Abstract
5-Enolpyruvylshikimate-3-phosphate synthase (EPSPS) catalyzes the transfer of a carboxyvinyl group from phosphoenolpyruvate (PEP) to shikimate-3-phosphate and in plants is the target of the herbicide glyphosate. EPSPSs with high catalytic efficiency and insensitivity to glyphosate are of microbial origin, including the enzyme from Agrobacterium strain CP4, in which insensitivity is conferred by an active site alanine. In the sequence context of plant EPSPSs, alanine in place of glycine at the equivalent position interferes with the binding of both glyphosate and PEP. We show here that iterative optimization of maize EPSPS containing the G101A substitution yielded variants on par with CP4 in terms of catalytic activity in the presence of glyphosate. The improvement relative to G101A alone was entirely due to reduction in Km for PEP from 333 to 18 μm, versus 9.5 μm for native maize EPSPS. A large portion of the reduction in Km was conferred by two down-sizing substitutions (L97C and V332A) within 8 Å of glyphosate, which together reduced Km for PEP to 43 μm. Although the original optimization was conducted with maize EPSPS, contextually homologous substitutions conferred similar properties to the EPSPSs of other crops. We also discovered a variant having the known glyphosate-desensitizing substitution P106L plus three additional ones that reduced the Km for PEP from 47 μm, observed with P106L alone, to 10.3 μm. The improvements obtained with both Ala101 and Leu106 have implications regarding glyphosate-tolerant crops and weeds.
Keywords: molecular evolution, enzyme inhibitor, protein engineering, biotechnology, plant biochemistry, 5-enolpyruvylshikimate-3-phosphate synthase, glyphosate, glyphosate resistance, herbicide resistance, plant EPSP synthase
Introduction
5-Enolpyruvylshikimate-3-phosphate synthase (EPSPS)5 (EC 2.5.1.19) catalyzes the transfer of a carboxyvinyl group from phosphoenolpyruvate (PEP) to shikimate-3-phosphate (S3P) (Fig. 1). The enzyme is inhibited by glyphosate, competitively with PEP (1, 2). The herbicidal efficacy of glyphosate against all plant species, its low cost, low mammalian toxicity, and benign environmental impact favor its use in crops that are endowed with a tolerance mechanism (3–7). Glyphosate-tolerant crops have been created by expressing glyphosate-insensitive bacterial EPSPS encoded on a transgene (8, 9).
Figure 1.
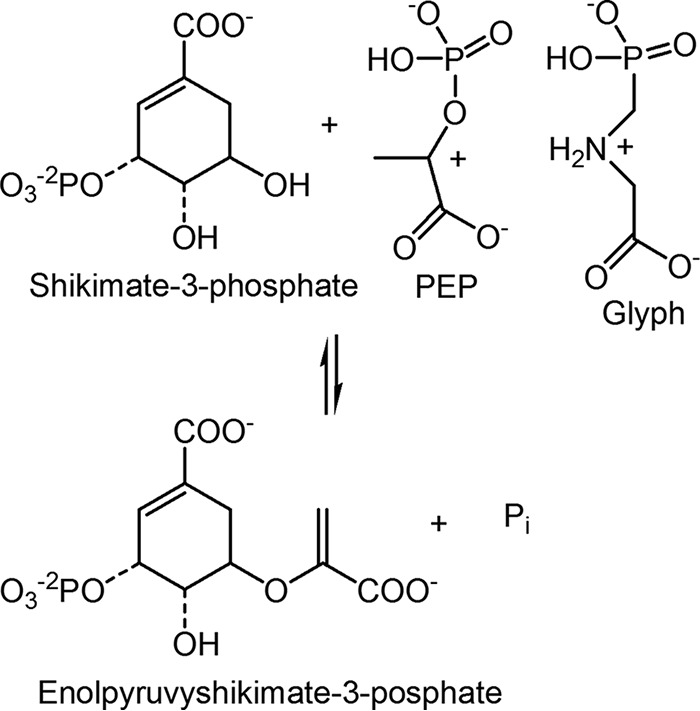
EPSPS reaction. The reaction is an addition/elimination in which an enzymic base deprotonates the 5-hydroxyl of S3P, allowing the electron pair to attack the oxocarbenium ion of PEP (shown to suggest the species mimicked by glyphosate), generated by the enzyme.
With gene editing facilitated by Cas9/CRISPR technology (10), it is theoretically possible to create glyphosate tolerance without a foreign transgene by introducing a set of mutations into the EPSPS gene that effectively desensitizes the enzyme to glyphosate while maintaining sufficient catalytic capacity. USDA, the American agency regulating genetically modified crops, has given “unregulated” status to many traits created with gene editing, without the usual lengthy and costly (∼$35 million) process required for a transgenic trait (48). Thus, gene editing could make glyphosate tolerance economically accessible for low volume crops or for staple crops in developing countries. In the latter case, the hand weeding prevalent in sub-Saharan Africa is typically far from optimal for preserving yield, is enormously time-consuming and often results in spinal deformation (11). However, enabling glyphosate tolerance by editing the native EPSPS gene will require a set of mutations that result in a much fitter enzyme than the plant variants currently known.
The known mutations in plant EPSPS and close homologs (termed Class I EPSPS) all involve modulations of active site Gly101 (numbering according to mature maize EPSPS (CAA44974.1, Fig. 2); 96 in Escherichia coli) that can create interference with the binding of glyphosate through one of its phosphonate oxygens (2). Changing Pro106 to glycine, serine, threonine, leucine, or alanine has the effect of moving the α carbon of Gly101 closer to glyphosate, causing moderately reduced affinity (12). These mutations were discovered by mutagenesis of bacterial genes (13) and are present in seven species of glyphosate-resistant weeds (14–16). Depending on the substitution, the degree of desensitization (increase in Ki) and whole plant resistance are proportional and are in the range of 2–9-fold (14, 15, 17).
Figure 2.

Amino acid sequence of the variant termed “native maize EPSPS” as expressed in vector pHD2114. The sequence shown is the reference for all position numbers mentioned in the text. The actual expressed protein has an N-terminal extension consisting of MGHHHHHHHHHHSSGHIEGRHM.
A second glyphosate-desensitized EPSPS variant is a double-mutant maize enzyme in which threonine at position 102 is changed to isoleucine in concert with the P106S mutation. The enzyme, termed TIPS, is highly desensitized to glyphosate, but whereas its Km for phosphoenolpyruvate is nearly normal, it has only 5% of the kcat of the native enzyme, as shown here and in earlier studies (18, 19). Like the Pro106 mutations, the TIPS mutations exert their effect by shifting Gly101 closer to the glyphosate-binding site (18). Although catalytic capacity for TIPS EPSPS is insufficient if the mutations are created by natural mutagenesis and gene editing (see “Discussion”), sufficiently high transgenic expression can result in competitive performance, as in GA21 maize (20). Very recently, a mutation resulting in the same threonine (position 102) being changed to serine was shown to account for glyphosate resistance in a tropical weed (21). We here provide the first kinetic characterization of the variant, which shows it to be no fitter than P106S and thus unsuitable for gene editing.
The other known de-sensitizing substitution is alanine for glycine at the equivalent of our maize position 101, first reported with the enzyme from a glyphosate-resistant strain of Klebsiella (22). Alanine occurs naturally at this position in EPSPS from Agrobacterium strain CP4, the enzyme present in RoundupReadyTM crops (8, 9). The CP4 enzyme is highly insensitive to inhibition by glyphosate and has 11–15% of the catalytic efficiency (kcat/Km) of the plant enzyme (23, 24). Plant EPSPS with the G101A mutation has similar insensitivity to CP4 but only 1.4% of the catalytic efficiency of the native plant enzyme, largely due to the 40-fold increase in Km for PEP imposed by the additional methyl group (23, 25). Although the structures of the CP4 and E. coli enzymes show that they share the same structural-fold and topology (26), they share only 23% identity. In contrast, the E. coli enzyme shares >50% identity with plant EPSPS. This divergence in sequence homology was the basis for designating CP4 and homologous microbial EPSPSs as Class II. The divergent amino acid sequence of CP4 has been thought to provide the structural context for an optimal spatial location of the alanine methyl group that results in interference with the longer glyphosate but not with PEP.
The G101A mutation has not been found in a glyphosate-resistant weed, likely due to its poor affinity for PEP. We here show that with 17 or more additional mutations, the enzyme from maize, a Class I EPSPS, can be adapted to accommodate the G101A mutation, resulting in kinetic parameters equal to or better than those of CP4. The variants are no closer in homology to CP4 than is the native enzyme.
