Abstract
SOX2 is a dose-dependent master stem cell protein that controls the self-renewal and pluripotency or multipotency of embryonic stem (ES) cells and many adult stem cells. We have previously found that SOX2 protein is monomethylated at lysine residues 42 and 117 by SET7 methyltransferase to promote SOX2 proteolysis, whereas LSD1 and PHF20L1 act on both methylated Lys-42 and Lys-117 to prevent SOX2 proteolysis. However, the mechanism by which the methylated SOX2 protein is degraded remains unclear. Here, we report that L3MBTL3, a protein with the malignant-brain-tumor (MBT) methylation–binding domain, is required for SOX2 proteolysis. Our studies showed that L3MBTL3 preferentially binds to the methylated Lys-42 in SOX2, although mutation of Lys-117 also partially reduces the interaction between SOX2 and L3MBTL3. The direct binding of L3MBTL3 to the methylated SOX2 protein leads to the recruitment of the CRL4DCAF5 ubiquitin E3 ligase to target SOX2 protein for ubiquitin-dependent proteolysis. Whereas loss of either LSD1 or PHF20L1 destabilizes SOX2 protein and impairs the self-renewal and pluripotency of mouse ES cells, knockdown of L3MBTL3 or DCAF5 is sufficient to restore the protein levels of SOX2 and rescue the defects of mouse ES cells caused by LSD1 or PHF20L1 deficiency. We also found that retinoic acid–induced differentiation of mouse ES cells is accompanied by the enhanced degradation of the methylated SOX2 protein at both Lys-42 and Lys-117. Our studies provide novel insights into the mechanism by which the methylation-dependent degradation of SOX2 protein is controlled by the L3MBTL3–CRL4DCAF5 ubiquitin ligase complex.
Keywords: protein methylation, ubiquitin ligase, stem cells, transcription regulation, pluripotency, CRL4DCAF5, L3MBTL3, SOX2, proteolysis
Introduction
The SRY-box2 protein, SOX2, is a master transcriptional factor that regulates the self-renewal and pluripotency or multipotency of ES cells2 and many adult stem cells, including neural stem cells (1–5). It is also one of the key factors to convert somatic cells to the induced pluripotent stem cells (5). SOX2 is a dose-dependent transcription factor that regulates the self-renewal and differentiation of ES cells and other stem cells in development (6). Even at the four-cell embryonic stage, heterogeneous binding of SOX2 to target genes defines the first cell fate decision (7). In ES cells, SOX2 activates its own transcription, as well as that of OCT4 and NANOG, to form a feed-forward core stem cell transcriptional circuitry for self-renewal and pluripotency of ES cells (8). An increase of SOX2 by 2-fold or less promotes development into ectoderm and mesoderm lineages, whereas loss or reduction of SOX2 induces differentiation into endoderm and trophectoderm lineages (9–13). In fetal development, SOX2 expression is also required for the formation of fetal endodermal and ectodermal tissues, such as the nervous system, lens epithelium, and anterior foregut endoderm (1, 14). SOX2 is also critical in progenitor cells of many adult tissues, including brain, retina, trachea, and the epithelium of the cervix, tongue, and testes, allowing for tissue regeneration and repair (14).
Lysine methylation is a major protein modification, and extensive investigation of histone modifications has revealed that site-specific lysine methylation is a key mechanism that regulates chromatin structure and gene expression (15). Emerging evidence indicates that many nonhistone proteins, such as SOX2, p53, DNMT1, E2F1, ERα, NF-κB/RelA, SMAD7, FOX3A, RB, GLI3, LIN28A, and STAT3, are monomethylated on specific lysine residues by methyltransferase SET7 (SETD7, KMT7, SET7/9, or SET9) (16–25). A novel function of these methylation events in a group of proteins, such as SOX2, DNMT1, E2F1, NF-κB/RelA, SMAD7, FOX3A, and STAT3, by SET7 is to trigger the ubiquitin-dependent proteolysis of the methylated proteins (16–18, 22, 23, 25). We have recently found that SOX2 protein is quite unusual because it contains two lysine residues, Lys-42 and Lys-117, that are monomethylated by SET7 (25). Methylation of either Lys-42 or Lys-117 is sufficient to trigger the ubiquitin-dependent degradation of SOX2 (25). The methylation-dependent SOX2 degradation is dynamically regulated by a demethylase, LSD1 (KMD1A), that is capable of removing the methyl group from both methylated Lys-42 and Lys-117 to stabilize SOX2 protein (25). The methylated Lys-42 and Lys-117 in SOX2 are further protected by the binding of PHF20L1 (25), an MBT-Tudor domain–containing protein encoded by a putative oncogene that is amplified or overexpressed in aggressive breast cancers (26, 27), to prevent SOX2 degradation. The methylation of Lys-117 by SET7 is prevented by the AKT-dependent phosphorylation on threonine 116 (17). However, it remains unclear how the methylated SOX2 protein is targeted for proteolysis. We report here the identification of a methyl-binding protein that preferentially recognizes the methylated SOX2 to recruit specific ubiquitin E3 ligase complexes to target the methylated SOX2 protein for ubiquitin-dependent proteolysis. We found that this mechanism plays an important role in maintaining the self-renewal and pluripotency of mouse ES cells.
Results
Identification of SOX2-associated proteins
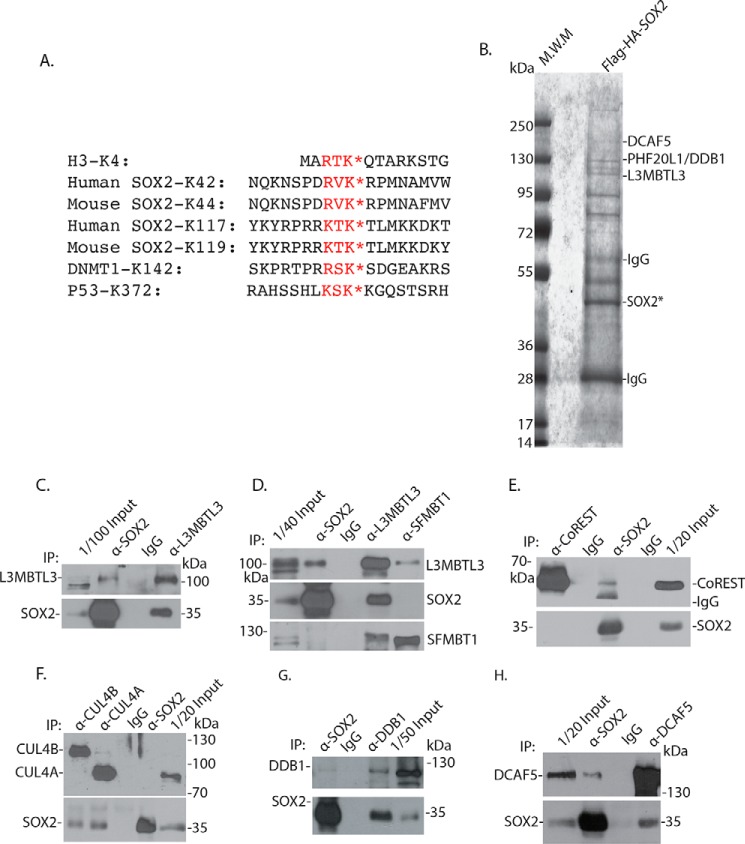
Our recent studies found that human SOX2 protein is methylated at both Lys-42 and Lys-117 by SET7 (Fig. 1A), and methylation of either Lys-42 or Lys-117 triggers SOX2 proteolysis in human teratocarcinoma PA-1 cells and mouse ES cells (25). A previous study has reported that the stability of the Lys-119–methylated murine SOX2 protein (human Lys-117 equivalent; Fig. 1A) is regulated by the WWP2 ubiquitin E3 ligase (Fig. 1A) (17). However, the Lys-42 methylation motif, 40RVK42, in SOX2 appears quite different from the methylated Lys-117 motif, 115KTK117, in SOX2, because it contains an arginine (Arg-40) and a valine (Val-41) preceding the methylated Lys-42, which corresponds to a lysine (Lys-115) and a threonine (Lys-116) in the Lys-117 methylation motif of SOX2 (Fig. 1A) (17, 25). In addition, Val-41 in the Lys-42 methylation motif is also unusual because a serine or threonine is usually present at this position in the highly conserved consensus methylation motif, (R/K)(S/T)K, in other SET7 substrate proteins such as that of lysine 4 (H3-K4) in histone H3, lysine 142 (K142) in DNMT1, and lysine 372 (K372) in p53 (Fig. 1A) (18, 22, 23). It also remains unclear whether regions outside the Lys-42 or Lys-117 methylation motifs, which are quite different between these two methylation degron regions, are also required for the degradation of the methylated SOX2 protein (Fig. 1A).
Figure 1.
Identification of SOX2-associated proteins. A, the conserved lysine residues (K*) methylated by SET7 in a methylation motif with the (R/K)(S/T)K* consensus sequences in histone H3, SOX2, DNMT1, and p53. B, isolated SOX2 protein complexes contain L3MBTL3, PHF20L1, DDB1, and DCAF5 proteins (see Table 1 for additional information). The triple FLAG– and triple HA–SOX2 protein was stably expressed in PA-1 cells, and the 3XFLAG-3XHA–SOX2 protein complexes were isolated from PA-1 cells by sequential anti-FLAG and anti-HA antibody affinity resins. The proteins associated with the isolated SOX2 complexes were separated in protein gel, excised, and trypsinized, and derivative peptides were fractionated by nanoliter LC and identified by MS. The positions of some proteins associated with the SOX2 complex are shown. The observed proteins are labeled according to the molecular weight markers/standard (M.W.M). C, endogenous L3MBTL3 interacts with SOX2. L3MBTL3 and SOX2 were immunoprecipitated (IP) from mouse ES cells and detected by Western blotting analyses as indicated. D, endogenous SFMBT1 interacts with L3MBTL3. SOX2, L3MBTL3, and SFMBT1 were immunoprecipitated from mouse ES cells, and their interactions were examined by Western blotting analyses as indicated. E, CoREST interacts with SOX2. CoREST and SOX2 were immunoprecipitated from mouse ES cells, and their interaction was examined by Western blotting analyses. F, endogenous CUL4A and CUL4B interact with SOX2. SOX2, CUL4A, and CUL4B were immunoprecipitated from mouse ES cells, and their interactions were determined by Western blotting analyses as indicated. G, endogenous DDB1 interacts with SOX2. DDB1 and SOX2 were immunoprecipitated from mouse ES cells, and their interactions were analyzed by Western blotting analyses as indicated. H, SOX2 and DCAF5 were immunoprecipitated from mouse ES cells, and their interaction was examined by Western blotting analyses as indicated.
