Abstract
Microbial formate–nitrite transporter-type proteins (FNT) exhibit dual transport functionality. At neutral pH, electrogenic anion currents are detectable, whereas upon acidification transport of the neutral, protonated monoacid predominates. Physiologically, FNT-mediated proton co-transport is vital when monocarboxylic acid products of the energy metabolism, such as l-lactate, are released from the cell. Accordingly, Plasmodium falciparum malaria parasites can be killed by small-molecule inhibitors of PfFNT. Two opposing hypotheses on the site of substrate protonation are plausible. The proton relay mechanism postulates proton transfer from a highly conserved histidine centrally positioned in the transport path. The dielectric slide mechanism assumes decreasing acidity of substrates entering the lipophilic vestibules and protonation via the bulk water. Here, we defined the transport mechanism of the FNT from the amoebiasis parasite Entamoeba histolytica, EhFNT, and also show that BtFdhC from Bacillus thuringiensis is a functional formate transporter. Both FNTs carry a nonprotonatable amide amino acid, asparagine or glutamine, respectively, at the central histidine position. Despite having a nonprotonatable residue, EhFNT displayed the same substrate selectivity for larger monocarboxylates including l-lactate, a low substrate affinity as is typical for FNTs, and, strikingly, proton motive force–dependent transport as observed for PfFNT harboring a central histidine. These results argue against a proton relay mechanism, indicating that substrate protonation must occur outside of the central histidine region, most likely in the vestibules. Furthermore, EhFNT is the sole annotated FNT in the Entamoeba genome suggesting that it could be a putative new drug target with similar utility as that of the malarial PfFNT.
Keywords: transporter, membrane transport, membrane biophysics, proton motive force, lactic acid, protozoan, malaria, Bacillus, electron microscopy (EM), liposome, acetate, Entamoeba histolytica, formate, proton transfer
Introduction
Bacterial and protozoal formate–nitrite transporters (FNT)3 are key elements in processes with high biotechnological, agrarian and pharmaceutical value (1, 2). In particular, uptake of formate/H+ via the FNT family member FocA for subsequent conversion by formate hydrogen lyase is the basis for gaseous hydrogen production in bacteria (3, 4). The bacterial FNT isoform NirC facilitates the import of nitrite providing a route for nitrogen assimilation by chemical reduction of the ingested nitrite to ammonium (5, 6). NirC, thus, contributes to the global biogeochemical nitrogen cycle. Eukaryotic FNTs carry substrate selectivity filters that are wider in diameter permitting efficient transport of larger monocarboxylates, such as l-lactate (7, 8). The viability and virulence of human-pathogenic malaria parasites depends on the swift release of l-lactate/H+ via its single FNT-type monocarboxylate transporter, PfFNT (2). Recently discovered PfFNT inhibitors killed the parasites in vitro and validated PfFNT as a novel antimalarial drug target (9). Despite such prominent potential for exploitation, basic questions about the structure–function relationships of FNTs and their transport mechanism remain unclear.
Based on electrophysiology studies, FNTs were initially classified as anion channels for weak acid substrates, such as formate or nitrite (10). Because slight acidification of the assay media blocked the electrogenic anion transport, a pH-gating mechanism was assumed (11). Contrary to the electrophysiology approach, monitoring of the total transport of anionic and neutral acid substrate by using a radiolabel showed increasing uptake rates with steeper proton gradients (2, 12). The transport rates in the acidic range exceeded anion permeability at neutral pH by more than 1 order of magnitude. Therefore, FNTs exhibit a dual functionality, with a basic anion conductance, and transport of the protonated neutral acid form; the latter predominates at acidic pH.
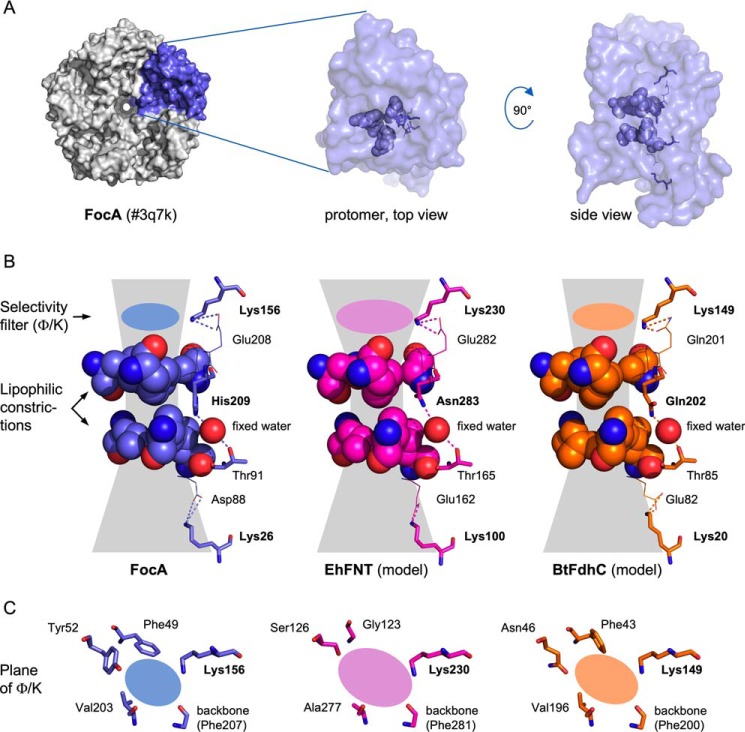
Two different sites and mechanisms for the neutralizing proton transfer onto the substrate anion have been proposed. A proton relay hypothesis was formulated when first crystal structures of the pentameric FNT proteins became available (1). The aquaporin-like fold of the individual substrate-conducting protomers is characterized by two lipophilic constrictions that sandwich a highly conserved histidine residue (His-209 in FocA; Fig. 1A, and Fig. 1B, left panel) (4). It was postulated that the central histidine carries a proton, which is transferred to the entering substrate anion (1). Re-protonation of the histidine would occur by a proton transfer chain from the exiting substrate via a conserved threonine residue (Thr-91 in FocA, Fig. 1) and a fixed water molecule. Theoretical studies questioned whether the lipophilic protein environment would permit a protonated, charged histidine for energetic reasons (13, 14), and further calculated that a substrate anion/histidine-H+ ion pair may be too stable to allow passage (14). We favor a dielectric slide mechanism based on lysine residues that are symmetrically positioned in the two funnel-like vestibules at the cytoplasmic and periplasmic/extracellular FNT entrance sites (Lys-26 and Lys-156 in FocA; Fig. 1) (12, 15). Their positive electrostatic field attracts substrate anions into an increasingly lipophilic environment reducing substrate acidity along its way. At a certain point a proton would be transferred from the aqueous bulk, and the neutralized substrate could pass the lipophilic constrictions. This view was derived from experimental evidence, such as pH and heavy water effects, or mutational exchange of lysine, all leading to specific modulations of the transport properties (12).
Figure 1.
