Abstract
The amphibian Xenopus laevis has long been used as a model for studying vertebrate cell and developmental biology largely due to the easiness to manipulate this system in vivo and in vitro. While most of the developmental studies have been on Xenopus embryogenesis, considerable efforts have been made to understand its metamorphosis, a process mimicking postembryonic development in mammals when many organs mature into their adult forms in the presence of high levels of thyroid hormone (T3). Amphibian metamorphosis is totally dependent on T3 and offers a number of advantages for experimental analyses compared to the late stage, uterus-enclosed mammalian embryos. Earlier studies on metamorphosis in Xenopus laevis have revealed dual functions of T3 receptors (TR) during premetamorphic development and metamorphosis as well as important roles of TR-interacting corepressors and coactivators during these two periods, respectively. The development of gene-editing technologies that functions in amphibians in recent years has made possible for the first time to study function of endogenous TRs, especially in the highly related diploid anuran species Xenopus tropicalis. Here, we first review the current mechanistic understanding of the regulation of metamorphosis by T3 and TR, and then describe a detailed method to use TALEN to knock out TRα for studying its role in gene regulation by T3 in vivo and Xenopus development.
Keywords: Xenopus laevis, Xenopus tropicalis, Metamorphosis, Gene-editing, Thyroid hormone receptor, Postembryonic development, TALEN
1. Introduction
Amphibian undergoes biphasic development with its embryogenesis producing a free-living larva, which later metamorphoses into the adult form [1]. Anuran amphibians undergo the most dramatic metamorphosis, with essentially every single organ/tissue changed during this process. This process bears strong similarity to the so-called postembryonic development in mammals, a period around birth when many organs/tissues are matured into the adult form, resembling the organ remodeling during anuran metamorphosis [1, 2]. Furthermore, both postembryonic development in mammals and anuran metamorphosis are characterized by peak levels of plasma thyroid hormone (T3) [1, 2]. Importantly, T3 is the causative agent of amphibian metamorphosis. Blocking synthesis of endogenous T3 leads to the formation of giant tadpoles that cannot metamorphose while exogenous T3 can induce premetamorphic tadpoles to undergo precocious metamorphosis prior to the synthesis of endogenous T3 [1, 2]. Furthermore, most, if not all, organs are genetically predetermined to undergo specific organ autonomous changes, making it possible to study anuran metamorphosis even in organ cultures. These properties together with the recent advancement in genetic tools for amphibian studies make anuran metamorphosis one of the best models to study the in vivo function of T3 and its underlying molecular mechanisms. Below, we summarize some of the studies on Xenopus laevis and Xenopus tropicalis, two highly related and best studied anuran species.
1.1. Dual Roles of Thyroid Hormone Receptors (TRs) in Xenopus Metamorphosis
T3 can exert its effect through both genomic and non-genomic pathways. It can regulate gene transcription through its nuclear receptors, TRs, while its non-genomic effects are believed to be mediated by cell surface and cytoplasmic binding proteins, including the small fractions of TRs present in the cytoplasm [3, 4, 5, 6, 7, 8, 9, 10, 11, 12]. TRs are known to function mainly as heterodimers with 9-cis retinoic acid receptors (RXRs) on T3-inducible genes. TR/RXR heterodimers are nuclear proteins that bind constitutively to thyroid hormone response elements (TREs) in chromatin to repress or activate target gene expression in the absence or presence of T3, respectively.
Earlier studies have shown that zygotic transcription of TR, especially TRα , occurs well before the synthesis and secretion of endogenous T3. TRα and RXRα genes are expressed at high levels by stage 45 when tadpole feeding begins while plasma T3 is detectible only by stage 54, the onset of metamorphosis [13, 14, 15, 16]. This and the ability of TR to repress or activate target gene in a T3-dependent manner have led to a dual function model for TR during frog development [17, 18]. That is, between stage 45, when tadpole feeding begins, and stage 54, the onset of metamorphosis, TRs are present mainly in the unliganded form due to only very low levels of T3, if any, in the tadpole plasma . The unliganded TR/RXR heterodimers repress T3 inducible genes to prevent precocious metamorphosis. After stage 54, the rising concentration of endogenous T3 leads to the binding of T3 to TR, and the liganded TR/RXR then activate the target genes to promote metamorphosis.
1.2. Analyses of TR Function via Overexpression during Xenopus laevis Development
Functional analysis of frog TRs initially came from microinjection studies. Overexpression of TR and RXR by microinjecting their mRNAs into fertilized eggs led to repression of endogenous target genes in the resulting embryos, and when the embryos were treated with exogenous T3, the target genes were activated, leading to embryonic defects and lethality [19]. RXR was found to be important for these effects, suggesting that TR functions as heterodimers with RXR in vivo during Xenopus development.
The development of the reliable sperm-mediated transgenic approach [20] made it possible to study the effect of overexpressed mutant TRs during Xenopus development. Several different laboratories analyzed the effect of overexpressing a dominant negative TR that had a C-terminal deletion to prevent T3 binding, leading to a constitutively unliganded TR that represses T3-inducible genes. Overexpression of such a dominant negative TR by either transgenesis in whole animals or in vivo gene transfer in the tail inhibited target gene induction by T3 and metamorphic transformation in Xenopus laevis [21, 22, 23, 24, 25]. Mechanistically, the transgenic dominant negative TR was found to compete against endogenous TR for binding to endogenous target genes, consequently preventing the regulation of the genes by T3 [25]. Thus, TR is required to mediate the effects of T3 on target gene induction and metamorphosis of different organs/tissues.
