ssDNA molecules. These data represent the mean for all molecules tested (n = 51); the solid line represents a fit to the binding data, and the error bars represent S.D. of all individual ssDNA molecules. G, association rates (kon) for each observed Dmc1–ssDNA molecule (n = 51). The error bars represent S.D. of the data.
To complete the kinetic mechanism of binding, we next tested the association rate (kon) for Hop2–Mnd1 binding to Dmc1–ssDNA. Having measured the Kd and koff, we first calculated the expected kon for Hop2–Mnd1 using the equation Kd = kon/koff. This calculation yielded a value for kon,calc of 10.4 × 107 m/s−1. We next measured the experimentally observed kon,obs value by monitoring the binding of Hop2–Mnd1–mCherry to Dmc1–ssDNA filaments in real time (Fig. 4E). We then fit the mean integrated Hop2–Mnd1–mCherry fluorescence signal intensity for all Dmc1–ssDNA molecules tested (n = 51) to an association equation (see “Experimental procedures”; note that the calculated decay rate was used as part of the fitting equation (Fig. 4F)). The data fit yielded an experimental kon,obs value of 8.5 × 107 ± 1.5 m/s with an r2 value of 0.91 (Fig. 4F). Additionally, we fit each individual Dmc1–ssDNA molecule with the same equation and found no deviation from the averaged mean (Fig. 4G). Thus, we found good agreement between the calculated and observed values for the association rate constants describing Hop2–Mnd1–mCherry binding to the Dmc1–ssDNA complexes. Our findings are most consistent with a model where the ∼12 nm Kd value for Hop2–Mnd1–mCherry association with the Dmc1–ssDNA filaments arises from a fast on-rate coupled to a relatively slow off-rate.
Punctate binding of Hop2–Mnd1 to recombinase filaments
We had shown previously that Rad51 and Dmc1 can form segregated filaments bound to the same ssDNA molecule when reconstituted in vitro (48). This observation is in good agreement with previous microscopy studies that suggested that Rad51 and Dmc1 form closely spaced but nonoverlapping filaments in vivo (18). The segregation of the two recombinases within our in vitro mixed recombinase filaments is readily revealed in experiments using a GFP-tagged version of Hed1, which is a meiosis-specific Rad51–binding protein (49–51).
As indicated above, we detected binding of Hop2–Mnd1–mCherry to Dmc1–ssDNA but did not observe any Hop2–Mnd1–mCherry binding to Rad51–ssDNA (Fig. 2D). Therefore, we next asked whether we could detect the Dmc1 subsections of mixed recombinase filaments using Hop2–Mnd1–mCherry as a Dmc1-specific binding factor. For these experiments, we reconstituted mixed recombinase filaments at a 3:1 or 1:1 ratio of Rad51 to Dmc1 and then injected 30 nm Hop2–Mnd1–mCherry into the sample chambers. The images in Fig. 5A show examples of wide-field microscope images of recombinase ssDNA molecules at each ratio of recombinase tested after incubation with 30 nm Hop2–Mnd1–mCherry. As shown above, 30 nm Hop2–Mnd1–mCherry was sufficient to uniformly coat Dmc1–ssDNA filaments (Fig. 2A). However, at a 3:1 ratio of Rad51 to Dmc1, we observed that Hop2–Mnd1–mCherry did not uniformly coat the presynaptic complexes but instead exhibited a more punctate binding pattern (Fig. 5A). We interpreted the punctate Hop2–Mnd1–mCherry binding patterns as reflecting segregation of Rad51 and Dmc1 into spatially distinct nucleoprotein filaments bound to the same ssDNA molecule, as we had observed previously from experiments using Hed1–GFP (48).
Figure 5.
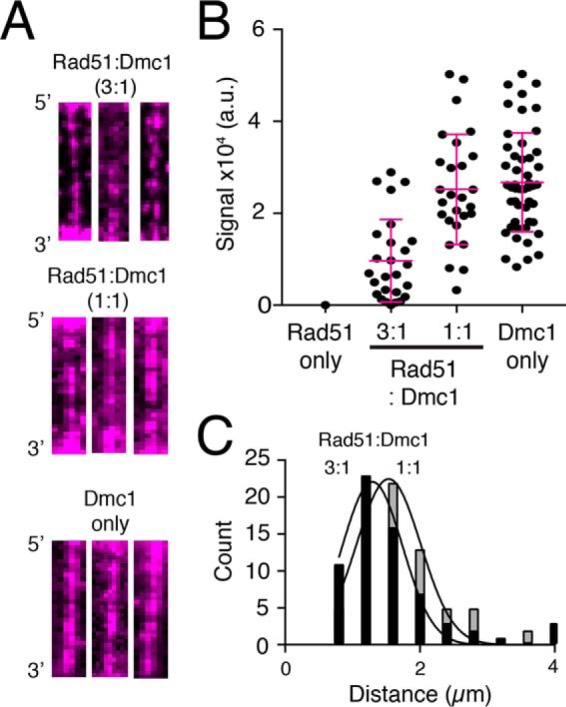
Hop2–Mnd1 binds to mixed recombinase filaments containing both Rad51 and Dmc1. A, wide-field TIRFM images of recombinase ssDNA filaments bound by 30 nm Hop2–Mnd1–mCherry (magenta) for filaments composed of 3:1 Rad51: Dmc1, 1:1 Rad51: Dmc1, and Dmc1 only, as indicated. B, distribution of integrated Hop2–Mnd1–mCherry fluorescence signal intensity values (a.u., arbitrary units) from individual ssDNA molecules for experiments conducted with Rad51 only, 3:1 Rad51:Dmc1, 1:1 Rad51:Dmc1, or Dmc1 only, as indicated. The error bars represent S.D. of the data. C, histogram showing the distribution of distances between adjacent Hop2–Mnd1–mCherry foci bound to mixed recombinase filaments composed of 3:1 Rad51:Dmc1 and 1:1 Rad51:Dmc1 ratios. The data fit a Gaussian distribution, and the mean distance between adjacent Hop2–Mnd1–mCherry foci was determined from the fit.
We also measured the Hop2–Mnd1–mCherry fluorescence signal intensity for data collected at the different ratios of Rad51 to Dmc1. The integrated fluorescence signal intensity of Hop2–Mnd1–mCherry bound to presynaptic complexes prepared at a 3:1 ratio of Rad51 to Dmc1, as expected, was significantly lower (∼50%) than the integrated signal intensity for presynaptic complexes prepared with just Dmc1 (Fig. 5B). This observation suggests that the presence of Rad51 within these mixed recombinase filaments reduces the availability of potential binding sites for Hop2–Mnd1–mCherry. Together, these results are most consistent with a model where Hop2–Mnd1 can bind to the Dmc1 portions of the mixed filaments but does not spread appreciably into the Rad51 subsections of the filaments.