Structural comparison of FocA and models of EhFNT and BtFdhC. A, shown is a FocA pentamer (PDB 3q7k) as well as the top and side views of the chain A protomer. In the protomer, key residues of the transduction path are highlighted. B, details of the transduction paths in side view. The residues of the hydrophobic constriction sites are shown space-filled. The central position holding a highly conserved histidine (FocA), or the alternative asparagine (EhFNT) or glutamine (BtFdhC) are depicted as sticks. The sphere indicates the position of the fixed water between the central position and the conserved threonine (sticks). The symmetrically placed, conserved lysine residues in the vestibules are shown as sticks. The planes of the Φ/K selectivity filters are marked by colored ovals. C, top down views on the Φ/K selectivity filter regions.
Previous attempts to replace the central histidine by mutation led to nonfunctional FNTs indicating its essential role (10). However, the loss of function prevented conclusions on how the histidine is involved in substrate transport. Is the contribution purely structural or is the histidine part of the proton transfer mechanism?
Here, we describe FNTs from the causative agent of human amoebiasis Entamoeba histolytica (EhFNT) (16) and from the Bacillus cereus group (B. cereus, Bacillus anthracis, and Bacillus thuringiensis; BtFdhC) (17, 18) that carry natural replacements of the central histidine by asparagine (position 283) or glutamine (position 202), respectively (Fig. 1B, middle and right panels). Amide amino acid side chains are incapable of transferring protons, which enabled us to elucidate the role of the central histidine position in FNT substrate transport. Furthermore, EhFNT is annotated as the sole monocarboxylate transporter encoded in the E. histolytica genome, thus, representing a novel putative drug target similar to PfFNT of malaria parasites (2).
Results
Amide amino acids are found as rare natural replacements of the central histidine
Analysis of 2206 individual FNT protein sequences deposited in the dbFNT database (19) showed an almost invariant conservation of the central histidine residue. In 27 instances (1.2%), the histidine was replaced by an amide amino acid. EhFNT represents a group of 6 sequences carrying an asparagine instead of histidine, and BtFdhC is one of 21 group members with a glutamine replacing the histidine. Central amide amino acid-containing FNTs have not been experimentally characterized before.
Based on the prototypical FocA crystal structure (PDB number 3q7k) (11), we generated structure models of EhFNT and BtFdhC using SwissModel (20) (for a sequence alignment, see Fig. S1). Although EhFNT and BtFdhC carry 4 and 5 histidine residues (Fig. S1), respectively, none can substitute for the central histidine because of their locations outside of the transduction path. In the models, asparagine and glutamine can well replace the central histidine without producing steric clashes (Fig. 1B, middle and right panels). Furthermore, and probably importantly, the amide moieties maintain hydrogen bonds to the canonical fixed water molecule (1, 4). Threonine acting as a second hydrogen bond partner to the water (12) is conserved in EhFNT and BtFdhC. Both FNTs further carry two symmetrically placed, anion-attracting lysine residues in the vestibules above and below the lipophilic constriction sites. The substrate selectivity filter (lipophilic/lysine region, Φ/K) of EhFNT holds small amino acid side chains (Gly, Ser, Ala), whereas BtFdhC harbors larger residues (Phe, Asn, Val) (Fig. 1C, middle and right panels). As a result, the diameter of the filter in EhFNT is wider and may permit passage of larger substrates such as l-lactate; this layout follows a previously noted scheme according to which eukaryotic FNTs have wider selectivity filters than prokaryotic ones (7).
Cell-free expression delivers functional EhFNT and BtFdhC proteins
Initially, we attempted to produce EhFNT and BtFdhC in a well-established Saccharomyces cerevisiae yeast system (2, 7–9, 12). However, the yeast cells produced fragmented (EhFNT) or little amounts of protein (BtFdhC), and transport functionality was absent. Therefore, we switched to an Escherichia coli S30 extract-based cell-free system, which we successfully employed before for the production of PfFNT (21–23). Based on prior findings that terminally truncated FNT variants are more stable (8), we used an N-terminally shortened version of EhFNT (Δ1–90) for setting up cell-free expression. We employed EhFNT and BtFdhC expression constructs with green fluorescent protein (GFP) fusions at the C termini as folding indicators (21) and identified Brij78 as a well-suited detergent for solubilization during the cell-free translation reaction (Fig. 2, A and B). The laborious procedure and small sample yields made us focus mainly on the characterization of EhFNT in the following experiments. In the cell-free system, EhFNT turned out to be stable even with the full-length N terminus. Hence, we decided to produce EhFNT without GFP fusion, yet kept a factor Xa/His tag at the C terminus for affinity purification (Fig. 2C; the purification of BtFdhC is shown in Fig. S2).
Figure 2.
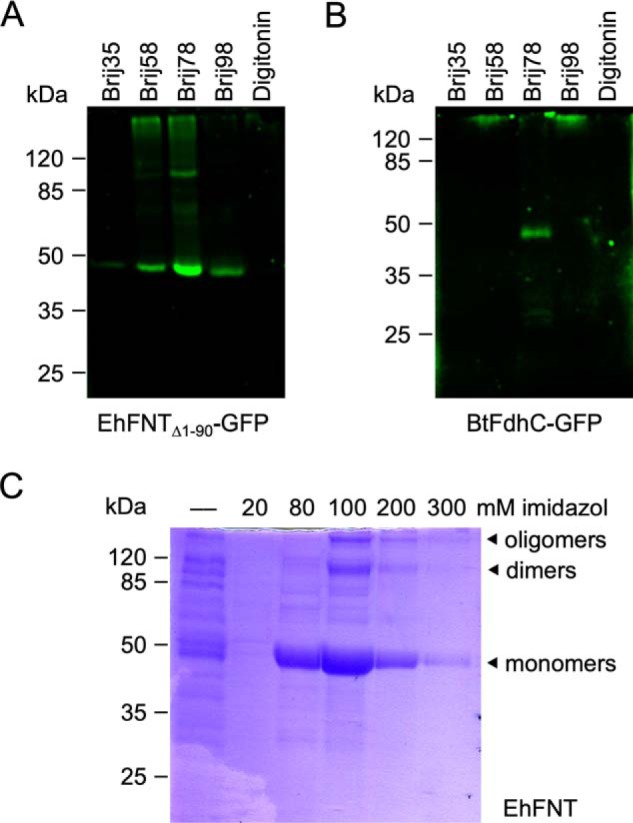
Cell-free production and purification of EhFNT and BtFdhC. A and B show detergent screens with C-terminal GFP fusions of EhFNT (first 90 amino acids truncated) and BtFdhC. C, Ni2+-affinity purification of full-length EhFNT (without GFP) via a C-terminal His6 tag. The elution fractions contained partially SDS-resistant EhFNT dimers and oligomers.
