Abstract
Homologous recombination (HR)-directed DNA double-strand break (DSB) repair enables template-directed DNA repair to maintain genomic stability. RAD51 recombinase (RAD51) is a critical component of HR and facilitates DNA strand exchange in DSB repair. We report here that treating triple-negative breast cancer (TNBC) cells with the fatty acid nitroalkene 10-nitro-octadec-9-enoic acid (OA-NO2) in combination with the antineoplastic DNA-damaging agents doxorubicin, cisplatin, olaparib, and γ-irradiation (IR) enhances the antiproliferative effects of these agents. OA-NO2 inhibited IR-induced RAD51 foci formation and enhanced H2A histone family member X (H2AX) phosphorylation in TNBC cells. Analyses of fluorescent DSB reporter activity with both static-flow cytometry and kinetic live-cell studies enabling temporal resolution of recombination revealed that OA-NO2 inhibits HR and not nonhomologous end joining (NHEJ). OA-NO2 alkylated Cys-319 in RAD51, and this alkylation depended on the Michael acceptor properties of OA-NO2 because nonnitrated and saturated nonelectrophilic analogs of OA-NO2, octadecanoic acid and 10-nitro-octadecanoic acid, did not react with Cys-319. Of note, OA-NO2 alkylation of RAD51 inhibited its binding to ssDNA. RAD51 Cys-319 resides within the SH3-binding site of ABL proto-oncogene 1, nonreceptor tyrosine kinase (ABL1), so we investigated the effect of OA-NO2–mediated Cys-319 alkylation on ABL1 binding and found that OA-NO2 inhibits RAD51–ABL1 complex formation both in vitro and in cell-based immunoprecipitation assays. The inhibition of the RAD51–ABL1 complex also suppressed downstream RAD51 Tyr-315 phosphorylation. In conclusion, RAD51 Cys-319 is a functionally significant site for adduction of soft electrophiles such as OA-NO2 and suggests further investigation of lipid electrophile–based combinational therapies for TNBC.
Keywords: homologous recombination, breast cancer, drug action, drug discovery, DNA repair, signal transduction, cysteine-mediated cross-linking, homologous recombination, nitro fatty acid, Rad51, triple negative breast cancer, nitroalkene, PARP1
Introduction
Fatty acid nitroalkenes are endogenously detectable products of nitric oxide and nitrite-dependent metabolic and inflammatory reactions with unsaturated fatty acids. By virtue of their electrophilic nature, fatty acid nitroalkenes mediate post-translational modifications of hyper-reactive nucleophilic cysteine thiols in proteins, including NF-κB, Keap1, peroxisome proliferator–activated receptor γ, and HSF-1, thus modulating protein structure and function and mediating pleiotropic cytoprotective and anti-inflammatory signaling responses (1, 2). Emerging evidence has revealed that triple-negative breast cancer cell growth, migration, and invasion are suppressed by treatment with the electrophilic nitro-fatty acid derivative nitro-oleic acid (10-nitro-octadec-9-enoic acid (OA-NO2)4) through modulation of NF-κB signaling, whereas nontumorigenic breast epithelial cells are resistant to the effects of OA-NO2 because of more intact mechanisms for maintaining redox homeostasis. (3). We further explored the effects of fatty acid nitroalkenes on triple-negative breast cancer (TNBC) cell growth in concert with the clinically relevant antineoplastic DNA-damaging agents doxorubicin, cisplatin, olaparib, and γ-irradiation (IR).
The multitude of exogenously and endogenously stimulated DNA-damaging events requires that DNA damage be vigilantly detected and efficiently repaired. Several DNA repair mechanisms have been identified that ameliorate deleterious genomic perturbations such as direct reversal, mismatch repair, nucleotide excision repair, base excision repair, and double-stranded break (DSB) repair (4). DNA DSBs are particularly pathogenic as the loss of genomic material and mutations promote genomic variability and disequilibrium. There are two main pathways to repair DSBs: nonhomologous end joining (NHEJ) and HR. Although NHEJ is faster and more frequently used, HR repair mechanisms maintain the highest fidelity of the genome. HR repair protects cells from the deleterious genomic instability caused by DSB by correcting for genetic material loss through homologous template searches that maintain the genomic landscape (5). RAD51 is a critical component of HR that facilitates the homology search and strand exchange to repair DSBs (6). RAD51 and the structurally similar proteins XRCC2, XRCC3, RAD51B, RAD51C, RAD51D, DMC1, and SWSAP1 all work in concert to promote HR (7). Consequently, reductions in RAD51 and paralog activity are linked with carcinogenesis (8).
Although RAD51 is essential for high-fidelity repair of DSB to maintain genomic homeostasis, overexpression of RAD51 in cancer can also have detrimental consequences. The extent of RAD51 overexpression is correlated with breast cancer tumor grade, and RAD51 overexpression has been identified in TNBC cell lines and metastatic patient samples (9, 10). Overexpression of RAD51 inhibits chemotherapeutic efficacy in cancer patients by rendering cancer cells more resistant to DNA-damaging agents. Responses to neoadjuvant chemotherapy are inversely correlated with BRCA1, γH2AX, and RAD51 foci before treatment, as well as the numbers of RAD51 foci following treatment (11, 12).
We report that combination treatments of OA-NO2 with the antineoplastic agents doxorubicin, cisplatin, IR, and olaparib enhance the antiproliferative effect of these DNA-damaging therapeutic strategies in TNBC cells. OA-NO2 was identified to suppress IR-induced RAD51 foci formation, inhibit RAD51 binding to ssDNA, decrease HR, induce phosphorylation of Ser-139 H2AX (γH2AX), disrupt RAD51–ABL heterodimerization, and decrease RAD51 Tyr-315 phosphorylation. These observations reinforce the concept that reactive species induce genomic perturbations in part via the disruption of HR and reveal a novel therapeutic strategy: that redox-derived soft electrophiles sensitize cancer cells to DNA-directed therapeutic strategies such as IR, cisplatin, and doxorubicin.
