Abstract
Disruption of circadian rhythms has been implicated in an increased risk for cancer development. The Period2 (Per2) gene encodes one of the major components of the mammalian circadian clock, which plays a key role in controlling the circadian rhythms in physiology and behavior. PER2 has also been reported to suppress the malignant transformation of cells, but its role in the regulation of cancer susceptibility to chemotherapeutic drugs remains unclear. In this study, we found that oncogene-transformed embryonic fibroblasts prepared from Per2-mutant (Per2m/m) mice, which are susceptible to both spontaneous and radiation-induced tumorigenesis, were resistant against common chemotherapeutic drugs and that this resistance is associated with up-regulation of the aldehyde dehydrogenase 3a1 (Aldh3a1) gene. Co-expression of the oncogenes H-rasV12 and SV40 large T-antigen induced malignant transformation of both WT and Per2m/m cells, but the cytotoxic effects of the chemotherapeutic agents methotrexate, gemcitabine, etoposide, vincristine, and oxaliplatin were significantly alleviated in the oncogene-transformed Per2m/m cells. Although introduction of the two oncogenes increased the expression of Aldh3a1 in both WT and Per2m/m cells, the ALDH3A1 protein levels in the Per2m/m cells were ∼7-fold higher than in WT cells. The elevated ALDH3A1 levels in the oncogene-transformed Per2m/m cells were sufficient to prevent chemotherapeutic drug–induced accumulation of reactive oxygen species. Consequently, shRNA-mediated suppression of Aldh3a1 expression relieved the chemoresistance of the Per2m/m cells. These results suggest a role for mutated PER2 in the development of multiple drug resistance and may inform therapeutic strategies for cancer management.
Keywords: clock gene, chemoresistance, anticancer drug, chromatin modification, reactive oxygen species (ROS), Aldh3a1, Period2, chemotherapy, oncogene, circadian rhythm, cancer, aldehyde dehydrogenase
Introduction
The rotation of the Earth with a period length of about 24 h has led to the evolution of an endogenous timing system within a large number of species, the circadian clock, which allows organisms to adapt their physiological and behavioral functions to anticipatory changes in their environment. In mammals, circadian rhythms in physiological functions are generated by a molecular oscillator driven by a transcriptional–translational feedback loop consisting of negative and positive regulators (1). The gene products of Clock and Bmal1 (also known as Arntl) form a heterodimer that acts as a positive transcription factor to activate the transcription of the Period (Per) and cryptochrome (Cry) genes. Once the PER and CRY proteins have reached a critical concentration, they act as negative transcription factors to attenuate CLOCK/BMAL1-mediated transactivation (2, 3). Rev-erbα (known as Nrd1d1) is also activated by CLOCK/BMAL1 and transrepressed by PER and CRY, resulting in circadian oscillation in the expression of Rev-erbα (4). In turn, REV-ERBα periodically represses Bmal1 transcription, thereby interconnecting the positive and negative loops (5). Like the mechanism of Rev-erbα transcription, clock genes, which comprise the core oscillation loop, control rhythmic RNA and protein abundance (6–8) and also allow organisms to synchronize their physiological and behavioral functions to anticipatory changes in their environment.
Because the expression of up to 10% of genes has been suggested to be under the control of the circadian clock (9), it should not come as a surprise that disruptions in the circadian clock system lead to the onset of various diseases. In fact, several epidemiological analyses and laboratory animal studies have revealed a relationship between disruptions in circadian rhythms and cancer development. For example, long-term shift workers are at an increased risk of developing breast, prostate, colon, and endometrial cancers, as well as non-Hodgkin lymphoma (10–12). These epidemiological findings are supported by animal studies in which repetitive changes in the light–dark cycle are found to facilitate the growth of implanted tumors (13, 14). Furthermore, genetic ablation of the circadian clock gene also enhances the tumorigenesis in the laboratory animals. PER2 is an essential component of mammalian circadian clock (15). Mice with a mutated Per2 gene (Per2m/m) are predisposed to spontaneous as well as radiation-induced tumor development (16). We also demonstrated previously that embryonic fibroblasts prepared from Per2m/m mice were susceptible to transformation induced by the co-expression of H-rasV12 and SV40 large T-antigen (SV40LT), and the oncogene-transformed Per2m/m cells have a high tumor formation potential (17). However, the role of the Per2 gene in the regulation of cellular chemosensitivity remains unclear.
In this study, we found that the cytotoxic effects of common chemotherapeutic drugs were diminished in oncogene-transformed Per2m/m cells. Expression of the aldehyde dehydrogenase 3a1 (Aldh3a1) gene in Per2m/m cells was remarkably increased by the introduction of oncogenes, and potent elevation of its enzymatic activity attenuated the cytotoxicity of chemotherapeutic drugs. Collectively, the results of the present study suggest a role for PER2 in the development of multiple drug resistance and offer new insights into therapeutic strategies for the treatment of cancers.
Results
Oncogene-transformed Per2m/m cells resist the cytotoxicity of chemotherapeutic drugs
We previously reported the preparation of oncogene-transformed WT and Per2m/m cells that were infected concomitantly with retrovirus vectors expressing H-rasV12 and SV40LT (17). The expression of mRNAs for these oncogenes was detected on day 3 after infection, and they were equally expressed in both types of cells (17). The concomitant introduction of H-rasV12 and SV40LT significantly enhanced the anchorage-independent growth of WT and Per2m/m cells (17). Therefore, we used these cells to investigate the role of the Per2 gene in the regulation of susceptibility of cells to chemotherapeutic drugs, methotrexate (MTX),3 gemcitabine (GEM), etoposide (VP-16), oxaliplatin (L-OHP), and vincristine (VCR).
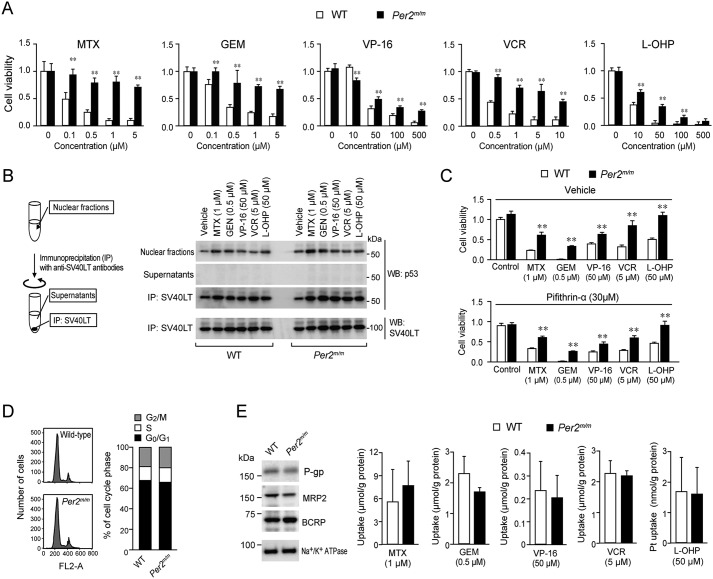
The viability of oncogene-transformed WT cells was dose-dependently decreased by treatment with all five types of chemotherapeutic drugs (Fig. 1A). The IC50 values of WT cells to MTX, GEM, VP-16, VCR, and L-OHP were 0.12, 0.27, 26.02, 0.39, and 4.32 μm, respectively. Similar dose-dependent decreases in viability were observed when oncogene-transformed Per2m/m cells were treated with MTX, GEM, VP-16, VCR, and L-OHP, but the cytotoxic effect of all five chemotherapeutic drugs on Per2m/m cells was attenuated as compared with those on WT cells. We prepared oncogene-transformed cells three times. In every preparation, the chemosensitivity of Per2m/m cells was lower than that of WT cells.
Figure 1.