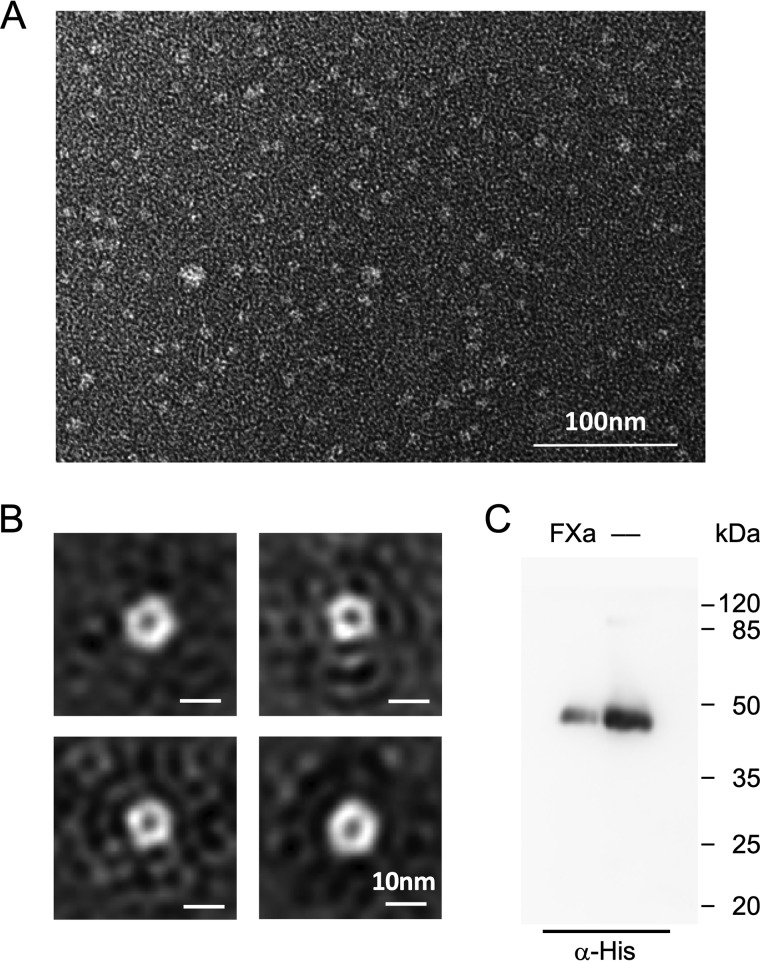
To test for proper homopentamer formation in the absence of a lipid membrane, we analyzed cell-free produced, Brij78-solubilized, and affinity-purified EhFNT by negative-stain transmission EM. The preparations contained even distributions of particles (Fig. 3A). Single-particle class averaging indicated a pentameric EhFNT structure throughout (Fig. 3B). Because the formation of homopentamers hinges on intact protein–protein interaction interfaces, it is indicative of a generally correct fold of the individual EhFNT protomers.
Figure 3.
Oligomeric state of detergent-solubilized EhFNT and orientation of reconstituted EhFNT in proteoliposomes. A, transmission EM EhFNT solubilized by Brij78. B, class sum images of averaged particles indicate pentameric protein homocomplexes. C, the orientation of EhFNT in proteoliposomes was probed by the accessibility and cleavage of a factor Xa protease site situated upstream of the C-terminal His6 tag. Loss of the His6 tag results in reduced signal intensity in the Western blotting using an anti-His antibody (lane labeled with FXa) compared with a sample without protease treatment (—).
We then reconstituted EhFNT into proteoliposomes for transport assays. The factor Xa site at the N terminus allowed us to determine the EhFNT orientation in the membrane (23). This protease cleavage site is accessible for the factor Xa enzyme only if the EhFNT termini are oriented toward the buffer outside of the proteoliposomes. We were able to cleave off the major portion of the C-terminal His10 tags via the factor Xa site, and found a 80:20 preference for the outside-in orientation, i.e. with the termini pointing into the extra-liposomal buffer (Fig. 3C). This preference in orientation in proteoliposomes is shared with the malaria parasite's PfFNT (23). Hence, transport of substrate via EhFNT into the proteoliposome lumen mainly corresponds to the efflux direction in E. histolytica cells.
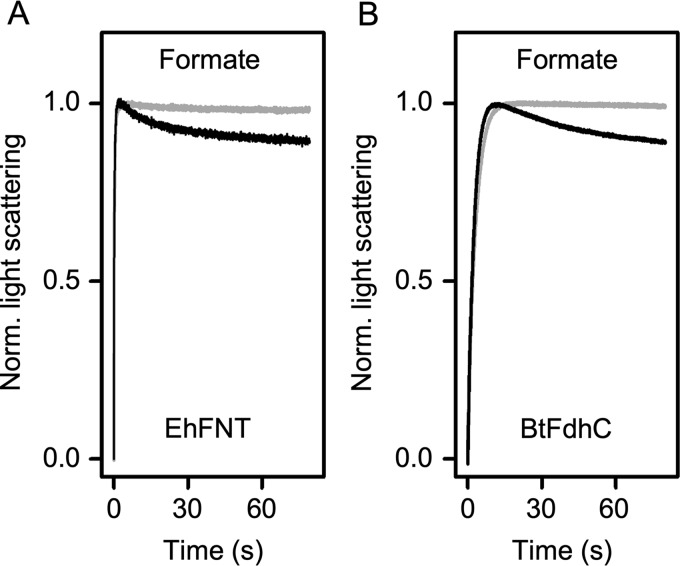
Next, we tested transport functionality of EhFNT proteoliposomes by abruptly subjecting them to 200 mm external formate using a stopped-flow light-scattering device (24–27). Under these hypertonic conditions, the proteoliposomes as well as the liposomes without protein shrank in a first, rapid phase (<5 s) by osmotic release of water across the lipid membrane (Fig. 4A, initial increase in light scattering). Protein-free liposomes were impermeable for formate and maintained the same level of light scattering after the initial rise in intensity (>5 s). The proteoliposomes, however, regained volume by the uptake of formate via EhFNT and a secondary influx of water (Fig. 4A, decrease in light scattering after 5 s). Changes in light scattering thus reflect substrate transport in an indirect fashion. Nevertheless, the assay is suitable to determine basic transport properties of the reconstituted FNT proteins. We also tested for functionality of reconstituted BtFdhC and confirmed formate transport (Fig. 4B). Despite the replacement of the central histidine by asparagine or glutamine, respectively, EhFNT and BtFdhC displayed functionality as formate transporters.
Figure 4.
Transport functionality of EhFNT (A) and BtFdhC (B) in proteoliposomes. The proteoliposomes (black traces) were abruptly challenged with hypertonic formate gradients in a stopped-flow device. An increase in light scattering intensity indicates liposome shrinking due to rapid water efflux following the osmotic gradient. The subsequent decrease in light scattering derives from transport of formate into the proteoliposomes and accompanying secondary water influx. Traces from empty control liposomes are shaded gray. 6–9 traces were averaged for each condition.