As indicated above, T3 has both genomic and non-genomic effects. To determine whether TR is sufficient to mediate the metamorphic effects of T3, a transgenic study with a dominant positive TR was carried out. This dominant positive TR consisted of the dominant negative TR above fused to the N-terminus a strong viral transcription activation protein, VP16 [26]. This mutant TR could not bind to T3 but functioned as a constitutive activator on T3 target genes due to the presence of the VP16 moiety. To prevent likely precocious induction of metamorphosis by the expression of the dominant positive TR, it was placed under the control of a heat shock-inducible promoter for transgenesis. When transgenic tadpoles and their wild-type siblings reached premetamorphic stages, they were subjected to heat shock treatment to induce the transgene expression. This enabled the transgenic but not the wild-type tadpoles to metamorphose just like premetamorphic tadpoles after T3 treatment [26]. Mechanistically, the dominant positive TR was shown to bind to endogenous target genes and specifically activate their expression [26]. Furthermore, when the intestine of such transgenic premetamorphic tadpoles was cultured in vitro , heat shock treatment also led to intestinal metamorphosis [27]. Thus, TR is sufficient to mediate the effects of T3 on individual organs during metamorphosis. These findings further demonstrate that the non-genomic effects of T3 are not necessary for amphibian metamorphosis.
1.3. Determining the Roles of Endogenous TR in Xenopus tropicalis
While molecular and transgenic studies have provided strong evidence to support the dual functions of TR during amphibian development, the roles of endogenous TRs had eluded researchers until recently. With the development of first TALEN and then CRISPR-nuclease mediated knockout/knockdown technologies that can effectively edit endogenous genes in both the pseudo-tetraploid Xenopus laevis and diploid Xenopus tropicalis, it becomes possible to study endogenous genes during Xenopus development [28, 29, 30, 31, 32, 33]. Using the TALEN technology, we and the Buchholz’s laboratory initially independently knocked down the endogenous TRα [34, 35, 36, 37]. Xenopus tropicalis was chosen for such studies due to its diploid nature. Both groups microinjected mRNAs encoding a TALEN nuclease targeting TRα into fertilized eggs and studied the effect on the resulting animals, the Fo generation knockdown animals, with similar conclusions [34, 35, 36, 37]. In our study, we generated a TALEN targeting exon 3, encoding part of the DNA binding domain of Xenopus tropicalis TRα (Fig. 1) [35]. Microinjecting the mRNA encoding the left and right arms of the TALEN into fertilized egg prior to the first cell division led to the expression of the TALEN in all cells of the early stage embryos and the resulting Fo tadpoles had genetic alterations at the TRα target site with about 90% efficiency by the time when feeding began (stage 45, 3 days of age) [35]. Such high efficiency suggested that we had nearly a complete knockout of the TRα gene and could study the phenotype in the Fo animals. Indeed, the findings in the Fo knockdown animals were subsequently confirmed by studies on complete TRα knockout animals derived from germ line breeding of the knockdown animals [38, 39].
Fig. 1.
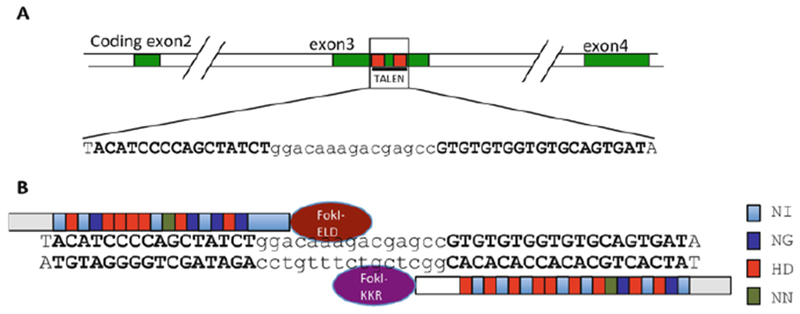
Targeting TRα with TALEN in Xenopus tropicalis embryos. (a) Schematic diagram of the TRα gene showing the TALEN target site. The protein coding exons were shown as numbered, colored boxes. The TALEN-recognized sequences were shown as red boxes (top) or bold letters (below). (b) Schematic representation of the TRα TALEN and the targeted DNA sequence in the TRα gene. The four types of RVDs (repeat variable di-residues) recognizing nucleotide A, G, T, or C were depicted in different colors, respectively. The left arm contained the FokI-ELD nuclease and the right arm contained the FokI-KKR nuclease, which, when both arms bind to their respective binding sites, heterodimerize to form the functional nuclease to make a double-stranded break in the intervening sequence. See [35] for more details
Using either Fo knockdown or complete knockout animals, both the groups found that TRα knockdown/knockout led to precocious limb development (Fig. 2) [34, 35, 36, 37, 38, 39]. Stage 54 is considered to be the onset of metamorphosis and is determined based on hindlimb morphology. Based on this, both groups observed that the TRα knockdown/knockout tadpoles initiated metamorphosis at younger age, with concurrent upregulation (derepression) of several well-known T3-inducible genes [35, 36, 38, 39]. In addition, the TRα knockdown/knockout tadpoles had reduced response to exogenous T3 treatment in terms of both gene regulation and morphological changes, indicating that TRα is important both for controlling the timing of the onset of metamorphosis and for mediating the response to T3. Consistently, when the knockdown/knockout animals were allowed to undergo natural metamorphosis, it was found that TRα knockdown/knockout animals took much longer time to develop from stage 54, the onset of metamorphosis, to stage 58, the early metamorphic climax. Furthermore, the studies on total TRα knockout animals also led to several novel observations [38, 39]. First, TRα knockout appears to have no gross effect on embryogenesis. Second and surprisingly, TRα knockout does not prevent the animal from completing metamorphosis or affect the total developmental time from fertilization to the end of metamorphosis as judged based on total resorption of the tail. Finally, TRα is important for temporal coordination of organ-specific transformations.
Fig. 2.