Results
Mutagenesis, maize native EPSPS
Because the use of degenerate oligonucleotides for saturation mutagenesis theoretically can access all 19 changes at each position, we began by searching for single desensitizing mutations in native maize EPSPS that may have been missed by earlier methodology. Saturation mutagenesis was performed and positional libraries screened as described under “Experimental procedures.” No new desensitizing mutations were found. The only ones found that showed appreciable desensitization were the known mutations at Pro106 (further evaluated below). However, many neutral or slightly beneficial mutations were identified.
Combinatorial shuffling, initial library
The mutations discovered were used to construct a combinatorial library designed to explore combinations of functional diversity at the known desensitizing positions (G101A, T102(IALGV), and P106(SGLVQWA)) in novel sequence contexts provided by the newly identified neutral mutations; in all, 43 substitutions at 29 positions. The library was synthesized entirely from oligonucleotides, using the technique of synthetic shuffling (27). The vector DNA of the library was transformed into our BL21(DE3) Tuner-AroA knockout strain and the cells were plated onto minimal medium with glyphosate (see Fig. S1 for details on library design, host cells, screening conditions, and advancement criteria). The variant forms of EPSPS from the 184 colonies that grew were evaluated by enzyme assay and the best of those were subjected to substrate saturation kinetic analysis.
Three variants each included one of the previously known mutations or pair of mutations that reduce sensitivity of EPSPS to inhibition by glyphosate. Variant P106L+3 has leucine substituted for proline at position 106 and three additional mutations (A100S, E301S, and A337S). Alone, the P106L mutation raised Ki for glyphosate 60-fold, but also raised Km for PEP 5-fold (Table 1). The three additional mutations served to lower Km for PEP from 47 μm, seen with P106L alone, to 10.3 μm with 60% retention of Ki. The overall result was 30-fold improved fitness ((kcat/Km) × Ki) compared with native maize EPSPS and 7-fold improved relative to P106S.
Table 1.
Kinetic parameters of maize EPSPS with mutations found in glyphosate-resistant weeds
Values represent mean ± S.E. for at least 4 sets of rate measurements. For assay procedure and analysis, see “Experimental procedures.” Values for Km were obtained by varying one substrate, with the other present at saturation (10 times its Km) and therefore are apparent Km. Values shown for kcat are those obtained from substrate saturation with PEP, which in no case differed significantly from those obtained with S3P.
| kcat | Km PEP | kcat/Km | Ki | Km S3P | kglya | (kcat/Km) × Kib | |
|---|---|---|---|---|---|---|---|
| min−1 | μm | μm | min−1 | ||||
| Zm nativec | 1630 ± 14 | 9.5 ± 0.3 | 172 | 0.066 ± 0.003 | 13.2 ± 0.6 | <lodd | 11 |
| Zm-P106Se | 1540 ± 12 | 11.5 ± 0.5 | 134 | 0.33 ± 0.02 | 15.4 ± 0.4 | 2.3 ± 0.07 | 44 |
| Zm-P106Lf | 1760 ± 10 | 47.0 ± 1.1 | 37.5 | 3.94 ± 0.17 | 27.6 ± 0.7 | 5.7 ± 0.17 | 148 |
| Zm-P106L+3g7 | 1450 ± 22 | 10.3 ± 0.7 | 140 | 2.34 ± 0.18 | 17.5 ± 0.6 | 10.1 ± 0.1 | 329 |
| Zm-TIPS+2h | 105 ± 1.0 | 16.2 ± 0.7 | 6.5 | 731 ± 36 | 27.5 ± 0.7 | 25.5 ± 0.4 | 4740 |
| Zm-T102Si | 1600 ± 13 | 30.9 ± 1.0 | 51.8 | 0.69 ± 0.02 | 28.8 ± 0.6 | 2.0 ± 0.09 | 35.7 |
| Ha-T102Sj | 1800 ± 13 | 30.4 ± 0.8 | 59.1 | 0.75 ± 0.03 | 21.5 ± 0.5 | 2.4 ± 0.08 | 44.3 |
a Enzyme turnover (min−1) at 30 μm PEP and S3P, 1 mm glyphosate (see “Results” for rational as a fitness parameter).
b Calculated with Km for PEP.
c Native Z. mays EPSPS having the amino acid sequence described in the legend to Fig. 2.
d lod; limit of detection calculated as Blank + 3xSD of blank. The value is 1.5 min−1.
e A mutation found in multiple species of glyphosate-resistant weeds, here created in maize EPSPS.
f A mutation found in several species of glyphosate-resistant weeds, here created in maize EPSPS.
g Variant captured from the initial combinatorial library (see “Results”), having P106L and three additional mutations (A100S, E301S, A337S).
h Variant captured from the initial combinatorial library, having T102I, P106S and two other mutations (T277G, A337S).
i A mutation found in a glyphosate-resistant population of T. procumbens, here created in maize EPSPS.
j A mutation found in a glyphosate-resistant population of T. procumbens, here created in H. annuus (sunflower) EPSPS.
Along with two other mutations (T277G, A337S), variant TIPS+2 has the T102I and P106S (TIPS) mutations present in the GA21 maize transformation event (20). Kinetic analysis indicated a high level of insensitivity to glyphosate (Ki), near native affinity for PEP (Km) but only ∼5% of the native kcat (Table 1), reflecting results obtained previously for TIPS (18, 19).
Variant G101A+3 has alanine substituted for glycine at position 101, plus three other substitutions (E301S, E390G, V437R) and was the starting point for the optimization scheme (Fig. S1) that resulted in the fittest glyphosate-desensitized variants (see below).
P106L and T102I-P106S variants could not be improved
P106L+3 and TIPS+2 were each subjected to a cycle of saturation mutagenesis and combinatorial shuffling, attempting to improve the Ki of P106L+3 or to improve the kcat of TIPS+2. Neither attempt was successful (not shown). We found no mutation(s) that could work in concert with P106L (and presumably P106S or -A) to enable any further desensitization. Nor did we find any mutation(s) that could compensate for the low kcat imposed by the TIPS mutations.
Very recently, glyphosate resistance in a population of Tridax procumbens (Tribe Heliantheae) was attributed to a change from threonine to serine at position 102 (21). We created the T102S mutation in the sequence contexts of maize and Helianthus annuus (XP_022017499.1) EPSPS. The mutation had the effect of increasing Ki by 10-fold, but also elevating Km for PEP by 3-fold (Table 1). We made no attempt to optimize this variant because it was unknown at the outset of our work.
Further optimization of maize EPSPS-G101A
In the context of the maize enzyme, G101A is highly insensitive to glyphosate, but has 35-fold elevated Km for PEP relative to native EPSPS (Table 2), confirming earlier results with Class I EPSPS (22, 23, 28). However, unlike the situation with variants containing P106L or the TIPS mutations, G101A was amenable to improvement through iterative cycles of diversity generation and combinatorial shuffling. The process is shown schematically in Fig. S1. For every application of saturation mutagenesis, at no position was the amino acid fixed. Rather, for each position, all 20 options were created in the sequence context of the current fittest variant and subjected to selection at the prevailing level of stringency. By not fixing the backbone, detrimental mutations can change to a different amino acid or revert to the native one.
Table 2.