Differences in the sensitivity to chemotherapeutic drugs between oncogene-transformed WT and Per2m/m cells. A, cells were treated with MTX, GEM, VP-16, VCR, or L-OHP at the indicated concentrations. After treatment for 48 h, cell viability was determined by an ATP luminescent assay. Values are means ± S.D. (error bars) (n = 3–4). **, p < 0.01 significantly different from WT cells. B, oncogene-transformed cells were treated with chemotherapeutic drugs for 24 h. Thereafter, cells were lysed and then subjected to immunoprecipitation with anti-SV40LT antibodies. The amounts of p53 and SV40LT in cell lysates, supernatants, and immunocomplexes were detected by Western blotting (WB). C, influence of p53 inhibitor pifithrin-α on the sensitivity of oncogene-transformed WT and Per2m/m cells to chemotherapeutic drugs. Cells were treated with chemotherapeutic drugs at the indicated concentrations in the presence or absence of pifithrin-α (30 μm). Cell viability was assessed 48 h after the initiation of drug treatment. Values are means ± S.D. (n = 4). **, p < 0.01 significantly different from WT cells. D, the analysis of cell-cycle distribution of oncogene-transformed WT and Per2m/m cells. There was no significant difference in the distribution of cell-cycle phase between the two genotypes (G2/M-phase, 22.01 ± 3.99% for WT and 23.02 ± 6.95% for Per2m/m; S-phase, 12.61 ± 4.26% for WT and 14.26 ± 2.78% for Per2m/m; G0/G1-phase, 65.38 ± 4.59% for WT and 62.73 ± 4.56% for Per2m/m; n = 4, means ± S.D.). E, left panel shows the expression profiles of P-gp, MRP2, and BCRP in oncogene-transformed WT and Per2m/m cells. Na+/K+-ATPase was used for membrane protein-loading control. The results shown are representative of three independent experiments. The right panels show intracellular accumulation of chemotherapeutic drugs in oncogene-transformed WT and Per2m/m cells. Concentrations of drugs were determined until 2 h after the addition of the drugs into the medium. Values shown are means ± S.D. (n = 3–4).
p53 acts as a universal sensor of genotoxic stress and plays a critical role in chemotherapeutic drug-induced apoptotic cell death (18, 19). However, SV40LT-transduced cells are immortalized by inactivation of p53 through protein–protein interaction (20). After treatment with chemotherapeutic drugs, p53 protein was accumulated in the nuclear fraction of oncogene-introduced WT and Per2m/m cells. The results of an immunoprecipitation analysis revealed that the greatest amounts of p53 protein in WT and Per2m/m cells were precipitated together with SV40LT (Fig. 1B), suggesting that p53 is unlikely to be involved in the chemotherapeutic drug–induced decrease in the viability of cells that were infected with H-rasV12 and SV40LT. In fact, treatment of oncogene-transformed WT and Per2m/m cells with 30 μm pifithrin-α, an inhibitor of p53-mediated transcription, was also unable to modulate their chemosensitivity (Fig. 1C). Because 30 μm pifithrin-α is sufficient to enhance the chemosensitivity of several types of cancer cell lines (21–23), SV40LT seemed to inactivate p53 in oncogene-transformed WT and Per2m/m cells.
The sensitivity of cells to chemotherapeutic drugs is also thought to be dependent on cell-cycle phase, but comparison of flow cytometry histograms from oncogene-transformed WT and Per2m/m cells revealed no significant difference in the cell-cycle distribution between the genotypes (Fig. 1D). These results suggest that apoptotic process and cell-cycle phase are unlikely to contribute to the diminished chemosensitivity of Per2m/m cells.
Elevated expression of several ABC transporters is often associated with multidrug resistance (24, 25). However, the levels of P-glycoprotein (P-gp), multidrug resistance–associated protein-2 (MRP2), and breast cancer–resistant protein (BCRP) in oncogene-transformed Per2m/m cells were comparable with those expressed in WT cells (Fig. 1E, left). Intracellular accumulation of MTX, GEM, VP-16, VCR, and L-OHP was also not significantly different between WT and Per2m/m cells (Fig. 1E, right panels), suggesting that the tolerance of the oncogene-transformed Per2m/m cells to the chemotherapeutic drugs is not due to the function of efflux transporters.
Up-regulation of Aldh3a1 in oncogene-transformed Per2m/m cells
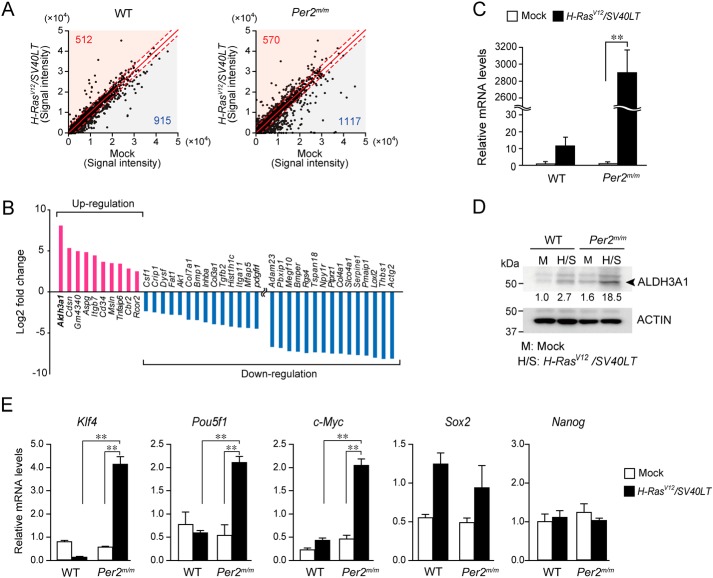
To investigate the underlying mechanism of the chemoresistance of oncogene-transformed Per2m/m cells, we carried out microarray analysis to identify the gene regulating the susceptibility of Per2m/m cells to chemotherapeutic drugs. After concomitant introduction of H-rasV12 and SV40LT, 1,427 genes were induced or repressed in WT cells (Fig. 2A). Similarly, expression of 1,687 genes in Per2m/m cells were altered by oncogenic stimuli. A total of 73 genes in oncogene-transformed Per2m/m cells were differentially expressed as compared with those in WT cells (Table S1). Of these differentially expressed genes, the expression of 10 genes in oncogene-transformed Per2m/m cells was higher than that in WT cells, whereas the expression of 63 genes was lower in oncogene-transformed Per2m/m cells. Among the differentially regulated genes, we identified Aldh3a1 as the up-regulated gene in oncogene-transformed Per2m/m cells with the greatest -fold change (Fig. 2B). Elevated expression of Aldh3a1 mRNA in oncogene-transformed Per2m/m cells was also confirmed by RT-qPCR analysis (Fig. 2C). The results of Western blot analysis revealed that ALDH3A1 levels in Per2m/m cells were ∼7-fold higher than in WT cells (Fig. 2D). High ALDH activity is often detected in cells with stemlike properties (26). Our previous study demonstrated that oncogene-transformed Per2m/m cells have potent tumor formation ability (17). Indeed, the expression levels of known cancer stemness markers (Kruppel-like factor 4 (Klf4); POU domain, class 5, transcription factor 1 (Pou5f1); and c-Myc) in oncogene-transformed Per2m/m cells were significantly higher than those in WT cells (p < 0.01, respectively; Fig. 2E), confirming that oncogene-transformed Per2m/m cells have stemlike properties.
Figure 2.
Enhanced expression of Aldh3a1 gene in oncogene-transformed Per2m/m cells. A, microarray analysis of oncogene introduced WT and Per2m/m cells. Up-regulated or down-regulated genes in WT and Per2m/m cells with concomitant introduction of oncogenes H-rasV12 and SV40LT. The criteria for up-regulated genes were set at a z-score of 2.0 or more and a ratio of 3-fold or more; and the criteria for down-regulated genes were set at a z-score of −2.0 or less and a ratio of one-third or less. The numbers at the top left in the graphs indicate up-regulated genes, and those at the bottom right indicate down-regulated genes. B, differentially expressed genes between oncogene-introduced WT and Per2m/m cells in microarray analysis. The full transcriptome data have been deposited in NCBI GEO (accession number GSE113242). C, elevation of Aldh3a1 mRNA levels in oncogene-transformed Per2m/m cells. Values shown are means ± S.D. (error bars) (n = 3). **, p < 0.01, significantly different between two groups. D, elevation of ALDH3A1 protein levels in oncogene-transformed Per2m/m cells. The results shown are representative of three independent experiments. The band intensity was plotted by normalizing to actin, and the mean value of basal levels was set as 1.0. The values are plotted in the photographs of Western blotting. E, the expression levels of Klf4, Pou5f1, c-Myc, Sox2, and Nanog in WT and Per2m/m cells before and after oncogenic transformation. Data were normalized by actin mRNA levels. Values show the mean ± S.D. (n = 3–5). **, p < 0.01, significantly different between the two groups.
Elevated ALDH3A1 attenuates the cytotoxic effects of chemotherapeutic drugs in oncogene-transformed Per2m/m cells through the prevention of H2O2 accumulation
High ALDH activity in cancer cells is often relevant to their resistance against chemotherapy (27). Therefore, we further focused on this enzyme and investigated its role in the regulation of cellular chemosensitivity.