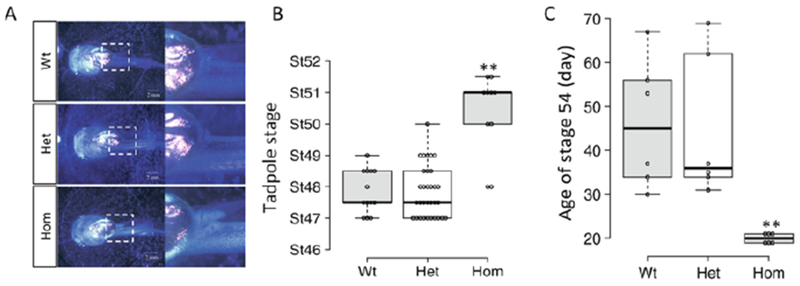
TRα regulates metamorphic timing. (a) TRα knockout tadpoles develop faster. One representative animal is shown for each of the three genotypes: wild type, heterozygous, and homozygous TRα knockout siblings. Dashed boxes in the left panel were enlarged and shown in the right panel. Note that the homozygous TRα knockout tadpole had significantly larger and more advanced hindlimb buds. (b) TRα knockout tadpoles develop into more advanced stages. The stage of individual sibling tadpoles in the three genotypes was plotted with the median (darker solid line) for each genotype shown in the figure. Note that homozygous TRα knockout tadpole reached a median stage of 51 while the wild-type and TRα heterozygous knockout tadpoles reached a median stage of 47.5. (c) TRα knockout tadpoles initiate metamorphosis, i.e., reaching stage 54, at a younger age. The age in days for each tadpole to reach stage 54 was plotted with the median (darker solid line) for each genotype shown in the figure. “**” indicates significant difference between homozygous knockout and the other groups (P < 0.01). See [38] for more details
Thus, the gene-editing technologies make it possible to investigate TR function during postembryonic development. Furthermore, the ability to introduce TALEN into fertilized eggs and the high efficiency of TALEN-mediated mutagenesis also enable one to determine the function of TR in Fo generation animals within a few weeks after micro-injecting of the TALEN mRNAs, providing a fast and powerful tool to study gene function in vivo.
In addition, TALEN-mediated mutations in certain genes may lead to lethal phenotype during early embryogenesis. This makes it difficult to study the functions of such genes during postembryonic development. However, it is possible to microinject mRNAs/proteins into fertilized eggs to rescue the embryonic defect due to the gene alteration, leading to the formation of free-feeding tadpoles. As the mRNA or protein injected into the eggs only last a few days, one can use the resulting rescued animals to study the effect of the knockout on subsequent tadpole development and metamorphosis.
2. Materials
2.1. Animal
Adult Xenopus tropicalis, Nasco (Wisconsin).
2.2. Devices for Microinjections
2.3. Plasmids and Constructs
2.4. Enzymes
T4 DNA ligase (include 10× T4 DNA ligase buffer).
Restriction endonuclease BsaI (New England Biolabs).
Restriction endonuclease Esp31 (Fermentas or Fisher).
Restriction endonuclease AgeI (New England Biolabs).
Restriction endonuclease EcoRI (New England Biolabs).
Restriction endonuclease NotI (New England Biolabs).
PrimeSTAR HS DNA Polymerase (Clontech) (Note that any high fidelity Taq DNA polymerase would be fine).
GoTaq® Green Master Mix (Promega).
2.5. Chemicals and Reagents
10 mM ATP.
SOC medium: 3.6 g/l glucose, 0.19 g/l potassium chloride, 0.58 g/l sodium chloride, 20 g/l tryptone, and 5 g/L yeast extract (Quality Biologicals, MD).
Human chorionic gonadotropin (HCG, Novarel).
20 mg/ml X-gal in DMSO.
10 mg/ml IPTG.
LB broth: 10 g/l sodium chloride, 10 g/l tryptone, and 5 g/l yeast extract (Quality Biologicals, MD).
10 mg/ml tetracycline stock solution.
50 mg/ml spectinomycin stock solution.
100 mg/ml ampicillin stock solution.
1× Marc’s Modified Ringers (MMR): 100 mM NaCl, 2 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 5 mM HEPES, pH 7.5.
0.1× MMR.
0.1× MMR/6% Ficoll.
Ficoll (Sigma).
Cysteine.
Agarose.
1× TAE buffer for DNA electrophoresis.
dNTPs, individual solutions or mix of equal concentration for each, for PCR reactions.
Mineral oil (for microinjectors).
2.6. Primers
-
Primers for sequencing during TALEN assembly:
-
○
pCR8_F1: 5′-TTGATGCCTGGCAGTTCCCT-3′
-
○
pCR8_R1: 5′-CGAACCGAACAGGCTTATGT-3′
-
○
TAL_F1: 5′-TTGGCGTCGGCAAACAGTGG-3′
-
○
TAL_R2: 5′-GGCGACGAGGTGGTCGTTGG-3′
-
○
TAL_Seq: 5′-CATCGCGCAATGCACTGAC-3′
-
○
TAL_R3: 5′-GGCTCAGCTGGGCCACAATG-3′
-
○
-
Primers for modifying TALEN backbone constructs:
-
○
Forward: 5′-CTTGGTCTCACCGGCACCATGGCTCCAAAGAAGAAG-3′ (AgeI-compatible end by BsaI digestion)
-
○
Reverse: 5′- CTTGGTCTCCAATTCGTAATACGACTCACTATAGTTC-3′ (EcoRI-compatible end by BsaI digestion)
-
○
-
Primers for detecting TRα mutations due to TALEN expression [35]:
-
○
Forward primer F: 5′-ATTGGGTTGGTTGTGGGTCG-3′
-
○
Reverse primer R: 5′-TAAGGTGGGGGCAGTACAGG-3′
-
○
Forward primer f: 5′-ATGTTCCATGAGACGCCCCT-3′
-
○
Forward primer f1: 5′-CCAGCTATCTGGACAAAGAC-3′
-
○
2.7. Other Materials
RNA transcription kit, Ambion mMessage mMachine.
Plasmid DNA Miniprep kit (Qiagen).
PCR Purification kit (Qiagen).
DNA Gel Extraction kit (Qiagen).
pCR TOPO-TA Cloning kit (Invitrogen).
Chemically Competent cells TOP10 (Invitrogen).
LB plates with 10 mg/l tetracycline, 50 mg/l spectinomycin, or 50 mg/l ampicillin, respectively.
3. Methods
3.1. TALEN Assembly
-
1.
Design of TALEN target
Obtain Xenopus tropicalis TRα gene sequence and intron/exon structure from frog genome database (http://www.xenbase.org/entry/ (http://www.xenbase.org/entry/)).
Choose the sequence encompassing the exon 2 to exon 4 for TALEN target screening.
Screen for TALEN targets via a web-based TALEN-targeter tool (https://talent.cac.cornell.edu/node/add/talen (https://talent.cac.cornell.edu/node/add/talen)).