Results
OA-NO2 inhibits TNBC cell growth, RAD51 foci formation, and sensitivity to ionizing radiation
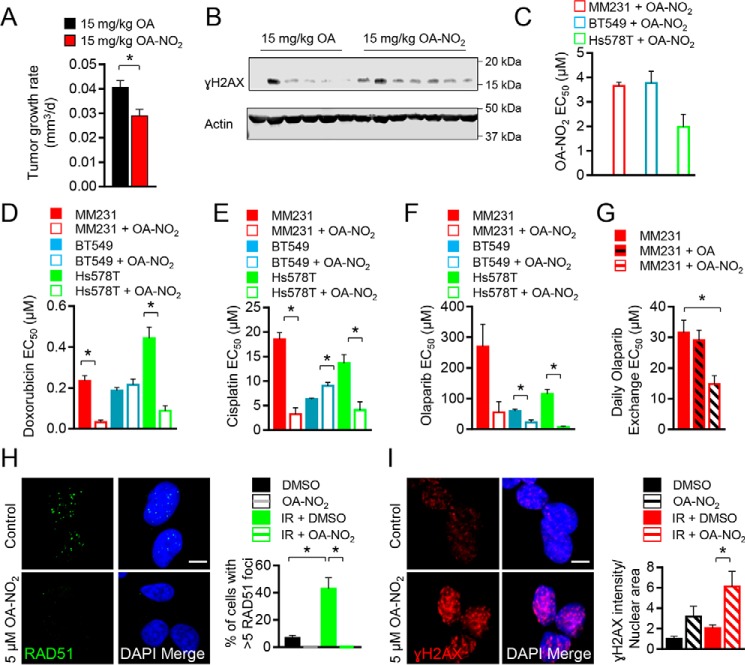
Current data indicate that OA-NO2 inhibits multiple aspects of TNBC epithelial cell, but not nontumorigenic breast epithelial cell, NF-κB signaling, by alkylating functionally significant thiols in (a) the inhibitor of NF-κB subunit kinase β, thus limiting downstream IκKα phosphorylation and (b) the NF-κB RelA protein, thus preventing DNA binding and promoting RelA polyubiquitination and proteasomal degradation (3). This motivated assessing whether OA-NO2 could also enhance TNBC DNA damage in vivo. MDA-MD-231 cells were implanted into the mammary gland of mice, and when tumors reached a volume of 100 mm3, mice were treated with 15 mg/kg nonelectrophilic fatty acid oleic acid (OA) or OA-NO2 by gavage for 4 weeks. Mice treated with OA-NO2 had significantly decreased tumor growth rates when compared with OA-treated controls (Fig. 1A and Fig. S1A). Probing tumor levels of the DNA damage biomarker γH2AX by immunoblotting showed that OA-NO2–treated mice displayed higher levels of γH2AX (Fig. 1B). Densitometric quantification of tumoral γH2AX/β-actin protein levels in OA- and OA-NO2–treated mice showed increased γH2AX in OA-NO2–treated mice. This response became statistically significant after Grubbs outlier detection and elimination of OA–treated mouse 2 (Fig. S1, B and C). The orthotopic tumor in OA mouse 2 was the largest tumor in the study, with necrosis potentially causing enhanced γH2AX levels.
Figure 1.
OA-NO2 inhibits TNBC cell growth, RAD51 foci formation, and sensitivity to ionizing radiation. A, MDA-MB-231 (MM231) cells (0.5 × 106) were orthotopically injected into 6-week-old mice, and mice were gavaged with 15 mg/kg OA (black) or OA-NO2 (red) for 4 weeks when tumors reached a volume of 100 mm3. Values indicate average, and error bars represent S.E.; n = 6–7 per group. B, tumoral γH2AX was increased in OA-NO2–treated mice compared with OA control mice as assessed by immunoblotting (n = 6–7 per group). C, MDA-MB-231 (red), BT549 (blue), or Hs578T (green) cells were treated with increasing concentrations of OA-NO2, and relative growth was measured by quantifying luminescent ATP levels (CellTiter-Glo). EC50 values indicate average, and error bars represent S.E.; n = 3. D–F, MDA-MB-231 (red), BT549 (blue), or Hs578T (green) cells were treated with increasing concentrations of doxorubicin, cisplatin, or olaparib ± OA-NO2 and measured as above. G, MDA-MB-231 (red) cells were treated with increasing concentrations of olaparib daily + vehicle, OA, or OA-NO2 and measured as above. H and I, OA-NO2 diminished RAD51 foci formation (green) and increased γH2AX (red) in MDA-MB-231 cells following irradiation with 5 Gy. Merged samples include 4′,6-diamidino-2-phenylindole (DAPI)-stained nuclei (blue). Cells on 16-well coverslips were dosed with 5 Gy and then treated with 5 μm OA-NO2 or vehicle for 6 h prior to immunofluorescence processing. The average percentages of cells with five or more foci from confocal z-stacked images are indicated from 0- or 5-Gy samples, and error bars represent. * = p < 0.05.
The TNBC growth-inhibitory effects of OA-NO2 were then evaluated in combination with DNA-damaging agents. The cell lines MDA-MB-231, BT-549, and Hs578T were treated with OA-NO2 daily for 3 days, and relative cell numbers were quantified by measuring the ATP-dependent luminescence signal generated using Ultra-Glo luciferase with the substrate luciferin. The EC50 values for growth inhibition of TNBC cells ranged from 1.98 ± 0.52 (Hs578T) to 3.78 ± 0.48 μm (BT-549) with MDA-MB-231 cells displaying an EC50 value of 3.66 ± 0.14 μm (Fig. 1C and Fig. S1D). We next tested the DNA-damaging agents doxorubicin and cisplatin in combination with daily treatment with 2 μm OA-NO2. OA-NO2 enhanced growth inhibition of doxorubicin in MDA-MB-231 and Hs578T cells, by 7- and 5-fold, respectively (Fig. 1D and Fig. S1E). The growth of BT-549 cells was not affected. Cotreatment with OA-NO2 and cisplatin showed a similar trend for MDA-MB-231 and Hs578T cells, which displayed increased growth inhibition by 6- and 3-fold, respectively, whereas growth inhibition of BT-549 cells was suppressed 1.4-fold (Fig. 1E and Fig. S1F). A subset of TNBC cells are sensitive to PARP inhibition and display a BRCAness phenotype in the presence of WT BRCA1 (14), so the PARP-1 inhibitor olaparib was evaluated to determine whether a combination treatment with OA-NO2 enhanced potency. MDA-MB-231, Hs578T, and BT-549 cells all displayed enhanced growth inhibition when olaparib was combined with OA-NO2 by 5-, 17-, and 3-fold, respectively (Fig. 1F and Fig. S1G). To specifically show that olaparib treatment in combination with OA-NO2, but not OA, affected proliferation and to evaluate the effect of daily olaparib media exchanges, MDA-MB-231 cell proliferation was quantified in dose-response assays. Daily administration of olaparib in combination with OA did not significantly alter the EC50 of olaparib in MDA-MB-231 cells. In contrast, olaparib in combination with OA-NO2 significantly inhibited growth (Fig. 1G). Thus, standard TNBC chemotherapeutic drugs as well as targeted PARP-1 inhibition exhibited enhanced antiproliferative effects when coadministered with OA-NO2 in TNBC cells.