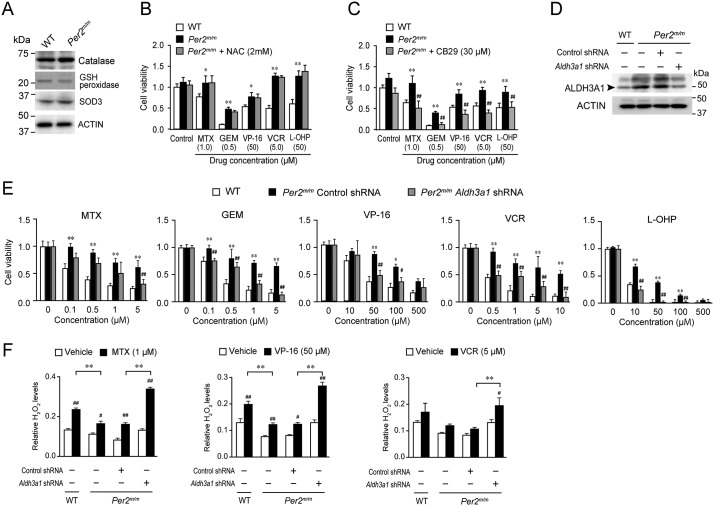
The expressions of antioxidant degradation enzymes, catalase, GSH peroxidase, and superoxide dismutase-3 (SOD3), were not significantly different between WT and Per2m/m cells (Fig. 3A). Furthermore, N-acetylcysteine (NAC), an antioxidant precursor to GSH, had a negligible effect on the chemosensitivity of oncogene-transformed Per2m/m cells (Fig. 3B). In contrast to these observations, treatment of oncogene-transformed Per2m/m cells with 30 μm CB29, a selective ALDH3A1 inhibitor, significantly restored their sensitivity to MTX, GEM, VP-16, VCR, and L-OHP (p < 0.01, respectively; Fig. 3C).
Figure 3.
Down-regulation of ALDH3A1 relieves the resistance of oncogene-transformed Per2m/m cells against chemotherapeutic drugs. A, the expression profiles of catalase, GSH peroxidase, and SOD3 in oncogene-transformed WT and Per2m/m cells. The results shown are representative of three independent experiments. B, effects of NAC, a GSH precursor, on the sensitivity of oncogene-transformed Per2m/m cells to chemotherapeutic drugs. Cells were treated with 1 μm MTX, 0.5 μm GEM, 50 μm VP-16, 5 μm VCR, or 50 μm L-OHP at the indicated concentrations in the presence or absence of NAC (2 mm). After treatment with each drug for 48 h, cell viability was determined by an ATP luminescent assay. Values are means ± S.D. (error bars) (n = 4). **, p < 0.01, significantly different from WT cells. C, effects of selective ALDH3A1 inhibitor CB29 on the sensitivity of oncogene-transformed Per2m/m cells to chemotherapeutic drugs. Cells were treated with chemotherapeutic drugs in the presence or absence of CB29 (30 μm). Cell viability was assessed 48 h after the initiation of drug treatment. Values are means ± S.D. (n = 4). **, p < 0.01, significantly different from WT cells. ##, p < 0.01, significantly different from Per2m/m cells without treatment with CB29. D, down-regulation of ALDH3A1 in oncogene-transformed Per2m/m cells by infection with shRNA-expressing vectors. E, effects of chemotherapeutic drugs on viability of Aldh3a1 down-regulated oncogene-transformed Per2m/m cells. Cell viability was assessed 48 h after the initiation of drug treatment. Values shown are means ± S.D. (n = 3–4). **, p < 0.01; *, p < 0.05, significantly different from WT cells. ##, p < 0.01; #, p < 0.05, significantly different from control shRNA-transfected oncogene-transformed Per2m/m cells (Per2m/m Control shRNA). F, influence of down-regulation of ALDH3A1 on the chemotherapeutic drug–induced production of ROS in oncogene-transformed Per2m/m cells. Values shown are means ± S.D. (n = 3). **, p < 0.01, significantly different between the two groups. ##, p < 0.01; #, p < 0.05, significantly different from the vehicle-treated group.
To further investigate the role of ALDH3A1 in the regulation of cellular chemosensitivity, we prepared oncogene-transformed Per2m/m cells with down-regulated expression of ALDH3A1. Infection of oncogene-transformed Per2m/m cells with retrovirus vectors expressing shRNA against Aldh3a1 caused a reduction of its protein levels (Fig. 3D). Down-regulation of ALDH3A1 in oncogene-transformed Per2m/m cells also restored their susceptibility to the chemotherapeutic drugs, with their susceptibilities becoming similar to those observed in oncogene-transformed WT cells (Fig. 3E). These results suggest that elevated expression of ALDH3A1 in oncogene-transformed Per2m/m cells attenuates the cytotoxicity of chemotherapeutic drugs.
Chemotherapeutic drug-induced DNA damage ultimately causes cell death via enhanced production of reactive oxygen species (ROS) (28). High ALDH activity protects cells from the cytotoxic effect of chemotherapeutic drugs through degradation of ROS (29). After treatment with 1 μm MTX, 50 μm VP-16, or 5 μm VCR, H2O2 accumulated in both oncogene- transformed WT and Per2m/m cells (Fig. 3F), but the accumulation of H2O2 in oncogene-transformed Per2m/m cells was lower than that in WT cells. Down-regulation of ALDH3A1 in oncogene-transformed Per2m/m cells restored the chemotherapeutic drug-induced ROS accumulation (Fig. 3F).
Histone modifications in Aldh3a1 gene of oncogene-transformed Per2m/m cells
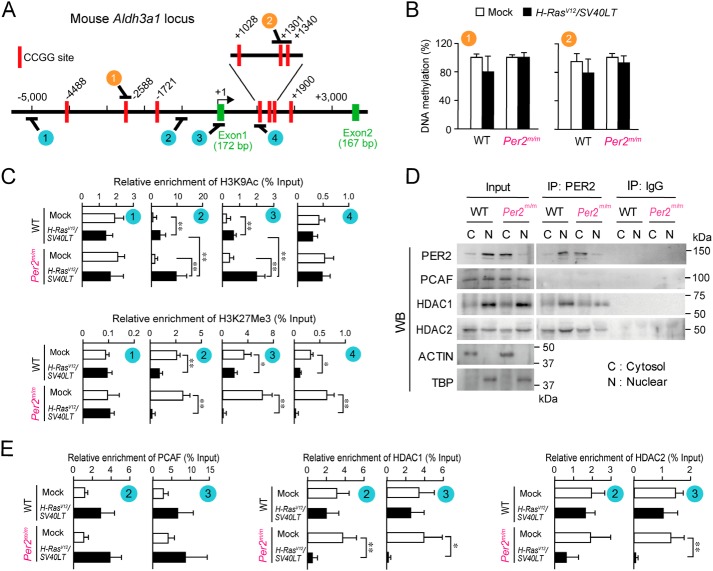
A large number of genes was induced or repressed by concomitant introduction of H-rasV12 and SV40LT (Fig. 2A). Alterations of gene expression in oncogenic cells are often associated with epigenetic modifications (30). To investigate the underlying mechanism of the up-regulation of Aldh3a1 gene in oncogene- transformed Per2m/m cells, we assessed the DNA methylation status of Aldh3a1. The upstream region of the mouse Aldh3a1 gene was retrieved by using an on-line genome browser hosted by the University of California, Santa Cruz (http://genome.ucsc.edu/index.html)4 (49). Although there were no significant CpG islands within the 5,000 bp up- and downstream from the transcription start site of the mouse Aldh3a1 gene, several 5′-CCGG-3′ sequences were located in the up- and downstream regions of the mouse Aldh3a1 gene (Fig. 4A). Because the methylation status of 5′-CCGG-3′ sequences around the transcriptional start site of the human ALDH3A1 is relevant to its expression levels (31), we investigated the methylation status of these sites in the mice by a methylation-sensitive amplification polymorphism method using isoschizomers, HpaII and MspI. However, no significant difference in the DNA CCGG methylation was detected between oncogene-introduced WT and Per2m/m cells (Fig. 4B). The methylation status of 5′-CCGG-3′ sequences within Aldh3a1 in oncogene-transformed Per2m/m cells was also not significantly different from those in mock-transformed cells, confirming that changes in the status of promoter DNA methylation are unlikely to be associated with the elevation of Aldh3a1 expression in oncogene-introduced Per2m/m cells.
Figure 4.
Epigenetic modifications of histone H3 on the mouse Aldh3a1 in oncogene-transformed Per2m/m cells. A, schematic representation of the mouse Aldh3a1 gene. The numbers indicate the distance (in bp) from the transcription start site (+1). Red rectangles, CCGG sites. The circled numbers (orange circles and blue circles) indicate the location on the gene where each of the different primer sets localize for analysis of DNA methylation and ChIP studies. B, methylation rate of DNA CCGG sites on Aldh3a1 in WT and Per2m/m cells. After digestion with isoschizomers, purified DNA was quantified by qPCR using primer sets that recognize the different regions indicated in the schematic in A (orange circles). Values shown are means ± S.D. (error bars) (n = 3). C, ChIP analysis for oncogene-induced changes in H3K9Ac and H3K27me3 enrichment across Aldh3a1 in WT and Per2m/m cells. Immunopurified DNA was quantified by qPCR using primer sets that recognize the different regions indicated in the schematic in A (blue circles). Values shown are means ± S.D. (n = 6). **, p < 0.01; *, p < 0.05, significantly different between the two groups. D, PER2 co-precipitated with HDAC1 and HDAC2. Cytosolic and nuclear extracts from oncogene-transformed WT and Per2m/m cells were immunoprecipitated with antibodies against PER2 or mouse IgG. Immune complexes generated by each antibody were subjected to Western blotting (WB). The results shown are representative of three independent experiments. E, ChIP analysis for oncogene-induced changes in recruitment of PCAF, HDCA1, and HDAC2 on Aldh3a1 in WT and Per2m/m cells. Immunopurified DNA was quantified by qPCR using primer sets that recognize the different regions indicated in the schematic in A (blue circles). Values shown are means ± S.D. (n = 4). **, p < 0.01; *, p < 0.05, significantly different between the two groups.