EhFNT exhibits substrate selectivity and affinity typical for eukaryotic FNTs
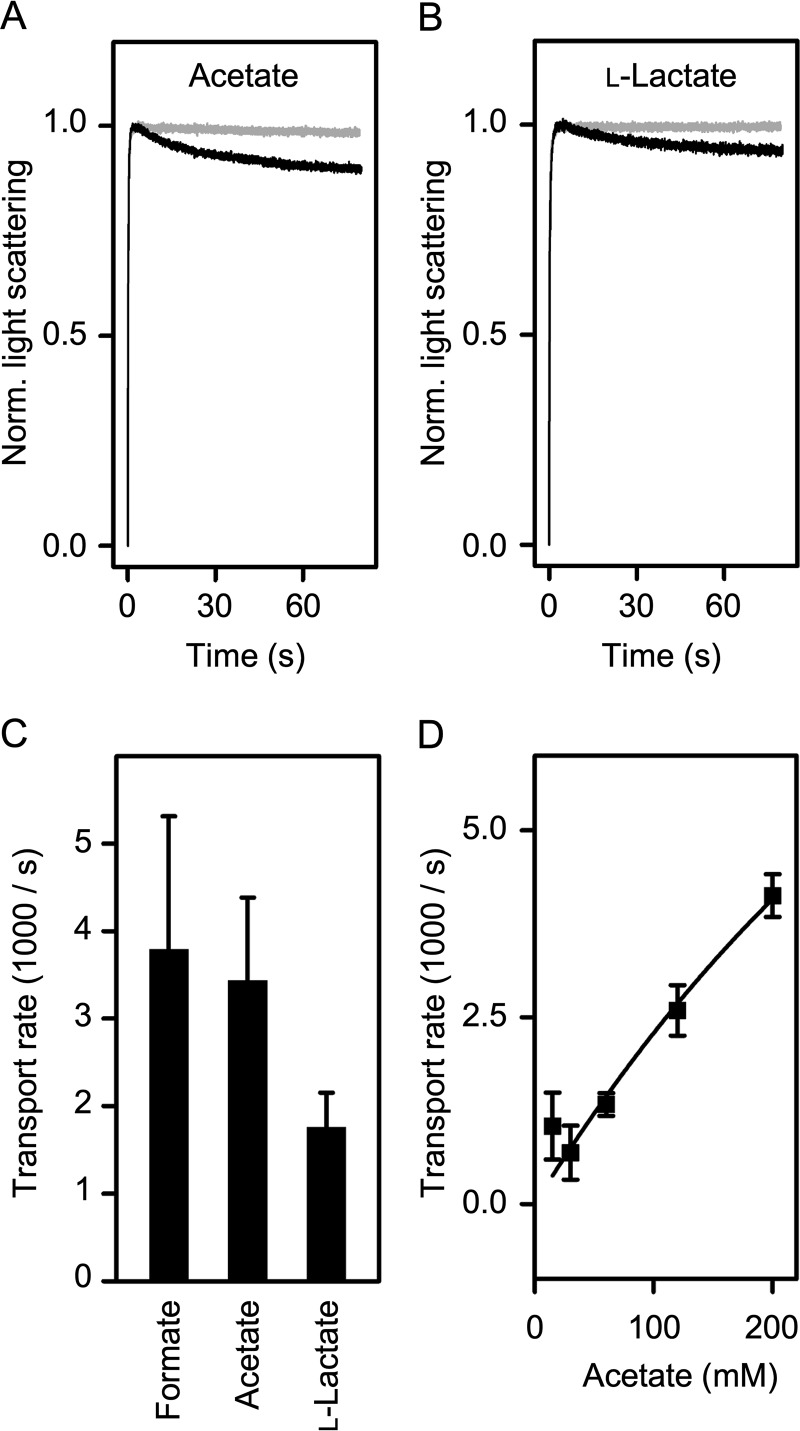
The composition of the Φ/K selectivity filter of EhFNT (Fig. 1C) suggests that the transporter may accommodate larger monocarboxylate substrates than formate (molecular mass 45 Da). Indeed, using the stopped-flow approach at a 200 mm hyperosmotic substrate gradient, we identified acetate (60 Da) and l-lactate (90 Da) as additional substrates of EhFNT (Fig. 5, A and B). Curve fitting yielded equal and half-transport rates, respectively, compared with formate (Fig. 5C). Inorganic chloride anions were excluded.
Figure 5.
Substrate selectivity and affinity of EhFNT. A, transport of acetate via EhFNT (black trace) and empty liposomes (gray). B, transport of l-lactate via EhFNT (black trace) and empty liposomes (gray). C, comparison of EhFNT transport rates for formate, acetate, and l-lactate at 200 mm inward gradients. D, dependence of EhFNT transport rates on substrate concentration. Shown are the rates of acetate transport at concentrations in the range 15–200 mm. The error bars denote S.E. from three independent replicates.
FNTs are further characterized by the lack of a strongly interacting substrate-binding site. As a consequence, saturation of transport is mild even at high millimolar substrate concentrations (2, 10, 12). We challenged EhFNT containing proteoliposomes with increasing acetate gradients in the range of 15–200 mm (original trace data are shown in Fig. S3). The determined influx rates increased almost linearly with the acetate concentration in the buffer (Fig. 5D). Even at 200 mm acetate, transport was far from being saturated. Together, despite replacement of the central histidine neither substrate selectivity nor substrate binding intensity appeared altered in EhFNT.
EhFNT substrate transport is driven by the proton motive force
Chemically, the imidazole side chain of histidine is a weak base. In free aqueous solution, its pKa is 6, meaning that at a pH of 6 half of the histidine side chains become protonated and capable of transferring these protons to other molecules. Vice versa, unprotonated histidine side chains can accept a proton. The pKa of acids and bases depends on the hydrophobicity or permittivity of the surrounding media. Generally, pKa values are difficult to obtain or predict for sites within a protein where access to the bulk water is limited or absent (28). The central region of the FNT transport path is strictly separated from the bulk water by two lipophilic constriction sites (Fig. 1). The energy barriers at the lipophilic constrictions for charged substrate anions are high (14), which led to the assumption that neutralization of the substrate anion by protonation is a necessity to pass these constrictions (1). Its close proximity and general proton transfer capability rendered the central histidine a candidate site for the proton transfer. We favor an alternative mechanism for substrate protonation via the bulk water situated in the vestibule regions at the cytoplasmic and periplasmic/extracellular entrance sites of the FNT transporters (12, 15).
EhFNT, due to its nonprotonatable asparagine at the central histidine position, is a suitable and natural candidate for a decisive experiment regarding the substrate protonation mechanism. For comparison, we produced and reconstituted PfFNT from the malaria parasite carrying a central histidine. Earlier, we found pH dependence of PfFNT transport in yeast, i.e. in the influx direction (2, 12). The preferential outside-in orientation of EhFNT (this study) and PfFNT (23) in the proteoliposomes now allowed us to directly analyze the effect of a proton motive force on transport in the physiological direction (29).