To determine the mechanism by which the Lys-42–methylated SOX2 protein is targeted for degradation, we tried to isolate the SOX2 protein complexes and used the MS-based analysis to interrogate SOX2-associated proteins (28). A triple FLAG tag and a triple HA tag (3XFLAG-3XHA) was fused to the N terminus of the human SOX2 protein, and this tagged SOX2 protein was stably expressed in human teratocarcinoma PA-1 cells that also express endogenous SOX2 (25). The 3XFLAG-3XHA–SOX2 protein complexes were isolated by sequential anti-FLAG and anti-HA antibody affinity chromatography (Fig. 1B) (28). The SOX2-associated proteins were identified by an Obitrap XL MS system coupled with an Easy nanoliter LC (29).
Using this approach, we have identified many peptides derived from SOX2 protein (Fig. 1B and Table 1). Our MS analyses of the SOX2 protein complexes also revealed the presence of peptides derived from PHF20L1 (Fig. 1B and Table 1), a protein that we have shown to bind to the methylated SOX2 and protect SOX2 protein from the methylation-dependent proteolysis (25). We also identified a putative methyl-binding protein (Fig. 1B and Table 1), L3MBTL3, which contains the three tandem-repeated malignant-brain-tumor (MBT) domain that is capable of binding to mono- or dimethylated lysine residues in histones (30, 31). L3MBTL3 is mutated in medulloblastoma and is further implicated in other pathological disorders, such as multiple sclerosis, insulin resistance, prostate cancer, and breast cancer (30, 32–35). Mouse null mutation of L3MBTL3 causes late embryonic lethality (36). Our recent studies revealed that L3MBTL3 is critical in regulating the protein stability of DNMT1 when it is methylated by SET7 (29). L3MBTL3 is thus a good candidate for the methylated SOX2 proteolysis, although there are substantial differences between the Lys-42 methylation motif and the surrounding region in SOX2 and that of the methylated Lys-142 in DNMT1 (Fig. 1A) (25, 29). To verify the interaction, we tested whether endogenous L3MBTL3 protein interacts with SOX2 by using independent immunocoprecipitation and Western blotting analyses. Our analyses confirmed that the endogenous L3MBTL3 protein associated with SOX2 in mouse ES cells (Fig. 1, C and D).
Table 1.
Partial list of peptides identified by mass spectrometry
The 3XFLAG-3XHA–tagged Sox2 protein complex was purified by anti-FLAG antibody affinity chromatography from PA-1 cells by anti-FLAG affinity chromatography, eluted with 3XFLAG peptide, and repurified with anti-HA affinity chromatography. The proteins in the 3XFLAG-3XHA–tagged Sox2 protein complexes were fractionated in protein SDS gel. Protein bands were excised, trypsinized, and fractionated on nanoliter liquid chromatography. The peptides were identified by mass spectrometry analyses in an Orbitrap XL mass spectrometry system with the Protein Discovery software. Three independent mass spectrometry analyses were performed for the identity of a specific peptide/protein in repeated purification samples. Only one preparation sample is shown.
| Protein identification | Number of peptides | NCBI accession number |
|---|---|---|
| SOX2 | 47 | NM_003106.3 |
| SOX21 | 23 | NM_007084.3 |
| DDB1 | 18 | NM_001923.4 |
| PHF20L1 | 14 | NM_016018.4 |
| DCAF5 | 13 | NM_003861.2 |
| L3MBTL3 | 10 | NM_032438.3 |
| LSD1 (KDM1A) | 9 | NM_001009999.2 |
| HDAC1 | 8 | NM_004964.2 |
| CUL4B | 8 | NM_003588.3 |
| SOX1 | 7 | NM_005986.2 |
| SOX3 | 5 | NM_005634.2 |
| OCT4 | 5 | NM_002701.5 |
| CUL4A | 4 | NM_001008895.3 |
| CAND1 | 3 | NM_018448.4 |
| CSN1 | 2 | NM_001330541.1 |
| CSN7A | 2 | NM_001164095.2 |
L3MBTL3 was previously isolated as an accessory component of polycomb repressive complex 1 (PRC1), which also contains LSD1, CoREST, and SFMBT1 (37, 38). Our examination indicates that CoREST, a protein that is required for LSD1 demethylation of the mono- and dimethylated lysine 4 in histone H3 (H3-K4 in Fig. 1A), also binds to SOX2 (Fig. 1E). This is consistent with our previous studies indicating that LSD1 also binds to SOX2 to remove the methyl group from the monomethylated Lys-42 and Lys-117 in human SOX2 protein and that loss of CoREST reduced the SOX2 protein levels, similar to that of LSD1 knockdown (25, 39). These studies suggest that LSD1 and CoREST may act together to demethylate SOX2. However, whereas our examination found that SFMBT1 interacts with L3MBTL3, we could not detect significant interaction between endogenous SOX2 and SFMBT1 proteins (Fig. 1D).
Our analysis also revealed the presence of peptides from DDB1 and DCAF5, components of the CRL4DCAF5 ubiquitin E3 ligase complexes (Fig. 1B and Table 1) (29). Our independent immunoprecipitation and Western blotting analyses in mouse ES cells confirmed that the components of the CRL4DCAF5, such as CUL4A or CUL4B, DDB1, and DCAF5, are associated with endogenous SOX2 protein (Fig. 1 (F–H) and Fig. S1A).
L3MBTL3 is involved in regulating the protein stability of SOX2 in a methylated lysine–dependent manner
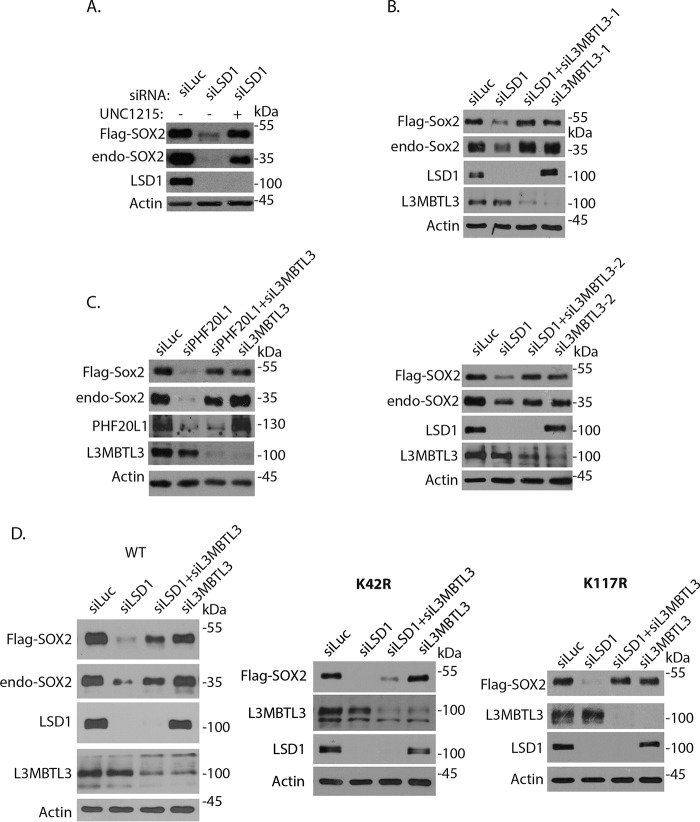
To determine whether L3MBTL3 regulates SOX2 at the protein level, we examined the effects of its inactivation on the FLAG-SOX2 protein that is stably and ectopically expressed under the retroviral LTR promoter control in PA-1 cells (Fig. 2) (25). We initially tested the effect of UNC1215, a reported chemical inhibitor of L3MBTL3, on SOX2 protein stability in PA-1 cells at low concentration (<1 μm), although high concentrations (about 20 μm) of this compound were shown to inhibit other methyl-binding proteins, such as PHF20L1 (27, 40–42). Whereas loss of LSD1, a demethylase that removes the methyl groups from the methylated SOX2 protein (25), destabilizes both endogenous and ectopically expressed SOX2 proteins in PA-1 cells, treatment of LSD1-deficient PA-1 cells with 1 μm UNC1215 restored the levels of both endogenous and ectopically expressed SOX2 proteins (Fig. 2A). These observations suggest that L3MBTL3 might regulate SOX2 protein stability.
Figure 2.
L3MBTL3 selectively regulates the stability of Lys-42–methylated SOX2 protein. A, PA-1 cells stably expressing FLAG-SOX2 under the retroviral LTR promoter control (pMSCV-puro) were transfected with 50 nm luciferase (control) and LSD1 siRNAs for 48 h. The cells were then treated with 1 μm UNC1215 for 10 h, and FLAG-tagged and endogenous SOX2 proteins were analyzed by Western blotting. B, L3MBTL3 regulates the protein stability of SOX2. PA-1 cells stably expressing FLAG-SOX2 were transfected with 50 nm luciferase (control) siRNA, LSD1 siRNA, LSD1 plus two independent L3MBTL3 siRNAs, and two independent L3MBTL3 siRNAs for 48 h. The FLAG-SOX2 and endogenous SOX2 proteins, LSD1, L3MBTL3, and actin (loading control) were analyzed by Western blotting. C, loss of L3MBTL3 stabilizes SOX2 protein in PHF20L1-deficient cells. PA-1 cells stably expressing FLAG-SOX2 were transfected with 50 nm luciferase (control), PHF20L1, PHF20L1 + L3MBTL3, and L3MBTL3 siRNAs for 48 h. The FLAG-SOX2 and endogenous SOX2 proteins and the indicated proteins were analyzed by Western blotting. D, analysis of K42R and K117R SOX2 mutants. PA-1 cells stably expressing the FLAG-SOX2 WT and the K42R and K117R mutants were transfected with 50 nm luciferase (control), LSD1, L3MBTL3 + LSD1, and L3MBTL3 siRNAs for 48 h. The FLAG-SOX2, K42R and K117R mutants, endogenous SOX2, and indicated proteins were analyzed by Western blotting.
We used the FLAG-tagged SOX2 protein to further verify whether L3MBTL3 is involved in SOX2 protein regulation. Whereas knockdown of LSD1 by specific siRNAs in PA-1 cells destabilized both FLAG-SOX2 and endogenous SOX2 proteins, we found that co-knockdown of L3MBTL3 by two independent siRNAs led to the stabilization of both FLAG-tagged and endogenous SOX2 proteins in LSD1-deficient PA-1 cells (Fig. 2B). We have previously shown that loss of PHF20L1, a protein that binds to the methylated SOX2 protein to prevent its proteolysis, also destabilized the methylated SOX2 protein (Fig. 2C) (25). Our studies indicate that loss of L3MBTL3 can stabilize both the destabilized endogenous and ectopically expressed SOX2 proteins in PHF20L1-deficient PA-1 cells (Fig. 2C). These results indicate that L3MBTL3 is involved in regulating the protein stability of FLAG-SOX2, which is sensitive to the loss of both LSD1 and PHF20L1.