The acetylation and methylation status of specific lysine residues on histone H3 is also involved in the regulation of the expression for numerous genes (32). Next, we investigated whether histone modifications were induced in the Aldh3a1 gene after the introduction of oncogenes. To achieve this, ChIP analysis was performed on WT and Per2m/m cells to identify the enrichment of an active histone mark (histone H3 lysine 9 acetylation (H3K9Ac)) and repressive histone mark (trimethylation of histone H3 lysine 27 (H3K27me3)) in the upstream and downstream regions of the transcriptional start site of Aldh3a1 gene. The presence of H3K9Ac and H3K27me3 in the promoter region of Aldh3a1 was confirmed by qPCR analysis, which revealed that proximal to the transcriptional start site of Aldh3a1 gene in oncogene-introduced Per2m/m cells was enrichment of the active histone mark H3K9Ac and depletion of the repressive mark H3K27me3 (Fig. 4C). Although a similar pattern of histone modifications was also observed in oncogene-transformed WT cells, the active mark H3K9Ac was significantly enriched in oncogene-introduced Per2m/m cells (p < 0.01). Because H3K9 acetylation is implicated in chromatin remodeling to promote efficient gene transcription, the histone modifications in oncogene-transformed Per2m/m cells appeared to be involved in the elevated expression of Aldh3a1.
Alleviation of HDAC1 and HDAC2 recruitment to Aldh3a1 gene in oncogene-transformed Per2m/m cells
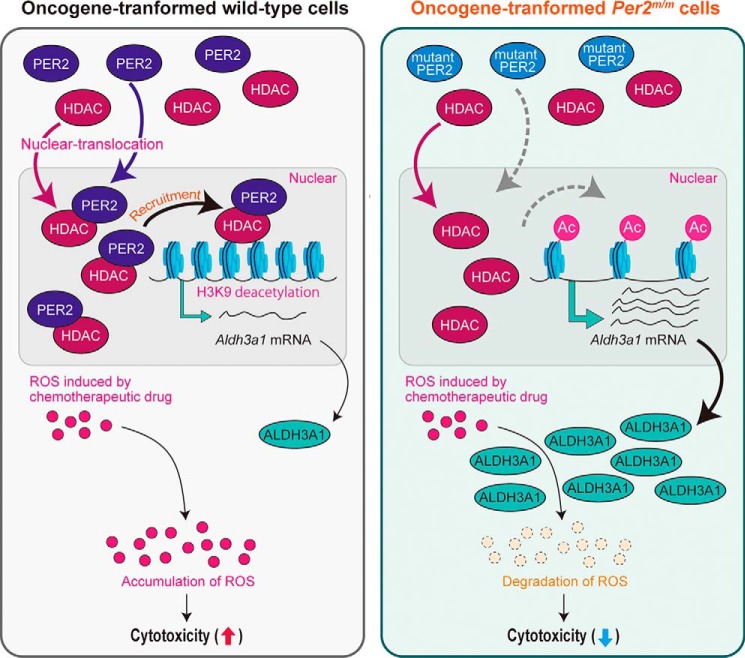
Because the acetylation state of histone H3K9 was different between oncogene-transformed WT and Per2m/m cells, we investigated whether PER2 interacted with histone modification enzymes that are associated with acetylation of H3K9. In the nuclear fraction of oncogene-transformed WT cells, PER2 was co-immunoprecipitated with HDAC1 and HDAC2, which are known as PER2-associated enzymes (33, 34) exhibiting deacetylation activity toward H3K9 (Fig. 4D). Per2m/m cells harbor a deletion of 87 amino acids from the PAS (PER-ARNT-SIM) domain of the PER2 protein (15). Mutated PER2 protein (missing residues 348–434) exhibits reduced translocation into the nucleus and instead accumulates in the cytoplasm (35). As reported previously, the level of the mutated PER2 protein remained lower in the nuclear fraction of oncogene-transformed Per2m/m cells (Fig. 4D, Input). Although obvious amounts of HDAC1 and HDAC2 were presented in nuclear fraction of both oncogene-transformed WT and Per2m/m cells (Fig. 4D, Input), the amounts of immunoprecipitated HDAC1 and HDCA2 in the nuclear fraction of oncogene-transformed Per2m/m cells were lower than those in WT cells (Fig. 4D, IP: PER2). Furthermore, the binding amounts of both HDACs around the transcriptional start site of the Aldh3a1 gene were significantly decreased in oncogene-transformed Per2m/m cells (Fig. 4E), although recruiting p300/CBP-associated factor (PCAF) was comparable between WT and Per2m/m cells. Taken together, these data support a model in which PER2 acts as a repressor of oncogene-induced Aldh3a1 expression through recruitment of HDACs to the promoter region. Dysfunction of PER2 appears to allow H3K9 acetylation, therefore leading to enhancement of oncogene-induced expression of ALDH3A1 (Fig. 5).
Figure 5.
Possible mechanism underlying the resistance of oncogene-transformed Per2m/m cells against chemotherapeutic drugs. Oncogenic stimuli induce the expression of Aldh3a1 in both WT and Per2m/m cells accompanied by histone H3 modifications. In oncogene-transformed Per2m/m cells, PER2 (mutated PER2 protein) fails to recruit HDACs on Aldh3a1, resulting in its enhanced expression. Elevated levels of ALDH3A1 contribute to the resistance against chemotherapeutic drugs through the enhancement of ROS degradation.
Discussion
Recent accumulating evidence has established a significant role of circadian genes in the regulation of cell-cycle progression and DNA damage response (36, 37). In mammals, dysfunction of the circadian machinery has been implicated in carcinogenesis as well as its recurrence (38). Although Per2 has a critical role in controlling the malignancy of cancers (17, 34), our results also showed a mechanism regulating the resistance of oncogene-transformed Per2m/m cells against the cytotoxicity of chemotherapeutic drugs. The development of chemoresistance was associated with up-regulation of the Aldh3a1 gene, accompanied by histone H3 modification.
Negative correlation between the expression levels of PER2 and tumor malignancies has been reported for breast cancers (34). Down-regulation of PER2 promotes the malignancy of human breast cancer cell lines by enhancing invasion and activating expression of epithelial–mesenchymal transition genes. The retrospective analysis of patients with breast cancer also indicates that low expression of PER2 is associated with poor prognosis. The role of clock genes in controlling the sensitivity of cancer cells to chemotherapeutic drugs has been demonstrated by the overexpression of Bmal1 (39), loss of Cry1/2 (40), and down-regulation of Per2 (41). The modulation of chemosensitivity of cancer cells by clock genes is closely related to their ability to regulate cell-cycle progression and apoptosis. The products of several clock genes and/or clock-controlled genes regulate the expression of genes responsible for cell-cycle progression and DNA damage response (36, 37). However, our present results indicated that the chemosensitivity of oncogene-transformed Per2m/m cells was unlikely to be dependent on alteration of the apoptotic process and cell-cycle phase. p53 acts as a universal sensor of genotoxic stress and partially mediates ROS-induced cell death (42). However, the results of an immunoprecipitation analysis revealed that the greatest amounts of p53 protein in Per2m/m cells as well as WT cells were precipitated together with SV40LT, even when cells were treated with chemotherapeutic drugs. Furthermore, the distribution of the cell-cycle phase of oncogene-introduced Per2m/m cells was not significantly different from that of WT cells. Therefore, the underlying mechanism of the development of chemoresistance of oncogene-transformed Per2m/m cells appeared to be distinct from findings in previous reports describing clock gene–deficient cells (39–41).