For experiments performed at a 1:1 ratio of Rad51 to Dmc1, some of the presynaptic complexes (∼40–50%) still displayed punctate Hop2–Mnd1–mCherry binding patterns; however, we also observed presynaptic complexes that appeared to be more uniformly coated with Hop2–Mnd1 (Fig. 5A). This is consistent with an assembly reaction in which the formation of Dmc1 filaments is slightly favored over Rad51 even at equimolar concentration.
We also found that integrated Hop2–Mnd1–mCherry signal intensity for experiments conducted at a 1:1 ratio of Rad51 to Dmc1 was somewhat lower, albeit not significantly different from values obtained from experiments performed with just Dmc1 (Fig. 5B). However, this observation is also consistent with our previously reported observation that Hed1 staining reaches a relative minimum at a 1:1 ratio of Rad51 to Dmc1 within the context of mixed filaments (48). In addition, it is also likely that a 1:1 mixture of Rad51:Dmc1 may retain as many Hop2–Mnd1 binding sites as the Dmc1 only filaments, especially given the substoichiometric binding of Hop2–Mnd1–mCherry to Dmc1 only filaments (e.g. at 30 nm Hop2–Mnd1–mCherry, we expect only one Hop2–Mnd1 heterodimer to bind/22 molecules of Dmc1 (see Fig. 3E)).
Finally, for molecules with spatially resolvable fluorescent foci, we measured the distance between adjacent Hop2–Mnd1–mCherry foci bound to presynaptic complexes reconstituted at 3:1 or 1:1 Rad51:Dmc1 ratios. This analysis revealed mean foci-to-foci distances of 1.2 ± 0.54 and 1.5 ± 0.49 μm, respectively (Fig. 5C). In this case, we interpreted these foci-to-foci distances to reflect the average center-to-center distance between adjacent Dmc1 filaments. The ∼1.5 μm foci-to-foci value for filaments prepared at a 1:1 ratio of Rad51:Dmc1 distance is comparable with the previously reported foci-to-foci value of ∼1.7 μm for Hed1–GFP binding to mixed recombinase filaments prepared at a 1:1 ratio of Rad51:Dmc1. Note that we cannot make a comparison of Hed1–GFP foci-to-foci distances for 3:1 Rad51 to Dmc1 because Hed1–GFP completely coats the presynaptic complexes under these conditions (48).
Spatial distribution of Hed1–GFP and Hop2–Mnd1–mCherry
We had shown previously that Hed1–GFP binds only to Rad51 filaments, and here we provided evidence that Hop2–Mnd1–mCherry associates exclusively with Dmc1 filaments. Both findings are consistent with prior studies of Hed1 and Hop2–Mnd1 recombinase binding specificities (48, 49). Together, these findings suggest that during meiosis, when both Hed1 and Hop2–Mnd1 are present, each protein should associate with distinct subsections of the meiotic presynaptic complex: Hed1 should bind to the Rad51 subsections of the filaments, and Hop2–Mnd1 should bind to the Dmc1 subsections.
To test this prediction, we next performed two-color ssDNA curtain assays using presynaptic complexes reconstituted at a 1:1 ratio of Rad51:Dmc1 and examined the binding of both Hed1–GFP and Hop2–Mnd1–mCherry. Under these conditions, we observed examples where the Hop2–Mnd1–mCherry and Hed1–GFP signals were well-resolved from one another, and we also observed examples where the fluorescent signals from the two proteins appeared to overlap one another (Fig. 6, A–C). A large number of ssDNA molecules exhibited readily observed tracts of both Hop2–Mnd1–mCherry and Hed1–GFP, and in many cases these foci were clearly separated from one another (cf. Fig. 5A and Fig. 6, A–C). We interpreted these nonoverlapping fluorescent signals from Hop2—Mnd1—mCherry and Hed1–GFP as reflecting the formation of spatially distinct Dmc1 and Rad51 filaments, respectively.
Figure 6.
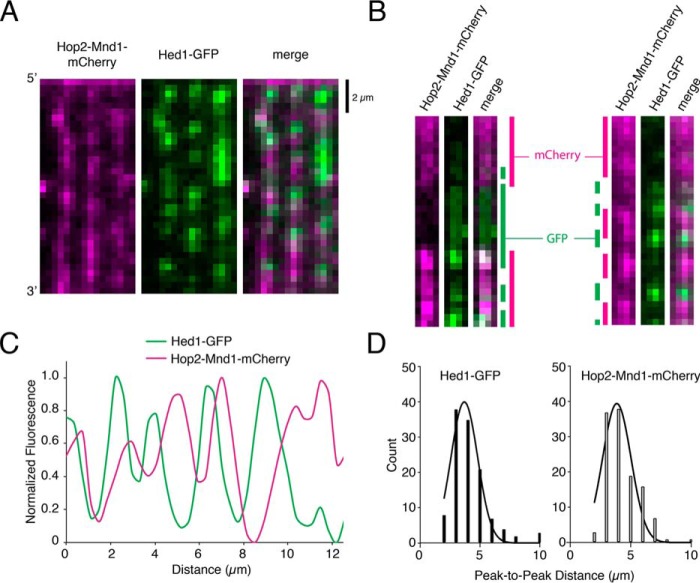
Hop2–Mnd1–mCherry and Hed1–GFP bind to separate sections of the mixed recombinase filaments. A, wide-field image of recombinase ssDNA molecules prepared at a 1:1 ratio of Rad51 to Dmc1 and bound by both 30 nm Hop2–Mnd1–mCherry (magenta) and 30 nm Hed1–GFP (green). B, typical examples of single ssDNA molecules bound by 1:1 Rad51:Dmc1 filaments followed by the addition of Hop2–Mnd1–mCherry (magenta) and Hed1–GFP (green). In these examples, the locations of Hop2–Mnd1 and Hed1 are highlighted by color-coded bars shown at the sides of the images. C, a typical normalized fluorescent signal trace showing the spatial distributions of Hop2–Mnd1–mCherry and Hed1–GFP bound to an individual ssDNA molecule that was preassembled with a 1:1 ratio of Rad51 to Dmc1. D, histograms showing the peak-to-peak distances between either Hed1–GFP foci (left panel, n = 125) or Hop2–Mnd1–mCherry foci (right panel, n = 115) for ssDNA molecules (n = 125) bound by a 1:1 ratio of Rad51 to Dmc1. The mean distance between adjacent fluorescent foci bound to the same ssDNA molecules was determined from a Gaussian fit to the data.