Our previous studies revealed that SOX2 is methylated at both Lys-42 and Lys-117 by SET7, and the methylation at either Lys-42 or Lys-117 is sufficient to induce SOX2 protein degradation (25). We tested whether L3MBTL3 regulates the SOX2 protein through the methylated Lys-42, Lys-117, or both. Because SET7 cannot methylate arginine residues, we converted either Lys-42 or Lys-117 to arginine (K42R or K117R) in SOX2 to determine whether L3MBTL3 acts through the methylated Lys-42 or Lys-117 in SOX2 (25). The FLAG-SOX2 WT and K42R or K117R mutant constructs were ectopically expressed in PA-1 cells as FLAG-tagged protein (Fig. 2D) (25). Although we often observed that loss of L3MBTL3 can fully rescue the reduced levels of FLAG-SOX2 protein in these experiments (Fig. 2, A–C), we also found in some experiments that knockdown of L3MBTL3 caused partial rescue of down-regulated FLAG-SOX2 protein in LSD1-deficient cells (Fig. 2D, left). For the mutant SOX2 proteins, we have repeatedly found that knockdown of L3MBTL3 usually stabilized the K117R SOX2 mutant in LSD1 knockdown cells (Fig. 2D), whereas loss of L3MBTL3 either completely lacked the ability to rescue or only partially restored the destabilized K42R protein levels in LSD1-deficient cells (Fig. 2D, middle and right; also see Fig. S1B). Because the K117R mutant of SOX2 still contains an intact Lys-42 residue, whereas K42R has an intact Lys-117 for the methylation-dependent degradation of SOX2 protein (25), these studies suggest that loss of L3MBTL3 preferentially affects the protein stability of the Lys-42–methylated SOX2 protein, but L3MBTL3 deficiency has lesser effects on the Lys-117–methylated SOX2 protein in LSD1-deficient PA-1 cells. Because previous studies showed that the methylation of Lys-117 by SET7 is prevented if Thr-116 of SOX2 is phosphorylated by AKT and because the methylated Lys-117 in SOX2 is also reported to be targeted by ubiquitin E3 ligases such as WWP2 for SOX2 degradation (17), the variable rescuing effects of L3MBTL3 knockdown on SOX2 WT and Lys-42 mutant proteins may be in part affected by the levels of AKT and WWP2 activities in different experiments for the Lys-117–methylated SOX2 protein.
L3MBTL3 regulates the protein levels of SOX2 in mouse ES cells
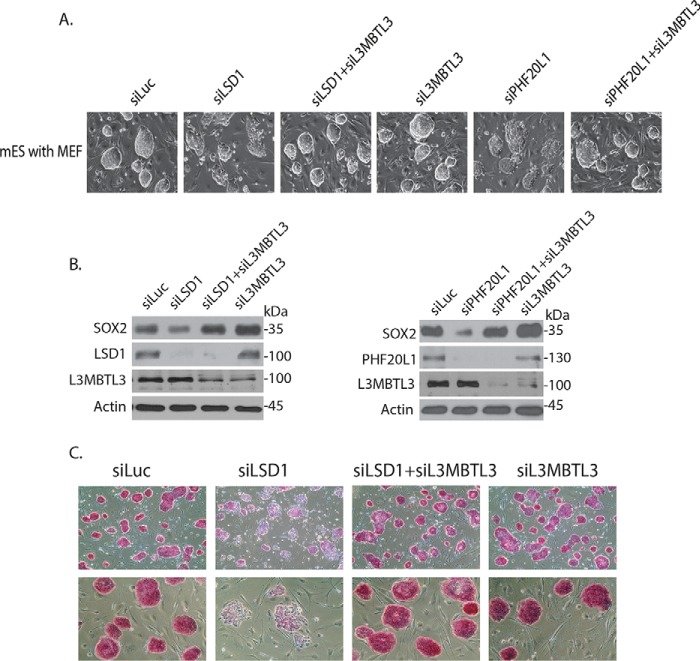
Because L3MBTL3 binds to SOX2 protein in mouse ES cells (Fig. 1, C and D), we further examined whether L3MBTL3 regulates the protein levels of endogenous SOX2 in mouse ES cells. We found that knockdown of L3MBTL3 by the specific siRNA alone often caused detectable increased levels of SOX2 protein in moue ES cells (Fig. 3B). Whereas the siRNA-mediated LSD1 knockdown reduced the level of SOX2 protein, co-knockdown of L3MBTL3 and LSD1 using the specific siRNAs restored the level of endogenous SOX2 protein in LSD1-deficient mouse ES cells (Fig. 3B), consistent with our observation in PA-1 cells (Fig. 2). Our studies also revealed that the reduced level of endogenous SOX2 protein in PHF20L1 knockdown mouse ES cells was also recovered by co-knockdown of L3MBTL3 and PHF20L1 by specific siRNAs (Fig. 3B). These studies indicate that L3MBTL3 regulates the SOX2 protein level in mouse ES cells.
Figure 3.
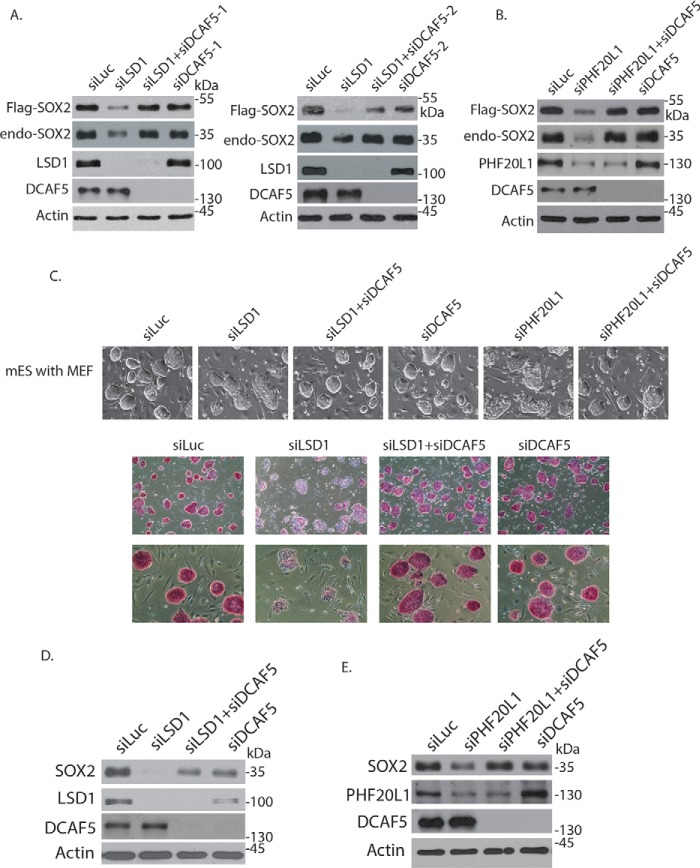
Loss of L3MBTL3 stabilizes SOX2 protein and restores self-renewal and pluripotency in LSD1- or PHF20L1-deficient mouse ES cells. A, mouse ES cells grown on mitotically inactivated mouse embryonic fibroblasts (MEF) were transfected with 50 nm siRNAs of luciferase, LSD1, LSD1 + L3MBTL3, L3MBTL3, PHF20L1, and PHF20L1 + L3MBTL3 for 48 h. Cells were examined, and cell images were acquired with a Nikon ECLIPSE Ti-S microscope equipped with NIS-Elements BR version 3.1 software. B, the proteins in Fig. 3A were analyzed by Western blotting with anti-SOX2, LSD1, PHF20L1, L3MBTL3, and actin antibodies. C, mouse ES cells in Fig. 3A were stained with AP, and cell images were acquired.
In mouse ES cells, loss of LSD1 or PHF20L1 not only reduced the SOX2 protein level, but also blocked the self-renewal of mouse ES cells and impaired their pluripotency, with cells assuming a flat morphology that is usually associated with differentiation (Fig. 3A) (25). Whereas pluripotent mouse ES cells are positively stained by alkaline phosphatase (AP), a well-established pluripotent stem cell marker, knockdown of LSD1 led to the loss of AP-positive ES colonies due to the induction of cellular differentiation (Fig. 3C) (17, 43). However, co-knockdown of L3MBTL3 and LSD1 or PHF20L1 with specific siRNAs not only restored the SOX2 protein level, but also reestablished the self-renewal and AP-positive staining of pluripotent mouse ES cells in LSD1- or PHF20L1-deficient mouse ES cells (Fig. 3, A–C), indicating that the stabilization of SOX2 protein after loss of L3MBTL3 is sufficient to prevent the differentiation of LSD1- or PHF20L1-deficient mouse ES cells. These observations also suggest that the SOX2 protein is the main target of LSD1 and PHF20L1 in mouse ES cells.