The ALDH3 family includes enzymes able to oxidize medium-chain aliphatic and aromatic aldehydes (43). These enzymes also have noncatalytic activities, including antioxidant function and some structural roles. ALDH3A1 is highly expressed in the stomach, lung, keratinocytes, and cornea, but poorly detected in normal liver (43). High ALDH3A1 expression and activity have been correlated with cell proliferation, resistance against aldehydes derived from lipid peroxidation, and resistance against the cytotoxic effects of drugs; therefore, the activity is also used as a marker for cancer stemlike cells (44, 45). In fact, the expression levels of cancer stemness markers Klf4, Pou5f1, and c-Myc were significantly increased in oncogene-introduced Per2m/m cells. Our previous study also demonstrated that oncogene-transformed Per2m/m cells show high tumor formation and increased anchorage-independent growth activity (17). Therefore, these malignant phenotypes may also be due to high ALDH3A1 expression. Chemotherapeutic drug–induced DNA damage ultimately causes cell death via enhancing the production of ROS (28). Due to antioxidant function (29), elevated expression of ALDH3A1 in oncogene-transformed Per2m/m cells appeared to contribute to their resistance against chemotherapeutic drugs through preventing ROS accumulation. This notion was also supported by the present finding that down-regulation of ALDH3A1 in oncogene-transformed Per2m/m cells restored the chemotherapeutic drug-induced accumulation of H2O2. The expressions of other H2O2 degradation enzyme were not significantly different between WT and Per2m/m cells. Therefore, enhanced expression of ALDH3A1 appeared to be involved in the development of chemoresistance of oncogene-transformed Per2m/m cells.
The expression of rat ALDH3 is induced by polycyclic aromatic hydrocarbons, chlorinated compounds, or the activation of the aryl carbon receptor (46). Peroxisome proliferator–activated receptor γ also negatively regulates the expression of human ALDH3A1 (47). Therefore, the orphan receptor agonist is able to suppress ALDH3A1 expression. Oncogene transformation of cells often causes irreversible changes in gene expression, leading to rapid proliferation and high invasiveness. Such alterations of gene expression in oncogene-transformed cells are thought to be associated with epigenetic modifications (30). Although methylation of CCGG sites around the transcriptional start site of Aldh3a1 was not significantly changed by the introduction of oncogenes, acetylation and trimethylation of histone H3 were modified in oncogene-transformed cells. As compared with WT cells, acetylation of H3K9 on Aldh3a1 gene was enriched in oncogene-transformed Per2m/m cells. H3K9 acetylation is implicated in chromatin remodeling to promote efficient gene transcription, suggesting a potential underlying cause of enhanced Aldh3a1 expression in oncogene-transformed Per2m/m cells. Actually, we prepared oncogene-transformed cells three times, and although the expression levels of ALDH3A1 protein were ∼2-fold different in each preparation, the dehydrogenase levels in oncogene-transformed Per2m/m cells were 7–12-fold higher than those in WT cells in every preparation.
PER2 is capable of interacting with several histone modification enzymes, such as HCAC1, HDAC2, SIN3-HDAC, EZH2, SUZ12, and SUV39H (33, 34). In this study, we also observed the interaction of PER2 with HDAC1 and HDAC2 in nucleus of WT cells. HDACs catalyze H3K9Ac deacetylation, resulting in gene silencing (48). Mutated PER2 protein, which is produced in Per2m/m cells, also interacted with HDACs in the cytoplasm, but the difficulties in nuclear translocation of the mutated PER2 protein appeared to prevent the recruitment of HDACs around the transcriptional start site of the Aldh3a1 gene. This may have allowed the enrichment of H3K9 acetylation, which accounted for the enhanced ALDH3A1 expression in oncogene-transformed Per2m/m cells. However, we were unable to clarify how PER2 protein directs deacetylation activity of HDACs toward the Aldh3a1 gene. In addition to the Aldh3a1 gene, decreased deacetylation of H3K9 may also modify the expression of genes in oncogene-transformed Per2m/m cells (Fig. 2A), whose transcriptional activity is highly dependent on the acetylation state of H3K9. Further studies are required to investigate the role of PER2 in the regulation of histone modification during oncogene transformation.
Because the basal mechanism of the circadian clock is well-conserved in many mammalian species, PER2 is assumed to function in human cells in a manner similar to what we observed in murine cells. The present results suggest a newly discovered role for PER2 in the regulation of susceptibility of cancer cells to chemotherapeutic drugs and may contribute to the development of new strategies in the treatment of cancer.
Experimental procedures
Treatment of animals and cells
Per2m/m mice in an ICR background and WT ICR mice were maintained under a standardized light and dark cycle. All animal experiments were reviewed and approved by the Animal Care and Use Committee of Kyushu University (Fukuoka, Japan). Fibroblasts were prepared from littermate embryos of WT or Per2m/m mice using standard techniques. The cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Sigma-Aldrich) supplemented with 10% fetal bovine serum and 0.5% penicillin-streptomycin. For oncogene transformation, embryonic cells were infected with 1 × 106 cfu/ml of retroviral vectors (pBABE-puro retroviral vector, RTV-001-PURO, Cell Biolabs, Inc., San Diego, CA) expressing H-rasV12 and SV40LT (Clontech). Transgene-expressing cells were selected with 2 μg/ml puromycin (Wako Chemical, Osaka, Japan). The preparation of oncogene-transformed cells of each genotype was performed three times.
To down-regulate ALDH3A1 expression, cells were infected with lentiviral vectors expressing shRNA against the mouse Aldh3a1 gene (pGFP-C-shAldh3a1 Lenti Vector; Origene Technologies, Inc., Rockville, MD). After infection, cells were maintained in DMEM. GFP-expressing cells were selected by sorting using FACS (BD Biosciences). Down-regulation of ALDH3A1 was confirmed by Western blotting.
Determination of cell viability
Cells were seeded at a density of 1 × 103 cells/well in 200 μl of DMEM in 96-well culture plates. After incubation for 24 h at 37 °C, the cells were treated with MTX, GEM, VP-16, VCR, or L-OHP at the indicated concentrations. Cells were also treated concomitantly with 30 μm pifithrin-α (Wako Chemical), 2 mm NAC (Sigma-Aldrich), or 30 μm CB29 (Merck, Darmstadt, Germany). The viability of the cells was determined by an ATP luminescent assay using a Cell Titer-Glo Luminescent Cell Visibility Assay Kit (Promega, Madison, WI).
Immunoprecipitation
Nuclear fractions were prepared from cells after treatment with MTX, GEM, VP-16, VCR, L-OHP, or vehicle for 24 h. The nuclear fractions were diluted (∼1 mg/ml) in 25 mm Tris-HCl, pH 8.0, 137 mm NaCl, 2.7 mm KCl, and 1% Triton X-100 supplemented with protease inhibitor mixtures and were then subjected to immunoprecipitation with anti-SV40LT antibodies (sc-58665, Santa Cruz Biotechnology, Inc.). After centrifugation, the amounts of p53 and SV40LT in supernatants and immune complexes were assessed by Western blotting. We also assessed the amounts of p53 in nuclear fractions as input.
Cell-cycle analysis
Single-cell suspension was prepared, and cells were incubated with 0.05 mg/ml propidium iodide for specific DNA staining. The samples were analyzed on an EPICS Elite flow cytometer (Beckman Coulter, Inc.). The total number of cells analyzed from each sample was 10,000.
Western blotting
Nuclear and cytosolic fractions of cells were prepared using a LysoPure nuclear and cytoplasmic extractor kit (Wako Chemicals). Denatured samples containing 20 or 40 μg of each protein fraction were separated by SDS-PAGE and then transferred from the gels to polyvinylidene difluoride membranes. The membranes were incubated with primary antibodies against p53 (1:3,000, sc-6243; Santa Cruz Biotechnology), SV40LT (1:1,000, sc-58665; Santa Cruz Biotechnology), P-gp (1:3,000, C219; Thermo Fisher Scientific), BCRP (1:3,000, BXP-53, sc-58224; Santa Cruz Biotechnology), MRP2 (1:3,000, sc-5770; Santa Cruz Biotechnology), Na+/K+ ATPase (1:3,000, ab76020; Abcam, Cambridge, UK), ALDH3A1 (1:3,000, ab76976; Abcam), catalase (1:3,000, ab16731; Abcam), GSH peroxidase (1:3,000, ab22604; Abcam), SOD3 (1:3,000, ab90258; Abcam), PCAF (1:3,000, ab176316), histone deacetylase-1 (HDAC1; 1:3,000, ab7028), HDAC2 (1:3,000, ab7029), TATA-binding protein (1:3,000, ab51841), or β-actin (1:3,000, sc-1616; Santa Cruz Biotechnology). Specific antigen–antibody complexes were visualized using horseradish peroxidase–conjugated secondary antibodies and a Chemi-Lumi One assay kit (Nacalai Tesque, Kyoto, Japan). Blot images were scanned using an Image Quant LAS4000 system (GE Healthcare).