In addition to the well-resolved Hop2–Mnd1–mCherry and Hed1–GFP fluorescent foci described above, we also observed many examples of overlap between Hop2–Mnd1–mCherry and Hed1–GFP fluorescent signals (Fig. 6, A–C). This finding was not completely unexpected, as we anticipated that presynaptic complexes prepared at a 1:1 ratio of Rad51 to Dmc1 likely comprised a broad distribution of Rad51 and Dmc1 filament lengths. In this scenario, it is possible, if not likely, that there exist many small homotypic Rad51 and homotypic Dmc1 filaments that would not be optically resolved. The resolution of our observations was ∼1000 nucleotides of ssDNA/pixel, and we cannot as yet define the spatial distribution of proteins below this resolution limit. However, the fact that large isolated homotypic filaments were also observed supports the idea of homotypic, instead of heterotypic, filaments of Dmc1 and Rad51.
For the nonoverlapping fluorescent foci, we measured the peak-to-peak distances between Hop2–Mnd1–mCherry and Hed1—GFP–labeled filaments to estimate the relative size of each filament type. This analysis revealed a distance of 1.6 ± 0.54 μm between adjacent Hed1–GFP foci and a distance of 1.45 ± 0.49 μm between adjacent Hop2–Mnd1–mCherry, corresponding to a length of ∼3600 nucleotides or ∼1200 recombinase monomers (Fig. 6, D and E). These observations support the conclusions that Rad51 and Dmc1 form homotypic filaments and that Hop2–Mnd1 is recruited to Dmc1, whereas Hed1 is recruited to Rad51 in the context of mixed meiotic filaments.
Discussion
In this study we used single-molecule imaging to investigate the binding properties of S. cerevisiae Hop2–Mnd1 during its interactions with presynaptic complexes comprising either Dmc1–ssDNA, Rad51–ssDNA, or mixtures of both recombinases. Our work revealed that Hop2–Mnd1 binds with high affinity to Dmc1–ssDNA and exhibits binding kinetics that are characterized by a rapid rate of association and a slower rate of dissociation. We saw no evidence of Hop2–Mnd1 interactions with Rad51–ssDNA, consistent with prior reports indicating that S. cerevisiae Hop2–Mnd1 is specific for Dmc1 (40). Finally, we have provided evidence that Hop2–Mnd1 binds specifically to Dmc1 even in the context of mixed meiotic filaments. These observations provide new mechanistic insights into how Hop2–Mnd1 associates with the presynaptic complex during homologous recombination and suggest a model wherein meiotic presynaptic complexes are composed of distinct subsets of recombination factors whose spatial distribution is defined by the underlying distribution of segregated Rad51 and Dmc1 filaments.
Our work provides detailed insights into the kinetics of Hop2–Mnd1 association with Dmc1 filaments. These results indicate that Hop2–Mnd1 binds very quickly to Dmc1–ssDNA filaments, exhibiting an experimentally measured association rate constant of kon = 8.5 × 107 m−1/s−1. Hop2–Mnd1 dissociates more slowly, exhibiting a dissociation rate constant of koff = 0.009 ± 0.0004 s−1, which corresponds to a half-life of ∼1.3 min. Although this could be considered a relatively long half-life, we note that it is much shorter than other components of the presynaptic complex that we measured in our assays. For instance, Dmc1, Rad51, Hed1, Rad52, and Rad54 do not exhibit any evidence of appreciable dissociation over the course of a 2-h observation periods (27, 49, 74, 75). We speculated that the more rapid turnover observed for Hop2–Mnd1 may reflect its function in promoting dsDNA capture by the Dmc1–ssDNA presynaptic complex (see below).
In the context of mixed meiotic filaments, we observed that Hop2–Mnd1 specifically associated with Dmc1 regions of the presynaptic filaments and showed a significant degree of isolation from the Rad51-specific–binding protein Hed1. There was some overlap between Hop2–Mnd1 and Hed1; however the fact that large segregated regions of the filament were optically resolvable suggests that regions of overlapping signal may simply represent areas containing shorter Dmc1 and Rad51 filaments that are not readily resolved by optical microscopy.
The biological importance of our findings lies in how meiosis-specific proteins are sorted in the context of mixed meiotic filaments. In this case, Hed1 and Hop2–Mnd1 are anticipated to exist within spatially distinct subregions of the meiotic presynaptic complex by virtue of the underlying spatial segregation of Rad51 and Dmc1 into separate filaments (Fig. 7A). Hed1 blocks the binding of Rad54 to Rad51, thus down-regulating the strand exchange activity of Rad51 (49–52, 76, 77). Rad54 has been implicated in numerous processes during mitotic HR, including the homology search, chromatin remodeling, strand invasion, and removal of Rad51 from heteroduplex dsDNA upon completion of strand invasion (4, 78–80). Thus, Hed1-mediated down-regulation of Rad54 during meiosis may affect any or all of these processes. As indicated above, Hop2–Mnd1 is thought to promote dsDNA capture by Dmc1, thus enhancing the ability of Dmc1 to conduct the homology search (40, 56, 65, 67). Taken together, these findings lead to a model in which recombinases that have otherwise very similar biochemical activities can be selectively activated or deactivated by the presence of the appropriate recombinase-specific regulatory proteins (Fig. 7B). Although our work focuses on S. cerevisiae as a model system, it also seems plausible that a similar mechanism involving the self-segregation of the Rad51 and Dmc1 recombinases, and the binding of recombinase-specific proteins, may impact the organization of meiotic presynaptic complexes in other eukaryotes.
Figure 7.