Kinetic parameters of variants at each turn of the optimization cycle of maize EPSPS with G101A as the key desensitizing mutation
For details on data collection and presentation, see the legend to Table I.
| kcat | Km PEP | kcat/Km | Ki | Km S3P | kglya | (kcat/Km) × Kib | |
|---|---|---|---|---|---|---|---|
| min−1 | μm | μm | min−1 | ||||
| Zm native | 1630 ± 14 | 9.5 ± 0.3 | 172 | 0.066 ± 0.003 | 13.2 ± 0.6 | <lodc | 11 |
| G101A+3d | 824 ± 15 | 347 ± 6.8 | 2.4 | 1430 ± 137 | 153 ± 3.9 | 17.7 ± 0.6 | 3,400 |
| G101A | 1000 ± 35 | 333 ± 26 | 3.0 | 1930 ± 40 | 84.0 ± 2.5 | 25.7 ± 0.2 | 5,780 |
| H6 | 397 ± 4 | 25.1 ± 0.9 | 15.8 | 989 ± 31 | 14.0 ± 0.5 | 95.5 ± 2.0 | 15,700 |
| C1 | 517 ± 7 | 18.9 ± 0.5 | 27.4 | 449 ± 48 | 11.4 ± 0.5 | 104 ± 1.6 | 12,300 |
| D2 | 414 ± 38 | 14.5 ± 2.2 | 28.6 | 935 ± 66 | 11.3 ± 0.3 | 119 ± 2.0 | 26,700 |
| D2–67 | 530 ± 17 | 14.4 ± 0.6 | 36.8 | 945 ± 98 | 10.9 ± 0.4 | 146 ± 2.0 | 34,800 |
| D2–124 | 631 ± 5 | 18.1 ± 0.7 | 34.9 | 893 ± 53 | 11.1 ± 0.7 | 175 ± 2.5 | 31,130 |
| D2c-A5 | 741 ± 6 | 18.1 ± 0.6 | 40.9 | 839 ± 40 | 12.6 ± 1.4 | 189 ± 3.1 | 34,350 |
| CP4e | 411 ± 5 | 15.5 ± 0.4 | 26.5 | 1970 ± 276 | 5.2 ± 0.5 | 176 ± 1.6 | 52,240 |
| 101A-97Cf | 477 ± 3.5 | 93.0 ± 2.7 | 5.1 | 646 ± 30 | 36.7 ± 1.0 | 79.3 ± 2.2 | 3,300 |
| 101A-332Af | 764 ± 10 | 123 ± 6.6 | 6.2 | 2390 ± 224 | 52.5 ± 3.1 | 47.8 ± 0.5 | 14,800 |
| 101A-401Gf | 222 ± 2.3 | 291 ± 9.0 | 0.8 | 2970 ± 183 | 91.8 ± 4.0 | 23.4 ± 1.9 | 2,290 |
| ACAg | 408 ± 3.7 | 42.5 ± 1.8 | 9.6 | 1470 ± 96 | 31.4 ± 1.1 | 117 ± 1.8 | 14,100 |
| A5-A101Gh | 135 ± 0.8 | 3.9 ± 0.2 | 34.7 | 0.17 ± 0.01 | 11.0 ± 0.6 | 0.2 ± 0.08 | 5.9 |
a Enzyme turnover (min−1) at 30 μm PEP and S3P, 1 mm glyphosate (see “Results” for rational as a fitness parameter).
b Calculated with Km for PEP,
c lod; limit of detection calculated as Blank + 3xSD of blank. The value is 1.5 min−1.
d Variant captured from the initial combinatorial library, having G101A plus three other mutations (E301S, E390G, V437R).
e EPSPS from Agrobacterium sp. strain CP4, present in RoundUp Ready crops.
f Variants of maize EPSPS having only the two mutations shown.
g Variant having G101A, L97C, and V332A. ACA, G101A-L98C, and G101A-V332A were accessed commercially by gene synthesis.
h D2c-A5 with Ala101 changed back to glycine.
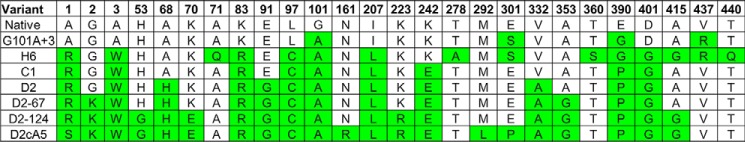
Although G101A+3 was our starting point, the process was effectively the progressive addition of mutations to G101A. The kinetic parameters for G101A+3 are poorer than those of G101A (Table 2), suggesting that one or more of its three additional mutations (E301S, E390G, V437R) was detrimental. Interestingly, all three were eliminated in the next phase of optimization. H6 and C2 were variants captured from a combinatorial library of diversity generated in the context of G101A+3, having 15 and 16 mutations, respectively. To fully explore the diversity present in H6 and C2, a small library was constructed in which the amino acids at their variable positions were toggled with the native amino acid. This allowed all positions encompassed by H6 and C2 to acquire a mutation or revert to the native amino acid, thus creating all possible combinations of the mutations present in the two variants and eliminating nonessential or deleterious ones. Although not resulting in significant improvement relative to H6 (the better of the two parents), the procedure yielded variant C1, which had kinetic parameters similar to those of H6 (Table 2) but only 9 mutations compared with the 15 present in H6. Five of the six substitutions eliminated from H6 by the backcross procedure (including the three from G101A+3) did not reappear in subsequent cycles of saturation mutagenesis and combinatorial shuffling (Fig. 3). From variant C1 on, with one exception (R1S in D2c-A5), the process was the progressive addition of new mutations to the previous best backbone sequence.
Figure 3.
Amino acid substitutions present in variants in the progressive optimization of maize EPSPS for activity in the presence of glyphosate. Numbers are the amino acid positions in the reference sequence (Fig. 2). Amino acids that differ from those in the native enzyme are highlighted.
About the stage of variant C1, we created a parameter for predicting performance in the treated plant that would be a more rapid and informative representation of fitness than (kcat/Km) × Ki. Although useful for assessing the intrinsic potential for activity in the presence of a competitive inhibitor (29), (kcat/Km) × Ki is inadequate for predicting the reaction velocity under the conditions of the application (plants sprayed with glyphosate) because it omits concentrations of substrates and inhibitor, factors that are not intrinsic to the enzyme, but on which the reaction rate depends. Therefore, libraries subsequent to variant C1 in Fig. S1 were evaluated with a single rate measurement designed to take all factors in the rate equation for competitive inhibition into account. The concentrations of PEP and S3P were set as nearly as possible (30 μm, limited by the sensitivity of our assay) to the presumed intracellular concentrations of 10–15 μm, the approximate values of Km for both PEP and S3P for the native enzyme (Table 1). Glyphosate was included at 1 mm, a concentration attainable in tissues, especially meristems, receiving metabolite flow from treated leaves (30). The pH (7.0), ionic strength (100 mm KCl) and co-solvent concentration (5% ethylene glycol) were also intended to mimic in vivo conditions (31, 32). The unit for the parameter is reaction rate (μm min−1) per enzyme concentration (μm), or min−1, describing the enzyme turnover under application conditions, which we abbreviate as “kgly.” Although we continued to determine individual kinetic parameters for key variants, kgly was adopted as the one parameter needed both for mediums throughput screening and for ultimate evaluation of fitness.
The progressive increase in kgly is shown graphically in Fig. 4. The largest step in the progression was the 4-fold increase found in the first combinatorial library involving G101A. Variants from H6 on show dramatically reduced Km for PEP, with some variants falling within 1.5-fold of the native value (Table 2). Most of the insensitivity to glyphosate conferred by the G101A mutation was retained, with Ki values clustering around 900 μm for all variants but C1. Optimization culminated with variants D2-124 and D2c-A5, with 18 and 21 mutations, respectively.
Figure 4.
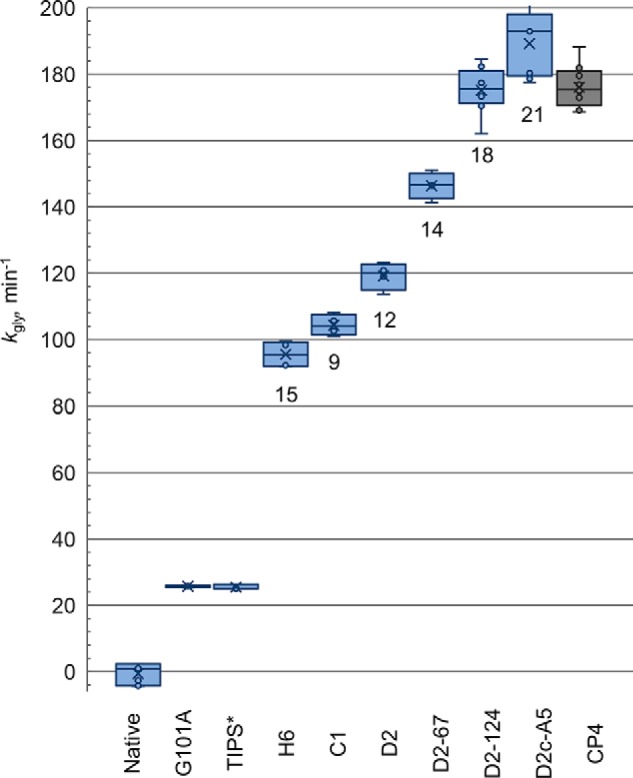
Progressive fitness resulting from optimization of maize EPSPS with the G101A mutation. Values are mean ± S.E. of 3–6 rate measurements, taken from Table 2. kgly is enzyme turnover, min−1, under simulated in vivo application conditions (30 μm PEP, 30 μm S3P, and 1 mm glyphosate; for rationale, see “Results”). Numbers below the box indicates the number of amino acid changes relative to native maize EPSPS. The reduction from 15 to 9 occurred in the H6-C2-native backcross library (see text). *TIPS, maize EPSPS with the T102I and P106S mutations plus an additional N444G mutation at the C terminus. TIPS* and CP4 are included for reference.