L3MBTL3 preferentially binds to the methylated Lys-42 in SOX2
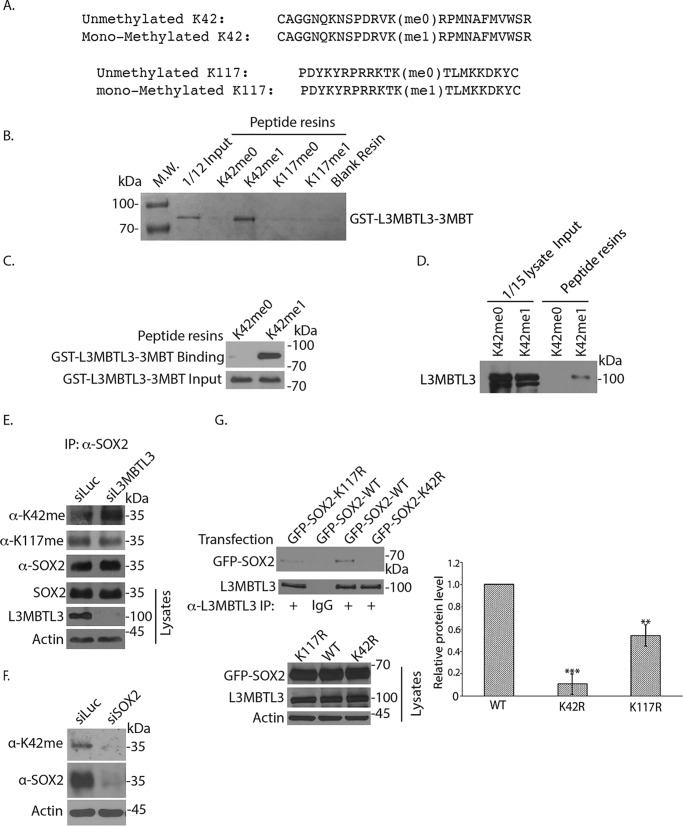
L3MBTL3 contains an MBT domain that contains three MBT tandem repeats (3MBT) at its N-terminal half that can bind to the mono- and dimethylated lysine residues (29, 31, 41). To further determine whether the 3MBT region of L3MBTL3 can directly and preferentially recognize the methylated Lys-42 or Lys-117 in SOX2, we tested the affinity of the recombinant GST-L3MBTL3–3MBT fusion protein, which contains the glutathione S-transferase (GST) fused to the three tandem-repeated MBT region of L3MBTL3, toward pairs of synthetic SOX2 peptides that contain either the monomethylated Lys-42 or monomethylated Lys-117 and their cognate un-methylated control peptides in vitro (Fig. 4A) (25, 27). The recombinant GST-L3MBTL3–3MBT protein, purified from bacteria, were incubated with these peptides, which were immobilized by covalent disulfide linkage to Sulfolink-Sepharose resins through cysteine residues at the end of each peptide (25). Our studies repeatedly revealed that the recombinant GST-L3MBTL3–3MBT protein preferentially binds to the monomethylated Lys-42 peptide resins, as compared with the unmethylated cognate peptide (Fig. 4, B and C). However, GST-L3MBTL3–3MBT did not appear to have a significant preference for the monomethylated Lys-117 peptide and its cognate unmethylated peptide under our assay conditions (Fig. 4, A and B). We also repeatedly found that incubation of the methylated and unmethylated Lys-42 peptide resins with PA-1 cell lysates detected the preferential binding of endogenous L3MBTL3 to the methylated Lys-42 peptide (Fig. 4D and Fig. S1B), as compared with that of unmethylated Lys-42 cognate peptide. These studies indicate that L3MBTL3 directly binds to the monomethylated Lys-42 in SOX2.
Figure 4.
L3MBTL3 preferentially binds to the methylated Lys-42 (K42) in SOX2. A, two pairs of synthetic SOX2 peptides were immobilized onto Sulfolink-Sepharose through the cysteine residues at the ends of the peptides. One pair of SOX2 peptides contains the monomethylated Lys-42 (Kme1) or the cognate peptide with unmethylated Lys-42 (Kme0); another pair contains the monomethylated Lys-117 (Kme1) or cognate unmethylated Lys-117 (Kme0) as indicated. B, L3MBTL3 directly interacts with the monomethylated Lys-42 peptide. The K42me0, K42me1, K117me0, and K117me1 peptide resins were preincubated with 5 μg of GST for 2 h to block the background. The resins were then incubated with 1.5 μg of GST-L3MBTL3–3MBT recombinant protein for 4 h at 4 °C and then washed. The GST-L3MBTL3–3MBT protein associated with various resins was separated in protein gel and visualized by Coomassie Blue staining. C, the K42me0 and K42me1 peptide resins were incubated with GST-L3MBTL3–3MBT recombinant protein as in B. The proteins associated with the resins were blotted with anti-L3MBTL3 antibodies. D, endogenous L3MBTL3 preferentially binds to the monomethylated Lys-42 peptide. The monomethylated (Kme1) or cognate unmethylated Lys-42 (Kme0) peptide resins were incubated with 250 μg of PA-1 cell lysates for 4 h. L3MBTL3 binding to the peptide resins was analyzed by the anti-L3MBTL3 Western blotting. E, loss of L3MBTL3 caused the accumulation of methylated Lys-42 in SOX2. PA-1 cells were transfected with 50 nm luciferase (control) and L3MBTL3 siRNAs for 48 h. SOX2 were immunoprecipitated (IP) by SOX2 antibodies and Western blotted with affinity-purified anti-methylated Lys-42, anti-methylated Lys-117, and SOX2 antibodies. F, PA-1 cells were transfected with 50 nm luciferase (control) and SOX2 siRNAs for 48 h. SOX2 and Lys-42–methylated SOX2 protein were Western blotted with affinity-purified anti-methylated Lys-42 and SOX2 antibodies in cell lysates. G, GFP-tagged SOX2 (WT), K42R, and K117R mutant SOX2 expression constructs were transfected into 293 cells. The L3MBTL3 complexes were immunoprecipitated by anti-L3MBTL3 antibodies and Western blotted by anti-SOX2 antibodies. The proteins bands were quantified and normalized to the WT SOX2 band. The quantifications are represented by a bar graph with mean and S.D. (error bars) from replicated experiments. The p value of K117R or K42R bands to the WT SOX2 band was calculated by independent Student's t test (**, p < 0.01; ***, p < 0.001). Experiments were repeated with the same conclusion (see Fig. S1C).
L3MBTL3 binding to SOX2 is abolished by K42R mutation but only partially reduced by K117R mutation in vivo
We have previously developed specific peptide antibodies against the monomethylated Lys-42 or monomethylated Lys-117 in SOX2 (25). Our studies revealed that knockdown of L3MBTL3 led to the accumulation of the methylated Lys-42 in SOX2, but the level of methylated Lys-117 SOX2 protein was not significantly affected (Fig. 4E). The specificity of the affinity-purified anti-monomethylated Lys-42 antibody was confirmed by the down-regulation of total SOX2 and the methylated Lys-42 SOX2 protein by treatment of specific SOX2 siRNA in PA-1 cells (Fig. 4F).
To further characterize the interaction between L3MBTL3 and SOX2, we also examined L3MBTL3 binding to the transfected GFP fusion of WT SOX2, K42R, and K117R mutant proteins in 293 cells (Fig. 4G). Our studies repeatedly indicate that whereas GFP-SOX2 can bind to L3MBTL3, the K42R mutant abolished the interaction between SOX2 and L3MBTL3 (Fig. 4G and Fig. S1C). Our experiments, however, also repeatedly revealed that the binding of L3MBTL3 to SOX2 was slightly reduced by the K117R mutation (Fig. 4G and Fig. S1C). Our studies thus indicate that L3MBTL3 preferentially recognizes and directly binds to the monomethylated Lys-42 in SOX2 using the purified L3MBTL3–3MBT protein in vitro, and its binding to SOX2 is abolished if Lys-42 is mutated. However, our transfection studies also provided evidence that L3MBTL3 may interact with the methylated Lys-117 in SOX2 weakly in vivo, as the K117R mutant also slightly reduced the interaction between L3MBTL3 and SOX2 in multiple transfection-based assays (Fig. 4G and Fig. S1C). It is possible that our failure to detect the weak interaction between recombinant L3MBTL3–3MBT and monomethylated Lys-117 peptide might be due to our stringent in vitro binding conditions (Fig. 4B).
Our in vivo and in vitro binding analyses are consistent with our analysis of L3MBTL3 knockdown effects on the K42R and K117R mutants, which indicate that the K117R mutant still retains full response to L3MBTL3 deficiency, whereas the K42R mutant either completely or partially abolishes the rescuing effects of L3MBL3 knockdown to restore SOX2 levels in LSD1-deficient cells (Fig. 2D and Fig. S1B). These studies indicate that L3MBTL3 preferentially regulates the Lys-42 methylation–dependent degradation of SOX2, although it also weakly binds the methylated Lys-117 and partially controls the methylated Lys-117–dependent SOX2 degradation. However, it is possible that additional ubiquitin ligases, such as WWP2, are involved to recognize and target the methylated Lys-117 in SOX2 for degradation (17).
DCAF5 is required for the methylated SOX2 degradation
Because the methylated SOX2 protein is degraded by ubiquitin-dependent proteolysis (25), we also looked for potential ubiquitin E3 ligase that may function to target the methylated SOX2 protein for degradation. Our MS data and our independent immunocoprecipitation and Western blot analysis have shown that the CRL4DCAF5 ubiquitin ligase complex, consisting of CUL4A or CUL4B and DDB1, and DCAF5 interact with the endogenous SOX2 protein in mouse ES and PA-1 cells (Fig. 1 (B and F–H), Fig. S1A, and Table 1). These studies suggest that CRL4DCAF5 may be involved in the degradation of the methylated SOX2 protein in the mouse ES cells.
Because DCAF5 is a specific subunit of CRL4 ubiquitin ligase complex (29, 44, 45), we tried to determine whether it is involved in SOX2 degradation on the ectopically and stably expressed FLAG-SOX2 protein in PA-1 cells (Fig. 5, A and B). We found that knockdown of DCAF5 by two independent siRNAs led to the stabilization of the FLAG-SOX2 protein in LSD1- or PHF20L1-deficient cells (Fig. 5, A and B). These studies indicated that DCAF5 is required for the protein stability of the methylated SOX2 protein. We also tested whether DCAF5 regulates endogenous SOX2 protein in mouse ES cells. Knockdown of DCAF5 with specific siRNAs repeatedly restored the SOX2 protein level and rescued the defects on self-renewal and pluripotency in LSD1 knockdown mouse ES cells (Fig. 5, C and D). In addition, loss of DCAF5 also reestablished the levels of SOX2 protein and the self-renewal of mouse ES cell colonies in PHF20L1 knockdown mouse ES cells (Fig. 5, C and E). These studies revealed that DCAF5 is involved in regulating SOX2 protein levels in LSD1- or PHF20L1-deficient mouse ES cells.
Figure 5.
DCAF5 regulates SOX2 in mouse embryonic stem cells and PA-1 cells. A, PA-1 cells stably expressing FLAG-SOX2 were transfected with 50 nm luciferase (control) siRNA, LSD1 siRNA, LSD1 + two independent DCAF5 siRNAs, and two independent DCAF5 siRNAs alone for 48 h. The FLAG-SOX2 and endogenous SOX2 proteins and the indicated proteins were analyzed by Western blotting. B, PA-1 cells stably expressing FLAG-SOX2 were transfected with 50 nm luciferase (control), PHF20L1, PHF20L1 + DCAF5, and DCAF5 siRNAs for 48 h. The FLAG-SOX2 and endogenous SOX2 proteins and the indicated proteins were analyzed by Western blotting. C, mouse ES cells grown on mitotically inactivated mouse embryonic fibroblasts (MEF) were transfected with 50 nm siRNAs of luciferase, LSD1, LSD1 + DCAF5, DCAF5, PHF20L1, and PHF20L1 + DCAF5 for 48 h. Top panels, cells were examined, and cell images were acquired. Bottom panels, the mouse ES cells transfected with 50 nm siRNAs of luciferase, LSD1, LSD1 + DCAF5, and DCAF5 were stained with AP, and cell images were acquired. D and E, the proteins in C were analyzed by Western blotting with anti-SOX2, LSD1, PHF20L1, DCAF5, and actin antibodies as indicated.