The heightened tumor γH2AX levels in vivo and sensitization of TNBC cells to DNA-damaging agents, especially in the context of olaparib-induced responses, led to further exploration of DNA damage repair modulation by OA-NO2. As olaparib sensitivity is a hallmark of HR-deficient cells (15), whether OA-NO2 impacted DNA repair by HR was evaluated. To specifically probe DNA double-strand break repair, MDA-MB-231 cells were challenged with 5 Gy of IR, and RAD51 foci were quantified. The treatment of breast cancer cells with OA-NO2 inhibited RAD51 foci formation as reflected by (a) the number of cells with more than five foci and (b) responses of vehicle-treated cells following IR (Fig. 1H). Cell cycle analysis of MDA-MB-231 cells confirmed that no significant changes to the cell cycle occurred that might indirectly alter RAD51 foci formation (Fig. S1H). Evaluation of nuclear γH2AX staining to probe for DNA damage of MDA-MB-231 cells in the presence or absence of 5 Gy of IR found that OA-NO2 significantly increased nuclear γH2AX localization in irradiated MDA-MB-231 cells compared with vehicle controls, indicating increases in DSBs and overall DNA damage (Fig. 1I). Increasing concentrations of OA-NO2 also enhanced breast cancer cell death in a clonogenic assay following irradiation with 2 Gy of IR (Fig. S1I). Evaluation of the DNA-damaging effects of OA and OA-NO2 on nontransformed MCF10A cells following 5 Gy of IR found that nuclear γH2AX staining was only increased in MDA-MB-231 TNBC cells treated with OA-NO2 and not MCF10A cells (Fig. S2, A and B).
OA-NO2 decreases HR but not NHEJ efficiency
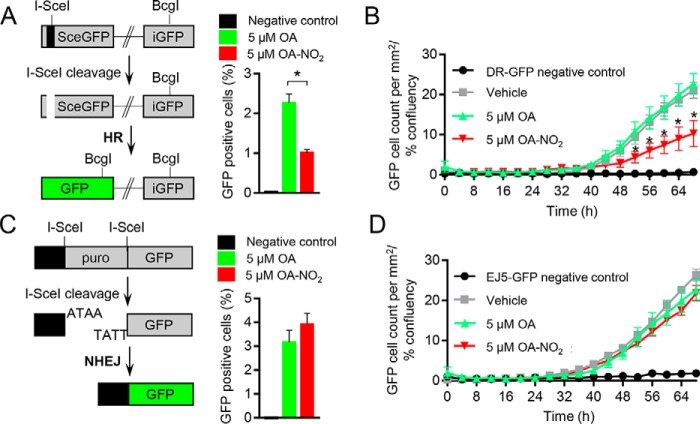
OA-NO2–dependent effects on HR DNA repair were further investigated by utilizing a direct repeat GFP (DR-GFP) reporter assay. This analysis quantifies intracellular recombination of an integrated cDNA cassette of two tandem nonfluorescent GFP constructs, following introduction of an I-SceI cleavage to the system, by measuring the fluorescent GFP protein that is produced following successful recombination (16). Daily OA-NO2 treatment of U2OS cells harboring the DR-GFP construct revealed that after I-SceI transfection, the number of GFP-positive cells was significantly decreased by 2-fold when compared with native OA or vehicle control after 48 h (Fig. 2A and Fig. S3A). A novel strategy was used to measure the kinetics of changes in HR in live cells by using automated fluorescence microscopy to track the emergence of GFP-positive cells over time in monolayers, as opposed to making static measurements of detached cells using flow cytometry. To account for changes in cell density over time, cell confluence was measured following treatment of DR-GFP U2OS cells with vehicle, 5 μm OA or OA-NO2 every 4 h for 3 days (Fig. S3B). The emergence of GFP-positive cells subsequent to I-SceI–induced cleavage was quantified and normalized to cell confluence over time to compare OA-NO2–treated with OA-treated and untreated cells. Daily administration of 5 μm OA-NO2 decreased the number of GFP-positive cells by 2-fold over 68 h when compared with controls (Fig. 2B). The impact of OA-NO2 on suppression of DSB repair through both the HR and NHEJ pathways was examined by utilizing an EJ5-GFP NHEJ reporter assay, which separates GFP cDNA from a transcriptional promoter with a puromycin resistance gene flanked by two I-SceI cleavage sites (17). In contrast to the effects seen by DR-GFP–mediated HR measurements, EJ5-GFP U2OS cells showed no effect of OA-NO2 on NHEJ. This was indicated by an absence of changes in the number of GFP-positive cells following I-SceI cleavage after 48 h by flow cytometric analysis or over 68 h by live-cell fluorescence microscopy (Fig. 2, C and D, and Fig. S3C).
Figure 2.
Inhibition of homologous recombination but not nonhomologous end joining by OA-NO2. A and B, U2OS cells containing the HR reporter construct DR-GFP were transfected with an I-SceI plasmid and treated with vehicle (gray), 5 μm OA (green), or 5 μm OA-NO2 (red). Negative control cells did not have I-SceI present. Values indicate average, and error bars represent S.E.; n ≥ 3. A, the number of GFP-positive cells was detected by flow cytometry at 48 h. B, emergence of GFP-positive cells over 68 h was quantified using live-cell fluorescence microscopy. GFP-positive cell counts were normalized to cell confluence and compared. C and D, U2OS cells containing the NHEJ reporter construct EJ5-GFP were transfected with an I-SceI plasmid and treated with vehicle (gray), 5 μm OA (green), or 5 μm OA-NO2 (red). Negative control cells did not have I-SceI present. Values indicate average, and error bars represent S.E.; n = 3. C, the number of GFP-positive cells were detected by flow cytometry at 48 h. D, emergence of GFP-positive cells over 68 h was quantified using live-cell fluorescence microscopy. GFP-positive cell counts were normalized to cell confluence and compared.