Large contribution from two near-active site substitutions
Examination of a model in which the maize amino acid sequence was threaded onto PDB code 1G6S (E. coli EPSPS) provided clues as to how substitutions outside the active site could result in the dramatic reduction in Km for PEP from 333 μm in G101A to 18 μm in variants D2-124 and D2c-A5. Three downsizing substitutions occurred that were within 9 Å of glyphosate. L97C and D401G became incorporated in variant H6, and V332A in D2 (Fig. S1 and Fig. 3). In combination with G101A, D401G had little if any effect on Km for either PEP or S3P relative to G101A alone and was detrimental to kcat (Table 2). In contrast, when L97C and V332A were each tested in combination with G101A, each reduced Km for PEP relative to G101A alone by half or more with no impact on kcat (Table 2). The two together with G101A resulted in a Km for PEP of 47 μm (variant ACA, Table 2), accounting for half of the improvement in kcat/Km of the best variant, D2c-A5, relative to G101A. As judged by kgly, the triple mutant ACA was 63% as fit as the best variant.
Mutations in maize are transferable to EPSPS from other plant species
The high degree of conservation among plant EPSPS (identity at 66.4% of the positions with consensus at 95% of positions among 11 species) suggests that the mutations defined in the maize background would have a similar effect in EPSPS from other crop species. To map the D2-124 substitutions to EPSPS from other crop species, we used ClustalW to align native maize EPSPS, maize variant D2-124, and the native sequences from the target species. We used the ChloroP Prediction Server (http://www.cbs.dtu.dk/services/ChloroP/)6 (47) to approximate the N terminus of the mature proteins. Nucleotide sequences were optimized for expression in E. coli and synthesized commercially. The synthetic genes were cloned into pHD2114 and expressed, purified, and analyzed. In most but not all cases, the substitutions defined in maize endowed a high level of fitness (kgly) in the alternative plant EPSPS (Table 3). In the case of wheat, the Km for PEP was close to that of the maize enzyme (18.9 versus 18.1 μm for maize), but the substitutions conferred a deficiency in kcat (223 min−1 versus 631 min−1 for maize). In the other case of inefficient translation, the result was the opposite. The kcat of soybean D2-124 was the same as maize D2-124, but the Km for PEP was 5-fold higher, indicating that the D2-124 substitutions in the sequence context of the soy enzyme were only partially effective in accommodating the G101A substitution.
Table 3.
Efficiency of translation of mutations from maize variant D2-124 to EPSPS from other crop species
The 18 mutations present in maize variant D2–124 were mapped onto the amino acid sequence of the predicted mature form of EPSPS from the species shown (see “Results” for details). Proteins were expressed, purified and evaluated as described under “Experimental procedures” and the legend to Table I.
| Speciesa | kglyb | kcat | Km PEP | Ki |
|---|---|---|---|---|
| min−1 | μm | |||
| Maize | 175 ± 2.5 | 631 ± 5 | 18.1 ± 0.7 | 893 ± 53 |
| Rice | 174 ± 3.1 | 707 ± 17 | 22.3 ± 2.7 | 893 ± 89 |
| Sorghum | 172 ± 8.5 | 846 ± 2.9 | 20.7 ± 1.7 | 404 ± 44 |
| Sunflower | 155 ± 7.7 | 381 ± 7.0 | 18.0 ± 1.8 | 918 ± 37 |
| Grapevine | 144 ± 11 | 652 ± 8.1 | 30.4 ± 1.7 | 1150 ± 98 |
| Cotton | 143 ± 8.4 | 413 ± 2.7 | 15.3 ± 0.6 | 940 ± 85 |
| Cassava | 133 ± 6.3 | 688 ± 8.6 | 24.7 ± 1.5 | 1400 ± 228 |
| Soybean | 114 ± 4.1 | 618 ± 4.8 | 90.3 ± 2.3 | 3550 ± 374 |
| Wheat | 102 ± 6.1 | 223 ± 4.3 | 18.9 ± 1.7 | 820 ± 119 |
a Species, accession: Z. mays, CAA44974.1; Oryza sativa, AF413082.1; Sorghum bicolor, XM_002436379.2; H. annuus, XM_022161807.1; Vitis vinifera, NC_012021.3; Gossypium hirsutum, UniProt A7Y7Y2; Manihot esculenta, XM_021758443.1; Glycine max, XM_003516991.3; T. aestivum, ACH72672.1.
b kgly is the enzyme turnover (min−1) at 30 μm PEP and S3P, 1 mm glyphosate (see “Results” for rational as a fitness parameter).
Discussion
No novel mutations
Extensive mutagenesis with a method (oligonucleotides with the “NNK” degenerate codon usage) theoretically capable of capturing all changes at all positions revealed no new mechanisms for desensitizing plant EPSPS to glyphosate. All known mutations function directly or indirectly at the contact between Gly101 (position 96 in E. coli) and a phosphonate oxygen of glyphosate. We tentatively conclude that the other amino acids with contacts to glyphosate in the E. coli structure (PDB 1G6S) (2), namely, Lys22, Asn94, Arg124, Gln171, Glu341, Arg344, Arg386, and Lys411, corresponding to Lys24, Asn99, Arg131, Gln180, Glu359, Arg362, Arg404, and Lys429 in our maize model, are too important for binding or catalysis to be mutated or have no means through which their spatial positions can be adjusted to improve discrimination between PEP and glyphosate.
Improved context for P106L, implications for weed resistance
In screening our first combinatorial library, we found variants representing all three of the previously known mechanisms by which Class I EPSPS (e.g. plant, E. coli) can be desensitized to inhibition by glyphosate. Variant P106L+3 has leucine substituted for proline at position 106 plus three other mutations. Substitutions at position 106 that reduce glyphosate affinity are present in seven species of glyphosate-resistant weeds (14, 15). Although Pro106 is not directly involved in molecular interaction with PEP or glyphosate, mutations at 106 have the effect of moving other amino acids, particularly Gly101, sufficiently closer toward the phosphonate end of glyphosate to moderately impede its binding (12). Although the P106S mutation did not perturb kinetic parameters, P106L caused a 5-fold increase in Km for PEP and doubled Km for S3P (Table 1) (also, see Refs. 12 and 33). P106L conferred 60-fold reduced sensitivity (elevated Ki) to glyphosate compared with native EPSPS and 12-fold versus P106S (Table 1). Until recently, only P106S and P106T were known to confer resistance without involvement of another mechanism (14), leading to conjecture that the high Km for PEP imposed by P106L precluded its emergence as a resistance mechanism. However, two weed species where resistance was due to P106L recently have been reported (15, 16). Interestingly, in our variant P106L+3, Km for PEP is restored nearly to normal, yielding intrinsic fitness ((kcat/Km) × Ki) that is 30-fold improved over native maize EPSPS and a kgly value 4-fold improved over P106S (Table 1). Were the P106L mutation to occur in a plant having the right sequence context, a plant with minimal fitness penalty and proportionally greater resistance factor (effective dose of glyphosate relative to WT) than that conferred by P106S would be expected.
T102S, the recently reported third mutation in glyphosate-resistant weeds
Our established methods presented the opportunity to evaluate this variant, recently reported as the basis for glyphosate resistance in T. procumbens, of the Tribe Heliantheae (21). In the sequence context of either maize or H. annuus (sunflower), the mutation had the effect of elevating Ki 10-fold while elevating Km for PEP 3-fold (Table 1). These results are supportive of the prediction, based on molecular dynamic simulations in a homology model, that the mutation increases the affinity for PEP relative to that of glyphosate (21). Compared with P106S, the combined effects of the T102S mutation (greater desensitization to glyphosate and reduced affinity for PEP) leaves the two with about equal fitness as measured by kgly (Table 1).