L3MBTL3 interacts with DCAF5 to cooperatively target the methylated SOX2 protein for polyubiquitination
To investigate whether DCAF5 is involved in the regulation of the methylated SOX2 proteolysis, we transfected and expressed FLAG-tagged DCAF5 protein in the presence or absence of SET7 to determine whether SET7 has any effects on the interaction between SOX2 and DCAF5 in PA-1 cells. We found that SET7 expression stimulated the interaction between SOX2 and DCAF5, suggesting that the interaction between these two proteins may be mediated through SET7-dependent methylation (Fig. 6A). Consistent with our observation that DCAF5 regulates the stability of methylated SOX2 protein, our studies showed that whereas the transfected GFP-SOX2 (WT) interacts with DCAF5, the K42R mutant of SOX2 is defective to associate with DCAF5 (Fig. 6B). Because L3MBTL3 is preferentially involved in regulating the protein stability of the methylated SOX2 protein, we examined whether L3MBTL3 interacts with DCAF5 to regulate the methylated SOX2 degradation. Our studies revealed that endogenous DCAF5 interacts with L3MBTL3 in mouse ES cells (Fig. 6C). This is consistent with our previous studies indicating that L3MBLT3 recruits DCAF5 and its associated CRL4 core complex to the methylated lysine residues, such as the methylated Lys-142 in DNMT1, to target the methylated proteins for degradation (29). However, whereas we found that L3MBTL3 interacts with DCAF5, SFMBT1, and CoREST, we could not detect significant interactions between DCAF5 and SFMBT1 or CoREST (Fig. 6C). To determine whether L3MBTL3 recruits DCAF5 to interact with SOX2, we knocked down L3MBTL3 and found that loss of L3MBTL3 significantly reduced the interaction between SOX2 and DCAF5 (Fig. 6D), indicating that L3MBTL3 is required for the interaction between DCAF5 and SOX2. In addition, we found that expression of L3MBTL3 and DCAF5 promoted the polyubiquitination of SOX2 protein, but not that of the K42R mutant SOX2 protein in vivo, in the presence of SET7 and other components of CRL4 core complexes (Fig. 6E), indicating that L3MBTL3 cooperates with the CRL4DCAF5 ubiquitin ligase complex to promote the polyubiquitination-dependent proteolysis of the methylated SOX2 protein.
Figure 6.
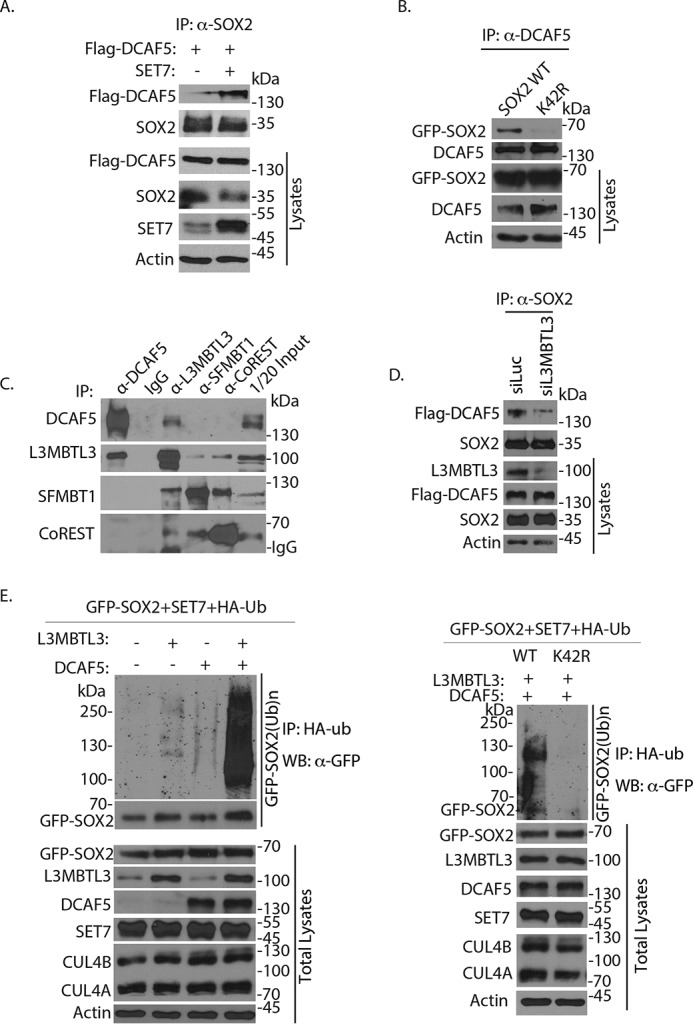
Regulation of methylated SOX2 protein by DCAF5. A, the interaction between SOX2 and DCAF5 is enhanced by SET7 expression. The FLAG-DCAF5– and SET7–expressing constructs were transfected into PA-1 cells for 48 h and treated with proteasome inhibitor MG132 (5 μg/ml) for last 6 h, and interactions between SOX2 and FLAG-DCAF5 were analyzed by coimmunoprecipitation (IP) and Western blotting analyses. B, DCAF5 poorly binds to the K42R mutant of SOX2. GFP-tagged SOX2 (WT) and its K42R mutant expression constructs were transfected into 293 cells. The DCAF5 protein was immunoprecipitated and Western blotted by anti-SOX2 antibodies. C, L3MBTL3 and DCAF5 interact in mouse ES cells. Endogenous L3MBTL3, DCAF5, SFMBT1, and CoREST were immunoprecipitated from mouse ES cells, and their interactions were detected by Western blotting analyses using specific antibodies as indicated. D, the interaction between SOX2 and DCAF5 is reduced after loss of L3MBTL3. PA-1 cells stably expressing FLAG-DCAF5 were transfected with 50 nm luciferase and L3MBTL3 siRNAs for 48 h. The SOX2 complexes were immunoprecipitated by anti-SOX2 antibodies and Western blotted with anti-FLAG-DCAF5 and SOX2 antibodies. E, L3MBTL3 and CRL4DCAF5 target SOX2 for polyubiquitination in vivo. Left, GFP-SOX2–expressing constructs were cotransfected into 293 cells together with vectors expressing HA-tagged ubiquitin (HA-ub), SET7, CUL4A, CUL4B, and DDB1, in the presence or absence of L3MBTL3- and DCAF5-expressing constructs as indicated. Proteins were immunoprecipitated with anti-HA antibodies and Western blotted (WB) with the anti-GFP-SOX2 antibodies as indicated. Right, the K42R SOX2 mutant abolishes the L3MBTL3–CRL4DCAF5–mediated SOX2 polyubiquitination in vivo; the same as the left panel except GFP-SOX2 (WT)– and K42R SOX2 mutant–expressing constructs were used.
Methylated Sox2 protein is a critical target of LSD1 in mouse ES cells
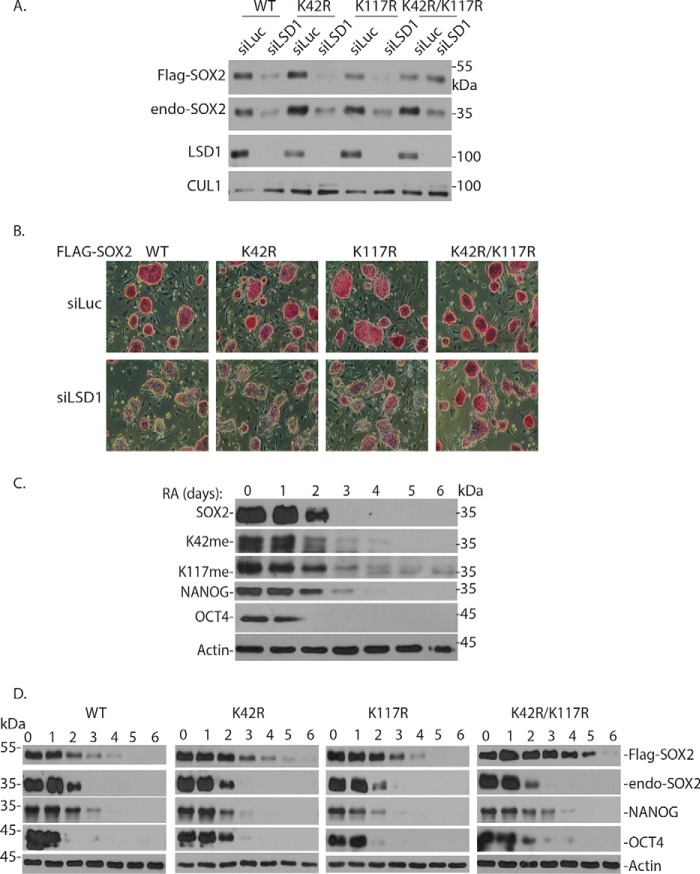
To examine how SOX2 is regulated in mouse ES cells, we used a retroviral vector (pMSCV-puro) to stably express the FLAG-tagged WT, K42R, K117R, and K42R/K117R SOX2 proteins in mouse ES cells after puromycin selection (Fig. 7A) (29). Knockdown of LSD1 in mouse ES cells caused the down-regulation of FLAG-SOX2 (WT) and single K42R or K117R mutant proteins and loss of pluripotency of mouse ES cells by AP staining (Fig. 7, A and B). However, the double K42R/K117R mutant SOX2 protein was relatively resistant to LSD1 knockdown, and the ES cells expressing the K42R/K117R mutant substantially retained their pluripotency and self-renewal after loss of LSD1 (Fig. 7, A and B), indicating that the SOX2 protein methylated at both Lys-42 and Lys-117 is a critical target of LSD1 to maintain the self-renewal and pluripotency of mouse ES cells.
Figure 7.
Expression of the K42R/K117R Sox2 mutant partially suppresses the effects of LSD1 knockdown and retinoic acid–induced ES differentiation. A, the 3XFLAG-3XHA–tagged WT-SOX2 and K42R, K117R, and K42R/K117R mutant SOX2–expressing constructs (pMSCV-Puro) were stably expressed in mouse ES cells. The ES cells were transfected with 50 nm siRNAs of luciferase and LSD1 for 48 h, and LSD1 knockdown effects on ectopically expressed FLAG-SOX2 (WT), K42R, K117R, and K42R/K117R mutant proteins were analyzed by Western blotting. B, cells in 7A were stained with AP, and cell images were acquired. C, retinoic acid–mediated ES differentiation induces SOX2 protein degradation. Mouse ES cells were cultured on gelatin-coated dishes at a density of 0.3 × 106 cells/10-cm dish. Twenty-four hours after plating (day 0 time point). LIF was removed, and retinoic acid was added at a concentration of 5 μm. Cells were collected daily for 6 days for analyzing SOX2, methylated SOX2, OCT4, NANOG, and actin proteins by Western blot analyses. D, mouse ES cells expressing FLAG-tagged WT SOX2 and the K42R, K117R, and K42R/K117R mutant SOX2 proteins were seeded on 0.1% gelatin-coated dishes in ES culture medium without LIF for 24 h. They were then treated with 5 μm RA (day 0 time), and cells were harvested at various time points (in days) as indicated. The levels of FLAG-tagged Sox2 and endogenous SOX2, endogenous Lys-42– and Lys-117–methylated SOX2, OCT4, NANOG, and actin were analyzed by Western blotting with specific antibodies.