OA-NO2 targets RAD51 Cys-319 and decreases RAD51 phosphorylation
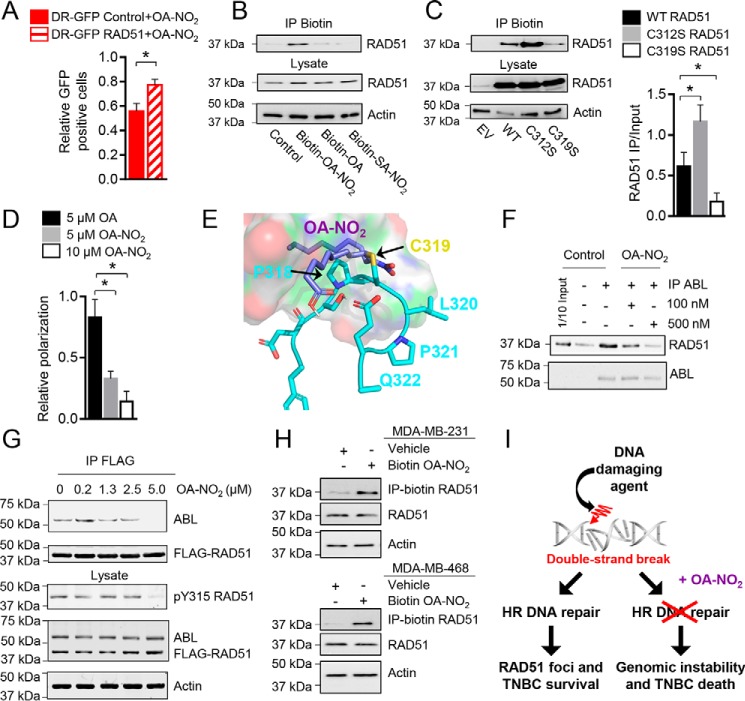
Inhibition of IR-induced RAD51 foci formation and DR-GFP HR reporter functionality by OA-NO2 was further studied by testing whether overexpression of the critical HR protein RAD51 in the HR reporter cells could rescue the effects of OA-NO2. HR activity, as measured by the percentage of GFP-positive cells relative to OA control treatment, was significantly increased in U2OS DR-GFP reporter cells stably overexpressing RAD51 treated with 5 μm OA-NO2 compared with control reporter cells (Fig. 3A and Fig. S4A). Protein structural data (Protein Data Bank (PDB) code 1N0W) show that Cys-319 is a solvent-exposed nucleophile within the RAD51 C terminus (Fig. S4B) that is susceptible to reaction with RI-1, a reagent also having Michael acceptor qualities (18). Moreover, fluorophore adduction of Cys-319 disrupts RAD51 filament formation in vitro (19). It was hypothesized that OA-NO2 would react with RAD51 Cys-319. Indeed, biotin-OA-NO2, but not the nonelectrophilic biotin-OA and biotin-10-nitro-octadecanoic acid (SA-NO2), supported affinity precipitation of RAD51 from cell lysates with streptavidin-labeled beads (Fig. 3B). Comparing RAD51 C312S or C319S mutant reaction with biotin-OA-NO2 revealed a preferential reaction of OA-NO2 with Cys-319 (Fig. 3C). RAD51 C312S and RAD51 WT controls were readily affinity-precipitated by biotin-OA-NO2 as opposed to when RAD51 C319S was expressed in mutant cells. Of note, the RAD51 C312S mutant displayed enhanced precipitation of OA-NO2, which may reflect interruption of a disulfide bond between RAD51 Cys-312 and Cys-319 or another intracellular protein that obscures Cys-319. The ability of OA-NO2 to specifically disrupt RAD51 binding from DNA was probed by quantifying changes in fluorescence polarization of an Alexa Fluor 488–conjugated single-stranded oligonucleotide in vitro. OA-NO2, but not OA, decreased the relative polarization of RAD51 in the presence of ATP and DNA (Fig. 3D). Control experiments found that OA and OA-NO2 did not cause nonspecific effects through fluorophore quenching to decrease fluorescence polarization (Fig. S4C). Computational analysis revealed that OA-NO2 alkylation of RAD51 Cys-319 is further stabilized by hydrophobic interactions with Pro-318 of RAD51 and hydrogen bonding with Glu-322 (Fig. 3E).
Figure 3.
OA-NO2 binds to RAD51 at Cys-319 and blocks ABL heterodimerization. A, U2OS cells containing the HR reporter construct DR-GFP were stably transfected with a control or RAD51 overexpression plasmid, and the cells were investigated as above with 5 μm OA-NO2 in control (red) or RAD51-overexpressing cells (red striped). B, OA-NO2 binds RAD51 in vitro. Purified RAD51 protein was incubated with control, biotinylated OA, OA-NO2, or SA-NO2 for 1 h; precipitated with streptavidin-coated agarose; and then detected by immunoblotting. C, OA-NO2 binds to RAD51 Cys-319 in cells expressing RAD51. 293T cells expressing WT or cysteine mutant RAD51 were incubated with biotinylated OA-NO2 for 1 h, precipitated, and detected. Three independent experiments were quantified and analyzed by one-way analysis of variance. *, p < 0.05. D, Alexa Fluor 488–conjugated DNA was incubated with purified RAD51, ATP, and 5 μm OA (black), 5 μm OA-NO2 (gray), or 10 μm OA-NO2 (white), and fluorescence polarization was quantified and normalized to a control lacking ATP. E, molecular modeling of RAD51 (blue) and OA-NO2 (purple). Binding of OA-NO2 with the Cys-319 residue (gold) of RAD51 is predicted to be further stabilized by hydrophobic interactions with Pro-318 and possible hydrogen bonding with Glu-322 of RAD51. F, OA-NO2 disrupts ABL binding to RAD51 in vitro. Purified RAD51 and ABL core proteins were incubated with OA-NO2 at 0, 100, or 500 nm for 1 h, and ABL was precipitated. The amount of bound RAD51 was detected by immunoblotting. G, OA-NO2 disrupts RAD51 and ABL interactions by IP and reduces RAD51 Tyr-315 phosphorylation. 293T cells were transfected with FLAG-RAD51 and ABL core protein and then treated with OA-NO2 for 1 h. RAD51 interactions with ABL and phosphorylated RAD51 Tyr-315 were probed by IP and immunoblotting. H, OA-NO2 binds RAD51 in MDA-MB-231 or MDA-MB-468 cells. Cells were incubated with biotinylated OA-NO2, and then lysates were precipitated with streptavidin-coated agarose and detected by immunoblotting. I, OA-NO2 decreases HR and causes genomic instability and death in TNBC cells. All values indicate average, and error bars represent S.E.; n = 3.