Combinations involving Thr102 and Pro106 could not be further improved
Variant TIPS+2 contains the T102I and P106S (TIPS) mutations present in the GA21 maize transformation event (20). The variant had a high level of insensitivity to glyphosate while retaining near native affinity for PEP but with only ∼5% of the native kcat (Table 1), confirming earlier reports with TIPS variants (18, 19). The low catalytic efficiency of TIPS EPSPS evidently makes it unsuitable as a resistance mechanism except at high-level transgenic expression. Stepwise acquisition of both T102I and P106S mutations was documented in a population of Eleusine indica (19). However, out of a population of 193 individuals, only 1.6% were homozygous for TIPS. The highest frequency allelic combination was TIPS/P106S, suggesting that the normal catalytic capacity contributed from the P106S allele was more important for fitness than having the second allele encode a highly insensitive, but catalytically deficient enzyme. In controlled experiments, the same group later confirmed the extreme fitness penalty imposed by the TIPS mutations (34). Similarly, unacceptable agronomics were observed in cassava edited to create the TIPS or TIPA mutations, unless highly expressed by a constitutive viral promoter (35, 36). Furthermore, homozygosity for TIPS where the T0 rice plants were produced by CRISPR/Cas9 gene editing could not be found among 93 T2 plants analyzed, and was suggested to be lethal (37). Our attempt to improve kcat in variants containing the T102I and P106S mutations was unsuccessful. The narrow range of parameters seen among the 32 variants analyzed (i.e. those that supported colony growth under selection conditions) suggests that the properties conferred by the T102I and P106S mutations are not significantly influenced by the sequence context but rather, are strongly determinant for a narrow range of kinetic parameters (data not shown).
Class II properties from an optimized Class I EPSPS
Only variant G101A+3, having alanine substituted for glycine at position 101, was amenable to further optimization. G101A was associated with a 30,000-fold increase in Ki, but also with a 35-fold increase in Km for PEP (Table 2), confirming earlier results (23, 25, 28, 38, 39). Alanine is present naturally at the homologous position in the Class II EPSPS from Agrobacterium sp. strain CP4. CP4 EPSPS exhibited a high degree of insensitivity to glyphosate but with a Km for PEP of just 15.5 μm (Table 2), in good agreement with the 12 μm obtained earlier (23). Comparison of the crystal structures of CP4 ligated with S3P and glyphosate (PDB 2GGA) (24) and E. coli EPSPS with the contextually equivalent glycine changed to alanine, ligated with S3P and glyphosate (modeled (28)), indicate that the alanine methyl group in CP4 is positioned 0.3 Å further away from the phosphonate group of glyphosate than in the E. coli structure. Because PEP is shorter than glyphosate, it is hypothesized that the alanine methyl group in CP4 EPSPS is ideally positioned to interfere with binding of glyphosate but not PEP. Although there is only 24–26% homology between the CP4 enzyme and E. coli or maize EPSPS, structures of the CP4 and E. coli enzymes show that they share the same structural-fold and topology (2, 24). Presumably, the amino acid sequence of CP4 creates an overall structural context that places the alanine methyl group in its favorable position. The novel global sequence context that we arrived at resulted in Class 1 EPSPSs (D2-124 and D2c-A5) that match CP4 in catalytic proficiency in the presence of glyphosate, yet are no closer in sequence homology to CP4 than is the native maize EPSPS.
Structural rationale for the effect of substitutions outside the active site
We surmise that the new sequence context we arrived at through our process had a similar effect of increasing the distance between the Ala101 methyl and the phosphonate group of glyphosate. Two downsizing substitutions (L97C and V332A) when present together with G101A resulted in a Km for PEP of 47 μm (variant ACA, Table 2), accounting for 50% the improvement in kcat/Km of the best variant relative to G101A. Both are within 8 Å of glyphosate in our model of maize EPSPS threaded onto the E. coli enzyme (PDB 1G6S). Leu97 is in a loop that links a β-strand to helix-3, the key catalytic element harboring G101A. The bulky Leu97 side chain packs tightly against the helix, ∼6 Å from Ala101. Downsizing the side chain to cysteine would create a void that could create space for Ala101 to move outward from the PEP-glyphosate–binding site. The 18 remaining substitutions in D2c-A5 are remote from the active site, yet somehow transmit a collective effect through the structure, further optimizing the position of the methyl group so as to provide a further 2-fold improvement in kcat/Km while retaining robust insensitivity to glyphosate.
Further evidence that the active site of the best variant (D2c-A5) is enlarged is that when its Ala101 was changed back to glycine, the kinetic parameters obtained were very different from those of the native enzyme. As expected, sensitivity to glyphosate was almost completely restored. Surprisingly, however, Km for PEP fell from 18 to 3.9 μm (versus 9.5 μm for the native enzyme), while kcat fell to 20% of the optimized variant (Table 2). The effects on both kcat and Km for PEP can be rationalized by presuming that the re-modeled active site has become enlarged and that in the absence of the extra methyl group, is too large. We speculate that Km is lower in the methyl group-free optimized active site than in the native because in the native active site, PEP must expend some of its binding energy in binding to a relatively constricted pocket compared with that of the methyl group-free evolved variant, manifesting in a relatively higher Km. The negative effect on kcat of the expanded active site may be a consequence of the substrates being held in positions that are out of range for optimal reaction rate. A comparison of crystal structures of maize native and variant EPSPS may confirm this conjecture and provide further details of how global sequence context creates an active site appropriately re-modeled to accommodate the extra methyl group.
In addition to its 35-fold effect on affinity for PEP, the G101A mutation also reduced affinity for S3P, but only by 6-fold (Km = 84 μm versus 13.2 μm for native EPSPS, Table 2). Reduced affinity for S3P was also observed with the same mutation in the Klebsiella enzyme (38). Optimization resulted in the Km for S3P returning to near normal in parallel with the reduction in Km for PEP. We hypothesize that in the native sequence context, the G101A mutation does not directly interfere with S3P binding, but that the extra methyl group simply reduces the space available for S3P when PEP is bound.
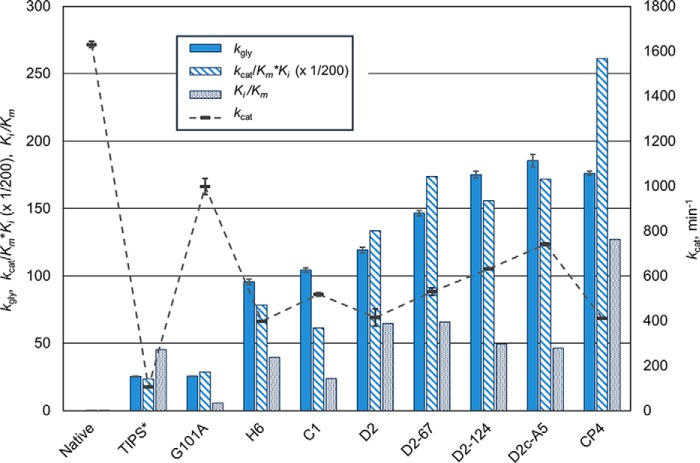
Fitness as quantified by kgly versus (kcat/Km) × Ki
The fitness parameter kgly, which takes into account anticipated concentrations of substrates and inhibitor as well as kinetic parameters, is intended to better capture enzyme fitness under the conditions of the application than (kcat/Km) × Ki. In fact, the two measures of fitness correlated rather well with a few exceptions (Fig. 5). CP4, with its very high Ki, displayed a disproportionately high (kcat/Km) × Ki. This is due to the greater impact of Ki on that parameter compared with its impact on the velocity equation for competitive inhibition, v = kcat [E] [S]/Km (1 + [I]/Ki + [S], which our kgly parameter seeks to represent. The relatively low kgly for CP4 given its outstanding selectivity (Ki/Km) is attributable to its low value for kcat. The converse, high kcat but poor selectivity (G101A), is likewise unsatisfactory. D2c-A5 incorporates the best combination of the parameters under optimization. Initially (H6), substitutions that rectified the high Km for PEP imposed by G101A were also detrimental to kcat. Improvement from D2 on occurred mainly by improving kcat while retaining the selectivity established earlier.
Figure 5.
Impact of improvements in kcat and selectivity (Ki/Km) on fitness of optimized maize EPSPS kgly is enzyme turnover, min−1, under simulated in vivo application conditions (30 μm PEP, 30 μm S3P, and 1 mm glyphosate; for rationale, see “Results”). Units for kcat and kcat/Km × Ki are also min−1. *TIPS, maize EPSPS with the T102I and P106S mutations plus an additional N444G mutation at the C terminus.