Select a pair of TALEN targets within the exon 3 [35] (see Note 3).
-
2.
Modification on TALEN backbone constructs
Golden Gate TALEN and TAL Effector Kit assemble both arms of a TALEN into a backbone vector which expresses TALEN containing the same FokI domain [40] (see Note 2). For assembling the two arms of a TALEN into a set of backbones such that one TALEN construct expresses TALEN containing a mutant FokI-ELD and the other TALEN construct expresses TALEN containing a mutant FokI-KKR in order to further increase the specificity [29, 30], the TALEN backbone constructs are modified to be compatible with the Golden Gate TALEN and TAL Effector Kit. The modified TALEN backbone constructs are based on two double-promoter transgenesis constructs, pDP(GFP)HG (with a green fluorescent protein gene driven by a γ-crystallin promoter) and the pDP(RFP)HG (with a red fluorescent protein gene driven by a γ-crystallin promoter), both of which can produce the TALEN mRNA from a SP6 promoter in vitro for microinjection or express the TALEN protein in transgenic animals from a heat shock-inducible promoter if desired [41] (see Note 4).
PCR-amplify the TAL- KKR and TAL- ELD coding regions from the corresponding pCS2+ vector-based constructs [29, 30] with the same set of primers (forward primer 5′-CTTGGTCTCACCGGCACCATGGCTCCAAAGAAGAAG-3′ and reverse primer 5′- CTTGGTCTCCAATTCGTAATACGACTCACTATAGTTC-3′).
Gel-purify the PCR fragments and digest with BsaI to produce AgeI and EcoRI compatible ends, respectively.
Digest the double-promoter transgenesis constructs pDP(GFP)HG (with a green fluorescent protein gene driven by a γ-crystallin promoter) and the pDP(RFP)HG (with a red fluorescent protein gene driven by a γ-crystallin promoter) with AgeI and EcoRI, respectively, and gel-purify the vector part.
Ligate the PCR amplicon of TAL-KKR containing AgeI and EcoRI-compatible ends with the pDP(RFP)HG vector bearing AgeI and EcoRI ends to make pDP(RFP)-TAL-KKR. Simultaneously, ligate the PCR amplicon of TAL-ELD containing AgeI and EcoRI-compatible ends with the pDP(GFP)HG vector bearing AgeI and EcoRI ends to make pDP(GFP)-TAL-ELD.
Transform the ligated products into competent E. coli TOP10 cells, plate the cells on LB plates containing 100 μg/ml ampicillin, and incubate at 37 °C overnight.
Pick up a couple of colonies each for bacteria growth in 5 ml of LB broth supplemental with ampicillin overnight and extract plasmid DNA from them.
Confirm the cloned TAL-KKR and TAL-ELD sequences by DNA sequencing.
-
3.
TALEN assembly into pDP(GFP)-TAL-ELD and pDP(RFP)-TAL-KKR constructs
Assemble the TALEN arms essentially following the instructions for Golden Gate TALEN and TAL Effector Kit (https://www.addgene.org/taleffector/goldengatev2/ (https://www.addgene.org/taleffector/goldengatev2/)) [40] except for the use of the TALEN backbone vectors.
Assemble the TALEN left arm (TALEN-L) and right arm (TALEN-R) into the double-promoter transgenesis constructs-based TALEN backbones pDP(GFP)-TAL-ELD and pDP(RFP)-TAL-KKR, respectively.
Confirm the TALEN-L and TALEN-R by DNA sequencing.
3.2. In Vitro Transcription
Linearize plasmid DNA with restriction enzyme NotI so that the SP6 RNA polymerase transcription would produce sense mRNA with a 3′-untranslated region followed by a poly(A)30 tail suitable for TALEN translation (see Note 5). Prepare enough linearized DNA (e.g., digest about 20μg) for at least several transcription reactions at 1 μg per reaction.
Confirm linearization and DNA purity by running a small aliquot on an agarose gel (see Note 6).
Purify the linearized TALEN constructs following the manufacturer’s instructions of a PCR Purification kit (see Note 7).
Transcribe RNA following directions of Ambion’s mMessage mMachine kit (see Note 8). One μg of the DNA was transcribed in vitro with SP6 RNA polymerase.
Remove template DNA, purify RNA and quantitate it (see Note 9).
RNA can be aliquoted and stored at −80 °C for months.
3.3. In Vitro Fertilization of Xenopus Eggs
Xenopus tropicalis males and females are primed with 20 U of chorionic gonadotropin (HCG) the evening prior to the day that ovulation is desired (see Note 10).
The primed frogs are then boost with a 200 U of HCG injection early next morning. The females can be injected at different intervals of 1~2 h apart so they begin to ovulate at different times to ensure constant supply of eggs within the whole day.
A Xenopus male frog is sacrificed when the first batch of females begin to ovulate, the testes are removed, and keep in 1× MMR on ice.
Macerate 1 testis in 0.3 ml of 1 × MMR to make sperm solution, transfer into a 1.5 ml microcentrifuge tube, and keep on ice. The sperm solution can be used throughout the day.
Eggs are expelled from ovulating females onto petri dishes with gentle squeezing. Remove the water from the petri dish and add 0.1 ml of the sperm solution for about 200 eggs and 0.9 ml of ddH2O.
After gentle splaying out of eggs into the sperm solution and 2-4 min incubation, the dishes are flooded with 0.1× MMR and incubated for an additional 15 min.
To remove the jelly coat, the fertilized eggs are washed with 3% cysteine, pH 8.0, until they started to touch each other (about 3-5 min), and then washed 4-6 times with 0.1× MMR before being transferred into fresh 0.1× MMR/6% Ficoll for microinjection (see Note 10).
3.4. Microinjection of TALEN mRNAs and Mutation Analysis
Place the fertilized eggs in an agar-coated plate and inject the mRNA into the cytoplasm of the fertilized eggs prior to the first division with a Drummond Nanojet II Auto-Nanoliter Microinjector.
For TALEN-targeted mutagenesis of TRα, equal amounts of TALEN-L and TALEN-R mRNA were mixed together and injected at 400 pg for each mRNA per egg (see Note 11).