Determination of intracellular concentrations of chemotherapeutic drugs
Cells were treated with 1 μm MTX, 0.5 μm GEM, 50 μm VP-16, 5 μm VCR, or 50 μm L-OHP. Intracellular concentrations of platinum were determined according to the amount of incorporated L-OHP, using inductively coupled plasma MS. After treatment with MTX, GEM, VP-16, and VCR, the cells were washed with PBS and collected in methanol containing an internal standard (aminopterin for MTX, 5-bromouracil for GEM, and teniposide for VP-16 and VCR). After centrifugation (12,000 rpm, 5 min, 4 °C), the methanol solution of MTX was analyzed without extraction, whereas the methanol solutions of GEM, VP-16, and VCR were extracted with ethyl acetate (Wako Chemical) and dissolved into the mobile phase. Concentrations of MTX, GEM, VP-16, and VCR in aliquots taken were measured by LC-MS/MS. A reversed-phase system column (Shim-pack XR-ODS, Shimadzu, Kyoto, Japan) was used for LC, and a liquid chromatograph mass spectrometer system (LCMS-8040, Shimadzu) was used for MS/MS. The multiple-reaction–monitoring transitions and the composition of the mobile phases are listed in Table 1. The amount of each drug was normalized to protein concentrations.
Table 1.
Multiple-reaction–monitoring transitions and the composition of the mobile phases for LC-MS/MS analysis
| Drugs (precursor ion → product ion (m/z)) | Internal standard (precursor ion → product ion (m/z)) | Mobile phase (flow rate (ml/min)) |
|---|---|---|
| MTX (455 → 308) | Aminopterin (441 → 294) | 1 mm CH3COONH4/acetonitrile = 4:1 (0.2) |
| GEM (264 → 112) | Bromouracil (189 → 42) | H2O/acetonitrile = 1:1 (0.2) |
| VP-16 (606 → 229) | Teniposide (674 → 382) | 1 mm CH3COONH4/methanol = 4:1 (0.2) |
| VCR (825 → 765) | Teniposide (674 → 382) | H2O/acetonitrile = 1:1 (0.2) |
Microarray gene expression analysis
The quality of the extracted RNA was analyzed using an Agilent 2100 Bioanalyzer (Agilent Technologies). The complementary RNA was amplified and labeled using a Low Input Quick Amp Labeling Kit (Agilent). Labeled complementary RNA was hybridized to a 44-K Agilent 60-mer oligo-microarray (Whole Mouse Genome Microarray Kit version 2.0). Probe level data were processed using the robust multiarray analysis algorithm to obtain data at the expression level. This produced a gene expression matrix consisting of 39,427 probe sets and six samples (two groups with three replications). To identify up-regulated or down-regulated genes, z-scores and ratios were calculated from the normalized signal intensities of each probe. The criteria for up-regulated genes were set at a z-score of 2.0 or more and a ratio of 3-fold or more. The criteria for down-regulated genes were set at a z-score of −2.0 or less and a ratio of 0.33 or less. The full data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database (accession number GSE113242).
Quantitative RT-PCR analysis
mRNA was extracted from WT or Per2m/m cells by using RNAiso (Takara Bio Inc., Otsu, Japan). The complementary DNA was synthesized by reverse transcription using the ReverTra Ace quantitative real-time PCR kit (Toyobo, Osaka, Japan). The real-time PCR assays were performed using THUNDERBIRDSYBR qPCR mix (Toyobo) and the LightCycler 96 system (Roche Applied Science). The sequences of the PCR primers are shown in Table 2.
Table 2.
Primer sets for PCR analysis of gene expression
| Genes | Primers |
|---|---|
| Mouse 18S ribosome | |
| Forward | 5′-CGGCTACCACATCCAAGGAA-3′ |
| Reverse | 5′-GCTGGAATTACCGCGGCT-3′ |
| Mouse Sox2 | |
| Forward | 5′-CCCACCTACAGCATGTCCTAC-3′ |
| Reverse | 5′-GCCTCGGACTTGACCACAG-3′ |
| Mouse Pou5f1 | |
| Forward | 5′-TGAGCCGTCTTTCCACCAGGC-3′ |
| Reverse | 5′-GGAAGCTTAGCCAGGTTCGAGGATC-3′ |
| Mouse Nanog | |
| Forward | 5′-TTTATTGGTGCCAGAGCAAACC-3′ |
| Reverse | 5′-GTCTCCAAAGCCTAGAGTTAAC-3′ |
| Mouse c-Myc | |
| Forward | 5′-ATGCCCCTCAACGTGAACTTC-3′ |
| Reverse | 5′-CGCAACATAGGATGGAGAGCA-3′ |
| Mouse Klf4 | |
| Forward | 5′-GGCGAGTCTGACATGGCTG-3′ |
| Reverse | 5′-GCTGGACGCAGTGTCTTCTC-3′ |
| Mouse β-actin | |
| Forward | 5′-ACTGTCGAGTCGCGTCC-3′ |
| Reverse | 5′-CGCAGCGATATCGTCATCCAT-3′ |
| Mouse Aldh3a1 | |
| Forward | 5′-AGAAGCCCCTGGCACTCTAT-3′ |
| Reverse | 5′-GCAAAGTGGGCACAGTGATG-3′ |
Measurement of H2O2
Cells were seeded at a density of 4 × 103 cells/well in 78 μl of DMEM in 96-well culture plates. Cells were treated with 1 μm MTX, 50 μm VP-16, or 5 μm VCR. The amount of hydrogen peroxide (H2O2) in the cells was determined using a ROS-Glo H2O2 assay kit (Promega), following the manufacturer's protocol. After treatment with the chemotherapeutic drugs, 20 μl of H2O2 substrate solution was added to the medium and further incubated for 3 h. After incubation, 100 μl of detection solution was added to each well of cells and incubated for 20 min at room temperature. The intensity of luminescence was measured for the amount of H2O2 in the cells. The H2O2 amounts were normalized to the number of cells in each well.
DNA methylation analysis
The methylation status of DNA CCGG sites in the upstream and downstream regions of mouse Aldh3a1 gene was measured using an EpiJet DNA methylation analysis kit (Thermo Fisher Scientific) according to the manufacturer's protocol. Briefly, genomic DNA (500 ng) was digested with HpaII and MspI, which are isoschizomers with different sensitivities to CpG methylation. When the internal CpG in the 5′-CCGG-3′ tetranucleotide sequence is methylated, cleavage with HpaII is blocked, but cleavage with MspI is unaffected. After digestion, the DNA was subjected to real-time PCR analysis using primers shown in Table 3. The status of DNA methylation was calculated based on Ct value using the following formula.
| (Eq. 1) |
Table 3.
Primer sets for PCR analysis of DNA methylation
The numbers indicate the distance from the putative transcription start site (+1).
| Genes | Primers |
|---|---|
| Mouse Aldh3a1 gene (from −2,678 to −2,507) | |
| Forward | 5′-AAATCTGTTCCTTCACATCATCAG-3′ |
| Reverse | 5′-TTAGCATCAGCCATGAGTTAGAAA-3′ |
| Mouse Aldh3a1 gene (from +1,265 to +1,435) | |
| Forward | 5′-CTTGAAGCTCTGGCTATCAGGAA-3′ |
| Reverse | 5′-CATGTTGACCCTTAGACTTCCTAC-3′ |
Immunoprecipitation analysis
One milligram of protein from the nuclear and cytosolic fractions were treated using a cross-link immunoprecipitation kit (Thermo Fisher Scientific). Three hundred microliters of the lysate was precleared with control protein A/G–agarose and then incubated at 4 °C for 12 h with protein A/G–agarose–binding anti-PER2 antibodies (1:500, sc-101105; Santa Cruz Biotechnology) or mouse IgG (1:100, sc-66931; Santa Cruz Biotechnology). After washing of the reactants multiple times, the immunoprecipitation lysates were denatured at 90 °C for 30 min with 0.1% Triton X-100, 1% SDS, 15% glycerol, 0.25 m Tris, and 5% 2-mercaptoethanol, separated by SDS-PAGE, and transferred to a polyvinylidene difluoride membrane. The immunoprecipitated proteins were detected by Western blotting.
ChIP analysis
Cells were treated with 4% paraformaldehyde for 20 min to cross-link the chromatin, and 250 mm glycine was added as a reaction-stopping agent. Cross-linked chromatin was sonicated on ice, and the nuclear fractions were obtained by centrifugation at 10,000 × g for 5 min. Acetylated or trimethylated lysine residues in histone H3 were immunoprecipitated using antibodies against H3K9Ac (1:500, ab10812; Abcam), H3K4Me3 (1:500; ab8898), H3K9Me3 (1:500; ab8580), H3K27Me3 (1:500; ab6002), PCAF (1:500; ab176316), HDAC1 (1:5000; ab7028), and HDAC2 (1:500; ab7029). DNA was purified using a DNA purification kit (Promega) and amplified by PCR for upstream and downstream regions of the mouse Aldh3a1. The PCR primer sequences are listed in Table 4. The quantitative reliability of PCR was evaluated by kinetic analysis of the amplified products to ensure that the signals were derived from only the exponential phase of amplification. As negative controls, ChIP was performed in the absence of antibody or in the presence of rabbit IgG.
Table 4.