Slow release of glyphosate from maize native EPSPS
Our value of 66 nm for Ki for the native enzyme is in accord with the 80 nm reported for EPSPS from Pisum sativum (40) and the 48 nm obtained with the E. indica enzyme (17). Although glyphosate has not been characterized as a “slow, tight-binding” inhibitor (41), its dissociation from an E·S3P·glyphosate complex was slow enough to be observed over a 40-s span using our continuous assay (Fig. S3). The slow release effectively removed a fraction of the enzyme from the reaction, causing the substrate saturation curves with and without glyphosate to resemble noncompetitive inhibition (Fig. S2). When the protocol was revised to avoid a preincubation with enzyme, glyphosate, and S3P, classic competitive inhibition was observed (Figs. S4 and S5). In view of nanomolar values for Ki and our observation of time-dependent dissociation, the rather high application rates of glyphosate (relative to, e.g. AHAS inhibiting herbicides) may have led to an underappreciation of glyphosate's potency as an inhibitor of plant EPSPS.
Previous attempts to use genome engineering to introduce known EPSPS mutations into crops to develop glyphosate-tolerant plants met with limited success unless a strong constitutive promoter was used to drive EPSPS expression (35, 36). It will be interesting to observe whether converting native maize EPSPS to an optimized variant by genome editing will result in commercial level trait efficacy and normal phenotype. If so, the facile transfer of properties obtained by mapping the mutations identified in this work onto EPSPS from other species should enable nontransgenic glyphosate tolerance in many diverse crops.
Experimental procedures
Reagents
S3P was prepared from cultures of Klebsiella pneumonia aroA- (ATCC 25597). Cells from a 500-ml culture grown in 2× YT were used to inoculate 6 liters of minimal medium augmented with 55 μm tyrosine, 60 μm phenylalanine, 25 μm tryptophan, 0.1 μm 4-aminobenzoate, and 0.1 μm 4-hydroxybenzoate (42). Accumulation of S3P was monitored by anion exchange HPLC. After 4 days shaking at 37 ºC, the concentration reached ∼1 mm. S3P was purified from the culture supernatant by anion exchange chromatography in ammonium bicarbonate at pH 7.3, with gradient elution up to 0.7 m. S3P was cleanly separated from phosphate, which eluted earlier. 2-Amino-6-mercapto-7-methylpurine ribonucleoside (MESG) was from Setareh Biotech, Eugene, OR. Glyphosate was from ORICO. All other reagents were from Sigma.
Vector for expressing plant EPSPS in E. coli
The amino acid sequence of mature Zea mays (maize) EPSPS was obtained from GenBankTM entry CAA44974.1. A nucleotide sequence was created to add an N-terminal methionine and optimize codon usage for expression in E. coli. The synthetic gene was supplied by a commercial vendor and was cloned into pHD2114, a derivative of pET16 E. coli expression vector that provides a T7 promoter driving expression of the protein (Fig. S6). The vector was modified to change the 6× N-terminal histidine tag to a 10× tag. Later, to reduce leaky expression and increase stringency, the origin of replication was altered to produce a lower copy number (see below, “Screening procedures”). The resulting coding region of the vector yields an expressed protein with the amino acid sequence shown in Fig. 2. CP4, the EPSPS derived from Agrobacterium sp. strain CP4 (9), was accessed by gene synthesis.
Saturation mutagenesis
Native or improved variants of EPSPS were subjected to saturation mutagenesis to discover novel mutations that reduce sensitivity to glyphosate. Libraries of substitutions for each position in the EPSPS polypeptide chain were created using NNK (where N represents a 25% mix each of adenine, thymine, guanine, and cytosine nucleotides; and K represents a 50% mix each of thymine and guanine nucleotides) as the degenerate codon for the position to be mutagenized. PCR mixtures contained a mutagenic forward primer (NNK codon flanked by 28 nucleotides matching with template at each side of the NNK) and a reverse primer that was the complement of the sequence preceding the forward primer, 28 nucleotides in length. To make circular dsDNA plasmids from the blunt ended PCR products, the products were digested with T4 polynucleotide kinase, T4 DNA ligase, and DpnI (to disrupt the parental DNA template). After desalting by ultrafiltration, the ligation products were ready for transformation and downstream applications.
Combinatorial library construction
The initial combinatorial library (neutral diversity identified in the native EPSPS plus known mutations at positions 101, 102, and 106) was constructed entirely from oligonucleotides, using the technique of synthetic shuffling (27). All other libraries were made by random toggling of desired substitutions into a specific sequence, termed the backbone, by the technique of semi-synthetic shuffling (27).
Screening procedures
We used a tiered screening cascade to identify the fittest variants in each library. At the outset of optimization beginning with native maize EPSPS, plasmid libraries of single or multiple mutations were transformed into BL21(DE3)-Tuner cells in which the native AroA gene was functionally deleted by phage transduction. The donor strain was JW0891 (CGSC), in which the AroA gene is disrupted with a kanamycin-resistant gene. Variant H6 (Fig. 3, Table 2) had a level of fitness that allowed growth on >10 mm glyphosate (at which the native EPSPS is completely inhibited), allowing its descendants to be expressed in standard BL21(DE3) cells. Cells were plated on minimal agar medium containing M9 salts, 2% glucose, 2 mm MgSO4, 0.1 mm CaCl2, and 10 mg/liter of thiamine, creating the requirement for a functional, plasmid-encoded EPSPS to enable colony growth. As fitness improved, amendments were added to increase the stringency of selection, as indicated in Fig. S1. Glyphosate was added up to 300 mm, its limit of solubility in agar. Polymyxin B nonapeptide was included at 1 mg/liter to increase membrane permeability to glyphosate (43). Betaine was added to provide an intracellular osmolyte to support growth in the presence of high concentrations of glyphosate (44). Later, the pH of the agar medium was reduced from 7.0 to 5.5, under the hypothesis that a larger fraction of glyphosate would be protonated and thus be more likely to be transported by proton cotransporters.
The plates were incubated at 37 ºC until colonies grew to a size conducive for automated picking. Saturation mutagenesis generates a small library of 19 variants at each position. For screening, 96 positional libraries were pooled, combining for roughly 2000 variants per pool, easily over-sampled by plating on selective medium. For combinatorial libraries, sufficient cell transformation reactions were performed to generate as many colony-forming units as theoretical unique members in the library, typically ∼106, all of which were plated and screened.
At the fitness level attained by variant D2, two more adaptations were made to increase stringency of the plate selection. The origin of replication in the expression vector was replaced with that from pSC101, which typically generates ∼5 copies of the plasmid instead of ∼20 (45). In the final stages, vector DNA encoding the libraries was transformed into BL21(DE3)-C41 cells. In that strain, mutations in the promoter of T7 RNA polymerase results in lower levels of induced, and presumably uninduced, expression (46).
EPSPS production and purification
Proteins were produced in E. coli strain BL21(DE3). Transformed cells were grown in Magic Medium (Thermo Fisher Scientific). After 4 h of growth at 37 ºC, cells were transferred to 16 ºC and grown another 48 h. Pelleted cells were lysed with BPER (Pierce) protein extraction reagent containing 25 mm Tris (to elevate pH to 7.8), 0.2 mg/ml of lysozyme, 1 mm DTT, protease inhibitor mixture (Sigma, bacterial mixture), and endonuclease. Insoluble cellular debris was removed by centrifugation. EPSPS protein was purified from the soluble protein solution by affinity chromatography on the nickel form of nitrilotriacetic acid resin (Qiagen). Protein concentration was measured by absorbance at 280 nm using the molar extinction coefficient calculated by Vector NTI for the amino acid sequence of each variant, including the additional N-terminal amino acids coded on the vector.
EPSPS assay
Activity was determined by quantifying release of Pi, coupled to reaction with MESG, catalyzed by purine-nucleoside phosphorylase (49). The absorbance change that occurs was monitored with a Spectramax plate reader (Molecular Devices) at 360 nm, where the extinction is 11,200 m−1 cm−1. Reaction mixtures contained 25 mm Hepes, pH 7, 100 mm KCl, 5% (v/v) ethylene glycol, 0.15 mm MESG, 1.5 unit/ml of purine nucleoside phosphorylase (Sigma N8264) and concentrations of EPSPS customized to result in linear initial reaction rates. In addition to these common components, reaction mixtures contained substrates and glyphosate as required by the fitness of the variant. To determine kinetic parameters with substrate saturation analysis, the varied substrate was present at eight concentrations typically ranging from zero (blank) to 800 μm, but as high as 3.2 mm for G101A, and the unvaried substrate was supplied at the saturating concentration of 10 times its Km. Five microliters of 60-fold concentrated stock solutions of the varied substrate were placed in the wells of the 96-well assay plate and reactions were started with the addition of 295 μl of a mixture of all other components. Reactions were observed at 360 nm for 1 min, taking readings at 4-s intervals.