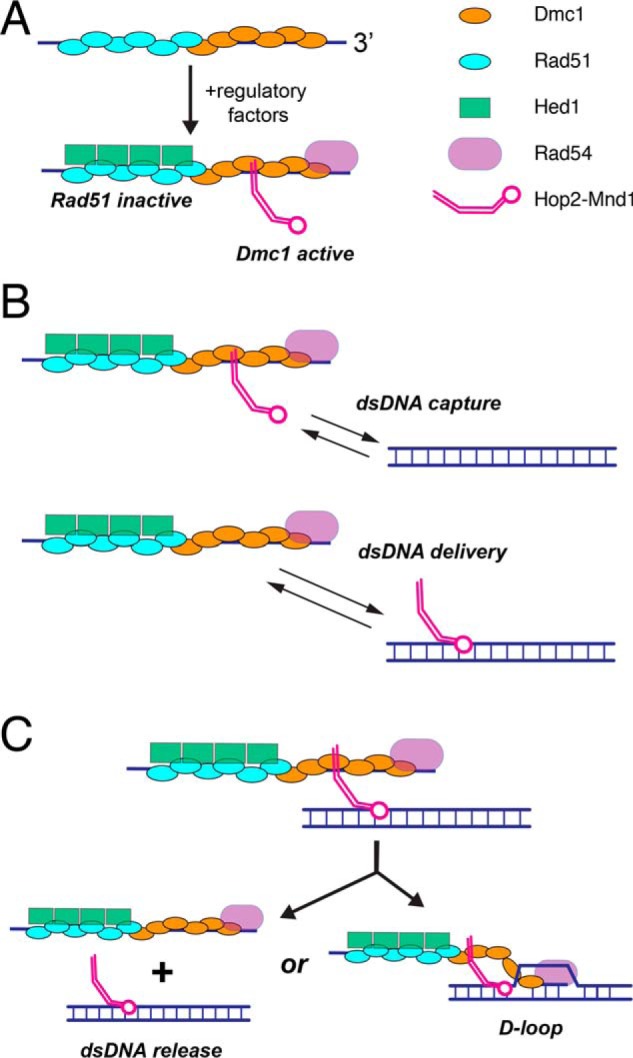
Models for Hop2–Mnd1 interactions with the meiotic presynaptic complex. A, proposed mechanism for regulating the activation of Dmc1 and the inactivation of Rad51 within meiotic presynaptic filaments through the combined actions of Hop2–Mnd1, Hed1, and Rad54. B, models describing possible roles of Hop2–Mnd1 in mediating the interactions between the meiotic presynaptic complex and potential dsDNA templates. In the dsDNA capture model, Hop2–Mnd1 is bound in cis to the Dmc1 subsections of the meiotic presynaptic complex and utilizes its dsDNA-binding domain to facilitate dsDNA capture. In the dsDNA delivery model, Hop2–Mnd1 is bound to the dsDNA and then delivers this dsDNA to the presynaptic complex. C, the presynaptic complex can promote D-loop formation when a homologous dsDNA is available. If the dsDNA substrate bound by Hop2–Mnd1 is not homologous to the Dmc1-bound ssDNA, then the turnover of Hop2–Mnd1 may allow for release of the nonhomologous DNA. Additional descriptions of these models are provided under “Discussion.”
Although many questions remain, Hop2–Mnd1 is believed to promote dsDNA capture by Dmc1 during the homology search (57, 60–62). In this model, Hop2–Mnd1 promotes homologous recombination by promoting dsDNA interactions with the Dmc1–ssDNA presynaptic complex, but the details of this mechanism remain unknown. In principle, Hop2–Mnd1 could function through a cis-acting mechanism, where Hop2–Mnd1 associates first with Dmc1–ssDNA and then allows for more efficient capture of dsDNA (Fig. 7B). Alternatively, dsDNA delivery by Hop2–Mnd1 could occur either in trans, where Hop2–Mnd1 associates first with dsDNA and then helps deliver the dsDNA to Dmc1–ssDNA (Fig. 7B). Our current work does not directly distinguish between these two models. However, our observation that the binding interaction of Hop2–Mnd1 with Dmc1–ssDNA is relatively short-lived (t½ ∼ 1.3 min) may reflect a requirement that the Dmc1–dsDNA complex be quickly released from Dmc1–ssDNA if the bound dsDNA is not an appropriate template for guiding DSB repair (Fig. 7C). Thus, the rapid release of Hop2–Mnd1 from the presynaptic complex may play a role in the Dmc1-mediated homology search mechanism by ensuring turnover of nonhomologous dsDNA substrates. An important next stage of this research will be to begin examining how Hop2–Mnd1 influences the interactions of Dmc1–ssDNA with dsDNA substrates.
Interestingly, yeast Hop2–Mnd1 is expressed only during meiosis and is restricted to interactions with the meiosis-specific recombinase Dmc1 (40). However, Hop2–Mnd1 in higher eukaryotes also supports Rad51-mediated HR reactions and may function during both mitotic and meiotic recombination. Thus, our studies of yeast Hop2–Mnd1 may also lead to new insights into recombination mechanisms from other organisms.
Experimental procedures
Proteins
S. cerevisiae RPA, Rad51, Dmc1, GFP–RPA, mCherry–RPA, GST–Hed1–mCherry, and GST–Hed1–GFP were all purified as described previously (27, 48, 49, 74). His6–Hop2–Mnd1 and His6–Hop2–Mnd1–mCherry were purified as follows. The pET-Duet plasmid, containing ORFs of Hop2 and Mnd1 plasmids, was transformed into E. coli Rosetta(DE3) cells (Novagen). Cell were grown to an OD of 0.6–0.8 at 37 °C, and cultures were then shifted to 16 °C and induced overnight with 0.3 mm isopropyl 1-thio-β-d-galactopyranoside. After overnight expression, cells were harvested and resuspended in 10 ml of cell lysis buffer (CLB) (25 mm Tris-Cl (pH 7.5),1 m KCl, 1 mm EDTA, 5 mm β-mercaptoethanol, 2 mm imidazole, protease inhibitor mixture (Roche, catalog No. 05892988001), 10% glycerol, and 1 mm phenylmethylsulfonyl fluoride) per liter of cell culture. Cells were lysed with lysozyme and sonication. The lysate was clarified by ultracentrifugation for 45 min at 95,000 × g. The clarified extract was incubated in-batch with cOmplete nickel resin (Roche, catalog No. 05893682001) for 1 h at 4 °C. The resin was then washed twice in 5 column volumes of CLB followed by elution in CLB plus 150 mm imidazole. The peak fractions were pooled and then loaded onto a Superdex 200 size exclusion column (120 ml) pre-equilibrated with SEC buffer (25 mm Tris-Cl (pH 7.5), 500 mm KCl, 1 mm EDTA, 5 mm β-mercaptoethanol, and 10% glycerol). The column was developed at 1 ml/min, and 1.5-ml fractions were collected. The protein eluted at a molecular mass of around 50,000 Da based on size standards, and peak fractions were pooled and quantified by absorbance at 280 nm; in the case of mCherry protein concentrations, they were quantified by measuring the absorbance of the chromophores at 587 nm (ϵ587 nm = 72,000 cm−1m−1). Samples were flash-frozen and stored at −80 °C.
TIRFM experiments
All experiments were conducted with a custom-built prism-type TIRF microscope (Nikon) equipped with a 488-nm sapphire laser (200 milliwatts, Coherent) and a 561-nm sapphire laser (200 milliwatts, Coherent) as described previously (48, 72, 73). For all two-color images, we used a custom-built shuttering system to avoid the bleed-through from the green into the red channel during image acquisition. With this system, images from the green (GFP) and the red (mCherry) channels are recorded independently. These recordings are offset by 100 ms, such that when one camera records the red channel image, the green laser is shuttered off, and vice versa (48, 49, 73, 81).