We monitored formate transport via EhFNT, PfFNT, and with plain liposomes at external pH values of 7.8, 6.8, and 4.8; the internal liposomal pH was 6.8 in all cases. EhFNT and PfFNT behaved almost identically throughout (Fig. 6, A and B). For both, the rates correlated with the steepness of the proton gradient. Transport increased in the acidic range (pH 4.8), and decreased below the detection limit of the assay when the proton gradient was reversed (pH 7.8) (Fig. 6C). However, the possibility remained that the different transport rates resulted from a simple pH effect, e.g. by altering the protonation status of the EhFNT protein or the substrate. To make sure that the proton gradient acts as a driving force of EhFNT formate transport, we prepared proteoliposomes with an acidic internal pH of 4.8 and exposed them to an external buffer that was adjusted to the same pH of 4.8 (Fig. 6C, inset), i.e. we generated acidic conditions, yet in the absence of a transmembrane proton gradient. Confirmatively, the transport rate decreased to the same level as in the assay with an equal internal and external pH of 6.8 (Fig. 6C). When we then established a ×100 inward proton gradient (external pH 2.8), the transport rate increased again.
Figure 6.
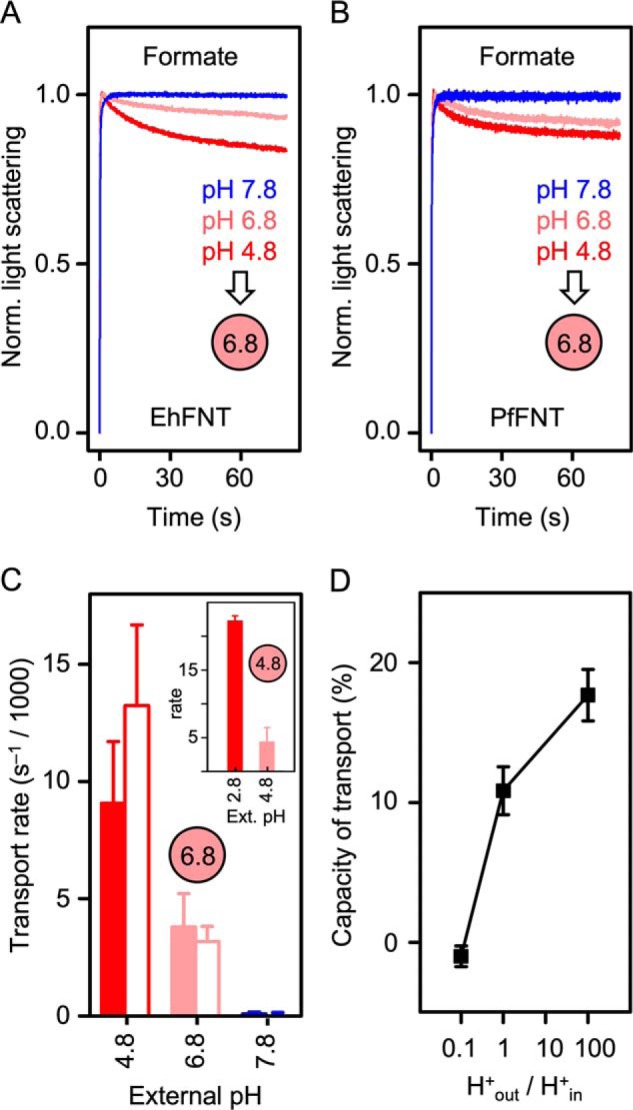
Effect of transmembrane proton gradients on the transport rates of EhFNT and PfFNT. A, EhFNT formate transport at external pH 4.8 (red), 6.8 (light red), and 7.8 (blue). B, formate transport via PfFNT at pH 4.8 (red), 6.8 (light red), and 7.8 (blue). The intra-liposomal pH (spheres) was 6.8 in all cases shown in A and B. C, formate transport rates of EhFNT (filled bars) and PfFNT (open bars) at different external pH conditions. The intra-liposomal pH was 6.8 or 4.8 (inset). D, transport capacity of formate into EhFNT proteoliposomes (% change of total light scattering intensity at 80 s time) in relationship to the steepness of the transmembrane proton gradient (H+out/H+in). The error bars indicate S.E. (n = 3).
Importantly, the capacity of transport, i.e. the total amount of substrate that was loaded into the liposomes, also increased with the proton gradient (Fig. 6D). If the substrate was transported only in its anionic form, an electrochemical potential would build up inside the liposomes. This accumulation of charge would rapidly limit the transport, which is probably the case at pH 7.8, i.e. at an outward proton gradient (H+out/H+in = 0.1). If, however, the protonated, neutral substrate is transported, this will leave the membrane potential undisturbed and result in higher transport capacity. The increase in EhFNT transport capacity with increasing proton concentrations (H+out/H+in ≥ 1), thus, indicates a shift in the transported substrate toward the protonated, neutral form.
These data show that independent of the presence or absence of a proton-accepting and donating central histidine, the FNT substrate undergoes protonation, and transport remains driven by the proton motive force. Consequently, pH-dependent substrate protonation must occur outside of the central histidine region, most likely in the vestibules.
Discussion
The major outcome of this work is the finding that in direct comparison EhFNT (central asparagine) and PfFNT (central histidine) exhibit highly similar transport properties, i.e. substrate selectivity, very low substrate affinity, substrate protonation, and pH dependence in the stopped-flow light-scattering assay. We further showed functionality of BtFdhC (central glutamine). Hence, FNTs carrying amide amino acids at the central histidine position function as typical FNT-type monocarboxylate transporters (7, 12).
Asparagine residues occupy a volume of 127.5 Å3, and glutamine of 149.4 Å3 in the protein interior (30), i.e. they are slightly less voluminous than histidine (159.3 Å3) explaining why these amino acid exchanges were sterically tolerated. In an earlier study, replacement of the central histidine by phenylalanine resulted in a nonfunctional FNT (10). At 193.5 Å3, the volume of phenylalanine is somewhat larger than histidine possibly coercing a conformational change in this central protein region that may affect substrate passage. Besides volume considerations, the physicochemical properties of the side chains must be taken into account as well. The imidazole moiety of the histidine side chain can act as an acceptor and donor of hydrogen bonds. This way, one of two interactions with the canonical fixed water molecule of the FNT protein structure is established (Fig. 1). The aromatic phenyl side chain of phenylalanine is incapable of forming hydrogen bonds. Therefore, a mutational exchange of histidine for phenylalanine may lead to the loss of the water molecule. The protein fold may collapse around the resulting gap, which would severely disturb the overall structure. FNTs have assumed the same fold as aquaporin water and uncharged solute channels even though the primary sequences are unrelated (4). Similar to the aquaporins, the conduction path of the FNTs is long and narrow forcing substrate molecules to pass in single file (15, 31). Even a slight kink in this tube-like structure, which could derive from loss of the fixed water, is likely to generate a section that is too narrow for the substrate to pass. We have found before that the second hydrogen bond partner of the fixed water molecule, i.e. a conserved threonine (Fig. 1), can be replaced by the hydroxyl amino acid serine, but not by an isosteric, yet hydrophobic valine (12). The dbFNT database (19) contains additional FNT sequences with asparagine as an alternative residue at the threonine position. Notably, asparagine is highly similar in volume (127.5 Å3) as threonine (120.0 Å3) (30) and equally capable of forming hydrogen bonds. Together, this suggests that the presence of a water molecule and fixation from two sides by hydrogen bonds is a must to obtain a functional FNT.