The TALEN mRNA-injected eggs along with some uninjected eggs from the same batch of in vitro fertilization were incubated side by side in a 25 °C incubator overnight.
Examine the embryos for potential phenotypes following TALEN mRNA injection (see Note 12).
Collect embryos for mutation rates evaluation starting from day 3 after the TALEN mRNA injection (see Note 13). To do so, three or more embryos from the same group are pooled together and lysed with the lysis buffer (10 mM Tris–HCl, 100 mM NaCl, 10 mM EDTA, and 0.5% SDS, pH 8.0) containing 8 μg/ml proteinase K at 56 °C overnight.
Precipitate the genomic DNA with isopropanol, wash with 70% ethanol, dry in air for 10 min., and dissolve the DNA in 10 mM Tris–HCl, pH 8.0.
Subject the genomic DNA to PCR amplification for a DNA fragment encompassing the 2 TALEN-targeted regions by using primers F and R [35].
Clone the PCR fragments into pCR TOPO-TA vector (see Note 14), transform into E. coli competent cells, plate the cells on LB plates containing 100 μg/ml ampicillin, and incubate at 37 °C overnight.
Screen the colonies for mutations by colony PCR using semi-nested primers f or f1 paired with R [35] (see Note 15).
Grow up the colonies containing mutant DNA as identified by PCR above for plasmid DNA purification and sequencing to determine the exact mutations (see Note 16)
If the mutation rate is high, grow up the remaining injected embryos for phenotypic analysis of the Fo knockdown animals, or use the Fo animals to produce total knockout animals for analysis (see [35, 38].
Acknowledgments
This work in the laboratory was supported by the Intramural Research Program of National Institute of Child Health and Human Development, National Institutes of Health.
4 Notes
Many instruments and approaches can be used to pull needles and inject RNA or DNA samples including simple hand pulled needles using a Bunsen Burner and forced air injectors using “house air” found in most laboratories, etc. The crucial criteria are to choose a needle and a method that allow for a small needle diameter (about 10 μm) to be pulled and a way to calibrate or control injection volumes (about 5–30 nl per injection).
There are several TALEN assembly tools available from different resources. TALEN constructs can be also custom-designed and made through commercial companies on the market. Golden Gate TALEN and TAL Effector Kit assembles both arms of a TALEN into a backbone vector which expresses TALEN containing the same FokI domain [40]. However, the two arms of a TALEN can be assembled into two separate backbones such that one TALEN construct expresses TALEN containing the mutant FokI-ELD and the other TALEN construct expresses TALEN containing the mutant FokI-KKR to further increase the specificity [29, 30]. FokI-ELD and FokI-KKR can form heterodimer to become active enzyme, but they do not form homodimer as the wild-type FokI itself [42].
Xenopus tropicalis TRα gene expresses two alternative splicing isoforms (TRα1 and TRα2) and both contain the DNA binding domain (DBD). The exon 3 encodes the start region of the DBD, thus mutation within this targeted region from TALEN-mediated mutagenesis would produce truncated protein lacking DBD and the ligand binding domain (LBD) or a TR with mutations in the DBD. In principle, TALEN targeted region should be as close to the translational start site of the gene as possible to disrupt protein function without worrying about potential side effects from a long truncated protein. Alternative splicing on mRNA or alternative translational start sites in the proximity of the TALEN target should also be taken into consideration when choosing an appropriate target region for TALEN.
TALEN-mediated mutations in certain genes may lead to lethal phenotype during early embryogenesis. Using inducible promoter to drive TALEN expression in transgenic animals may be an alternative approach to knock down a target gene at later developmental stages to investigate gene function. Our TALEN arms are also under the control of a heat shock-inducible promoter in the vectors. If they are used to generate transgenic tadpoles, the arms should not be expressed without heat shock in the transgenic animals, thus allowing the target gene to function normally. However, when the animals are treated with heat shocks at desired stages to induce TALEN expression, the target gene would be mutated in some cells, thus affecting development. As the eye-specific γ-crystallin promoter drives GFP expression from one TALEN construct for one of the arm and RFP expression from the TALEN construct for the other arm, one can easily identify animals expressing both arms for such conditional knockdown studies. On the other hand, while TALENs assembled in these backbone vectors work effectively in mRNA microinjection experiments [35, 43], their efficiency in transgenic studies remains to be determined.
When linearizing plasmids, avoid excess DNA sequence following the gene of interest (such as long multiple cloning sites found in some plasmids). This is because long untranslated sequences may form RNA secondary structures that inhibit translation. In addition, avoid linearizing DNA with restriction enzymes which leave 3′ overhangs (such as Pst I) as that may prime transcription from the inappropriate strand [44].
Use gel electrophoresis to confirm that the DNA is pure and completely linearized. Circular plasmid templates will generate extremely long, heterogeneous RNA transcripts because RNA polymerases are very processive.
The linearized DNA should be purified for high yield RNA production. Commercial DNA purification kits are effective and usually produce reproducible results in the following RNA production. The linearized DNA can be also purified through phenol/chloroform extraction and isopropanol precipitation; however, care must be taken to ensure that the process does not carry any inhibitory chemical residues to affect RNA synthesis efficiency from the RNA polymerase.
Though a commercial RNA transcription kit is optimized for producing translatable RNA, cost may prohibit the use of such a kit. Transcription can easily be performed with transcription components purchased individually. RNA polymerases, their appropriate buffers, nucleotides, m7G(5′)ppp(5′)G cap analogs, DNase, and other reagents are available from numerous companies. Care must be taken, however, to ensure that when these reagents are used together, the conditions are optimized for efficient transcription. This may take some trials and errors.
Once the transcription reaction is complete (following the removal of the DNA template with DNase), the RNA must be purified prior to its injection. LiCl precipitation is a good and quick method for removing unincorporated nucleotides and proteins. Phenol/chloroform extraction followed by ethanol precipitation is a more stringent method of purification, though care must be taken to remove all traces of phenol and chloroform prior to injection.
The in vitro fertilization for Xenopus tropicalis is essentially identical to that for Xenopus laevis. For details, see Xenopus fertilization protocols by [45].