Flow cells were constructed by deposition of chrome barriers on quartz microscope slides via electron beam lithography and thermal evaporation as described (82). Bilayers were prepared with 91.5% DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine), 0.5% biotinylated-PE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(cap-biotinyl)), and 8% mPEG 2000–DOPE (18:1 PEG 2000:1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000]). Bilayers were deposited through sequential injections of a lipid master mix in lipid buffer (20 mm Tris-Cl (pH 7.5) and 100 mm NaCl) as described (72, 73, 81, 82). The surface was then blocked with Dmc1 buffer (30 mm Tris-Cl (pH 7.5), 100 mm KCl, 5 mm MgCl2, 2 mm CaCl2, 0.2 mg/ml BSA, and 1 mm DTT) and conjugated to streptavidin to provide attachment points for tethering the biotinylated ssDNA. The ssDNA substrate was prepared by rolling circle replication using phi29 DNA polymerase with a biotinylated primer annealed to M13 circular ssDNA as a template (72, 75). The ssDNA was then deposited onto the bilayer, and the flow cell was attached to the microfluidic system.
Rad51 and Dmc1 filament assembly
Rad51 and Dmc1 filaments were prepared as described previously (48, 49). In brief, The ssDNA molecules were aligned along the diffusion barriers at a flow rate of 0.5 ml/min in Dmc1 buffer plus RPA (30 mm Tris-Cl (pH 7.5), 100 mm KCl, 5 mm MgCl2, 2 mm CaCl2, 0.2 mg/ml BSA, 1 mm DTT, and 0.1 nm RPA–GFP or RPA–mCherry). Once molecules were aligned, the flow rate was adjusted to 1.0 ml/min, and 0.5 ml of 7 m urea was injected into the flow cell to disrupt any remaining secondary structure; plus RPA–GFP or RPA–mCherry (0.1 nm) was then flushed through the sample chamber at 1.0 ml/min for 4 min. After 4 min, Dmc1 buffer plus ATP (30 mm Tris-Cl (pH 7.5), 100 mm KCl, 5 mm MgCl2, 2 mm CaCl2, 0.2 mg/ml BSA, 1 mm DTT, plus 2.5 mm ATP) was flushed through the sample chamber at 1.0 ml/min for 3 min. Rad51, Dmc1, or mixed ratios of the two were injected (2 μm) into the flow cell, buffer flow was terminated, and the reaction was incubated at 30 °C for 20 min to allow filament assembly as described (27, 48, 49, 72). The RPA fluorescence signal was then monitored to verify filament assembly. Following this 15-min incubation, free recombinase was flushed from the sample chamber with Dmc1 buffer plus ATP.
Data acquisition and analysis
All data were collected with 100-ms integration time. Image acquisition frequency was varied as required for each specific experiment, and the laser was shuttered between frames to minimizing photobleaching as described (48, 49, 81). Images collected using Nikon software were exported as individual TIFF images. TIFF stacks were imported into ImageJ (Fiji) as described (73, 81). For two-color imaging, the two channels were first corrected for stage drift and then merged into TIFF images, which were then converted to TIFF stacks. For each time course experiment, kymographs were generated from the TIFF image stacks by defining a 1-pixel-wide region of interest (ROI) along the axis of each individual ssDNA molecule, and these ROI were extracted from each image within the TIFF stack. All of the slices corresponding to one ssDNA molecule were then aligned to yield a kymograph representing the entire experimental time course; this process was repeated for each ssDNA molecule that was analyzed. For ssDNA intensity analysis, a 35-pixel ROI was taken from individual DNA molecules, and the pixel intensity was summed over the length of the DNA to generate an intensity value. A separate 35-pixel ROI was also taken from a region with no DNA molecules, and the intensity measured. This was subtracted from all DNA samples as background, and the resulting background-subtracted values were averaged. For kinetic measurements, the background was subtracted, and the data were normalized by setting the peak intensity to 1 and considering all other signals relative to 1. The data were then either fit by a single exponential decay curve or to an association curve of the form,
| (Eq. 1) |
where
| (Eq. 2) |
and
| (Eq. 3) |
Curve fitting was performed using the program GraphPad Prism 7. For association data, the fitting required the use of the measured koff value.
Author contributions
J. B. C. conceptualization; J. B. C. and Y. K. investigation; J. B. C. writing-original draft; J. B. C. and E. C. G. writing-review and editing; Y. K. methodology; P. S. and E. C. G. funding acquisition; E. C. G. supervision; E. C. G. project administration.
Acknowledgments
We thank members of the Greene and Sung laboratories for comments on the manuscript.
This research was supported by National Institutes of Health Grants R35 GM118026 (to E. C. G.), R01 ES007061, RO1 CA220123, and P30 CA054174 (to P. S.), and PO1 CA092584 (to E. C. G. and P. S.) and National Science Foundation (NSF) Grant MCB1154511 (to E. C. G.). The authors declare they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- HR
- homologous recombination
- DSB
- double-strand DNA break(s)
- RPA
- replication protein A
- TIRFM
- total internal reflection microscopy
- CLB
- cell lysis buffer
- ROI
- region of interest.