Data collection and analysis
All the data in Tables 1 and 2 were obtained from a single panel of purified enzymes prepared together and meeting the purity criterion of no additional bands on a Coomassie Blue-stained SDS gel loaded with 3 μg of each enzyme. The initial, linear reaction rate was captured using the first 6 or 7 readings. Data from runs in which a linear initial rate was not captured due to excessive enzyme concentration were excluded. Kinetic parameters for S3P were obtained by nonlinear regression analysis (GraphPad Prism) of at least four sets of rate measurements at 7 substrate concentrations, fitted to the Michaelis-Menten equation. For PEP, the procedure was repeated with one or two concentrations of glyphosate and the data were processed by nonlinear regression analysis fitted to the Michaelis-Menten equation for competitive inhibition.
Author contributions
Y. D., E. C. N., T. K. F., Y. T., and D. L. S. data curation; Y. D., E. C. N., J. L., T. K. F., M. S., and D. L. S. investigation; Y. D., E. C. N., J. L., T. K. F., Z. H., and D. L. S. methodology; Y. D., S. B., P. P., M. W. L., and D. L. S. project administration; Y. D., E. C. N., J. L., T. K. F., Y. T., S. B., M. S., E. B., Z. H., P. P., M. W. L., and D. L. S. writing-review and editing; J. L. validation; Y. T., S. B., E. B., P. P., M. W. L., and D. L. S. conceptualization; E. B., P. P., M. W. L., and D. L. S. supervision; Z. H. resources; Z. H. and D. L. S. visualization; D. L. S. writing-original draft.
Supplementary Material
Acknowledgments
We thank Jarred Oral for suggesting the use of BL-21(DE3)-C41 for higher stringency selection, and Dr. Steve Bass for coaching on P1 viral transduction for creating the AroA knockout E. coli strain.
The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S6.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- EPSPS
- 5-enolpyruvylshikimate-3-phosphate synthase
- PEP
- phosphoenolpyruvate
- PDB
- Protein Data Bank
- S3P
- shikimate-3-phosphate
- MESG
- 2-amino-6-mercapto-7-methylpurine ribonucleoside.
References
- 1. Boocock M. R., and Coggins J. R. (1983) Kinetics of 5-enolpyruvylshikimate-3-phosphate synthase inhibition by glyphosate. FEBS Lett. 154, 127–133 10.1016/0014-5793(83)80888-6 [DOI] [PubMed] [Google Scholar]
- 2. Schönbrunn E., Eschenburg S., Shuttleworth W. A., Schloss J. V., Amrhein N., Evans J. N., and Kabsch W. (2001) Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail. Proc. Natl. Acad. Sci. U.S.A. 98, 1376–1380 10.1073/pnas.98.4.1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Franz J. E., Mao M. K., and Sikorski J. A. (1997) Glyphosate: A unique global herbicide, American Chemical Society, Washington, D. C. [Google Scholar]
- 4. Solomon K. (2017) What is the problem with glyphosate? Outlooks on Pest Management 28, 173–174 10.1564/v28_aug_08 [DOI] [Google Scholar]
- 5. Duke S. O., and Powles S. B. (2008) Glyphosate: a once-in-a-century herbicide. Pest Manag. Sci. 64, 319–325 10.1002/ps.1518 [DOI] [PubMed] [Google Scholar]
- 6. Nielsen L. N., Roager H. M., Casas M. E., Frandsen H. L., Gosewinkel U., Bester K., Licht T. R., Hendriksen N. B., and Bahl M. I. (2018) Glyphosate has limited short-term effects on commensal bacterial community composition in the gut environment due to sufficient aromatic amino acid levels. Environ. Pollut. 233, 364–376 10.1016/j.envpol.2017.10.016 [DOI] [PubMed] [Google Scholar]
- 7. Tarone R. E. (2018) Conflicts of interest, bias, and the IARC Monographs Program. Regul. Toxicol. Pharmacol. 98, A1–A4 10.1016/j.yrtph.2018.09.005 [DOI] [PubMed] [Google Scholar]
- 8. Heck G. R., Armstrong C. L., Astwood J. D., Behr C. F., Bookout J. T., Brown S. M., Cavato T. A., DeBoer D. L., Deng M. Y., George C., Hillyard J. R., Hironaka C. M., Howe A. R., Jakse E. H., Ledesma B. E., et al. (2005) Development and characterization of a CP4 EPSPS-based, glyphosate-tolerant corn event. Crop Sci. 45, 329–339 10.2135/cropsci2005.0329 [DOI] [Google Scholar]
- 9. Padgette S. R., Kolacz K. H., Delanny X., Re D. B., AlVallee B. J., Tinius C. N., Rhodes W. K., Otero Y. I., Barry G. F., Eichholtz D. A., Peschke V. M., Nida D. L., Taylor N. B., and Kishore G. M. (1995) Development, identification, and characterization of a glyphosate-tolerant soybean line. Crop Sci. 35, 1451–1461 10.2135/cropsci1995.0011183X003500050032x [DOI] [Google Scholar]
- 10. Sander J. D., and Joung J. K. (2014) CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 32, 347–355 10.1038/nbt.2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gianessi L. P. (2013) The increasing importance of herbicides in worldwide crop production. Pest Manag. Sci. 69, 1099–1105 10.1002/ps.3598 [DOI] [PubMed] [Google Scholar]
- 12. Healy-Fried M. L., Funke T., Priestman M. A., Han H., and Schönbrunn E. (2007) Structural basis of glyphosate tolerance resulting from mutations of Pro-101 in Escherichia coli 5-enolpyruvylshikimate-3-phosphate synthase. J. Biol. Chem. 282, 32949–32955 10.1074/jbc.M705624200 [DOI] [PubMed] [Google Scholar]
- 13. Stalker D. M., Hiatt W. R., and Comai L. (1985) A single amino acid substitution in the enzyme 5-enolpyruvylshikimate-3-phosphate synthase confers resistance to the herbicide glyphosate. J. Biol. Chem. 260, 4724–4728 [PubMed] [Google Scholar]
- 14. Sammons R. D., and Gaines T. A. (2014) Glyphosate resistance: state of knowledge. Pest Manag. Sci. 70, 1367–1377 10.1002/ps.3743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ngo T. D., Krishnan M., Boutsalis P., Gill G., and Preston C. (2018) Target-site mutations conferring resistance to glyphosate in feathertop Rhodes grass (Chloris virgata) populations in Australia. Pest Manag. Sci. 74, 1094–1100 10.1002/ps.4512 [DOI] [PubMed] [Google Scholar]
- 16. Chen J., Huang H., Zhang C., Wei S., Huang Z., Chen J., and Wang X. (2015) Mutations and amplification of EPSPS gene confer resistance to glyphosate in goosegrass (Eleusine indica). Planta 242, 859–868 10.1007/s00425-015-2324-2 [DOI] [PubMed] [Google Scholar]
- 17. Baerson S. R., Rodriguez D. J., Tran M., Feng Y., Biest N. A., and Dill G. M. (2002) Glyphosate-resistant goosegrass: identification of a mutation in the target enzyme 5-enolpyruvylshikimate-3-phosphate synthase. Plant Physiol. 129, 1265–1275 10.1104/pp.001560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Funke T., Yang Y., Han H., Healy-Fried M., Olesen S., Becker A., and Schönbrunn E. (2009) Structural basis of glyphosate resistance resulting from the double mutation Thr-97 → Ile and Pro-101 → Ser in 5-enolpyruvylshikimate-3-phosphate synthase from Escherichia coli. J. Biol. Chem. 284, 9854–9860 10.1074/jbc.M809771200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu Q., Jalaludin A., Han H., Chen M., Sammons R. D., and Powles S. B. (2015) Evolution of a double amino acid substitution in the 5-enolpyruvylshikimate-3-phosphate synthase in Eleusine indica conferring high-level glyphosate resistance. Plant Physiol. 167, 1440–1447 10.1104/pp.15.00146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Spencer M., Mumm R. and Gwynn J. (March 21, 2000) Glyphosate resistant maize lines. U. S. Patent 6040497