Once injected, RNA stability (half-life) is dependent on a number of factors including transcript size, secondary structure, 5′ and 3′ UTR lengths and sequences, polyA tail lengths, and others. Most injected RNA (such as TR or RXR) can still be readily detectable after 10 h, though Northern or other analysis should be used to determine the half-life of the transcript used if needed. TALEN mRNA stability has not been measured, but the mutation rate is a useful endpoint for validating the TALEN mRNA injection. Injecting higher doses of RNA may produce a higher mutation rate but may also be toxic to the embryos. Typical nucleic acid toxicity phenotypes include gastrulation defects (ring embryos, sometimes called the spina bifida phenotype), kinked embryos (abnormal somitogenesis), anterio-dorsal deficiencies (small heads or eyes, lack of pigment cells) and death. Nucleic acids also become toxic at much lower concentrations if too big a volume is injected [44].
TRα knockdown may produce distinct phenotypes depending on the absence or presence of T3. Scoring (classification) of embryos must be achieved with readily visible criteria. Thus, axis length, body width, presence/size of eye or cement gland, and other finer details such as number and organization of somites can all be used to correlate with a treatment. Subsequently, more detailed molecular or biochemical analyses can be carried out. RNA can be isolated from embryos and Northern blot or RT-PCR analyses can be used to examine changes of expression in downstream genes. Soluble proteins can be isolated from embryos for Western analysis of various proteins of interest, or in gel mobility shift assays to examine the binding level of TR/RXR heterodimer to TREs. Finally, whole mount in situ hybridization analysis can be used to investigate gene expression, etc.
Upon TALEN mRNA injection, TALEN proteins would be produced and bind to the target sequences to cleave the DNA in between the two TALEN binding sites. Mutations could be introduced when the cells repair the DNA damage. The DNA break/repair cycle can continue as long as the TALEN binding sites are intact and the distance between the two TALEN binding sites is still suitable for the TALEN pair to form active FokI enzyme. The mutations would be mosaic in the Fo animals, and the mutation spectrum and rate should be evaluated for deciding if the animals are worthwhile to keep for germ-line breeding for the target gene knockout lines or phenotype analysis on the Fo animals. TALEN pairs that produce very low mutation rates, e.g., 10%, should be discarded.
If high-fidelity DNA polymerases are used for PCR amplification of the target DNA fragment, the fragment should be cloned into pCR Blunt II TOPO vector (Invitrogen) or similar cloning vectors for blunt-ended PCR fragments. If possible, high-fidelity DNA polymerase should be used for this step of PCR amplification to avoid potential random mutations introduced by Taq DNA polymerase during PCR amplification.
For colony-PCR, PCR amplification system based on high-fidelity DNA polymerase should be avoided to ensure no correction occurs on the primer in order to distinguish the wild-type target sequences and the mutant DNA sequences within the target.
A number of methods can be used to facilitate the identification of mutations caused by designer nucleases such as TALEN. The enzyme mismatch cleavage assays (MCAs) such as Surveyor [46] or T7 endonuclease I (T7E1) [47] mismatch cleavage assay are widely adapted to quantify mutation rates as a primary evaluation of targeted mutagenesis by designer nucleases. Other methods, such as High Resolution Melt Assay (HRMA) [48], droplet digital PCR (ddPCR) [49], have also been applied with varied benefits and disadvantages. PCR cloning and sequencing of the DNA sequences encompassing the TALEN targets would directly identify the mutation types and determine the mutation rates. For the purpose of gene knockdown or knockout, only the mutations that introduce out-of-frame mutations in the coding sequences of the target gene are desirable. For this purpose, the recently developed LacZ recovery/disruption assay [50] and direct visualization of out-of-frame mutation assay based on mCherry-GFP or LacZα-GFP fusion protein expression [51] would be valuable tools.
References
- 1.Shi Y-B (1999) Amphibian metamorphosis: from morphology to molecular biology. John Wiley & Sons, Inc., New York [Google Scholar]
- 2.Tata JR (1993) Gene expression during metamorphosis: an ideal model for post-embryonic development. BioEssays 15(4):239–248 [DOI] [PubMed] [Google Scholar]
- 3.Lazar MA (1993) Thyroid hormone receptors: multiple forms, multiple possibilities. Endocr Rev 14(2):184–193 [DOI] [PubMed] [Google Scholar]
- 4.Evans RM (1988) The steroid and thyroid hormone receptor superfamily. Science 240:889–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yen PM (2001) Physiological and molecular basis of thyroid hormone action. Physiol Rev 81(3):1097–1142 [DOI] [PubMed] [Google Scholar]
- 6.Davis PJ, Davis FB (1996) Nongenomic actions of thyroid hormone. Thyroid 6:497–504 [DOI] [PubMed] [Google Scholar]
- 7.Tsai MJ, O’Malley BW (1994) Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem 63:451–486 [DOI] [PubMed] [Google Scholar]
- 8.Buchholz DR, Paul BD, Fu L, Shi YB (2006) Molecular and developmental analyses of thyroid hormone receptor function in Xenopus laevis, the African clawed frog. Gen Comp Endocrinol 145(1): 1–19 [DOI] [PubMed] [Google Scholar]
- 9.Davis PJ, Davis FB, Cody V (2005) Membrane receptors mediating thyroid hormone action. Trends Endocrinol Metab 16(9):429–435 [DOI] [PubMed] [Google Scholar]