Cys-319 is located in the RAD51 C terminus within one of the two ABL-SH3–binding domains (amino acids 283–286 and 318–321) (20). In addition to RAD51 filament disruption, OA-NO2 inhibited heterodimerization of RAD51 and ABL. IP analysis revealed that purified RAD51 and catalytic ABL core (Src homology 2 (SH2), SH3, and kinase domains only) complex formation was abolished by OA-NO2 (Fig. 3F). ABL regulates RAD51 activity via sequential phosphorylation of RAD51 Tyr-54 and then Tyr-315 (21, 22). By transfecting FLAG-RAD51 and ABL core into 293T cells, the impact of OA-NO2 on RAD51–ABL complex formation and RAD51 Tyr-315 phosphorylation was examined. After treating cells with 0–5 μm OA-NO2 for 1 h, FLAG IP analysis revealed that OA-NO2 decreased the amount of ABL bound to RAD51 (Fig. 3G). Along with the inhibition of RAD51–ABL complex formation, RAD51 Tyr-315 phosphorylation was also inhibited by OA-NO2 in FLAG-RAD51– and ABL-expressing cells. To define whether OA-NO2 alkylates endogenous RAD51 in TNBC cells, biotin-OA-NO2 was added to MDA-MB-231 and MDA-MB-468 cells. Biotin-OA-NO2–RAD51 complex formation upon streptavidin precipitation was observed in lysates of both cell lines (Fig. 3H). Overall, these data reveal that OA-NO2 inhibited HR by forming adducts with RAD51 and possibly additional HR-related target proteins to enhance sensitivity to DNA-directed cancer therapies (Fig. 3I).
Discussion
Fatty acid nitroalkenes are endogenously produced products of nitric oxide– and nitrite-dependent nitration of unsaturated fatty acids. By virtue of kinetically rapid and reversible Michael addition, fatty acid nitroalkenes mediate the post-translational modification of susceptible Cys residues of proteins, in some cases modifying protein function and inducing signaling responses via pleiotropic mechanisms (1, 2, 23, 24). The present results indicate that OA-NO2 decreases the proliferation of TNBC cells, especially when coadministered with clinically relevant DNA-directed therapeutic agents (Fig. 1). OA-NO2 also amplified the induction of DSB through IR or I-SceI DNA cleavage by limiting IR-induced nuclear RAD51 foci formation and DNA recombination, specifically via inhibiting HR and not NHEJ (Figs. 1 and 2). The functionally significant Cys-319 of RAD51 was targeted by OA-NO2, but not by nonelectrophilic native and nitroalkane-substituted control fatty acids (Fig. 3). Cys-319 alkylation by OA-NO2 disrupted RAD51 dimerization with ABL and decreased ABL-induced phosphorylation of RAD51 Tyr-315 (Fig. 3). OA-NO2 may also target proteins beyond RAD51 that modulate HR.
The potent antiproliferative effect of olaparib, when administered in combination with OA-NO2 in TNBC cell lines, indicates the pharmacological induction of a BRCAness phenotype by OA-NO2 (Fig. 1). Suppression of HR-mediated DNA DSB repair by OA-NO2 is reflective of loss of function mutations in BRCA genes, which cause deficits in DNA repair capacity via impairment of HR. Breast cancer patients harboring BRCA1 loss of function mutations may also benefit from suppression of RAD51, as increased expression of RAD51 bypasses BRCA1 function and is a common feature of BRCA1-deficient breast tumors (25). Additional investigation of further OA-NO2 targets and actions, coupled with PARP inhibition in BRCA1-deficient breast cancer backgrounds, is thus warranted.
Although functional HR is important for maintaining genome stability, an enhancement of homology-directed DNA repair activities impedes chemotherapeutic and ionizing radiation treatments for cancer (6, 7). The elevated expression of RAD51 is positively correlated with breast cancer tumor grade and has been identified in several TNBC cell lines and metastatic patient samples (9, 10). Several studies have attempted to harness RAD51 inhibition to promote lethality in cancer cells. Inhibition of RAD51 with small-molecule inhibitors can sensitize cancer cells to chemotherapeutic agents or IR (e.g. DIDS (26), B02 (27, 28), RI-1 (18), and IBR2 (29)). For example, RI-1 was identified in a high-throughput screen to potentiate RAD51 filament formation and HR activity, fortuitously adducting Cys-319 (18). Unfortunately, RI-1 has multiple electrophilic centers in a complex biphenolic morpholino structure, thus inducing irreversible Michael addition and an incompatibility for in vivo applications due to toxicity.
The RAD51 Cys-319 represents an important functional site within the RAD51 protein. Homomultimeric RAD51 filaments interface surrounding the Cys-319 residue, which is located within an SH3 domain and nearby an ATPase domain (PDB code 1N0W) (30). Post-translational thiol modification or pharmacological targeting of Cys-319 may thus disrupt RAD51 function through multiple mechanisms.
It was previously reported that OA-NO2 more selectively targets TNBC thiols of the NF-κB signaling pathway, as opposed to nontumorigenic breast epithelium, due to the more effective mechanisms for maintaining redox homeostasis in normal breast epithelium (3). We now identify another specific target of electrophilic nitroalkenes, the Cys-319 of RAD51, that upon alkylation inhibits RAD51 binding to ssDNA (Fig. 3). Thus, the administration of synthetic homologs of endogenously occurring fatty acid nitroalkenes offers a viable option for inactivating RAD51. The clinical administration of intravenous and oral formulations of OA-NO2 (i.v. IND, 122583; oral IND, 124524) is safe, having cleared multiple Phase I and drug–drug interaction studies. An oral formulation of OA-NO2 is now in multicenter Phase II trials for treating chronic inflammatory-related diseases. The present results motivate further investigation into whether endogenously generated soft electrophiles or their exogenously administered synthetic homologs might play a role in modulating DNA repair and other signaling responses that improve treatment of drug-resistant cancers.
Experimental procedures
Cell culture and reagents
HEK 293T, MDA-MB-231, MDA-MB-468, Hs578T, and BT-549 cells (American Type Culture Collection) were cultured at 37 °C with 5% CO2 in Dulbecco's modified Eagle's medium (Gibco) supplemented with 5% FBS (HyClone), 100 units/ml penicillin, 100 mg/ml streptomycin (Gibco), nonessential amino acids (Gibco), and 2 mm L-glutamine (Gibco). Doxorubicin (Selleckchem), cisplatin (Sigma), or olaparib (Selleckchem) was dissolved in DMSO or N,N-dimethylformamide (cisplatin). OA-NO2 and biotinylated OA-NO2 were synthesized as described previously (24, 31). Pure OA-NO2 was diluted in DMSO and added to cells after solvation in assay medium. Relative cell numbers were compared by measuring the luminescence signal generated by ATP using the CellTiter-Glo (Promega) assay. Cells were plated in a 96-well plate at 5,000 (MDA-MB-231) or 6,600 (BT549 or Hs578T) cells/well. Cells were treated with doxorubicin, cisplatin, or olaparib at the indicated concentrations for 72 h in the presence or absence of 2 μm OA-NO2, which was replenished every 24 h.