Is the central histidine region including the fixed water and threonine involved in proton transfers to passing substrate molecules? Most likely not. Estimation of the acidity of the central histidine in its protonated form using the algorithms residue DEPTH (32) and propka (33) yielded pKa values below 1 or around 3, respectively. We found this range of values for all FNTs with available crystal data (4, 6, 11, 34, 35). Deviations of such scale from a pKa of 6 in aqueous solution suggest that a positive charge at the histidine due to protonation would not be supported. FNTs carrying central amide amino acids, therefore, fit well into the picture as amides are neither basic nor acidic, i.e. proton transfers are impossible. The strong lipophilicity at the FNT center in combination with full functionality and pH-dependent transport of amide amino acid FNTs call for substrate protonation in the vestibule regions by the dielectric slide mechanism (12, 15). The central, neutral histidine would assume a structural function as an element in the lining of the transport path and in the fixation of the water molecule via a hydrogen bond.
In addition to resolving the structural role of the central FNT histidine, our findings are of putative pharmaceutical value as well. The intestinal protist E. histolytica is an obligate fermenter (16). Enzymes of the tricarboxylic acid cycle are absent and it even lacks the mitochondrion organelle. The parasite draws energy from anaerobic glycolysis, and monocarboxylates, such as acetate, are among the fermentation products. This study showed that EhFNT efficiently transports such monocarboxylates in the efflux direction. As EhFNT is annotated as the sole FNT encoded in the Entamoeba genome, it might have a vital role in the energy metabolism of the parasite. The situation is quite similar to malaria parasites, which express PfFNT from a single FNT encoding gene (2). Drug-like inhibitors of PfFNT led to accumulation of the metabolic end product l-lactate and to acidification of the cytosol killing the parasites (9, 36). By making use of the preferential orientation of PfFNT in the proteoliposome membrane, we could now show pH-dependent transport via PfFNT (and EhFNT) in the physiological direction, i.e. outwards. There is a good chance that blockade of EhFNT may be equally detrimental for amoebiasis parasites as for malaria parasites.
The suitability of the current proteoliposome system for inhibitor screenings is limited, however. The sample size is small and the lipid bilayer is sensitive toward organic solvents. In particular, the solvent dimethyl sulfoxide (DMSO) is typically added at low concentrations to keep lipophilic, drug-like molecules in solution. The significance of amoebiasis for mortality in developing countries mainly due to systemic and liver complications warrants a search for cell-based EhFNT expression systems that can be employed for drug-screening programs.
Experimental procedures
DNA sequences and protein models
DNA and protein sequences were derived from AmoebaDB (EhFNT; gene ID EHI_198990) and the NCBI database (BtFdhC gene ID 2854446). The EhFNT ORF DNA was codon-optimized (Fig. S4) and synthesized (GenScript Biotech Corp.). BtFdhC was amplified by PCR from genomic B. thuringensis DNA (DMSZ-German Collection of Microorganisms and Cell Cultures). Protein structure data (Salmonella typhimurium FocA; PDB 3q7k) (11) were from the RCSB data bank. Sequence alignments were set using TeXshade (37). EhFNT and BtFdhC structure models were generated with SwissModel (20). Visualization of protein structures was done using the PyMOL Molecular Graphics System, Schrödinger, LLC.
Cell-free production of FNTs
Protein production was done in a continuous exchange cell-free system based on an E. coli S30 extract as described before (23). Briefly, EhFNT-DNA (SpeI/XhoI) and BtFdhC-DNA (BamHI/XhoI) were cloned into pIVEX2.3 yielding open reading frames encoding a factor Xa-digestion site followed by a His10 tag at the C-terminal end. For screening purposes the vector included a GFP encoding sequence between the factor Xa site and the His10 tag. All constructs were verified by DNA sequencing. The detergent Brij78 at 1.0% was optimal for the protein production and purification procedure. Large-scale protein production was carried out in 1-ml reaction volumes containing the E. coli S30-extract, plasmid DNA, and all ingredients required for the transcription and translation reactions, such as nucleotides, amino acids, t-RNAs, T7-polymerase, energy regenerating system (23), using Slide-A-Lyzer dialysis cassettes (Thermo Scientific) with 10-kDa cut-off. The cassettes were placed into 17 ml of feeding mix acting as a reservoir of the low molecular-weight components, and as dilutant for waste products. The reactions were kept at 30 °C in a shaking water bath for 20 h.
Affinity purification of FNT proteins, in-gel fluorescence, and Western blotting
The cell-free reaction was mixed with 300 μl of washed nickel-nitrilotriacetic acid beads slurry (50%) and purification buffer (20 mm HEPES, pH 8.0, 150 mm NaCl, 0.05% Brij78) to a final volume of 5 ml. The samples were rotated overnight at 4 °C. The resin was washed with 5 column volumes of purification buffer supplemented with 20 mm imidazole. His-tagged proteins were eluted in 2 column volumes of increasing imidazole concentration in the range of 80–500 mm. FNT-containing fractions were pooled and passed over a PD-Midi-Trap desalting column (GE Healthcare) concentrated using Amicon Ultra-4 Ultracel filter units (Millipore). Initially, we used 35-kDa cut-off filters (BtFdhC data), which we replaced by wider 100-kDa filters (EhFNT) because we noticed a general improvement in liposome membrane tightness probably due to better removal of empty detergent micelles. The protein samples were separated by SDS-PAGE using 12.5% polyacrylamide gels. GFP fusion constructs were visualized by in-gel fluorescence monitoring. For Western blotting, the proteins were electrotransferred to polyvinylidene difluoride membranes (Amersham Biosciences). Detection was done with a monoclonal mouse anti-penta-His antibody (1:5000 dilution in TBS with 5% milk powder and 0.1% TWEEN 20, M-TBS-T; Qiagen) and a polyclonal goat anti-mouse antibody carrying horseradish peroxidase (1:5000 in M-TBS-T; Jackson ImmunoResearch) using the ECL Plus system (GE Healthcare). Chemiluminescence as well as in-gel GFP fluorescence were documented with a Lumi-Imager F1 (Roche Applied Science).
Negative staining EM and class sum formation
Negative staining was performed as described before (38), yet with half-saturated uranyl-acetate as a staining reagent. Samples were inserted into a JEOL1400 Plus operating at 100 kV and images were taken on a TVIPS F416 digital camera binned to 2048 × 2048 pixel at 50,000 magnification. This results in a resolution of 4.58 Å/pixel. Images were analyzed using EMAN2 (39). Particles were selected using boxer, and class sum images were calculated using the reference-free class averaging as a tool of e2refine2d.py. Per class, 10–15 single particles were grouped and there was no symmetry imposed during image analysis.