- 10.Shi YB, Matsuura K, Fujimoto K, Wen L, Fu L (2012) Thyroid hormone receptor actions on transcription in amphibia: the roles of histone modification and chromatin disruption. Cell Biosci 2(1):42 10.1186/2045-3701-2-42 ( 10.1186/2045-3701-2-42). 2045-3701-2-42 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guigon CJ, Cheng SY (2009) Novel non-genomic signaling of thyroid hormone receptors in thyroid carcinogenesis. Mol Cell Endocrinol 308(1-2):63–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiroi Y, Kim HH, Ying H, Furuya F, Huang Z, Simoncini T, Noma K, Ueki K, Nguyen NH, Scanlan TS, Moskowitz MA, Cheng SY, Liao JK (2006) Rapid nongenomic actions of thyroid hormone. Proc Natl Acad Sci U S A 103(38):14104–14109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yaoita Y, Brown DD (1990) A correlation of thyroid hormone receptor gene expression with amphibian metamorphosis. Genes Dev 4(11):1917–1924 [DOI] [PubMed] [Google Scholar]
- 14.Wong J, Shi Y-B (1995) Coordinated regulation of and transcriptional activation by Xenopus thyroid hormone and retinoid X receptors. J Biol Chem 270:18479–18483 [DOI] [PubMed] [Google Scholar]
- 15.Wang X, Matsuda H, Shi Y-B (2008) Developmental regulation and function of thyroid hormone receptors and 9-cis retinoic acid receptors during Xenopus tropicalis metamorphosis. Endocrinology 149:5610–5618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leloup J, Buscaglia M (1977) La triiodothyronine: hormone de la métamorphose des amphibiens. CR Acad Sci 284:2261–2263 [Google Scholar]
- 17.Shi Y-B, Wong J, Puzianowska-Kuznicka M, Stolow M (1996) Tadpole competence and tissue-specific temporal regulation of amphibian metamorphosis: roles of thyroid hormone and its receptors. BioEssays 18:391–399 [DOI] [PubMed] [Google Scholar]
- 18.Sachs LM, Damjanovski S, Jones PL, Li Q, Amano T, Ueda S, Shi YB, Ishizuya-Oka A (2000) Dual functions of thyroid hormone receptors during Xenopus development. Comp Biochem Physiol B Biochem Mol Biol 126(2):199–211 [DOI] [PubMed] [Google Scholar]
- 19.Puzianowska-Kuznicka M, Damjanovski S, Shi Y-B (1997) Both thyroid hormone and 9-cis retinoic acid receptors are required to efficiently mediate the effects of thyroid hormone on embryonic development and specific gene regulation in xenopus laevis. Mol Cell Biol 17:4738–4749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amaya E, Kroll KL (1999) A method for generating transgenic frog embryos. Methods Mol Biol 97:393–414 [DOI] [PubMed] [Google Scholar]
- 21.Nakajima K, Yaoita Y (2003) Dual mechanisms governing muscle cell death in tadpole tail during amphibian metamorphosis. Dev Dyn 227:246–255 [DOI] [PubMed] [Google Scholar]
- 22.Schreiber AM, Brown DD (2003) Tadpole skin dies autonomously in response to thyroid hormone at metamorphosis. Proc Natl Acad Sci U S A 100:1769–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das B, Schreiber AM, Huang H, Brown DD (2002) Multiple thyroid hormone-induced muscle growth and death programs during metamorphosis in Xenopus laevis. Proc Natl Acad Sci U S A 99:12230–12235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schreiber AM, Das B, Huang H, Marsh-Armstrong N, Brown DD (2001) Diverse developmental programs of Xenopus laevis metamorphosis are inhibited by a dominant negative thyroid hormone receptor. Proc Natl Acad Sci U S A 98:10739–10744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buchholz DR, Hsia VS-C, Fu L, Shi Y-B (2003) A dominant negative thyroid hormone receptor blocks amphibian metamorphosis by retaining corepressors at target genes. Mol Cell Biol 23:6750–6758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buchholz DR, Tomita A, Fu L, Paul BD, Shi Y-B (2004) Transgenic analysis reveals that thyroid hormone receptor is sufficient to mediate the thyroid hormone signal in frog metamorphosis. Mol Cell Biol 24:9026–9037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hasebe T, Buchholz DR, Shi YB, Ishizuya-Oka A (2011) Epithelial-connective tissue interactions induced by thyroid hormone receptor are essential for adult stem cell development in the Xenopus laevis intestine. Stem Cells 29(1):154–161. https://doi.org/10.1002/stem.560 ( 10.1002/stem.560 10.1002/stem.560 ( https://doi.org/10.1002/stem.560 ) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo X, Zhang T, Hu Z, Zhang Y, Shi Z, Wang Q, Cui Y, Wang F, Zhao H, Chen Y (2014) Efficient RNA/Cas9-mediated genome editing in Xenopus tropicalis. Development 141(3):707–714. 10.1242/dev.099853 ( 10.1242/dev.099853) [DOI] [PubMed] [Google Scholar]
- 29.Lei Y, Guo X, Deng Y, Chen Y, Zhao H (2013) Generation of gene disruptions by transcription activator-like effector nucleases (TALENs) in Xenopus tropicalis embryos. Cell Biosci 3(1):21 10.1186/2045-3701-3-21 ( 10.1186/2045-3701-3-21). 2045-3701-3-21 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lei Y, Guo X, Liu Y, Cao Y, Deng Y, Chen X, Cheng CH, Dawid IB, Chen Y, Zhao H (2012) Efficient targeted gene disruption in Xenopus embryos using engineered transcription activator-like effector nucleases (TALENs). Proc Natl Acad Sci U S A 109:17484–17489. https://doi.org/10.1073/pnas.1215421109 ( 10.1073/pnas.1215421109 10.1073/pnas.1215421109 ( https://doi.org/10.1073/pnas.1215421109 ). 