OA-NO2 in vivo
Animals used for this study were approved by and conducted according to the guidelines of the University of Pittsburgh Institutional Animal Care and Use Committee (protocol 18093443). MDA-MB-231 cells (0.5 × 106) were injected into the mammary fat pad (left fourth gland) of 6-week-old female nude mice in a volume of 20 μl of sterile saline. When tumors reached an average volume of 100 mm3, mice were randomized into groups and administered vehicle (tricaprylin) + 15 mg/kg OA or vehicle + 15 mg/kg OA-NO2 every day by gavage (200 μl) for 4 weeks. The surgical procedure has been described previously (32).
Plasmids
The DR-GFP reporter and I-SceI pCAGGS plasmids were a kind gift from Prof. Maria Jasin (16, 33). pLVX-Neo-RAD51 was cloned by PCR amplification of RAD51 with PCR primers incorporating SpeI (5′) and BamHI (3′) restrictions sites. The PCR product was then ligated into the corresponding restrictions sites of pLVX-Neo (Clontech) and transformed into DH5α MAX Efficiency cells (Invitrogen). pLVX-Neo-RAD51 cysteine to serine mutant plasmids (137, 312, or 319) were produced using the QuikChange II site-directed mutagenesis kit (Agilent) using pLVX-Neo-RAD51 as a template.
DSB repair assays
Measurements of HR and NHEJ assays were performed as described previously (16, 34). HR activity was measured by counting GFP-positive cells by flow cytometry at the Magee-Womens Research Institute Flow Cytometry Core using a BD LSR II flow cytometer (BD Biosciences). RAD51-overexpressing cells were generated by stable transfection of pLVX-RAD51-IRES-Neo and selection with geneticin (Invitrogen).
Kinetic DSB repair assays
U2OS cells were prepared as above, but 5 h following compound treatment, cells were transferred into the Incucyte Zoom (Essen) live-cell imaging automated fluorescence microscope at 37 °C with 5% CO2. Cell confluence and green object count per mm2 were determined using Incucyte Zoom software. Green object count per field was normalized to cell confluence to correct for OA-NO2–induced effects on cell proliferation.
Immunostaining and imaging
To analyze RAD51 foci formation, 10,000 cells were plated to a CultureWell 16-well chambered coverglass (MIDSCI) coated with poly-l-lysine (Sigma) and incubated overnight in 5% FBS medium. Cells were then treated with OA-NO2 and irradiated (Gammacell 40 Exactor γ-Irradiator, Best Medical) with 5 Gy and incubated for 6 h. Cells were fixed with 10% formalin for 20 min at 4 °C and immunostained with RAD51 (Santa Cruz Biotechnology) or γH2AX (EMD Biosciences) antibody. z-stack images were acquired using a Nikon A1R confocal microscope with 60× oil objective, and acquisition was performed using NIS Elements software. Quantification of z-stacks and foci were completed using ImageJ software.
Cell cycle analysis
Cell cycle analysis was performed with propidium iodide on MDA-MB-231 cells treated with DMSO or 5 μm OA-NO2 (35). Samples were analyzed at the Magee-Womens Research Institute Flow Cytometry Core utilizing a BD LSR II flow cytometer.
Western blotting and immunoprecipitation
Cell lysates and immunoprecipitations were prepared as described previously (36). For immunoprecipitation analysis, one million HEK 293T cells were transiently transfected with FuGENE 6 (Promega) and 2 μg of pQCXIP (empty vector) or FLAG-RAD51 pQCXIP plasmids and precipitated with anti-FLAG M2 affinity gel (Sigma).
Biotinylated OA-NO2 affinity capture of RAD51
HEK 293T were transiently transfected with FuGENE 6 and 5 μg of RAD51-expressing vectors (WT, C312S, or C319S). Cells were treated 24 h later with 5 μm biotin-OA-NO2 or biotin-SA-NO2 for 1 h in 5% FBS medium. Cells were prepared as above. Precipitation of biotinylated OA-NO2 protein adducts was accomplished with 8 μl of streptavidin-agarose beads with 1 mg of total cell lysates incubated for 16 h at 4 °C. Detection of RAD51 was accomplished by immunoblotting with RAD51 antibody (1:2,000) with actin antibody (1:3,000) probed as a loading control.
Protein purification for in vitro RAD51-ABL binding assay
Recombinant His-tagged RAD51 in the pET21a vector was transformed into Escherichia coli BL21(DE3)pLysS cells (EMD Millipore) and purified as described previously (37).
DNA binding assays
Reactions were performed in black 96-well plates (Greiner) in 50-μl reaction volumes in 20 mm HEPES, pH 7.5, 10 mm MgCl2, 0.25 μm BSA, 2% glycerol, 30 mm NaCl, and 4% DMSO. Purified RAD51 protein (Abcam) and OA (negative control) or OA-NO2 were preincubated for 5 min at 25 °C. 2 mm ATP and 100 nm 5′-Alexa Fluor 488–conjugated ssDNA poly(dT) (Integrated DNA technologies) were added to the reaction and incubated for 90 min at 37 °C. DNA binding was measured using fluorescence polarization on a Tecan Spark 20 m (excitation/emission, 480 nm/535 nm). Compound fluorescence quenching was detected as above in the absence of RAD51 protein.
Molecular modeling
Structures for RAD51 (PDB code 1N0W (30) and OA-NO2 were aligned using PyMOL 1.7.1. The structure of OA-NO2 was generated using ChemDraw 15 (PerkinElmer Life Sciences) and converted to 3D structure using Open Babel version 2.3.1 (13).
Statistical analysis
Data represent the mean ± S.E. from three independent experiments unless otherwise noted. A p value <0.05 was considered statistically significant. Nonlinear curves were generated in GraphPad Prism 7.0 (GraphPad Software, La Jolla, CA) for statistical analysis. EC50 values and standard error were calculated from three independent experiments utilizing a nonlinear dose-response variable slope model. Significance was tested by one-way analysis of variance for multiple groups with Tukey post-test or by t test when groups were less than three. RAD51 foci number was analyzed with ImageJ. Nuclear boundaries were individually identified in more than 50 cells per treatment group in three independent experiments.
Author contributions
A. A., J. J. S., B. A. F., and C. A. N. conceptualization; A. A., J. J. S., C.-S. C. W., and C. A. N. data curation; A. A., J. J. S., D. N., C. J. C., and C. A. N. formal analysis; A. A. and J. J. S. visualization; A. A., J. J. S., S. R. W., and C. A. N. methodology; A. A., J. M. S., B. A. F., and C. A. N. writing-review and editing; J. J. S., C.-S. C. W., B. M. W., and C. A. N. investigation; J. J. S. writing-original draft; J. J. S. and C. A. N. project administration; S. R. W., Y. H., J. M. S., and C. A. N. resources; B. A. F. and C. A. N. funding acquisition; C. A. N. supervision; C. A. N. validation.