Primer sets for PCR analysis of chromatin immunoprecipitation
The numbers indicate the distance from the putative transcription start site (+1).
| Genes | Primers |
|---|---|
| Mouse Aldh3a1 gene (from −5,078 to −4,937) | |
| Forward | 5′-CTTTGTTGGAGGAAATGTGT-3′ |
| Reverse | 5′-CACTTCTGCATTTGGATCT-3′ |
| Mouse Aldh3a1 gene (from −1,068 to −857) | |
| Forward | 5′-TTGGTTACTGGAATGTGTG-3′ |
| Reverse | 5′-GCTCCAACATCATATCACAT-3′ |
| Mouse Aldh3a1 gene (from −38 to +160) | |
| Forward | 5′-AGAAGGGTCCTTAAATGTGTT-3′ |
| Reverse | 5′-CTCCCATCTAGAGACGAAG-3′ |
| Mouse Aldh3a1 gene (from +906 to +1,083) | |
| Forward | 5′-TATGGCGGTTTTCTGAAACT-3′ |
| Reverse | 5′-CGGAGTTACTGACCGTTG-3′ |
Statistical analysis
Statistical significance for differences among the groups was analyzed by Student's t tests or analysis of variance followed by Tukey–Kramer test. p < 0.05 was considered significant.
Author contributions
C. K., S. K., and S. O. designed the study and wrote the paper. C. K., S. K., and N. K. performed and analyzed the experiments shown in Figs. 1–4 and contributed to the preparation of the figures. K. H., T. A., and N. M. provided technical assistance. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Takashi Ogino and Yuya Yoshida for technical assistance with flow cytometry analysis. We are grateful for the technical support provided by the Research Support Center, Graduate School of Medical Sciences, Kyushu University.
This work was supported in part by KAKENHI Grants-in-aid for Scientific Research 16H02636 and 17H06262 (to S. O.) and 26670317 (to S. K.) from the Japan Society for the Promotion of Science (JSPS). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1.
The array data have been deposited in the NCBI Gene Expression Omnibus and are accessible through GEO Series accession number GSE113242.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- MTX
- methotrexate
- GEM
- gemcitabine
- VP-16
- etoposide
- L-OHP
- oxaliplatin
- VCR
- vincristine
- P-gp
- P-glycoprotein
- MRP2
- multidrug resistance–associated protein-2
- BCRP
- breast cancer–resistant protein
- qPCR
- quantitative PCR
- SOD3
- superoxide dismutase-3
- NAC
- N-acetylcysteine
- ROS
- reactive oxygen species
- H3K9
- histone H3 lysine 9
- H3K9Ac
- H3K9 acetylation
- H3K27me3
- histone H3 lysine 27 trimethylation
- DMEM
- Dulbecco's modified Eagle's medium
- PCAF
- p300/CBP-associated factor.
References
- 1. Reppert S. M., and Weaver D. R. (2002) Coordination of circadian timing in mammals. Nature 418, 935–941 10.1038/nature00965 [DOI] [PubMed] [Google Scholar]
- 2. Gekakis N., Staknis D., Nguyen H. B., Davis F. C., Wilsbacher L. D., King D. P., Takahashi J. S., and Weitz C. J. (1998) Role of the CLOCK protein in the mammalian circadian mechanism. Science 280, 1564–1569 10.1126/science.280.5369.1564 [DOI] [PubMed] [Google Scholar]
- 3. Kume K., Zylka M. J., Sriram S., Shearman L. P., Weaver D. R., Jin X., Maywood E. S., Hastings M. H., and Reppert S. M. (1999) mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98, 193–205 10.1016/S0092-8674(00)81014-4 [DOI] [PubMed] [Google Scholar]
- 4. Preitner N., Damiola F., Lopez-Molina L., Zakany J., Duboule D., Albrecht U., and Schibler U. (2002) The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110, 251–260 10.1016/S0092-8674(02)00825-5 [DOI] [PubMed] [Google Scholar]
- 5. Akashi M., and Takumi T. (2005) The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat. Struct. Mol. Biol. 12, 441–448 10.1038/nsmb925 [DOI] [PubMed] [Google Scholar]
- 6. Cheng M. Y., Bullock C. M., Li C., Lee A. G., Bermak J. C., Belluzzi J., Weaver D. R., Leslie F. M., and Zhou Q. Y. (2002) Prokineticin 2 transmits the behavioural circadian rhythm of the suprachiasmatic nucleus. Nature 417, 405–410 10.1038/417405a [DOI] [PubMed] [Google Scholar]
- 7. Maemura K., de la Monte S. M., Chin M. T., Layne M. D., Hsieh C. M., Yet S. F., Perrella M. A., and Lee M. E. (2000) CLIF, a novel cycle-like factor, regulates the circadian oscillation of plasminogen activator inhibitor-1 gene expression. J. Biol. Chem. 275, 36847–36851 10.1074/jbc.C000629200 [DOI] [PubMed] [Google Scholar]
- 8. Ripperger J. A., Shearman L. P., Reppert S. M., and Schibler U. (2000) CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 14, 679–689 [PMC free article] [PubMed] [Google Scholar]
- 9. Panda S., Antoch M. P., Miller B. H., Su A. I., Schook A. B., Straume M., Schultz P. G., Kay S. A., Takahashi J. S., and Hogenesch J. B. (2002) Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109, 307–320 10.1016/S0092-8674(02)00722-5 [DOI] [PubMed] [Google Scholar]
- 10. Masri S., Kinouchi K., and Sassone-Corsi P. (2015) Circadian clocks, epigenetics, and cancer. Curr. Opin. Oncol. 27, 50–56 10.1097/CCO.0000000000000153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lahti T. A., Partonen T., Kyyrönen P., Kauppinen T., and Pukkala E. (2008) Night-time work predisposes to non-Hodgkin lymphoma. Int. J. Cancer 123, 2148–2151 10.1002/ijc.23566 [DOI] [PubMed] [Google Scholar]
- 12. Kloog I., Haim A., Stevens R. G., and Portnov B. A. (2009) Global co-distribution of light at night (LAN) and cancers of prostate, colon, and lung in men. Chronobiol. Int. 26, 108–125 10.1080/07420520802694020 [DOI] [PubMed] [Google Scholar]
- 13. Filipski E., Innominato P. F., Wu M., Li X. M., Iacobelli S., Xian L. J., and Lévi F. (2005) Effects of light and food schedules on liver and tumor molecular clocks in mice. J. Natl. Cancer Inst. 97, 507–517 10.1093/jnci/dji083 [DOI] [PubMed] [Google Scholar]
- 14. Lee S., Donehower L. A., Herron A. J., Moore D. D., and Fu L. (2010) Disrupting circadian homeostasis of sympathetic signaling promotes tumor development in mice. PLoS One 5, e10995 10.1371/journal.pone.0010995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zheng B., Larkin D. W., Albrecht U., Sun Z. S., Sage M., Eichele G., Lee C. C., and Bradley A. (1999) The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature 400, 169–173 10.1038/22118 [DOI] [PubMed] [Google Scholar]
- 16. Fu L., Pelicano H., Liu J., Huang P., and Lee C. (2002) The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell 111, 41–50 10.1016/S0092-8674(02)00961-3 [DOI] [PubMed] [Google Scholar]
- 17. Katamune C., Koyanagi S., Shiromizu S., Matsunaga N., Shimba S., Shibata S., and Ohdo S. (2016) Different roles of negative and positive components of the circadian clock in oncogene-induced neoplastic transformation. J. Biol. Chem. 291, 10541–10550 10.1074/jbc.M115.706481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. El-Deiry W. S. (2003) The role of p53 in chemosensitivity and radiosensitivity. Oncogene 22, 7486–7495 10.1038/sj.onc.1206949 [DOI] [PubMed] [Google Scholar]
- 19. Fojo T. (2002) p53 as a therapeutic target: unresolved issues on the road to cancer therapy targeting mutant p53. Drug Resist. Updat. 5, 209–216 10.1016/S1368-7646(02)00119-X [DOI] [PubMed] [Google Scholar]
- 20. Zhu J. Y., Abate M., Rice P. W., and Cole C. N. (1991) The ability of Simian virus 40 large T antigen to immortalize primary mouse embryo fibroblasts cosegregates with its ability to bind to p53. J. Virol. 65, 6872–6880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guo X. L., Hu F., Zhang S. S., Zhao Q. D., Zong C., Ye F., Guo S. W., Zhang J. W., Li R., Wu M. C., and Wei L. X. (2014) Inhibition of p53 increases chemosensitivity to 5-FU in nutrient-deprived hepatocarcinoma cells by suppressing autophagy. Cancer Lett. 346, 278–284 10.1016/j.canlet.2014.01.011 [DOI] [PubMed] [Google Scholar]
- 22. Wehbe H., Henson R., Lang M., Meng F., and Patel T. (2006) Pifithrin-α enhances chemosensitivity by a p38 mitogen-activated protein kinase-dependent modulation of the eukaryotic initiation factor 4E in malignant cholangiocytes. J. Pharmacol. Exp. Ther. 319, 1153–1161 10.1124/jpet.106.109835 [DOI] [PubMed] [Google Scholar]