Retinoic acid–mediated differentiation induces the proteolysis of methylated Sox2 in mouse ES cells
When mouse ES cells are treated with retinoic acid (RA) and grown on the gelatin-coated culture dishes in the absence of leukemia-inhibitory factor (LIF), they rapidly undergo cellular differentiation (46). We found that RA-induced ES cell differentiation led to the rapid down-regulation of endogenous major stem cell proteins SOX2, OCT4, and NANOG within 2–3 days in mouse ES cells (Fig. 7C). Importantly, we also found that RA induced the degradation of the ectopically expressed FLAG-SOX2 WT, K42R, or K117R single mutant protein (Fig. 7D). However, the K42R/K117R double mutant of SOX2 protein was significantly more resistant to the RA-induced SOX2 degradation (Fig. 7D). Our studies revealed that RA-induced mouse ES cell differentiation enhanced the degradation of SOX2 protein, which depends on the methylation of both Lys-42 and Lys-117 in SOX2 protein.
Discussion
We have recently reported that methylated SOX2 protein at either Lys-42 or Lys-117 is targeted for degradation in mouse ES cells (25). Our current investigation indicates that the protein stability of the methylated SOX2 protein is regulated by the methylation-dependent CRL4DCAF5 ubiquitin E3 ligase complex containing L3MBTL3 for ubiquitin-dependent proteolysis, as schematically illustrated in Fig. 8. We found that L3MBTL3 preferentially recognizes the monomethylated Lys-42 in SOX2 to recruit the CRL4DCAF5 ubiquitin E3 ligase complex to target SOX2 for ubiquitin-dependent degradation. Our evidence also suggests that L3MBTL3 may have relatively weak affinity to the methylated Lys-117 in SOX2 to regulate SOX2 protein degradation (Figs. 2D and 4G and Fig. S1 (B and C)). Our current studies are consistent with our recent findings that L3MBTL3 and the CRL4DCAF5 ubiquitin E3 ligase complex regulates the stability of the methylated DNMT1 and E2F1 proteins (29). Taken together, our studies indicate that the function of L3MBTL3 and CRL4DCAF5 ubiquitin E3 ligase complex is to regulate the protein degradation of a set of methylated nonhistone proteins, including the methylated SOX2, DNMT1, and E2F1 (29). The different methylation motifs in these L3MBTL3-regulated proteins suggest that L3MBTL3 may tolerate substantial variation in the methylated lysine motifs (29).
Figure 8.
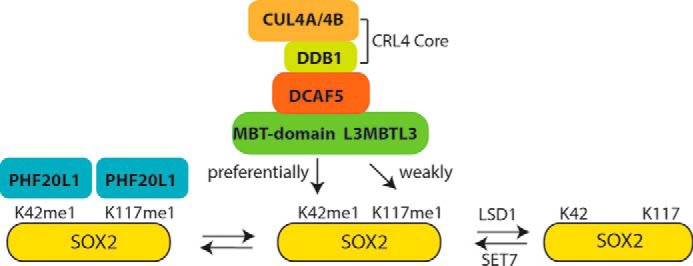
A model for the degradation of the methylated SOX2 protein. Shown is a model for the regulation of the methylated SOX2 protein by the L3MBTL3–CRL4DCAF5 ubiquitin E3 ligase complex. The SOX2 protein is methylated at Lys-42 (K42) and Lys-117 (K117) by SET7, and the methylated SOX2 is protected by LSD1, which removes the methyl group, and by PHF20L1, which binds to the methylated SOX2 at both Lys-42 and Lys-117. L3MBTL3 preferentially binds to the methylated Lys-42 and weakly interacts with methylated Lys-117 in SOX2. The binding of L3MBTL3 leads to the recruitment of the CRL4DCAF5 ubiquitin E3 ligase complex to target SOX2 protein for ubiquitin-dependent proteolysis.
It is important to note that the human L3MBTL3 gene is mutated in medulloblastoma and is also implicated in pathological disorders, such as multiple sclerosis, insulin resistance, prostate cancer, and breast cancer (30, 32–35). The human DCAF5 gene is located in 14q24.1, the deletion of which is associated with various disorders, including breast cancer susceptibility, leukemia (including lymphoblastic and chronic lymphocytic leukemia), congenital heart defects, brachydactyly, and intellectual disability (47–50). Because SOX2 is a master stem cell protein in mouse and human ES cells, induced pluripotent stem cells, and many adult stem cells, and overexpression of SOX2 protein is associated with many different types of cancers (6, 51), our studies suggest that the L3MBTL3-dependent CRL4DCAF5 pathway may play an important role in controlling the stability of SOX2 protein in various embryonic stem cells, adult stem cells, and cancer cells. Further investigation of the function and regulation of the L3MBTL3-dependent CRL4DCAF5 pathway in development and various human diseases, including cancers, is warranted.
Experimental procedures
Cell culture
Human ovarian carcinoma PA-1 and embryonic kidney 293 cells were purchased from American Type Cell Collection (ATCC). Mouse embryonic stem cells (CMTI-2, strain C57/BL6J, passage 11) were obtained from Millipore-Sigma. PA-1 cells were cultured in minimum essential medium (MEM), and 293 cells were in Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% fetal bovine serum and penicillin/streptomycin (25, 39). Mouse embryonic stem cells were cultured on the mitomycin C–treated mouse fibroblast feeder layer in knockout DMEM and knockout serum replacement, supplemented with LIF, GlutaMax, β-mercaptoethanol, MEM nonessential amino acid solution, and penicillin/streptomycin (all from Life Technologies) (25). All cells have been recently authenticated and tested based on protein markers such as SOX2 or Oct4 in mouse embryonic stem cells and PA-1 cells and p53 or p21 in 293 cells (39). All cells were negative for the mycoplasma tests. Alkaline Phosphatase Staining kit II was purchased from Reprocell, and mouse ES cells were stained according to the manufacturer's protocol. For stable expression, human SOX2 and its mutant cDNAs were cloned into the retroviral pMSCV-3XFLAG-3XHA vector (Clontech) to construct the N-terminal tagged 3XFLAG-3XHA–SOX2 fusion proteins (25). Transient transfection for recombinant viral packaging was conducted in 293 cells using the calcium phosphate method (29). The recombinant viruses were collected from cell culture media of transfected cells and were used to infect PA-1 cells. Cells with stable expression of targeted proteins were selected by the puromycin resistance (25). Expression of SOX2 protein was confirmed by Western blotting analyses. Lipofectamine 2000 was used for other transient expression constructs into 293 cells, such as the expression of the GFP-SOX2, DCAF5, and L3MBTL3 for in vivo polyubiquitination analyses (39).
Antibodies and immunological analysis
Anti-LSD1 (A300-215A), L3MBTL3 (A302-852), SOX2 (SOX2 (A301-741A), SET7 (A301-747A), CoREST (A300-130A), SFMBT1 (A303-221A), CUL4A (A300-739A), CUL4B (A303-863A), and DDB1 (A300-462A) antibodies were purchased from Bethyl Laboratories; anti-PHF20L1 (HPA028417) was from Sigma; and anti-actin (sc-1616) antibodies were from Santa Cruz Biotechnology, Inc. The anti-monomethylated Lys-42 and monomethylated Lys-117 SOX2 peptide antibodies, anti-CUL4A, -CUL4B, -CUL1, -FLAG, -HA, and -GST antibodies were described before (25, 29). A DCAF5 peptide (MKRRAGLGGSMRSVVGFLSQRGLHC) was synthesized at ABI Scientific. The peptides were used to raise rabbit polyclonal antibodies after coupling these peptides to keyhole limpet hemocyanin (25, 29). For direct Western blotting, the cells were washed with PBS and directly lysed in the 2× SDS sample buffer (4% SDS, 100 mm Tris, pH 6.8, and 20% glycerol) (25). Protein concentrations were measured by the protein assay dye (Bio-Rad) and equalized before the addition of 0.2% bromphenol blue and freshly added 10% β-mercaptoethanol and boiled for 15 min. Usually three replicate repeats of siRNA-based knockdown experiments were conducted to evaluate the effects on target proteins. Each set of protein samples in an siRNA-based knockdown experiment was normalized by total proteins to the luciferase siRNA control and further analyzed by Western blot analysis in three replicates to normalize protein levels to the protein-loading controls, such as actin, to evaluate knockdown efficiencies, effects on target proteins, and operational variables.
Methylated peptide binding assays
The monomethylated Lys-42 (CAGGNQKNSPDRVK(me1)RPMNAFMVWSR) and cognate unmethylated peptides and the monomethylated Lys-117 (PDYKYRPRRKTK(me1)TLMKKDKYC) and cognate unmethylated peptides of SOX2 were synthesized at ABI Scientific. They were covalently coupled to the Sulfolink-coupled resins (Thermo Fisher Scientific) through the disulfide bond between the C-terminal cysteine of the peptides and the resins (25). The GST was fused in frame to the N terminus of the full-length L3MBTL3 in pGEXKG, and the recombinant GST fusion proteins were expressed in Escherichia coli and purified by GSH-Sepharose (GE Healthcare) (25). For peptide-binding assays, 25–30 μl of peptide-coupled resins were prewashed with the binding buffer (0.1% NP-40, 50 mm Tris-Cl, pH 7.5, 150 mm NaCl), and the resins were preblocked with 5 μg of GST protein at room temperature for 2 h. The peptide-coupled resins were then incubated with 1.5 μg each of GST or the indicated GST fusion proteins in the binding buffer overnight at 4 °C (25, 29). The beads were extensively washed (4–5 times) with PBS, and the proteins associated with the resins were analyzed by Western blotting with anti-GST antibodies (25, 29). In the case of cell lysates as the source of endogenous L3MBTL3, PA-1 cells were lysed with the NP-40–containing lysis buffer, nuclear fractions were cleared, and 250–500 μg of soluble cell lysate proteins were used for each peptide resin–binding assay (25).