Supplementary Material
Acknowledgments
We thank Dr. M. Jasin (Memorial Sloan Kettering Cancer Center, New York, NY) for providing HR plasmids and Dr. P. Grover and Dr. T. Smithgall (University of Pittsburgh, Pittsburgh, PA) for providing purified RAD51 and ABL protein for the in vitro binding assay.
This work was supported by National Institutes of Health Grants R01-CA131350 and P30-CA047904 (to the UPMC Hillman Cancer Center and D. N.), T32-EB009403 (to B. M. W.), R01-CA197506 (to J. M. S), and R01-HL058115, R01-HL64937, P30-DK072506, and P01-HL103455 (to B. A. F.); Basic Biology of Aging, University of Pittsburgh (to C. A. N.); University of Pittsburgh Department of Pharmacology and Chemical Biology and William C. de Groat predoctoral fellowships (to A. A.); and a Cotswold Foundation postdoctoral fellowship (to J. J. S.). B. A. F. and C.-S. C. W. acknowledge an interest in Complexa, Inc. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supporting methods and Figs. S1–S4.
- OA-NO2
- 10-nitro-octadec-9-enoic acid
- HR
- homologous recombination
- DSB
- double-strand break
- TNBC
- triple-negative breast cancer
- IR
- γ-irradiation
- H2AX
- H2A histone family member X
- NHEJ
- nonhomologous end joining
- ssDNA
- single-stranded DNA
- SH
- Src homology
- OA
- oleic acid
- PARP
- poly(ADP-ribose) polymerase
- Gy
- grays
- DR-GFP
- direct repeat GFP
- SA-NO2
- 10-nitro-octadecanoic acid
- IP
- immunoprecipitation
- DIDS
- 4,4′-diisothiocyanostilbene-2,2′-disulfonic acid
- IND
- Investigational New Drug
- FBS
- fetal bovine serum.
References
- 1. Schopfer F. J., Cipollina C., and Freeman B. A. (2011) Formation and signaling actions of electrophilic lipids. Chem. Rev. 111, 5997–6021 10.1021/cr200131e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Delmastro-Greenwood M., Freeman B. A., and Wendell S. G. (2014) Redox-dependent anti-inflammatory signaling actions of unsaturated fatty acids. Annu. Rev. Physiol. 76, 79–105 10.1146/annurev-physiol-021113-170341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Woodcock C. C., Huang Y., Woodcock S. R., Salvatore S. R., Singh B., Golin-Bisello F., Davidson N. E., Neumann C. A., Freeman B. A., and Wendell S. G. (2018) Nitro-fatty acid inhibition of triple-negative breast cancer cell viability, migration, invasion, and tumor growth. J. Biol. Chem. 293, 1120–1137 10.1074/jbc.M117.814368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Curtin N. J. (2012) DNA repair dysregulation from cancer driver to therapeutic target. Nat. Rev. Cancer 12, 801–817 10.1038/nrc3399 [DOI] [PubMed] [Google Scholar]
- 5. Moynahan M. E., and Jasin M. (2010) Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol. 11, 196–207 10.1038/nrm2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Prakash R., Zhang Y., Feng W., and Jasin M. (2015) Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb. Perspect. Biol. 7, a016600 10.1101/cshperspect.a016600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Godin S. K., Sullivan M. R., and Bernstein K. A. (2016) Novel insights into RAD51 activity and regulation during homologous recombination and DNA replication. Biochem. Cell Biol. 94, 407–418 10.1139/bcb-2016-0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen J., Morrical M. D., Donigan K. A., Weidhaas J. B., Sweasy J. B., Averill A. M., Tomczak J. A., and Morrical S. W. (2015) Tumor-associated mutations in a conserved structural motif alter physical and biochemical properties of human RAD51 recombinase. Nucleic Acids Res. 43, 1098–1111 10.1093/nar/gku1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maacke H., Opitz S., Jost K., Hamdorf W., Henning W., Krüger S., Feller A. C., Lopens A., Diedrich K., Schwinger E., and Stürzbecher H. W. (2000) Over-expression of wild-type Rad51 correlates with histological grading of invasive ductal breast cancer. Int. J. Cancer 88, 907–913 [DOI] [PubMed] [Google Scholar]
- 10. Wiegmans A. P., Al-Ejeh F., Chee N., Yap P. Y., Gorski J. J., Da Silva L., Bolderson E., Chenevix-Trench G., Anderson R., Simpson P. T., Lakhani S. R., and Khanna K. K. (2014) Rad51 supports triple negative breast cancer metastasis. Oncotarget 5, 3261–3272 10.18632/oncotarget.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Graeser M., McCarthy A., Lord C. J., Savage K., Hills M., Salter J., Orr N., Parton M., Smith I. E., Reis-Filho J. S., Dowsett M., Ashworth A., and Turner N. C. (2010) A marker of homologous recombination predicts pathologic complete response to neoadjuvant chemotherapy in primary breast cancer. Clin. Cancer Res. 16, 6159–6168 10.1158/1078-0432.CCR-10-1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Asakawa H., Koizumi H., Koike A., Takahashi M., Wu W., Iwase H., Fukuda M., and Ohta T. (2010) Prediction of breast cancer sensitivity to neoadjuvant chemotherapy based on status of DNA damage repair proteins. Breast Cancer Res. 12, R17 10.1186/bcr2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. O'Boyle N. M., Banck M., James C. A., Morley C., Vandermeersch T., and Hutchison G. R. (2011) Open Babel: an open chemical toolbox. J. Cheminform. 3, 33 10.1186/1758-2946-3-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sharma P. (2018) Update on the treatment of early-stage triple-negative breast cancer. Curr. Treat. Options Oncol. 19, 22 10.1007/s11864-018-0539-8 [DOI] [PubMed] [Google Scholar]
- 15. Robert M., Frenel J. S., Gourmelon C., Patsouris A., Augereau P., and Campone M. (2017) Olaparib for the treatment of breast cancer. Expert Opin. Investig. Drugs 26, 751–759 10.1080/13543784.2017.1318847 [DOI] [PubMed] [Google Scholar]
- 16. Moynahan M. E., Pierce A. J., and Jasin M. (2001) BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell 7, 263–272 10.1016/S1097-2765(01)00174-5 [DOI] [PubMed] [Google Scholar]