References
- 1. Hunter N. (2015) Meiotic recombination: The essence of heredity. Cold Spring Harb. Perspect. Biol. 7, a016618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bernstein K. A., Gangloff S., and Rothstein R. (2010) The RecQ DNA helicases in DNA repair. Annu. Rev. Genet. 44, 393–417 10.1146/annurev-genet-102209-163602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Prakash R., Zhang Y., Feng W., and Jasin M. (2015) Homologous recombination and human health: The roles of BRCA1, BRCA2 and associated proteins. Cold Spring Harb. Perspect. Biol. 7, a016600 10.1101/cshperspect.a016600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Heyer W. D., Ehmsen K. T., and Liu J. (2010) Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 44, 113–139 10.1146/annurev-genet-051710-150955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sung P., and Klein H. (2006) Mechanism of homologous recombination: Mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 7, 739–750 10.1038/nrm2008 [DOI] [PubMed] [Google Scholar]
- 6. Symington L. S., Rothstein R., and Lisby M. (2014) Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics 198, 795–835 10.1534/genetics.114.166140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. San Filippo J., Sung P., and Klein H. (2008) Mechanism of EUkaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257 10.1146/annurev.biochem.77.061306.125255 [DOI] [PubMed] [Google Scholar]
- 8. Haber J. E. (2012) Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics 191, 33–64 10.1534/genetics.111.134577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Keeney S., Giroux C. N., and Kleckner N. (1997) Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384 10.1016/S0092-8674(00)81876-0 [DOI] [PubMed] [Google Scholar]
- 10. Keeney S. (2008) Spo11 and the Formation of DNA Double-strand breaks in meiosis. Genome Dyn. Stab. 2, 81–123 10.1007/7050_2007_026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Symington L. S. (2014) End resection at double-strand breaks: Mechanism and regulation. Cold Spring Harb. Perspect. Biol. 6, a016436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Symington L. S. (2016) Mechanism and regulation of DNA end resection in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 51, 195–212 10.3109/10409238.2016.1172552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mimitou E. P., Yamada S., and Keeney S. (2017) A global view of meiotic double-strand break end resection. Science 355, 40–45 10.1126/science.aak9704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cejka P., Cannavo E., Polaczek P., Masuda-Sasa T., Pokharel S., Campbell J. L., and Kowalczykowski S. C. (2010) DNA end resection by Dna2–Sgs1–RPA and its stimulation by Top3–Rmi1 and Mre11–Rad50–Xrs2. Nature 467, 112–116 10.1038/nature09355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen H., Lisby M., and Symington L. S. (2013) RPA coordinates DNA end resection and prevents formation of DNA hairpins. Mol. Cell 50, 589–600 10.1016/j.molcel.2013.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Niu H., Chung W. H., Zhu Z., Kwon Y., Zhao W., Chi P., Prakash R., Seong C., Liu D., Lu L., Ira G., and Sung P. (2010) Mechanism of the ATP-dependent DNA end resection machinery from S. cerevisiae. Nature 467, 108–111 10.1038/nature09318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brown M. S., and Bishop D. K. (2014) DNA Strand exchange and RecA homologs in meiosis. Cold Spring Harb. Perspect. Biol. 7, a016659 10.1101/cshperspect.a016659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brown M. S., Grubb J., Zhang A., Rust M. J., and Bishop D. K. (2015) Small Rad51 and Dmc1 complexes often co-occupy both Ends of a meiotic DNA double strand break. PLoS Genet. 11, e1005653 10.1371/journal.pgen.1005653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sheridan S., and Bishop D. K. (2006) Red-Hed regulation: Recombinase Rad51, though capable of playing the leading role, may be relegated to supporting Dmc1 in budding yeast meiosis. Genes Dev. 20, 1685–1691 10.1101/gad.1447606 [DOI] [PubMed] [Google Scholar]
- 20. Bishop D. K. (1994) RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell 79, 1081–1092 10.1016/0092-8674(94)90038-8 [DOI] [PubMed] [Google Scholar]
- 21. Kowalczykowski S. C. (2015) An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 7, a016410 10.1101/cshperspect.a016410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sheridan S. D., Yu X., Roth R., Heuser J. E., Sehorn M. G., Sung P., Egelman E. H., and Bishop D. K. (2008) A comparative analysis of Dmc1 and Rad51 nucleoprotein filaments. Nucleic Acids Res. 36, 4057–4066 10.1093/nar/gkn352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Conway A. B., Lynch T. W., Zhang Y., Fortin G. S., Fung C. W., Symington L. S., and Rice P. A. (2004) Crystal structure of a Rad51 filament. Nat. Struct. Mol. Biol. 11, 791–796 10.1038/nsmb795 [DOI] [PubMed] [Google Scholar]
- 24. Xu J., Zhao L., Xu Y., Zhao W., Sung P., and Wang H. W. (2017) Cryo-EM structures of human recombinase RAD51 filaments in the catalysis of DNA strand exchange. Nat. Struct. Mol. Biol. 24, 40–46 10.1038/nsmb.3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen Z., Yang H., and Pavletich N. P. (2008) Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature 453, 489–494 10.1038/nature06971 [DOI] [PubMed] [Google Scholar]
- 26. Hunter N., and Kleckner N. (2001) The single-end invasion: An asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell 106, 59–70 10.1016/S0092-8674(01)00430-5 [DOI] [PubMed] [Google Scholar]
- 27. Qi Z., Redding S., Lee J. Y., Gibb B., Kwon Y., Niu H., Gaines W. A., Sung P., and Greene E. C. (2015) DNA sequence alignment by microhomology sampling during homologous recombination. Cell 160, 856–869 10.1016/j.cell.2015.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Greene E. C. (2016) DNA sequence alignment during homologous recombination. J. Biol. Chem. 291, 11572–11580 10.1074/jbc.R116.724807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Renkawitz J., Lademann C. A., and Jentsch S. (2014) Mechanisms and principles of homology search during recombination. Nat. Rev. Mol. Cell Biol. 15, 369–383 10.1038/nrm3805 [DOI] [PubMed] [Google Scholar]
- 30. Zickler D., and Kleckner N. (2015) Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb. Perspect. Biol. 7, a016626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chang H. Y., Liao C. Y., Su G. C., Lin S. W., Wang H. W., and Chi P. (2015) Functional relationship of ATP hydrolysis, presynaptic filament stability, and homologous DNA pairing activity of the human meiotic recombinase DMC1. J. Biol. Chem. 290, 19863–19873 10.1074/jbc.M115.666289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cox M. M. (2007) Motoring along with the bacterial RecA protein. Nat. Rev. Cell Biol. 8, 127–138 10.1038/nrm2099 [DOI] [PubMed] [Google Scholar]