Preparation of proteoliposomes
We followed a previously described protocol (Holm-Bertelsen16). Briefly, 25 mg of E. coli polar lipids were dissolved in 1 ml of chloroform, evaporated under a nitrogen stream, and fully dried under vacuum while rotating. The resulting thin lipid film was hydrated for 1–2 h at room temperature in liposome buffer (EhFNT: 5 mm HEPES, 5 mm MES, pH 6.8, 100 mm KCl, 2 mm β-mercaptoethanol, 200 mm sucrose; BtFdhC: 20 mm HEPES, pH 6.8) to yield a final lipid concentration of 50 mg ml−1. The samples were shock-frozen in liquid nitrogen and thawed before sonication to obtain multilamellar liposomes. 100 μg of purified FNT protein were added at a protein–lipid ratio of 1:50. By diluting into 25 volumes of liposome buffer the protein was forced to reconstitute into proteoliposomes. Multilamellar proteoliposomes were harvested by ultracentrifugation (140,000 × g for 45 min), resuspended in 1 ml of liposome buffer, and passed through a LiposoFast extruder (Avestin) with 0.2-μm pore diameter 21 times. The obtained unilamellar proteoliposomes were analyzed for integration of EhFNT by density gradient centrifugation. The directionality of the integrated EhFNT was assayed by treatment with factor Xa protease (Qiagen). Therefore, 48 μg of EhFNT protein integrated in proteoliposomes were supplemented with 1 mm CaCl2, and treated with 15 units of factor Xa enzyme or were left untreated and kept at room temperature for 9 h. These reaction conditions were chosen, because they led to full digestion of 48 μg of solubilized, nonintegrated EhFNT protein. Samples were precipitated by adding trichloric acid and then analyzed via Western blotting. The band intensities of digested and undigested EhFNT protein were compared.
Functional transport assays using proteoliposomes
Substrate transport into (proteo)liposomes was monitored at 20 °C by changes in 524 nm light scattering using a stopped flow device (SFM-2000, Bio-Logic Science Instruments). Liposome suspensions were rapidly mixed with an equivalent volume of liposome buffer generating external pH 4.8, 6.8, or 7.8, and supplemented with 30 to 400 mm sodium salts of formate, acetate, l-lactate, chloride, or glycerol, yielding 15 to 200 mm gradients, respectively. The internal pH of the (proteo)liposomes was 6.8 or 4.8, respectively. Shrinkage of the (proteo-) liposomes increased the light scattering signal intensity, whereas swelling led to a decrease in light scattering. The obtained signal traces were averaged (n = 3–9) and normalized. Transport rates were calculated from double-exponential fitting (Bio-Logic software).
Author contributions
F. H., P. A., B. H., L. M. P., and E. B. validation; F. H., P. A., B. H., and E. B. investigation; F. H., P. A., B. H., and E. B. visualization; F. H., P. A., B. H., and L. M. P. methodology; F. H., P. A., B. H., and L. M. P. writing-review and editing; P. A. resources; E. B. conceptualization; E. B. funding acquisition; E. B. writing-original draft; E. B. project administration.
Supplementary Material
Acknowledgments
We thank B. Henke and A. Fuchs for technical assistance. We further thank Z3 from SFB877 for support of the Electron Microscopic Unit of the Anatomical Institute.
This work was supported by German Research Foundation Deutsche Forschungsgemeinschaft (DFG) Research Grant Be2253/8–1 (to E. B.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S4.
- FNT
- formate–nitrite transporter
- GFP
- green fluorescent protein
- Φ/K
- lipophilic lysine region
- PDB
- Protein Data Bank.
References
- 1. Lü W., Du J., Schwarzer N. J., Wacker T., Andrade S. L., and Einsle O. (2013) The formate/nitrite transporter family of anion channels. Biol. Chem. 394, 715–727 [DOI] [PubMed] [Google Scholar]
- 2. Wu B., Rambow J., Bock S., Holm-Bertelsen J., Wiechert M., Soares A. B., Spielmann T., and Beitz E. (2015) Identity of a Plasmodium lactate/H+ symporter structurally unrelated to human transporters. Nat. Commun. 6, 6284 10.1038/ncomms7284 [DOI] [PubMed] [Google Scholar]
- 3. Sawers R. G. (2005) Formate and its role in hydrogen production in Escherichia coli. Biochem. Soc. Trans. 33, 42–46 [DOI] [PubMed] [Google Scholar]
- 4. Wang Y., Huang Y., Wang J., Cheng C., Huang W., Lu P., Xu Y.-N., Wang P., Yan N., and Shi Y. (2009) Structure of the formate transporter FocA reveals a pentameric aquaporin-like channel. Nature 462, 467–472 10.1038/nature08610 [DOI] [PubMed] [Google Scholar]
- 5. Jia W., Tovell N., Clegg S., Trimmer M., and Cole J. (2009) A single channel for nitrate uptake, nitrite export and nitrite uptake by Escherichia coli NarU and a role for NirC in nitrite export and uptake. Biochem. J. 417, 297–304 10.1042/BJ20080746 [DOI] [PubMed] [Google Scholar]
- 6. Lü W., Schwarzer N. J., Du J., Gerbig-Smentek E., Andrade S. L., and Einsle O. (2012) Structural and functional characterization of the nitrite channel NirC from Salmonella typhimurium. Proc. Natl. Acad. Sci. U.S.A. 109, 18395–18400 10.1073/pnas.1210793109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wiechert M., Erler H., Golldack A., and Beitz E. (2017) A widened substrate selectivity filter of eukaryotic formate-nitrite transporters enables high-level lactate conductance. FEBS J. 284, 2663–2673 10.1111/febs.14117 [DOI] [PubMed] [Google Scholar]
- 8. Erler H., Ren B., Gupta N., and Beitz E. (2018) The intracellular parasite Toxoplasma gondii harbors three druggable FNT-type formate and l-lactate transporters in the plasma membrane. J. Biol. Chem. 293, 17622–17630 10.1074/jbc.RA118.003801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Golldack A., Henke B., Bergmann B., Wiechert M., Erler H., Blancke Soares A., Spielmann T., and Beitz E. (2017) Substrate-analogous inhibitors exert antimalarial action by targeting the Plasmodium lactate transporter PfFNT at nanomolar scale. PLoS Pathog. 13, e1006172 10.1371/journal.ppat.1006172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lü W., Du J., Schwarzer N. J., Gerbig-Smentek E., Einsle O., and Andrade S. L. (2012) The formate channel FocA exports the products of mixed-acid fermentation. Proc. Natl. Acad. Sci. U.S.A. 109, 13254–13259 10.1073/pnas.1204201109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lü W., Du J., Wacker T., Gerbig-Smentek E., Andrade S. L., and Einsle O. (2011) pH-dependent gating in a FocA formate channel. Science 332, 352–354 10.1126/science.1199098 [DOI] [PubMed] [Google Scholar]