1215421109 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakayama T, Fish MB, Fisher M, Oomen-Hajagos J, Thomsen GH, Grainger RM (2013) Simple and efficient CRISPR/Cas9-mediated targeted mutagenesis in Xenopus tropicalis. Genesis 51(12):835–843. https://doi.org/10.1002/dvg.22720 ( 10.1002/dvg.22720 10.1002/dvg.22720 ( https://doi.org/10.1002/dvg.22720 ) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blitz IL, Biesinger J, Xie X, Cho KW (2013) Biallelic genome modification in F(o) Xenopus tropicalis embryos using the CRISPR/Cas system. Genesis 51(12):827–834. https://doi.org/10.1002/dvg.22719 ( 10.1002/dvg.22719 10.1002/dvg.22719 ( https://doi.org/10.1002/dvg.22719 ) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang F, Shi Z, Cui Y, Guo X, Shi YB, Chen Y (2015) Targeted gene disruption in Xenopus laevis using CRISPR/Cas9. Cell Biosci 5:15 https://doi.org/10.1186/s13578-015-0006-1 ( 10.1186/s13578-015-0006-1 10.1186/s13578-015-0006-1 ( https://doi.org/10.1186/s13578-015-0006-1 ) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yen PM (2015) Unliganded TRs regulate growth and developmental timing during early embryogenesis: evidence for a dual function mechanism of TR action. Cell Biosci 5:8 https://doi.org/10.1186/2045-3701-5-8 ( 10.1186/2045-3701-5-8 10.1186/2045-3701-5-8 ( https://doi.org/10.1186/2045-3701-5-8 ) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wen L, Shi YB (2015) Unliganded thyroid hormone receptor alpha controls developmental timing in Xenopus tropicalis. Endocrinology 156:721–734. https://doi.org/10.1210/en.2014-1439 ( 10.1210/en.2014-1439 10.1210/en.2014-1439 ( https://doi.org/10.1210/en.2014-1439 ) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi J, Suzuki KI, Sakuma T, Shewade L, Yamamoto T, Buchholz DR (2015) Unliganded thyroid hormone receptor alpha regulates developmental timing via gene repression as revealed by gene disruption in Xenopus tropicalis. Endocrinology 156:735–744. https://doi.org/10.1210/en.2014-1554 ( 10.1210/en.2014-1554 10.1210/en.2014-1554 ( https://doi.org/10.1210/en.2014-1554 ) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sachs LM (2015) Unliganded thyroid hormone receptor function: amphibian metamorphosis got TALENs. Endocrinology 156(2):409–410. https://doi.org/10.1210/en.2014-2016 ( 10.1210/en.2014-2016 10.1210/en.2014-2016 ( https://doi.org/10.1210/en.2014-2016 ) [DOI] [PubMed] [Google Scholar]
- 38.Wen L, Shibata Y, Su D, Fu L, Luu N, Shi Y-B (2017) Thyroid hormone receptor α controls developmental timing and regulates the rate and coordination of tissue specific metamorphosis in Xenopus tropicalis. Endocrinology 158(6):1985–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi J, Ishizuya-Oka A, Buchholz DR (2017) Growth, development, and intestinal remodeling occurs in the absence of thyroid hormone receptor alpha in tadpoles of Xenopus tropicalis. Endocrinology 158(6):1623–1633 [DOI] [PubMed] [Google Scholar]
- 40.Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF (2011) Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39:e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu L, Buchholz D, Shi YB (2002) Novel double promoter approach for identification of transgenic animals: a tool for in vivo analysis of gene function and development of gene-based therapies. Mol Reprod Dev 62(4):470–476 [DOI] [PubMed] [Google Scholar]
- 42.Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J, Xia DF, Miller JC, Urnov FD, Gregory PD, Holmes MC (2011) Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods 8(1):74–79. https://doi.org/10.1038/nmeth.1539 ( 10.1038/nmeth.1539 10.1038/nmeth.1539 ( https://doi.org/10.1038/nmeth.1539 ) [DOI] [PubMed] [Google Scholar]
- 43.Wen L, Fu L, Guo X, Chen Y, Shi YB (2015) Histone methyltransferase Dot1L plays a role in postembryonic development in Xenopus tropicalis. FASEB J 29:385–393. https://doi.org/10.1096/fj.14-252171 ( 10.1096/fj.14-252171 10.1096/fj.14-252171 ( https://doi.org/10.1096/fj.14-252171 ) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vize PD, Melton DA, hemmati-Brivanlou A, Harland RM (1991) Assays for gene function in developing Xenopus embryos. Methods Cell Biol 36:367–387 [DOI] [PubMed] [Google Scholar]
- 45.Wu M, Gerhart J (1991) Raising Xenopus in the laboratory. Methods Cell Biol 36:3–18 [DOI] [PubMed] [Google Scholar]
- 46.Qiu P, Shandilya H, D’Alessio JM, O’Connor K, Durocher J, Gerard GF (2004) Mutation detection using surveyor nuclease. BioTechniques 36(4):702–707 [DOI] [PubMed] [Google Scholar]
- 47.Vouillot L, Thelie A, Pollet N (2015) Comparison of T7E1 and surveyor mismatch cleavage assays to detect mutations triggered by engineered nucleases. G3 (Bethesda) 5(3):407–415. https://doi.org/10.1534/g3.114.015834 ( 10.1534/g3.114.015834 10.1534/g3.114.015834 ( https://doi.org/10.1534/g3.114.015834 ) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dahlem TJ, Hoshijima K, Jurynec MJ, Gunther D, Starker CG, Locke AS, Weis AM, Voytas DF, Grunwald DJ (2012) Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet 8(8):e1002861 https://doi.org/10.1371/journal.pgen.1002861 ( 10.1371/journal.pgen.1002861 10.1371/journal.pgen.1002861 ( https://doi.org/10.1371/journal.pgen.1002861 ) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sedlak RH, Liang S, Niyonzima N, De Silva Feelixge HS, Roychoudhury P, Greninger AL, Weber ND, Boissel S, Scharenberg AM, Cheng A, Magaret A, Bumgarner R, Stone D, Jerome KR (2016) Digital detection of endonuclease mediated gene disruption in the HIV provirus. Sci Rep 6:20064 https://doi.org/10.1038/srep20064 ( 10.1038/srep20064 10.1038/srep20064 ( https://doi.org/10.1038/srep20064 ) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hisano Y, Ota S, Arakawa K, Muraki M, Kono N, Oshita K, Sakuma T, Tomita M, Yamamoto T, Okada Y, Kawahara A (2013) Quantitative assay for TALEN activity at endogenous genomic loci. Biol Open 2(4):363–367. https://doi.org/10.1242/bio.20133871 ( 10.1242/bio.20133871 10.1242/bio.20133871 ( https://doi.org/10.1242/bio.20133871 ) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fu L, Wen L, Luu N, Shi YB (2016) A simple and efficient method to visualize and quantify the efficiency of chromosomal mutations from genome editing. Sci Rep 6:35488 https://doi.org/10.1038/srep35488 ( 10.1038/srep35488 10.1038/srep35488 ( https://doi.org/10.1038/srep35488 ) [DOI] [PMC free article] [PubMed] [Google Scholar]