- 33. Lee J. Y., Terakawa T., Qi Z., Steinfeld J. B., Redding S., Kwon Y., Gaines W. A., Zhao W., Sung P., and Greene E. C. (2015) Base triplet stepping by the Rad51/RecA family of recombinases. Science 349, 977–981 10.1126/science.aab2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bzymek M., Thayer N. H., Oh S. D., Kleckner N., and Hunter N. (2010) Double Holliday junctions are intermediates of DNA break repair. Nature 464, 937–941 10.1038/nature08868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Morrical S. W. (2015) DNA-pairing and annealing processes in homologous recombination and homology-directed repair. Cold Spring Harb. Perspect. Biol. 7, a016444 10.1101/cshperspect.a016444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Matos J., and West S. C. (2014) Holliday junction resolution: Regulation in space and time. DNA Repair 19, 176–181 10.1016/j.dnarep.2014.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Anand R. P., Lovett S. T., and Haber J. E. (2013) Break-induced DNA replication. Cold Spring Harb. Perspect. Biol. 5, a010397 10.1101/cshperspect.a010397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mitchel K., Lehner K., and Jinks-Robertson S. (2013) Heteroduplex DNA position defines the roles of the Sgs1, Srs2, and Mph1 helicases in promoting distinct recombination outcomes. PLoS Genet. 9, e1003340 10.1371/journal.pgen.1003340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lin Z., Kong H., Nei M., and Ma H. (2006) Origins and evolution of the recA/RAD51 gene family: Evidence for ancient gene duplication and endosymbiotic gene transfer. Proc. Natl. Acad. Sci. U.S.A. 103, 10328–10333 10.1073/pnas.0604232103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chan Y. L., Brown M. S., Qin D., Handa N., and Bishop D. K. (2014) The third exon of the budding yeast meiotic recombination gene HOP2 is required for calcium-dependent and recombinase Dmc1-specific stimulation of homologous strand assimilation. J. Biol. Chem. 289, 18076–18086 10.1074/jbc.M114.558601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chi P., Kwon Y., Moses D. N., Seong C., Sehorn M. G., Singh A. K., Tsubouchi H., Greene E. C., Klein H. L., and Sung P. (2009) Functional interactions of meiotic recombination factors Rdh54 and Dmc1. DNA Repair 8, 279–284 10.1016/j.dnarep.2008.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sung P. (1994) Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science 265, 1241–1243 10.1126/science.8066464 [DOI] [PubMed] [Google Scholar]
- 43. Cloud V., Chan Y. L., Grubb J., Budke B., and Bishop D. K. (2012) Dmc1 catalyzes interhomolog joint molecule formation in meiosis with Rad51 and Mei5-Sae3 as accessory factors. Science 337, 1222–1225 10.1126/science.1219379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bishop D. K. (2012) Rad51, the lead in mitotic recombinational DNA repair, plays a supporting role in budding yeast meiosis. Cell Cycle 11, 4105–4106 10.4161/cc.22396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hong E. L., Shinohara A., and Bishop D. K. (2001) Saccharomyces cerevisiae Dmc1 protein promotes renaturation of single-strand DNA (ssDNA) and assimilation of ssDNA into homologous super-coiled duplex DNA. J. Biol. Chem. 276, 41906–41912 10.1074/jbc.M105563200 [DOI] [PubMed] [Google Scholar]
- 46. Busygina V., Gaines W. A., Xu Y., Kwon Y., Williams G. J., Lin S. W., Chang H. Y., Chi P., Wang H. W., and Sung P. (2013) Functional attributes of the S. cerevisiae meiotic recombinase Dmc1. DNA Repair 12, 707–712 10.1016/j.dnarep.2013.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee M. H., Chang Y. C., Hong E. L., Grubb J., Chang C. S., Bishop D. K., and Wang T. F. (2005) Calcium ion promotes yeast Dmc1 activity via formation of long and fine helical filaments with single-stranded DNA. J. Biol. Chem. 280, 40980–40984 10.1074/jbc.M505896200 [DOI] [PubMed] [Google Scholar]
- 48. Crickard J. B., Kaniecki K., Kwon Y., Sung P., and Greene E. C. (2018) Spontaneous self-segregation of Rad51 and Dmc1 DNA recombinases within mixed recombinase filaments. J. Biol. Chem. 293, 4191–4200 10.1074/jbc.RA117.001143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Crickard J. B., Kaniecki K., Kwon Y., Sung P., Lisby M., and Greene E. C. (2018) Regulation of Hed1 and Rad54 binding during maturation of the meiosis-specific presynaptic complex. EMBO J. 37, e98728 10.15252/embj.201798728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Busygina V., Saro D., Williams G., Leung W.-K., Say A. F., Sehorn M. G., Sung P., and Tsubouchi H. (2012) Novel attributes of Hed1 affect dynamics and activity of the Rad51 presynaptic filament during meiotic recombination. J. Biol. Chem. 287, 1566–1575 10.1074/jbc.M111.297309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Busygina V., Sehorn M. G., Shi I. Y., Tsubouchi H., Roeder G. S., and Sung P. (2008) Hed1 regulates Rad51-mediated recombination via a novel mechanism. Genes Dev. 22, 786–795 10.1101/gad.1638708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tsubouchi H., and Roeder G. S. (2006) Budding yeast Hed1 down-regulates the mitotic recombination machinery when meiotic recombination is impaired. Genes Dev. 20, 1766–1775 10.1101/gad.1422506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Niu H., Li X., Job E., Park C., Moazed D., Gygi S. P., and Hollingsworth N. M. (2007) Mek1 kinase is regulated to suppress double-strand break repair between sister chromatids during budding yeast meiosis. Mol. Cell. Biol. 27, 5456–5467 10.1128/MCB.00416-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Callender T. L., Laureau R., Wan L., Chen X., Sandhu R., Laljee S., Zhou S., Suhandynata R. T., Prugar E., Gaines W. A., Kwon Y., Börner G. V., Nicolas A., Neiman A. M., and Hollingsworth N. M. (2016) Mek1 down regulates Rad51 activity during yeast meiosis by phosphorylation of Hed1. PLoS Genet. 12, e1006226 10.1371/journal.pgen.1006226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu Y., Gaines W. A., Callender T., Busygina V., Oke A., Sung P., Fung J. C., and Hollingsworth N. M. (2014) Down-regulation of Rad51 activity during meiosis in yeast prevents competition with Dmc1 for repair of double-strand breaks. PLoS Genet. 10, e1004005 10.1371/journal.pgen.1004005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pezza R. J., Petukhova G. V., Ghirlando R., and Camerini-Otero R. D. (2006) Molecular activities of meiosis-specific proteins Hop2, Mnd1, and the Hop2-Mnd1 complex. J. Biol. Chem. 281, 18426–18434 10.1074/jbc.M601073200 [DOI] [PubMed] [Google Scholar]