- 17. Bennardo N., Cheng A., Huang N., and Stark J. M. (2008) Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 4, e1000110 10.1371/journal.pgen.1000110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Budke B., Logan H. L., Kalin J. H., Zelivianskaia A. S., Cameron McGuire W., Miller L. L., Stark J. M., Kozikowski A. P., Bishop D. K., and Connell P. P. (2012) RI-1: a chemical inhibitor of RAD51 that disrupts homologous recombination in human cells. Nucleic Acids Res. 40, 7347–7357 10.1093/nar/gks353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Modesti M., Ristic D., van der Heijden T., Dekker C., van Mameren J., Peterman E. J., Wuite G. J., Kanaar R., and Wyman C. (2007) Fluorescent human RAD51 reveals multiple nucleation sites and filament segments tightly associated along a single DNA molecule. Structure 15, 599–609 10.1016/j.str.2007.04.003 [DOI] [PubMed] [Google Scholar]
- 20. Slupianek A., Dasgupta Y., Ren S. Y., Gurdek E., Donlin M., Nieborowska-Skorska M., Fleury F., and Skorski T. (2011) Targeting RAD51 phosphotyrosine-315 to prevent unfaithful recombination repair in BCR-ABL1 leukemia. Blood 118, 1062–1068 10.1182/blood-2010-09-307256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Popova M., Shimizu H., Yamamoto K., Lebechec M., Takahashi M., and Fleury F. (2009) Detection of c-Abl kinase-promoted phosphorylation of Rad51 by specific antibodies reveals that Y54 phosphorylation is dependent on that of Y315. FEBS Lett. 583, 1867–1872 10.1016/j.febslet.2009.04.044 [DOI] [PubMed] [Google Scholar]
- 22. Subramanyam S., Ismail M., Bhattacharya I., and Spies M. (2016) Tyrosine phosphorylation stimulates activity of human RAD51 recombinase through altered nucleoprotein filament dynamics. Proc. Natl. Acad. Sci. U.S.A. 113, E6045–E6054 10.1073/pnas.1604807113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Baker P. R., Schopfer F. J., Sweeney S., and Freeman B. A. (2004) Red cell membrane and plasma linoleic acid nitration products: synthesis, clinical identification, and quantitation. Proc. Natl. Acad. Sci. U.S.A. 101, 11577–11582 10.1073/pnas.0402587101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baker P. R., Lin Y., Schopfer F. J., Woodcock S. R., Groeger A. L., Batthyany C., Sweeney S., Long M. H., Iles K. E., Baker L. M., Branchaud B. P., Chen Y. E., and Freeman B. A. (2005) Fatty acid transduction of nitric oxide signaling: multiple nitrated unsaturated fatty acid derivatives exist in human blood and urine and serve as endogenous peroxisome proliferator-activated receptor ligands. J. Biol. Chem. 280, 42464–42475 10.1074/jbc.M504212200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Martin R. W., Orelli B. J., Yamazoe M., Minn A. J., Takeda S., and Bishop D. K. (2007) RAD51 up-regulation bypasses BRCA1 function and is a common feature of BRCA1-deficient breast tumors. Cancer Res. 67, 9658–9665 10.1158/0008-5472.CAN-07-0290 [DOI] [PubMed] [Google Scholar]
- 26. Lamont K. R., Hasham M. G., Donghia N. M., Branca J., Chavaree M., Chase B., Breggia A., Hedlund J., Emery I., Cavallo F., Jasin M., Rüter J., and Mills K. D. (2013) Attenuating homologous recombination stimulates an AID-induced antileukemic effect. J. Exp. Med. 210, 1021–1033 10.1084/jem.20121258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang F., and Mazin A. V. (2014) A small molecule inhibitor of human RAD51 potentiates breast cancer cell killing by therapeutic agents in mouse xenografts. PLoS One 9, e100993 10.1371/journal.pone.0100993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Alagpulinsa D. A., Ayyadevara S., and Shmookler Reis R. J. (2014) A small-molecule inhibitor of RAD51 reduces homologous recombination and sensitizes multiple myeloma cells to doxorubicin. Front. Oncol. 4, 289 10.3389/fonc.2014.00289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhu J., Zhou L., Wu G., Konig H., Lin X., Li G., Qiu X. L., Chen C. F., Hu C. M., Goldblatt E., Bhatia R., Chamberlin A. R., Chen P. L., and Lee W. H. (2013) A novel small molecule RAD51 inactivator overcomes imatinib-resistance in chronic myeloid leukaemia. EMBO Mol. Med. 5, 353–365 10.1002/emmm.201201760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pellegrini L., Yu D. S., Lo T., Anand S., Lee M., Blundell T. L., and Venkitaraman A. R. (2002) Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature 420, 287–293 10.1038/nature01230 [DOI] [PubMed] [Google Scholar]
- 31. Cui T., Schopfer F. J., Zhang J., Chen K., Ichikawa T., Baker P. R., Batthyany C., Chacko B. K., Feng X., Patel R. P., Agarwal A., Freeman B. A., and Chen Y. E. (2006) Nitrated fatty acids: endogenous anti-inflammatory signaling mediators. J. Biol. Chem. 281, 35686–35698 10.1074/jbc.M603357200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dunphy K. A., Tao L., and Jerry D. J. (2010) Mammary epithelial transplant procedure. J. Vis. Exp. 1849 10.3791/1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Richardson C., Moynahan M. E., and Jasin M. (1998) Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 12, 3831–3842 10.1101/gad.12.24.3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gunn A., and Stark J. M. (2012) I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods Mol. Biol. 920, 379–391 10.1007/978-1-61779-998-3_27 [DOI] [PubMed] [Google Scholar]
- 35. Pozarowski P., and Darzynkiewicz Z. (2004) Analysis of cell cycle by flow cytometry. Methods Mol. Biol. 281, 301–311 10.1385/1-59259-811-0:301 [DOI] [PubMed] [Google Scholar]
- 36. Hopkins B. L., Nadler M., Skoko J. J., Bertomeu T., Pelosi A., Shafaei P. M., Levine K., Schempf A., Pennarun B., Yang B., Datta D., Bucur O., Ndebele K., Oesterreich S., Yang D., et al. (2018) A peroxidase peroxiredoxin 1-specific redox regulation of the novel FOXO3 microRNA target let-7. Antioxid. Redox Signal. 28, 62–77 10.1089/ars.2016.6871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grover P., Shi H., Baumgartner M., Camacho C. J., and Smithgall T. E. (2015) Fluorescence polarization screening assays for small molecule allosteric modulators of ABL kinase function. PLoS One 10, e0133590 10.1371/journal.pone.0133590 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.