- 21. Li J., Peng Q., Han H., Nyporko A., Kulynych T., Yu Q., and Powles S. (2018) Glyphosate resistance in Tridax procumbens via a novel EPSPS Thr-102-Ser substitution. J. Agric. Food Chem. 66, 7880–7888 10.1021/acs.jafc.8b01651 [DOI] [PubMed] [Google Scholar]
- 22. Sost D., and Amrhein N. (1990) Substitution of Gly-96 to Ala in the 5-enolpyruvylshikimate-3-phosphate synthase of Klebsiella pneumoniae results in a greatly reduced affinity for the herbicide glyphosate. Arch. Biochem. Biophys. 282, 433–436 10.1016/0003-9861(90)90140-T [DOI] [PubMed] [Google Scholar]
- 23. Sikorski J. A., and Gruys K. J. (1997) Understanding glyphosate's molecular mode of action with EPSP synthase: evidence favoring an allosteric inhibitor model. Acc. Chem. Res. 30, 2–8 10.1021/ar950122 [DOI] [Google Scholar]
- 24. Funke T., Han H., Healy-Fried M. L., Fischer M., and Schönbrunn E. (2006) Molecular basis for the herbicide resistance of Roundup Ready crops. Proc. Natl. Acad. Sci. U.S.A. 103, 13010–13015 10.1073/pnas.0603638103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Padgette S. R., Re D. B., Gasser C. S., Eichholtz D. A., Frazier R. B., Hironaka C. M., Levine E. B., Shah D. M., Fraley R. T., and Kishore G. M. (1991) Site-directed mutagenesis of a conserved region of the 5-enolpyruvylshikimate-3-phosphate synthase active site. J. Biol. Chem. 266, 22364–22369 [PubMed] [Google Scholar]
- 26. Pollegioni L., Schonbrunn E., and Siehl D. (2011) Molecular basis of glyphosate resistance-different approaches through protein engineering. FEBS J. 278, 2753–2766 10.1111/j.1742-4658.2011.08214.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ness J. E., Kim S., Gottman A., Pak R., Krebber A., Borchert T. V., Govindarajan S., Mundorff E. C., and Minshull J. (2002) Synthetic shuffling expands functional protein diversity by allowing amino acids to recombine independently. Nat. Biotechnol. 20, 1251–1255 10.1038/nbt754 [DOI] [PubMed] [Google Scholar]
- 28. Eschenburg S., Healy M. L., Priestman M. A., Lushington G. H., and Schönbrunn E. (2002) How the mutation glycine96 to alanine confers glyphosate insensitivity to 5-enolpyruvyl shikimate-3-phosphate synthase from Escherichia coli. Planta 216, 129–135 10.1007/s00425-002-0908-0 [DOI] [PubMed] [Google Scholar]
- 29. Lu J., Dong Y., Ng E. C., and Siehl D. L. (2017) Novel form of the Michaelis-Menten equation that enables accurate estimation of (kcat/KM)*KI with just two rate measurements; utility in directed evolution. Protein Eng. Des. Sel. 30, 395–399 10.1093/protein/gzx012 [DOI] [PubMed] [Google Scholar]
- 30. Kirkwood R., Hetherington R., Reynolds T. L., and Marshall G. (2000) Absorption, localisation, translocation and activity of glyphosate in barnyardgrass (Echinochloa crus-galli (L) Beauv): influence of herbicide and surfactant concentration. Pest Manag. Sci. 56, 359–367 10.1002/(SICI)1526-4998(200004)56:4%3C359::AID-PS145%3E3.0.CO%3B2-S [DOI] [Google Scholar]
- 31. Kronzucker H. J., Szczerba M. W., and Britto D. T. (2003) Cytosolic potassium homeostasis revisited: 42K-tracer analysis in Hordeum vulgare L. reveals set-point variations in [K+]. Planta 217, 540–546 10.1007/s00425-003-1032-5 [DOI] [PubMed] [Google Scholar]
- 32. Cuin T. A., Miller A. J., Laurie S. A., and Leigh R. A. (2003) Potassium activities in cell compartments of salt-grown barley leaves. J. Exp. Bot. 54, 657–661 10.1093/jxb/erg072 [DOI] [PubMed] [Google Scholar]
- 33. Zhou M., Xu H., Wei X., Ye Z., Wei L., Gong W., Wang Y., and Zhu Z. (2006) Identification of a glyphosate-resistant mutant of rice 5-enolpyruvylshikimate 3-phosphate synthase using a directed evolution strategy. Plant Physiol. 140, 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Han H., Vila-Aiub M. M., Jalaludin A., Yu Q., and Powles S. B. (2017) A double EPSPS gene mutation endowing glyphosate resistance shows a remarkably high resistance cost. Plant Cell Environ. 40, 3031–3042 10.1111/pce.13067 [DOI] [PubMed] [Google Scholar]
- 35. Chauhan R. D., Hummel A., Cermak T., Starker C., Bart R., Voytas D., and Taylor N. (2017) Generation of glyphosate tolerant cassava plants through CRISPR/Cas9-mediated gene editing. In Vitro Cell Dev. Biol.—Animal 53, (Suppl. 1) S30–S38 [Google Scholar]
- 36. Hummel A. W., Chauhan R. D., Cermak T., Mutka A. M., Vijayaraghavan A., Boyher A., Starker C. G., Bart R., Voytas D. F., and Taylor N. J. (2018) Allele exchange at the EPSPS locus confers glyphosate tolerance in cassava. Plant Biotechnol. J. 16, 1275–1282 10.1111/pbi.12868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li J., Meng X., Zong Y., Chen K., Zhang H., Liu J., Li J., and Gao C. (2016) Gene replacements and insertions in rice by intron targeting using CRISPR-Cas9. Nat. Plants 2, 16139 10.1038/nplants.2016.139 [DOI] [PubMed] [Google Scholar]
- 38. Sost D., Schultz A., and Amrhein N. (1984) Characterization of a glyphosate-insensitive 5-enolpyruvylshikimic acid-3-phosphate synthase. FEBS Lett. 173, 238–242 10.1016/0014-5793(84)81054-6 [DOI] [Google Scholar]
- 39. Barry G. F., Kishore G. M., Padgette S. R., and Stallings W. C. (May 27, 1997) Glyphosate-tolerant 5-enolpyruvylshikimate-3-phosphate synthases. U. S. Patent 5633435
- 40. Mousdale D. M., and Coggins J. R. (1984) Purification and properties of 5-enolpyruvylshikimate 3-phosphate synthase from seedlings of Pisum sativum L. Planta 160, 78–83 10.1007/BF00392469 [DOI] [PubMed] [Google Scholar]
- 41. Morrison J. F. (1982) The slow-binding and slow, tight-binding inhibition of enzyme-catalysed reactions. Trends Biochem. Sci. 7, 102–105 10.1016/0968-0004(82)90157-8 [DOI] [Google Scholar]
- 42. Weiss U., Davis B. D., and Mingioli E. S. (1953) Aromatic biosynthesis X: identification of an early precursor as 5-dehydroquinic acid. J. Am. Chem Soc. 75, 5572–5576 10.1021/ja01118a028 [DOI] [Google Scholar]
- 43. Vaara M. (1992) Agents that increase the permeability of the outer membrane. Microbiol. Rev. 56, 395–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Record M. T. Jr., Courtenay E. S., Cayley D. S., and Guttman H. J. (1998) Responses of E. coli to osmotic stress: large changes in amounts of cytoplasmic solutes and water. Trends Biochem. Sci. 23, 143–148 10.1016/S0968-0004(98)01196-7 [DOI] [PubMed] [Google Scholar]
- 45. Manen D., and Caro L. (1991) The replication of plasmid pSC101. Mol. Microbiol. 5, 233–237 10.1111/j.1365-2958.1991.tb02103.x [DOI] [PubMed] [Google Scholar]
- 46. Schlegel S., Genevaux P., and de Gier J. W. (2015) De-convoluting the genetic adaptations of E. coli C41(DE3) in real time reveals how alleviating protein production stress improves yields. Cell Rep. 10, 1758–1766 10.1016/j.celrep.2015.02.029 [DOI] [PubMed] [Google Scholar]
- 47. Emanuelsson O., Nielsen H., and von Heijne G. (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984 10.1110/ps.8.5.978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Introduction of organisms and products altered or produced through genetic engineering which are plant pests or which there is reason to believe are plant pests. 7 C.F.R. §340.1 (2018) [Google Scholar]
- 49. Webb M. R. (1992) A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc. Natl. Acad. Sci. U.S.A. 89, 4884–4887 10.1073/pnas.89.11.4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.