- 57. Tsubouchi H., and Roeder G. S. (2002) The Mnd1 protein forms a complex with Hop2 to promote homologous chromosome pairing and meiotic double-strand break repair. Mol. Cell. Biol. 22, 3078–3088 10.1128/MCB.22.9.3078-3088.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pezza R. J., Voloshin O. N., Vanevski F., and Camerini-Otero R. D. (2007) Hop2/Mnd1 acts on two critical steps in Dmc1-promoted homologous pairing. Genes Dev. 21, 1758–1766 10.1101/gad.1562907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Leu J. Y., Chua P. R., and Roeder G. S. (1998) The Meiosis-specific Hop2 protein of S. cerevisiae ensures synapsis between homologous chromosomes. Cell 94, 375–386 10.1016/S0092-8674(00)81480-4 [DOI] [PubMed] [Google Scholar]
- 60. Gerton J. L., and DeRisi J. L. (2002) Mnd1p: An evolutionarily conserved protein required for meiotic recombination. Proc. Natl. Acad. Sci. U.S.A. 99, 6895–6900 10.1073/pnas.102167899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tsubouchi H., and Roeder G. S. (2003) The Importance of genetic recombination for fidelity of chromosome pairing in meiosis. Dev. Cell 5, 915–925 10.1016/S1534-5807(03)00357-5 [DOI] [PubMed] [Google Scholar]
- 62. Henry J. M., Camahort R., Rice D. A., Florens L., Swanson S. K., Washburn M. P., and Gerton J. L. (2006) Mnd1/Hop2 facilitates Dmc1-dependent interhomolog crossover formation in meiosis of budding yeast. Mol. Cell. Biol. 26, 2913–2923 10.1128/MCB.26.8.2913-2923.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gerton J. L., and Hawley R. S. (2005) Homologous chromosome interactions in meiosis: diversity amidst conservation. Nat. Rev. Genet. 6, 477–487 10.1038/nrg1614 [DOI] [PubMed] [Google Scholar]
- 64. Cho Nam W., Dilley Robert L., Lampson Michael A., and Greenberg Roger A.. Interchromosomal homology searches drive directional ALT telomere movement and synapsis. Cell 159, 108–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhao W., and Sung P. (2015) Significance of ligand interactions involving Hop2-Mnd1 and the RAD51 and DMC1 recombinases in homologous DNA repair and XX ovarian dysgenesis. Nucleic Acids Res. 43, 4055–4066 10.1093/nar/gkv259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pezza R. J., Voloshin O. N., Volodin A. A., Boateng K. A., Bellani M. A., Mazin A. V., and Camerini-Otero R. D. (2014) The dual role of HOP2 in mammalian meiotic homologous recombination. Nucleic Acids Res. 42, 2346–2357 10.1093/nar/gkt1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhao W., Saro D., Hammel M., Kwon Y., Xu Y., Rambo R. P., Williams G. J., Chi P., Lu L., Pezza R. J., Camerini-Otero R. D., Tainer J. A., Wang H. W., and Sung P. (2014) Mechanistic insights into the role of Hop2–Mnd1 in meiotic homologous DNA pairing. Nucleic Acids Res. 42, 906–917 10.1093/nar/gkt924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Peng M., Yang Z., Zhang H., Jaafar L., Wang G., Liu M., Flores-Rozas H., Xu J., Mivechi N. F., and Ko L. (2013) GT198 splice variants display dominant-negative activities and are induced by inactivating mutations. Genes Cancer 4, 26–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chen Y. K., Leng C. H., Olivares H., Lee M. H., Chang Y. C., Kung W. M., Ti S. C., Lo Y. H., Wang A. H., Chang C. S., Bishop D. K., Hsueh Y. P., and Wang T. F. (2004) Heterodimeric complexes of Hop2 and Mnd1 function with Dmc1 to promote meiotic homolog juxtaposition and strand assimilation. Proc. Natl. Acad. Sci. U.S.A. 101, 10572–10577 10.1073/pnas.0404195101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kang H. A., Shin H. C., Kalantzi A. S., Toseland C. P., Kim H. M., Gruber S., Peraro M. D., and Oh B. H. (2015) Crystal structure of Hop2–Mnd1 and mechanistic insights into its role in meiotic recombination. Nucleic Acids Res. 43, 3841–3856 10.1093/nar/gkv172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Moktan H., Guiraldelli M. F., Eyster C. A., Zhao W., Lee C. Y., Mather T., Camerini-Otero R. D., Sung P., Zhou D. H., and Pezza R. J. (2014) Solution structure and DNA-binding properties of the winged helix domain of the meiotic recombination HOP2 protein. J. Biol. Chem. 289, 14682–14691 10.1074/jbc.M114.548180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Qi Z., and Greene E. C. (2016) Visualizing recombination intermediates with single-stranded DNA curtains. Methods 105, 62–74 10.1016/j.ymeth.2016.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ma C. J., Steinfeld J. B., and Greene E. C. (2017) Single-stranded DNA curtains for studying homologous recombination. Methods Enzymol. 582, 193–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gibb B., Ye L. F., Kwon Y., Niu H., Sung P., and Greene E. C. (2014) Protein dynamics during presynaptic complex assembly on individual ssDNA molecules. Nat. Struct. Mol. Biol. 21, 893–900 10.1038/nsmb.2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gibb B., Ye L. F., Gergoudis S. C., Kwon Y., Niu H., Sung P., and Greene E. C. (2014) Concentration-dependent exchange of replication protein A on single-stranded DNA revealed by single-molecule imaging. PLoS One 9, e87922 10.1371/journal.pone.0087922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lao J. P., Cloud V., Huang C. C., Grubb J., Thacker D., Lee C. Y., Dresser M. E., Hunter N., and Bishop D. K. (2013) Meiotic crossover control by concerted action of Rad51-Dmc1 in homolog template bias and robust homeostatic regulation. PLoS Genet. 9, e1003978 10.1371/journal.pgen.1003978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Niu H., Wan L., Busygina V., Kwon Y., Allen J. A., Li X., Kunz R. C., Kubota K., Wang B., Sung P., Shokat K. M., Gygi S. P., and Hollingsworth N. M. (2009) Regulation of meiotic recombination via Mek1-mediated Rad54 phosphorylation. Mol. Cell 36, 393–404 10.1016/j.molcel.2009.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Heyer W. D., Li X., Rolfsmeier M., and Zhang X. P. (2006) Rad54: the Swiss Army knife of homologous recombination? Nucleic Acids Res. 34, 4115–4125 10.1093/nar/gkl481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wright W. D., and Heyer W. D. (2014) Rad54 functions as a Heteroduplex DNA pump modulated by its DNA substrates and Rad51 during D-loop formation in homologous recombination. Mol. Cell 53, 420–432 10.1016/j.molcel.2013.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Li X., Zhang X. P., Solinger J. A., Kiianitsa K., Yu X., Egelman E. H., and Heyer W. D. (2007) Rad51 and Rad54 ATPase activities are both required to modulate Rad51-dsDNA filament dynamics. Nucleic Acids Res. 35, 4124–4140 10.1093/nar/gkm412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. De Tullio L., Kaniecki K., and Greene E. C. (2018) Single-stranded DNA curtains for studying the Srs2 helicase using total internal reflection fluorescence microscopy. Methods Enzymol. 407–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Collins B. E., Ye L. F., Duzdevich D., and Greene E. C. (2014) DNA curtains: Novel tools for imaging protein–nucleic acid interactions at the single-molecule level. Methods Cell Biol. 123, 217–234 [DOI] [PubMed] [Google Scholar]