- 23. Xu G. W., Mymryk J. S., and Cairncross J. G. (2005) Pharmaceutical-mediated inactivation of p53 sensitizes U87MG glioma cells to BCNU and temozolomide. Int. J. Cancer 116, 187–192 10.1002/ijc.21071 [DOI] [PubMed] [Google Scholar]
- 24. Tian C., Ambrosone C. B., Darcy K. M., Krivak T. C., Armstrong D. K., Bookman M. A., Davis W., Zhao H., Moysich K., Gallion H., and DeLoia J. A. (2012) Common variants in ABCB1, ABCC2 and ABCG2 genes and clinical outcomes among women with advanced stage ovarian cancer treated with platinum and taxane-based chemotherapy: a gynecologic oncology group study. Gynecol. Oncol. 124, 575–581 10.1016/j.ygyno.2011.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cascorbi I. (2006) Role of pharmacogenetics of ATP-binding cassette transporters in the pharmacokinetics of drugs. Pharmacol. Ther. 112, 457–473 10.1016/j.pharmthera.2006.04.009 [DOI] [PubMed] [Google Scholar]
- 26. Duong H. Q., You K. S., Oh S., Kwak S. J., and Seong Y. S. (2017) Silencing of NRF2 reduces the expression of ALDH1A1 and ALDH3A1 and sensitizes to 5-FU in pancreatic cancer cells. Antioxidants (Basel) 6, 52 10.3390/antiox6030052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Awad O., Yustein J. T., Shah P., Gul N., Katuri V., O'Neill A., Kong Y., Brown M. L., Toretsky J. A., and Loeb D. M. (2010) High ALDH activity identifies chemotherapy-resistant Ewing's sarcoma stem cells that retain sensitivity to EWS-FLI1 inhibition. PLoS One 5, e13943 10.1371/journal.pone.0013943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Trachootham D., Alexandre J., and Huang P. (2009) Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach. Nat. Rev. Drug. Discov. 8, 579–591 10.1038/nrd2803 [DOI] [PubMed] [Google Scholar]
- 29. Raha D., Wilson T. R., Peng J., Peterson D., Yue P., Evangelista M., Wilson C., Merchant M., and Settleman J. (2014) The cancer stem cell marker aldehyde dehydrogenase is required to maintain a drug-tolerant tumor cell subpopulation. Cancer Res. 74, 3579–3590 10.1158/0008-5472.CAN-13-3456 [DOI] [PubMed] [Google Scholar]
- 30. Fujimoto A., Furuta M., Totoki Y., Tsunoda T., Kato M., Shiraishi Y., Tanaka H., Taniguchi H., Kawakami Y., Ueno M., Gotoh K., Ariizumi S., Wardell C. P., Hayami S., Nakamura T., et al. (2016) Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat. Genet. 48, 500–509 10.1038/ng.3547 [DOI] [PubMed] [Google Scholar]
- 31. Buro-Auriemma L. J., Salit J., Hackett N. R., Walters M. S., Strulovici-Barel Y., Staudt M. R., Fuller J., Mahmoud M., Stevenson C. S., Hilton H., Ho M. W., and Crystal R. G. (2013) Cigarette smoking induces small airway epithelial epigenetic changes with corresponding modulation of gene expression. Hum. Mol. Genet. 22, 4726–4738 10.1093/hmg/ddt326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rice J. C., and Allis C. D. (2001) Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr. Opin. Cell Biol. 13, 263–273 10.1016/S0955-0674(00)00208-8 [DOI] [PubMed] [Google Scholar]
- 33. Duong H. A., and Weitz C. J. (2014) Temporal orchestration of repressive chromatin modifiers by circadian clock Period complexes. Nat. Struct. Mol. Biol. 21, 126–132 10.1038/nsmb.2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hwang-Verslues W. W., Chang P. H., Jeng Y. M., Kuo W. H., Chiang P. H., Chang Y. C., Hsieh T. H., Su F. Y., Lin L. C., Abbondante S., Yang C. Y., Hsu H. M., Yu J. C., Chang K. J., Shew J. Y., et al. (2013) Loss of corepressor PER2 under hypoxia up-regulates OCT1-mediated EMT gene expression and enhances tumor malignancy. Proc. Natl. Acad. Sci. U.S.A. 110, 12331–12336 10.1073/pnas.1222684110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shearman L. P., Sriram S., Weaver D. R., Maywood E. S., Chaves I., Zheng B., Kume K., Lee C. C., van der Horst G. T., Hastings M. H., and Reppert S. M. (2000) Interacting molecular loops in the mammalian circadian clock. Science 288, 1013–1019 10.1126/science.288.5468.1013 [DOI] [PubMed] [Google Scholar]
- 36. Matsuo T., Yamaguchi S., Mitsui S., Emi A., Shimoda F., and Okamura H. (2003) Control mechanism of the circadian clock for timing of cell division in vivo. Science 302, 255–259 10.1126/science.1086271 [DOI] [PubMed] [Google Scholar]
- 37. Geyfman M., Kumar V., Liu Q., Ruiz R., Gordon W., Espitia F., Cam E., Millar S. E., Smyth P., Ihler A., Takahashi J. S., and Andersen B. (2012) Brain and muscle Arnt-like protein-1 (BMAL1) controls circadian cell proliferation and susceptibility to UVB-induced DNA damage in the epidermis. Proc. Natl. Acad. Sci. U.S.A. 109, 11758–11763 10.1073/pnas.1209592109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Climent J., Perez-Losada J., Quigley D. A., Kim I. J., Delrosario R., Jen K. Y., Bosch A., Lluch A., Mao J. H., and Balmain A. (2010) Deletion of the PER3 gene on chromosome 1p36 in recurrent ER-positive breast cancer. J. Clin. Oncol. 28, 3770–3778 10.1200/JCO.2009.27.0215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zeng Z. L., Luo H. Y., Yang J., Wu W. J., Chen D. L., Huang P., and Xu R. H. (2014) Overexpression of the circadian clock gene Bmal1 increases sensitivity to oxaliplatin in colorectal cancer. Clin. Cancer Res. 20, 1042–1052 10.1158/1078-0432.CCR-13-0171 [DOI] [PubMed] [Google Scholar]
- 40. Lee J. H., and Sancar A. (2011) Circadian clock disruption improves the efficacy of chemotherapy through p73-mediated apoptosis. Proc. Natl. Acad. Sci. U.S.A. 108, 10668–10672 10.1073/pnas.1106284108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Momma T., Okayama H., Saitou M., Sugeno H., Yoshimoto N., Takebayashi Y., Ohki S., and Takenoshita S. (2017) Expression of circadian clock genes in human colorectal adenoma and carcinoma. Oncol. Lett. 14, 5319–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu B., Chen Y., and St Clair D. K. (2008) ROS and p53: a versatile partnership. Free Radic. Biol. Med. 44, 1529–1535 10.1016/j.freeradbiomed.2008.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marchitti S. A., Brocker C., Stagos D., and Vasiliou V. (2008) Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin. Drug Metab. Toxicol. 4, 697–720 10.1517/17425255.4.6.697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ma I., and Allan A. L. (2011) The role of human aldehyde dehydrogenase in normal and cancer stem cells. Stem Cell Rev. 7, 292–306 10.1007/s12015-010-9208-4 [DOI] [PubMed] [Google Scholar]
- 45. Burchiel S. W., Thompson T. A., Lauer F. T., and Oprea T. I. (2007) Activation of dioxin response element (DRE)-associated genes by benzo(a)pyrene 3,6-quinone and benzo(a)pyrene 1,6-quinone in MCF-10A human mammary epithelial cells. Toxicol. Appl. Pharmacol. 221, 203–214 10.1016/j.taap.2007.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xie Y. Q., Takimoto K., Pitot H. C., Miskimins W. K., and Lindahl R. (1996) Characterization of the rat class 3 aldehyde dehydrogenase gene promoter. Nucleic Acids Res. 24, 4185–4191 10.1093/nar/24.21.4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Muzio G., Trombetta A., Maggiora M., Martinasso G., Vasiliou V., Lassen N., and Canuto R. A. (2006) Arachidonic acid suppresses growth of human lung tumour A549 cells through down-regulation of ALDH3A1 expression. Free Radic. Biol. Med. 40, 1929–1938 10.1016/j.freeradbiomed.2006.01.020 [DOI] [PubMed] [Google Scholar]
- 48. Chen H. P., Zhao Y. T., and Zhao T. C. (2015) Histone deacetylases and mechanisms of regulation of gene expression. Crit. Rev. Oncog. 20, 35–47 10.1615/CritRevOncog.2015012997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., Zahler A. M., and Haussler D. (2002) The human genome browser at UCSC. Genome Res. 6, 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.