Retinoic acid–induced differentiation of mouse embryonic stem cells
Mouse embryonic stem cells (CMTI-2, passage 14) were cultured on 0.1% gelatin-coated culture dishes in knockout DMEM and Knockout Serum Replacement, supplemented with 103 units of LIF, GlutaMax, β-mercaptoethanol, MEM nonessential amino acid solution, and penicillin/streptomycin (25). For differentiation assays (46), cells were plated at a density of 0.3 × 106 cells/10-cm dish. Twenty-four hours after plating (day 0 time point), LIF was removed, and RA (Sigma) was added at a concentration of 5 μm. Cell samples were collected daily for 6 days for protein analysis.
Immunoaffinity purification and MS analysis
Cell lysates from 50 dishes (150 mm × 25 mm) of PA-1 cells stably expressing the 3XFLAG- and 3XHA-SOX2 were employed for immunoaffinity purification using 1 ml of the anti-FLAG M2 affinity gel (Sigma, A2220) (29). The proteins in the 3XFLAG-3XHA–SOX2 immunocomplexes were eluted by incubating with 3XFLAG peptide (Sigma). The eluted complexes were loaded onto 0.5 ml of anti-HA antibody–agarose, and bound proteins were eluted with an SDS-containing buffer. The proteins were separated on an SDS-polyacrylamide gel, and protein bands were excised and trypsinized. Tryptic peptides derived from each gel slice were separated by an on-line C18 (New Objective, IntegraFritTM column, ProteoPepTMC18) nano-flow reversed-phase LC (Easy nano-LC) connected to an LTQ Orbitrap XL mass spectrometer (Thermo Scientific). The LC eluent was directly nanosprayed into the LTQ Orbitrap XL mass spectrometer as described previously (29).
Transfection, siRNAs, and oligonucleotides
DharmaFECT 1 transfection reagent (Dharmacon) was used for transfection of siRNAs into PA-1 and mouse embryonic stem cells (25). Oligofectamine (Life Technologies) was used for transfection of siRNAs into PA-1 or 293 cells for siRNA-mediated knockdown analysis (25, 29). Typically, a 50 nm concentration of each siRNA or their combinations were transfected into target cells for 48 h, and cells were directly lysed in the SDS or NP-40 lysis buffers (25). The knockdown effects were examined by Western blotting with specific antibodies. For verification of the effects of new proteins, usually two or three independent siRNAs were designed to examine the knockdown efficiency and the consequences of knockdown on target proteins. The siRNAs for humans are as follows: LSD1 (also mouse), GGAAGAAGAUAGUGAAAAC; PHF20L1, TGGGGTTGATGGTGCTGAA; DCAF5-1, GCUGCAGAAACCUCUACAA; DCAF5-2, ATCACCAACTTCTGACATA; L3MBTL3-1, GATGCAGATTCTCCTGATA; L3MBTL3-2, GGTACCAACTGCTCAAGAA; mouse L3MBTL3, GCTGAGGTTTGTGGATATA; mouse DCAF5, GGATGTGTTTGCTCATGAA; mouse PHF20L1, UCCAGCUUCAGGGAAUAAA; human and mouse SOX2, CGCUCAUGAAGAAGGAUAA. All siRNAs were synthesized from GE Dharmacon.
Statistical information
Experiments were usually performed with at least three independent repeats (biological replicates) to ensure the results. Statistical plot analyses were performed using Microsoft Excel. Protein bands were quantified using ImageJ. To quantify protein loading in each Western blot analysis of a set of protein samples, the same protein samples were analyzed with three repeated loading experiments (technical replicates). For cell-based assays, triplicated repeats in the same set of cells (technical replicates) were measured, and the experiments were usually repeated three times independently with independently cultured cells (biological replicates). Quantitative data are expressed by bar graphs, with mean and S.D. (error bars) from independent replicates. For siRNA-mediated knockdown experiments, statistically significant differences or variations between means of double and single knockdowns were normalized to the luciferase siRNA control and compared using a two-tailed paired Student's t test. The data in all figures met the assumption of normal distribution for tests. Different data sets were considered to be statistically significant when the p value was <0.05 (*), 0.01 (**), or 0.001 (***) (29).
Author contributions
H. Z. conceived the work, organized approaches, designed and supervised all research activities, analyzed results, and wrote the manuscript; C. Z. organized and conducted most experiments; F. Leng made significant contributions. J. Y. and H. Z. performed the mass spectrometry–based proteolysis and data analysis; L. S., S. A., N. H., L. L., D. Q., and F. Lu assisted in some experiments. H. S. assisted with mouse ES cells. All authors read and agreed on the manuscript drafted by H. Z.
Supplementary Material
This work was supported by National Institutes of Health Grants R15NS096694 (to H. S.) and R15GM131255 (to H. Z.) and a grant from the Cancer Research Gift Fund to the College of Sciences at the University of Nevada, Las Vegas (to H. S. and H. Z.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Fig. S1.
- ES cell
- embryonic stem cell
- MBT
- malignant brain tumor
- AP
- alkaline phosphatase
- GST
- glutathione S-transferase
- LIF
- leukemia-inhibitory factor
- MEM
- minimum essential medium
- DMEM
- Dulbecco's modified Eagle's medium
- me1
- monomethylated
- me0
- unmethylated
- NP-40
- Nonidet P-40
- RA
- retinoic acid.
References
- 1. Driessens G., and Blanpain C. (2011) Long live sox2: sox2 lasts a lifetime. Cell Stem Cell 9, 283–284 10.1016/j.stem.2011.09.007 [DOI] [PubMed] [Google Scholar]
- 2. Masui S., Nakatake Y., Toyooka Y., Shimosato D., Yagi R., Takahashi K., Okochi H., Okuda A., Matoba R., Sharov A. A., Ko M. S., and Niwa H. (2007) Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat. Cell Biol. 9, 625–635 10.1038/ncb1589 [DOI] [PubMed] [Google Scholar]
- 3. Episkopou V. (2005) SOX2 functions in adult neural stem cells. Trends Neurosci. 28, 219–221 10.1016/j.tins.2005.03.003 [DOI] [PubMed] [Google Scholar]
- 4. Rizzino A., and Wuebben E. L. (2016) Sox2/Oct4: a delicately balanced partnership in pluripotent stem cells and embryogenesis. Biochim. Biophys. Acta 1859, 780–791 10.1016/j.bbagrm.2016.03.006 [DOI] [PubMed] [Google Scholar]
- 5. Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., and Yamanaka S. (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- 6. Sarkar A., and Hochedlinger K. (2013) The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell 12, 15–30 10.1016/j.stem.2012.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goolam M., Scialdone A., Graham S. J. L., Macaulay I. C., Jedrusik A., Hupalowska A., Voet T., Marioni J. C., and Zernicka-Goetz M. (2016) Heterogeneity in Oct4 and Sox2 targets biases cell fate in 4-cell mouse embryos. Cell 165, 61–74 10.1016/j.cell.2016.01.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G., Gifford D. K., Melton D. A., Jaenisch R., and Young R. A. (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 10.1016/j.cell.2005.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Adachi K., Suemori H., Yasuda S. Y., Nakatsuji N., and Kawase E. (2010) Role of SOX2 in maintaining pluripotency of human embryonic stem cells. Genes Cells 15, 455–470 [DOI] [PubMed] [Google Scholar]
- 10. Boer B., Kopp J., Mallanna S., Desler M., Chakravarthy H., Wilder P. J., Bernadt C., and Rizzino A. (2007) Elevating the levels of Sox2 in embryonal carcinoma cells and embryonic stem cells inhibits the expression of Sox2:Oct-3/4 target genes. Nucleic Acids Res. 35, 1773–1786 10.1093/nar/gkm059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kopp J. L., Ormsbee B. D., Desler M., and Rizzino A. (2008) Small increases in the level of Sox2 trigger the differentiation of mouse embryonic stem cells. Stem Cells 26, 903–911 10.1634/stemcells.2007-0951 [DOI] [PubMed] [Google Scholar]
- 12. Chew J. L., Loh Y. H., Zhang W., Chen X., Tam W. L., Yeap L. S., Li P., Ang Y. S., Lim B., Robson P., and Ng H. H. (2005) Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol. Cell Biol. 25, 6031–6046 10.1128/MCB.25.14.6031-6046.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fong H., Hohenstein K. A., and Donovan P. J. (2008) Regulation of self-renewal and pluripotency by Sox2 in human embryonic stem cells. Stem Cells 26, 1931–1938 10.1634/stemcells.2007-1002 [DOI] [PubMed] [Google Scholar]
- 14. Arnold K., Sarkar A., Yram M. A., Polo J. M., Bronson R., Sengupta S., Seandel M., Geijsen N., and Hochedlinger K. (2011) Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell 9, 317–329 10.1016/j.stem.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Klose R. J., Kallin E. M., and Zhang Y. (2006) JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 7, 715–727 10.1038/nrg1945 [DOI] [PubMed] [Google Scholar]
- 16. Elkouris M., Kontaki H., Stavropoulos A., Antonoglou A., Nikolaou K. C., Samiotaki M., Szantai E., Saviolaki D., Brown P. J., Sideras P., Panayotou G., and Talianidis I. (2016) SET9-mediated regulation of TGF-β signaling links protein methylation to pulmonary fibrosis. Cell Rep. 15, 2733–2744 10.1016/j.celrep.2016.05.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fang L., Zhang L., Wei W., Jin X., Wang P., Tong Y., Li J., Du J. X., and Wong J. (2014) A methylation-phosphorylation switch determines Sox2 stability and function in ESC maintenance or differentiation. Mol. Cell 55, 537–551 10.1016/j.molcel.2014.06.018 [DOI] [PubMed] [Google Scholar]
- 18. Zhang X., Wen H., and Shi X. (2012) Lysine methylation: beyond histones. Acta Biochim. Biophys. Sin. (Shanghai) 44, 14–27 10.1093/abbs/gmr100 [DOI] [PubMed] [Google Scholar]