- 12. Wiechert M., and Beitz E. (2017) Mechanism of formate-nitrite transporters by dielectric shift of substrate acidity. EMBO J. 36, 949–958 10.15252/embj.201695776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lv X., Liu H., Ke M., and Gong H. (2013) Exploring the pH-dependent substrate transport mechanism of FocA using molecular dynamics simulation. Biophys. J. 105, 2714–2723 10.1016/j.bpj.2013.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Atkovska K., and Hub J. S. (2017) Energetics and mechanism of anion permeation across formate-nitrite transporters. Sci. Rep. 7, 12027 10.1038/s41598-017-11437-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wiechert M., and Beitz E. (2017) Formate-nitrite transporters: monoacids ride the dielectric slide. Channels 11, 365–367 10.1080/19336950.2017.1329999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Loftus B., Anderson I., Davies R., Alsmark U. C., Samuelson J., Amedeo P., Roncaglia P., Berriman M., Hirt R. P., Mann B. J., Nozaki T., Suh B., Pop M., Duchene M., Ackers J., et al. (2005) The genome of the protist parasite Entamoeba histolytica. Nature 433, 865–868 10.1038/nature03291 [DOI] [PubMed] [Google Scholar]
- 17. Radnedge L., Agron P. G., Hill K. K., Jackson P. J., Ticknor L. O., Keim P., and Andersen G. L. (2003) Genome differences that distinguish Bacillus anthracis from Bacillus cereus and Bacillus thuringiensis. Appl. Environ. Microbiol. 69, 2755–2764 10.1128/AEM.69.5.2755-2764.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Challacombe J. F., Altherr M. R., Xie G., Bhotika S. S., Brown N., Bruce D., Campbell C. S., Campbell M. L., Chen J., Chertkov O., Cleland C., Dimitrijevic M., Doggett N. A., Fawcett J. J., Glavina T., et al. (2007) The complete genome sequence of Bacillus thuringiensis Al Hakam. J. Bacteriol. 189, 3680–3681 10.1128/JB.00241-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mukherjee M., Vajpai M., and Sankararamakrishnan R. (2017) Anion-selective formate/nitrite transporters: taxonomic distribution, phylogenetic analysis and subfamily-specific conservation pattern in prokaryotes. BMC Genomics 18, 560 10.1186/s12864-017-3947-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T. G., Bertoni M., Bordoli L., and Schwede T. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 10.1093/nar/gku340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Müller-Lucks A., Bock S., Wu B., and Beitz E. (2012) Fluorescent in situ folding control for rapid optimization of cell-free membrane protein synthesis. PLoS ONE 7, e42186 10.1371/journal.pone.0042186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Müller-Lucks A., Gena P., Frascaria D., Altamura N., Svelto M., Beitz E., and Calamita G. (2013) Preparative scale production and functional reconstitution of a human aquaglyceroporin (AQP3) using a cell free expression system. N. Biotechnol. 30, 545–551 10.1016/j.nbt.2013.03.007 [DOI] [PubMed] [Google Scholar]
- 23. Holm-Bertelsen J., Bock S., Helmstetter F., and Beitz E. (2016) High-level cell-free production of the malarial lactate transporter PfFNT as a basis for crystallization trials and directional transport studies. Protein Expr. Purif. 126, 109–114 10.1016/j.pep.2016.06.008 [DOI] [PubMed] [Google Scholar]
- 24. Wu B., Song J., and Beitz E. (2010) Novel channel-enzyme fusion proteins confer arsenate resistance. J. Biol. Chem. 285, 40081–40087 10.1074/jbc.M110.184457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Song J., Almasalmeh A., Krenc D., and Beitz E. (2012) Molar concentrations of sorbitol and polyethylene glycol inhibit the Plasmodium aquaglyceroporin but not that of E. coli: involvement of the channel vestibules. Biochim. Biophys. Acta 1818, 1218–1224 10.1016/j.bbamem.2012.01.025 [DOI] [PubMed] [Google Scholar]
- 26. Song J., Baker N., Rothert M., Henke B., Jeacock L., Horn D., and Beitz E. (2016) Pentamidine is not a permeant but a nanomolar inhibitor of the Trypanosoma brucei aquaglyceroporin-2. PLoS Pathog. 12, e1005436 10.1371/journal.ppat.1005436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rothert M., Rönfeldt D., and Beitz E. (2017) Electrostatic attraction of weak monoacid anions increases probability for protonation and passage through aquaporins. J. Biol. Chem. 292, 9358–9364 10.1074/jbc.M117.782516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stanton C. L., and Houk K. N. (2008) Benchmarking pKa prediction methods for residues in proteins. J. Chem. Theory Comput. 4, 951–966 10.1021/ct8000014 [DOI] [PubMed] [Google Scholar]
- 29. Marchetti R. V., Lehane A. M., Shafik S. H., Winterberg M., Martin R. E., and Kirk K. (2015) A lactate and formate transporter in the intraerythrocytic malaria parasite, Plasmodium falciparum. Nat. Commun. 6, 6721 10.1038/ncomms7721 [DOI] [PubMed] [Google Scholar]
- 30. Harpaz Y., Gerstein M., and Chothia C. (1994) Volume changes on protein folding. Structure 2, 641–649 10.1016/S0969-2126(00)00065-4 [DOI] [PubMed] [Google Scholar]
- 31. Murata K., Mitsuoka K., Hirai T., Walz T., Agre P., Heymann J. B., Engel A., and Fujiyoshi Y. (2000) Structural determinants of water permeation through aquaporin-1. Nature 407, 599–605 10.1038/35036519 [DOI] [PubMed] [Google Scholar]
- 32. Tan K. P., Nguyen T. B., Patel S., Varadarajan R., and Madhusudhan M. S. (2013) Depth: a web server to compute depth, cavity sizes, detect potential small-molecule ligand-binding cavities and predict the pKa of ionizable residues in proteins. Nucleic Acids Res. 41, W314–W321 10.1093/nar/gkt503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Olsson M. H. M., Søndergaard C. R., Rostkowski M., and Jensen J. H (2011) PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 7, 525–537 10.1021/ct100578z [DOI] [PubMed] [Google Scholar]
- 34. Waight A. B., Love J., and Wang D. N. (2010) Structure and mechanism of a pentameric formate channel. Nat. Struct. Mol. Biol. 17, 31–37 10.1038/nsmb.1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Czyzewski B. K., and Wang D. N. (2012) Identification and characterization of a bacterial hydrosulphide ion channel. Nature 483, 494–497 10.1038/nature10881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hapuarachchi S. V., Cobbold S. A., Shafik S. H., Dennis A. S., McConville M. J., Martin R. E., Kirk K., and Lehane A. M. (2017) The malaria parasite's lactate transporter PfFNT is the target of antiplasmodial compounds identified in whole cell phenotypic screens. PLoS Pathog. 13, e1006180 10.1371/journal.ppat.1006180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Beitz E. (2000) TeXshade: shading and labeling of multiple sequence alignments using LaTeX2e. Bioinformatics 16, 135–139 10.1093/bioinformatics/16.2.135 [DOI] [PubMed] [Google Scholar]
- 38. Arnold P., Himmels P., Weiß S., Decker T. M., Markl J., Gatterdam V., Tampé R., Bartholomäus P., Dietrich U., and Dürr R. (2014) Antigenic and 3D structural characterization of soluble X4 and hybrid X4-R5 HIV-1 Env trimers. Retrovirology 11, 42 10.1186/1742-4690-11-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tang G., Peng L., Baldwin P. R., Mann D. S., Jiang W., Rees I., and Ludtke S. J. (2007) EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 10.1016/j.jsb.2006.05.009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.