- 19. Kim S. K., Lee H., Han K., Kim S. C., Choi Y., Park S. W., Bak G., Lee Y., Choi J. K., Kim T. K., Han Y. M., and Lee D. (2014) SET7/9 methylation of the pluripotency factor LIN28A is a nucleolar localization mechanism that blocks let-7 biogenesis in human ESCs. Cell Stem Cell 15, 735–749 10.1016/j.stem.2014.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fu L., Wu H., Cheng S. Y., Gao D., Zhang L., and Zhao Y. (2016) Set7 mediated Gli3 methylation plays a positive role in the activation of Sonic Hedgehog pathway in mammals. eLife 5, e15690 10.7554/eLife.15690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang J., Hevi S., Kurash J. K., Lei H., Gay F., Bajko J., Su H., Sun W., Chang H., Xu G., Gaudet F., Li E., and Chen T. (2009) The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 41, 125–129 10.1038/ng.268 [DOI] [PubMed] [Google Scholar]
- 22. Estève P. O., Chang Y., Samaranayake M., Upadhyay A. K., Horton J. R., Feehery G. R., Cheng X., and Pradhan S. (2011) A methylation and phosphorylation switch between an adjacent lysine and serine determines human DNMT1 stability. Nat. Struct. Mol. Biol. 18, 42–48 10.1038/nsmb.1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kontaki H., and Talianidis I. (2010) Lysine methylation regulates E2F1-induced cell death. Mol. Cell 39, 152–160 10.1016/j.molcel.2010.06.006 [DOI] [PubMed] [Google Scholar]
- 24. Estève P. O., Chin H. G., Benner J., Feehery G. R., Samaranayake M., Horwitz G. A., Jacobsen S. E., and Pradhan S. (2009) Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 106, 5076–5081 10.1073/pnas.0810362106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang C., Hoang N., Leng F., Saxena L., Lee L., Alejo S., Qi D., Khal A., Sun H., Lu F., and Zhang H. (2018) LSD1 demethylase and the methyl-binding protein PHF20L1 prevent SET7 methyltransferase-dependent proteolysis of the stem-cell protein SOX2. J. Biol. Chem. 293, 3663–3674 10.1074/jbc.RA117.000342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jiang Y., Liu L., Shan W., and Yang Z. Q. (2016) An integrated genomic analysis of Tudor domain-containing proteins identifies PHD finger protein 20-like 1 (PHF20L1) as a candidate oncogene in breast cancer. Mol. Oncol. 10, 292–302 10.1016/j.molonc.2015.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Estève P. O., Terragni J., Deepti K., Chin H. G., Dai N., Espejo A., Corrêa I. R. Jr., Bedford M. T., and Pradhan S. (2014) Methyllysine reader plant homeodomain (PHD) finger protein 20-like 1 (PHF20L1) antagonizes DNA (cytosine-5) methyltransferase 1 (DNMT1) proteasomal degradation. J. Biol. Chem. 289, 8277–8287 10.1074/jbc.M113.525279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yin F., Lan R., Zhang X., Zhu L., Chen F., Xu Z., Liu Y., Ye T., Sun H., Lu F., and Zhang H. (2014) LSD1 regulates pluripotency of embryonic stem/carcinoma cells through histone deacetylase 1-mediated deacetylation of histone H4 at lysine 16. Mol. Cell Biol. 34, 158–179 10.1128/MCB.00631-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leng F., Yu J., Zhang C., Alejo S., Hoang N., Sun H., Lu F., and Zhang H. (2018) Methylated DNMT1 and E2F1 are targeted for proteolysis by L3MBTL3 and CRL4(DCAF5) ubiquitin ligase. Nat. Commun. 9, 1641 10.1038/s41467-018-04019-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bonasio R., Lecona E., and Reinberg D. (2010) MBT domain proteins in development and disease. Semin. Cell Dev. Biol. 21, 221–230 10.1016/j.semcdb.2009.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nady N., Krichevsky L., Zhong N., Duan S., Tempel W., Amaya M. F., Ravichandran M., and Arrowsmith C. H. (2012) Histone recognition by human malignant brain tumor domains. J. Mol. Biol. 423, 702–718 10.1016/j.jmb.2012.08.022 [DOI] [PubMed] [Google Scholar]
- 32. Northcott P. A., Nakahara Y., Wu X., Feuk L., Ellison D. W., Croul S., Mack S., Kongkham P. N., Peacock J., Dubuc A., Ra Y. S., Zilberberg K., McLeod J., Scherer S. W., Sunil Rao J., et al. (2009) Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat. Genet. 41, 465–472 10.1038/ng.336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kar S. P., Beesley J., Amin Al Olama A., Michailidou K., Tyrer J., Kote-Jarai Z., Lawrenson K., Lindstrom S., Ramus S. J., Thompson D. J., ABCTB Investigators, Kibel A. S., Dansonka-Mieszkowska A., Michael A., Dieffenbach A. K., et al. (2016) Genome-wide meta-analyses of breast, ovarian, and prostate cancer association studies identify multiple new susceptibility loci shared by at least two cancer types. Cancer Discov. 6, 1052–1067 10.1158/2159-8290.CD-15-1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Andlauer T. F., Buck D., Antony G., Bayas A., Bechmann L., Berthele A., Chan A., Gasperi C., Gold R., Graetz C., Haas J., Hecker M., Infante-Duarte C., Knop M., Kümpfel T., et al. (2016) Novel multiple sclerosis susceptibility loci implicated in epigenetic regulation. Sci. Adv. 2, e1501678 10.1126/sciadv.1501678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lotta L. A., Gulati P., Day F. R., Payne F., Ongen H., van de Bunt M., Gaulton K. J., Eicher J. D., Sharp S. J., Luan J., De Lucia Rolfe E., Stewart I. D., Wheeler E., Willems S. M., Adams C., et al. (2017) Integrative genomic analysis implicates limited peripheral adipose storage capacity in the pathogenesis of human insulin resistance. Nat. Genet. 49, 17–26 10.1038/ng.3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Arai S., and Miyazaki T. (2005) Impaired maturation of myeloid progenitors in mice lacking novel Polycomb group protein MBT-1. EMBO J. 24, 1863–1873 10.1038/sj.emboj.7600654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang J., Bonasio R., Strino F., Kluger Y., Holloway J. K., Modzelewski A. J., Cohen P. E., and Reinberg D. (2013) SFMBT1 functions with LSD1 to regulate expression of canonical histone genes and chromatin-related factors. Genes Dev. 27, 749–766 10.1101/gad.210963.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lin S., Shen H., Li J. L., Tang S., Gu Y., Chen Z., Hu C., Rice J. C., Lu J., and Wu L. (2013) Proteomic and functional analyses reveal the role of chromatin reader SFMBT1 in regulating epigenetic silencing and the myogenic gene program. J. Biol. Chem. 288, 6238–6247 10.1074/jbc.M112.429605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang X., Lu F., Wang J., Yin F., Xu Z., Qi D., Wu X., Cao Y., Liang W., Liu Y., Sun H., Ye T., and Zhang H. (2013) Pluripotent stem cell protein Sox2 confers sensitivity to LSD1 inhibition in cancer cells. Cell Rep. 5, 445–457 10.1016/j.celrep.2013.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. James L. I., Korboukh V. K., Krichevsky L., Baughman B. M., Herold J. M., Norris J. L., Jin J., Kireev D. B., Janzen W. P., Arrowsmith C. H., and Frye S. V. (2013) Small-molecule ligands of methyl-lysine binding proteins: optimization of selectivity for L3MBTL3. J. Med. Chem. 56, 7358–7371 10.1021/jm400919p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. James L. I., Barsyte-Lovejoy D., Zhong N., Krichevsky L., Korboukh V. K., Herold J. M., MacNevin C. J., Norris J. L., Sagum C. A., Tempel W., Marcon E., Guo H., Gao C., Huang X. P., Duan S., et al. (2013) Discovery of a chemical probe for the L3MBTL3 methyllysine reader domain. Nat. Chem. Biol. 9, 184–191 10.1038/nchembio.1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang Q., Yu M., Ma Y., Zhang X., Zhang H., Li S., Lan R., and Lu F. (2018) PHF20L1 antagonizes SOX2 proteolysis triggered by the MLL1/WDR5 complexes. Lab. Invest. 98, 1627–1641 10.1038/s41374-018-0106-8 [DOI] [PubMed] [Google Scholar]
- 43. Adamo A., Sesé B., Boue S., Castaño J., Paramonov I., Barrero M. J., and Izpisua Belmonte J. C. (2011) LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat. Cell Biol. 13, 652–659 10.1038/ncb2246 [DOI] [PubMed] [Google Scholar]
- 44. Jin J., Arias E. E., Chen J., Harper J. W., and Walter J. C. (2006) A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell 23, 709–721 10.1016/j.molcel.2006.08.010 [DOI] [PubMed] [Google Scholar]
- 45. Higa L. A., and Zhang H. (2007) Stealing the spotlight: CUL4-DDB1 ubiquitin ligase docks WD40-repeat proteins to destroy. Cell Div. 2, 5 10.1186/1747-1028-2-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ivanova N., Dobrin R., Lu R., Kotenko I., Levorse J., DeCoste C., Schafer X., Lun Y., and Lemischka I. R. (2006) Dissecting self-renewal in stem cells with RNA interference. Nature 442, 533–538 10.1038/nature04915 [DOI] [PubMed] [Google Scholar]
- 47. Oehl-Jaschkowitz B., Vanakker O. M., De Paepe A., Menten B., Martin T., Weber G., Christmann A., Krier R., Scheid S., McNerlan S. E., McKee S., and Tzschach A. (2014) Deletions in 14q24.1q24.3 are associated with congenital heart defects, brachydactyly, and mild intellectual disability. Am. J. Med. Genet. A 164A, 620–626 10.1002/ajmg.a.36321 [DOI] [PubMed] [Google Scholar]
- 48. Lee P., Fu Y. P., Figueroa J. D., Prokunina-Olsson L., Gonzalez-Bosquet J., Kraft P., Wang Z., Jacobs K. B., Yeager M., Horner M. J., Hankinson S. E., Hutchinson A., Chatterjee N., Garcia-Closas M., Ziegler R. G., et al. (2012) Fine mapping of 14q24.1 breast cancer susceptibility locus. Hum. Genet. 131, 479–490 10.1007/s00439-011-1088-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xu W., Lu X., Kim Y., Luo Y., Martin M., Mulvihill J. J., and Li S. (2008) Deletion of 14q24.1 approximately q24.3 in a patient with acute lymphoblastic leukemia: a hidden chromosomal anomaly detected by array-based comparative genomic hybridization. Cancer Genet. Cytogenet. 185, 43–46 10.1016/j.cancergencyto.2008.04.007 [DOI] [PubMed] [Google Scholar]
- 50. Reindl L., Bacher U., Dicker F., Alpermann T., Kern W., Schnittger S., Haferlach T., and Haferlach C. (2010) Biological and clinical characterization of recurrent 14q deletions in CLL and other mature B-cell neoplasms. Br. J. Haematol. 151, 25–36 10.1111/j.1365-2141.2010.08299.x [DOI] [PubMed] [Google Scholar]
- 51. Weina K., and Utikal J. (2014) SOX2 and cancer: current research and its implications in the clinic. Clin. Transl. Med. 3, 19 10.1186/2001-1326-3-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.